Abstract
Rare genetic variants have recently been studied for genome-wide associations with human complex diseases. Existing rare variant methods are based on the hypothesis-testing framework that predefined variant sets need to be tested separately. The power of those methods is contingent upon accurate selection of variants for testing, and frequently, common variants are left out for separate testing. In this article, we present a novel Bayesian method for simultaneous testing of all genome-wide variants across the whole frequency range. The method allows for much more flexible grouping of variants and dynamically combines them for joint testing. The method accounts for correlation among variant sets, such that only direct associations with the disease are reported, whereas indirect associations due to linkage disequilibrium are not. Consequently, the method can obtain much improved power and flexibility and simultaneously pinpoint multiple disease variants with high resolution. Additional covariates of categorical, discrete, and continuous values can also be added. We compared our method with seven existing categories of approaches for rare variant mapping. We demonstrate that our method achieves similar power to the best methods available to date when testing very rare variants in small SNP sets. When moderately rare or common variants are included, or when testing a large collection of variants, however, our method significantly outperforms all existing methods evaluated in this study. We further demonstrate the power and the usage of our method in a whole-genome resequencing study of type 1 diabetes.
Keywords: N brare variant, disease association mapping, Bayesian graphical model
WITH recent advances in sequencing technologies (Shendure and Ji 2008), genome-wide association studies (GWAS) for complex diseases have included both rare and structural variants for association mapping. Individuals’ genomes carry many more rare as opposed to common variants in the human population. Rare variants are more likely to be the mutations under selection and their effects could potentially explain a portion of the missing heritability in complex diseases (Bodmer and Bonilla 2008; Schork et al. 2009). Identifying disease-associated rare variants at a large scale, however, is statistically challenging. Evaluating the effects of individual rare mutations to the disease risk is powerless due to their low frequency in the population, for which tens of thousands or even hundreds of thousands of individuals may be needed to obtain sufficient statistical power.
An alternative approach is to simultaneously test the effects of multiple rare variants to accumulate sufficient statistical power in limited samples. Many burden tests (Morgenthaler and Thilly 2007; Li and Leal 2008; Pan 2009; Madsen and Browning 2009; Price et al. 2010; Morris and Zeggini 2010; Han and Pan 2010; Zawistowski et al. 2010) have been developed following this type of approach, which test the cumulative effects of an entire set of SNPs. Burden tests are powerful when most rare variants under testing have the same direction of effects to the disease risk, but are otherwise less effective when some of the variants have opposite effects and/or if most of the variants under testing are not contributing to the disease risk (Neale et al. 2011; Basu and Pan 2011). Depending on how minor alleles are accumulated into a set, some burden tests could also produce seriously inflated false positives and lose power due to potential correlation among variants. To overcome the limitation of burden tests, random-effect variational methods (Wu et al. 2011; Lin and Tang 2011) have been recently proposed to detect association of rare variants with opposite effects allowing most variants in a set to have near zero or no effects. The random-effect models achieve power by evaluating the variance of the estimated disease effects of multiple rare variants, but not testing the mean effect size. Complementary to the burden tests, the variational effect models are powerful when the effects of rare variants have opposite signs and are small, but do not perform as well when most variants in a set have effects in the same direction. Given that we do not know which of the two scenarios is more likely in human complex diseases, or if both occur simultaneously, the two types of approaches were recently merged to improve power (Lee et al. 2012).
Despite a large number of rare variant testing methods developed to date, almost all of them perform hypothesis testing on predefined SNP sets. There are several limitations of such approaches. First, only the predefined sets of SNPs can be tested for associations, whereas improperly defined sets containing too many nondisease variants will result in loss of power. Second, each test is carried out independently without accounting for the correlation between tests. Tests are correlated not only because of linkage disequilibrium (LD) among SNPs, but also because the SNP sets under testing may overlap. SNP sets are often defined based on biological knowledge (e.g., genes and pathways), the sizes of which vary considerably. There are substantial overlaps in gene/pathway annotations, yet few existing methods can handle overlapping or nested tests properly. Third, many methods work well only for rare variants. It requires ad hoc choices of how “rare” a variant needs to be to be tested. The variants with minor allele frequency (MAF) above a threshold will be left out without testing, or tested separately, which reduces power. Most current rare variant methods follow the single-“variant”-test paradigm that was conventionally used in GWAS, except that a variant is now a set of SNPs rather than a single SNP. As a result, current rare variant methods inherit the same drawbacks as those experienced in single SNP tests.
To tackle the above-mentioned limitations for rare variant mapping, we propose a new approach based on statistical variable selection, i.e., a joint model for selecting “variables” from all rare and common variants. The new method is generalized from our previously developed method called BEAM3 (Zhang 2011) for common variants. In the new method, a variable can be either a single SNP or a set of SNPs. The new method retains most advantages of the joint modeling approaches for GWAS and simultaneously works for rare variants. In particular:
The method alleviates the need of accurate preselection of SNP sets for rare variant testing by dynamically grouping the predefined SNP sets for joint testing.
The method handles correlation among sets of SNPs via Bayesian graphical models, such that indirect disease associations purely due to correlation with “true” disease variants are filtered out. Unlike typical solutions that model the dependence structure of all variables, our approach handles dependence implicitly and locally. While the former is computationally prohibitive in GWAS, the latter can satisfactorily resolve the dependence issue with drastically reduced computation time.
With dependence accounted for, the method pinpoints the most likely subsets of disease variants within local regions, such that the method achieves much improved mapping resolution than existing tools.
The method handles both common and rare SNPs without arbitrary separation. We allow SNPs to be represented in both forms of a single unit (to test the effect of itself) and as part of a group of SNPs (to test its group effect jointly with other SNPs). More generally, the method is flexible in that it allows the users to group SNPs with overlaps. Our method analyzes all SNP sets and their combinations in a joint Bayesian probabilistic model, where the best SNP sets associated with the disease are automatically selected and the multiplicity issue is handled via Bayesian priors.
