Abstract
Regular meiotic chromosome segregation requires sister centromeres to mono-orient (orient to the same pole) during the first meiotic division (meiosis I) when homologous chromosomes segregate, and to bi-orient (orient to opposite poles) during the second meiotic division (meiosis II) when sister chromatids segregate. Both orientation patterns require cohesion between sister centromeres, which is established during meiotic DNA replication and persists until anaphase of meiosis II. Meiotic cohesion is mediated by a conserved four-protein complex called cohesin that includes two structural maintenance of chromosomes (SMC) subunits (SMC1 and SMC3) and two non-SMC subunits. In Drosophila melanogaster, however, the meiotic cohesion apparatus has not been fully characterized and the non-SMC subunits have not been identified. We have identified a novel Drosophila gene called sisters unbound (sunn), which is required for stable sister chromatid cohesion throughout meiosis. sunn mutations disrupt centromere cohesion during prophase I and cause high frequencies of non-disjunction (NDJ) at both meiotic divisions in both sexes. SUNN co-localizes at centromeres with the cohesion proteins SMC1 and SOLO in both sexes and is necessary for the recruitment of both proteins to centromeres. Although SUNN lacks sequence homology to cohesins, bioinformatic analysis indicates that SUNN may be a structural homolog of the non-SMC cohesin subunit stromalin (SA), suggesting that SUNN may serve as a meiosis-specific cohesin subunit. In conclusion, our data show that SUNN is an essential meiosis-specific Drosophila cohesion protein.
Keywords: Drosophila melanogaster, meiosis, chromosome, cohesion, centromere, Sisters Unbound (SUNN)
MEIOSIS is a specialized cell division that generates haploid gametes from diploid precursor cells and is essential for sexual reproduction. Segregation of chromosomes during meiosis occurs in two stages called meiosis I and meiosis II that follow a single round of DNA replication. During meiosis I, homologs pair and orient toward opposite poles of the spindle (bi-orient) with sister centromeres oriented toward the same pole (mono-oriented). As a result, homologous chromosomes segregate to opposite poles at the onset of anaphase I in a reductional segregation pattern. In meiosis II, as in mitosis, the sister centromeres are bi-oriented and sister chromatids segregate to opposite poles at the onset of anaphase II, a pattern referred to as equational segregation (Page and Hawley 2003; Petronczki et al. 2003).
In most eukaryotes, pairing of homologs during meiosis I is facilitated and reinforced by synapsis and recombination. Synapsis involves formation of elaborate zipper-like structures, called synaptonemal complexes (SCs), which hold homologs tightly together during prophase I. SCs are composed of the tightly paired sister chromatid axes of the two homologs, known as axial elements (AEs) before synapsis or as lateral elements (LEs) after synapsis, cross-linked by multiple transverse filament (TF) proteins. Synapsis initiates at a limited number of discrete sites of homolog alignment during zygotene and subsequently spreads until SCs form continuous chromosome-length structures during pachytene (Page and Hawley 2004). Most synapsis initiation sites appear to be in the euchromatin but in some eukaryotes, including yeast, several plant species, and female Drosophila, synapsis also initiates at centromeres and is preceded by homologous and/or non-homologous pairing of centromeres (Khetani and Bickel 2007; Stewart and Dawson 2008; Takeo et al. 2011; Tanneti et al. 2011). Recombination overlaps temporally with synapsis and involves programmed formation and repair of double-strand breaks, resulting in high levels of exchange (crossing over) between homologous chromatids. After SC disassembly at the end of pachytene, homolog crossovers, stabilized by cohesion between sister chromatid arms, serve as stable interhomolog linkers known as chiasmata (Page and Hawley 2003; Kleckner 2006). Some eukaryotes achieve stable homolog pairing and regular homolog segregation during meiosis I without SCs or recombination. In Drosophila males, which lack meiotic recombination, stable homolog connections are provided by a male-specific “homolog conjunction complex” that serves as a functional substitute for chiasmata and is removed at anaphase I (Thomas et al. 2005).
