Abstract
Several helicases function during repair of double-strand breaks and handling of blocked or stalled replication forks to promote pathways that prevent formation of crossovers. Among these are the Bloom syndrome helicase BLM and the Fanconi anemia group M (FANCM) helicase. To better understand functions of these helicases, we compared phenotypes of Drosophila melanogaster Blm and Fancm mutants. As previously reported for BLM, FANCM has roles in responding to several types of DNA damage in preventing mitotic and meiotic crossovers and in promoting the synthesis-dependent strand annealing pathway for repair of a double-strand gap. In most assays, the phenotype of Fancm mutants is less severe than that of Blm mutants, and the phenotype of Blm Fancm double mutants is more severe than either single mutant, indicating both overlapping and unique functions. It is thought that mitotic crossovers arise when structure-selective nucleases cleave DNA intermediates that would normally be unwound or disassembled by these helicases. When BLM is absent, three nucleases believed to function as Holliday junction resolvases—MUS81-MMS4, MUS312-SLX1, and GEN—become essential. In contrast, no single resolvase is essential in mutants lacking FANCM, although simultaneous loss of GEN and either of the others is lethal in Fancm mutants. Since Fancm mutants can tolerate loss of a single resolvase, we were able to show that spontaneous mitotic crossovers that occur when FANCM is missing are dependent on MUS312 and either MUS81 or SLX1.
Keywords: mitotic recombination, helicase, Holliday junction resolvase, Fanconi anemia
HELICASES are best known as enzymes that separate the strands of duplex nucleic acids, but many DNA repair helicases process more complex structures to direct repair pathways toward specific outcomes (reviewed in Brosh 2013). The Bloom syndrome helicase (BLM) has activities that promote disassembly of D loops and double-Holliday junction (dHJ) intermediates (Karow et al. 2000; Van Brabant et al. 2000; Wu and Hickson 2003). These activities prevent formation of crossovers during repair of DNA double-strand breaks (DSBs) (reviewed in Andersen and Sekelsky 2010). Disassembly of a D loop is a key step in the noncrossover synthesis-dependent strand annealing (SDSA) pathway. Drosophila BLM plays an important role in SDSA during gap repair, most likely by promoting D-loop disassembly after repair synthesis (Adams et al. 2003; McVey et al. 2004b). Likewise, the Saccharomyces cerevisiae ortholog Sgs1 generates noncrossovers during meiotic DSB repair and mitotic gap repair (De Muyt et al. 2012; Mitchel et al. 2013). If D loops are not disassembled, repair may proceed to generate a dHJ intermediate. BLM collaborates with topoisomerase 3α and other proteins to disassemble dHJs into noncrossover products in vitro, a process termed “dissolution” (Wu and Hickson 2003). Evidence for dHJ dissolution in vivo comes from meiotic return-to-growth experiments and mitotic gap repair assays in budding yeast (Dayani et al. 2011; Mitchel et al. 2013). If a dHJ is not dissolved, it must be resolved by structure-selective endonucleases (resolvases), which may generate reciprocal crossover products.
FANCM helicase also prevents crossing over (reviewed in Whitby 2010). FANCM mutations in humans cause Fanconi anemia (FA), a hereditary disorder characterized by developmental abnormalities, bone marrow failure, and cancer predisposition (reviewed in Soulier 2011). Cells from FA patients exhibit heightened sensitivity to agents that cause DNA interstrand crosslinks (ICLs), suggesting a defect in ICL repair (reviewed in Kim and D’Andrea 2012). FANCM is thought to function during an early step in the FA repair pathway, perhaps in damage recognition and recruitment of additional FA proteins. FANCM also has functions outside of the FA pathway. Anticrossover functions for FANCM and its orthologs have been observed in several contexts. Spontaneous sister-chromatid exchange is elevated in mouse embryonic fibroblasts and chicken DT40 cells that lack FANCM (Mosedale et al. 2005; Bakker et al. 2009). Orthologs in S. cerevisiae (Mph1) and Schizosaccharomyces pombe (Fml1) promote noncrossover outcomes during mitotic DSB repair (Sun et al. 2008; Prakash et al. 2009; Mazón and Symington 2013; Mitchel et al. 2013), and meiotic crossovers are elevated in Arabidopsis FANCM mutants and S. pombe fml mutants (Crismani et al. 2012; Lorenz et al. 2012). Like BLM, FANCM and its orthologs can branch-migrate HJs and disassemble D loops (Gari et al. 2008; Sun et al. 2008; Prakash et al. 2009). FANCM is not thought to not be capable of catalyzing dissolution; hence, it has been proposed that the D-loop disassembly activity of Fml1 and Mph1 promotes SDSA, thereby preventing formation of dHJs and resolution of these into crossovers (Sun et al. 2008; Prakash et al. 2009; Mitchel et al. 2013).
Biochemical and genetic studies have identified several likely nuclear HJ resolvases (reviewed in Schwartz and Heyer 2011). Among these, GEN1/Yen1 appears to have the greatest selectivity for HJs, but also has activity on 5′ flaps and replication fork-like structures (Ip et al. 2008). Genetic studies in vertebrate cells, budding yeast, and Caenorhabditis elegans have failed to identify a major function for GEN1 or its orthologs in generating meiotic or mitotic crossovers (Blanco et al. 2010; Ho et al. 2010; Tay and Wu 2010; Wechsler et al. 2011; Agostinho et al. 2013; Saito et al. 2013). In budding yeast, Yen1 functions are revealed in mus81 yen1 double mutants, leading to the hypothesis that Yen1 functions as a backup to the Mus81–Mms4 endonuclease (Blanco et al. 2010; Ho et al. 2010; Tay and Wu 2010; Wechsler et al. 2011).
Mus81–Mms4/Eme1 and its orthologs have important roles in generating meiotic and mitotic crossovers in several organisms (Boddy et al. 2001; De Los Santos et al. 2003; Berchowitz et al. 2007; Ho et al. 2010; Wechsler et al. 2011). Although this enzyme was reported to cut HJs (Boddy et al. 2001; Chen et al. 2001), studies with recombinant protein found that intact HJs are not a good substrate, but nicked HJs, D loops, and 3′ flaps are (Ehmsen and Heyer 2008). This apparent paradox suggested models in which crossovers are generated by cleavage of a D loop (Whitby 2005) or a structure with nicked HJs (Osman et al. 2003; Schwartz and Heyer 2011). Another solution suggested by recent studies is that Mus81–Mms4/Eme1 functions as an HJ resolvase together with the Slx1 nuclease, with Slx1 making the first nick and Mus81 then cutting the nicked HJ (Castor et al. 2013; Garner et al. 2013; Wyatt et al. 2013). Human SLX1 was previously shown to have HJ resolution activity in vitro; this activity is dependent on SLX4, a scaffolding protein that also interacts with MUS81–EME1 (Fekairi et al. 2009; Muñoz et al. 2009; Svendsen et al. 2009). Thus, it is proposed that SLX4 coordinates the activities of SLX1 and MUS81–EME1 to coordinately resolve HJs.