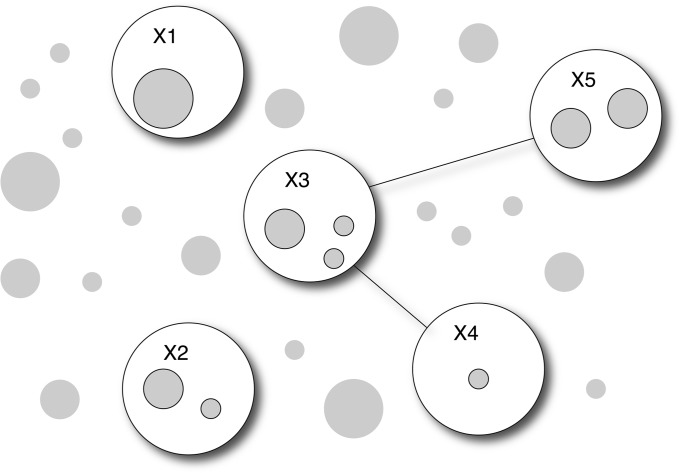
An illustration of the framework of our method is shown in Figure 1. Each shaded circle in the figure represents a user-defined set of SNPs. Each set of SNPs may contain one or multiple SNPs. Each SNP may also appear in multiple sets. The size of each shaded circle is proportional to the number of SNPs contained in the set. Existing approaches are designed to test each set of SNPs separately without considering their joint distributions. In contrast, we model the joint distribution of all SNP sets and use Bayesian graphical models to identify the best combinations of SNP sets for testing their joint associations with the disease. In Figure 1, some SNP sets are selected to form nodes (open circles) in the graph if they are associated with the disease. The connectivity between nodes suggests joint disease association between pairs of nodes. For the example in Figure 1, SNP sets in nodes X1 and X2 are associated with the disease independently, SNP sets in the pairs of nodes (X3, X4) and (X3, X5) are associated with the disease jointly, and the association of SNP sets in X4 and X5 are conditionally independent given X3. Using a Monte Carlo Markov chain (MCMC) algorithm, we dynamically explore combinations of SNP sets to maximize the power of association mapping, and we learn the disease association structure via Bayesian graphs.
Figure 1.
Illustration of the model principle. Predefined SNP sets (shaded circles) are in different sizes. A graph is used to identify SNP sets (included in nodes, open circle) that are associated with the disease, either by itself or jointly with other SNP sets. Connectivity between nodes further combines selected SNP sets from pairs of nodes for joint testing. The SNP sets to be selected in the graph and the graph structures are both learned from the data.
Our method evaluates the association between a SNP set and the disease traits using a Bayesian regression model. In a regression framework, covariates such as environmental factors, individual factors, and population structure components can be incorporated to account for confounding effects. Different from conventional regression models, our regression inversely models the distribution of genotype data on the disease traits. This inverse modeling has several advantages: (1) genotype distribution is relatively simple to model, whereas disease traits may follow any distribution; (2) direct associations with the disease can be effectively distinguished from indirect associations (due to SNP correlation) by explicitly modeling the joint distribution of genotypes at multiple SNPs; and (3) we can detect genetic effects on the variation or high-order moments of the disease traits by including interaction terms, and we can also include multivariate traits.
Using data from the 1000 Genomes project (1000 Genomes Project Consortium 2010), we performed extensive simulation studies to evaluate the power of our method compared to six sets of existing rare variant methods (Pan 2009; Price et al. 2010; Neale et al. 2011; Wu et al. 2011; Lee et al. 2012; Ionita-Laza et al. 2013) and a single SNP test method (Conneely and Boehnke 2007). We show that the new method performs similarly to the best rare variant methods when testing only a handful number of very rare variants. The method, however, performs better and sometimes substantially so, when moderately rare or common variants are included and/or when the disease variants are not randomly distributed in a predefined genomic interval for testing. In addition, when testing associations in a large collection of variants, which is the common scenario in practice, our method substantially outperforms existing methods. Beyond set-based tests, the new method further reveals the locations of the most likely disease variants via Bayesian variable selection. We demonstrate an application of our method to a whole-genome resequencing data set generated in our laboratory from 97 type 1 diabetes (T1D) patients, where we handled sample stratifications and identified novel T1D loci.
Materials and Methods
The general model framework
Let Y = (Y1,…,Yp) denote the disease data measured on p traits, where Yj for j = 1,…, p is a n-dim vector containing data in n individuals. Since we use Y as independent variables, it may take any measurements such as discrete, continuous, and categorical. Let X = (X1,…, XL) denote the genotype data at L SNP sets (each SNP set may contain one or more SNPs; i.e., each Xi could be a matrix by itself), and Z = (Z1,…,Zm) denote additional m covariates to be adjusted for their confounding effects. Again, each Xl (l = 1,…, L) and Zk (k = 1,…, m) contain data in n individuals. Our task is to identify a subset of SNPs in X that are directly associated (not due to LD with other genotyped SNPs) with at least one trait in Y, conditioning on Z. Let XA and XU denote a nonoverlapping partition of SNPs, such that XA include the SNPs directly associated with Y, XU denote the remaining SNPs, and X = {XA, XU}. Let A denote the partition. We write the joint probability
| (1) |
The second equality in (1) is due to our definition that XU is not associated with Y conditioning on XA and Z (but XU could be marginally associated with Y due to its LD with XA). This is a major distinction between our method and existing ones, as the latter do not distinguish the two.
Our goal is to identify the partition A, which relies solely on Pr(XU|XA,Z,A)Pr(XA|Y,Z,A)Pr(A), which can be rewritten as
| (2) |
Note that Equation 2 is proportional to the odds of XA against Y conditioning on Z, along with a prior distribution of partition A. We dropped A from the condition of probability function Pr(X|Z) because the partition is irrelevant without disease information Y. Therefore, we need only to compute the term within the brackets in (2) to identify the partition A.
When the size of A is large, directly modeling Pr(XA|Y,Z,A) and Pr(XA|Z,A) as multivariate distributions can be powerless due to the quickly increasing size of model parameters. Instead, we use undirected acyclic graphs (Zhang 2011) to reduce model complexities. Let GA = (NA, EA) and GA′ = (NA′, EA′) denote the graphical structures of XA with and without Y, respectively, where a node (N) denotes one or a set of SNPs in XA, and an edge (E) denotes “interactions” (joint association) between the two nodes. We augment Pr(XA|Y,Z,A) in (2) to
| (3) |
where “a in NA” denotes the enumeration of all nodes, and a∼a′ in EA denotes the enumeration of all edges. Similarly, we write Pr(XA|Z,A) in (2) as
| (4) |
The difference between (3) and (4) is that the graphs are different, and we sum over in (4) as it is in the denominator of the odds in (2). Plugging (3) and (4) back into (2) [replace Pr(XA|Y,Z,A) in (2) by Pr(GA,XA|Y,Z,A) in (3)], we use MCMC to learn the SNP partition A as well as the graph GA, which we call a disease graph that further details the joint association structures of the identified disease SNPs.
In this study, we choose simple prior distributions for convenience. We assign each SNP set with equal probability (p) to be included in XA, and hence the prior distribution of the size of A is Binomial(L, p). We use a Pitman–Yor process (Pitman and Yor 1997) to further partition SNP sets in XA into nodes in GA (and GA′), with strength parameter 0.5. We use independent Bernoulli priors with probability 0.5 to indicate the presence of an edge between each pair of nodes, with constraint that the graph must be acyclic (for simplicity, we ignore the difference in normalizing constants due to this constraint, and such omission can be regarded as part of a prior setup).