Proper chromosome segregation at both meiotic divisions, as well as in mitosis, requires cohesion between sister chromatids provided by conserved four-protein complexes called cohesins. The mitotic cohesin complex is composed of SMC1, SMC3, SCC1/MCD1/RAD21 (henceforth called RAD21), and SCC3/stromalin/SA (henceforth called SA). SMC1, SMC3, and RAD21 form a tripartite ring structure that is thought to embrace the newly formed sister chromatids during S phase. Cleavage of the RAD21 subunit by the conserved protease Separase at anaphase releases cohesion and allows sister chromatids to segregate to the poles. SA is an all α-helical protein that binds to RAD21. It is essential for cohesion but its precise role in cohesion remains unclear. Meiotic cohesins are similar in composition to mitotic cohesin but frequently contain one or more paralogous meiosis-specific subunits that replace their mitotic counterparts. Most such paralogs are restricted to fairly narrow taxonomic lineages and have specialized functions, but REC8 replaces RAD21 in most meiotic cohesins and is required for nearly all meiotic chromosome interactions in most eukaryotes (Lee and Orr-Weaver 2001; Nasmyth 2001; Nasmyth and Haering 2009).
In meiosis I, cohesin is abundant all along the chromosome axes but arm and centromere cohesion play distinct roles in meiosis. During meiosis I, arm cohesion stabilizes the chiasmata that provide resistance to poleward forces required for homologs to biorient on the meiosis I spindle. Release of arm cohesion at the onset of anaphase I, by separase-mediated cleavage of REC8, destabilizes chiasmata and serves as the triggering event for homolog segregation (Petronczki et al. 2003; Nasmyth and Haering 2009). Arm cohesins also play important roles in synapsis and recombination during meiosis I although it remains unclear to what extent those roles are related to arm cohesion (Nasmyth 2001; Brar et al. 2009; Nasmyth and Haering 2009). Cohesion between sister centromeres enables sister chromatids to biorient on the meiosis II spindle and is preserved until a second round of separase activation at anaphase II cleaves centromeric cohesins and triggers sister chromatid separation. Preservation of centromeric cohesins during anaphase I is mediated by shugoshins, which are centromere proteins that inhibit separase cleavage of cohesin (Watanabe 2005; Clift and Marston 2011).
Centromere cohesion is also required during meiosis I, to enable sister centromeres to mono-orient. Mono-orientation is thought to require a “side-by-side” alignment of sister centromeres (rather than the “back-to-back” alignment characteristic of mitosis or meiosis II) enabling them to form a functionally single kinetochore that binds microtubules from only one pole (Hauf and Watanabe 2004). In Schizosaccharomyces pombe, this specialized centromere orientation entails establishing cohesion within the kinetochore-forming centromere core domain and requires both REC8 cohesin and a specialized meiosis-specific centromere protein called Moa1 (Watanabe and Nurse 1999; Yokobayashi and Watanabe 2005). In S. cerevisiae, both cohesin and a meiosis I-specific centromere complex called monopolin are required for regular mono-orientation (Toth et al. 2000). In several higher eukaryotes, mutations in rec8 or other cohesion genes have also been found to disrupt mono-orientation (Klein et al. 1999; Pasierbek et al. 2001; Cai et al. 2003; Wang et al. 2003; Chelysheva et al. 2005; Golubovskaya et al. 2006; Severson et al. 2009). However, no specific mono-orientation factors have been identified in higher eukaryotes and the mechanism of mono-orientation remains unclear.
Drosophila has been a major model for meiotic studies for more than a century. However, insight into the mechanism and roles of cohesion in Drosophila meiosis has been hampered by limited data on the composition of the meiotic cohesion apparatus. Recent findings have pointed to meiotic roles for the cohesin SMC proteins. SMC1 localizes to centromeres during meiosis in both sexes and persists on centromeres until anaphase II in male meiosis (Khetani and Bickel 2007; Yan et al. 2010). Both SMC1 and SMC3 localize to LEs in female prophase I and loss of either protein completely ablates formation of LEs and SCs (Khetani and Bickel 2007; Tanneti et al. 2011; Yan and McKee 2013). However, as yet there is only indirect evidence for roles of SMC1 and SMC3 in arm or centromere cohesion. Moreover, the non-SMC subunits of meiotic cohesins in Drosophila remain poorly characterized. There are no published reports relevant to a meiotic role of SA. Although RAD21 was recently shown to localize to lateral elements in female meiosis and to be required for SC stability, neither premature cleavage or degradation of RAD21 during prophase I nor mutation of its Separase cleavage sites had any effect on sister chromatid cohesion or on chromosome segregation at either meiotic division (Urban et al. 2014). In addition, unlike all other characterized eukaryotes, Drosophila appears to lack a true REC8 homolog. The Drosophila genome does encode a meiosis-specific RAD21 paralog, C(2)M, that localizes to LEs and is required for synapsis, SC formation, and normal levels of recombination (Manheim and McKim 2003). However, C(2)M does not form centromeric foci and is not required for either centromere or arm cohesion during the meiotic division stages in female meiosis or for any aspect of male meiosis (Manheim and McKim 2003; Heidmann et al. 2004) .