Not surprisingly, helicase and resolvase gene mutations often show genetic interactions. In S. cerevisiae, sgs1 mutations are synthetically lethal with mutations in mus81 or mms4 and with mutations in slx1 or slx4 (Kaliraman et al. 2001; Fricke and Brill 2003). Lethality of sgs1 mus81 mutants is suppressed by mutations that prevent recombination, but lethality of sgs1 slx1 (or slx4) is not, suggesting different causes for the lethality.
The above discussion hints at the inherent functional complexity between BLM, FANCM, and resolvases. We have sought to tease apart some of this complexity through genetic studies in the model metazoan Drosophila melanogaster. Mitotic crossovers are highly elevated in Drosophila Blm mutants, and these mutants have defects in SDSA and meiotic recombination (Adams et al. 2003; McVey et al. 2004a,b; Kohl et al. 2012). Blm mutations are synthetically lethal with mutations in mus81, mus312 (encodes the ortholog of Slx4), Slx1, or Gen (Trowbridge et al. 2007; Andersen et al. 2009, 2011). As in yeast, different double mutants have different phenotypes that reveal different functional overlaps.
We describe here characterization of Drosophila Fancm mutants and comparison to Blm mutants. We show that FANCM has roles in preventing both mitotic and meiotic crossovers, independent of its function in the FA pathway, although the mitotic crossover frequency is lower in Fancm mutants than in Blm mutants. Similarly, Fancm mutants have a defect in SDSA repair of a gap, but it is significantly less severe than the defect in Blm mutants. Unlike Blm mutations, Fancm mutations are not synthetically lethal with single resolvase gene mutations; however, some combinations of multiple resolvase mutations are lethal to Fancm mutants. Finally, we show that spontaneous mitotic crossovers that occur in the absence of FANCM are dependent on MUS312 and either MUS81 or SLX1.
Materials and Methods
Drosophila stocks
Fly stocks were maintain at 25° with standard medium. Mutants were heteroallelic or hemizygous for null alleles (Table 1). Mutations in Fancm (CG7922) were found by TILLING (Cooper et al. 2008). Fancm0693 is T-to-C substitution at 3R: 27,905,053 that generates a nonsense mutation (L78ter). In experiments reported here, Fancm mutants were Fancm0693/Df(3R)ED6058. FanclLL00701 is an insertion of a PBac{SAstopDsRed} element into the boundary between the first intron and second exon between codons 37 and 38 (Schuldiner et al. 2008). The stock from the Drosophila Genetic Resource Center (Kyoto) had three P elements inserted onto other locations on the same chromosome arm; we removed these by recombination before doing experiments with FanclLL00701. Alleles of other genes are listed in Table 1.
Table 1. Mutations used in this study.
| Allele | Type | Reference |
|---|---|---|
| BlmN1 | Deletion | McVey et al. (2007) |
| BlmD2 | Nonsense | Kusano et al. (2001) |
| Gen5997 | Frameshift | Andersen et al. (2011) |
| mei-9a | Missense | Yıldız et al. (2004) |
| mus312D1 | Nonsense | Yıldız et al. (2002) |
| mus312Z1973 | Nonsense | Yıldız et al. (2002) |
| mus81Nhe | Frameshift | Trowbridge et al. (2007) |
| slx1F93I | Missense | Andersen et al. (2011) |
| spn-A057 | Missense | Staeva-Vieira et al. (2003) |
| spn-A093A | Nonsense | Staeva-Vieira et al. (2003) |
Sensitivity assays
Sensitivity to DNA-damaging agents was determined as in Yıldız et al. (2002). For HN2 and MMS, 250 μl of an aqueous solution at the indicated concentrations was added to the medium on which larvae were feeding. For ionizing radiation (IR), vials with larvae were exposed to gamma rays in an irradiator with 145 Ci of 137Cs. Adults were counted daily from day 10 (after parents were first placed in vials) until day 18. Most treatments had at least three technical replicates (treatments on different days), each with 10 biological replications (different vials). Vials with <20 total progeny in either the untreated or the treated brood were discarded. Relative survival was calculated for each vial as the ratio between mutant and control flies in the treated vial, normalized to the same ratio in the untreated vial. To estimate absolute survival, we compared the number of control progeny in treated and untreated vials. For low doses, these were 81% (HN2), 82% (MMS), and 89% (IR). This reduced recovery is probably because flies in untreated vials were allowed to mate and lay eggs for 3 days, whereas flies in treated vials were allowed to lay eggs for only 2 days. Normalizing to these numbers, the highest doses had 81% (HN2), 56% (MMS), and 47% (IR) absolute survival. Statistical analyses were done in Prism 6 (GraphPad). For treatments that involved more than two genotypes, a Kruskal–Wallace test with a Dunn post-test was done. P-values reported are corrected for multiple comparisons. Treatments that involved only two genotypes were compared using an unpaired t-test.
Mitotic crossover assay
Mitotic crossovers in the male germline were measured as in McVey et al. (2007), using the genetic markers st and Sb. Each vial, each with a single male, was counted as a separate biological replicate. Vials with <20 progeny were discarded. Statistical analyses were done in Prism 6 (GraphPad). For treatments that involved more than two genotypes, a Kruskal–Wallace test with a Dunn post-test was done. P-values reported are corrected for multiple comparisons.
Meiotic crossover and nondisjunction assays
To measure meiotic crossovers, virgin females of the genotype net dppd-ho dp b pr cn /+; Fancm0693/Df(3R)ED6058 (or wild-type control) were collected and aged 1–4 days and then crossed to net dppd-ho dp b pr cn males. Progeny were counted and scored for each marker from day 10 to 14 after the cross was set up. To measure nondisjunction of the X chromosome, virgin females were crossed to males carrying Dp(1;Y)BS, a Y chromosome carrying the BS dominant marker. Normal progeny were females with wild-type eyes and males with Bar eyes; nondisjunctional progeny were females with Bar eyes (XXY progeny from XX ova or XY sperm) and males with wild-type eyes (XO progeny from nullo-X ova or nullo-XY sperm).
SDSA assay
The P{wa} assay was done as described previously (McVey et al. 2004a), using the CyO, H{w+, Δ2-3} transposase source. Because Df(3R)ED6058 has a w+ allele associated with the deletion, the deletion chromosome was marked with Sb, and only Sb+ progeny were scored for eye color. A control was done with Sb Df(3R)ED6058/+ males; the results were not different from previous controls that did not have this Df chromosome.
Data archiving
Raw data have been deposited in the Carolina Digital Repository with the digital object identifier 10.15139/S3159M.