From (2), the model parameters to be updated by MCMC include the SNP partition A and the disease graph GA (nodes and edges). We show in the next section that additional parameters for modeling the data distribution can be analytically integrated out. As a result, after random initialization, our MCMC algorithm works by iteratively adding/removing a SNP set in/out of XA one at a time and, simultaneously, updating GA by adding/removing a SNP set in/out of a graph node and adding/removing an edge between two nodes. All these updating procedures are done via standard Gibbs samplers derived from (2), details of which are omitted here but can be found in Zhang (2011). Finally, enumerating all graphs in (4) can be time consuming for large size of A. For quick computation, we provide the users with an option to enumerate only a subset of graphs that shares the same structure as that of GA except for the current SNP set to be updated; i.e., we enumerate only {: = GA,-i} in (4), where SNP set i is the set to be added/removed from XA. This is an approximate solution that does not yield the correct posterior distribution from (2). Empirically, however, we found that this option produces very similar results to that produced by the full model when the signals are not extremely strong (as is the case in GWAS), but it results in a dramatic reduction in computing time (Zhang 2011).
The rationale underlying our approach is to evaluate whether a SNP set i (Xi) should be added into the disease partition A, given those SNP sets already included in A and the covariates Z. This is done by comparing probabilities (3) and (4) for Xi_ during MCMC, where (3) represents the probability that Xi is associated with Y conditioning on current XA and Z, and (4) represents not associated. Note that (3) is a more complex model (with more parameters) than (4) due to Y. Via Bayesian priors, therefore, (3) tends to be smaller than (4) when Xi is not associated with Y, and thus Xi tends not to be included in the disease partition A. Also note that SNP dependence is modeled via Bayesian graphical models in (4), which accounts for LD. As a result, our method is able to distinguish direct disease association from indirect association due to SNP correlation.
Bayesian regression for a set of variants
A major difference between the new method and the original BEAM3 algorithm lies in the definition of the probability functions Pr(Xa|GA,Y,Z,A) in (3) and Pr(Xa|GA,Z,A) in (4). In BEAM3, we used a saturated multinomial distribution to describe the genotypes in SNP set Xa, which works powerfully for common SNPs, but not so much for rare variants because there are many more rare variants to be tested together. It is also analytically complicated to incorporate continuous values of Y and covariates Z in a multinomial distribution. In the new method, therefore, we model Pr(Xa|GA,Y,Z,A) and Pr(Xa|GA,Z,A) by a multivariate regular Bayesian regression function. Using conjugate priors, it is analytically easy to compute and straightforward to incorporate any forms of disease traits Y and covariates Z without model parameter estimation.
Let Xa denote a (n × q) response matrix of genotype data, where n denotes the total number of individuals and q denotes the number of SNPs to be tested in a set. By default, Xa contains the minor allele counts (0, 1, 2) per individual per SNP. Alternatively, a dummy coding for the three genotypes can be used. Let Y be a (n × p) predictor matrix of disease traits. Let Z be a (n × m) matrix of covariates. Without loss of generality, we assume that Xa, Y, and Z are all column centered. Our regression model assumes that
where B denotes a (p × q) matrix representing the effects of Y on SNPs in Xa, C denotes a (m × q) matrix representing the effects of covariates, and Σ denotes a (q × q) covariance matrix of noise.
Since our interest is only to identify SNP partitions, whereas SNP effects can always be estimated in postanalysis, we analytically integrate out the parameters (B, C, Σ). We assume that B follows a matrix normal distribution MN(0, HB, Σ), C follows another matrix normal distribution MN(0, HC, Σ), and Σ follows an inverse Wishart distribution IW(Ψ, ν). Here, HB = diag(c/q, q), HC = diag(h, m), Ψ = Ιq, ν = θ/2+1 denote fixed hyperparameters. By default, we choose c = 0.01 and h = 1000. A small value of c penalizes on large magnitude of the effects of disease Y, and a large value of h allows any magnitude of the effects of covariates Z.
Let H = diag(HB, HC) denote a (p + m) × (p + m) diagonal block matrix carrying HB and HC along its diagonal, U = (Y, Z) denote a n × (p + m) matrix with Y and Z combined. Following standard procedures, it is straightforward to show the following conditional probability function with model parameters (except for hyperparameters) integrated out
| (5) |
where Γq(.) denotes a multivariate Gamma function.
Using (5), we can further obtain the null function Pr(Xa|GA,Z) by removing Y from U and modify the dimension parameters for matrices accordingly. Finally, we plug (5) back into (3) and (4), which completes the new model.
Our choice of the Gaussian regression form in (5) is mainly due to its analytical simplicity. Not only is it convenient to model SNP correlation and include covariates, but also its closed-form marginal probability functions enable practical computation of genome-wide data sets without estimating continuous parameters. The novelty of our method does not lie in Equation 5. Instead, it lies in our joint modeling approach defined in (2), the usage of Bayesian graphical models in (3) and (4) for implicit modeling of SNP correlation, and the Bayesian approach for integrating common and rare variants via a joint probabilistic framework for testing their marginal and joint effects. To our best knowledge, no other methods have been able to achieve the same goals and simultaneously being computationally feasible for large data sets.
Construction of SNP sets
When testing a single set of SNPs, Equation 5 can synergize information of the disease effects in both directions, which is similar to the existing variational methods. Our model further dynamically explores combinations of SNP sets for joint testing via MCMC. This is a variable selection procedure that is unique compared to hypothesis testing. When a pathway involves many genes, it is unclear whether it will be more powerful to test all genes in the pathway or to focus on a subset of genes. Using our method, the users can define each gene or a subset of genes as a variable and then let the method explore combinations of SNP sets for the most powerful association mapping. Each gene often carries several SNPs, both common and rare. It is unclear what is the best cutoff for “rare” variants to be tested together. In our method, the users can define a SNP as a variable by itself and simultaneously group the SNP with others as a set. Our method then automatically evaluates the individual effect and the group effect of the same SNP simultaneously to maximize power.
The new method allows three ways to define sets of SNPs ahead of the analysis:
As used by current rare variant methods, the users can define SNP sets in genic regions based on biological knowledge.
The users can input two cutoffs a1 ≤ a2 and a parameter d to define SNP sets, particularly for intergenic regions. For SNPs whose MAF > a1, we define the SNP as a variable by itself. For SNPs whose MAF < a2, we group them together if they are within d SNPs away. Since a1 ≤ a2, SNPs with MAF between a1 and a2 will be evaluated for both single and group effects. This approach creates a buffer that effectively alleviates the need for a hard and ad hoc threshold for defining common and rare variants. To the extremes, when a1 = a2 = 0, all SNPs will be tested individually; when a1 = a2 = 1, all SNPs will be tested in sets; and when a1 = 0, a2 = 1, all SNPs will be tested for both individual and group effects. By default, we let a1 = 0.005, a2 = 0.05, d = 30.
The users can ask the method to hierarchically split large SNP sets into smaller sets. For a predefined SNP set containing many SNPs, we introduce k additional SNP sets that are subsets of the original SNP set. If some of the k new SNP sets still contain too many SNPs (greater than a user-specified threshold), we split them further. As a result, the sets of SNPs to be analyzed will include (i) the original large SNP set; (ii) the k subsets; and (iii) additional smaller subsets split hierarchically. This creates new SNP sets in different sizes to be tested for association, which increase the chance for the true disease variants to be properly covered and detected with improved power.
