Abstract
Homology-dependent exchange of genetic information between DNA molecules has a profound impact on the maintenance of genome integrity by facilitating error-free DNA repair, replication, and chromosome segregation during cell division as well as programmed cell developmental events. This chapter will focus on homologous mitotic recombination in budding yeast Saccharomyces cerevisiae. However, there is an important link between mitotic and meiotic recombination (covered in the forthcoming chapter by Hunter et al. 2015) and many of the functions are evolutionarily conserved. Here we will discuss several models that have been proposed to explain the mechanism of mitotic recombination, the genes and proteins involved in various pathways, the genetic and physical assays used to discover and study these genes, and the roles of many of these proteins inside the cell.
Keywords: recombination, repair, budding yeast
IN the course of this review, we will touch on many of the genes and processes conserved between mitosis and meiosis. Indeed, early studies in yeast and other fungi showed that mitotic recombination exhibited many of the same properties of meiotic recombination. For example, gene conversion, the nonreciprocal transfer of genetic information (see below), is sometimes associated with exchange (i.e., crossover). Heteroduplex DNA, which is detected by the failure to repair mismatches between genetically distinct DNA molecules, is indicative of strand exchange and is often found at or near sites of crossovers. Importantly, the unrepaired mismatched sequences segregate after the next round of DNA replication and can be seen as sectored colonies, similar to postmeiotic segregation observed by tetrad analysis.
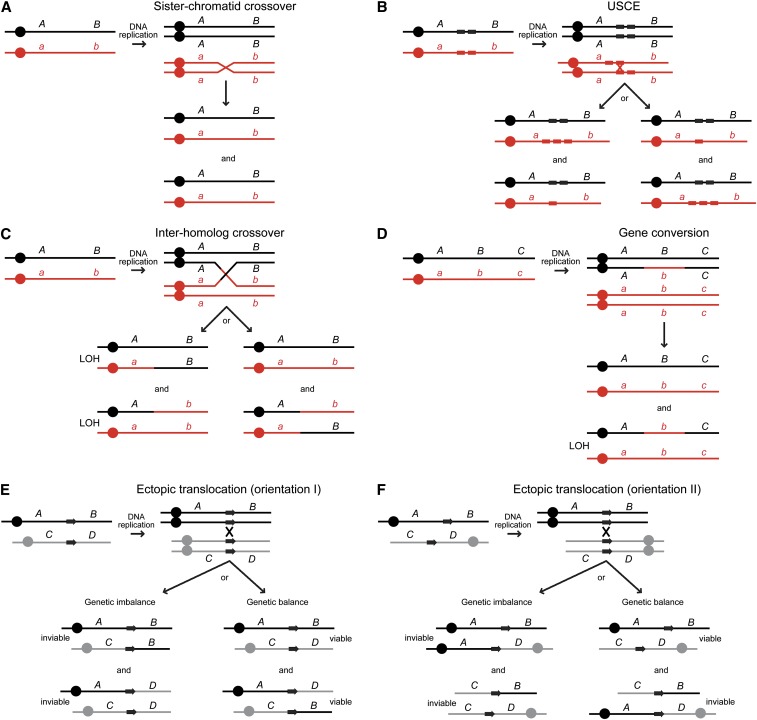
For simplicity, homologous recombination (HR) is minimally defined as the repair of DNA lesions using homologous sequences. During S phase and G2, in both haploid and diploid cells, repair of the damage uses the unbroken sister chromosome as the homologous sequence (Figure 1A). Such repair is the main role of mitotic recombination and it can lead to genetic consequences. When sister chromatid repair is accompanied by a crossover, it results in sister-chromatid exchange (SCE). If the repair event occurs between misaligned repetitive sequences in a tandem array, it results in an unequal SCE (USCE) (Figure 1B). In diploids, repair can also be templated from the unbroken homologous chromosome and if associated with a crossover, the exchange can lead to loss of heterozygosity (LOH) (Figure 1C). Mitotic gene conversion results when there is a nonreciprocal transfer of genetic information from one chromosome to the other during the repair event (Figure 1D). DNA repair from homologous sequences at nonallelic positions, called ectopic recombination, can lead to deletions, inversions, translocations, and acentric or dicentric chromosomes if repair is associated with a crossover (Figure 1, E and F).
Figure 1.
Genetic outcomes of homologous recombination. The letters A/a and B/b indicate heteroalleles. Circles indicate centromeres. Colors red and black indicate homologous chromosomes in diploid cells. (A) Sister-chromatid crossover. A crossover between sister chromatids results in two genetically identical cells. (B) Unequal sister-chromatid exchange (USCE). Within repetitive sequence elements (boxes), a crossover between misaligned repeats results in repeat copy number expansion and contraction. (C) Interhomolog crossover. A crossover between homologs leads to loss of hetorozygosity (LOH), if the recombinant molecules segregate to different cells in the ensuing cell division. (D) Gene conversion. A nonreciprocal genetic exchange between homologs leads to LOH in one of the resulting cells. (E) Productive ectopic translocation. A crossover between homologous sequences (boxes with arrows) with the same orientation relative to the centromere (circles) on different chromosomes in gray and black results in a productive ectopic translocation. Cosegregation of the recombinant molecules results in genetically balanced cells, shown on the right. Segregation of the recombinant molecules to different cells, shown on the left, leads to lethality if the regions represented by B and D are essential. (F) Nonproductive ectopic translocation. If the recombining sequences have opposite orientation with respect to the centromere, the reciprocal translocation results in inviable dicentric and acentric chromosomes.
Most of our attention focuses on the repair of double-strand breaks (DSBs); however, the exact nature of the initiating spontaneous lesion is unknown. Indeed nicks can be processed into single-stranded DNA (ssDNA) gaps or DSBs as the result of ligation failure from the previous round of DNA replication or during the repair of damaged or misincorporated nucleotides via processes such as nucleotide excision repair (NER), mismatch repair (MMR), base excision repair (BER), or transcription-coupled repair (TCR). Nicks can also be formed after the failed catalysis of Top1 covalently attached to DNA. Upon subsequent replication, these protein-bound nicks can also become DSBs. Reactive oxygen species (ROS), cellular metabolism, and exogenous damage from ultraviolet light or gamma-irradiation can also produce nicks. Similarly, collapsed and stalled replication forks can lead to structures that can be processed into DSBs. Finally, DNA ends are produced when gamma-irradiation breaks both strands or when telomeres are uncapped due to problems in assembling the shelterin complex. In some cases, simple ligation of the ends, so-called nonhomologous end joining (NHEJ), results in repair that may or may not be error-free. For example, in a G1 cell, NHEJ is likely the preferred repair choice. However, given the many different sources of DNA lesions, it is clear that one of the central questions in the regulation of recombination is how the cell “determines” how to repair a DSB—NHEJ or HR. Among some of the issues the cell must confront are the necessity to interpret whether it is haploid or diploid (controlled by the MAT locus), where it is in the cell cycle (controlled by CDK), what kind of processing the DNA ends need [controlled in part by the Mre11–Rad50–Xrs2 (MRX) complex] and the chromatin environment of the DNA lesion. All of this information must be integrated for the appropriate repair decision to be made. Understanding this integration will clarify how all of the pathways that impact recombination are intertwined to lead to repair of a DNA lesion and a viable cell.
Many of the genes described in this chapter that affect genetic recombination were originally identified by their requirement to repair radiation-induced DNA damage. For example, most of the genes of the RAD52 epistasis group are ionizing radiation (IR) sensitive, while RAD1 and RAD10 are ultraviolet light sensitive (Game and Cox 1971; Game and Mortimer 1974). In addition, genes have been identified that are sensitive to other types of DNA damaging agents, such as methyl methanesulfonate (MMS), camptothecin (CPT), 4-nitroquinoline 1-oxide (4-NQO), etc. Over the years, genes involved in many pathways have been shown to affect genetic recombination. Some of these genes were discovered in mutation analyses that looked for effects on recombination and repair assays, while others were identified when the yeast gene disruption library was systematically tested for gross chromosomal rearrangements (Huang and Koshland 2003; Smith et al. 2004), mitotic crossing over (Andersen et al. 2008), sensitivity of polyploidy (Storchova et al. 2006), Rad52 foci (Alvaro et al. 2007), doxorubicin sensitivity (Westmoreland et al. 2009), sensitivity to R-loops (Gomez-Gonzalez et al. 2011), fragility of triplex structure-forming GAA/TTC (Zhang et al. 2012), stability of quasipalindromes (Zhang et al. 2013), and many more. Bioinformatic analyses have also revealed many genes involved in genome stability (Putnam et al. 2012). Furthermore, screens for hyperrecombination (hyper-rec) uncovered genes involved in a multitude of pathways (Aguilera and Klein 1988; Keil and McWilliams 1993; Scholes et al. 2001).
Finally, it is worth mentioning that most assays that have been constructed to identify genes that affect recombination were applied without knowing the precise recombination pathway being assayed. By isolating mutations that affect that pathway, we can start to understand the mechanism of the assay. At the same time, the precise function of the gene is refined by understanding its role in the assay. This “yin–yang” situation makes the study of homologous recombination so challenging.
II. Mechanisms of Recombination
A. Models for DSB-initiated homologous recombination
DSB repair and synthesis-dependent strand annealing models:
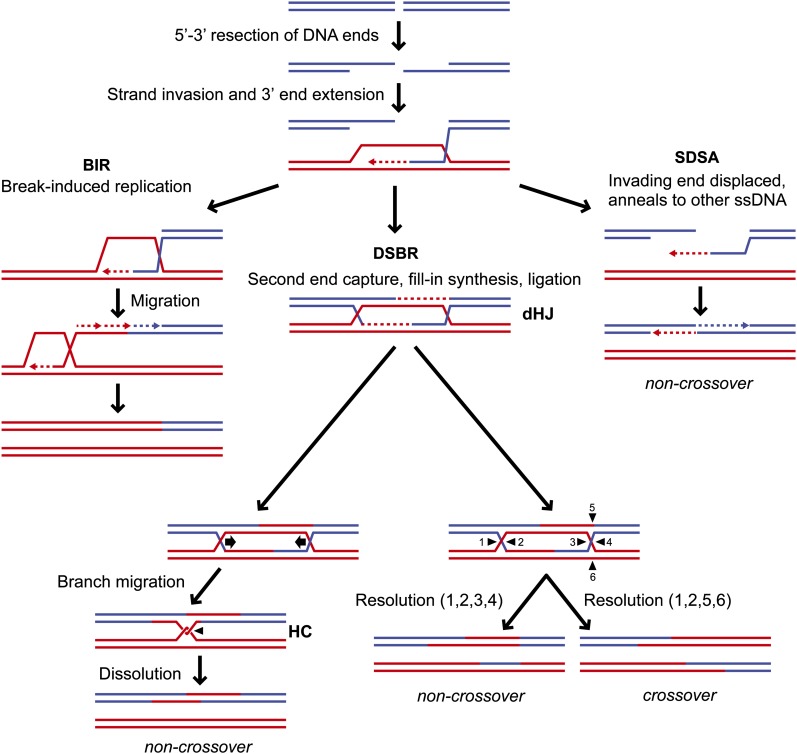
The DSB repair (DSBR) model was first proposed to explain the mechanism of plasmid gap repair and is currently the most accepted model to rationalize the association of crossing over with gene conversion during homologous recombination (Orr-Weaver et al. 1981; Szostak et al. 1983). In this model, the 5′ ends at the DSB are degraded to yield 3′ ssDNA tails, one of which invades a homologous double-stranded DNA (dsDNA) to form a displacement loop (D-loop) and is used to prime DNA synthesis, templated by the donor duplex (Figure 2). The 3′-terminated strand at the other side of the break anneals to the displaced strand from the donor duplex and primes a second round of leading strand synthesis. After ligation of the newly synthesized DNA to the resected 5′ strands, a double Holliday junction intermediate (dHJ) is generated. To segregate the recombinant duplexes, the HJs must be removed, which can occur by the activity of a helicase and topoisomerase to produce only noncrossover (NCO) products (dissolution), or by endonucleolytic cleavage (resolution). Cutting the inner strands of both HJs yields NCO products, whereas cleavage of the inner strands of one HJ and the outer strands of the other generates crossovers (COs). Heteroduplex DNA (hDNA), which is formed by pairing one strand from one duplex with a complementary strand from the other duplex, is a hallmark of homologous recombination and can be formed during the initial strand invasion and/or by second end capture. Repair of mismatches present in hDNA can give rise to gene conversion or restoration to the original sequence (Boiteux and Jinks-Robertson 2013).
Figure 2.
Models for homology-dependent DSB repair. Recombinational repair of a DSB is initiated by 5′ to 3′ resection of the DNA end(s). The resulting 3′ single-stranded end(s) invades an intact homologous duplex (in red) to prime leading strand DNA synthesis. For one-ended breaks, a migrating D-loop is established to facilitate break-induced replication (BIR) to the end of the chromosome and the complementary strand is synthesized by conservative replication. For two-ended breaks, the classical double-strand break repair (DSBR) model predicts that the displaced strand from the donor duplex pairs with the 3′ ssDNA tail at the other side of the break and primes a second round of leading strand synthesis. After ligation of the newly synthesized DNA to the resected 5′ strands, a double Holliday junction intermediate (dHJ) is generated. The dHJ can be either dissolved by branch migration into a hemicatenane (HC) leading to noncrossover (NCO) products or resolved by endonucleolytic cleavage to produce NCO (positions 1, 2, 3, and 4) or CO (positions 1, 2, 5, and 6) products. In mitotic cells, the invading strand is often displaced after limited synthesis and the nascent complementary strand anneals with the 3′ single-stranded tail of the other end of the DSB and after fill-in synthesis and ligation generate exclusively NCO products (synthesis-dependent strand annealing, SDSA).
While the DSBR model explains many properties of meiotic recombination, mitotic recombination generally shows a lower association of crossing over with gene conversion than observed during meiotic recombination. This observation led to two variations of the DSBR model, the synthesis-dependent strand annealing (SDSA) and migrating D-loop models, to explain the lower incidence of associated COs during mitotic DSB repair (Nassif et al. 1994; Ferguson and Holloman 1996; Paques et al. 1998). The SDSA model proposes that both 3′ ssDNA tails invade the homologous duplex(es) and after limited DNA synthesis are displaced by DNA helicases; the nascent complementary strands anneal and after fill-in synthesis and ligation generate exclusively NCO products (Nassif et al. 1994). The migrating D-loop model proposes that only one of the two 3′ ssDNA tails invades the homologous DNA duplex and after limited DNA synthesis is dissociated and anneals to the 3′ ssDNA tail at the other side of the DSB (Ferguson and Holloman 1996). Gap filling and ligation yields only NCO products (Figure 2). SDSA is the acronym generally used to refer to both models.
Break-induced replication:
Break-induced replication (BIR) is a recombination-dependent replication process that results in the nonreciprocal transfer of DNA from the donor to recipient chromosome. For repair by BIR, a single end of a DSB invades a homologous duplex DNA and initiates replication to the chromosome end (Kraus et al. 2001; Llorente et al. 2008) (Figure 2). As BIR from one of the two ends of a DSB would result in extensive loss of heterozygosity (LOH), it suggests BIR is suppressed when DSBs have two homologous ends in order for repair to occur by a more conservative HR mechanism. Indeed, the frequency of BIR at an endonuclease-induced DSB is <1%, but can be substantially higher if gene conversion is prevented by limiting homology to one side of the break (Malkova et al. 1996, 2005; Bosco and Haber 1998). The initial steps of BIR appear to be similar to DSBR and SDSA in requiring end resection, homologous pairing, and strand invasion (Davis and Symington 2004). Recent studies identified a migrating D-loop intermediate during BIR and demonstrated that synthesis is conservative (Donnianni and Symington 2013; Saini et al. 2013a), suggesting the “lagging” strand initiates on the nascent strand extruded from the trailing end of the D-loop. BIR can occur by several rounds of strand invasion, DNA synthesis, and dissociation, resulting in chromosome rearrangements when dissociation and reinvasion occur within dispersed repeated sequences (Smith et al. 2007; Ruiz et al. 2009). Thus, the highly mutagenic nature of BIR could contribute to genome evolution and disease development in humans.
Single-strand annealing and microhomology-mediated end joining:
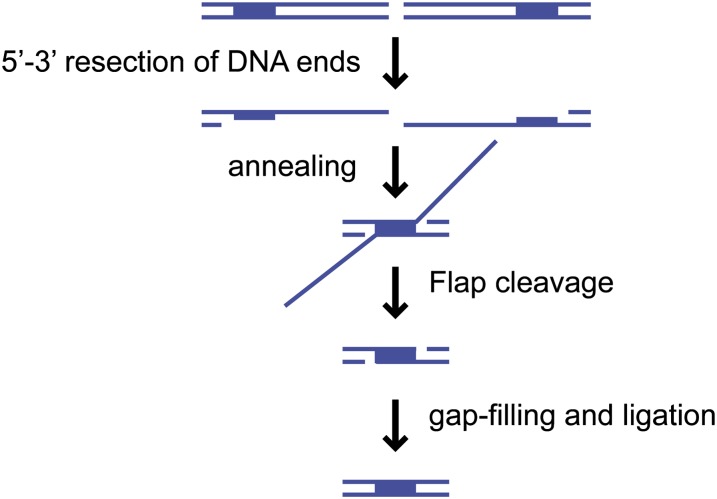
The single-strand annealing (SSA) mechanism has been most extensively studied in the context of repair of an induced DSB formed between direct repeats (Paques and Haber 1999). SSA might also be responsible for spontaneous deletions between direct repeats, but other mechanisms could operate. SSA efficiently repairs DSBs formed between repeats of >200 bp, but the frequency drops significantly for repeats of <50 bp (Sugawara et al. 2000). After resection of the DSB ends, the 3′ ssDNA tails can anneal when resection is sufficient to reveal complementary single-stranded regions corresponding to the repeats (Figure 3). Following annealing of the complementary ssDNA, heterologous flaps are formed if the repeats are separated from the break site by unique sequence. The flaps are removed by nucleases prior to gap filling and ligation (Fishman-Lobell et al. 1992; Ivanov and Haber 1995). Microhomology-mediated end joining (MMEJ) applies to an end joining mechanism that, like SSA, involves end resection and annealing between short (5–25 nt) direct repeats flanking a DSB (Ma et al. 2003; Decottignies 2007; Villarreal et al. 2012; Deng et al. 2014). These mechanisms are always mutagenic because they result in deletions, and MMEJ may be responsible for gross chromosome rearrangements (GCRs) that exhibit microhomologies at the junctions (Putnam et al. 2005).
Figure 3.
Single-strand annealing. Repair of a DSB flanked by direct repeat sequences (boxes) can occur by single-strand annealing (SSA), if 5′ to 3′ resection is allowed to progress past the repeats. The complementary single-stranded repeats can anneal, leaving heterologous flaps to be removed by the Rad1–Rad10 endonuclease, before gap-filling and ligation completes the repair thereby deleting one of the repeats and the intervening sequence.
B. Proteins involved in homologous recombination
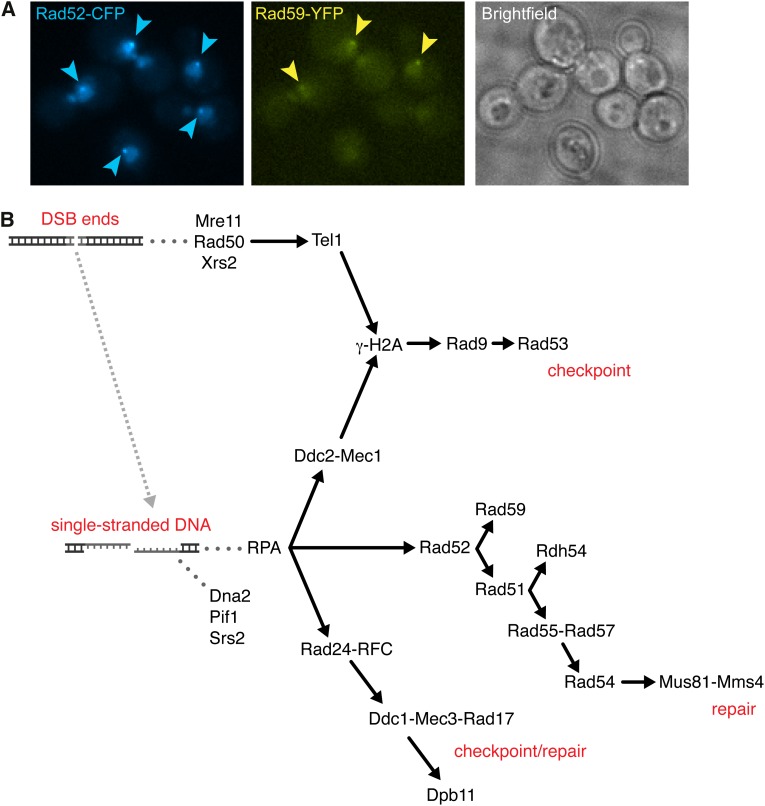
Here we summarize the activities of proteins known to function at discrete steps of homology-dependent repair, as determined by biochemical analysis using defined DNA substrates and from the phenotypes of mutants in genetic and physical assays (for a list of human homologs and associated human diseases see Table 1). Physical assays, such as the MAT switching system, have been particularly useful to identify DNA intermediates formed during DSB repair and the genetic control of discrete steps in HR (Haber 2012). MAT switching is initiated by HO endonuclease cleavage of a specific site at the MAT locus and repair occurs by gene conversion using one of the transcriptionally silent donor cassettes, HML or HMR. The system has been adapted for studies of DSB repair by placing the HO gene under the control of the GAL1 promoter to synchronously induce HO in a population of cells by addition of galactose to the growth medium (Jensen and Herskowitz 1984). The HO recognition site can be inserted at other genomic sites unrelated to MAT to induce recombination (Nickoloff et al. 1986). I-SceI, a site-specific endonuclease responsible for intron mobility in yeast mitochondria, has also been used to initiate DSB-induced recombination in the yeast nuclear genome by expressing an engineered version from the GAL1 promoter (Plessis et al. 1992). The advantage of both systems is that 50–100% of the target sequences are cut within 1 hr following induction of the nuclease, and, by taking DNA samples at different times after DSB formation, intermediates in the process, for example, resected DNA ends and strand invasion intermediates, can be identified by Southern blot hybridization and/or PCR methods.
Table 1. Evolutionary conservation of homologous recombination proteins and examples of related human diseases.
| Functional class | S. cerevisiae | H. sapiens | Associated disease(s) | References |
|---|---|---|---|---|
| End resection | Mre11-Rad50-Xrs2 | MRE11-RAD50-NBS1 | Nijmegen breakage syndrome; AT-like disorder | (Varon et al., 1998) |
| Sae2 | CtIP | |||
| Exo1 | EXO1 | Colorectal cancer | (Yang et al., 2014) | |
| Dna2-Sgs1-Top3-Rmi1 | DNA2-BLM-TOP3-RMI1-RMI2 | Bloom syndrome | (Kaneko and Kondo, 2004) | |
| Adaptors | Rad9 | 53BP1, MDC1 | Breast cancer | (Bartkova et al., 2007; Rapakko et al., 2007) |
| - | BRCA1 | Breast cancer | (Petrucelli et al., 2010) | |
| Checkpoint signaling | Tel1 | ATM | Ataxia-telangiectasia | (Gatti et al., 2001) |
| Mec1-Ddc2 | ATR-ATRIP | Seckel syndrome | (O'Driscoll et al., 2003) | |
| Rad53 | CHK2 | |||
| Rad24-RFC | RAD17-RFC | |||
| Ddc1-Mec3-Rad17 | RAD9-HUS1-RAD1 | |||
| Dpb11 | TOPBP1 | Breast cancer | (Karppinen et al., 2006) | |
| Single-stranded DNA binding | Rfa1-Rfa2-Rfa3 | RPA1-RPA2-RPA3 | ||
| Single-strand annealing | Rad52 | RAD52 | ||
| Rad59 | - | |||
| Mediators | Rad52 | - | ||
| - | BRCA2-PALB2 | Breast cancer | (Petrucelli et al., 2010) | |
| Strand exchange | Rad51 | RAD51 | Breast cancer | (Nissar et al., 2014; Sun et al., 2011) |
| Rad54 | RAD54A, RAD54B | |||
| Rdh54 | - | |||
| Rad51 paralogs | Rad55-Rad57 | RAD51B-RAD51C-RAD51D-XRCC2-XRCC3 | Breast cancer | (Silva et al., 2010; Vuorela et al., 2011) |
| Psy3-Csm2-Shu1-Shu2 | RAD51D-XRCC2-SWS1 | |||
| Anti-recombinases | Srs2 | FBH1, PARI | ||
| Mph1 | FANCM | Fanconi Anemia | (Meetei et al., 2005) | |
| - | RTEL1 | Hoyeraal-Hreidarsson syndrome | (Ballew et al., 2013) | |
| Resolvases and nucleases | Mus81-Mms4 | MUS81-EME1 | ||
| Slx1-Slx4 | SLX1-SLX4 | Fanconi Anemia | (Crossan et al., 2011; Kim et al., 2011; Stoepker et al., 2011) | |
| Yen1 | GEN1 | |||
| Rad1-Rad10 | XPF-ERCC1 | Xeroderma pigmentosum | (Gregg et al., 2011) |
See also http://www.malacards.org.