Results
Sensitivity of Fancm mutants to DNA-damaging agents
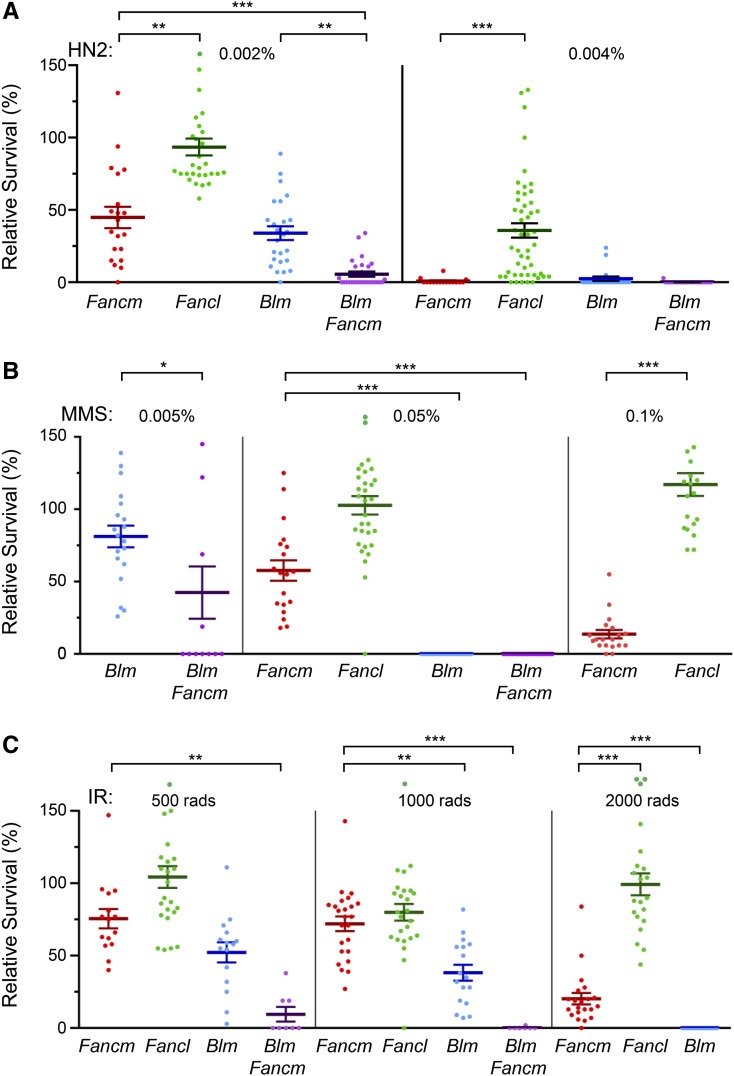
To investigate functions of Drosophila FANCM that are independent of the FA pathway, we compared phenotypes of Fancm mutants to those of Fancl mutants because FANCL is an essential component of the FA pathway but has no other known roles in DNA repair or recombination. Given the central function of the FA pathway in responding to ICLs, we first assayed sensitivity to a crosslinking agent, the nitrogen mustard mechlorethamine (HN2). Fancl mutants are hypersensitive to a high dose of HN2 (Figure 1A), consistent with a previous study that showed hypersensitivity to cross-linking agents after RNA interference knockdown of FANCL (Marek 2006). Fancm mutants were significantly more sensitive than Fancl mutants at this dose and were also hypersensitive to a lower dose of HN2, at which Fancl mutants were not hypersensitive.
Figure 1.
Comparison of sensitivities of Fancm, Fancl, and Blm mutants. Plots show survival of the indicated mutants relative to control flies in the same vial after exposure to (A) the nitrogen mustard mechloramine (HN2), (B) MMS, or (C) IR. Survival of control flies did not appear to be reduced at the lower doses used here, but the highest doses reduced survival of control flies by ∼19% (HN2), 46% (MMS), and 53% (IR) (see Materials and Methods). Each dot represents one vial. Heavy bars are means; error bars are standard error of the mean. n = (left to right) HN2: 20, 30, 25, 28 | 16, 49, 20, 23; MMS: 26, 19 | 22, 32, 24, 28 | 19, 20; IR: 15, 25, 15, 8 | 24, 25, 17, 28 | 21, 21, 25. Statistical comparisons were done for Fancm compared to each other genotype, and Fancm Blm double mutants were compared to Blm single mutants: *P < 0.05; **P < 0.01, ***P < 0.001 (corrected for multiple comparisons; see Materials and Methods); all statistically significant comparisons are indicated. Seven of the 625 data points, all from Fancl vials, are off the scale and are not shown (values in parentheses): 0.002 HN2 (1.92), 0.05% MMS (1.97), 0.1% MMS (1.84, 1.84, and 1.80), and 500 rad IR (1.92 and 1.76).
The greater sensitivity of Fancm mutants suggests that FANCM has an FA-independent role in responding to ICLs or to another type of damage induced by HN2. Like most cross-linking agents, HN2 can induce mono-adducts and intrastrand cross-links in addition to ICLs (Wijen et al. 2000). We therefore assayed sensitivity to MMS, which generates mono-adducts but not cross-links (Beranek 1990). Fancl mutants were not hypersensitive to MMS at the doses assayed, but Fancm mutants showed significant hypersensitivity to a high dose (Figure 1B). Studies in other model organisms revealed functions for FANCM orthologs in DSB repair pathways (Sun et al. 2008; Prakash et al. 2009; Crismani et al. 2012; Lorenz et al. 2012; Mazón and Symington 2013; Mitchel et al. 2013), so we also measured sensitivity to IR. Fancm mutants were hypersensitive to IR, but Fancl mutants were not, suggesting an FA-independent role for FANCM in DSB repair (Figure 1C).
Given the functional similarities between FANCM and BLM, we compared sensitivity between Fancm and Blm mutants. Blm mutants had about the same severity of hypersensitivity to HN2 as Fancm mutants, but were significantly more sensitive to MMS and to IR (Figure 1). Blm Fancm double mutants are fully viable in the absence of exogenous damage, but were more sensitive to MMS and HN2 than either single mutant. This suggests the existence of separate FANCM-dependent and BLM-dependent pathways for responding to base adduct damage and possibly to ICLs. Double mutants also appear to be more sensitive to IR than single mutants, but the difference between Blm and Blm Fancm is not statistically significant. This suggests that FANCM participates in a subset of the BLM-dependent responses to DSBs (e.g., one of multiple branches that converge on or diverge from a BLM-dependent step). Vial-to-vial variation is often large in whole-animal assays such as this, especially at doses where survival is low, and this may have prevented us from detecting some real differences. If Blm Fancm double mutants are actually more sensitive to IR than Blm single mutants, it would suggest that FANCM and BLM contribute to different repair mechanisms, although this would not preclude overlap in the same mechanism.