In summary, our method allows for greater flexibility than existing methods in defining SNP sets, testing combination of SNP sets, evaluating both individual and group effects, and testing both common and rare variants without hard cutoffs. Also, SNP sets may overlap, where the correlation among overlapping SNP sets is accounted for via probabilities.
Data simulation
We used the phased haplotype data from the 1000 Genomes project to generate simulated case control data sets in this study. Using individuals with European origins, we generated new haplotypes as mosaic combinations of the 1000 Genomes haplotypes, with recombination rate 1 per 100 kb. We then generated new individuals by randomly pairing the new haplotypes. The data of each new individual contained genotypes at L consecutive SNPs in a randomly chosen region. Among the L SNPs, we randomly selected x SNPs as the disease variants. For a given disease model specified in Results, we then generated cases and controls from the new individuals according to the genotypes at the x selected SNPs.
Results
Simulation study in small data sets
We first performed simulation studies to evaluate the power of our method (implemented in BEAM3) compared to seven categories of existing methods on SNP sets that are small enough (a few hundreds of SNPs) such that a single test can be performed on all SNPs together. The methods we compared with include (1) SKAT (Wu et al. 2011), SKAT-O (Lee et al. 2012), and SKAT-C (Ionita-Laza et al. 2013), which are kernel regression methods, and SKAT-C combines effects of both rare and common variants; (2) MultiVar, a standard multivariate score test (Wald test); (3) SSU and SSUw (Pan 2009), unweighted and weighted sum of squared scores; (4) Common, a standard regression assuming same effect of all variants; (5) Single, a single SNP test reporting minimum P-value adjusted by multiple testing corrections (Conneely and Boehnke 2007); (6) C-alpha (Neale et al. 2011), a homogeneity test of a set of Binomial proportions; and (7) VT1-4 (Price et al. 2010), a regression method subject to variable allele-frequency thresholds using four different criteria. These methods employ very different approaches and are good representatives of the current rare variant mapping algorithms. All methods are capable of detecting effects in opposite directions.
We simulated case control data sets under four disease models. Each data set contains 1000 cases and 1000 controls at 300 SNPs. Among the 300 SNPs, 15 (5%) are selected as disease variants. All models assume additive effects of the disease variants, and the effect size of each disease variant is given by λ = p−0.1747− 1 (Wu et al. 2011), where p denotes the MAF of the disease variant. For p = 0.3, 0.03, and 0.003, the effect size is 0.23, 0.84, 1.76, respectively, which mimic the effect sizes observed in genome-wide association studies for complex diseases. The four disease models differ by the locations of disease variants and the directions of effects. In model 1 and model 2, we assume a uniform distribution of disease variants among the 300 SNPs, while in model 3 and model 4 we assume a clustered distribution. That is, in model 3 and model 4, the 15 disease variants are distributed within two nonoverlapping SNP clusters. Each cluster contains 30 SNPs, carrying 25% disease variants each, yet the overall percentage of disease variants among the 300 SNPs is still 5%. For the directions of effects, in models 1 and 3, we assumed independent and random directions of effects with probability 0.5 each, while in models 2 and 4, we assumed that all disease variants have positive effects to the disease risk.
To evaluate how each method performs with respect to the rareness of variants, the MAFs of the 300 SNPs in each data set was bounded above by 0.01, 0.05, and 0.5, respectively, representing very rare, moderately rare, and common + rare data sets. In the 1000 Genomes data, there are ∼33% SNPs in each category of MAF <0.01, between (0.01, 0.05), and >0.05, respectively. For each MAF bound and for each disease model, we simulated 1000 data sets to evaluate power. To control type I error rate, we did not use the P-values provided by the original methods, because the P-values provided by C-alpha and VT were seriously inflated, and the asymptotic P-values of MultiVar were too conservative. Instead, we ran 200,000 permutations in each scenario to obtain empirical P-values of all methods. For BEAM3, we set a1 = 0.005 and a2 = 0.05, such that SNPs with MAF >0.005 forms its own SNP set, and SNPs with MAF <0.05 form groups with other SNPs within d = 15 SNPs. The test statistic for BEAM3 is the sum of posterior probabilities of disease association over all SNP sets in each data set, and P-value is calculated by comparing with the statistics obtained from permuted data.
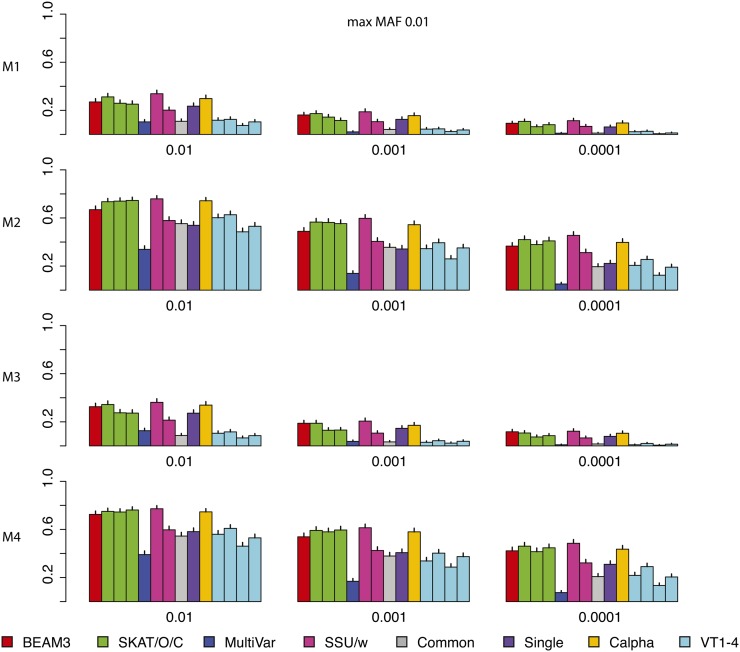
Figure 2 shows the power comparison of all methods on data sets with maximum MAF 0.01. In these data sets, only the very rare variants are included. We observed that SKAT, SSU and Calpha all performed similarly with the best power in all scenarios. Our method (BEAM3), in comparison, achieved similar power in most cases. Overall, models 2 and 4 with all positive effects are detected by all methods more easily than by models 1 and 3 with opposite effects. The multivariate regression score test (MultiVar) performed the worst in all scenarios, whereas the common effect method (Common) performed poorly for models 1 and 3, because the effects were in opposite directions. The variable threshold approach (VT) performed poorly too; particularly, its power was worse than the single SNP test (Single) in all scenarios.
Figure 2.
Comparison on data sets with MAF bounded at 0.01. x: significance. y: power for four models.