DNA end resection:
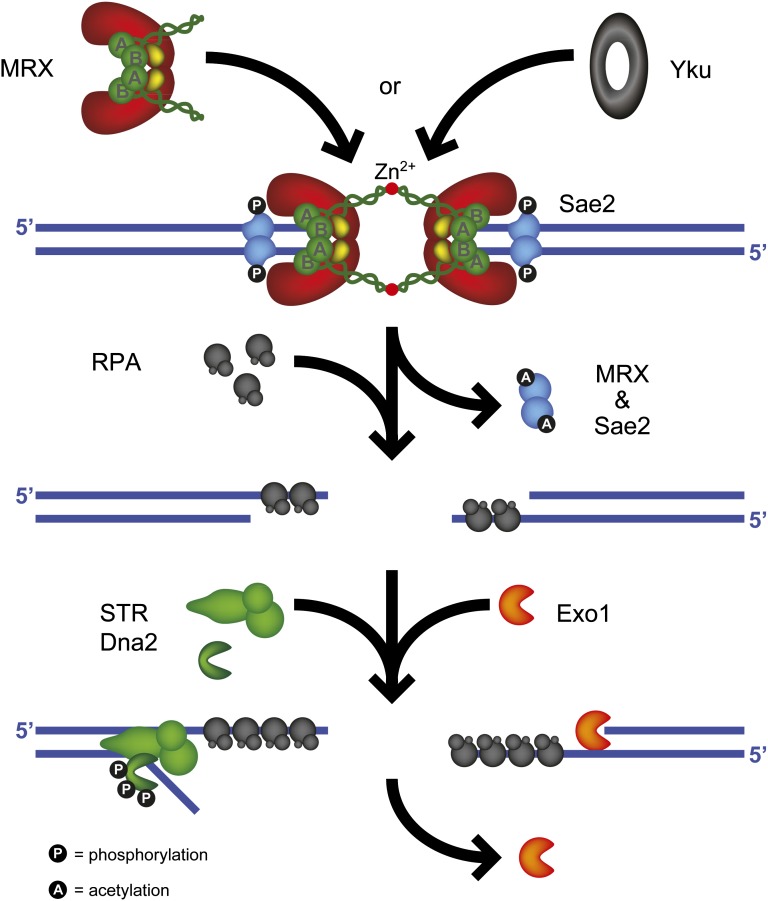
Homology-dependent DSB repair initiates by nucleolytic degradation of the 5′ strands to yield 3′ ssDNA tails, a process referred to as 5′–3′ resection (reviewed by Mimitou and Symington 2009) (Figure 4). The RAD50 and XRS2 genes were initially implicated in controlling end resection because the mutants show a marked delay in the initiation of resection at endonuclease-induced DSBs; however, recombination products are still produced, but with delayed kinetics and lower yield than wild-type cells (Ivanov et al. 1994). Since recombination is not completely defective, it is puzzling why mre11, rad50, and xrs2 mutants exhibit extreme sensitivity to DNA damaging agents and could be due to other functions of the proteins, such as in telomere maintenance, processing of ends with adducts, and nonhomologous end joining repair. The Mre11, Rad50, and Xrs2 proteins form a stable heterotrimeric complex (MRX) that plays structural and catalytic roles in the initiation of end resection (Mimitou and Symington 2009). Mre11 contains conserved phosphoesterase motifs that are essential for 3′–5′ dsDNA exonuclease and ssDNA endonuclease activities in vitro (Furuse et al. 1998; Usui et al. 1998; Moreau et al. 1999; Trujillo and Sung 2001). Rad50 has a similar domain organization to the structural maintenance of chromosomes (SMC) family of proteins (Alani et al. 1989; Lammens et al. 2011; Lim et al. 2011; Williams et al. 2011). The integrity of the long coiled-coil domains, as well as the conserved Cys-X-X-Cys dimerization motif (Rad50 hook) at the apex of the coiled-coil domains, is important for all functions of the MRX complex, suggesting tethering of DNA ends or sister chromatids via MR binding and dimerization is critical for repair (De Jager et al. 2001; Hopfner et al. 2002; Wiltzius et al. 2005; Hohl et al. 2011). Mre11 binds to the base of the Rad50 coiled-coils forming a “head” region composed of the Mre11 nuclease and Rad50 ATPase domain that together provides DNA binding and end processing activities that are regulated by ATP binding and hydrolysis (Lammens et al. 2011; Lim et al. 2011; Williams et al. 2011). Interaction with Xrs2 is required for translocation of Mre11 to the nucleus, and Xrs2 is also thought to regulate the MR complex and to mediate DNA damage signaling via interactions with Tel1 and Sae2 (Nakada et al. 2003; Tsukamoto et al. 2005; Lloyd et al. 2009; Schiller et al. 2012).
Figure 4.
Resection of DSB ends. Resection of DSB ends progress 5′ to 3′ and in two steps. First, MRX and Sae2 catalyze short-range resection of ∼100 nt. The initial resection by MRX and Sae2 is particularly important for cleaning up “dirty” ends harboring chemical adducts, secondary structures, or covalently attached proteins. The Yku complex inhibits initial end resection by competing with MRX for binding to ends. Second, extensive resection for up to 50 kb is catalyzed by Exo1 and/or STR–Dna2. Phosphorylation of Sae2 at serine 267 is required for resection. Sae2 is degraded upon acetylation. Dna2 nuclear localization and recruitment to DSBs require its phosphorylation at threonine 4 and serines 17 and 237 (see text for details).
MRX can act as an endonuclease with Sae2 to cleave oligonucleotides from the 5′ strands resulting in short (∼100 nucleotides) 3′ ssDNA tails, or it can promote resection indirectly by recruitment of the Exo1 and/or Dna2 nucleases (Mimitou and Symington 2008; Zhu et al. 2008; Shim et al. 2010). The Mre11 nuclease activity and Sae2 are essential to remove covalent adducts, such as Spo11 (which forms a covalent attachment to 5′ ends to initiate meiotic recombination) or hairpin caps, from DNA ends, but not for resection of endonuclease-induced DSBs (Mimitou and Symington 2009). There is a delay of ∼30 min before resection of an HO-induced DSB initiates in the sae2 mutant and this results in an increased frequency of NHEJ repair (Lee et al. 2008; Mimitou and Symington 2008; Deng et al. 2014). Sae2 exhibits endonuclease activity in vitro that is stimulated by MRX, but unlike Mre11 has no obvious nuclease motifs (Lengsfeld et al. 2007). Which of these two nucleases, or if both, contributes to the initiation of resection is currently unknown. A class of separation-of-function rad50 alleles, rad50S, confers similar phenotypes to the sae2 and mre11 nuclease-defective mutants (Alani et al. 1990). One attractive model for resection that has emerged from studies of meiotic recombination is for MRX and Sae2 to incise the 5′ strand at a distance from the end, followed by bidirectional resection from the nick using the Mre11 3′–5′ exonuclease and Exo1 5′–3′ exonuclease (Zakharyevich et al. 2010; Garcia et al. 2011). This model rationalizes how the Mre11 3′–5′ exonuclease participates in end resection and how Exo1 can overcome the block imposed by Ku binding at DNA ends.
Extensive resection is catalyzed by the 5′–3′ dsDNA exonuclease, Exo1, or by the combined action of the Sgs1 helicase and Dna2 endonuclease (Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008) (Figure 4). The Sgs1 interacting partners Top3 and Rmi1 are also required for end resection, but their roles appear to be structural rather than catalytic (Zhu et al. 2008; Niu et al. 2010). In vitro reconstitution of the Sgs1-Dna2 reaction revealed an essential role for the heterotrimeric replication protein A (RPA, Rfa1–Rfa2–Rfa3) to stimulate Sgs1 unwinding and Dna2 cleavage of the 5′ strand (Cejka et al. 2010a; Niu et al. 2010). Depletion of RPA from cells at the time of HO cleavage results in a block to extensive resection and failure to recruit Dna2 to DSBs. RPA is also required to prevent formation of secondary structures within the 3′ ssDNA tails that can generate hairpin capped ends or can be degraded by MRX and Sae2 (Chen et al. 2013). Although Exo1 or Sgs1–Dna2 can directly process free DNA ends with no covalent adducts, they are unable to remove end-blocking lesions, such as Spo11. One possible reason for the greatly delayed initiation of resection observed for the mre11, rad50, or xrs2 mutants is the poor recruitment of Exo1 and Sgs1–Dna2 in the absence of the MRX complex. The other possible reason is the presence of Ku, a heterodimeric dsDNA end binding protein essential for NHEJ, which inhibits Exo1-mediated resection and is greatly enriched at DSBs in the absence of the MRX complex (Ivanov et al. 1994; Clerici et al. 2008; Mimitou and Symington 2010; Nicolette et al. 2010; Shim et al. 2010). Indeed, the resection defect of the mre11 mutant is suppressed by elimination of Ku or overexpression of EXO1 (Bressan et al. 1999; Tsubouchi and Ogawa 2000; Moreau et al. 2001; Lewis et al. 2002). Similarly, the resection defect of the sae2 mutant is suppressed by yku70 in an Exo1-dependent manner (Limbo et al. 2007; Mimitou and Symington 2010).
The rate of extensive end resection is ∼4 kb/hr and can remove up to 50 kb of DNA if there is no donor sequence to template repair of the DSB (Zhu et al. 2008). However, extensive resection is unlikely to be necessary for HR, and even the short 3′ ssDNA tails resulting from MRX–Sae2-dependent cleavage in the absence of Exo1 and Sgs1–Dna2 are sufficient for Rad51-dependent recombination (Mimitou and Symington 2008; Zhu et al. 2008). Although extensive resection is not required for HR, resection of >10 kb is needed to activate the DNA damage checkpoint (Gravel et al. 2008; Zhu et al. 2008; Roberts et al. 2012). Elimination of MRX, the Mre11 nuclease or Sae2 in the exo1 sgs1 background results in a complete block to end resection and lethality (Mimitou and Symington 2008).
Homologous pairing and strand invasion:
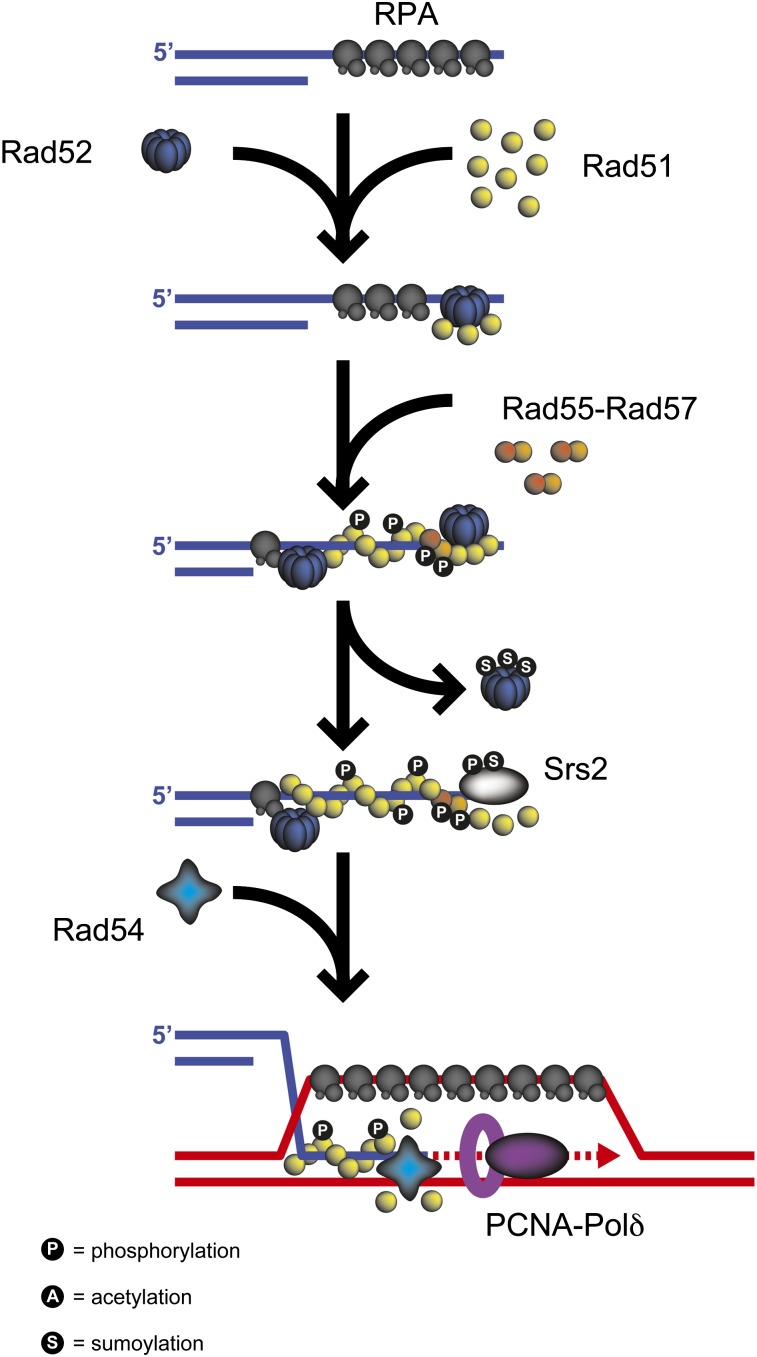
The critical step of HR is pairing between the ssDNA formed by end resection and one strand of the donor duplex to form hDNA, a reaction catalyzed by the Rad51/RecA family of recombinases (Shinohara et al. 1992; Symington 2002). Although Rad51 can catalyze DNA strand exchange under certain conditions in vitro, efficient exchange requires the activity of several other proteins, including RPA, Rad52, Rad54, Rad55, and Rad57 in vivo. Rad51 binds to ssDNA and dsDNA in an ATP-dependent fashion to form right-handed helical nucleoprotein filaments that can span thousands of nucleotides (Ogawa et al. 1993; Sung 1994; Sehorn et al. 2004; Sheridan et al. 2008; Ferrari et al. 2009). The contour length of DNA within the nucleoprotein filaments is extended by 50% relative to B-form DNA. Although Rad51 binds dsDNA in vitro, only the Rad51–ssDNA filament is active for homologous pairing and strand exchange. Rad51 has two DNA binding sites, one of which is required for high-affinity DNA binding and filament formation, and the second is required for homologous pairing (Cloud et al. 2012). Alone, Rad51 exhibits weak strand exchange activity that is stimulated by RPA, but this stimulation only occurs if Rad51 is allowed to nucleate on single-stranded DNA prior to the addition of RPA (Sung 1994, 1997a; Sugiyama et al. 1997). RPA is thought to stimulate Rad51 binding to ssDNA by removing secondary structures to allow assembly of a contiguous Rad51 nucleoprotein filament. If RPA and Rad51 are added simultaneously to ssDNA then RPA inhibits the reaction. This inhibition can be overcome by the addition of mediator proteins, such as Rad52 (Sung 1997a; New et al. 1998; Shinohara and Ogawa 1998). RPA also stimulates Rad51-mediated strand transfer by sequestering the displaced ssDNA that can inhibit the pairing reaction (Eggler et al. 2002).
Rad51 mediators:
Rad51 mediators include proteins that enable loading of Rad51 on RPA-coated ssDNA, stabilize Rad51 nucleoprotein filaments and/or promote strand exchange by Rad51 (Sung et al. 2003). Rad52 interacts directly with both RPA and Rad51 and is thought to mediate Rad51 filament assembly by delivering Rad51 to RPA-bound ssDNA where it binds cooperatively to DNA, displacing RPA (Sung 1997a; New et al. 1998; Krejci et al. 2002; Sugiyama and Kowalczykowski 2002) (Figure 5). These in vitro studies are consistent with Rad52 being required for Rad51 recruitment to an HO-induced DSB in vivo as measured by chromatin IP and fluorescence microscopy (Sugawara et al. 2003; Lisby et al. 2004). Rad52 interaction with Rad51 occurs through a domain in the nonconserved C-terminal region of the protein (Milne and Weaver 1993; Krejci et al. 2002). Mutants expressing Rad52 C-terminal truncations that remove the Rad51 interacting domain exhibit intermediate IR sensitivity as compared to wild type and rad52 mutants, and the IR sensitivity is suppressed by overexpression of RAD51 or by deleting SRS2, which encodes a helicase that disrupts Rad51–ssDNA complexes in vitro (Boundy-Mills and Livingston 1993; Milne and Weaver 1993; Kaytor et al. 1995). An acidic region of Rad52, encompassing residues 308–311 is required for interaction with RPA and for DNA damage resistance (Plate et al. 2008).
Figure 5.
Rad51 filament dynamics. During Rad51-catalyzed strand invasion, Rad52 mediates the loading of Rad51 onto RPA-coated ssDNA to facilitate formation of a Rad51 nucleoprotein filament. The Rad51 filament is further stabilized by Rad55–Rad57. In contrast, the Srs2 helicase counteracts the Rad51 mediators by displacing Rad51 from ssDNA to disrupt toxic recombination intermediates. Similarly, Rad54 can displace Rad51 from dsDNA to allow loading of PCNA–Polδ at the 3′ end of the invading strand. Phosphorylation of Rad51 at serine 192 is required for ATP hydrolysis and DNA binding. Rad55 is phosphorylated at serines 2, 8, and 14. Sumoylation of Rad52 at lysines 43, 44, and 253 mediates its dissociation from ssDNA. Phosphorylation and sumoylation of Srs2 have pro- and antirecombination functions, respectively (see text for details).
Rad55 and Rad57 are referred to as Rad51 paralogs because they share ∼20% identity to the Rad51/RecA core region (Kans and Mortimer 1991; Lovett 1994). Rad55 and Rad57 interact to form a stable heterodimer and Rad55 also interacts with Rad51 (Hays et al. 1995; Johnson and Symington 1995; Sung 1997b). The Rad55-Rad57 complex is implicated as a mediator of Rad51 filament assembly because it alleviates the RPA inhibition to Rad51-catalyzed strand exchange in vitro (Sung 1997b). Rad51 recruitment to an HO-induced DSB occurs more slowly in the absence of Rad55, consistent with a role for Rad55–Rad57 in Rad51 filament formation or stabilization (Sugawara et al. 2003). The IR sensitivity of rad55 and rad57 mutants is partially bypassed by overexpression of RAD51, or by RAD51 gain-of-function alleles, such as rad51-I345T, that encode proteins with higher affinity for DNA than wild-type Rad51 (Hays et al. 1995; Johnson and Symington 1995; Fortin and Symington 2002). Deletion of SRS2 also suppresses the IR sensitivity of rad55 and rad57 mutants (Fung et al. 2009). In vitro studies show the Rad55–Rad57 complex stabilizes Rad51 nucleoprotein filaments and counteracts Srs2-mediated displacement of Rad51 from ssDNA (Liu et al. 2011).
The Psy3, Csm2, Shu1, and Shu2 proteins (collectively referred to as the Shu proteins) are thought to function in early HR because mutation of any of the SHU genes suppresses the top3 slow growth defect and sgs1 HU sensitivity, similar to rad51, rad55, and rad57 mutations (Shor et al. 2002, 2005). The shu mutants are not sensitive to IR, but do exhibit sensitivity to MMS, an alkylating agent that stalls replication, and show increased mutation rates dependent on the translesion synthesis (TLS) DNA polymerase, Polζ (Huang et al. 2003; Shor et al. 2005), which is consistent with a role in error-free postreplicative repair (Ball et al. 2009). The shu mutations partially suppress the accumulation of sister-chromatid joint molecules in the sgs1 mutant, suggesting they function specifically in early HR events to fill gaps during replication (Mankouri et al. 2007). The Shu proteins appear to antagonize the activity of the Srs2 antirecombinase, possibly through Rad51 filament stabilization (Bernstein et al. 2011). The Shu proteins interact to form a complex and structural studies of the Psy3–Csm2 subcomplex indicate structural similarity to a Rad51 dimer, raising the possibility that the Shu complex is incorporated into the Rad51 filament, as suggested for Rad55–Rad57 (Liu et al. 2011; Tao et al. 2012; Sasanuma et al. 2013). The Psy3–Csm2 complex exhibits DNA binding in vitro, with a preference for forked and 3′ overhang DNA substrates, and is able to stabilize Rad51 binding to ssDNA (Godin et al. 2013; Sasanuma et al. 2013).
Single-strand annealing:
Rad52 also promotes annealing of ssDNA in vitro (Mortensen et al. 1996; Shinohara et al. 1998) and this activity is likely important for capture of the second end in DSBR and SDSA and for the SSA mechanism of recombination (Sugawara and Haber 1992; Sugiyama et al. 2006; Lao et al. 2008). However, Rad52 is not required for MMEJ unless the microhomologies are >14 bp in length (Villarreal et al. 2012). This Rad51-independent function of Rad52 may explain the greater defect in most recombination assays reported for rad52 mutants compared to rad51, rad54, rad55, and rad57 mutants (Symington 2002). RPA is inhibitory to ssDNA annealing in vitro; however, Rad52 is able to overcome this inhibition (Sugiyama et al. 1998). In vivo, RPA prevents annealing between microhomologies that are too short to be annealed by Rad52 and prevents SSA in the absence of Rad52 (Smith and Rothstein 1999; Villarreal et al. 2012; Deng et al. 2014). Rad59, a protein with homology to the N-terminal DNA binding domain of Rad52 (Bai and Symington 1996), interacts with Rad52 and augments the strand-annealing activity (Petukhova et al. 1999; Sugawara et al. 2000; Davis and Symington 2001; Wu et al. 2006). The N-terminal domain of Rad52 is required for strand annealing and multimerization of Rad52 and is essential for Rad52 function in vivo (Kagawa et al. 2002; Mortensen et al. 2002; Singleton et al. 2002).
DNA translocases:
Rad54 and Rdh54/Tid1, members of the Swi2/Snf2 family of chromatin remodeling proteins, stimulate homologous pairing by Rad51 in vitro (Petukhova et al. 1999, 2000). Rad54 stimulates D-loop formation between ssDNA and a homologous supercoiled plasmid, but also promotes dissociation of D-loop structures. The dissociation function is decreased when long ssDNA substrates are used or when duplex regions flank the invading ssDNA (Wright and Heyer 2014). Rad54 mediates chromatin remodeling and is able to promote Rad51-dependent D-loop formation on chromatinized templates (Alexiadis and Kadonaga 2002; Alexeev et al. 2003; Jaskelioff et al. 2003). Rad54 and Rdh54 exhibit dsDNA-specific ATPase activity and translocate on dsDNA to generate unconstrained negative and positive supercoils (Mazin et al. 2000; Van Komen et al. 2000). Strand separation at unwound regions is expected to facilitate the search for homology between dsDNA and the incoming Rad51 nucleoprotein filament. Interestingly, Rad54 enables Rad51-dependent D-loop formation between ssDNA and linear duplex DNA, a reaction not observed for the Escherichia coli RecA protein (Wright and Heyer 2014). Since homologous pairing between recipient and donor sequences is still observed in vivo in rad54 mutants, it has been suggested that the primary function of Rad54 is postsynaptic rather than during the search for homology (Sugawara et al. 2003). Indeed, Rad54 is important to remove Rad51 from the end of strand invasion intermediates to permit access to DNA polymerases to extend the invading end (Li and Heyer 2009) (Figure 5). Rad54 and Rdh54 displace Rad51 from dsDNA and this could be important to remove unproductive association of Rad51 with dsDNA during presynapsis, thus increasing the pool of Rad51 available for HR, for turnover of Rad51 upon completion of recombination, or to uncover the 3′ end of paired intermediates to allow initiation of DNA synthesis (Solinger et al. 2002; Holzen et al. 2006; Shah et al. 2010). Overexpression of RAD51 in the rdh54 mutant results in the formation of toxic Rad51 foci on undamaged chromatin, suggesting Rdh54 is the major translocase to remove Rad51 in undamaged cells, whereas Rad54 acts on damaged chromatin (Shah et al. 2010).
Srs2 is a DNA helicase that plays both positive and negative roles in recombination. Srs2 mutants show elevated levels of spontaneous recombination, but in some DSB-induced recombination assays, the recovery of recombinants is reduced (Rong et al. 1991; Vaze et al. 2002). In vitro, Srs2 translocates on ssDNA, displacing Rad51 (Krejci et al. 2003; Veaute et al. 2003). The current view is that Srs2 prevents initiation of recombination events presynaptically by disrupting the Rad51 nucleoprotein filament, and this activity could be important to prevent unwanted recombination at replication forks, where Srs2 colocalizes with PCNA (Pfander et al. 2005; Burgess et al. 2009). A direct interaction of Srs2 with PCNA may also regulate DNA synthesis during HR to suppress formation of crossover products (Burkovics et al. 2013). Although Srs2 is able to disrupt D-loop intermediates in vitro, it is less effective than Mph1, and the pattern of hDNA products recovered from plasmid gap repair in the srs2 mutant is not consistent with a simple D-loop dissociation mechanism (Sebesta et al. 2011; Mitchel et al. 2013).
DNA synthesis during HR:
DNA synthesis is essential to extend the 3′ end within the D-loop and is likely to be required after strand displacement to fill gaps adjacent to the annealed sequences, replacing the nucleotides lost by end resection. Genetic studies suggest these two phases of DNA synthesis may use different polymerases. The reversion frequency of a marker located 300 bp from an HO cut site was higher during DSB repair than during normal growth, and the mutagenesis was largely DNA Polζ dependent (Holbeck and Strathern 1997; Rattray et al. 2002). Polζ-dependent mutagenesis of nearby genes has also been reported for other recombinogenic initiating lesions, such as inverted repeats, GAA repeats, and interstitial telomere repeats (Shah et al. 2012; Aksenova et al. 2013; Saini et al. 2013b; Tang et al. 2013). In contrast, another study demonstrated that the mutagenic DNA synthesis associated with gap repair is independent of the TLS polymerases, Polζ or Polη (Hicks et al. 2010). The DNA synthesized in the context of the D-loop appears to be carried out by DNA Polδ operating at much lower fidelity and processivity than during S-phase synthesis (Maloisel et al. 2008; Hicks et al. 2010). Consistent with these findings, the Polδ complex is able to extend D-loop intermediates generated in vitro by Rad51 and Rad54 (Li and Heyer 2009). It is possible that long tracts of ssDNA formed by end resection are subject to base modification and gap-filling synthesis by DNA Polζ causing mutations in sequences close to the DSB, but not in the context of D-loop synthesis.
Conditional alleles of essential replication genes have been used to determine the role of replication proteins during DSBR by physical monitoring of MAT switching. Gene conversion of the MAT locus is independent of ORC, the Cdc7–Dbf4 kinase, the MCM complex, Cdc45, DNA Polα, and Okazaki fragment processing proteins, but requires PCNA, Dpb11, and either Polδ or Polε (Wang et al. 2004; Germann et al. 2011; Hicks et al. 2011). In contrast to gene conversion repair of a two-ended DSB, BIR requires lagging strand as well as leading strand synthesis (Lydeard et al. 2007). The extensive DNA synthesis associated with BIR needs the nonessential subunit of the Polδ complex, Pol32, and is also compromised by the pol3-ct mutation, which affects stability of the Polδ complex (Lydeard et al. 2007; Deem et al. 2008; Payen et al. 2008; Smith et al. 2009; Brocas et al. 2010). The Pif1 helicase is required for BIR and in vitro studies show Pif1 functions with the Polδ complex to extend the 3′ end of a Rad51-generated D-loop (Saini et al. 2013a; Wilson et al. 2013). Pif1 facilitates extensive DNA synthesis by liberating the newly synthesized ssDNA to establish a migrating D-loop, in agreement with the current model for BIR synthesis (Figure 2). All three replicative DNA polymerases are required for BIR, but the need for Polε occurs later than for Polα and Polδ (Lydeard et al. 2007).