Meiotic crossovers are elevated in some regions of the genome when FANCM is absent
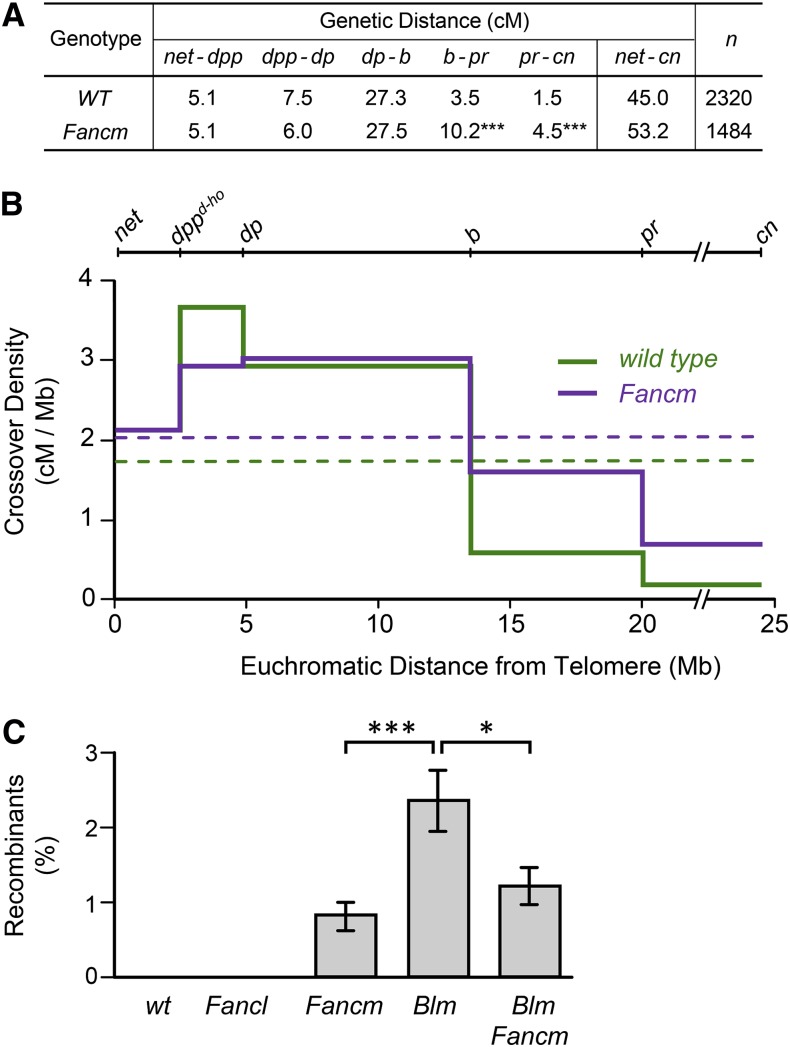
S. pombe Fml1 and Arabidopsis FANCM suppress crossovers during meiotic recombination (Crismani et al. 2012; Knoll et al. 2012; Lorenz et al. 2012), and Fml1 and S. cerevisiae Mph1 suppress crossovers in vegetative cells (Sun et al. 2008; Prakash et al. 2009; Mazón and Symington 2013). We therefore assayed both meiotic and mitotic crossovers in Drosophila Fancm mutants. Meiotic crossovers were scored in five adjacent intervals spanning the tip of 2L to the base of 2R, a region comprising ∼20% of the genome. The genetic distance across this region increased from 45.0 cM in wild-type females to 53.2 cM (118% of wild type) in Fancm mutants (Figure 2A; P < 0.0001). The increase is restricted to the two centromere-proximal intervals, each of which has about a threefold increase in crossovers compared to wild type (Figure 2, A and B). Double crossovers (DCOs) were also significantly more frequent in Fancm mutants: There were 44 DCOs among 2320 progeny (1.9%) from wild-type females, compared to 79 DCOs among 1484 progeny (5.3%) from Fancm (P < 0.0001). When progeny with multiple crossovers (DCOs and a small number with triple crossovers) are excluded, the crossover rates are not significantly different across the entire region assayed (P = 0.1061), but remain significantly elevated in the two proximal intervals (P < 0.0001 in each case; no significant differences in other intervals). Thus, in Fancm mutants, meiotic crossovers are elevated, but in only a subset of the genome.
Figure 2.
Meiotic and mitotic crossover elevation in Fancm mutants. (A) Genetic distances, in centimorgans, are given for five adjacent intervals on chromosome 2 (***P < 0.0001); other intervals were not significantly different (P = 0.9647, 0.0776, and 0.9406). (B) Meiotic crossover density. Data from (A) were graphed as crossover density, in centimorgans per megabase pair (Mb). The markers used are shown above the graph. Hash marks between pr and cn indicate the position of the centromere and pericentric heterochromatin (∼16 Mb, not counted in distances shown). Solid lines depict density in each interval; dashed lines are mean density across the entire region. (C) Mitotic crossovers in the male germline. Bars show mean percentage of progeny that were recombinant between st and Sb. Error bars are standard error of the mean. No crossovers were detected in wild-type (wt) or Fancl mutant males. Fancm, Blm, and Blm Fancm were each significantly different from wild-type and Fancl (P < 0.01 for each comparison). The difference between Fancm and Blm Fancm was not significant (P > 0.99). *P < 0.0294; ***P < 0.0001. P-values reported have been adjusted for multiple comparisons (see Materials and Methods). n = (left to right) 40, 41, 46, 39, and 30.
This elevation does not appear to have any negative impact on chromosome segregation. In an assay for meiotic nondisjunction of the X chromosome, we detected one case of nondisjunction among 1698 progeny of wild-type females and one among 1592 progeny of Fancm mutant females (P = 0.9636 by χ2 test).
Spontaneous mitotic crossovers are elevated when FANCM is absent
We assayed spontaneous mitotic crossovers in the male germline because there are no meiotic crossovers in males (Morgan 1912). We scored crossovers between the visible markers st and Sb, which are separated by >36 Mbp (∼20% of the genome). Crossovers were not detected among progeny of wild-type males or Fancl mutant males, but were significantly elevated in Fancm mutant males (Figure 2C). In the same assay, the crossover frequency in Blm mutants is about threefold higher than in Fancm mutants. The rate in Blm Fancm double mutants was not significantly different from that in Fancm single mutants but was significantly lower than in Blm single mutants. A straightforward interpretation of this result is that FANCM functions upstream of BLM in a pathway that prevents mitotic crossovers; however, this is likely to be an oversimplification, given the multiple functions of these enzymes and the possibility of partial overlap in function (see Discussion).