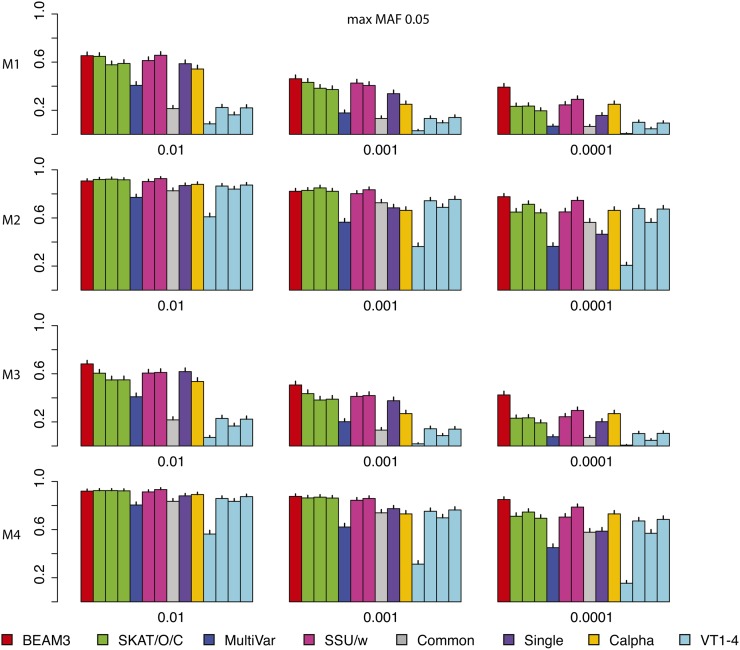
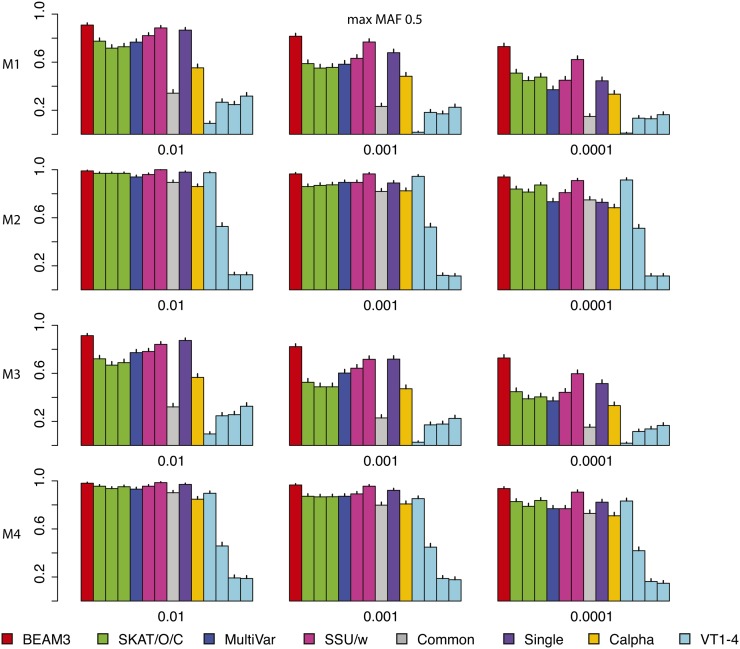
Figure 3 and Figure 4 show the power comparisons on data sets with maximum MAF 0.05 and 0.5, respectively, which included not only very rare variants, but also moderately rare and common variants, respectively. It is seen that the powers of all methods increased as more common variants were included. For all models and at all significant levels, BEAM3 performed consistently and sometimes substantially better than the others. Again, SKAT, SSU, and Calpha performed similarly in all scenarios, and they obtained better power than the remaining methods in most cases. It is worth mentioning that, apart from Single, our method is the only approach that can reveal the locations of disease variants within each data set. We have also performed additional simulation studies with each data set carrying 30% disease variants, for which we observed similar results (supporting information File S1). In summary, our method performed similarly or better than existing methods when testing on a small set of SNPs, particularly when moderately rare and common SNPs were included. Even for the very rare SNPs, our method still performed competitively to the best methods, but we further pinpointed disease variants within each set.
Figure 3.
Comparison on data sets with MAF bounded at 0.05. x: significance. y: power for four models.
Figure 4.
Comparison on data sets with MAF bounded at 0.5. x: significance. y: power for four models.
Simulation study in large data sets
We next evaluated the power of our method on larger data sets containing 1000 cases and 1000 controls at 10,000 SNPs with maximum MAF <0.05. In this case, no existing methods can perform a single test on all SNPs simultaneously, but they have to split the SNPs into subsets and perform multiple tests. Based on the small data results, we compared only our method with SKAT, because SKAT performed similarly to SSU and C-alpha and was one of the best among all methods. In each data set, we simulated 50 disease variants equally partitioned into 5 groups (10 disease variants per group). The disease variants in each group were randomly distributed within either a 5- or a 50-kb region, with equal probability. Also, the disease variants in a group either have 50% opposite directions of effects or have positive effects, with 50% chance each. The effect sizes were determined in the same way as before. Since we cannot run SKAT to test all SNPs together, we partitioned each data set into equal-sized windows containing M SNPs per window and we ran SKAT in each window separately. We tested SKAT for M = 10, 25, 50, 100, 200, respectively. We ran our method on the entire data set with a1 = 0.005, a2 = 0.05, and d = 30 (number of SNPs per set for those with MAF <a2). We also used the hierarchical splitting strategy to split each 30-SNP set into k = 4 subsets and kept both for analysis. Again, we used permutation P-value to control type I errors. For SKAT, the P-value from each M SNP window was used as the statistic to obtain data-wide significance thresholds. For BEAM3, the posterior probability of disease association from each predefined SNP set was used as the statistic to obtain data-wide significance thresholds.
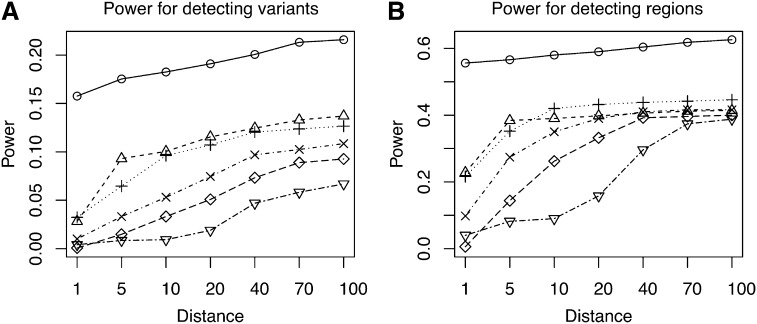
We performed two different power comparisons: (a) the power for detecting a disease variant and (b) the power for detecting a disease region (one of the five disease groups). To calculate power, we first identified the significant SNP sets reported by each method at data-wide significance level 0.01. For each significant SNP set, we then identified its nearest true disease variant to the center of the SNP set. The disease variant (or the region it belonged to) was counted as detected if the distance (in number of SNPs) between the two was within a threshold.
Figure 5 shows the results obtained from 100 simulated data sets of 10,000 SNPs each (MAF < 0.05). We observed that BEAM3 performed considerably and consistently better in terms of detecting and localizing disease variants and disease regions, compared to SKAT using any window size. The performance of SKAT varied considerably for different window sizes. Additional simulation studies of even larger data sets containing 100,000 SNPs showed similar results (supporting information, File S1). In practice, the best window size is never known for hypothesis-testing methods. It is likely that both rare and common variants affect the disease risks, either independently or jointly. A flexible method like ours is thus strongly desirable.