Resolution of recombination intermediates:
Resolution of D-loop intermediates by displacement of the extended invading strand is the primary mode of DSB repair in mitotic cells, at least for events initiated by endonucleases (Mitchel et al. 2010) (Figure 2). The Mph1 helicase dissociates Rad51-generated D-loop intermediates in vitro and mutants show increased levels of COs during DSB-induced recombination, in agreement with a role in promoting SDSA repair (Sun et al. 2008; Prakash et al. 2009; Tay et al. 2010; Sebesta et al. 2011; Lorenz et al. 2012; Mazon and Symington 2013; Mitchel et al. 2013). The increased COs observed in the mph1 mutant are dependent on MUS81 (Mazon and Symington 2013). Joint molecules, detected by two-dimensional agarose gel electrophoresis, accumulate transiently in the absence of MPH1, and persist at high levels in the mph1mus81 double mutant (Mazon and Symington 2013). Dissociation of D-loop intermediates is expected to have a negative impact on BIR and indeed overexpression of MPH1 reduces the frequency of BIR, while the mph1 mutant exhibits an increased BIR frequency (Luke-Glaser and Luke 2012; Stafa et al. 2014). The srs2 mutant also exhibits increased levels of COs associated with DSB-induced recombination, but, as noted above, the pattern of hDNA products observed is not consistent with D-loop dissociation.
The DSBR model predicts the formation of a dHJ intermediate, which must be resolved for segregation of the recombinant duplexes. The Sgs1–Top3–Rmi1 complex can resolve dHJ intermediates in vitro by a process called dissolution (Wu and Hickson 2003; Cejka et al. 2010b). The helicase activity of Sgs1 branch migrates the constrained HJs and the topoisomerase activity of Top3 is thought to remove the supercoils between the two HJs eventually leading to NCO products (Figure 2). Consistent with the in vitro studies, sgs1 and top3 mutants show increased levels of COs associated with spontaneous recombination and DSB-induced gene conversion (Wallis et al. 1989; Watt et al. 1996; Ira et al. 2003). Furthermore, joint molecules (JMs) containing a dHJ intermediate are detected at higher levels during DSB-induced interhomolog recombination in sgs1 diploid cells than in wild type (Bzymek et al. 2010).
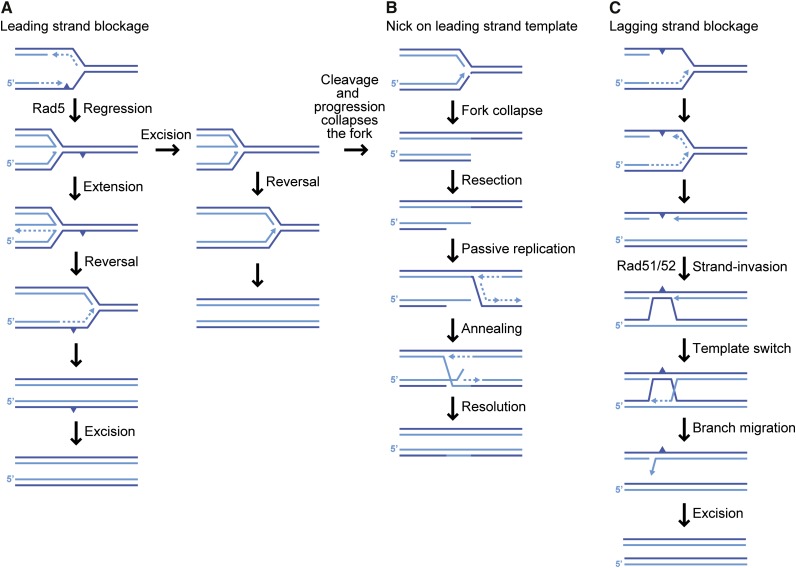
X-shaped JMs are detected in the highly repetitive ribosomal DNA (rDNA) locus during S-phase and their abundance increases when DNA Polα/primase is limiting (Zou and Rothstein 1997). Replication-dependent X-structures at unique sequences that are independent of Rad51 and Rad52 have been found in unperturbed cells. These structures are thought to be hemicatenanes due to their physical properties and resistance to cleavage by HJ resolvases in vitro (Lopes et al. 2003). Late forming replication-dependent X-structures accumulate in the absence of the Sgs1 helicase when cells are treated with MMS and in this case require HR functions, as well as the template-switching branch of postreplication repair for their formation, suggesting they are formed at ssDNA gaps as a means to bypass lesions (Figure 6) (Liberi et al. 2005; Branzei et al. 2008).
Figure 6.
Recombination at replication forks. Parental strands are shown in dark blue and nascent strands in light blue. Polymerase-blocking DNA lesion indicated by a filled triangle. (A) Error-free bypass of leading strand blockage. The replication fork stalled at a DNA lesion on the leading strand template may be regressed in a Rad5-dependent manner to expose the lesion for excision repair after which the regressed fork is reversed and replication resumed. Alternatively, leading strand synthesis may transiently switch templates within the regressed fork. Upon fork reversal and reanneling of the extended leading strand to its parental template, the DNA lesion is bypassed and can subsequently be repaired by excision repair. (B) Fork collapse and rescue by passive replication. Fork collapse may result if the replication fork encounters a nick on the leading strand template or if a regressed fork is endonucleolytically cleaved to form a one-ended DSB, which is most often rescued by passive replication from an adjacent replication fork that can anneal to the end and be resolved into two intact sister chromatids. (C) Error-free bypass of lagging strand blockage. Lagging strand synthesis can be completed by postreplicative recombination to reestablish strand continuity at the lesion using the nascent sister chromatid as a template. The remaining lesion on the parental lagging strand can subsequently be removed by excision repair.
The alternative means to remove HJ-containing intermediates is through endonucleolytic cleavage. Several structure-selective nucleases (Mus81–Mms4 heterodimer, Yen1 and Slx1–Slx4 heterodimer) have been shown to cleave branched DNA structures, including HJs, in vitro (Boddy et al. 2001; Kaliraman et al. 2001; Fricke and Brill 2003; Ip et al. 2008). Interestingly, mus81, mms4, slx1, and slx4 mutations were all identified on the basis of synthetic lethality with sgs1 (Mullen et al. 2001). Because Mus81–Mms4 (Eme1 in Schizosaccharomyces pombe) exhibits higher cleavage activity on D-loops and nicked HJs than intact HJs, Mus81–Mms4/Eme1 most likely processes an early strand exchange intermediate, prior to ligation to form a dHJ intermediate, to generate crossover products (Kaliraman et al. 2001; Osman et al. 2003; Mazon and Symington 2013; Mukherjee et al. 2014). Ho et al. (2010) found a significant decrease in the formation of DSB-induced CO products between homologs in the mus81 diploid and a greater decrease in the mus81yen1 double mutant, suggesting Mus81–Mms4 is the primary activity to resolve recombination intermediates with Yen1 serving as a back up function. The partial redundancy between these activities is also observed for DNA damage sensitivity, but Yen1 is not able to counteract the lethality of the mus81sgs1 mutant unless it is constitutively activated (Blanco et al. 2010; Ho et al. 2010; Tay et al. 2010; Matos et al. 2013). Surprisingly, COs between ectopic repeats are only reduced by 50% in the mus81yen1 double mutant (Agmon et al. 2011). Persistent JMs containing a single HJ (sHJ) connecting the ectopic sequences were identified in the mus81yen1 mutant, and their formation, as well as generation of CO products, was dependent on RAD1 (Mazon et al. 2012; Mazon and Symington 2013). However, Rad1–Rad10 has no apparent role in the formation of COs between chromosome homologs (Mazon et al. 2012). Rad1–Rad10 is proposed to facilitate ectopic CO formation by cleaving the leading edge of the captured D-loop at the heterology boundary creating a substrate for subsequent cleavage by Mus81–Mms4 or Yen1 (Mazon et al. 2012).
In contrast to mus81, the synthetic lethality of slx1 or slx4 with sgs1 is not suppressed by rad51 mutation, suggesting the lethality might be due to problems other than, or in addition to, unresolved recombination intermediates (Fabre et al. 2002; Bastin-Shanower et al. 2003). In vitro, the yeast Slx1–Slx4 complex preferentially cleaves 5′ flap structures; however, the human SLX1–SLX4 complex is reported to cleave intact HJs (Schwartz and Heyer 2011). The slx1 mutant has no obvious defect in the formation of mitotic or meiotic crossovers, however, in an slx4 mutant, spontaneous mitotic crossovers are reduced (Ho et al. 2010; De Muyt et al. 2012; Zakharyevich et al. 2012; Gritenaite et al. 2014).
Removal of heterologous flaps that can form during strand invasion or following strand annealing by the SDSA and SSA models requires the Rad1–Rad10 endonuclease, which cuts branched DNA structures at the transition between dsDNA and ssDNA (Fishman-Lobell and Haber 1992; Ivanov and Haber 1995; Mazon et al. 2012). The flap cleaving activity of Rad1–Rad10 requires Slx4, but not Slx1, and the mismatch repair proteins, Msh2 and Msh3 (Fishman-Lobell and Haber 1992; Ivanov and Haber 1995; Sugawara et al. 1997; Flott et al. 2007). Saw1, which was identified in a genome-wide screen for SSA defects, binds specifically to 3′ flap structures in vitro and recruits Rad1–Rad10 to ssDNA flaps in vivo (Li et al. 2013, 2008). Although Rad1–Rad10 is generally considered to be essential for heterologous flap removal, in an assay that detects chromosomal translocations formed by SSA, the rad1 defect was suppressed by rad51 (Manthey and Bailis 2010). This result raises the possibility that Rad51 binds to unrepaired ssDNA flaps and prevents access to nucleases other than Rad1–Rad10.
III. Genetic Assays of Mitotic Recombination
In this section, we describe the various assays that have been used over the years to both define mitotic recombination as well as to aid in the isolation of mutations in genes that affect the process. Further insight has been gained by the introducing of site-specific lesions in these assays, such as inserting the HO-cut site or site-specific nicking site (Nickoloff et al. 1986; Galli and Schiestl 1998; Cortes-Ledesma and Aguilera 2006; Nielsen et al. 2009). Concomitant expression of these nucleases has allowed researchers to induce DSBs and nicks to evaluate their role in many of these assays.
A. Allelic recombination in diploids
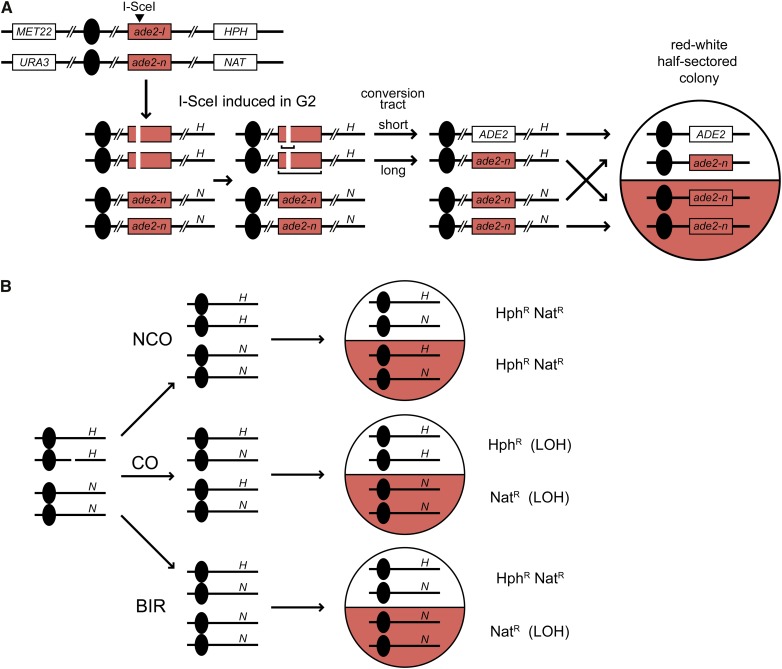
Allelic recombination refers to events that occur at allelic positions between homologous chromosomes. As shown in Figure 1C and Figure 7A, most recombination events occur in the G2 phase of the cell cycle and, although there is a strong preference for recombination between sister chromatids, numerous genetic studies have documented recombination between homologs. Conversion events can be selected by generation of a functional copy of a gene from different mutant alleles (heteroalleles). Use of heterozygous markers centromere (CEN) proximal to the recombining locus allows identification of associated crossovers by LOH, though half of the potential CO events are not detected because of random segregation of chromatids at mitosis (Figure 1C and Figure 7) (Chua and Jinks-Robertson 1991; Ho et al. 2010). The rate of spontaneous gene conversion is generally ∼10−6 events/cell/generation and 10–20% of events are associated with COs (Haber and Hearn 1985). Allelic recombination is stimulated by several orders of magnitude if cells are irradiated with IR or UV (Manney and Mortimer 1964; Esposito and Watsgaff 1981), or if one of the recombining loci has the recognition sequence for the HO or I-SceI rare-cutting endonucleases (Nickoloff et al. 1999; Palmer et al. 2003; Mozlin et al. 2008). The frequency of recombinants is sufficiently high when induced by a targeted DSB to analyze unselected events and to distinguish between a CO and BIR (Malkova et al. 1996; Ho et al. 2010) (Figure 7B).
Figure 7.
Assay for heteroallelic recombination. Nonfunctional ade2-I and ade2-n heteroalleles and wild-type ADE2 give rise to red and white colonies, respectively. (A) I-SceI-induced heteroallelic recombination. A DSB induced by the I-SceI endonuclease in the ade2-I allele in G2 can be repaired from the intact homolog by short-tract or long-tract gene conversion to give rise to ADE2 and ade2-n, respectively. Red-white half-sectored colonies are indicative of a recombination event that occurred in the first generation after plating. Markers MET22 and URA3 on the other side of the centromeres (filled circles) facilitate the scoring of chromosome nondisjunction events. Markers HPH and NAT adjacent to the ade2 locus facilitate the scoring of CO events. (B) Scoring CO, NCO, and BIR events associated with gene conversion. Genotyping of red-white half-sectored colonies with respect to the HPH (H) and NAT (N) markers described in panel A allows the distinction of CO and BIR events as reciprocal and nonreciprocal LOH, respectively. The remaining events are NCOs.
Because spontaneous gene conversion events occur at low frequency and only one recombinant daughter cell is selected, reciprocal COs cannot be distinguished from BIR. Petes and colleagues developed a clever genetic assay to detect spontaneous reciprocal crossovers that occur between CEN5 and the CAN1 locus (Barbera and Petes 2006). The rate of spontaneous reciprocal exchange is 4 × 10−5 within this 120-kb interval. By using haploid strains with 0.5% sequence divergence to create the diploid, single nucleotide polymorphisms (SNPs) were used to map the site of exchange between CEN5 and CAN1 and determine the length of the conversion tracts associated with crossovers (Lee et al. 2009; Lee and Petes 2010). Unlike meiotic recombination, which occurs at nonrandom positions, the mitotic crossovers were evenly distributed and the median length of conversion tracts was ∼12 kb, much longer than meiotic conversion tracts (Lee et al. 2009). Surprisingly, many of the conversion tracts associated with spontaneous crossovers showed 4:0 segregations of the heterozygous markers or hybrid tracts consisting of 3:1 adjacent to 4:0 segregations. Such events are best explained by the presence of a DSB in a G1 cell, replication of the broken chromosome and repair of the two broken sisters from the nonsister chromatids in G2, where one event would have to be associated with a CO to generate a sectored colony (Esposito 1978; Lee and Petes 2010).
The rate of spontaneous gene conversion is reduced >20-fold in rad51 and rad52 mutants, and by 5- to 10-fold in rad54 and rdh54 mutants as compared to wild type (Bai and Symington 1996; Klein 1997; Petukhova et al. 1999; Mortensen et al. 2002). In the absence of Rad52, aberrant products are recovered that are associated with chromosome loss and are thought to occur by a half crossover mechanism (Haber and Hearn 1985; Coic et al. 2008). Mutation of MRE11, RAD50, or XRS2 results in higher rates of allelic recombination in diploids, but a slight reduction in the recombination rate between repeats (Alani et al. 1990; Ajimura et al. 1993; Rattray and Symington 1995). Cohesin loading at DSBs and stalled replication forks is dependent on MRX, suggesting recombinogenic lesions are channeled from sister chromatids to homologs in the absence of this complex (Strom et al. 2004; Unal et al. 2004; Tittel-Elmer et al. 2012). Allelic recombination is also higher in rad59 diploids compared to wild type (Bai and Symington 1996). Rad59 physically interacts with the RSC chromatin-remodeling complex, which is required for cohesin loading at DSBs, and is important for sister chromatid recombination (Oum et al. 2011); thus, Rad59 may function to promote sister-chromatid recombination and in its absence, lesions might be channeled to nonsisters as suggested for the MRX complex.
B. Recombination between dispersed repeats
Ectopic or nonallelic homologous recombination (NAHR) refers to events occurring between homologous sequences located at nonallelic positions (Figure 1, E and F). Naturally occurring repeats (for example, the rDNA array and Ty elements) and artificial duplications have been used as substrates for ectopic recombination, mainly in haploid cells. The repeated sequences can be located on different chromosomes or within the same chromosome. Closely spaced repeats in the same orientation (tandem or nontandem) or inverted relative to each other generally exhibit higher rates of recombination than repeats present on different chromosomes (dispersed) (Liefshitz et al. 1995). To a first approximation, the rate of spontaneous recombination between dispersed repeats and its genetic control are more similar to allelic recombination than to direct or inverted repeat recombination, suggesting heterologous chromosomes interact as frequently as homologous chromosomes in mitotic cells (Lichten and Haber 1989). However, global analyses of the yeast genome three-dimensional organization by chromosome conformation capture indicate that some loci are more prone to contact than others and these restricted chromosome territories influence the frequency of ectopic recombination (Burgess and Kleckner 1999; Duan et al. 2010; Agmon et al. 2013). Reciprocal exchange between dispersed repeats yields chromosome translocations that can only be detected if the orientation of the repeats with respect to their centromeres is the same. About 10–50% of gene conversion events between dispersed repeats have an associated crossover (Jinks-Robertson and Petes 1986; Liefshitz et al. 1995; Robert et al. 2006). However, the frequency of associated COs is generally less for DSB-induced ectopic recombination (Inbar and Kupiec 1999; Ira et al. 2003).
Direct-repeat recombination:
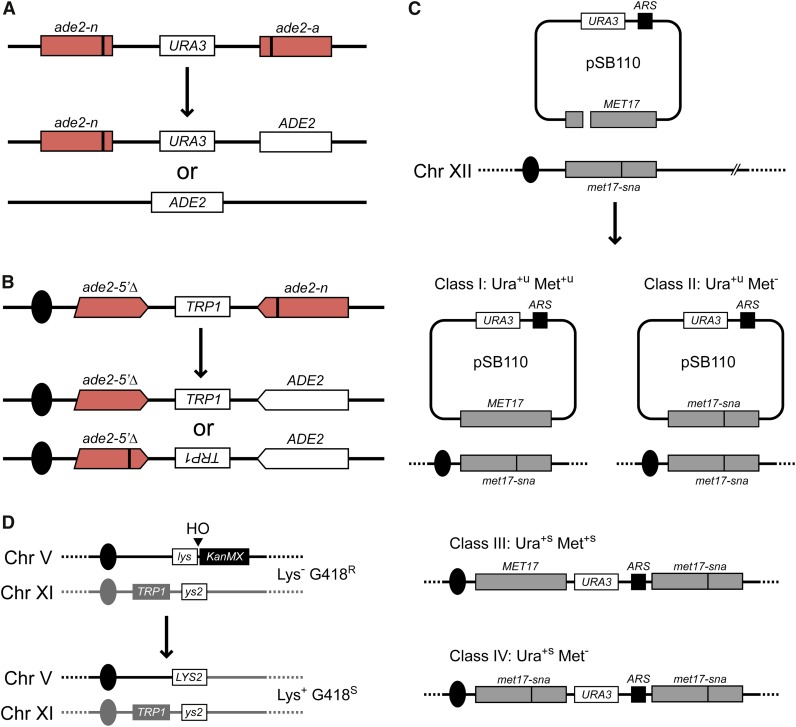
Gene conversion between direct repeats can be detected using heteroalleles of a selectable gene, and can occur by intrachromatid or sister-chromatid interactions (Figure 8A). Gene conversion between misaligned sister chromatids can generate a triplication or deletion on one chromatid while retaining the direct repeat on the other, whereas a crossover between misaligned sister chromatids generates triplication and deletion products (Klein 1988) (Figure 1B). Intrachromatid gene conversion associated with a crossover generates a chromosomal deletion and an episomal circular product. Although deletions are formed at high frequency between direct repeats, the reciprocal product is only associated with ∼7% of deletions, suggesting they arise primarily by a nonconservative mechanism (Schiestl et al. 1988; Santos-Rosa and Aguilera 1994).
Figure 8.
Genetic assays. (A) Direct-repeat recombination. Spontaneous homologous recombination between ade2-n and ade2-a alleles can occur by gene conversion to produce Ade+ Ura+ cells or by SSA to produce Ade+ Ura− cells (Fung et al. 2009). (B) Inverted repeat recombination. Inverted repeat recombination assays exclusively Ade+ recombinants arising from gene conversion since SSA will not produce viable recombinants. CO and NCO events will lead to inversion and noninversion of the TRP1 marker, respectively (Mott and Symington 2011). (C) Plasmid gap repair assay. The efficiency of plasmid–chromosome recombination, crossover frequency, and conversion tract length is assayed by transformation of the gapped pSB110 plasmid into yeast containing the chromosomal met17-sna mutant allele in which a SnaBI site is eliminated 216 bp downstream of the gap in the plasmid (Symington et al. 2000). When the plasmid gap is repaired by noncrossover gene conversion, the result is unstable (u) Ura+ transformants, which will be Met+ (class I) or Met− (class II), depending on the absence or presence of co-conversion of the met17-sna mutation, respectively. If the gene conversion event is associated with a crossover, the result is a stable (s) Ura+ phenotype (classes III and IV). ARS, autonomously replicating sequence. (D) Break-induced replication. In this assay, BIR is initiated by induction of an HO-mediated DSB adjacent to a 3′ truncated lys2 gene (lys) on chromosome V. The lys fragment has 2.1 kb of homology to a 5′ truncation of lys2 (ys2) close to the telomere on chromosome XI (Donnianni and Symington 2013), which serves as a donor for BIR. BIR results in deletion of the KanMX gene and all nonessential genes telomere proximal to the HO cut site and loss of G418 resistance (G418R). The strain has the MATa-inc allele to prevent cleavage at the endogenous HO cut site.
Gene conversion events between direct repeats that maintain the intervening sequence require RAD51, RAD54, RAD55, and RAD57, whereas deletions arise independently of these genes, consistent with their formation by SSA (McDonald and Rothstein 1994; Liefshitz et al. 1995; Petukhova et al. 1999). By contrast, rad52 mutants exhibit reduced frequencies of conversion and deletion events (Klein 1988; Thomas and Rothstein 1989b) likely due to the role of Rad52 as a Rad51 mediator and as a strand annealing protein, respectively.
The ribosomal genes in yeast naturally occur as a multiple tandem array (150–200 copies). Recombination within and between the repeats is important to maintain homogeneity of the cluster and is restricted to mitosis (Petes and Botstein 1977). Repeat homeostasis of rDNA is measured by determining the repeat length of the native array, which reveals either gain or loss of repeats. Recombination between repeats can also generate rDNA circles and the accumulation of rDNA circles in mother cells has been implicated in cellular aging (Sinclair and Guarente 1997; Kobayashi 2008). Due to their repetitive nature, rDNA have been excellent substrates for studying recombination. The insertion of selectable and counterselectable markers allows measurements for the effects of mutations on recombination outcomes—Rad51 is necessary for both deletion and duplication of the marker and Rad52 is necessary for duplication (Szostak and Wu 1980; Gangloff et al. 1996). In addition, the multiple copies of rDNA permit the detection of recombination intermediates after two-dimensional gel electrophoresis (Brewer and Fangman 1988; Zou and Rothstein 1997). Circles of rDNA are formed at high rates in mutants that are defective in DNA topoisomerases Top1 and Top2, indicating that regulating DNA topology is important for the stability of the array (Christman et al. 1988; Kim and Wang 1989). In addition, mutation of Top3, a eukaryotic type IA topoisomerase, also leads to increased rDNA instability (Wallis et al. 1989).
Importantly, to maintain sequence homogeneity, there is a genetic system in place that ensures recombination between the rDNA repeats. This system, controlled by the Fob1 replication block protein, is thought to help avoid collisions between transcription of rDNA and the bidirectional DNA synthesis that can be potentially initiated in every repeat (Takeuchi et al. 2003). At each repeat, Fob1 binds and inhibits DNA synthesis in one direction so that there are very few replication/transcription collisions. However, it is likely that the accumulation of blocked forks actually increases the amount of recombination that takes place naturally within this array, since in the absence of Fob1 protein, spontaneous recombination is reduced fivefold (Defossez et al. 1999). The increased recombination between the repeats stimulates homogenization of this multiple tandem array. The recent finding that spontaneous chromosomal fragile sites in yeast are enriched for motifs that correlate with paused replication forks supports the view that the Fob1 sites are playing this role in the rDNA array (Song et al. 2014).
Inverted-repeat recombination:
Substrates with inverted repeats were designed to avoid formation of recombinants by SSA (Figure 8B). Gene conversion between inverted repeats retains the original configuration, whereas intrachromatid gene conversion associated with a CO or long tract conversion between misaligned sister chromatids results in inversion of the intervening sequence (Rothstein et al. 1987; Rattray and Symington 1994; Chen et al. 1998). Spontaneous recombination between inverted repeats is highly dependent on RAD52, but reduced only 5- to 10-fold in the rad51 mutant (Dornfeld and Livingston 1992; Rattray and Symington 1994; Gonzalez-Barrera et al. 2002). DSB-induced recombination between chromosomal inverted repeats is reduced by >1000-fold in the rad51 mutant, but plasmid-borne inverted repeats exhibit less of a requirement for RAD51 (Gonzalez-Barrera et al. 2002; Ira and Haber 2002; Rattray et al. 2002). RAD59 is required for spontaneous RAD51-independent events, suggesting they occur by a strand annealing mechanism, possibly by template switching during DNA replication (Bai and Symington 1996; Gonzalez-Barrera et al. 2002; Mott and Symington 2011). The DSB-induced RAD51-independent events in the plasmid context might occur by BIR and SSA or by some other type of nonconservative event (Kang and Symington 2000; Ira and Haber 2002).