FANCM has a modest role in SDSA
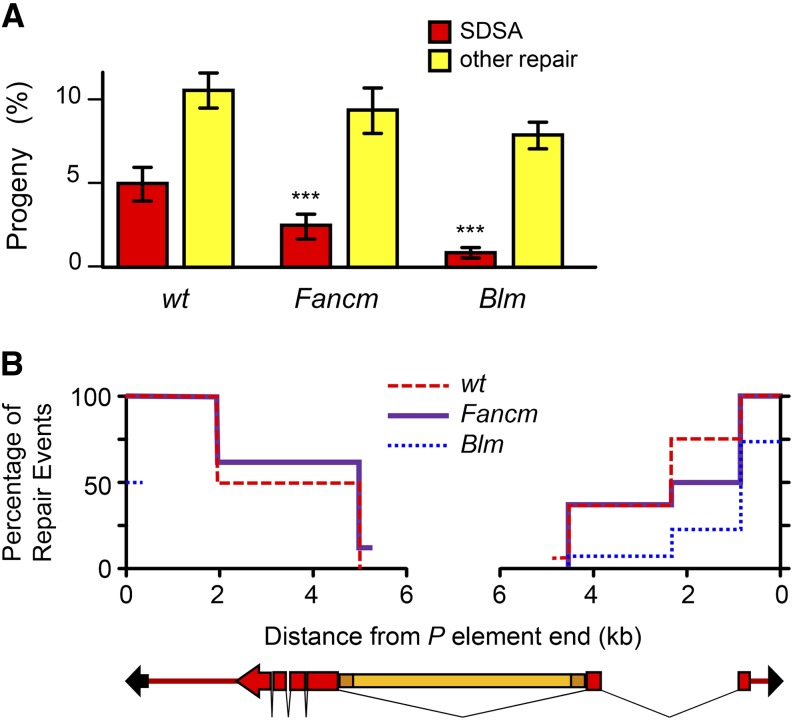
Hypersensitivity to IR and elevated mitotic crossovers suggest a role for FANCM in DSB repair, independent of its role in the FA pathway. It has been proposed that S. pombe Fml1 and S. cerevisiae Mph1 promote SDSA by disassembling D loops (Sun et al. 2008; Prakash et al. 2009; Mitchel et al. 2013), but this hypothesis has not been tested directly using an assay specific for SDSA. We used a gap repair assay in which products of SDSA can be distinguished from other types of repair (Adams et al. 2003; McVey et al. 2004a). A gap is generated by excision of a P{wa} element from the male X chromosome. This element carries the apricot allele of the white gene (wa), in which a copia retrotransposon is inserted into an intron, resulting in orange eye color instead of the wild-type red color (Figure 3B). Excision leaves a DSB that is repaired using the sister chromatid as a template. Since the sister still has an intact P{wa} element, this amounts to gap repair. Repair by two-ended SDSA can result in annealing between the long terminal repeats (LTRs) at the ends of copia, giving a product with only one LTR instead of an entire copia. Progeny that inherit this product have red eyes. This red-eyed class was decreased by ∼50% in Fancm mutants compared to controls (Figure 3A), revealing a reduced ability to complete repair by SDSA. In Blm mutants, the decrease in SDSA is significantly more severe (Figure 3A) (McVey et al. 2007). Other types of repair result in loss of white gene function, which we recover as progeny with yellow eyes. In wild-type flies this is almost exclusively aborted SDSA in which there is templated synthesis from one or both ends of the gap followed by joining through an alternative end-joining pathway (Adams et al. 2003; McVey et al. 2004a,c; Chan et al. 2010). Molecular analyses of these repair products shows that synthesis tracts are significantly shorter in Blm mutants than in wild-type flies (Adams et al. 2003), but did not reveal any differences between wild-type and Fancm mutants (Figure 3B).
Figure 3.
SDSA defects in Fancm mutants. (A) Gap repair outcomes. Bars show mean fraction of flies in each repair class: red bars indicate repair by SDSA, and yellow bars indicate other types of repair (usually aborted SDSA followed by end joining). Of the remaining flies, most came from cells that did not experience an excision event, although a small percentage may also be from the repair of the entire gap by SDSA. Blm data are from McVey et al. (2007). Error bars are standard error of the mean. n = 85 for Fancm and 45 for control. ***P < 0.001 compared to wild type. (B) Kaplan–Meier graph showing the amount of synthesis from each end of the P element in non-SDSA progeny (those with yellow eyes). PCR was done on sons of these progeny to detect synthesis from the left end at 5 bp, 1.7 kb, and 5.2 kb from the cut site and from the right end at 5 bp, 920 bp, 2.4 kb, and 4.6 kb from the cut site. Blm data are from Adams et al. (2003); the left end was analyzed only at 5 bp in that study. The drawing at the bottom represents the P{wa} element: black arrows, P-element ends; red, white gene (boxes, exons; lines, introns); orange, copia element (dark, long terminal repeats). n = 16 (Fancm), 83 (wild type), and 147 (Blm).
Both Fancm and Blm mutants have elevated spontaneous mitotic crossovers and a decreased ability to complete SDSA repair of a gap, but the defects in Blm mutants are more severe in both assays. Since Fancm is epistatic to Blm for mitotic crossovers and SDSA is thought to be an important pathway in crossover avoidance during DSB repair, we asked whether Fancm is epistatic to Blm in our SDSA assay. However, we were unable to generate any Blm Fancm double-mutant males carrying both the P{wa} element and transposase. This appeared to be due to recombination defects, since flies that also lacked the strand exchange protein Rad51 (spn-A mutants), and therefore are incapable of initiating recombination, do survive infrequently.
Spontaneous mitotic crossovers in the absence of FANCM require the scaffolding protein MUS312 and either MUS81 or SLX1
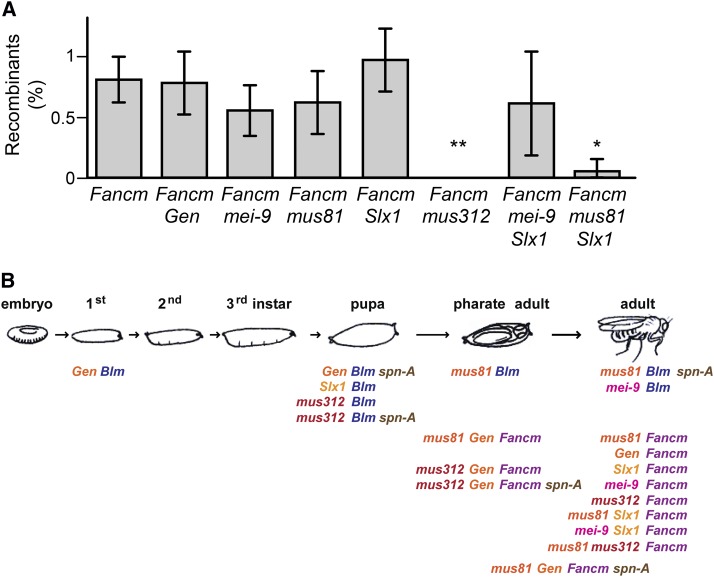
To better understand how spontaneous mitotic crossovers in Fancm mutants are generated, we asked whether any HJ resolvases are required. We made double mutants between Fancm and the genes encoding each of the catalytic subunits of the putative resolvases: Gen, mus81, Slx1, and mei-9. There was no significant difference in crossover frequency in any of these double mutants relative to Fancm single mutants (Figure 4A). However, crossovers were completely eliminated in double mutants with mus312 (Figure 4A). MUS312 is a scaffolding protein that interacts physically and functionally with both MEI-9 (the Drosophila ortholog of Rad1/XPF) and SLX1 (Yıldız et al. 2002; Andersen et al. 2009), so we considered the possibility that SLX1 and MEI-9 have redundant roles in generating these mitotic crossovers in Fancm mutants. This does not appear to be the case because mei-9; Slx1 Fancm triple mutants have the same crossover rate as Fancm single mutants and both double mutants (Figure 4A). Orthologs of MUS312 from vertebrates (SLX4) and C. elegans (HIM-18) interact physically with MUS81 (Fekairi et al. 2009; Muñoz et al. 2009; Svendsen et al. 2009; Saito et al. 2013). A similar interaction has not been reported for the Drosophila proteins, and we failed to detect such an interaction in yeast two-hybrid assays (J. R. LaRocque and J. Sekelsky, personal communication). Nevertheless, we asked whether MUS81 and SLX1 might be acting redundantly to make mitotic crossovers in Fancm mutants. Crossovers were nearly absent in mus81; Slx1 Fancm triple mutants (Figure 4A), suggesting that MUS81 and SLX1 indeed act redundantly to make these crossovers and that both nucleases require MUS312 for this function.