Figure 5.
Power comparison between BEAM3 and SKAT: (A) Power for detecting variants and (B) power for detecting regions. Distance: maximum allowed number of SNPs between the center of a reported significant SNP set (data-wide P-value 0.01) and the nearest true disease variant, such that the true variant is counted toward power. SKAT: in the parentheses shows the number of SNPs per set. o: BEAM3; Δ: SKAT(10); +: SKAT(25); x: SKAT(50); ⋄: SKAT(100); ∇: SKAT(200)
Type 1 diabetes resequencing data
We applied BEAM3 to a whole-genome resequencing (WGS) data generated in our laboratory on blood-derived DNA from 97 T1D patients. The samples were processed using SOLiD5500 sequencers. For sequence alignment, variant calling, and annotation, we employed our parallel read mapping and variant-calling pipeline, using Burrow Wheeler alignment (BWA) (Li and Durbin 2009, 2010), BOWTIE (Langmead et al. 2009), and SAMtools (Li et al. 2009) to call SNVs and indels. The output is in sequence alignment/mapping (SAM) format we compressed into a binary format (BAM). The BAM files that passed quality control (QC), including proportion of mappable reads and number of unique start sites, were subsequently used for downstream analysis. The average number of reads for the WGS data were 6–8× (i.e., low coverage) of high-quality sequence data following standard QC procedures.
Because no controls are sequenced in this study, we downloaded 85 unrelated CEU samples, whose origins are the closest to the T1D patients in this study, from the 1000 Genomes project. We retained only the SNPs that appeared in both the T1D samples and the CEU controls. We removed SNPs with >10% missing values and those with significant Hardy–Weinberg disequilibrium (P-value <10−6). We imputed missing genotypes by sampling within each SNP and we removed nonpolymorphic SNPs. The final data set contained 97 cases and 85 controls at 2.93 million SNPs.
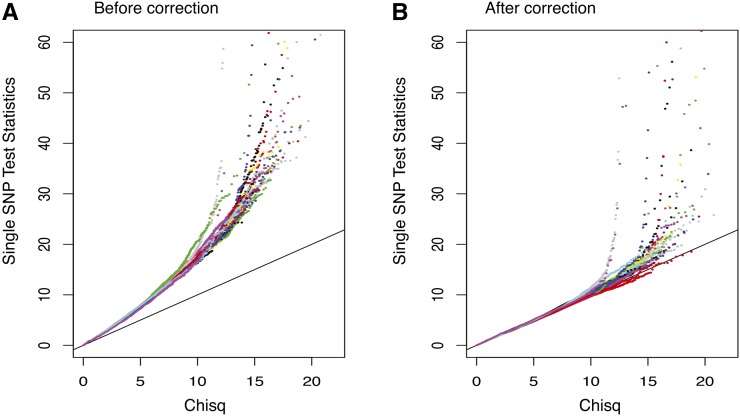
Since the cases and controls are generated by different protocols, we observed sample stratification. As shown in Figure 6A, single SNP test statistics are inflated genome-wide. We therefore decorrelated samples as follows: (1) we calculated a covariance matrix V of the 182 individuals using SNPs whose absolute correlation with the disease status is <99 percentile; (2) we decomposed V = LL′ by Cholesky decomposition; and (3) we calculated new “genotypes” by Xnew = X(L′)-1. As a result, the new “genotypes” are decorrelated under Normality assumption. Figure 6B shows that the single SNP test statistics are “correct” after this adjustment.
Figure 6.
QQ-plot of single SNP test statistics (A) before and (B) after correcting for stratification.
After correcting for sample stratification, we ran our method with a1 = 0.05, a2 = 0.1, d = 30 with hierarchical splitting (k = 4). Due to computational constraints, we applied our method on each chromosome separately. We ran our method four times on each chromosome independently and then summarized the posterior probabilities of associations by averaging. We reported in Table 1 the top 12 detected T1D loci whose posterior probabilities were >0.25. Based on genome-wide permutations, our criteria yielded P-value <10−7. Among the 12 detected loci, 7 had at least two SNPs within 500 kb showing single-SNP test P-value <10−5.
Table 1. Loci detected in the T1D resequencing data set.
| Detected locia | Assoc. prob.b | Multi- assoc.c | Nearest genesd |
|---|---|---|---|
| chr1:24281021–24388679 | 1.00 (1.00) | Yes | SRSF10, MYOM3, PNRC2 |
| chr3:625496–705294 | 1.00 (1.00) | Yes | AK126307 (CNTN6) |
| chr4:190436494–190538640 | 0.82 (2.06) | Yes | (DUX2, DUX4L4, FRG2, FRG1, TUBB4Q) |
| chr5:17479389–17622489 | 0.99 (0.99) | No | (LOC401177, BASP1, BC028204) |
| chr6:67706682–67780247 | 0.98 (0.98) | No | None, but has strong Pol2 signal in K562 and DNA methylation. |
| chr7:159100528–159126143 | 0.66 (0.66) | Yes | (VIPR2) |
| chr9:68435907–68800509 | 1.00 (1.00) | No | LOC100132352, AK096159, LOC642236 |
| chr10:127556783–127650870 | 0.85 (0.86) | No | FANK1, DHX32 |
| chr16:70813237–71062992 | 0.29 (0.67) | Yes | HYDIN, VAC14 |
| chr19:55268017–55332829 | 0.98 (0.98) | Yes | KIR2DL1-4, KIR3DL1, KIR2DL4, KIR2DS4 |
| chr20:20109354–20184868 | 1.00 (1.00) | No | C20orf26 (CRNKL1) |
| chr21:14946923–15168502 | 0.43 (0.43) | No | POTED, DQ590589, DQ591735 (C21orf15, DQ586768) |
| chr22:18622054–18923349 | 0.89 (1.14) | Yes | GGT3P, DGCR6, PRODH, AK302545, BC112340, BC051721, AL117485, DQ786190, AK129567, USP18 |
Positions are in hg19 coordinates.
Maximum posterior probability within the interval and the sum of posterior probabilities of all SNPs in the interval are in parentheses (italics indicate a significant difference between the two).
Whether or not the interval carries more than one SNP with marginal P-value <10−5.
Genes overlap with the interval, and genes that do not overlap with the interval but are within the 500-kb neighborhood are in the parentheses. Genes in italic type are discussed in the main text.