Naturally occurring inverted repeats, such as Ty elements and delta sequences, are also substrates for spontaneous recombination that can lead to genome instability and gross chromosomal rearrangements (Rothstein et al. 1987; Argueso et al. 2008; Casper et al. 2009; Paek et al. 2009; Chan and Kolodner 2011). Many of these events are dependent on DSB repair pathways; however, notably, fusion of inverted repeats that lead to chromosome rearrangements are replication dependent (Paek et al. 2009). The rearrangements involving Ty elements and delta sequences are almost completely dependent on the RAD51 and RAD52 gene products (Rothstein et al. 1987; Liefshitz et al. 1995).
C. Plasmid gap repair
Transformation-based assays using plasmids linearized in vitro have been used extensively to study the mechanism and genetic control of DSB repair (Orr-Weaver et al. 1981, 1983; Bartsch et al. 2000; Mitchel et al. 2010; Tay et al. 2010). A plasmid containing a DSB or double-stranded gap within sequences that have homology to a chromosomal locus are introduced to cells by transformation and repair of the DSB or gap is templated by the homologous chromosomal sequence. In most assays, a second marker on the plasmid is used to select for transformants. In some respects, plasmid–chromosome recombination resembles ectopic recombination between dispersed repeats because both involve limited homology. If the plasmid contains no origin of replication, then only CO recombinants (integration of the plasmid at the chromosomal locus) are recovered; however, if the plasmid has an origin to allow stable maintenance as an episome then both NCO and CO products can be detected (Figure 8C). Use of a CEN ARS vector restricts events to NCOs (Bartsch et al. 2000). The frequency of COs recovered from an ARS-containing plasmid varies between assays, ranging from 20 to 50%, significantly higher than observed for DSB-induced chromosomal ectopic recombination (Orr-Weaver and Szostak 1983; Inbar and Kupiec 1999; Bartsch et al. 2000; Ira et al. 2003; Welz-Voegele and Jinks-Robertson 2008). Use of a plasmid substrate with SNPs located ∼100 bp apart to detect hDNA intermediates that persist in a mismatch repair defective background revealed that most NCO products formed by SDSA and only a few events were diagnostic of dHJ dissolution. Furthermore, the CO products were most consistent with resolution of a sHJ intermediate (Mitchel et al. 2010).
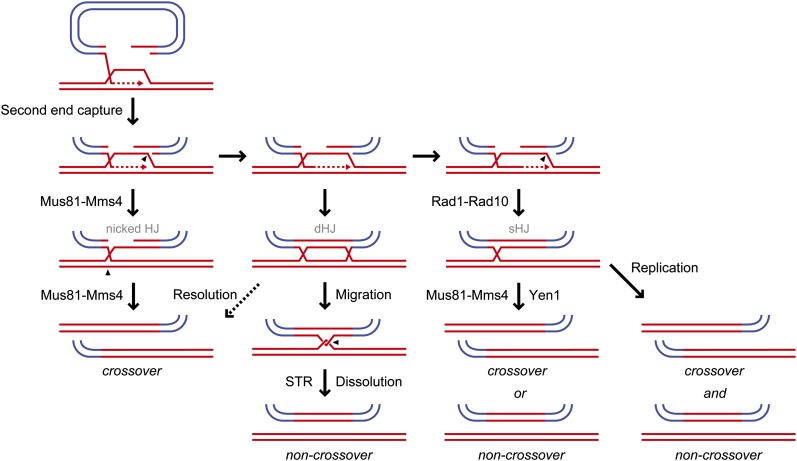
Early studies showed an essential role for RAD52 in plasmid gap repair, and subsequently, rad51, rad55, and rad57 mutations were shown to reduce the frequency of gap repair by >50-fold (Orr-Weaver et al. 1981; Bartsch et al. 2000). Elimination of RAD1 reduces integration of an ARS-containing plasmid by 5- to 10-fold, whereas mus81 and yen1 mutations do not decrease integration. These confusing data were rationalized by studies showing the RAD1-dependent accumulation of a sHJ intermediate between ectopic sequences in the mus81yen1 mutant leading to the hypothesis that Rad1–Rad10 clips the D-loop intermediate when it encounters the heterology barrier creating a sHJ intermediate linking the plasmid to the chromosome; replication through the sHJ would then generate CO and NCO products (Mazon et al. 2012; Mazon and Symington 2013). This model explains the pattern of hDNA observed in CO recombinants and also the high frequency of plasmid integration (Figure 9).
Figure 9.
Model for the role of endonucleases in the resolution of recombination intermediates. Invasion by a 3′ end of a gapped vector into a chromosomal donor sequence generates a D-loop, which is extended by DNA synthesis. Initially, second end capture results in a structure that is a potential substrate for Mus81–Mms4 cleavage to produce a nicked HJ and subsequently a CO upon further cleavage. If, on the other hand, the captured D-loop is gap filled and ligated, a dHJ is formed, which in most cases is converted to a hemicatene and dissolved by STR to yield a NCO, but could also be resolved by Mus81–Mms4 to produce a CO or NCO. Alternatively, if resection and DNA synthesis proceed beyond the heterology boundary, the D-loop can branch migrate to create a region of single-stranded DNA adjacent to the branch point, which could be cleaved by Rad1–Rad10 to generate a single HJ (sHJ). The sHJ can be resolved by either Mus81–Mms4 or Yen1 cleavage to produce a CO or NCO, or converted to a CO and a NCO product during the next S phase.
D. Assays for BIR
BIR is most easily studied by creating a DSB where just one of the two ends can undergo homology-dependent strand invasion. Telomeres are a natural source of one-ended DSBs and maintenance of telomeres in the absence of telomerase provides a convenient genetic assay for BIR (see Wellinger and Zakian 2012 for review). Cells senesce in the absence of telomerase but survivors can arise by Rad52- and Pol32-dependent recombination (Lundblad and Blackburn 1993; Lydeard et al. 2007). Two pathways for generation of survivors have been defined: type I survivors are due to amplification of the Y′ subtelomeric repeats, have very short telomere repeat tracts, and are dependent on Rad51, Rad52, Rad54, Rad57, and Pol32; type II survivors have long heterogeneous telomere tracts and require the MRX complex, Pol32, Rad52, Rad59, and Sgs1 for their formation (see Wellinger and Zakian 2012).
To force repair of a chromosome-internal DSB by BIR, most assays restrict homology to only one side of the DSB to prevent gene conversion repair. Several systems have been developed using HO to create a chromosomal DSB, and a transformation-based system utilizing linear plasmid vectors has also been described (Morrow et al. 1997; Bosco and Haber 1998; Davis and Symington 2004; Malkova et al. 2005; Lydeard et al. 2007; Donnianni and Symington 2013). Malkova and colleagues use a haploid strain disomic for chromosome III to study BIR (Malkova et al. 2005; Deem et al. 2011; Saini et al. 2013a). The MATa-inc allele, which is refractory to HO cleavage, is present on one homolog and the HO-induced DSB on the other is forced to repair by BIR due to deletion of homology on the CEN-distal side of the DSB. Heterozygous markers present on both chromosome arms are used to differentiate between BIR, chromosome loss, and half COs. Two systems to study BIR in haploids make use of truncated partially overlapping fragments of the CAN1 or LYS2 gene to regenerate a wild-type copy of the gene by BIR (Lydeard et al. 2007; Donnianni and Symington 2013). The recipient cassette has an HO cut site between at the border of homology and a selectable marker located in a nonessential region of chromosome V; the donor cassette is located on another chromosome. After induction of the DSB, the recipient sequence invades the donor copying to the end of the chromosome, and the nonessential sequences CEN-distal to the DSB are lost (Figure 8D). Although BIR was originally reported to be very slow by physical monitoring assays, when the donor is close to the telomere and only 15–20 kb of DNA needs to be synthesized, the efficiency and kinetics are similar to other ectopic recombination assays (Jain et al. 2009; Donnianni and Symington 2013). Furthermore, cells synchronously released from G1 show a higher BIR efficiency than cells arrested in G2/M at the time of DSB induction.
BIR has also been proposed to explain Fob1-stimulated recombination in rDNA (Kobayashi et al. 1998). Strand invasion events occurring at the matching repeat on the sister chromatid preserves copy number while those occurring at unmatched repeats cause rDNA copy number changes. Cohesin restricts recombination to matched repeats and is regulated by transcription of non-coding RNA sequences near the replication fork block (Kobayashi and Ganley 2005). Transcription of these RNAs is regulated by the Sir2 histone deacetylase, explaining the increased rates of rDNA recombination observed in sir2 mutant cells (Gottlieb and Esposito 1989; Kobayashi and Ganley 2005).
Although most of the genetic requirements for BIR are similar to other recombination reactions, POL32, which encodes a nonessential subunit of DNA Polδ, and PIF1 are required for BIR but not for short tract gene conversion (Lydeard et al. 2007; Deem et al. 2008; Smith et al. 2009; Saini et al. 2013a; Wilson et al. 2013; Stafa et al. 2014). Interestingly, half crossovers are recovered at high frequency from pol3-ct, pol32, and pif1 mutants (Deem et al. 2008; Smith et al. 2009; Saini et al. 2013a; Wilson et al. 2013), consistent with defective extension of the invading strand followed by cleavage of the D-loop intermediate. Half crossover products are also elevated in checkpoint mutants in agreement with the role of the DNA damage checkpoint in suppressing activation of structure-selective nucleases (Vasan et al. 2014).
IV. The Nature of the Recombinogenic DNA Lesion
This section describes the current thinking on the nature of the DNA lesion that results in defined recombination events. Many of our views on the exact kind of lesion that occurs have been influenced by studies of defined site-specific recombination events and the analysis of recombination mutants. Mating type switching is the “poster child” for this kind of study, having contributed greatly over the years to our understanding of the process.
A. Induction of recombination by DSBs and nicks
As described above, DSBs made by endonucleases serve as potent initiators of recombination. Furthermore, treatment of cells with ionizing radiation or radiomimetic drugs stimulates mitotic recombination. Testing the role of nicks as recombination initiators is more difficult because they can be healed by direct ligation, and any stimulation observed could be due to conversion to a DSB during replication (Figure 6). Insertion of the bacteriophage f1 gene II nick site between trp1 and his3 heteroalleles was shown to stimulate interchromosomal recombination when the gene II protein was expressed. Interestingly, there was a bias favoring conversion of the marker to the 5′ side of the nick site, in contrast to DSB initiated events that stimulate bidirectionally from the break site (Strathern et al. 1991). Conversion of the nick to a one-ended DSB during S-phase could account for the directionality of gene conversion observed. Aguilera and colleagues have shown a minimal HO site of 21 bp is cut by HO on one strand more frequently than on both strands, and the resulting nicks are converted to DSBs as cells transition through S-phase (Cortes-Ledesma and Aguilera 2006). Although the frequency of cutting is lower than at the optimal HO cut site, it is sufficient to detect intermediates by physical methods. This system has proven extremely useful to study replication-associated DSBs and their repair by sister-chromatid recombination (Gonzalez-Barrera et al. 2003; Cortes-Ledesma and Aguilera 2006).
Studies with DNA damaging agents are used to gain insight in how particular lesions behave in different assays (reviewed in Kupiec 2000). For example, γ-rays and UV induce mainly DSBs or single-stranded gaps (Ma et al. 2013), respectively, while CPT leads to covalently bound topoisomerase I to the 3′ phosphate end at a nick (Pommier 2009). Replication of the unremoved adduct leads to a DSB in the next round of replication. In addition, the absence of topoisomerases I, II, or III themselves leads to increased recombination, especially in the rDNA multiple tandem array as discussed above. In these cases, it is thought that the absence of the topoisomerase causes topological problems such as catenanes and hemicatenates resulting in broken chromosomes during mitosis.
B. Replication-coupled recombination
Most spontaneous recombination is thought to occur during DNA replication, when the replisome encounters various challenges, such as reduced dNTP levels, DNA adducts on the template strand, DNA secondary structures, or tightly bound proteins that can stall the replication fork, and these impediments occasionally cause fork collapse resulting in a DSB (reviewed in Aguilera and Gomez-Gonzalez 2008). Fork collapse is thought to result from the replication fork running into a transient nick on the template strand, by fork reversal (pairing of the nascent strands) leading to the formation of Holliday junction or “chicken-foot” structure that can be cut by a HJ endonuclease or direct cleavage of the stalled fork by structure-selective nucleases, such as Mus81–Mms4 or Slx1–Slx4 (Figure 6). Fork regression requires Rad5 to facilitate either immediate excision repair or limited extension of the leading strand using the nascent lagging strand as a template (Sogo et al. 2002; Cotta-Ramusino et al. 2005; Blastyak et al. 2007), known as “template switching” (Higgins et al. 1976), followed by fork reversal and postreplicative excision repair (Figure 6A). Instead of fork reversal, the regressed fork may be cleaved, causing fork collapse (reviewed in Atkinson and McGlynn 2009). The one-ended DSB generated by fork collapse could be rescued by replication from an adjacent origin (Figure 6B). DNA adducts that block progression of the replicative polymerases result in ssDNA gaps on both lagging and leading strands, which can be acted on by TLS polymerases or by recombination (Figure 6C).
Most recombinogenic lesions formed during S-phase are expected to be repaired by sister-chromatid recombination; however, some lesions must be repaired by a nonsister to account for the increased interchromosomal recombination observed for mutants with replication defects or after treatment of cells with agents that stall replication, such as UV. These events are initiated from either ssDNA gaps or DSBs. Fabre and colleagues have argued against DSBs as the spontaneous recombination initiating lesion on the grounds that rad52yku70 double mutants (deficient for both HR and NHEJ) are viable, yet the srs2sgs1 double mutant is inviable but rescued by loss of HR function, suggesting lethal spontaneous recombination intermediates occur at high frequency (Fabre et al. 2002). Furthermore, certain rad52 hypomorphic alleles confer high sensitivity to IR but are hyper-rec for spontaneous interchromosomal recombination (Lettier et al. 2006). Recently, whole chromosome analysis has revealed that single-stranded gaps are the intermediates of recombinational repair after UV irradiation (Ma et al. 2013). On the other hand, the pattern of conversion tracts associated with spontaneous mitotic crossovers is most compatible with initiation of recombination by a DSB present in a G1 cell (Lee and Petes 2010). Furthermore, analysis of conversion tracts associated with UV-induced mitotic crossovers showed they were similar to spontaneous events and those resulting from gamma-irradiation of G1 diploids (St Charles et al. 2012; Yin and Petes 2013).
C. Fragile sites and noncanonical structures
Fragile sites were first defined in mammalian cells as sequences that show gaps and breaks following inhibition of DNA synthesis and are associated with hotspots for genome rearrangements. Reducing the levels of DNA Polα or Polδ created a fragile site on yeast chromosome III detected by chromosome rearrangements between a pair of inverted Ty elements and other Ty elements located on other chromosomes (Lemoine et al. 2005, 2008). DSBs were detected at the inverted Ty elements when replication was compromised, analogous to mammalian fragile sites (Lemoine et al. 2005). Moreover, this fragile site experiences a high frequency of spontaneous BIR events leading to LOH on the right arm of chromosome III (Rosen et al. 2013).
Fragile sites are usually associated with DNA sequences that are difficult to replicate and prone to form secondary structures, such as certain trinucleotide repeats (TNRs), AT rich sequences, inverted repeats, and sequences with the potential to form G quadruplexes. Moreover, genome-wide mapping of fragile sites revealed a nonrandom distribution correlating with motifs that pause DNA replication forks, including replication-termination sites and binding sites for the helicase Rrm3 (Song et al. 2014). Insertion of inverted Alu elements, but not Alu direct repeats, was shown to stimulate ectopic recombination by 1000-fold in an MRX- and Sae2-dependent manner (Lobachev et al. 2002). A chromosomal DSB induced by the inverted Alu elements was detected by PFGE and shown to be hairpin capped in the absence of the MRX complex, the Mre11 nuclease, or Sae2. It was postulated that the Alu elements extrude into a cruciform, and the base, resembling a HJ, is cleaved by a structure-selective nuclease converting it into two hairpin-capped ends that must be opened by MRX and Sae2 to initiate recombination.
TNRs that are capable of forming hairpins when present in ssDNA show orientation-dependent replication fork stalling, increased chromosome fragility, and contractions and expansions when present on the lagging strand template (Freudenreich et al. 1997; Miret et al. 1998). Insertion of a long TNR tract between CEN5 and CAN1 was shown to stimulate mitotic crossovers by 30-fold, but the local stimulation was much greater and the majority of crossovers and conversion tracts were close to the TNR tract (Tang et al. 2011).
G-quadruplex (G4) DNA induces recombination by interfering with both replication and transcription. Interestingly, G4-induced recombination is observed only when the G-rich strand is the template for leading strand synthesis (Lopes et al. 2011) or when transcription of the G-rich DNA is oriented with the C-rich strand as the transcription template (Kim and Jinks-Robertson 2011). The effect of transcription orientation was enhanced in the absence of the type IB topoisomerase Top1, possibly due to enhanced R-loop formation (Kim and Jinks-Robertson 2011). Pif1 unwinds G4 structures in vitro and prevents replication fork stalling and DNA breakage at G4 motifs in vivo (Paeschke et al. 2011), which likely explains its suppression of recombination triggered by G-quadruplex forming tandem repeats (Ribeyre et al. 2009). Finally, Mre11 binds and cleaves G4 DNA in vitro (Ghosal and Muniyappa 2005).
D. Transcription-stimulated recombination
Early studies searching for hotspots of genetic recombination showed that promiscuous transcription by RNA polymerase I stimulates mitotic recombination (Keil and Roeder 1984; Voelkel-Meiman et al. 1987). In addition, a high level of transcription by RNA polymerase II resulted in increased repeat recombination (Thomas and Rothstein 1989a,b; Saxe et al. 2000). Topological changes induced by transcription may be responsible for creating recombinogenic lesions since mutations in topoisomerase I and II also lead to increased recombination especially in rDNA (Christman et al. 1988; Kim and Wang 1989; Wallis et al. 1989; El Hage et al. 2010). Studies of the THO/TREX complex indicate that an increased frequency of R-loop formation during transcription is likely the cause of transcription-stimulated recombination (Huertas and Aguilera 2003). In many cases, increased recombination can be suppressed by overexpressing RNaseH, which preferentially removes the RNA from the DNA–RNA hybrid (Huertas and Aguilera 2003; El Hage et al. 2010). Interestingly, it was recently reported that the Rad51 protein is involved in the formation of RNA–DNA hybrids and that Srs2 normally counteracts their potential for genome instability (Wahba et al. 2013). Although the precise lesion involved in stimultating recombination is not known, multiple lines of evidence suggest that the intermediate is a DSB (reviewed in Aguilera and Garcia-Muse 2012). Recently, the DNA damage checkpoint has been linked to transcription-associated R-loops that impede DNA replication (Bermejo et al. 2011). It is thought that highly transcribed genes associate with the nuclear periphery to aid in RNA export. When a replication fork is encountered head on with the transcribed gene, the resulting collision collapses the fork. This action activates the DNA damage checkpoint to release the transcription/replication unit from the nuclear pore to allow relief of topological stress.
V. Cell Biology of Recombination
Most recombination proteins can be expressed as functional fusions to genetically encoded fluorescent proteins such as GFP and mCherry (Lisby et al. 2004; Silva et al. 2012), which allows for the dynamic redistribution of these proteins to be monitored in real-time at the single-cell level during homologous recombination.
A. Recombination foci
Most homologous recombination proteins are recruited in many copies to the site of DNA damage during repair. The high local concentration of recombination proteins at the site of DNA damage can be visualized by fluorescence microscopy after immunostaining or by GFP-tagging of the proteins (Lisby et al. 2004; Eckert-Boulet et al. 2011; Silva et al. 2012). For example, a single DNA DSB is sufficient for the formation of a prominent focus containing 600–2100 molecules of Rad52 yielding a ≥50-fold higher local concentration of Rad52 at the DSB relative to the diffuse nuclear distribution in undamaged cells (Lisby et al. 2003b). Although the minimum number of Rad52 molecules required for mediating a single strand invasion is currently unknown, the high local concentration of recombination proteins within these foci may allow constitutively expressed proteins to be active only at the site of DNA damage, and therefore prevent untimely recombination or assembly of recombination complexes at undamaged DNA.
Recombination foci are highly dynamic in their protein composition and localization. Foci can assemble and disassemble within minutes. However, studies of Rad51, Rad52, and Rad54 foci in mammalian cells indicate that the residence time of individual molecules may vary between proteins and even subpopulations of proteins within foci (Essers et al. 2002). So far the dynamics of proteins within individual recombination foci has not been studied in yeast.
Although focus formation of recombination and checkpoint proteins is a useful tool for monitoring the cellular response to DSBs and a single DSB is sufficient to trigger focus formation (Lisby et al. 2003b), it is likely that some recombination events go undetected by this methodology. For example, recombinational restart of stalled replication forks, some sister chromatid events and intramolecular recombination may be too fast or require too few molecules of recombination proteins to be detected by current techniques.
B. Choreography of focus formation
HR starts with the recruitment of MRX, which binds directly to DNA ends (Chen et al. 2001; Hopfner et al. 2001; Lisby et al. 2004; Mimitou and Symington 2010). The two ends of a DSB are held together by a mechanism that is partially dependent on MRX and Sae2 (Chen et al. 2001; Lisby et al. 2003a; Kaye et al. 2004; Lobachev et al. 2004; Clerici et al. 2005). For this reason, the two ends of a DSB give rise to a single Mre11 focus rather than two foci. Further, the MRX complex interacts with the Tel1 kinase and is required for its recruitment to foci at all phases of the cell cycle (Nakada et al. 2003; Lisby et al. 2004) (Figure 10). The Tel1 kinase phosphorylates histone H2A, which is a chromatin mark specific for damaged DNA in most eukaryotes (Rogakou et al. 1998, 1999; Redon et al. 2003). Importantly, the modification of chromatin by H2A phosphorylation facilitates binding of the checkpoint adaptor Rad9 to sites of DNA damage likely through a dual interaction of its BRCT domains with H2A-S129P and its Tudor domain with histone H3 methylated at lysine 79 (H3-K79Me) leading to subsequent recruitment and activation of Rad53 (Giannattasio et al. 2005; Javaheri et al. 2006; Toh et al. 2006; Grenon et al. 2007; Hammet et al. 2007; Germann et al. 2011). Notably, Rad53 foci are faint and transient, which is consistent with the notion from mammalian cells that Rad53/CHK2 must redistribute from the site of DNA damage upon phosphorylation to mediate a pannuclear checkpoint response (Lukas et al. 2003).
Figure 10.
Choreography of HR focus assembly. (A) Focus formation of HR proteins. The high local concentration of Rad52 and Rad59 at DSBs induced by treatment with 200 µg/ml zeocin for 2 hr at 25°. Strain NEB110-25B is a MATa haploid containing RAD52-CFP and RAD59-YFP. Arrowheads mark foci. (B) Order of assembly of HR proteins at foci. Proteins are recruited from the left to right starting with MRX binding at DSB ends and later replaced by proteins recruited to ssDNA at resected DSBs.
The proteins involved in resection have very different focal appearances giving clues to their function and regulation. Mre11 and Sae2 form prominent foci at all phases of the cell cycle in response to DSBs (Lisby et al. 2004; Barlow et al. 2008). Dna2 shuttles between the nucleus and cytoplasm in a cell-cycle-dependent manner, residing in the cytoplasm during G1 phase and relocalizing to the nucleus in S/G2 upon phosphorylation by Cdc28 (CDK) (Kosugi et al. 2009; Chen et al. 2011). Dna2 forms Rad52-colocalizing foci after DSB formation (Zhu et al. 2008). Sgs1 is a low abundance nuclear protein, which forms foci in S/G2/M (Frei and Gasser 2000 and M. Wagner, personal communication). Exo1 levels are cell cycle regulated gradually increasing through G1 and peaking in late S/G2 phase before it is degraded in anaphase (M. Lisby, unpublished data). Resection is accompanied by the dissociation of MRX, Sae2, and Tel1 from the DSB and binding of RPA to the 3′ single-stranded overhangs (Figure 10) (Lisby et al. 2004; Barlow et al. 2008). The intensity of Rfa1 foci can be used to estimate the extent of resection. This approach was used to demonstrate at the single-cell level that the rate of DSB end resection increases at the G1–S transition (Barlow et al. 2008). RPA is necessary for recruiting a number of checkpoint and HR proteins including the Dna2, Mec1–Ddc2/Lcd1, Rad24–RFC and 9–1–1 (Ddc1–Mec3–Rad17) complexes. Notably, Tel1 and Mec1 have many of the same phosphorylation targets, including histone H2A. As a consequence, Tel1-dependent checkpoint signaling is likely replaced by Mec1-dependent signaling upon resection of DSB ends. Consistent with a functional crosstalk between the Ddc2–Mec1 and 9–1–1 complexes during checkpoint signaling, there is a reported requirement for the 9–1–1 complex to stabilize Ddc2 foci in irradiated G1 cells (Barlow et al. 2008). Further, in S and G2 phases, the 9–1–1 complex and the Cdc28 kinase both contribute to the stabilization of DNA damage-induced Ddc2 foci (Barlow et al. 2008), demonstrating how checkpoint signaling is coordinated with cell cycle phase. The multifunctional Dpb11 protein is recruited to foci by the 9–1–1 complex, reflecting its role in mediating the DNA damage checkpoint through activation of the Mec1 kinase (Puddu et al. 2008; Germann et al. 2011). In contrast, the DNA replication and recombination functions of Dpb11 are independent of focus formation (Germann et al. 2011).