Figure 4.
Phenotypes of Fancm mutants lacking one or more resolvases. (A) Mitotic crossovers in Fancm mutants lacking various resolvases. Error bars are standard error of the mean. Statistical significance was determined relative to Fancm single mutants (*P = 0.0298; **P = 0.0083). P-values reported have been adjusted for multiple comparisons (see Materials and Methods). n = (left to right) 46, 18, 18, 22, 17, 23, 20, and 22. (B) Lethality and viability of mutants lacking a helicase and one or more resolvases. At the top is a drawing of the developmental life cycle of Drosophila. Various genotypes lacking one of the anticrossover helicases (BLM or FANCM) and one or more of the putative resolvases are listed below the stage at which they die. Those listed below the adult live to adulthood. Life cycle stages and Blm mutant results are modified from Andersen et al. (2011).
Simultaneous loss of FANCM and multiple resolvases is lethal
In Drosophila, each of the three putative mitotic HJ resolvases (MUS81–MMS4, GEN, and MUS312–SLX1) are essential when BLM is absent (Trowbridge et al. 2007; Andersen et al. 2009, 2011). The experiments described above show that none of the resolvases are essential when FANCM is absent and that at least males are fertile (we did not assay female fertility since several of these resolvases are required for meiotic recombination and their absence causes high levels of nondisjunction and low fecundity). However, synthetic lethality was observed when certain combinations of resolvases were removed (Figure 4B). As in budding yeast (Blanco et al. 2010; Ho et al. 2010; Tay and Wu 2010), Drosophila MUS81–MMS4 and GEN have a partially redundant or compensatory relationship (Andersen et al. 2011). Simultaneous loss of both MUS81–MMS4 and GEN is lethal in Fancm mutants (Figure 4B). The mus81; Gen Fancm triple mutants survive to the pharate adult stage, which is the same stage at which mus81; Blm double mutants die but much later than lethality of Gen Blm double mutants, which only survive until the second instar larval stage. Preventing recombination partially suppresses mus81; Blm and Gen Blm lethality: mus81; Blm spn-A mutants are semiviable (∼70% live to adulthood), and Gen Blm spn-A mutants survive to the pupal stage (Trowbridge et al. 2007; Andersen et al. 2011). To ask whether mus81; Gen Fancm inviability is similarly due to recombination defects, we made mus81; Gen Fancm spn-A quadruple mutants. A few quadruple mutants did survive to adulthood (4, compared to 84 expected), and these had rough eyes and cuticle defects suggestive of high rates of cell death during development. Gen mus312 Fancm triple mutants also died as pharate adult pupae; mutating spn-A had no apparent effect on this lethality (Figure 4B).
Discussion
Comparison of Drosophila Fancm and Fancl mutants for hypersensitivity to DNA-damaging agents (Figure 1) indicates that, as in fungi and plants, Drosophila FANCM has functions outside of the FA pathway. Among the agents that we tested, Fancl mutants were hypersensitive to only the cross-linking agent HN2, consistent with a primary or sole function for FANCL in the FA pathway. Fancm mutants were more sensitive to HN2 and, unlike Fancl mutants, were hypersensitive to the alkylating agent MMS and to ionizing radiation. Among these FA-independent roles, we focus on functions that may prevent crossovers.
FANCM in meiotic recombination
Drosophila Fancm mutants have a significant elevation in both meiotic and mitotic crossovers (Figure 2). Interestingly, the increase in meiotic crossovers was observed only in the two most proximal intervals, each of which had threefold more crossovers than wild-type females. Elevated meiotic crossovers have also been reported for Arabidopsis FANCM mutants, but in this case the elevation seems to be genome-wide (Crismani et al. 2012). The significance and cause of the elevation in Drosophila being restricted to only the two proximal intervals is unknown. One of these (pr–cn) includes the centromere and pericentric heterochromatin. It seems unlikely that the crossovers that we recovered occurred in heterochromatic regions since these are normally devoid of DSBs (Jang et al. 2003), but we did not directly determine whether the crossovers between pr and cn were in the euchromatic or heterochromatic portion of this interval.
Based on immunolocalization of meiotic recombination proteins, Knoll et al. (2012) hypothesized that Arabidopsis FANCM suppresses crossovers produced by MUS81, which is usually responsible for only 10–15% of meiotic crossovers in normal meiosis (Berchowitz et al. 2007). In Drosophila, most meiotic crossovers are generated by a complex whose catalytic subunit is MEI-9, which is orthologous to XPF/Rad1 (Sekelsky et al. 1995). No role in generating meiotic crossovers has been detected for MUS81 (Trowbridge et al. 2007), so it will be interesting to determine whether the extra meiotic crossovers in Fancm mutants are dependent on MEI-9, MUS81, or another resolvase or combination of resolvases.
FANCM in synthesis-dependent strand annealing
Mitotic crossovers are elevated in the germlines of Fancm mutant males (Figure 2). Previous studies found elevated mitotic crossovers in S. pombe fml1 mutants and in S. cerevisiae mph1 mutants (Sun et al. 2008; Prakash et al. 2009; Mazón and Symington 2013; Mitchel et al. 2013). It is important to note that these studies in fungi involved enzymatic induction of DSBs, whereas the crossovers that we measured are spontaneous. Our experiments do not provide insight into the sources of these crossovers. FANCM may direct repair of spontaneous lesions toward noncrossover outcomes, or loss of FANCM may cause an increased incidence of some lesions, such as a DSB, that might be precursors to crossovers.
Mph1 and Fml1 have been proposed to promote DSB repair through the noncrossover SDSA pathway by disrupting D loops (Sun et al. 2008; Prakash et al. 2009; Tay et al. 2010; Mitchel et al. 2013). Hypersensitivity to ionizing radiation and elevated mitotic crossover frequency are consistent with Drosophila FANCM having a function in SDSA. In our gap repair assay for SDSA, we detected a 50% reduction in progeny with the phenotype diagnostic of completed SDSA repair, confirming a role for FANCM in this process (Figure 3). BLM/Sgs1 also has anticrossover functions during DSB repair. Although this is usually discussed in terms of the dHJ dissolution function, a function in SDSA is also apparent in the finding that Sgs1 generates meiotic noncrossovers (De Muyt et al. 2012) and that Drosophila Blm mutants are severely compromised in the P{wa} gap repair assay for SDSA (Adams et al. 2003). Molecular and genetic analysis of gap repair products from Blm mutants revealed that synthesis tracts are significantly shorter than in wild-type males, and deletions into flanking DNA sequences are significantly more frequent (Adams et al. 2003; McVey et al. 2007). It is thought that repair of the large gap in the P{wa} assay requires multiple cycles of strand exchange, synthesis, and D-loop disassembly (McVey et al. 2004a). This led to a model in which BLM is required for D-loop disassembly, and in the absence of BLM these structures are cut by a structure-selective endonuclease, leading to flanking deletions. This hypothesis raised the question of why there are synthesis tracts at all in Blm mutants. We propose that BLM-topoisomerase 3α is essential for disassembling D loops only after lengthy synthesis, but that FANCM can disassemble shorter D loops. Although some features of this model are attractive, it does not explain why there is an SDSA defect in Fancm mutants. It is also possible that both FANCM and BLM promote SDSA by disassembling D loops, but that they act at different stages of male germline development.