Curiously, the best-characterized meiotic cohesion genes in Drosophila are two genes with no apparent homology to any of the cohesins: orientation disruptor (ord) and sisters on the loose (solo) (Bickel et al. 1996; Yan et al. 2010). Mutations in both genes cause premature loss of sister centromere cohesion, accompanied by absence of centromeric SMC1 foci, leading to very high frequencies of both homolog and sister chromatid non-disjunction (NDJ) (Miyazaki and Orr-Weaver 1992; Bickel et al. 1997; Balicky et al. 2002; Yan et al. 2010). Both ORD and SOLO co-localize with SMC1 on centromeres in both sexes and persist there until anaphase II in male meiosis, disappearing simultaneously with SMC1 (Balicky et al. 2002; Khetani and Bickel 2007; Yan et al. 2010). Consistent with a cohesin-related role, SOLO was recently shown to reciprocally coimmunoprecipitate with SMC1 from ovary extracts (Yan and McKee 2013). These findings have led to suggestions that, despite their lack of sequence homology to cohesins, ORD and SOLO might be functional homologs of REC8 (Khetani and Bickel 2007; Yan et al. 2010; Yan and McKee 2013).
Here we describe a third Drosophila-specific, meiosis-specific cohesion gene, sunn (sisters unbound), with properties remarkably similar to those of ord and solo. sunn mutations cause high levels of both meiosis I and meiosis II NDJ in both sexes. In male meiosis, SUNN localizes primarily to centromeres until anaphase II and is required for centromeric cohesion, for mono-orientation of sister centromeres during meiosis I and for stable centromere recruitment of SMC1 and SOLO. In female meiosis, SUNN also localizes to centromeres during prophase I and is required for centromere pairing and cohesion during pachytene. These data identify SUNN as a major component of the meiotic cohesion apparatus in Drosophila. Although no sequence homologs of SUNN were identified in genome searches, structure-based bioinformatic analysis revealed similarity between SUNN and the Drosophila cohesin subunit stromalin (SA), suggesting SUNN’s possible role as a meiosis-specific cohesin subunit.
Materials and Methods
Fly stocks and Drosophila culturing
sunn mutations were obtained from the Zuker-3 (Z3) collection of EMS mutagenized third chromosomes (Koundakjian et al. 2004). The Z3- lines used in this study were identified in a screen for loss of paternal 4th chromosomes (Wakimoto et al. 2004). Dp (1:1) scv1 was obtained from Kim McKim (Rutgers University). All of the chromosome 3 deficiency stocks and compound chromosome stocks used in the crosses were obtained from the Bloomington Stock Center (Indiana University). Details about markers and special chromosomes can be found in FlyBase and the Bloomington Stock Center webpage (http://flystocks.bio.indiana.edu/).
Flies were cultured at 22° on a food mix containing cornmeal, malt, corn syrup, yeast, and propionic acid (antifungal agent) and crosses were set and maintained at 22°. Progeny from the cross were scored between 14 and 21 days after the cross was set.