In S and G2 phases, RPA facilitates the recruitment of Rad52 to resected DSBs, likely via a direct physical interaction (Hays et al. 1995; Lisby et al. 2001, 2004; Plate et al. 2008). The recruitment is independent of DNA replication and requires B-type cyclin/Cdc28 activity (Barlow and Rothstein 2009). However, the cell cycle regulation of Rad52 focus formation can be circumvented at high doses of ionizing radiation at which Rad52 also forms foci in G1 phase, although it is unknown if these foci are productive for recombination (Lisby et al. 2003a). Rad52 interacts with the Rad51 recombinase and Rad59 to recruit these proteins to foci (Milne and Weaver 1993; Davis and Symington 2003; Lisby et al. 2004). In addition, Rad59 also requires Rad52 for its nuclear accumulation (Lisby et al. 2004). However, recent data indicate that Rad59 has Rad52-independent functions, indicating that some Rad59 enters the nucleus in a rad52 mutant (Coic et al. 2008; Pannunzio et al. 2008; Pannunzio et al. 2012). Rad51 foci form, but are dimmer in the absence of Rad55–Rad57, consistent with the role of the Rad51 paralogs to stabilize Rad51 filaments. Interestingly, Rad55 focus formation requires Rad51 (Lisby et al. 2004; Fung et al. 2009). Formation of DNA damage-induced foci by Rad54 requires both Rad55–Rad57 and Rad51, suggesting that Rad54 recruitment to the site of DNA damage requires Rad51 nucleoprotein filament formation (Lisby et al. 2004). Interestingly, the Rad54 homolog, Rdh54, is recruited both to DSBs and to the kinetochore, although the functional significance of this dual localization is unknown. The recruitment of Rdh54 to DSBs is Rad52- and Rad51 dependent, while its localization to the kinetochore is independent of the recombination machinery. Interestingly, Rad54, which does not localize to the kinetochore in wild-type cells, localizes to the kinetochore in an rdh54 mutant (Lisby et al. 2004), possibly explaining some of the functional redundancy between these two proteins (Shinohara et al. 1997).
Additional proteins that are recruited to recombination foci include the Pif1 helicase, which forms Rad52-colocalizing foci (Wagner et al. 2006), and the Srs2 helicase and antirecombinase, which is recruited to two distinct classes of foci (Burgess et al. 2009). During S phase, Srs2 is recruited to sumoylated PCNA speckles, and in late S/G2 Srs2 is recruited to recombination foci marked by Rad52. The recruitment of Srs2 to recombination foci is independent of its SUMO-interacting motif (Burgess et al. 2009). The Mus81–Mms4 structure-selective endonuclease forms foci, which are largely dependent on Rad54, consistent with Mus81–Mms4 acting downstream of the strand-invasion step of homologous recombination (Matulova et al. 2009). The SUMO-targeted ubiquitin ligase Slx5–Slx8 forms foci that partially overlap with Rad52 and Rad9 foci in response to DNA damage (Cook et al. 2009).
C. DSB dynamics and recombination centers
Work in both haploid and diploid yeast cells has found that, after DNA damage, the volume of the nucleus explored by the broken chromosome more than doubles from that seen in the absence of DSBs (Dion et al. 2012; Mine-Hattab and Rothstein 2012). The dynamics of unbroken chromosomes also increase depending on the number of DSBs (Mine-Hattab and Rothstein 2012; Seeber et al. 2013). The pairing of the homologs in diploid cells takes ∼20 min before the repair center disassembles and the loci separate again (Mine-Hattab and Rothstein 2012). These studies suggest that increased chromosomal mobility facilitates the homology search, which is otherwise restricted by the proximity of donor and recipient loci (Agmon et al. 2013). Genetics and cell biological studies, in both haploid and diploid yeast cells, showed that increased DNA mobility depends on the Rad51 recombinase (Dion et al. 2012; Mine-Hattab and Rothstein 2012). In haploid yeast, increased mobility also depends on Rad54 and two checkpoint proteins, Rad9 and Mec1 (Dion et al. 2012; Seeber et al. 2013). In a sae2 mutant, which has delayed appearance of single-stranded DNA, increased chromosome mobility is also delayed (Mine-Hattab and Rothstein 2012). In contrast to the increased mobility observed after IR or enzymatically induced DSBs, spontaneous Rad52 foci are constrained, which may reflect recombination between sister chromatids in the context of DNA replication (Dion et al. 2013).
The mobilization of DSBs may also allow multiple DSBs to interact. In fact, it has been shown that multiple DSBs in the same cell often come together at a single Rad52 focus (Lisby et al. 2003b). These recombinational repair centers are observed in both haploid and diploid cells. The aggregation of multiple DSBs takes place subsequent to recognition by the MRX complex and prior to recruitment of Rad52, which is indicated by the observation that cells exposed to 40 krad of ionizing radiation (equivalent to 20 DSBs per haploid cell) initially exhibit up to 20 Mre11 foci within 5 min, which transitions to 1–2 Rad52 foci within 20–30 min (M. Lisby and R. Rothstein, unpublished data). Most likely multiple DSBs are held together by the same scaffolding processes that hold together the two ends of a single DSB, but the molecular components of the scaffold have not been fully described, although a partial dependency on Sae2 and the MRX complex for tethering ends has been reported (Chen et al. 2001; Lisby et al. 2003a; Kaye et al. 2004; Lobachev et al. 2004; Clerici et al. 2005). Thus, the tethering of DNA ends may facilitate DSB repair but at the same time pose a risk for translocation between clustered DSBs.
D. Nuclear compartments
Some regions of the genome are more susceptible to deleterious recombination including repetitive elements such as the centromeres, telomeres, and Ty elements, and the highly transcribed rDNA and tRNA genes. Untimely recombination at these loci is prevented by compartmentalization of the nucleus into regions that suppress recombination and regions that allow or even stimulate recombination. The most prominent example is the nucleolus from which late-acting recombination and checkpoint proteins such as RPA, Rad52, Rad51, Rad59, Rad55, Rad24–RFC, 9–1–1 (Ddc1), Ddc2, and Rad9 are largely excluded even in the absence of DNA damage (Torres-Rosell et al. 2007). Although DSBs in the rDNA are initially recognized by the MRX complex within the nucleolus and resected (Torres-Rosell et al. 2007), they are only bound by Rad52 and downstream factors after exiting the nucleolus. The relocalization of rDNA breaks from the nucleolus to the nucleoplasm requires the Smc5–Smc6 complex and SUMO modification of Rad52 (see below), and mutants that disrupt the Smc5–Smc6 complex or prevent Rad52 sumoylation lead to Rad52 focus formation inside the nucleolus and rDNA instability (Torres-Rosell et al. 2007).
Similar to the rDNA, telomeres are compartmentalized. Telomeres associate into 6–8 clusters (Gotta et al. 1996), which are largely refractory to recombination and the DNA damage checkpoint response in general (Khadaroo et al. 2009; Ribeyre and Shore 2012). Importantly, all telomeres are generally in the vicinity of the nuclear envelope over long periods of time (Hediger et al. 2002). A component of the nuclear envelope in Saccharomyces cerevisiae is the SUN domain protein Mps3 (Antoniacci et al. 2007; Bupp et al. 2007). Several studies indicate that Mps3 is involved in anchoring telomeres at the nuclear envelope and shielding telomeres against spontaneous recombination (Antoniacci et al. 2007; Bupp et al. 2007; Schober et al. 2009) (reviewed in Taddei and Gasser 2012). Nevertheless, anchoring of telomeres at the nuclear periphery is essential for efficient DSB repair in subtelomeric DNA (Therizols et al. 2006).
In contrast to the nucleolus and telomere clusters, homologous recombination appears to be enhanced in the vicinity of nuclear pore complexes (NPCs). Persistent DSBs, collapsed replication forks, and eroded telomeres relocalize to NPCs, which stimulates recombinational repair at those loci (Nagai et al. 2008; Khadaroo et al. 2009). In an independent study, unrepaired DSBs were found to be enriched at the nuclear periphery in an Mps3-dependent but NPC-independent manner (Oza et al. 2009). Further, it was reported that spontaneous gene conversion is enhanced in a Nup84- and Slx8-dependent manner by tethering of a donor sequence to the nuclear pore complex (NPC) (Nagai et al. 2008). It was suggested that desumoylation of repair proteins by the SUMO-specific protease Ulp1, which associates with the NPC (Takahashi et al. 2000), could be responsible for the observed stimulation of gene conversion (Nagai et al. 2008) (see below).
VI. Regulation of Homologous Recombination
Homologous recombination is tightly regulated according to the type of DNA lesion, cell cycle phase, ploidy, and other environmental and development cues. The regulation of HR serves to ensure that the HR machinery does not interfere with DNA transactions such as transcription and replication on undamaged chromosomes and to fine tune the fidelity of repair. In budding yeast, the regulation of HR takes place mainly at the transcriptional level and by post-translational modification of HR proteins, while there is so far little evidence for post-transcriptional regulation.
A. Transcriptional regulation of homologous recombination
Many recombination proteins are constitutively expressed with some exceptions where expression is regulated with cell cycle or in response to DNA damage. Presumably, a constitutive basal expression of many recombination and checkpoint proteins allows for a rapid response to DNA damage. DNA damage-induced genes with relevance to mitotic recombination are the ribonucleotide reductase genes RNR1–4, which are required for reduction of ribonucleotides to their corresponding 2′-deoxyribonucleotides (dNTPs) necessary for DNA synthesis during recombination, the DNA damage checkpoint genes RAD53 and MEC1, which form a positive feedback loop to increase their own expression, and the recombination genes RFA1, RFA2, RFA3, RAD50, SRS2, RAD54, and RAD51 which are also induced in a MEC1-dependent manner (Cole et al. 1987; Elledge and Davis, 1989, 1990; Yagle and McEntee 1990; Basile et al. 1992; Kiser and Weinert 1996; Jelinsky and Samson 1999; Vallen and Cross 1999; Gasch et al. 2001; Mercier et al. 2001; Benton et al. 2006) (reviewed in Fu et al. 2008). In addition, many recombination genes exhibit cell cycle regulated expression, which peaks in late G1 to early S phase (Basile et al. 1992; Spellman et al. 1998 and reviewed in Mathiasen and Lisby 2014), although the functional importance of this regulation remains to be determined.
B. Regulation of homologous recombination by post-translational modifications
Homologous recombination proteins are acted upon by most known post-translational modifications (PTMs) including phosphorylation, ubiquitylation, sumoylation, and acetylation, which are all reversible. Since the PTMs and their consequences are context dependent, this section will review the regulation by PTMs during DNA double-strand break repair and during recombinational restart of DNA replication (Table 2), while the regulation of specialized recombination events such as alternative lengthening of telomeres is described elsewhere (Wellinger and Zakian 2012). The following section will focus on the PTMs, for which a biological function has been described.
Table 2. Regulation of HR proteins by post-translational modifications.
| Target | PTM | Modifier(s) | Function | References |
|---|---|---|---|---|
| Sae2 | S267P | Cdc28 | Promotes resection | Aylon et al. (2004); Huertas et al. (2008); Ira et al. (2004); Zierhut and Diffley (2008) |
| P | Tel1/Mec1 | Activation through solubilization | Baroni et al. (2004); Fu et al. (2014) | |
| Ac | ? | Degradation | Robert et al. (2011) | |
| Dna2 | T4P, S17P, S237P | Cdc28 | Promotes long-range resection | Chen et al. (2011) |
| S17P | Cdc28 | Nuclear localization | Kosugi et al. (2009) | |
| Rad55 | S2P, S8P, S14P | Mec1 | ? | Bashkirov et al. (2000); Herzberg et al. (2006) |
| Rad51 | S192P | Mec1 | ATPase regulation | Flott et al. (2011) |
| Srs2 | P | CDC28 | Inhibits displacement of Rad51 | Saponaro et al. (2010) |
| K1081S, K1089S, K1142S | ? | Promotes displacement of Rad51 | Saponaro et al. (2010) | |
| Rad52 | K43S, K44S, K253S | Siz2 | Inhibits DNA binding and annealing of ssDNA | Altmannova et al. (2010); Sacher et al. (2006) |
| Ub | Slx5–Slx8 | ? | (Ii et al. (2007) | |
| Slx4 | T113P | Mec1/Tel1 | Activation of Rad1-Rad10 nuclease | Toh et al. (2010) |
| P | Mec1/Tel1 | Interaction with Dpb11 | Ohouo et al. (2010) | |
| Mms4 | P | CDC28, Cdc5 | Activation in M phase | Loog and Morgan (2005); Matos et al. (2011) |
| Yen1 | P | CDC28, Cdc5 | Inhibition outside of anaphase | Loog and Morgan (2005); Matos et al. (2011) |
| S655P, S679P | CDC28 | Nuclear localization | Eissler et al. (2014); Kosugi et al. (2009) | |
| Rad1 | K32S | Siz1, Siz2 | Inhibition of DNA binding | Sarangi et al. (2014) |
| PCNA | K127S, K164S | ? | Binding of Srs2 | Hoege et al. (2002); Papouli et al. (2005) |
| K164Ub | Rad6–Rad18 | Promotes translesion synthesis | Hoege et al. (2002) | |
| K164Ubn | Rad5–Ubc13–Mms2 | Promotes error-free lesion bypass by template switching | Blastyak et al. (2007); Hoege et al. (2002) | |
| Rtt107 | P | Mec1 (Slx4) | Replication restart | Roberts et al. (2006) |
| Mcm2–7 | S164P, S170P | Mec1 (Mrc1) | Replication restart | Randell et al. (2010); Stead et al. (2012) |
P, phosphorylation. Ac, acetylation. S, sumoylation. Ub, ubiquitylation. Parentheses indicate partial dependency. ?, unknown.
Regulation of DSB repair by PTMs:
The initiating step of recombinational repair of a DSB end, resection to produce 3′ single-stranded overhangs, is regulated by cyclin-dependent kinase Cdc28 (CDK). Inhibition of Cdc28 using an analog-sensitive allele of CDC28 or by overexpression of Sic1, an inhibitor of Cdc28, results in greatly reduced end resection (Aylon et al. 2004; Ira et al. 2004). As a consequence, resection of DSBs induced in G1 cells is greatly reduced compared with cycling or G2-arrested cells. Resection is regulated by PTMs at multiple levels. Initially, binding of the Ku complex to DSB ends blocks resection and its binding is inhibited by Cdc28 during S and G2 phase of the cell cycle (Clerici et al. 2008). G1 cells deficient for Ku show greater recruitment of Mre11 to an endonuclease-induced DSB and increased resection. As a consequence, HR can occur in G1 in yku mutants (Zhang et al. 2009; Trovesi et al. 2011). Overexpression of Exo1 is also able to overcome the inhibition to resection in G1 cells, consistent with other studies showing Ku is a barrier to Exo1-mediated end resection. The inhibitory effect on end resection was observed to a lesser extent in the dnl4 (ligase IV deficient) mutant, suggesting the end binding function of Ku and ligation both contribute to protecting ends from degradation in G1 (Clerici et al. 2008; Zierhut and Diffley 2008). Interestingly, inhibition of CDK in G2 yku80 cells fails to block short-range resection, similar to the situation in G1 yku80 cells, and activation of CDK in G1 by overexpression of Clb2 restores both initiation and extensive resection (Clerici et al. 2008). Together, these results suggest that Ku (and to a lesser extent NHEJ) is the primary rate-limiting factor for the initiation of end resection in G1 by competing with MRX and Exo1 for end binding.
Resection is positively regulated by phosphorylation of Sae2 at serine 267 by Cdc28 during S and G2 phases (Huertas et al. 2008). Mutation of this site to a nonphosphorylatable residue, S267A, phenocopies sae2, including hypersensitivity to camptothecin, defective sporulation, reduced hairpin-induced recombination, impaired DSB processing, persistent Mre11 foci, and delayed Rad52 recruitment. Sae2 is phosphorylated at additional (S/T)Q motifs by Mec1 and Tel1 in reponse to DNA damage and mutation of these phosphorylation sites also impairs DNA repair (Baroni et al. 2004). These phosphorylation events activate Sae2 through a transition from an insoluble oligomeric state to active monomers/dimers, which allow the protein to be recruited to sites of DNA damage (Fu et al. 2014). Further, the stability of Sae2 is regulated by acetylation and treatment with the histone deacetylase inhibitor valproic acid causes accumulation of acetylated Sae2 and degradation of Sae2 (Robert et al. 2011). The mechanism for acetylation of Sae2 remains to be determined. Extensive resection is promoted in S and G2 phases by Cdc28-dependent phosphorylation of Dna2 at threonine 4 and serines 17 and 237, which is required for its recruitment to DSBs (Chen et al. 2011). Further, Dna2 shuttles from the cytoplasm to the nucleus upon phosphorylation on serine 17 by Cdc28 (Kosugi et al. 2009). Interestingly, resection in G2 is more dependent on MRX than in cycling cells, suggesting replication forks could serve to recruit Exo1 and/or STR–Dna2 in lieu of the MRX complex, possibly through RPA, which interacts directly with Dna2 (Bae et al. 2003; Chen et al. 2013). Loss of Rad9 can partially bypass the Cdc28 requirement for resection, suggesting that Rad9 could also be a target of the Cdc28-dependent regulation of resection (Lazzaro et al. 2008).
The nucleolytically produced 3′ single-stranded DNA ends are bound by RPA (Alani et al. 1992). The recruitment of Rad52 to ssDNA by RPA in S/G2 phase requires Cdc28 activity (Alabert et al. 2009; Barlow and Rothstein 2009), however the responsible phosphorylation sites have not been identified. Rad55 is subject to Mec1-dependent phosphorylation on serines 2, 8, and 14 in response to MMS or an HO-induced DSB (Bashkirov et al. 2000; Herzberg et al. 2006), and a rad55-S2,8,14A mutant exhibits reduced survival after DNA damage although the underlying mechanism remains to be established. The Rad51 recombinase itself is phosphorylated in a Mec1-dependent manner at serine 192 in response to DNA damage (Flott et al. 2011). Biochemical analysis indicates that serine 192 is required for Rad51 ATP hydrolysis and DNA binding, whereas mutation of serine 192 does not interfere with Rad51 multimerization. Srs2 is inhibited by Cdc28 phosphorylation at multiple sites to allow Rad51-dependent DSB repair via SDSA by controlling turnover of Srs2 at the invading strand (Saponaro et al. 2010).
Some aspects of recombination are also likely to be regulated by sumoylation of RPA, Rad52, Rad59, and Srs2 (Sacher et al. 2006; Burgess et al. 2007; Ohuchi et al. 2008; Saponaro et al. 2010; Cremona et al. 2012; Psakhye and Jentsch 2012). For Rad52, it was shown that sumoylation on lysines 43, 44, and 253 inhibits its ssDNA binding and annealing activities without affecting its interaction with Rad51 and RPA (Altmannova et al. 2010), and sumoylation of Rad52 protects it from proteasomal degradation (Sacher et al. 2006). Moreover, stimulating Rad52 sumoylation by overexpression of the Siz2 SUMO ligase or by fusing SUMO to the C terminus of Rad52 suppressed the DNA damage sensitivity of srs2 cells (Esta et al. 2013). As a consequence, sumoylation of Rad52 improves the fidelity of recombinational repair by shifting DSB repair from SSA to gene conversion (Sacher et al. 2006; Altmannova et al. 2010). Sumoylation of Srs2 on lysines 1081, 1089, and 1142 appears to inhibit recombinational repair as mutation of these lysines to arginine partially suppresses the DNA repair defect of a Cdc28-phosphorylation deficient srs2 mutant (Saponaro et al. 2010), suggesting that phosphorylation and sumoylation may have counteracting effects on Srs2 activity. Furthermore, sumoylation of Srs2 decreases its interaction with sumoylated PCNA and at the same time sumoylated PCNA inhibits Srs2 sumoylation (Kolesar et al. 2012). This observation suggests that sumoylation could make Srs2 association to the replication fork more dynamic. The effects of RPA and Rad59 sumoylation on recombination remain to be established in yeast. However, in human cells, RPA1 sumoylation facilitates recruitment of Rad51 to DNA damage-induced foci to initiate DNA repair through homologous recombination (Dou et al. 2010). It has also been suggested that desumoylation of repair proteins by the SUMO-specific protease Ulp1, which associates with the NPC (Takahashi et al. 2000), is responsible for the stimulation of spontaneous gene conversion observed at a locus artificially tethered to the nuclear envelope or to the NPC (Nagai et al. 2008). This notion is supported by changes in sumoylation patterns of RPA, Rad52, and Rad59 observed in nucleoporin mutants and in slx8 (Burgess et al. 2007; Palancade et al. 2007).
The role of Rad1–Rad10 in processing heterologous flaps in collaboration with Slx4 requires phosphorylation of Slx4 on threonine 113 by Mec1 or Tel1 (Toh et al. 2010). Rad1 is sumoylated at lysine 32, which decreases the affinity of the Rad1–Rad10 for DNA without affecting its other activities, suggesting that Rad1 sumoylation promotes its disengagement from DNA after nuclease cleavage (Sarangi et al. 2014). Mus81–Mms4 and Yen1 activity are restricted to the G2/M transition and anaphase, respectively, by Cdc5- and Cdc28-dependent phosphorylation (Loog and Morgan 2005; Matos et al. 2011; Matos et al. 2013; Saugar et al. 2013). Premature activation of Mus81–Mms4 using a phosphomimetic Mms4 allele or by untimely activation of Cdc5 increases crossover-associated recombination events (Matos et al. 2013; Szakal and Branzei 2013). Yen1 is inactivated by Cdc28-dependent phosphorylation and activated at anaphase by the Cdc14 phosphatase (Blanco et al. 2014; Eissler et al. 2014). Further, to provide another level of regulation, Yen1 relocalizes from the cytoplasm to the nucleus upon phosphorylation on serines 655 and 679 by Cdc28 in G2/M phase (Kosugi et al. 2009; Blanco et al. 2014; Eissler et al. 2014).
Regulation of recombinational restart of replication by PTMs:
A range of DNA lesions may cause stalling of DNA replication. Replication fork blockage activates the replication checkpoint, which is responsible for slowing of S-phase and cell cycle progression, down-regulation of late origin firing, activation of DNA repair proteins, and stabilization of replication forks (reviewed in Friedel et al. 2009). The replication checkpoint is mediated by the Mec1 kinase and its downstream effector kinase Rad53. In the absence of Mec1 or Rad53, stalled replication forks collapse and the replisome dissociates (Tercero and Diffley 2001; Cobb et al. 2003), which is likely due to the failure to phosphorylate functional targets at the replication fork such as Mrc1, Pol31, Rtt107, Dbf4, and Pol1 (Osborn and Elledge 2003; Roberts et al. 2006; Chen et al. 2010; Randell et al. 2010). In contrast to active or stalled replication forks, which are refractory to recruitment of Rad52 (Lisby et al. 2004; Alabert et al. 2009), collapsed replication forks readily recruit Rad52 into foci.
PCNA is the master regulator of DNA damage tolerance pathways at the replication fork. During S phase and in response to replication stress by hydroxyurea, PCNA is sumoylated at lysines 127 and 164 (Hoege et al. 2002; Papouli et al. 2005). Sumoylated PCNA is bound by the Srs2 helicase (Papouli et al. 2005), which acts as an antirecombinase by displacing Rad51 from single-stranded DNA (Krejci et al. 2003; Veaute et al. 2003). Accordingly, Rad51 is enriched at the replication fork in a nonsumoylatable pol30-K127,164R mutant (Papouli et al. 2005). In contrast, when DNA damage such as UV- or MMS-induced lesions are encountered by the replication fork, PCNA is first monoubiquitylated at lysine 164 by the Rad6–Rad18 pathway (Hoege et al. 2002), which promotes translesion synthesis by a number of error-prone polymerases (reviewed in Finley et al. 2012). Sumoylation and ubiquitylation of PCNA are independent processes (Papouli et al. 2005). Lysine 164 of PCNA can be further modified by K63-linked polyubiquitylation through the Rad5–Ubc13–Mms2 pathway (Hoege et al. 2002), which facilitates error-free repair of lesions on the leading strand template by fork regression and template switching using the Rad5 helicase (Blastyak et al. 2007), while Rad52 mediates error-free repair of lesions on the lagging strand template (Prakash 1981; Zhang and Lawrence 2005; Gangavarapu et al. 2007) (Figure 6). However, the majority of error-free lesion bypass was reported to be RAD52 independent (Zhang and Lawrence 2005).
The role of Mec1-dependent phosphorylation during the restart of stalled replication forks is still poorly understood. Mec1-dependent phosphorylation of Slx4 facilitates assembly of an Rtt107–Slx4–Dpb11 complex at stalled forks (Ohouo et al. 2010). Further, Slx4-dependent phosphorylation of Rtt107 by Mec1 is critical for replication restart after alkylation damage (Roberts et al. 2006). Possibly, the Rtt107–Slx4–Dpb11 complex acts as a scaffold for the assembly of additional fork stabilizing and repair factors such as the Smc5–Smc6–Mms21 SUMO ligase (Ohouo et al. 2010; Leung et al. 2011). The Smc5–Smc6–Mms21 complex was shown to promote sister-chromatid junction-mediated intra-S repair (Branzei et al. 2006; De Piccoli et al. 2006; Sollier et al. 2009), although the relevant sumoylation targets remain to be identified. Finally, Mrc1 facilitates Mec1 phosphorylation of the S/T-Q motifs of chromatin-bound Mcm2–7 during S phase to facilitate replication restart during replication stress (Randell et al. 2010; Stead et al. 2012).
The Slx5–Slx8 (SUMO-targeted ubiquitin E3 ligase) is localized to replication foci and is important for suppressing recombination during DNA replication (Burgess et al. 2007). This may be explained by the observation that deletion of SLX5–SLX8 results in reduced levels of Rad52, Rad59, and RPA sumoylation. In the case of Rad52, its sumoylation inhibits recombination (Sacher et al. 2006; Altmannova et al. 2010). In vitro, Rad52 and Rad57 are targets of the ubiquitylation activity of Slx5–Slx8 (Ii et al. 2007).
C. Role of chromatin in controlling mitotic recombination
Several lines of evidence suggest that homologous recombination is controlled by modification of chromatin structure (Chai et al. 2005; Shim et al. 2005; Tsukuda et al. 2005; Kent et al. 2007; Van Attikum et al. 2007; Sinha et al. 2009; Sinha and Peterson 2009; Tsukuda et al. 2009; Chen et al. 2012; Costelloe et al. 2012; Adkins et al. 2013). Similarly, capping of telomeres is likely a major barrier for recombination at telomere sequences either by inhibiting recombination proteins or by preventing resection of telomeres (Grossi et al. 2001; Dubois et al. 2002).