Resolvases in generating mitotic crossovers in Fancm mutants
Spontaneous mitotic crossovers that occur in the absence of FANCM require the MUS312 nuclease scaffold protein and either MUS81 or SLX1 (Figure 3A). One simple interpretation is that FANCM acts at an early stage to direct repair down a noncrossover pathway (e.g., SDSA). In the absence of FANCM, an HJ-containing intermediate is generated, and it is the resolution of this intermediate by MUS81 or SLX1 that generates a crossover. Recent in vitro experiments suggest that vertebrate MUS81 and SLX1 collaborate to resolve HJs, with SLX1 making an initial nick and MUS81 making a second nick and both being coordinated by the MUS312 ortholog SLX4 (Castor et al. 2013; Garner et al. 2013; Wyatt et al. 2013). Our result is more consistent with MUS81 and SLX1 having redundant functions. If Drosophila MUS81 and SLX1 work together to resolve HJs, then it must not be loss of this activity that prevents crossovers when FANCM is absent.
The different phenotypes of mus81 and Slx1 mutants indicate that each enzyme has unique functions, but this does not preclude redundant or codependent functions (Trowbridge et al. 2007; Andersen et al. 2009). Both S. cerevisiae and human MUS81–EME1 and SLX4–SLX1 cut flap and replication fork structures in vitro (Fricke and Brill 2003; Ehmsen and Heyer 2008; Wyatt et al. 2013). It is possible that, rather than working late on a HJ intermediate, both MUS312–MUS81–MMS4 or MUS312–SLX1 can generate DSBs by cutting aberrant replication fork structures that would normally be processed by FANCM. Some of these DSBs may then be repaired through a pathway that results in a crossover with the homologous chromosome. Given the anticrossover roles of BLM during DSB repair, however, we might expect a synergistic effect on mitotic crossover frequency in Blm Fancm double mutants. Instead, we saw a crossover frequency more similar to that of Fancm single mutants. One weakness of this crossover assay is that we cannot detect complete failure of repair since this would likely result in apoptosis or spermatocyte defects. Blm Fancm males did not produce fewer progeny than single-mutant males, but we would not have been able to detect reductions of similar magnitude as the mitotic crossover frequency (∼2% in Blm single mutants).
We also found that mus81 Gen Fancm and Gen mus312 Fancm mutants are inviable. In the former of these genotypes, inviability may be explained by functional overlap between MSU81–MMS4 and GEN. S. cerevisiae Mus81–Mms4 and Yen1 exhibit partial redundancy, with Yen1 appearing to function primarily as a backup to Mus81–Mms4 (Blanco et al. 2010; Ho et al. 2010; Tay and Wu 2010). Redundancy has also been observed between Drosophila MUS81–MMS4 and GEN, although the relationship appears to be reversed, with Gen mutants having more severe phenotypes than mus81 mutants (Andersen et al. 2011). Inviability in mus81 Gen Fancm triple mutants is weakly suppressed by preventing recombination. This suggests that either the death of mus81 Gen Fancm mutants is only partially due to defects in recombination or that the alternatives to RAD51-mediated recombination are also detrimental.
No functional overlaps have been reported for GEN and MUS312–SLX1 or their orthologs. Triple mutants that lack GEN, MUS312, and FANCM may be inviable because of an accumulation of damage, some resulting from loss of GEN and FANCM and some resulting from loss of MUS312–SLX1 and FANCM. This lethality may also be in part due to loss of both GEN and those MUS81 activities that require MUS312 or some more complex interaction involving all three nucleases.
Concluding remarks
In summary, our analysis shows that Drosophila Fancm mutants have similar phenotypes to Blm mutants in several assays. These defects are generally less severe in Fancm mutants and more severe in Blm Fancm double mutants. This suggests that FANCM and BLM have overlapping functions and that these helicases are partially redundant or that either can partially compensate for loss of the other; however, several of the phenotypes that we assayed, such as viability after treatment with DNA-damaging agents, viability of flies carrying mutations in multiple genes, and mitotic crossing over, are fairly crude genetic readouts that might have several underlying causes. Thus, more detailed mechanistic studies will be necessary to tease apart the different cellular functions of BLM, FANCM, and the HJ resolvases.
Acknowledgments
We thank members of the Sekelsky lab for critical reading of the manuscript and the Bloomington Drosophila Stock Center for stocks. C.M.R. was supported by a Summer Undergraduate Research Fellowship from the University of North Carolina Office of Undergraduate Research. K.P.K. was supported in part by award T32 GM007092 from the National Institute of General Medical Sciences (NIGMS). This research was supported by grants from NIGMS under award nos. R01 GM061252 and R01 GM099890. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Communicating editor: S. K. Sharan
Literature Cited
- Adams M. D., McVey M., Sekelsky J., 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267. [DOI] [PubMed] [Google Scholar]
- Agostinho A., Meier B., Sonneville R., Jagut M., Woglar A., et al. , 2013. Combinatorial regulation of meiotic Holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 9: e1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S. L., Sekelsky J., 2010. Meiotic vs. mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic vs. mitotic DSB repair are reflected in different pathway usage and different outcomes. BioEssays 32: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S. L., Bergstralh D. T., Kohl K. P., Larocque J. R., Moore C. B., et al. , 2009. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol. Cell 35: 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S. L., Kuo H. K., Savukoski D., Brodsky M. H., Sekelsky J., 2011. Three structure-selective endonucleases are essential in the absence of BLM helicase in Drosophila. PLoS Genet. 7: e1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker S. T., Van De Vrugt H. J., Rooimans M. A., Oostra A. B., Steltenpool J., et al. , 2009. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum. Mol. Genet. 18: 3484–3495. [DOI] [PubMed] [Google Scholar]
- Beranek D. T., 1990. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 231: 11–30. [DOI] [PubMed] [Google Scholar]
- Berchowitz L. E., Francis K. E., Bey A. L., Copenhaver G. P., 2007. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M. G., Matos J., Rass U., Ip S. C., West S. C., 2010. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair (Amst.) 9: 394–402. [DOI] [PubMed] [Google Scholar]
- Boddy M. N., Gaillard P. H., McDonald W. H., Shanahan P., Yates J. R., III, et al. , 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548. [DOI] [PubMed] [Google Scholar]
- Brosh R. M., Jr, 2013. DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 13: 542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castor D., Nair N., Declais A. C., Lachaud C., Toth R., et al. , 2013. Cooperative control of Holliday junction resolution and DNA repair by the SLX1 and MUS81–EME1 nucleases. Mol. Cell 52: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. H., Yu A. M., McVey M., 2010. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6: e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. B., Melchionna R., Denis C. M., Gaillard P. H., Blasina A., et al. , 2001. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8: 1117–1127. [DOI] [PubMed] [Google Scholar]
- Cooper J. L., Till B. J., Henikoff S., 2008. Fly-TILL: reverse genetics using a living point mutation resource. Fly (Austin) 2: 300–302. [DOI] [PubMed] [Google Scholar]
- Crismani W., Girard C., Froger N., Pradillo M., Santos J. L., et al. , 2012. FANCM limits meiotic crossovers. Science 336: 1588–1590. [DOI] [PubMed] [Google Scholar]