Measuring NDJ
The methods for measuring male NDJ are explained in the Results section and in the legends to Table 1 and Table 2. To measure female NDJ, Dp(1;1)scv1, y .y+/y; sunn/Df females were crossed singly to two males of the genotype YSX.YL, In(1)EN, y B/Y. Regular segregation yields B females and B+ males. Progeny from diplo-X and nullo-X- non-disjunctional eggs are B+ females and yellow body y B males, respectively. The B+ daughters carry two maternal X chromatids and were classified as resulting from sister chromatid NDJ if they were y. Dp(1;1)scv1 has a duplication of the tip of the X chromosome on the right arm that carries the dominant y+ marker. Both X chromosomes have recessive y alleles at the native locus near the tip of XL. There is no recombination between the duplicated y+ allele and the X centromere so a NDJ female lacking both copies of the y+ allele is expected to carry two sister centromeres.
Table 1. Sex chromosome NDJ in sunn mutant males.
| Sperm genotype | ||||||
|---|---|---|---|---|---|---|
| Paternal genotype | X | Y | XY | O | na | %NDJb |
| sunnZ3-1956/Dfc | 337 | 353 | 177 | 337 | 1204 | 42.7 |
| sunnZ3-5839/Dfc | 388 | 416 | 181 | 419 | 1404 | 42.7 |
| sunnZ3-4085/Dfc | 344 | 353 | 158 | 400 | 1255 | 44.5 |
| sunnZ3-5839/sunnZ3-5839 | 286 | 266 | 92 | 311 | 955 | 42.2 |
| Total sunn | 1355 | 1388 | 608 | 1467 | 4818 | 43.1 |
| Gamete frequency (%) | 28.1 | 28.8 | 12.6 | 30.4 | — | — |
| Dfc/+ (WT) | 803 | 735 | 0 | 2 | 1540 | 0.1 |
w/BSYy+ males with the indicated third chromosome genotype for sunn were each crossed to two y w females. The dominant BS marker causes Bar eyes and was used to determine whether progeny inherited the Y chromosome.
Total number of progeny scored.
%NDJ = 100 × (XY+ O)/n.
Df (3L) ED4470.
Table 2. Sister chromatid vs. homolog NDJ in sunn mutant males.
| Sperm genotype | ||||||||
|---|---|---|---|---|---|---|---|---|
| X | Y | XY | XX | O | ||||
| Progeny phenotype | ||||||||
| Paternal genotype | w B+ ♂ | su-wa BS ♀ | w BS ♂ | w B+ ♀ | su-wa B+ ♀ | na | %NDJb | %sisc |
| sunnZ3-5839/Df | 388 | 234 | 270 | 64 | 265 | 1221 | 54.3 | 32.2 |
| sunnZ3-1956/Df | 366 | 265 | 223 | 56 | 279 | 1189 | 51.6 | 33.4 |
| sunnZ3-4085/Df | 256 | 188 | 138 | 50 | 264 | 896 | 56.0 | 42.0 |
| Total sunn | 1010 | 687 | 631 | 170 | 808 | 3306 | 53.8 | 35.0 |
| Gamete frequency (%) | 30.6 | 20.8 | 19.1 | 5.1 | 24.4 | — | — | — |
| Df/+ (WT) | 730 | 489 | 0 | 1 | 1 | 1221 | 0.3 | 1 |
w/BSYy+ males with the indicated third chromosome genotype for sunn were each crossed to two C(1)RM, y2 su (wa) wa/O females. These females produce only diplo-X and nullo-X eggs and permit recovery of viable progeny derived exclusively from sister chromatid NDJ sperm (XX), homolog NDJ sperm (XY), and nullo-XY (O) sperm (which result from both types of NDJ). The diplo-X eggs yield viable progeny when fertilized by Y or nullo-XY (O) sperm. These progeny exhibit a suppressed white-apricot (light brown) eye color (su-wa) caused by the su (wa) and wa alleles on C(1)RM. The nullo-X eggs yield viable progeny when fertilized by X, XY, XX, or XXY sperm. These progeny all have white eyes because of the null w allele carried on the paternal X chromosome. Progeny were classified by sperm genotype as described above in column labels.
Total number of progeny scored.
% NDJ = 100 × ((2 × XX) + XY + O)/ n.
% sister chromatid NDJ = 100 × (2 × XX)/ ((2 × XX) + XY).