One of the principal and evolutionarily conserved chromatin marks associated with DNA damage that expose DNA ends or single-stranded regions is the phosphorylation of histone H2A on serine 129 by the Tel1 and Mec1 kinases (Rogakou et al. 1999; Downs et al. 2000; Shroff et al. 2004). An hta-S129A mutant is sensitive to DNA damage-inducing agents such as phleomycin, camptothecin, and methyl methanesulfonate (Redon et al. 2003; Downs et al. 2004). The modification of chromatin by H2A phosphorylation occurs preferentially at unresected DSB ends and in G1 phase, whereas the recruitment of chromatin modifiers NuA4, SWR1, RSC, SWI/SNF, and INO80 occurs in G2/M and correlates with homologous recombination (Downs et al. 2004; Morrison et al. 2004; Van Attikum et al. 2004; Bennett et al. 2013). The NuA4 complex contains an associated histone acetyltransferase, which targets histone H4 for acetylation and is important for DNA repair (Choy and Kron 2002; Downs et al. 2004). Histone H3-K56 acetylation, which is formed transiently by Rtt109 during DNA replication, is important for sister-chromatid repair of DSBs arising during replication (Munoz-Galvan et al. 2013). Other histone acetyltransferases, such as Gcn5 and Hat1, also contribute to the wave of chromatin acetylation that follows DSB formation (Qin and Parthun 2002; Tamburini and Tyler 2005). A number of histone deacetylases including Rpd3, Hda1, Sir2, and Hst1 are also recruited to sites of DNA damage presumably to remove DNA damage-induced chromatin marks after completion of repair (Robert et al. 2011; Tamburini and Tyler 2005). The acetylation marks may serve to stabilize the SWR1 complex at DSBs via binding of its Bdf1 subunit through its double bromodomain (Kobor et al. 2004). SWR1 mediates deposition of the histone variant Htz1 (H2A.Z) in place of H2A in chromatin (Kobor et al. 2004; Mizuguchi et al. 2004; Morillo-Huesca et al. 2010). The effects of these chromatin modifications are not fully understood, but it has been reported that deposition of Htz1 into chromatin is important for DSB end resection (Kalocsay et al. 2009). The checkpoint adaptor Rad9 is recruited to sites of DNA damage likely through a dual interaction of its BRCT domains with H2A-S129P and its Tudor domain with histone H3 methylated at lysine 79 (H3-K79Me) (Giannattasio et al. 2005; Javaheri et al. 2006; Toh et al. 2006; Hammet et al. 2007; Germann et al. 2011). Finally, H2A-S129 phosphorylation is also required for loading cohesins at a defined DSB (Unal et al. 2004).
ATP-dependent chromatin remodeling is equally important for efficient homologous recombination especially in heterochromatin (Sinha and Peterson 2009; Sinha et al. 2009). The RSC complex is one of the earliest factors recruited to a DSB along with the MRX and Ku complexes (Shim et al. 2005), but RSC also appears to have a later role in recombination following synapsis (Chai et al. 2005). The RSC complex produces a histone-free region of a few hundred nucleotides immediately adjacent to a DSB to promote binding of the MRX and Ku complexes and to facilitate resection (Kent et al. 2007; Shim et al. 2007; Adkins et al. 2013). The later role of RSC in recombination may be linked to its interaction with Rad59 and/or its involvement in loading of cohesin at DNA breaks (see below) (Oum et al. 2011). In contrast, the SWI/SNF chromatin-remodeling complex was found to play a role at or preceding the strand-invasion step of HR (Chai et al. 2005). The Fun30 nucleosome-remodeling factor is important for extensive resection by Exo1 and Sgs1–Dna2 (Chen et al. 2012; Costelloe et al. 2012; Eapen et al. 2012). A fourth ATP-dependent chromatin remodeling factor, Rad54, enhances DNA strand invasion by Rad51 on chromatin substrates in vitro (Alexiadis and Kadonaga 2002), and further plays a crucial role in vivo for the initiation of DNA synthesis after strand invasion likely by enhancing the accessibility to DNA within nucleosomal arrays (Jaskelioff et al. 2003; Sugawara et al. 2003; Ceballos and Heyer 2011). Rad54 interacts directly with histone H3 and its chromatin-remodeling activity is stimulated by Rad51 (Jaskelioff et al. 2003; Kwon et al. 2007).
D. Role of cohesin in regulating mitotic recombination
Cohesin is loaded and maintained at double-strand breaks independent of global DNA replication and is required for efficient sister-chromatid recombination (Sjogren and Nasmyth 2001). Loading of cohesin at DSBs requires H2A-S129P, Mre11, and Scc2, a component of the cohesin loading machinery (Strom et al. 2004; Unal et al. 2004), and is further aided by the RSC chromatin remodeler and Rad59 (Oum et al. 2011). Replication-independent cohesion is induced genome-wide by the Eco1 acetyltransferase, which targets Smc3 in response to DNA damage (Strom et al. 2007; Unal et al. 2007; Heidinger-Pauli et al. 2009). The establishment of damage-induced cohesion by cohesin acetylation is further aided by Ctf4, Ctf18, Tof1, Csm3, Chl1, and Mrc1 (Borges et al. 2013). In contrast, cohesion must be relieved locally by separase in order for efficient resection of DSBs during postreplicative repair (McAleenan et al. 2013). Another structural maintenance of chromosomes (SMC) factor, which is loaded at DSBs by Scc2 is the Smc5–Smc6 complex in a manner dependent on the BRCT domain-containing protein Rtt107/Esc4 (Lindroos et al. 2006; Leung et al. 2011). The Smc5–Smc6 complex is required for MMS-induced recombination and DSB repair (Onoda et al. 2004; De Piccoli et al. 2006).
E. Ploidy/aneuploidy
Mitotic recombination is more efficient in heterozygous MATa/MATα diploids than in homozygous MATa/MATa or MATα/MATα cells, both spontaneously and after UV irradiation (Friis and Roman 1968; Hopper et al. 1975; Esposito and Watsgaff 1981). In diploid yeast, NHEJ is severely disabled through the repression of NEJ1, a key component of NHEJ, by the transcriptional repressor, Mata1–Matα2 (Heude and Fabre 1993; Frank-Vaillant and Marcand 2001). As expected, diploids are more radioresistant than haploids due to the extra copy of the genome (Mortimer 1958). However, a further increase in ploidy leads to increased radiosensitivity and reliance on homologous recombination for survival (Storchova et al. 2006). In response to replication stress, haploid cells use the Rad6-dependent pathways that resume stalled forks, whereas diploid cells use homologous recombination (Li and Tye 2011). Indeed, the DNA damage sensitivity of rad6 and rad18 mutants is suppressed by mating-type heterozygosity in a RAD52-dependent manner (Yan et al. 1995). The IR sensitivity of rad55 and rad57 mutants is also suppressed by MAT heterozygosity by an unknown mechanism. Although the mechanism was reported to be due to loss of NHEJ, in other strain backgrounds rad55yku70 or rad55dnl4 mutants retain IR sensitivity (Valencia-Burton et al. 2006; Fung et al. 2009). Mutation of SRS2 suppresses the IR sensitivity of rad55 and rad57 mutants consistent with the model that Srs2 and Rad55–Rad57 have opposing roles in Rad51 nucleoprotein filament stability (Fung et al. 2009; Liu et al. 2011). SRS2 transcript levels vary in response to ploidy and might contribute to the MAT heterozygosity suppression of rad6, rad18, rad55, and rad57 mutants. The only recombination gene that is clearly regulated at the transcriptional level by mating type is RDH54. There is a Mata1–Matα2 binding site within the RDH54 promoter and Rdh54 protein level is reduced fivefold in diploids compared with haploids (de Godoy et al. 2008; Galgoczy et al. 2004). Surprisingly, the phenotype of rdh54 mutants is most apparent in diploids (lethality with rad54 and srs2), suggesting the reduced levels are nevertheless required for HR (Klein 1997).
VII. Postscript
The study of mitotic recombination in yeast has progressed immensely since the last time a chapter in the precursor of the YeastBook was written (Petes et al. 1991). Much has been learned about the many genes that act to preserve genome stability through genetics, biochemistry, and cell biology. However, there is still important work to be done. The in vitro reconstitution of many of the fundamental reactions has not yet been achieved. In addition, the emergence of single molecule techniques for defining biochemical reactions will greatly expand our detailed understanding of the steps for each of these reactions. Genetic analysis of combinations of mutants will be needed to push the boundaries to define new gene and pathway interactions. The underlying cell biology of these genes and pathways must also be examined in detail. Finally, high throughput methods should be applied to more processes, which promises a greater depth of understanding of the relationships between the genetic, biochemical, and cell biological processes that are involved in this basic cellular function.
Acknowledgments
We thank Tom Petes, Peter Philippsen, Eric Bryant, Patrick Sung, and members of his laboratory and Patrick Ruff for insightful comments on the manuscript. We are grateful to the following funding agencies for financial support: Danish Council for Independent Research (FNU: 12-127136), the Villum Kann Rasmussen Foundation and the European Research Council (ERCStG, no. 242905) to M.L., and National Institutes of Health (GM50237 and GM67055 to R.R. and GM41784 and GM94386 to L.S.S.).
Footnotes
Available freely online through the author-supported open access option.
Communicating editor: J. Boeke
Literature Cited
- Adkins N. L., Niu H., Sung P., Peterson C. L., 2013. Nucleosome dynamics regulates DNA processing. Nat. Struct. Mol. Biol. 20: 836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agmon N., Liefshitz B., Zimmer C., Fabre E., Kupiec M., 2013. Effect of nuclear architecture on the efficiency of double-strand break repair. Nat. Cell Biol. 15: 694–699. [DOI] [PubMed] [Google Scholar]
- Agmon N., Yovel M., Harari Y., Liefshitz B., Kupiec M., 2011. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. 39: 7009–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A., Klein H. L., 1988. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics 119: 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera, A., and B. Gomez-Gonzalez, 2008 Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. 9: 204–217. [DOI] [PubMed]
- Aguilera A., Garcia-Muse T., 2012. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46: 115–124. [DOI] [PubMed] [Google Scholar]
- Ajimura M., Leem S. H., Ogawa H., 1993. Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics 133: 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksenova A. Y., Greenwell P. W., Dominska M., Shishkin A. A., Kim J. C., et al. , 2013. Genome rearrangements caused by interstitial telomeric sequences in yeast. Proc. Natl. Acad. Sci. USA 110: 19866–19871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C., Bianco J. N., Pasero P., 2009. Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J. 28: 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Subbiah S., Kleckner N., 1989. The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics 122: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Padmore R., Kleckner N., 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61: 419–436. [DOI] [PubMed] [Google Scholar]
- Alani E., Thresher R., Griffith J. D., Kolodner R. D., 1992. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J. Mol. Biol. 227: 54–71. [DOI] [PubMed] [Google Scholar]
- Alexeev A., Mazin A., Kowalczykowski S. C., 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 10: 182–186. [DOI] [PubMed] [Google Scholar]
- Alexiadis V., Kadonaga J. T., 2002. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 16: 2767–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmannova V., Eckert-Boulet N., Arneric M., Kolesar P., Chaloupkova R., et al. , 2010. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res. 38: 4708–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro D., Lisby M., Rothstein R., 2007. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 3: e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M. P., Nelson Z. W., Hetrick E. D., Gottschling D. E., 2008. A genetic screen for increased loss of heterozygosity in Saccharomyces cerevisiae. Genetics 179: 1179–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniacci L. M., Kenna M. A., Skibbens R. V., 2007. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle 6: 75–79. [DOI] [PubMed] [Google Scholar]
- Argueso J. L., Westmoreland J., Mieczkowski P. A., Gawel M., Petes T. D., et al. , 2008. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl. Acad. Sci. USA 105: 11845–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J., McGlynn P., 2009. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 37: 3475–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y., Liefshitz B., Kupiec M., 2004. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23: 4868–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K. H., Kim H. S., Bae S. H., Kang H. Y., Brill S., et al. , 2003. Bimodal interaction between replication-protein A and Dna2 is critical for Dna2 function both in vivo and in vitro. Nucleic Acids Res. 31: 3006–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Symington L. S., 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10: 2025–2037. [DOI] [PubMed] [Google Scholar]
- Ball L. G., Zhang K., Cobb J. A., Boone C., Xiao W., 2009. The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol. Microbiol. 73: 89–102. [DOI] [PubMed] [Google Scholar]
- Ballew B. J., Yeager M., Jacobs K., Giri N., Boland J., et al. , 2013. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Hum. Genet. 132: 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera M. A., Petes T. D., 2006. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103: 12819–12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow J. H., Rothstein R., 2009. Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase. EMBO J. 28: 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow J. H., Lisby M., Rothstein R., 2008. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol. Cell 30: 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroni E., Viscardi V., Cartagena-Lirola H., Lucchini G., Longhese M. P., 2004. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol. Cell. Biol. 24: 4151–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J., Horejsi Z., Sehested M., Nesland J. M., Rajpert-De Meyts E., et al. , 2007. DNA damage response mediators MDC1 and 53BP1: constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene 26: 7414–7422. [DOI] [PubMed] [Google Scholar]
- Bartsch S., Kang L. E., Symington L. S., 2000. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 20: 1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov V. I., King J. S., Bashkirova E. V., Schmuckli-Maurer J., Heyer W. D., 2000. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol. Cell. Biol. 20: 4393–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basile G., Aker M., Mortimer R. K., 1992. Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol. Cell. Biol. 12: 3235–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin-Shanower S. A., Fricke W. M., Mullen J. R., Brill S. J., 2003. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 23: 3487–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett G., Papamichos-Chronakis M., Peterson C. L., 2013. DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat. Commun. 4: 2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton M. G., Somasundaram S., Glasner J. D., Palecek S. P., 2006. Analyzing the dose-dependence of the Saccharomyces cerevisiae global transcriptional response to methyl methanesulfonate and ionizing radiation. BMC Genomics 7: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R., Capra T., Jossen R., Colosio A., Frattini C., et al. , 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146: 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein K. A., Reid R. J., Sunjevaric I., Demuth K., Burgess R. C., et al. , 2011. The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol. Biol. Cell 22: 1599–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M. G., Matos J., Rass U., Ip S. C., West S. C., 2010. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair (Amst.) 9: 394–402. [DOI] [PubMed] [Google Scholar]
- Blanco M. G., Matos J., West S. C., 2014. Dual control of Yen1 nuclease activity and cellular localization by Cdk and Cdc14 prevents genome instability. Mol. Cell 54: 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A., Pinter L., Unk I., Prakash L., Prakash S., et al. , 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell 28: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M. N., Gaillard P. H., McDonald W. H., Shanahan P., Yates J. R., 3rd, et al. , 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Jinks-Robertson S., 2013. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics 193: 1025–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges V., Smith D. J., Whitehouse I., Uhlmann F., 2013. An Eco1-independent sister chromatid cohesion establishment pathway in S. cerevisiae. Chromosoma 122: 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G., Haber J. E., 1998. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150: 1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boundy-Mills K. L., Livingston D. M., 1993. A Saccharomyces cerevisiae RAD52 allele expressing a C-terminal truncation protein: activities and intragenic complementation of missense mutations. Genetics 133: 33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., et al. , 2006. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522. [DOI] [PubMed] [Google Scholar]
- Branzei D., Vanoli F., Foiani M., 2008. SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920. [DOI] [PubMed] [Google Scholar]
- Bressan D. A., Baxter B. K., Petrini J. H., 1999. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 7681–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L., 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643. [DOI] [PubMed] [Google Scholar]
- Brocas C., Charbonnier J. B., Dherin C., Gangloff S., Maloisel L., 2010. Stable interactions between DNA polymerase delta catalytic and structural subunits are essential for efficient DNA repair. DNA Repair (Amst.) 9: 1098–1111. [DOI] [PubMed] [Google Scholar]
- Bupp J. M., Martin A. E., Stensrud E. S., Jaspersen S. L., 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179: 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R. C., Lisby M., Altmannova V., Krejci L., Sung P., et al. , 2009. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J. Cell Biol. 185: 969–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R. C., Rahman S., Lisby M., Rothstein R., Zhao X., 2007. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell. Biol. 27: 6153–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S. M., Kleckner N., 1999. Collisions between yeast chromosomal loci in vivo are governed by three layers of organization. Genes Dev. 13: 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkovics P., Sebesta M., Sisakova A., Plault N., Szukacsov V., et al. , 2013. Srs2 mediates PCNA-SUMO-dependent inhibition of DNA repair synthesis. EMBO J. 32: 742–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M., Thayer N. H., Oh S. D., Kleckner N., Hunter N., 2010. Double Holliday junctions are intermediates of DNA break repair. Nature 464: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper, A. M., P. W. Greenwell, W. Tang, and T. D. Petes, 2009 Chromosome aberrations resulting from double-strand DNA breaks at a naturally occurring yeast fragile site composed of inverted ty elements are independent of Mre11p and Sae2p. Genetics 183: 423–439, 421SI–426SI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos S. J., Heyer W. D., 2011. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim. Biophys. Acta 1809: 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., et al. , 2010a DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467: 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., Plank J. L., Bachrati C. Z., Hickson I. D., Kowalczykowski S. C., 2010b Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 17: 1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai B., Huang J., Cairns B. R., Laurent B. C., 2005. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19: 1656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. E., Kolodner R. D., 2011. A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet. 7: e1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Umezu K., Kolodner R. D., 1998. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell 2: 9–22. [DOI] [PubMed] [Google Scholar]
- Chen H., Lisby M., Symington L. S., 2013. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell 50: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Trujillo K., Ramos W., Sung P., Tomkinson A. E., 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8: 1105–1115. [DOI] [PubMed] [Google Scholar]
- Chen S. H., Albuquerque C. P., Liang J., Suhandynata R. T., Zhou H., 2010. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J. Biol. Chem. 285: 12803–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Niu H., Chung W. H., Zhu Z., Papusha A., et al. , 2011. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat. Struct. Mol. Biol. 18: 1015–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., D. Cui, A. Papusha, X. Zhang, C. D. Chu et al., 2012 The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489: 576–580. [DOI] [PMC free article] [PubMed]
- Choy J. S., Kron S. J., 2002. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell. Biol. 22: 8215–8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M. F., Dietrich F. S., Fink G. R., 1988. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell 55: 413–425. [DOI] [PubMed] [Google Scholar]
- Chua P., Jinks-Robertson S., 1991. Segregation of recombinant chromatids following mitotic crossing over in yeast. Genetics 129: 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M., Mantiero D., Lucchini G., Longhese M. P., 2005. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 280: 38631–38638. [DOI] [PubMed] [Google Scholar]
- Clerici M., Mantiero D., Guerini I., Lucchini G., Longhese M. P., 2008. The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 9: 810–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud V., Chan Y. L., Grubb J., Budke B., Bishop D. K., 2012. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337: 1222–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J. A., Bjergbaek L., Shimada K., Frei C., Gasser S. M., 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22: 4325–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coic E., Feldman T., Landman A. S., Haber J. E., 2008. Mechanisms of Rad52-independent spontaneous and UV-induced mitotic recombination in Saccharomyces cerevisiae. Genetics 179: 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G. M., Schild D., Lovett S. T., Mortimer R. K., 1987. Regulation of RAD54- and RAD52-lacZ gene fusions in Saccharomyces cerevisiae in response to DNA damage. Mol. Cell. Biol. 7: 1078–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook C. E., Hochstrasser M., Kerscher O., 2009. The SUMO-targeted ubiquitin ligase subunit Slx5 resides in nuclear foci and at sites of DNA breaks. Cell Cycle 8: 1080–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Ledesma F., Aguilera A., 2006. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep. 7: 919–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costelloe, T., R. Louge, N. Tomimatsu, B. Mukherjee, E. Martini et al., 2012 The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489: 581–584. [DOI] [PMC free article] [PubMed]
- Cotta-Ramusino C., Fachinetti D., Lucca C., Doksani Y., Lopes M., et al. , 2005. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol. Cell 17: 153–159. [DOI] [PubMed] [Google Scholar]
- Cremona C. A., Sarangi P., Yang Y., Hang L. E., Rahman S., et al. , 2012. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol. Cell 45: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossan G. P., van der Weyden L., Rosado I. V., Langevin F., Gaillard P. H. L., et al. , 2011. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nature Genet. 43: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Symington L. S., 2001. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Symington L. S., 2003. The Rad52-Rad59 complex interacts with Rad51 and replication protein A. DNA Repair (Amst.) 2: 1127–1134. [DOI] [PubMed] [Google Scholar]
- Davis A. P., Symington L. S., 2004. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 24: 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., et al. , 2008. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455: 1251–1254. [DOI] [PubMed] [Google Scholar]
- de Jager M., van Noort J., van Gent D. C., Dekker C., Kanaar R., et al. , 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8: 1129–1135. [DOI] [PubMed] [Google Scholar]
- De Muyt A., Jessop L., Kolar E., Sourirajan A., Chen J., et al. , 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G., Cortes-Ledesma F., Ira G., Torres-Rosell J., Uhle S., et al. , 2006. Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat. Cell Biol. 8: 1032–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies A., 2007. Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics 176: 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A., Barker K., Vanhulle K., Downing B., Vayl A., et al. , 2008. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics 179: 1845–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A., Keszthelyi A., Blackgrove T., Vayl A., Coffey B., et al. , 2011. Break-induced replication is highly inaccurate. PLoS Biol. 9: e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defossez P. A., Prusty R., Kaeberlein M., Lin S. J., Ferrigno P., et al. , 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3: 447–455. [DOI] [PubMed] [Google Scholar]
- Deng S. K., Gibb B., de Almeida M. J., Greene E. C., Symington L. S., 2014. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat. Struct. Mol. Biol. 21: 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V., Kalck V., Horigome C., Towbin B. D., Gasser S. M., 2012. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 14: 502–509. [DOI] [PubMed] [Google Scholar]
- Dion V., Kalck V., Seeber A., Schleker T., Gasser S. M., 2013. Cohesin and the nucleolus constrain the mobility of spontaneous repair foci. EMBO Rep. 14: 984–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnianni R. A., Symington L. S., 2013. Break-induced replication occurs by conservative DNA synthesis. Proc. Natl. Acad. Sci. USA 110: 13475–13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornfeld K. J., Livingston D. M., 1992. Plasmid recombination in a rad52 mutant of Saccharomyces cerevisiae. Genetics 131: 261–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H., Huang C., Singh M., Carpenter P. B., Yeh E. T., 2010. Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol. Cell 39: 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs J. A., Lowndes N. F., Jackson S. P., 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Downs J. A., Allard S., Jobin-Robitaille O., Javaheri A., Auger A., et al. , 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16: 979–990. [DOI] [PubMed] [Google Scholar]
- Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y. J., et al. , 2010. A three-dimensional model of the yeast genome. Nature 465: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBois M. L., Haimberger Z. W., McIntosh M. W., Gottschling D. E., 2002. A quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics 161: 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eapen V. V., Sugawara N., Tsabar M., Wu W. H., Haber J. E., 2012. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol. Cell. Biol. 32: 4727–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert-Boulet N., Rothstein R., Lisby M., 2011. Cell biology of homologous recombination in yeast. Methods Mol. Biol. 745: 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggler A. L., Inman R. B., Cox M. M., 2002. The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein A. J. Biol. Chem. 277: 39280–39288. [DOI] [PubMed] [Google Scholar]
- Eissler C. L., Mazon G., Powers B. L., Savinov S. N., Symington L. S., et al. , 2014. The cdk/cdc14 module controls activation of the yen1 holliday junction resolvase to promote genome stability. Mol. Cell 54: 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A., French S. L., Beyer A. L., Tollervey D., 2010. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W., 1989. DNA damage induction of ribonucleotide reductase. Mol. Cell. Biol. 9: 4932–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W., 1990. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 4: 740–751. [DOI] [PubMed] [Google Scholar]
- Esposito M., 1978. Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc. Natl. Acad. Sci. USA 75: 4436–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito M., Watsgaff J., 1981. Mechanisms of mitotic recombination, p. 29 in Molecular Biology of the Yeast Saccharomyces, edited by Strathern J., Jones E. W., Broach J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Essers J., Houtsmuller A. B., van Veelen L., Paulusma C., Nigg A. L., et al. , 2002. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21: 2030–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esta A., Ma E., Dupaigne P., Maloisel L., Guerois R., et al. , 2013. Rad52 sumoylation prevents the toxicity of unproductive Rad51 filaments independently of the anti-recombinase Srs2. PLoS Genet. 9: e1003833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F., Chan A., Heyer W. D., Gangloff S., 2002. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99: 16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson D. O., Holloman W. K., 1996. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc. Natl. Acad. Sci. USA 93: 5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S. R., Grubb J., Bishop D. K., 2009. The Mei5-Sae3 protein complex mediates Dmc1 activity in Saccharomyces cerevisiae. J. Biol. Chem. 284: 11766–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Ulrich H. D., Sommer T., Kaiser P., 2012. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192: 319–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell J., Haber J. E., 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258: 480–484. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J., Rudin N., Haber J. E., 1992. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 12: 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S., Alabert C., Toh G. W., Toth R., Sugawara N., et al. , 2007. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol. Cell. Biol. 27: 6433–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S., Kwon Y., Pigli Y. Z., Rice P. A., Sung P., et al. , 2011. Regulation of Rad51 function by phosphorylation. EMBO Rep. 12: 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin G. S., Symington L. S., 2002. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 21: 3160–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant M., Marcand S., 2001. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 15: 3005–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C., Gasser S. M., 2000. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 14: 81–96. [PMC free article] [PubMed] [Google Scholar]
- Freudenreich C. H., Stavenhagen J. B., Zakian V. A., 1997. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell. Biol. 17: 2090–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke W. M., Brill S. J., 2003. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 17: 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel A. M., Pike B. L., Gasser S. M., 2009. ATR/Mec1: coordinating fork stability and repair. Curr. Opin. Cell Biol. 21: 237–244. [DOI] [PubMed] [Google Scholar]
- Friis J., Roman H., 1968. The effect of the mating-type alleles on intragenic recombination in yeast. Genetics 59: 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q., Chow J., Bernstein K. A., Makharashvili N., Arora S., et al. , 2014. Phosphorylation-regulated transitions in an oligomeric state control the activity of the sae2 DNA repair enzyme. Mol. Cell. Biol. 34: 778–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Pastushok L., Xiao W., 2008. DNA damage-induced gene expression in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 32: 908–926. [DOI] [PubMed] [Google Scholar]
- Fung C. W., Mozlin A. M., Symington L. S., 2009. Suppression of the double-strand-break-repair defect of the Saccharomyces cerevisiae rad57 mutant. Genetics 181: 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Nagase Y., Tsubouchi H., Murakami-Murofushi K., Shibata T., et al. , 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 17: 6412–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgoczy D. J., Cassidy-Stone A., Llinas M., O’Rourke S. M., Herskowitz I., et al. , 2004. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 101: 18069–18074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli A., Schiestl R. H., 1998. Effects of DNA double-strand and single-strand breaks on intrachromosomal recombination events in cell-cycle-arrested yeast cells. Genetics 149: 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game J. C., Cox B. S., 1971. Allelism tests of mutants affecting sensitivity to radiation in yeast and a proposed nomenclature. Mutat. Res. 12: 328–331. [Google Scholar]