Among the top 12 ranked T1D loci, we found a few interesting genes. First, the locus chr19:55.27–55.33 Mb includes genes KIR2DL1–4, KIR3DL1, KIR2DL4, KIR2DS4. These are killer cell immunoglobulin-like receptor genes expressed in killer cells and subsets of T cells. They are subsets of HLA class I molecules and play important roles in regulation of the immune response. In addition, downregulation of KIRD3L1 has been shown to enhance inhibition of type 1 diabetes (Qin et al. 2011). Second, the locus chr5:17.48–17.62 Mb is 200 kb downstream of gene BASP1. This gene has been reported to promote apoptosis in diabetic nephropathy (Sanchez-Nino et al. 2010), and defective apoptosis is known to play an important role in type 1 diabetes (Hayashi and Faustman 2003). Incidentally, two other loci (chr10:127.56–127.65 Mb, chr22:18.62–18.92 Mb) also overlapped with genes (FANK1, DHX32, USP18) that are related to cell apoptosis. While FANK1 and DHX32 regulate T-cell apoptosis (Alli et al. 2007; Wang et al. 2011), USP18 is a key regulator of the interferon-driven gene network modulating pancreatic beta cell inflammation and apoptosis (Santin et al. 2012). Third, 3 (chr1:24.28–24.39 Mb, chr7:159.10–159.13 Mb; chr22:18.62–18.92) out of the 12 loci either overlapped or were near genes (PNRC2, VIPR2, DGCR6, PRODH) associated with obesity and type 2 diabetes. PNRC2 is a nuclear receptor coactivator that regulates energy expenditure and adiposity in mice (Zhou et al. 2008), which are the keys to understand obesity, insulin resistance, and type 2 diabetes. VIPR2, DGCR6 and PRODH are known to be significantly associated with schizophrenia (Vacic et al. 2011; Welsh et al. 2011; Liu et al. 2002), a mental disorder that is associated with decreased risk of type 1 diabetes (Juvonen et al. 2007) and increased risk of type 2 diabetes (Schoepf et al. 2012).
The human major histocompatibility complex (MHC) region is a well-known T1D locus. Our analysis did not capture this region at the genome-wide significance level due to several reasons. First, the sample size of our data set is small. The lead T1D SNP rs9268645 in MHC, reported by Barrett et al. (2009), has association P-value <<1e-100 from a metaanalysis combining two independent studies carrying over 10 thousands of individuals. This SNP is captured in our study, with MAF 0.47 in cases and 0.40 in controls. These MAFs were statistically the same as those observed in WTCCC T1D data set (Wellcome Trust Case Control Consortium 2007) (0.46 in cases and 0.40 in controls, respectively), but has insignificant P-value (>0.1) due to the small sample size. Should the sample size be 10000 with the same MAFs, its P-value will decrease to <1e-50. Second, there are many missing values due to low coverage sequencing. The lead T1D MHC SNP rs9273363 reported by Nejentsev et al. (2007) had P-value 1e-298, yet it was removed from our study because of 38% missingness (no reads). Should this SNP be retained in our study, but simply ignoring the missing values, we will obtain case MAF 0.63, control MAF 0.33, and P-value 2.2e-7 before adjusting for sample stratification. These MAFs are again similar to those observed in WTCCC T1D data set (case MAF 0.71, control MAF 0.30). Third, our data set has sample stratification problem due to lack of controls, which further reduces the power.
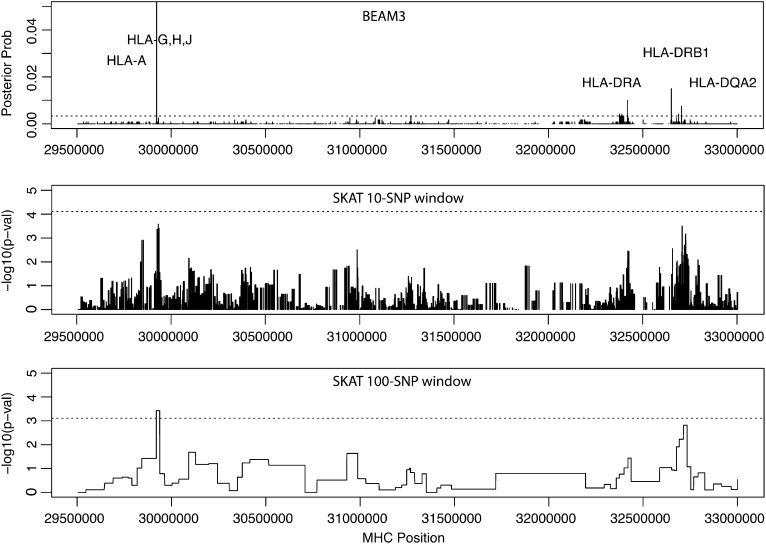
Given that we already know that MHC carries T1D variants, we applied our method to the MHC region (chr6:29.5 Mb–33 Mb, hg19, 6472 SNPs) specifically to localize “causative” T1D variants. The result is shown in Figure 7. Our method pinpointed four T1D loci with MHC-wide significance <0.05, including locus 29922754 bp in genes HLA-H, HLA-G, HLA-J and at 10 kb downstream of HLA-A, locus 32417825–32417891 bp at 3 kb downstream of HLA-DRA, locus 32651168–32651254 bp at 17 kb upstream of HLA–DRB1, and locus 32705193–32705276 bp at 2 kb upstream of HLA–DQA2. The latter three loci are all within the well-known HLA–DR–DQ genes in MHC class II complex. For comparison, we also ran SKAT in the same MHC region using two different window sizes: 10 and 100 SNPs, respectively. As shown in Figure 7, SKAT detected the HLA–H,G,J locus, but failed to yield significant P-values at the HLA–DR–DQ region.
Figure 7.
Posterior probability of T1D association by BEAM3 compared to P-values by SKAT using 10 and 100 SNPs per window in MHC region. Dashed lines indicate the MHC-wide 0.05 significance levels.
Discussion
In this article we introduced a powerful and flexible method for simultaneous testing of rare and common variants associated with complex diseases. Distinct from existing approaches, our method utilizes a joint statistical model to test all variants genome-wide simultaneously. The benefits are twofold: joint associations or cumulative effects of multiple variants are detectable with improved power by dynamic grouping of sets of variants; and our joint model accounts for correlation among variants, such that multiple disease variants within a local region can be detected and redundant associations due to LD are filtered out. As a consequence, we are able to define sets of variants that overlap with each other without concerning about multicolinearity among variants. A variant may be simultaneously present in the data set as a single variant by itself and as groups of variants with others. The new method will then evaluate the effects of the variant both as a single variant and as groups, and one shows that the most power will be automatically detected. This feature significantly alleviated the burden on the users to define sets of variants to be tested, which is often arbitrary. At the same time, the users can still design their favorable sets of variants for joint testing based on their biological knowledge. While it is unclear how much effects of rare variants contribute to the complex diseases, it is most likely that both common and rare variants are contributing to the disease risks to a different degree. We therefore believe that our method is more suitable to the current genome-wide association studies, where all genetic variants from sequencing studies are included in the analysis.