Notes: (1) Progeny with one or two copies of BSYy+ cannot be discriminated, so some progeny scored as derived from Y or XY sperm could have been YY or XYY. (2) In the %NDJ and %sis formulae, the XX sperm-derived progeny are doubled to account for the YY sperm-derived progeny, which cannot be discriminated from regular Y sperm-derived progeny and are poorly viable. (3) Two, seven, and two progeny derived from XXY non-disjunctional sperm were recovered from sunnZ3-5839/Df, sunnZ3-1956/Df, and sunnZ3-4085/Df hemizygotes, respectively (not shown in table).
Mapping and identification of sunn mutations
Mapping of sunn alleles was performed by deficiency complementation against the 3rd chromosome deficiency kit (Bloomington Stock Center) using the X-Y NDJ phenotype. sunn location was narrowed down to a critical region, 68C8–68D6, on chromosome arm 3L using the following chromosome deficiencies (deleted region in parenthesis)—Df(3L)vin6 (68C8–69A5), Df(3L)vin2 (67F2–68D6), Df(3L)vin3 (68C5–68E4), Df(3L)vin5 (68A2–69A1), Df(3L)vin4 (68B1–68F6), Df(3L)vin7 (68C8–69B5), Df(3L)ED4470 (68A6–68E1), and Df(3L)BK9 (68E2–69A1). Exons of candidate genes from the critical region were amplified by PCR using the genomic DNA of sunn mutants and sequenced (Cycle Sequencing Kit, Life Technologies) to identify single nucleotide polymorphisms (SNPs).
Generation of sunn cDNA clone and UAS–SUNN::Venus transgene
sunn cDNA was amplified from total ovary RNA of y w (yellow white) females using SuperscriptR III Reverse Transcriptase (Invitrogen) and Pfx polymerase (Invitrogen). Total RNA for reverse transcription was extracted using TRI-reagent (Sigma-Aldrich) and treated with DNase I (Invitrogen) before reverse transcription. sunn cDNA was amplified in two overlapping fragments, the first fragment stretching from the 1st exon to the 6th exon and the second fragment extending from the 6th exon to the 10th exon using Pfx polymerase (Invitrogen) and the following primers: First fragment forward, ATGGAATTTGTAAGCGCCATTTCGA and reverse, CAT CACACTCTGCTACTGAGTCAA; second fragment forward, GAATTGAGCCTTATTGCTGCGCAA and reverse, ATCAGTTAGATCTGTTGTATTATGAATAGTTTTAATCT. The two fragments were cloned separately into pJet 1.2/Blunt vector (Fermentas) using CloneJET PCR cloning kit (Fermentas) then ligated together into a pJet 1.2/Blunt vector using restriction sites common to the overlapping fragments. The cDNA was then transferred to pENTR4 (Invitrogen) and recombined into the Gateway P-element vector pPWV 1094 (Drosophila Genomics Resource Center), which contains a C-terminal Venus tag and UAS sequences, using GatewayR LR Clonase II Enzyme Mix (Invitrogen). The resulting construct was transformed into w1118 flies by BestGene.
Determining the 5′ and 3′ ends of Sunn mRNA using RACE
Total RNA was extracted from ovaries of y w (yellow white) females using TRI-reagent (Sigma Aldrich) and treated with DNase I (Invitrogen). RACE was performed using FirstChoice RLM-RACE kit (Ambion Inc). The length of the 5′ UTR determined by 5′ RLM-RACE was 69 bp. The 5′ UTR of CG32088 shown in FlyBase is 72 bp, longer by 3 bp at the 5′ end when compared to the sequence we determined. The 3′ UTR determined by 3′ RACE was found to be 75 bp long and expected to have the features that should be present in a 3′ UTR of the mRNA and the surrounding DNA sequence: a consensus polyadenylation sequence 10–25 bp upstream of the mRNA cleavage site and a conserved element located within 30 bp, downstream of the cleavage site (Retelska et al. 2006). The putative polyadenylation sites, AGUAAA and UAUAAA, are located 23 bp and 32 bp upstream of the cleavage site, respectively, and a U-rich downstream element is positioned 21 bp downstream of the cleavage site. The 3′ UTR for CG32088 shown in FlyBase is 105 bp long, but it lacks essential features of a 3′ UTR. Primer sequences used for 5′ RLM RACE and 3′ RACE are available upon request.