- Dayani Y., Simchen G., Lichten M., 2011. Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLoS Genet. 7: e1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Santos T., Hunter N., Lee C., Larkin B., Loidl J., et al. , 2003. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A., Jessop L., Kolar E., Sourirajan A., Chen J., et al. , 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmsen K. T., Heyer W. D., 2008. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 36: 2182–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S., Scaglione S., Chahwan C., Taylor E. R., Tissier A., et al. , 2009. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138: 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke W. M., Brill S. J., 2003. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 17: 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K., Décaillet C., Stasiak A. Z., Stasiak A., Constantinou A., 2008. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell 29: 141–148. [DOI] [PubMed] [Google Scholar]
- Garner E., Kim Y., Lach F. P., Kottemann M. C., Smogorzewska A., 2013. Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Reports 5: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. K., Mazon G., Lam A. F., Symington L. S., 2010. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell 40: 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip S. C., Rass U., Blanco M. G., Flynn H. R., Skehel J. M., et al. , 2008. Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361. [DOI] [PubMed] [Google Scholar]
- Jang J. K., Sherizen D. E., Bhagat R., Manheim E. A., McKim K. S., 2003. Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116: 3069–3077. [DOI] [PubMed] [Google Scholar]
- Kaliraman V., Mullen J. R., Fricke W. M., Bastin-Shanower S. A., Brill S. J., 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15: 2730–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow J. K., Constantinou A., Li J. L., West S. C., Hickson I. D., 2000. The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. USA 97: 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., D’Andrea A. D., 2012. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 26: 1393–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A., Higgins J. D., Seeliger K., Reha S. J., Dangel N. J., et al. , 2012. The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell 24: 1448–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K. P., Jones C. D., Sekelsky J., 2012. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338: 1363–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano K., Johnson-Schlitz D. M., Engels W. R., 2001. Sterility of Drosophila with mutations in the Bloom syndrome gene: complementation by Ku70. Science 291: 2600–2602. [DOI] [PubMed] [Google Scholar]
- Lorenz A., Osman F., Sun W., Nandi S., Steinacher R., et al. , 2012. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science 336: 1585–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek L. R., Bale A. E., 2006. Drosophila homologs of FANCD2 and FANCL function in DNA repair. DNA Repair (Amst) 5: 1317–1326. [DOI] [PubMed] [Google Scholar]
- Mazón G., Symington L. S., 2013. Mph1 and Mus81-Mms4 prevent aberrant processing of mitotic recombination intermediates. Mol. Cell 52: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Adams M. D., Staeva-Vieira E., Sekelsky J. J., 2004a Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167: 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Larocque J. R., Adams M. D., Sekelsky J. J., 2004b Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl. Acad. Sci. USA 101: 15694–15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Radut D., Sekelsky J., 2004c End-joining repair of double-strand breaks in Drosophila is largely DNA ligase IV-independent. Genetics 168: 2067–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Andersen S. L., Broze Y., Sekelsky J., 2007. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176: 1979–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K., Lehner K., Jinks-Robertson S., 2013. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet. 9: e1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. H., 1912. Complete linkage in the second chromosome of the male of Drosophila. Science 36: 719–720. [Google Scholar]
- Mosedale G., Niedzwiedz W., Alpi A., Perrina F., Pereira-Leal J. B., et al. , 2005. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol. 12: 763–771. [DOI] [PubMed] [Google Scholar]
- Muñoz I. M., Hain K., Declais A. C., Gardiner M., Toh G. W., et al. , 2009. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell 35: 116–127. [DOI] [PubMed] [Google Scholar]
- Osman F., Dixon J., Doe C. L., Whitby M. C., 2003. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12: 761–774. [DOI] [PubMed] [Google Scholar]
- Prakash R., Satory D., Dray E., Papusha A., Scheller J., et al. , 2009. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 23: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. T., Lui D. Y., Kim H.-M., Meyer K., Colaiácovo M. P., 2013. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet. 9: e1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O., Berdnik D., Levy J. M., Wu J. S., Luginbuhl D., et al. , 2008. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev. Cell 14: 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E. K., Heyer W. D., 2011. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120: 109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J. J., McKim K. S., Chin G. M., Hawley R. S., 1995. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141: 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier J., 2011. Fanconi anemia. Hematology Am. Soc. Hematol. Educ. Program 2011: 492–497. [DOI] [PubMed] [Google Scholar]
- Staeva-Vieira E., Yoo S., Lehmann R., 2003. An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 22: 5863–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Nandi S., Osman F., Ahn J. S., Jakovleska J., et al. , 2008. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell 32: 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen J. M., Smogorzewska A., Sowa M. E., O’Connell B. C., Gygi S. P., et al. , 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138: 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y. D., Wu L., 2010. Overlapping roles for Yen1 and Mus81 in cellular Holliday junction processing. J. Biol. Chem. 285: 11427–11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y. D., Sidebotham J. M., Wu L., 2010. Mph1 requires mismatch repair-independent and -dependent functions of MutSalpha to regulate crossover formation during homologous recombination repair. Nucleic Acids Res. 38: 1889–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge K., McKim K. S., Brill S., Sekelsky J., 2007. Synthetic lethality in the absence of the Drosophila MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics 176: 1993–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Brabant A. J., Ye T., Sanz M., German I. J., Ellis N. A., et al. , 2000. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry 39: 14617–14625. [DOI] [PubMed] [Google Scholar]
- Wechsler T., Newman S., West S. C., 2011. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature 471: 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby M. C., 2005. Making crossovers during meiosis. Biochem. Soc. Trans. 33: 1451–1455. [DOI] [PubMed] [Google Scholar]
- Whitby M. C., 2010. The FANCM family of DNA helicases/translocases. DNA Repair (Amst.) 9: 224–236. [DOI] [PubMed] [Google Scholar]
- Wijen J. P., Nivard M. J., Vogel E. W., 2000. The in vivo genetic activity profile of the monofunctional nitrogen mustard 2-chloroethylamine differs drastically from its bifunctional counterpart mechlorethamine. Carcinogenesis 21: 1859–1867. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I. D., 2003. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874. [DOI] [PubMed] [Google Scholar]
- Wyatt H. D., Sarbajna S., Matos J., West S. C., 2013. Coordinated actions of SLX1–SLX4 and MUS81–EME1 for Holliday junction resolution in human cells. Mol. Cell 52: 234–247. [DOI] [PubMed] [Google Scholar]
- Yıldız Ö., Majumder S., Kramer B. C., Sekelsky J., 2002. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol. Cell 10: 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yıldız Ö., Kearney H., Kramer B. C., Sekelsky J. J., 2004. Mutational analysis of the Drosophila DNA repair and recombination gene mei-9. Genetics 167: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]