Our simulation studies have demonstrated the superior power of the new method compared to existing rare variant mapping tools. In the small data study, we observed that our method performed similarly to the best existing methods when testing only the very rare variants (MAF <0.01). When more common variants were included, our method achieved better power and sometimes substantially so. In the large data study, our method performed substantially better than existing methods in terms of both power and mapping resolution. When applied to a whole-genome resequencing study of type 1 diabetes, we handled sample stratification and detected novel loci that are biologically relevant to T1D. We further demonstrated a fine mapping of T1D variants in the well-known MHC region, where we identified one locus in the HLA-G,H,J genes and three loci in the HLA–DR–DQ genes. In comparison, SKAT detected only one locus in MHC and its result is sensitive to window sizes. Many loci we detected involved common variants, which in part was due to the very limited sample size of the study, but also perhaps indicated that exclusively focusing on rare variants may not be the best strategy.
Our method can be directly applied to other types of data, such as copy-number variations and genomic/epigenetic data. In addition, our method can be used for QTL mapping, and covariates such as environmental factors can be included. Currently the method does not allow detection of SNP–environment interactions associated with the disease, but a simple modification can be added to allow detecting disease associated interactions between SNPs and covariates. The method can also take input of multiple traits simultaneously, such that SNPs associated with one or multiple disease traits can be detected. By inversely regressing SNPs on disease traits, we avoid modeling the distributions of disease traits. An interesting extension of the method is therefore to include kernels to detect nonlinear associations between SNPs and multiple disease traits. The URLs for data presented herein are as follows:
1000 Genomes: http://www.1000genomes.org/
Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/omim
Source code of BEAM3 can be found in Supporting Information, File S2.
Supplementary Material
Acknowledgments
Y.Z. is supported by National Institutes of Health grant R01-HG004718.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.167403/-/DC1.
Communicating editor: G. A. Churchill
Literature Cited
- 1000 Genomes Project Consortium, 2010 A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed]
- Alli Z., Chen Y., Wajid S. A., Al-Saud B., Abdelhaleem M., 2007. A role for DHX32 in regulating T-cell apoptosis. Anticancer Res. 27(1A): 373–377. [PubMed] [Google Scholar]
- Barrett J. C., Clayton D. G., Concannon P., Akolkar B., Cooper J. D., et al. , 2009. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 41(6): 703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S., Pan W., 2011. Comparison of statistical tests for disease association with rare variants. Genet. Epidemiol. 35: 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W., Bonilla C., 2008. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 40: 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conneely K. N., Boehnke M., 2007. So many correlated tests, so little time!: rapid adjustment of p values for multiple correlated tests. Am. J. Hum. Genet. 81: 1158–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Pan W., 2010. A data-adaptive sum test for disease association with multiple common or rare variants. Hum. Hered. 70: 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Faustman D. L., 2003. Role of defective apoptosis in type 1 diabetes and other autoimmune diseases. Recent Prog. Horm. Res. 58: 131–153. [DOI] [PubMed] [Google Scholar]
- Ionita-Laza, I., S. Lee, V. Makarov, J. D. Buxbaum, and X. Lin, 2013 Sequence kernel association tests for the combined effect of rare and common variants. Am. J. Hum. Genet. 92: 841–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juvonen H., Reunanen A., Haukka J., Muhonen M., Suvisaari J., et al. , 2007. Incidence of schizophrenia in a nationwide cohort of patients with type 1 diabetes mellitus. Arch. Gen. Psychiatry 64: 894–899. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Emond M. J., Bamshad M. J., Barnes K. C., Rieder M. J., et al. , 2012. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 91: 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Leal S. M., 2008. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16): 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. Y., Tang Z. Z., 2011. A general framework for detecting disease associations with rare variants in sequencing studies. Am. J. Hum. Genet. 89: 354–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Heath S. C., Sobin C., Roos J. L., Galke B. L., et al. , 2002. Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc. Natl. Acad. Sci. USA 99: 3717–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen B. E., Browning S. R., 2009. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 5: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenthaler S., Thilly W. G., 2007. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST). Mutat. Res. 615: 28–56. [DOI] [PubMed] [Google Scholar]
- Morris A. P., Zeggini E., 2010. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet. Epidemiol. 34: 188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale B. M., Rivas M. A., Voight B. F., Altshuler D., Devlin B., et al. , 2011. Testing for an unusual distribution of rare variants. PLoS Genet. 7: e1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejentsev S., Howson J. M., Walker N. M., Szeszko J., Field S. F., et al. , 2007. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 450(7171): 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., 2009. Asymptotic tests of association with multiple SNPs in linkage disequilibrium. Genet. Epi. 33: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman J., Yor M., 1997. The two-parameter Poisson–Dirichlet distribution derived from a stable subordinator. Ann. Probab. 25: 855–900. [Google Scholar]
- Price A. L., Kryukov G.V., de Bakker P. I. W., Purcell S. M., Staples J. et al, 2010. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 86: 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Wang Z., Du W., Lee W. H., Wu X., et al. , 2011. Killer cell Ig-like receptor (KIR) 3DL1 down-regulation enhances inhibition of type 1 diabetes by autoantigen-specific regulatory T cells. Proc. Natl. Acad. Sci. USA 108: 2016–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Niño M. D., Sanz A. B., Lorz C., Gnirke A., Rastaldi M. P., et al. , 2010. BASP1 promotes apoptosis in diabetic nephropathy. J. Am. Soc. Nephrol. 21: 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin I., Moore F., Grieco F. A., Marchetti P., Brancolini C., et al. , 2012. USP18 is a key regulator of the interferon-driven gene network modulating pancreatic beta cell inflammation and apoptosis. Cell Death Dis. 3: e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepf D., Potluri R., Uppal H., Natalwala A., Narendran P., et al. , 2012. Type-2 diabetes mellitus in schizophrenia: increased prevalence and major risk factor of excess mortality in a naturalistic 7-year follow-up. Eur. Psychiatry 27: 33–42. [DOI] [PubMed] [Google Scholar]
- Schork N. J., Murray S. S., Frazer K. A., Topol E. J., 2009. Common vs. rare allele hypotheses for complex diseases. Curr. Opin. Genet. Dev. 19: 212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J., Ji H., 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26: 1135–1145. [DOI] [PubMed] [Google Scholar]
- Vacic V., McCarthy S., Malhotra D., Murray F., Chou H. H., et al. , 2011. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature 471: 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Song W., Hu T., Zhang N., Miao S., et al. , 2011. Fank1 interacts with Jab1 and regulates cell apoptosis via the AP-1 pathway. Cell. Mol. Life Sci. 68: 2129–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium , 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh D. K., Craig D., Kelsoe J. R., Gershon E. S., Leal S. M., et al. , 2011. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature 471: 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. C., Lee S., Cai T., Li Y., Boehnke M., et al. , 2011. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawistowski M., Gopalakrishnan S., Ding J., Li Y., Grimm S., et al. , 2010. Extending rare-variant testing strategies: analysis of noncoding sequence and imputed genotypes. Am. J. Hum. Genet. 87: 604–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., 2011. A novel Bayesian graphical model for genome-wide multi-SNP association mapping. Genet. Epi. 36: 36–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Shen R., Ye J. J., Li Y., Tsark W., et al. , 2008. Nuclear Receptor Coactivator PNRC2 Regulates Energy Expenditure and Adiposity. J. Biol. Chem. 283: 541–553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.