Generation of Venus::SMC1 transgene
smc1 was PCR amplified from a smc1 cDNA clone using the following primers: forward, CACCATGACCGAAGAGGACGACG and reverse, TTACGTGTCCTCGAACGTTGTC. The product was cloned into pENTR/D-TOPO vector (Invitrogen) and the entry clone was recombined with Gateway P-element vector pPVW (1093) (Drosophila Genomics Research Center) using GatewayR LR Clonase II Enzyme Mix (Invitrogen). This vector contains an N-terminal Venus tag and UAS. The construct was transformed into w1118 flies (BestGene).
Testis immunostaining
Testes were dissected and fixed according to Cenci et al. (1994). Immunostaining was performed using the protocol described in Bonaccorsi (2000) with modifications. Testes were dissected in 1× phosphate buffered saline (PBS) (137 mM Nacl, 2.7 mM KCl, 4.3 mM Na2HPO4.7 H2O, 1.4 mM KH2PO4) and covered with Sigmacote (Sigma Aldrich) treated cover slips and frozen in liquid nitrogen. Cover slips were removed and slides were immersed in −20° ethanol for 20 min, followed by 10 min in PBS solution containing 4% formaldehyde. Slides were washed twice with PBT (PBS with 0.2% Triton X-100) and blocked with 1% BSA–PBT solution. Primary antibodies were diluted in 1% BSA–PBT solution and secondary antibodies were diluted in PBT solution. Primary antibody incubations were done for 12–16 hr at 4° and secondary antibody incubations were done for 1 hr at room temperature. Antibody incubations were followed by PBT washes and finally DAPI stain was incubated for 20 min followed by PBS washes and slide mounting using Vectashield (Vector Laboratories). For identifying centromere cohesion phenotypes, a rabbit anti-centromere identifier (CID) primary antibody (Active Motif) and Alexa Fluor 555, a donkey anti-rabbit IgG secondary antibody (H+L, Invitrogen) were used at 1:1000 dilutions. For the Venus::SMC1 and GFP–LacI localization experiments, native fluorescence was used to detect the tagged proteins. Slides were prepared according to the above protocol without the antibody staining steps. For the anti-tubulin/DAPI experiment, immunostaining was performed according to Thomas et al. (2005) using FITC-conjugated monoclonal anti-tubulin antibody (Sigma) at a 1:150 dilution. Meiosis I and meiosis II cells were discriminated on the basis of number of cells per cyst (16 or 32, respectively) and size of DAPI-stained masses. The criteria for meiosis I and II substages are described in Cenci et al. (1994).
Fluorescent in situ hybridization
Fluorescent in situ hybridization (FISH) experiments were performed according to Balicky et al. (2002) with modification (Thomas et al. 2005). The 359 bp satellite-repeat probe was amplified by PCR according to Thomas et al. (2005) and labeled using the Fluorescein-High Prime kit (Roche). The AATAC repeat probe was synthesized as a single-stranded oligonucleotide (IDT Biophysics) and labeled with Alexa Fluor 546 (Invitrogen) using terminal deoxynucleotidyl transferase (Promega).
Ovary immunostaining
Virgin females were placed in a food vial with yeast paste and males. After 2 days, their ovaries were dissected, fixed, and stained using the protocol described in Page and Hawley (2001). Slides were mounted using Prolong Gold Antifade reagent (Invitrogen). To determine centromeric clustering and cohesion phenotypes, rabbit anti-CID (Active Motif) and mouse anti-C(3)G (Scott Hawley, Stowers Institute for Medical Research) primary antibodies were used at 1:1000 dilutions. Alexa Fluor 488 donkey anti-rabbit IgG (H+L, Invitrogen) and Alexa Fluor 555 donkey anti-mouse IgG (H+L, Invitrogen) secondary antibodies were used at 1:1000 dilutions. For the CID spot counts, C(3)G positive cells in germaria and stage 2 were identified as oocytes/pro-oocytes. CID foci were counted to be part of an oocyte if they were within the C(3)G-stained and DAPI-stained boundary of the cell. [Note: C(3)G is a transverse filament protein that provides a useful marker of the SC (Page and Hawley 2004)]. For quantification, non-overlapping CID foci were counted as separate spots. To determine SMC1 localization to centromeres, guinea pig anti-SMC1 (Sharon Bickel, Dartmouth University) and rabbit anti-CID (Active Motif) primary antibodies were used at 1:2000 and 1:1000 dilutions, respectively. Alexa Fluor 488 goat anti-guinea pig IgG (H+L, Invitrogen) and Alexa Fluor 647 donkey anti-rabbit IgG (H+L, Invitrogen) were used as secondary antibodies at 1:1000 dilutions. Classification of oocyte stages was done according to Matthies (2000).