- Game J. C., Mortimer R. K., 1974. A genetic study of x-ray sensitive mutants in yeast. Mutat. Res. 24: 281–292. [DOI] [PubMed] [Google Scholar]
- Gangavarapu V., Prakash S., Prakash L., 2007. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 7758–7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., Zou H., Rothstein R., 1996. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 15: 1715–1725. [PMC free article] [PubMed] [Google Scholar]
- Garcia V., Phelps S. E., Gray S., Neale M. J., 2011. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479: 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Huang M., Metzner S., Botstein D., Elledge S. J., et al. , 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell 12: 2987–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti R. A., Becker-Catania S., Chun H. H., Sun X., Mitui M., et al. , 2001. The pathogenesis of ataxia-telangiectasia. Learning from a Rosetta Stone. Clin. Rev. Allergy Immunol. 20: 87–108. [DOI] [PubMed] [Google Scholar]
- Germann S. M., Oestergaard V. H., Haas C., Salis P., Motegi A., et al. , 2011. Dpb11/TopBP1 plays distinct roles in DNA replication, checkpoint response and homologous recombination. DNA Repair (Amst.) 10: 210–224. [DOI] [PubMed] [Google Scholar]
- Ghosal G., Muniyappa K., 2005. Saccharomyces cerevisiae Mre11 is a high-affinity G4 DNA-binding protein and a G-rich DNA-specific endonuclease: implications for replication of telomeric DNA. Nucleic Acids Res. 33: 4692–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M., Lazzaro F., Plevani P., Muzi-Falconi M., 2005. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 280: 9879–9886. [DOI] [PubMed] [Google Scholar]
- Godin S., Wier A., Kabbinavar F., Bratton-Palmer D. S., Ghodke H., et al. , 2013. The Shu complex interacts with Rad51 through the Rad51 paralogues Rad55-Rad57 to mediate error-free recombination. Nucleic Acids Res. 41: 4525–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B., Garcia-Rubio M., Bermejo R., Gaillard H., Shirahige K., et al. , 2011. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 30: 3106–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Barrera S., Garcia-Rubio M., Aguilera A., 2002. Transcription and double-strand breaks induce similar mitotic recombination events in Saccharomyces cerevisiae. Genetics 162: 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Barrera S., Cortes-Ledesma F., Wellinger R. E., Aguilera A., 2003. Equal sister chromatid exchange is a major mechanism of double-strand break repair in yeast. Mol. Cell 11: 1661–1671. [DOI] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H., et al. , 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134: 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S., Esposito R. E., 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56: 771–776. [DOI] [PubMed] [Google Scholar]
- Gravel S., Chapman J. R., Magill C., Jackson S. P., 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22: 2767–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg S. Q., Robinson A. R., Niedernhofer L. J., 2011. Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease. DNA Repair (Amst.) 10: 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenon M., Costelloe T., Jimeno S., O’Shaughnessy A., Fitzgerald J., et al. , 2007. Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast 24: 105–119. [DOI] [PubMed] [Google Scholar]
- Gritenaite D., Princz L. N., Szakal B., Bantele S. C., Wendeler L., et al. , 2014. A cell cycle-regulated Slx4-Dpb11 complex promotes the resolution of DNA repair intermediates linked to stalled replication. Genes Dev. 28: 1604–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi S., Bianchi A., Damay P., Shore D., 2001. Telomere formation by rap1p binding site arrays reveals end-specific length regulation requirements and active telomeric recombination. Mol. Cell. Biol. 21: 8117–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., 2012. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191: 33–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., Hearn M., 1985. RAD52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics 111: 7–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammet A., Magill C., Heierhorst J., Jackson S. P., 2007. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep. 8: 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays S. L., Firmenich A. A., Berg P., 1995. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. USA 92: 6925–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F., Dubrana K., Gasser S. M., 2002. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J. Struct. Biol. 140: 79–91. [DOI] [PubMed] [Google Scholar]
- Heidinger-Pauli J. M., Unal E., Koshland D., 2009. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol. Cell 34: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg K., Bashkirov V. I., Rolfsmeier M., Haghnazari E., McDonald W. H., et al. , 2006. Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol. Cell. Biol. 26: 8396–8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heude M., Fabre F., 1993. a/alpha-control of DNA repair in the yeast Saccharomyces cerevisiae: genetic and physiological aspects. Genetics 133: 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks W. M., Kim M., Haber J. E., 2010. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329: 82–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks W. M., Yamaguchi M., Haber J. E., 2011. Real-time analysis of double-strand DNA break repair by homologous recombination. Proc. Natl. Acad. Sci. USA 108: 3108–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Kato K., Strauss B., 1976. A model for replication repair in mammalian cells. J. Mol. Biol. 101: 417–425. [DOI] [PubMed] [Google Scholar]
- Ho C. K., Mazon G., Lam A. F., Symington L. S., 2010. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell 40: 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S., 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141. [DOI] [PubMed] [Google Scholar]
- Hohl M., Kwon Y., Galvan S. M., Xue X., Tous C., et al. , 2011. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat. Struct. Mol. Biol. 18: 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbeck S. L., Strathern J. N., 1997. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics 147: 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzen T. M., Shah P. P., Olivares H. A., Bishop D. K., 2006. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev. 20: 2593–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner K. P., Karcher A., Craig L., Woo T. T., Carney J. P., et al. , 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105: 473–485. [DOI] [PubMed] [Google Scholar]
- Hopfner K. P., Craig L., Moncalian G., Zinkel R. A., Usui T., et al. , 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418: 562–566. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Kirsch J., Hall B. D., 1975. Mating type and sporulation in yeast. II. Meiosis, recombination, and radiation sensitivity in an alpha-alpha diploid with altered sporulation control. Genetics 80: 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Koshland D., 2003. Chromosome integrity in Saccharomyces cerevisiae: the interplay of DNA replication initiation factors, elongation factors, and origins. Genes Dev. 17: 1741–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. E., Rio A. G., Nicolas A., Kolodner R. D., 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100: 11529–11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., Aguilera A., 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12: 711–721. [DOI] [PubMed] [Google Scholar]
- Huertas P., Cortes-Ledesma F., Sartori A. A., Aguilera A., Jackson S. P., 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ii T., Fung J., Mullen J. R., Brill S. J., 2007. The yeast Slx5-Slx8 DNA integrity complex displays ubiquitin ligase activity. Cell Cycle 6: 2800–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar O., Kupiec M., 1999. Homology search and choice of homologous partner during mitotic recombination. Mol. Cell. Biol. 19: 4134–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip S. C., Rass U., Blanco M. G., Flynn H. R., Skehel J. M., et al. , 2008. Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361. [DOI] [PubMed] [Google Scholar]
- Ira G., Haber J. E., 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22: 6384–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J. E., 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., et al. , 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov E. L., Haber J. E., 1995. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 2245–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov E. L., Sugawara N., White C. I., Fabre F., Haber J. E., 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 3414–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Sugawara N., Lydeard J., Vaze M., Tanguy Le Gac N., et al. , 2009. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 23: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskelioff M., Van Komen S., Krebs J. E., Sung P., Peterson C. L., 2003. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 278: 9212–9218. [DOI] [PubMed] [Google Scholar]
- Javaheri A., Wysocki R., Jobin-Robitaille O., Altaf M., Cote J., et al. , 2006. Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling. Proc. Natl. Acad. Sci. USA 103: 13771–13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky S. A., Samson L. D., 1999. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. USA 96: 1486–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. E., Herskowitz I., 1984. Directionality and regulation of cassette substitution in yeast. Cold Spring Harb. Symp. Quant. Biol. 49: 97–104. [DOI] [PubMed] [Google Scholar]
- Jinks-Robertson S., Petes T. D., 1986. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics 114: 731–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. D., Symington L. S., 1995. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol. 15: 4843–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa W., Kurumizaka H., Ishitani R., Fukai S., Nureki O., et al. , 2002. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol. Cell 10: 359–371. [DOI] [PubMed] [Google Scholar]
- Kaliraman V., Mullen J. R., Fricke W. M., Bastin-Shanower S. A., Brill S. J., 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15: 2730–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M., Hiller N. J., Jentsch S., 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell 33: 335–343. [DOI] [PubMed] [Google Scholar]
- Kaneko H., Kondo N., 2004. Clinical features of Bloom syndrome and function of the causative gene, BLM helicase. Expert Rev. Mol. Diagn. 4: 393–401. [DOI] [PubMed] [Google Scholar]
- Kang L. E., Symington L. S., 2000. Aberrant double-strand break repair in rad51 mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 20: 9162–9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kans J. A., Mortimer R. K., 1991. Nucleotide sequence of the RAD57 gene of Saccharomyces cerevisiae. Gene 105: 139–140. [DOI] [PubMed] [Google Scholar]
- Karppinen S. M., Erkko H., Reini K., Pospiech H., Heikkinen K., et al. , 2006. Identification of a common polymorphism in the TopBP1 gene associated with hereditary susceptibility to breast and ovarian cancer. Eur. J. Cancer 42: 2647–2652. [DOI] [PubMed] [Google Scholar]
- Kaye J. A., Melo J. A., Cheung S. K., Vaze M. B., Haber J. E., et al. , 2004. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 14: 2096–2106. [DOI] [PubMed] [Google Scholar]
- Kaytor M. D., Nguyen M., Livingston D. M., 1995. The complexity of the interaction between RAD52 and SRS2. Genetics 140: 1441–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil R. L., Roeder G. S., 1984. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell 39: 377–386. [DOI] [PubMed] [Google Scholar]
- Keil R. L., McWilliams A. D., 1993. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics 135: 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent N. A., Chambers A. L., Downs J. A., 2007. Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. J. Biol. Chem. 282: 27693–27701. [DOI] [PubMed] [Google Scholar]
- Khadaroo B., Teixeira M. T., Luciano P., Eckert-Boulet N., Germann S. M., et al. , 2009. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat. Cell Biol. 11: 980–987. [DOI] [PubMed] [Google Scholar]
- Kim N., Jinks-Robertson S., 2011. Guanine repeat-containing sequences confer transcription-dependent instability in an orientation-specific manner in yeast. DNA Repair (Amst.) 10: 953–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R. A., Wang J. C., 1989. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell 57: 975–985. [DOI] [PubMed] [Google Scholar]
- Kim Y., Lach F. P., Desetty R., Hanenberg H., Auerbach A. D., et al. , 2011. Mutations of the SLX4 gene in Fanconi anemia. Nature Genet. 43: 142–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser G. L., Weinert T. A., 1996. Distinct roles of yeast MEC and RAD checkpoint genes in transcriptional induction after DNA damage and implications for function. Mol. Biol. Cell 7: 703–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H. L., 1988. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics 120: 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H. L., 1997. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147: 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., 2008. A new role of the rDNA and nucleolus in the nucleus–rDNA instability maintains genome integrity. BioEssays 30: 267–272. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Ganley A. R., 2005. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581–1584. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Heck D. J., Nomura M., Horiuchi T., 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 12: 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor M. S., Venkatasubrahmanyam S., Meneghini M. D., Gin J. W., Jennings J. L., et al. , 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2: E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesar P., Sarangi P., Altmannova V., Zhao X., Krejci L., 2012. Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation. Nucleic Acids Res. 40: 7831–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Hasebe M., Tomita M., Yanagawa H., 2009. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 106: 10171–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus E., Leung W. Y., Haber J. E., 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98: 8255–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L., Song B., Bussen W., Rothstein R., Mortensen U. H., et al. , 2002. Interaction with Rad51 is indispensable for recombination mediator function of Rad52. J. Biol. Chem. 277: 40132–40141. [DOI] [PubMed] [Google Scholar]
- Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., et al. , 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309. [DOI] [PubMed] [Google Scholar]
- Kupiec M., 2000. Damage-induced recombination in the yeast Saccharomyces cerevisiae. Mutat. Res. 451: 91–105. [DOI] [PubMed] [Google Scholar]
- Kwon Y., Chi P., Roh D. H., Klein H., Sung P., 2007. Synergistic action of the Saccharomyces cerevisiae homologous recombination factors Rad54 and Rad51 in chromatin remodeling. DNA Repair (Amst.) 6: 1496–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens K., Bemeleit D. J., Mockel C., Clausing E., Schele A., et al. , 2011. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell 145: 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao J. P., Oh S. D., Shinohara M., Shinohara A., Hunter N., 2008. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol. Cell 29: 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro F., Sapountzi V., Granata M., Pellicioli A., Vaze M., et al. , 2008. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 27: 1502–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Zhang Y., Lee S. E., 2008. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature 454: 543–546. [DOI] [PubMed] [Google Scholar]
- Lee P. S., Petes T. D., 2010. From the Cover: mitotic gene conversion events induced in G1-synchronized yeast cells by gamma rays are similar to spontaneous conversion events. Proc. Natl. Acad. Sci. USA 107: 7383–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. S., Greenwell P. W., Dominska M., Gawel M., Hamilton M., et al. , 2009. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 5: e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F. J., Degtyareva N. P., Lobachev K., Petes T. D., 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120: 587–598. [DOI] [PubMed] [Google Scholar]
- Lemoine F. J., Degtyareva N. P., Kokoska R. J., Petes T. D., 2008. Reduced levels of DNA polymerase delta induce chromosome fragile site instability in yeast. Mol. Cell. Biol. 28: 5359–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld B. M., Rattray A. J., Bhaskara V., Ghirlando R., Paull T. T., 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28: 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettier G., Feng Q., de Mayolo A. A., Erdeniz N., Reid R. J., et al. , 2006. The role of DNA double-strand breaks in spontaneous homologous recombination in S. cerevisiae. PLoS Genet. 2: 1773–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung G. P., Lee L., Schmidt T. I., Shirahige K., Kobor M. S., 2011. Rtt107 is required for recruitment of the SMC5/6 complex to DNA double strand breaks. J. Biol. Chem. 286: 26250–26257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L. K., Karthikeyan G., Westmoreland J. W., Resnick M. A., 2002. Differential suppression of DNA repair deficiencies of Yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics 160: 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Dong J., Pan X., Oum J. H., Boeke J. D., et al. , 2008. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol. Cell 30: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Dong J., Eichmiller R., Holland C., Minca E., et al. , 2013. Role of Saw1 in Rad1/Rad10 complex assembly at recombination intermediates in budding yeast. EMBO J. 32: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Heyer W. D., 2009. RAD54 controls access to the invading 3′-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 37: 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. C., Tye B. K., 2011. Ploidy dictates repair pathway choice under DNA replication stress. Genetics 187: 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., et al. , 2005. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 19: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., Haber J. E., 1989. Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics 123: 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefshitz B., Parket A., Maya R., Kupiec M., 1995. The role of DNA repair genes in recombination between repeated sequences in yeast. Genetics 140: 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. S., Kim J. S., Park Y. B., Gwon G. H., Cho Y., 2011. Crystal structure of the Mre11-Rad50-ATPgammaS complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 25: 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O., Chahwan C., Yamada Y., de Bruin R. A., Wittenberg C., et al. , 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28: 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos H. B., Strom L., Itoh T., Katou Y., Shirahige K., et al. , 2006. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell 22: 755–767. [DOI] [PubMed] [Google Scholar]
- Lisby M., Rothstein R., Mortensen U. H., 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 98: 8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Antúnez de Mayolo A., Mortensen U. H., Rothstein R., 2003a Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle 2: 479–483. [PubMed] [Google Scholar]
- Lisby M., Mortensen U. H., Rothstein R., 2003b Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 5: 572–577. [DOI] [PubMed] [Google Scholar]
- Lisby M., Barlow J. H., Burgess R. C., Rothstein R., 2004. Choreography of the DNA damage response; spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713. [DOI] [PubMed] [Google Scholar]
- Liu J., Renault L., Veaute X., Fabre F., Stahlberg H., et al. , 2011. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479: 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B., Smith C. E., Symington L. S., 2008. Break-induced replication: What is it and what is it for? Cell Cycle 7: 859–864. [DOI] [PubMed] [Google Scholar]
- Lloyd J., Chapman J. R., Clapperton J. A., Haire L. F., Hartsuiker E., et al. , 2009. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell 139: 100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K. S., Gordenin D. A., Resnick M. A., 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193. [DOI] [PubMed] [Google Scholar]
- Lobachev K., Vitriol E., Stemple J., Resnick M. A., Bloom K., 2004. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 14: 2107–2112. [DOI] [PubMed] [Google Scholar]
- Loog M., Morgan D. O., 2005. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434: 104–108. [DOI] [PubMed] [Google Scholar]
- Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., et al. , 2011. G-quadruplex-induced instability during leading-strand replication. EMBO J. 30: 4033–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Liberi G., Foiani M., 2003. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol. Cell 12: 1499–1510. [DOI] [PubMed] [Google Scholar]
- Lorenz A., Osman F., Sun W., Nandi S., Steinacher R., et al. , 2012. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science 336: 1585–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett S. T., 1994. Sequence of the RAD55 gene of Saccharomyces cerevisiae: similarity of RAD55 to prokaryotic RecA and other RecA-like proteins. Gene 142: 103–106. [DOI] [PubMed] [Google Scholar]
- Lukas C., Falck J., Bartkova J., Bartek J., Lukas J., 2003. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 5: 255–260. [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S., Luke B., 2012. The Mph1 helicase can promote telomere uncapping and premature senescence in budding yeast. PLoS ONE 7: e42028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V., Blackburn E. H., 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73: 347–360. [DOI] [PubMed] [Google Scholar]
- Lydeard J. R., Jain S., Yamaguchi M., Haber J. E., 2007. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448: 820–823. [DOI] [PubMed] [Google Scholar]
- Ma J. L., Kim E. M., Haber J. E., Lee S. E., 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23: 8820–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Westmoreland J. W., Resnick M. A., 2013. Homologous recombination rescues ssDNA gaps generated by nucleotide excision repair and reduced translesion DNA synthesis in yeast G2 cells. Proc. Natl. Acad. Sci. USA 110: E2895–E2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A., Ross L., Dawson D., Hoekstra M. F., Haber J. E., 1996. Meiotic recombination initiated by a double-strand break in rad50Δ yeast cells otherwise unable to initiate meiotic recombination. Genetics 143: 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A., Naylor M. L., Yamaguchi M., Ira G., Haber J. E., 2005. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol. Cell. Biol. 25: 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloisel L., Fabre F., Gangloff S., 2008. DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol. Cell. Biol. 28: 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri H. W., Ngo H. P., Hickson I. D., 2007. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell 18: 4062–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manney T. R., Mortimer R. K., 1964. Allelic mapping in yeast by X-ray-induced mitotic reversion. Science 143: 581–583. [DOI] [PubMed] [Google Scholar]
- Manthey G. M., Bailis A. M., 2010. Rad51 inhibits translocation formation by non-conservative homologous recombination in Saccharomyces cerevisiae. PLoS ONE 5: e11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiasen D. P., Lisby M., 2014. Cell cycle regulation of homologous recombination in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 38: 172–184. [DOI] [PubMed] [Google Scholar]
- Matos J., Blanco M. G., Maslen S., Skehel J. M., West S. C., 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J., Blanco M. G., West S. C., 2013. Cell-cycle kinases coordinate the resolution of recombination intermediates with chromosome segregation. Cell Reports 4: 76–86. [DOI] [PubMed] [Google Scholar]
- Matulova P., Marini V., Burgess R. C., Sisakova A., Kwon Y., et al. , 2009. Cooperativity of Mus81·Mms4 with Rad54 in the resolution of recombination and replication intermediates. J. Biol. Chem. 284: 7733–7745. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mazin A. V., Bornarth C. J., Solinger J. A., Heyer W. D., Kowalczykowski S. C., 2000. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell 6: 583–592. [DOI] [PubMed] [Google Scholar]
- Mazon G., Symington L. S., 2013. Mph1 and Mus81-Mms4 prevent aberrant processing of mitotic recombination intermediates. Mol. Cell 52: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazon G., Lam A. F., Ho C. K., Kupiec M., Symington L. S., 2012. The Rad1-Rad10 nuclease promotes chromosome translocations between dispersed repeats. Nat. Struct. Mol. Biol. 19: 964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleenan A., Clemente-Blanco A., Cordon-Preciado V., Sen N., Esteras M., et al. , 2013. Post-replicative repair involves separase-dependent removal of the kleisin subunit of cohesin. Nature 493: 250–254. [DOI] [PubMed] [Google Scholar]
- McDonald J. P., Rothstein R., 1994. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics 137: 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei A. R., Medhurst A. L., Ling C., Xue Y., Singh T. R., et al. , 2005. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37: 958–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier G., Denis Y., Marc P., Picard L., Dutreix M., 2001. Transcriptional induction of repair genes during slowing of replication in irradiated Saccharomyces cerevisiae. Mutat. Res. 487: 157–172. [DOI] [PubMed] [Google Scholar]
- Milne G. T., Weaver D. T., 1993. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 7: 1755–1765. [DOI] [PubMed] [Google Scholar]
- Mimitou E. P., Symington L. S., 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E. P., Symington L. S., 2009. DNA end resection: many nucleases make light work. DNA Repair (Amst.) 8: 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E. P., Symington L. S., 2010. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 29: 3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine-Hattab J., Rothstein R., 2012. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 14: 510–517. [DOI] [PubMed] [Google Scholar]
- Miret J. J., Pessoa-Brandao L., Lahue R. S., 1998. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95: 12438–12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K., Zhang H., Welz-Voegele C., Jinks-Robertson S., 2010. Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol. Cell 38: 211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K., Lehner K., Jinks-Robertson S., 2013. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet. 9: e1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W. H., Sen S., et al. , 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348. [DOI] [PubMed] [Google Scholar]
- Moreau S., Ferguson J. R., Symington L. S., 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S., Morgan E. A., Symington L. S., 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159: 1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo-Huesca M., Clemente-Ruiz M., Andujar E., Prado F., 2010. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A. Z. PLoS One 5: e12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A. J., Highland J., Krogan N. J., Arbel-Eden A., Greenblatt J. F., et al. , 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119: 767–775. [DOI] [PubMed] [Google Scholar]
- Morrow D. M., Connelly C., Hieter P., 1997. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147: 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen U. H., Bendixen C., Sunjevaric I., Rothstein R., 1996. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. USA 93: 10729–10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen U. H., Erdeniz N., Feng Q., Rothstein R., 2002. A molecular genetic dissection of the evolutionarily conserved N terminus of yeast Rad52. Genetics 161: 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., 1958. Radiobiological and genetic studies on a polyploid series (haploid to hexaploid) of Saccharomyces cerevisiae. Radiat. Res. 9: 312–326. [PubMed] [Google Scholar]
- Mott C., Symington L. S., 2011. RAD51-independent inverted-repeat recombination by a strand-annealing mechanism. DNA Repair (Amst.) 10: 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozlin A. M., Fung C. W., Symington L. S., 2008. Role of the Saccharomyces cerevisiae Rad51 paralogs in sister chromatid recombination. Genetics 178: 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Wright W. D., Ehmsen K. T., Heyer W. D., 2014. The Mus81-Mms4 structure-selective endonuclease requires nicked DNA junctions to undergo conformational changes and bend its DNA substrates for cleavage. Nucleic Acids Res. 42: 6511–6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R., Kaliraman V., Ibrahim S. S., Brill S. J., 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Galvan S., Jimeno S., Rothstein R., Aguilera A., 2013. Histone H3K56 acetylation, Rad52, and non-DNA repair factors control double-strand break repair choice with the sister chromatid. PLoS Genet. 9: e1003237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M. B., Roberts T. M., et al. , 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D., Matsumoto K., Sugimoto K., 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17: 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif N., Penney J., Pal S., Engels W. R., Gloor G. B., 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New J. H., Sugiyama T., Zaitseva E., Kowalczykowski S. C., 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391: 407–410. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A., Chen E. Y., Heffron F., 1986. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc. Natl. Acad. Sci. USA 83: 7831–7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J. A., Sweetser D. B., Clikeman J. A., Khalsa G. J., Wheeler S. L., 1999. Multiple heterologies increase mitotic double-strand break-induced allelic gene conversion tract lengths in yeast. Genetics 153: 665–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolette M. L., Lee K., Guo Z., Rani M., Chow J. M., et al. , 2010. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol. 17: 1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen I., Bentsen I. B., Lisby M., Hansen S., Mundbjerg K., et al. , 2009. A Flp-nick system to study repair of a single protein-bound nick in vivo. Nat. Methods 6: 753–757. [DOI] [PubMed] [Google Scholar]
- Nissar, S., S. M. Baba, T. Akhtar, R. Rasool, Z. A. Shah et al., 2014 RAD51 G135C gene polymorphism and risk of colorectal cancer in Kashmir. Eur. J. Cancer Prev. 23: 264–268. [DOI] [PubMed]
- Niu H., Chung W. H., Zhu Z., Kwon Y., Zhao W., et al. , 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467: 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M., Ruiz-Perez V. L., Woods C. G., Jeggo P. A., Goodship J. A., 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 33: 497–501. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Yu X., Shinohara A., Egelman E. H., 1993. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science 259: 1896–1899. [DOI] [PubMed] [Google Scholar]
- Ohouo P. Y., Bastos de Oliveira F. M., Almeida B. S., Smolka M. B., 2010. DNA damage signaling recruits the Rtt107-Slx4 scaffolds via Dpb11 to mediate replication stress response. Mol. Cell 39: 300–306. [DOI] [PubMed] [Google Scholar]
- Ohuchi T., Seki M., Branzei D., Maeda D., Ui A., et al. , 2008. Rad52 sumoylation and its involvement in the efficient induction of homologous recombination. DNA Repair (Amst.) 7: 879–889. [DOI] [PubMed] [Google Scholar]
- Onoda F., Takeda M., Seki M., Maeda D., Tajima J., et al. , 2004. SMC6 is required for MMS-induced interchromosomal and sister chromatid recombinations in Saccharomyces cerevisiae. DNA Repair (Amst.) 3: 429–439. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., 1983. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. USA 80: 4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J., 1981. Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. USA 78: 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J., 1983. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 101: 228–245. [DOI] [PubMed] [Google Scholar]