Microscopy
All micrographs were obtained using an Axioplan microscope (Zeiss), which is equipped with a HBO 100 W mercury lamp. This microscope is fitted with a high-resolution charge-coupled device camera (Roper Industries). Metamorph software (Universal Imaging) was used to acquire pictures, pseudocolor them, and merge them together. For all immunostaining and FISH images, Z-series pictures were taken, deconvolved, and merged/stacked using sum algorithm. Images and figures were prepared using Adobe Photoshop (CS2), Adobe Illustrator, and Microsoft PowerPoint.
Results
Mutation of sunn causes homologous and sister chromatid NDJ in both male and female meiosis
Three alleles of sunn were identified in a screen of the Zuker-3 collection of EMS-treated third chromosomes for mutants that showed increased rates of fourth chromosome loss in male meiosis (Koundakjian et al. 2004; Wakimoto et al. 2004). Males hemizygous for each sunn allele and that carried a genetically marked Y chromosome (BsYy+) were tested for X and Y chromosome NDJ in crosses to chromosomally normal females. Progeny from XY and nullo-XY (O) sperm were recovered at frequencies of 42–45% in all three sunn mutants compared to <0.2% in wild-type (WT) controls (Table 1). A similar NDJ frequency was obtained in homozygous sunnZ3-5839 males (Table 1). Taken together, these data suggest, but do not prove, that all three sunn alleles are genetic null alleles.
The results in Table 1 show that sunn mutations cause high frequencies of homolog NDJ but do not address whether sunn mutations also cause sister chromatid NDJ. The diagnostic sperm class for NDJ of X sister chromatids is XX sperm, which yield inviable XXX progeny in crosses to chromosomally normal females. To detect XX sperm and compare the frequencies of homolog and sister chromatid NDJ, sunn males were crossed to females carrying an attached X chromosome [C(1)RM/O], which produce only diplo-X and nullo-X eggs in roughly equal proportions. In such crosses, all major sperm classes, including the XX, XY, and nullo-XY (O) NDJ classes, yield viable progeny in combination with one of the egg classes (see Table 2 legend for detailed explanation). Males hemizygous for the three sunn alleles produced XX, XY, and O non-disjunctional sperm at average frequencies of 5.1, 19.1, and 24.4%, respectively (Table 2), indicating that sunn mutations cause NDJ of both homologous and sister chromatids. The average NDJ frequency was 53.8% and the average relative frequency of sister chromatid NDJ out of total NDJ was 35%. In this assay, as in the previous one, differences among the three alleles were minor and insignificant. These results are consistent with random sex chromatid assortment through both meiotic divisions, as might result from loss of sister chromatid cohesion prior to meiosis I. Similar NDJ frequencies and patterns have been reported for null alleles of ord and solo (Miyazaki and Orr-Weaver 1992; Bickel et al. 1997; Yan et al. 2010). Mutation of sunn also causes high frequencies of both sister chromatid and homologous chromosome NDJ of the autosomal second (Supporting Information, Table S1) and fourth chromosomes (data not shown).
To determine whether sunn mutations also cause sex chromosome NDJ in female meiosis, sunn hemizygous females were crossed with males carrying a dominant Bar (B) mutation on their X chromosomes. The regular progeny from this cross are B females and B+ males; the NDJ progeny are B+ females and B males. The results showed that 56.9% of progeny from sunn females resulted from X-X NDJ compared to 0.1% in sibling WT control females (Table 3). Analysis of centromere-linked markers revealed that 25.4% of the B+ females carried two maternal sister chromatids and the remainder carried two maternal homologous chromatids, indicating that both homologous and sister chromatids nondisjoin in sunn females (Table 3). Thus, like ord and solo, sunn is required for proper chromosome segregation in both meiotic divisions in both sexes (Miyazaki and Orr-Weaver 1992; Bickel et al. 1997; Yan et al. 2010; Yan and McKee 2013).