- Osborn A. J., Elledge S. J., 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17: 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F., Dixon J., Doe C. L., Whitby M. C., 2003. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12: 761–774. [DOI] [PubMed] [Google Scholar]
- Oum J. H., Seong C., Kwon Y., Ji J. H., Sid A., et al. , 2011. RSC facilitates Rad59-dependent homologous recombination between sister chromatids by promoting cohesin loading at DNA double-strand breaks. Mol. Cell. Biol. 31: 3924–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P., Jaspersen S. L., Miele A., Dekker J., Peterson C. L., 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 23: 912–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek A. L., Kaochar S., Jones H., Elezaby A., Shanks L., et al. , 2009. Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes Dev. 23: 2861–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K., Capra J. A., Zakian V. A., 2011. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B., Liu X., Garcia-Rubio M., Aguilera A., Zhao X., et al. , 2007. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell 18: 2912–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S., Schildkraut E., Lazarin R., Nguyen J., Nickoloff J. A., 2003. Gene conversion tracts in Saccharomyces cerevisiae can be extremely short and highly directional. Nucleic Acids Res. 31: 1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannunzio N. R., Manthey G. M., Bailis A. M., 2008. RAD59 is required for efficient repair of simultaneous double-strand breaks resulting in translocations in Saccharomyces cerevisiae. DNA Repair (Amst.) 7: 788–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannunzio N. R., Manthey G. M., Liddell L. C., Fu B. X., Roberts C. M., et al. , 2012. Rad59 regulates association of Rad52 with DNA double-strand breaks. MicrobiologyOpen 1: 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli E., Chen S., Davies A. A., Huttner D., Krejci L., et al. , 2005. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19: 123–133. [DOI] [PubMed] [Google Scholar]
- Paques F., Haber J. E., 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Leung W. Y., Haber J. E., 1998. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 18: 2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C., Koszul R., Dujon B., Fischer G., 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 4: e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., Botstein D., 1977. Simple mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc. Natl. Acad. Sci. USA 74: 5091–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., Malone R. E., Symington L. S., 1991. Recombination in yeast, pp. 407–521 in The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis and Energetics, edited by Broach J. R., Pringle J. R., Jones E. W. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Petrucelli, N., M. B. Daly, and G. L. Feldman, 2010 Hereditary breast and ovarian cancer due to mutations in BRCA1 and BRCA2. Genet. Med. 12: 245–259. [DOI] [PubMed]
- Petukhova G., Stratton S. A., Sung P., 1999. Single strand DNA binding and annealing activities in the yeast recombination factor Rad59. J. Biol. Chem. 274: 33839–33842. [DOI] [PubMed] [Google Scholar]
- Petukhova G., Sung P., Klein H., 2000. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 14: 2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S., 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433. [DOI] [PubMed] [Google Scholar]
- Plate I., Hallwyl S. C., Shi I., Krejci L., Muller C., et al. , 2008. Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J. Biol. Chem. 283: 29077–29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessis A., Perrin A., Haber J. E., Dujon B., 1992. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics 130: 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., 2009. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 109: 2894–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash L., 1981. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 184: 471–478. [DOI] [PubMed] [Google Scholar]
- Prakash R., Satory D., Dray E., Papusha A., Scheller J., et al. , 2009. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 23: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I., Jentsch S., 2012. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151: 807–820. [DOI] [PubMed] [Google Scholar]
- Puddu F., Granata M., Di Nola L., Balestrini A., Piergiovanni G., et al. , 2008. Phosphorylation of the budding yeast 9–1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol. Cell. Biol. 28: 4782–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Allen-Soltero S. R., Martinez S. L., Chan J. E., Hayes T. K., et al. , 2012. Bioinformatic identification of genes suppressing genome instability. Proc. Natl. Acad. Sci. USA 109: E3251–E3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Pennaneach V., Kolodner R. D., 2005. Saccharomyces cerevisiae as a model system to define the chromosomal instability phenotype. Mol. Cell. Biol. 25: 7226–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Parthun M. R., 2002. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 22: 8353–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell J. C., Fan A., Chan C., Francis L. I., Heller R. C., et al. , 2010. Mec1 is one of multiple kinases that prime the Mcm2–7 helicase for phosphorylation by Cdc7. Mol. Cell 40: 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapakko K., Heikkinen K., Karppinen S. M., Erkko H., Winqvist R., 2007. Germline alterations in the 53BP1 gene in breast and ovarian cancer families. Cancer Lett. 245: 337–340. [DOI] [PubMed] [Google Scholar]
- Rattray A. J., Symington L. S., 1994. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics 138: 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A. J., Symington L. S., 1995. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics 139: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A. J., Shafer B. K., McGill C. B., Strathern J. N., 2002. The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics 162: 1063–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon C., Pilch D. R., Rogakou E. P., Orr A. H., Lowndes N. F., et al. , 2003. Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 4: 678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeyre C., Shore D., 2012. Anticheckpoint pathways at telomeres in yeast. Nat. Struct. Mol. Biol. 19: 307–313. [DOI] [PubMed] [Google Scholar]
- Ribeyre C., Lopes J., Boule J. B., Piazza A., Guedin A., et al. , 2009. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 5: e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert T., Dervins D., Fabre F., Gangloff S., 2006. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 25: 2837–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert T., Vanoli F., Chiolo I., Shubassi G., Bernstein K. A., et al. , 2011. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 471: 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S. A., Sterling J., Thompson C., Harris S., Mav D., et al. , 2012. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell 46: 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T. M., Kobor M. S., Bastin-Shanower S. A., Ii M., Horte S. A., et al. , 2006. Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol. Biol. Cell 17: 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M., 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273: 5858–5868. [DOI] [PubMed] [Google Scholar]
- Rogakou E. P., Boon C., Redon C., Bonner W. M., 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong L., Palladino F., Aguilera A., Klein H. L., 1991. The hyper-gene conversion hpr5–1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 127: 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen D. M., Younkin E. M., Miller S. D., Casper A. M., 2013. Fragile site instability in Saccharomyces cerevisiae causes loss of heterozygosity by mitotic crossovers and break-induced replication. PLoS Genet. 9: e1003817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R., Helms C., Rosenberg N., 1987. Concerted deletions and inversions are caused by mitotic recombination between delta sequences in Saccharomyces cerevisiae. [published erratum appears in Mol Cell Biol 1989 Aug;9(8):3592] Mol. Cell. Biol. 7: 1198–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz J. F., Gomez-Gonzalez B., Aguilera A., 2009. Chromosomal translocations caused by either pol32-dependent or pol32-independent triparental break-induced replication. Mol. Cell. Biol. 29: 5441–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M., Pfander B., Hoege C., Jentsch S., 2006. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 8: 1284–1290. [DOI] [PubMed] [Google Scholar]
- Saini N., Ramakrishnan S., Elango R., Ayyar S., Zhang Y., et al. , 2013a Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502: 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini N., Zhang Y., Nishida Y., Sheng Z., Choudhury S., et al. , 2013b Fragile DNA motifs trigger mutagenesis at distant chromosomal loci in saccharomyces cerevisiae. PLoS Genet. 9: e1003551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Aguilera A., 1994. Increase in incidence of chromosome instability and non-conservative recombination between repeats in Saccharomyces cerevisiae hpr1 delta strains. Mol. Gen. Genet. 245: 224–236. [DOI] [PubMed] [Google Scholar]
- Saponaro M., Callahan D., Zheng X., Krejci L., Haber J. E., et al. , 2010. Cdk1 targets Srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. PLoS Genet. 6: e1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangi P., Bartosova Z., Altmannova V., Holland C., Chavdarova M., et al. , 2014. Sumoylation of the Rad1 nuclease promotes DNA repair and regulates its DNA association. Nucleic Acids Res. 42: 6393–6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H., Tawaramoto M. S., Lao J. P., Hosaka H., Sanda E., et al. , 2013. A new protein complex promoting the assembly of Rad51 filaments. Nat. Commun. 4: 1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugar I., Vazquez M. V., Gallo-Fernandez M., Ortiz-Bazan M. A., Segurado M., et al. , 2013. Temporal regulation of the Mus81-Mms4 endonuclease ensures cell survival under conditions of DNA damage. Nucleic Acids Res. 41: 8943–8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe D., Datta A., Jinks-Robertson S., 2000. Stimulation of mitotic recombination events by high levels of RNA polymerase II transcription in yeast. Mol. Cell. Biol. 20: 5404–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Igarashi S., Hastings P. J., 1988. Analysis of the mechanism for reversion of a disrupted gene. Genetics 119: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller C. B., Lammens K., Guerini I., Coordes B., Feldmann H., et al. , 2012. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat. Struct. Mol. Biol. 19: 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H., Ferreira H., Kalck V., Gehlen L. R., Gasser S. M., 2009. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 23: 928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes D. T., Banerjee M., Bowen B., Curcio M. J., 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159: 1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E. K., Heyer W. D., 2011. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120: 109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebesta M., Burkovics P., Haracska L., Krejci L., 2011. Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Repair (Amst.) 10: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber A., Dion V., Gasser S. M., 2013. Checkpoint kinases and the INO80 nucleosome remodeling complex enhance global chromatin mobility in response to DNA damage. Genes Dev. 27: 1999–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehorn M. G., Sigurdsson S., Bussen W., Unger V. M., Sung P., 2004. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature 429: 433–437. [DOI] [PubMed] [Google Scholar]
- Shah K. A., Shishkin A. A., Voineagu I., Pavlov Y. I., Shcherbakova P. V., et al. , 2012. Role of DNA polymerases in repeat-mediated genome instability. Cell Reports 2: 1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P. P., Zheng X., Epshtein A., Carey J. N., Bishop D. K., et al. , 2010. Swi2/Snf2-related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth. Mol. Cell 39: 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan S. D., Yu X., Roth R., Heuser J. E., Sehorn M. G., et al. , 2008. A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res. 36: 4057–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim E. Y., Ma J. L., Oum J. H., Yanez Y., Lee S. E., 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell. Biol. 25: 3934–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim E. Y., Hong S. J., Oum J. H., Yanez Y., Zhang Y., et al. , 2007. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 27: 1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim E. Y., Chung W. H., Nicolette M. L., Zhang Y., Davis M., et al. , 2010. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29: 3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Ogawa T., 1992. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69: 457–470. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Ogawa T., 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391: 404–407. [DOI] [PubMed] [Google Scholar]
- Shinohara M., Shita-Yamaguchi E., Buerstedde J. M., Shinagawa H., Ogawa H., et al. , 1997. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147: 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A., Shinohara M., Ohta T., Matsuda S., Ogawa T., 1998. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells 3: 145–156. [DOI] [PubMed] [Google Scholar]
- Shor E., Gangloff S., Wagner M., Weinstein J., Price G., et al. , 2002. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics 162: 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E., Weinstein J., Rothstein R., 2005. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics 169: 1275–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R., Arbel-Eden A., Pilch D., Ira G., Bonner W. M., et al. , 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14: 1703–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S., Gallina I., Eckert-Boulet N., Lisby M., 2012. Live cell microscopy of DNA damage response in Saccharomyces cerevisiae. Methods Mol. Biol. 920: 433–443. [DOI] [PubMed] [Google Scholar]
- Silva, S. N., M. Tomar, C. Paulo, B. C. Gomes, A.P. Azevedo et al, 2010 Breast cancer risk and common single nucleotide polymorphisms in homologous recombination DNA repair pathway genes XRCC2, XRCC3, NBS1 and RAD51. Cancer Epidemiol. 34: 85–92. [DOI] [PubMed]
- Sinclair D. A., Guarente L., 1997. Extrachromosomal rDNA circles: a cause of aging in yeast. Cell 91: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Singleton M. R., Wentzell L. M., Liu Y., West S. C., Wigley D. B., et al. , 2002. Structure of the single-strand annealing domain of human RAD52 protein crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Proc. Natl. Acad. Sci. USA 99: 13492–13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M., Peterson C. L., 2009. Chromatin dynamics during repair of chromosomal DNA double-strand breaks. Epigenomics 1: 371–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M., Watanabe S., Johnson A., Moazed D., Peterson C. L., 2009. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell 138: 1109–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren C., Nasmyth K., 2001. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11: 991–995. [DOI] [PubMed] [Google Scholar]
- Smith C. E., Llorente B., Symington L. S., 2007. Template switching during break-induced replication. Nature 447: 102–105. [DOI] [PubMed] [Google Scholar]
- Smith C. E., Lam A. F., Symington L. S., 2009. Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase delta complex. Mol. Cell. Biol. 29: 1432–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J., Rothstein R., 1999. An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics 151: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Hwang J. Y., Banerjee S., Majeed A., Gupta A., et al. , 2004. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101: 9039–9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J. M., Lopes M., Foiani M., 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602. [DOI] [PubMed] [Google Scholar]
- Solinger J. A., Kiianitsa K., Heyer W. D., 2002. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell 10: 1175–1188. [DOI] [PubMed] [Google Scholar]
- Sollier J., Driscoll R., Castellucci F., Foiani M., Jackson S. P., et al. , 2009. The Saccharomyces cerevisiae Esc2 and Smc5–6 proteins promote sister chromatid junction-mediated intra-S repair. Mol. Biol. Cell 20: 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Dominska M., Greenwell P. W., Petes T. D., 2014. Genome-wide high-resolution mapping of chromosome fragile sites in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 111: E2210–E2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., et al. , 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Charles J., Hazkani-Covo E., Yin Y., Andersen S. L., Dietrich F. S., et al. , 2012. High-resolution genome-wide analysis of irradiated (UV and gamma-rays) diploid yeast cells reveals a high frequency of genomic loss of heterozygosity (LOH) events. Genetics 190: 1267–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafa A., Donnianni R. A., Timashev L. A., Lam A. F., Symington L. S., 2014. Template switching during break-induced replication is promoted by the Mph1 helicase in Saccharomyces cerevisiae. Genetics 196: 1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead B. E., Brandl C. J., Sandre M. K., Davey M. J., 2012. Mcm2 phosphorylation and the response to replicative stress. BMC Genet. 13: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoepker C., Hain K., Schuster B., Hilhorst-Hofstee Y., Rooimanst M. A., et al. , 2011. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nature Genet. 43: 138–141. [DOI] [PubMed] [Google Scholar]
- Storchova Z., Breneman A., Cande J., Dunn J., Burbank K., et al. , 2006. Genome-wide genetic analysis of polyploidy in yeast. Nature 443: 541–547. [DOI] [PubMed] [Google Scholar]
- Strathern J. N., Weinstock K. G., Higgins D. R., McGill C. B., 1991. A novel recombinator in yeast based on gene II protein from bacteriophage f1. Genetics 127: 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L., Lindroos H. B., Shirahige K., Sjogren C., 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16: 1003–1015. [DOI] [PubMed] [Google Scholar]
- Strom L., Karlsson C., Lindroos H. B., Wedahl S., Katou Y., et al. , 2007. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317: 242–245. [DOI] [PubMed] [Google Scholar]
- Sugawara N., Haber J. E., 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12: 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Kowalczykowski S. C., 2002. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 277: 31663–31672. [DOI] [PubMed] [Google Scholar]
- Sugawara N., Paques F., Colaiacovo M., Haber J. E., 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94: 9214–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Ira G., Haber J. E., 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20: 5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Wang X., Haber J. E., 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12: 209–219. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Zaitseva E. M., Kowalczykowski S. C., 1997. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 272: 7940–7945. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., New J. H., Kowalczykowski S. C., 1998. DNA annealing by Rad52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. USA 95: 6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Kantake N., Wu Y., Kowalczykowski S. C., 2006. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 25: 5539–5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Bai J., Chen F., Jin Y., Yu Y., et al. , 2011. RAD51 G135C polymorphism is associated with breast cancer susceptibility: a meta-analysis involving 22,399 subjects. Breast Cancer Res. Treat. 125: 157–161. [DOI] [PubMed] [Google Scholar]
- Sun W., Nandi S., Osman F., Ahn J. S., Jakovleska J., et al. , 2008. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell 32: 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265: 1241–1243. [DOI] [PubMed] [Google Scholar]
- Sung P., 1997a Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272: 28194–28197. [DOI] [PubMed] [Google Scholar]
- Sung P., 1997b Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11: 1111–1121. [DOI] [PubMed] [Google Scholar]
- Sung P., Krejci L., Van Komen S., Sehorn M. G., 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278: 42729–42732. [DOI] [PubMed] [Google Scholar]
- Symington L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Kang L. E., Moreau S., 2000. Alteration of gene conversion tract length and associated crossing over during plasmid gap repair in nuclease-deficient strains of Saccharomyces cerevisiae. Nucleic Acids Res. 28: 4649–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakal B., Branzei D., 2013. Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover. EMBO J. 32: 1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Wu R., 1980. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature 284: 426–430. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W., 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Taddei A., Gasser S. M., 2012. Structure and function in the budding yeast nucleus. Genetics 192: 107–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Mizoi J., Toh E. A., Kikuchi Y., 2000. Yeast Ulp1, an Smt3-specific protease, associates with nucleoporins. J. Biochem. 128: 723–725. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y., Horiuchi T., Kobayashi T., 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 17: 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini B. A., Tyler J. K., 2005. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 25: 4903–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Dominska M., Greenwell P. W., Harvanek Z., Lobachev K. S., et al. , 2011. Friedreich’s ataxia (GAA)n*(TTC)n repeats strongly stimulate mitotic crossovers in Saccharomyces cerevisae. PLoS Genet. 7: e1001270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Dominska M., Gawel M., Greenwell P. W., Petes T. D., 2013. Genomic deletions and point mutations induced in Saccharomyces cerevisiae by the trinucleotide repeats (GAA.TTC) associated with Friedreich’s ataxia. DNA Repair (Amst.) 12: 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Li X., Liu Y., Ruan J., Qi S., et al. , 2012. Structural analysis of Shu proteins reveals a DNA binding role essential for resisting damage. J. Biol. Chem. 287: 20231–20239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y. D., Sidebotham J. M., Wu L., 2010. Mph1 requires mismatch repair-independent and -dependent functions of MutSalpha to regulate crossover formation during homologous recombination repair. Nucleic Acids Res. 38: 1889–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J. A., Diffley J. F., 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557. [DOI] [PubMed] [Google Scholar]
- Therizols P., Fairhead C., Cabal G. G., Genovesio A., Olivo-Marin J. C., et al. , 2006. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 172: 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R., 1989a Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R., 1989b The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics 123: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittel-Elmer M., Lengronne A., Davidson M. B., Bacal J., Francois P., et al. , 2012. Cohesin association to replication sites depends on rad50 and promotes fork restart. Mol. Cell 48: 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh, G. W., A. M. O’Shaughnessy, S. Jimeno, I. M. Dobbie, M. Grenon et al., 2006 Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA Repair (Amst.) 5: 693–703. [DOI] [PubMed]
- Toh G. W., Sugawara N., Dong J., Toth R., Lee S. E., et al. , 2010. Mec1/Tel1-dependent phosphorylation of Slx4 stimulates Rad1-Rad10-dependent cleavage of non-homologous DNA tails. DNA Repair (Amst.) 9: 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J., Sunjevaric I., De Piccoli G., Sacher M., Eckert-Boulet N., et al. , 2007. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 9: 923–931. [DOI] [PubMed] [Google Scholar]
- Trovesi C., Falcettoni M., Lucchini G., Clerici M., Longhese M. P., 2011. Distinct Cdk1 requirements during single-strand annealing, noncrossover, and crossover recombination. PLoS Genet. 7: e1002263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo K. M., Sung P., 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 276: 35458–35464. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H., Ogawa H., 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell 11: 2221–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y., Mitsuoka C., Terasawa M., Ogawa H., Ogawa T., 2005. Xrs2p regulates Mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol. Biol. Cell 16: 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T., Fleming A. B., Nickoloff J. A., Osley M. A., 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438: 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T., Lo Y. C., Krishna S., Sterk R., Osley M. A., et al. , 2009. INO80-dependent chromatin remodeling regulates early and late stages of mitotic homologous recombination. DNA Repair (Amst.) 8: 360–369. [DOI] [PubMed] [Google Scholar]
- Unal E., Arbel-Eden A., Sattler U., Shroff R., Lichten M., et al. , 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16: 991–1002. [DOI] [PubMed] [Google Scholar]
- Unal E., Heidinger-Pauli J. M., Koshland D., 2007. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317: 245–248. [DOI] [PubMed] [Google Scholar]
- Usui T., Ohta T., Oshiumi H., Tomizawa J., Ogawa H., et al. , 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95: 705–716. [DOI] [PubMed] [Google Scholar]
- Valencia-Burton M., Oki M., Johnson J., Seier T. A., Kamakaka R., et al. , 2006. Different mating-type-regulated genes affect the DNA repair defects of Saccharomyces RAD51, RAD52 and RAD55 mutants. Genetics 174: 41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E. A., Cross F. R., 1999. Interaction between the MEC1-dependent DNA synthesis checkpoint and G1 cyclin function in Saccharomyces cerevisiae. Genetics 151: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Fritsch O., Gasser S. M., 2007. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 26: 4113–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Fritsch O., Hohn B., Gasser S. M., 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788. [DOI] [PubMed] [Google Scholar]
- Van Komen S., Petukhova G., Sigurdsson S., Stratton S., Sung P., 2000. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell 6: 563–572. [DOI] [PubMed] [Google Scholar]
- Varon R., Vissinga C., Platzer M., Cerosaletti K. M., Chrzanowska K. H., et al. , 1998. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93: 467–476. [DOI] [PubMed] [Google Scholar]
- Vasan S., Deem A., Ramakrishnan S., Argueso J. L., Malkova A., 2014. Cascades of genetic instability resulting from compromised break-induced replication. PLoS Genet. 10: e1004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaze M. B., Pellicioli A., Lee S. E., Ira G., Liberi G., et al. , 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10: 373–385. [DOI] [PubMed] [Google Scholar]
- Veaute X., Jeusset J., Soustelle C., Kowalczykowski S. C., Le Cam E., et al. , 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312. [DOI] [PubMed] [Google Scholar]
- Villarreal D. D., Lee K., Deem A., Shim E. Y., Malkova A., et al. , 2012. Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet. 8: e1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Keil R. L., Roeder G. S., 1987. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 48: 1071–1079. [DOI] [PubMed] [Google Scholar]
- Vuorela M., Pylkas K., Hartikainen J. M., Sundfeldt K., Lindblom A., et al. , 2011. Further evidence for the contribution of the RAD51C gene in hereditary breast and ovarian cancer susceptibility. Breast Cancer Res. Treat. 130: 1003–1010. [DOI] [PubMed] [Google Scholar]
- Wagner M., Price G., Rothstein R., 2006. The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae. Genetics 174: 555–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L., Gore S. K., Koshland D., 2013. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. eLife 2: e00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R., 1989. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58: 409–419. [DOI] [PubMed] [Google Scholar]
- Wang X., Ira G., Tercero J. A., Holmes A. M., Diffley J. F., et al. , 2004. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 6891–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt P. M., Hickson I. D., Borts R. H., Louis E. J., 1996. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger R. J., Zakian V. A., 2012. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 191: 1073–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz-Voegele C., Jinks-Robertson S., 2008. Sequence divergence impedes crossover more than noncrossover events during mitotic gap repair in yeast. Genetics 179: 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland T. J., Wickramasekara S. M., Guo A. Y., Selim A. L., Winsor T. S., et al. , 2009. Comparative genome-wide screening identifies a conserved doxorubicin repair network that is diploid specific in Saccharomyces cerevisiae. PLoS ONE 4: e5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. J., Williams R. S., Williams J. S., Moncalian G., Arvai A. S. et al, 2011. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 18: 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. A., Kwon Y., Xu Y., Chung W. H., Chi P., et al. , 2013. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature 502: 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltzius J. J., Hohl M., Fleming J. C., Petrini J. H., 2005. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 12: 403–407. [DOI] [PubMed] [Google Scholar]
- Wright W. D., Heyer W. D., 2014. Rad54 functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D loop formation. Mol. Cell 53: 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Hickson I. D., 2003. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874. [DOI] [PubMed] [Google Scholar]
- Wu Y., Sugiyama T., Kowalczykowski S. C., 2006. DNA annealing mediated by Rad52 and Rad59 proteins. J. Biol. Chem. 281: 15441–15449. [DOI] [PubMed] [Google Scholar]
- Yagle K., McEntee K., 1990. The DNA damage-inducible gene DIN1 of Saccharomyces cerevisiae encodes a regulatory subunit of ribonucleotide reductase and is identical to RNR3. Mol. Cell. Biol. 10: 5553–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y. X., Schiestl R. H., Prakash L., 1995. Mating-type suppression of the DNA-repair defect of the yeast rad6 delta mutation requires the activity of genes in the RAD52 epistasis group. Curr. Genet. 28: 12–18. [DOI] [PubMed] [Google Scholar]
- Yang R., Chen B., Pfutze K., Buch S., Steinke V., et al. , 2014. Genome-wide analysis associates familial colorectal cancer with increases in copy number variations and a rare structural variation at 12p12.3. Carcinogenesis 35: 315–323. [DOI] [PubMed] [Google Scholar]
- Yin Y., Petes T. D., 2013. Genome-wide high-resolution mapping of UV-induced mitotic recombination events in Saccharomyces cerevisiae. PLoS Genet. 9: e1003894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Ma Y., Tang S., Hwang P. Y., Boiteux S., et al. , 2010. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol. Cell 40: 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Tang S., Ma Y., Hunter N., 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lawrence C. W., 2005. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl. Acad. Sci. USA 102: 15954–15959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Shim E. Y., Davis M., Lee S. E., 2009. Regulation of repair choice: Cdk1 suppresses recruitment of end joining factors at DNA breaks. DNA Repair (Amst.) 8: 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Shishkin A. A., Nishida Y., Marcinkowski-Desmond D., Saini N., et al. , 2012. Genome-wide screen identifies pathways that govern GAA/TTC repeat fragility and expansions in dividing and nondividing yeast cells. Mol. Cell 48: 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Saini N., Sheng Z., Lobachev K. S., 2013. Genome-wide screen reveals replication pathway for quasi-palindrome fragility dependent on homologous recombination. PLoS Genet. 9: e1003979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G., 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C., Diffley J. F., 2008. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 27: 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Rothstein R., 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90: 87–96. [DOI] [PubMed] [Google Scholar]