Table 3. X chromosome NDJ in sunn mutant females.
| Progeny types | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Maternal genotype | DJa B♀ | DJa B+♂ | NDJb y+ B+♀ | NDJ(Sis)b,c y B+♀ | NDJb y B♂ | nd | %NDJe | %sisf | P/Fg |
| sunnZ3-5839/Df | 219 | 198 | 146 | 19 | 124 | 706 | 58.1 | 23.0 | 11.6 |
| sunnZ3-4085/Df | 264 | 174 | 136 | 19 | 123 | 716 | 55.9 | 24.5 | 10.9 |
| sunnZ3-1956/Df | 268 | 149 | 123 | 21 | 128 | 689 | 56.6 | 29.2 | 11.9 |
| Total sunn | 751 | 521 | 405 | 59 | 375 | 2111 | 56.9 | 25.4 | — |
| Gamete freq. (%) | 35.6 | 24.7 | 19.2 | 2.8 | 17.8 | — | — | — | — |
| Df/+ | 1033 | 879 | 0 | 0 | 1 | 1913 | 0.1 | NAh | 79.7 |
Dp(1;1)scv1, y .y+/y females with the above third chromosome genotypes were crossed with two YSX.YL, In(1)EN, y B/Y males.
DJ: progeny from normal (disjunctional) eggs.
NDJ: progeny from NDJ eggs. The B+ daughters result from diplo-X eggs and the y B sons from nullo-X eggs.
NDJ(Sis): The y B+ daughters derive from diplo-X eggs carrying two sister chromatids lacking the y+ centromere marker, so represent sister chromatid NDJ only. The other two NDJ categories reflect a mix of sister chromatid and homolog NDJ.
n: total number of progeny counted.
%NDJ = 100 × 2 (NDJ)/ (n + NDJ).
%sis = % sister chromatid NDJ = 2 × (y B+♀)/(y B+♀ + y+ B+♀).
P/F (progeny/female) = average number of progeny a single female produces when crossed to two males. hNot applicable.
sunn mutations disrupt sister chromatid cohesion during male meiosis
For an in-depth study of the NDJ mechanism in sunn mutants, we surveyed chromosome and nuclear morphology throughout male meiosis by staining spermatocytes with DAPI to label chromosomes and with a α-tubulin antibody to label spindles. WT male meiosis I occurs synchronously in interconnected cysts of 16 primary spermatocytes derived from a single germline stem cell. Although axial elements and synaptonemal complexes are absent and the chromosomes are decondensed, Drosophila spermatocytes traverse a series of prophase I substages, labeled S1–S6, during which the chromosomes undergo distinctive changes, the most prominent of which are the separation of the four bivalents into distinct nuclear territories near the end of stage S2 and their condensation during stage S6 to form four compact and roughly spherical bivalents. The bivalents then congress during prometaphase I to form a tight metaphase I bundle and segregate reductionally at anaphase I to form daughter nuclei with equal staining intensity. After a brief prophase II, meiosis II univalents recondense, congress, and segregate equationally at anaphase II, yielding cysts of 64 spermatids with round nuclei of uniform size (Cenci et al. 1994).
Despite the high rates of meiosis I NDJ in genetic crosses, DAPI-stained sunn spermatocytes appeared remarkably normal during meiosis I (Figure S1A). As in WT, three large DAPI-stained territories, corresponding to the X-Y, 2nd and 3rd chromosome bivalents, were present throughout mid- and late-prophase I, indicating that homolog pairing and territory formation are intact in sunn mutants. Sometimes, a small fourth territory is observed in WT and sunn mutants, which corresponds to 4th chromosome bivalent. The territories condensed into compact “blobs” by prometaphase I, congressed normally and segregated at anaphase I to form daughter nuclei that in most cases appeared to contain roughly equal amounts of chromatin. However, prematurely separated sister chromatids were common during and after anaphase I, and meiosis II was chaotic in sunn mutants. We frequently observed single chromatids during prometaphase II and metaphase II, defective metaphase II congression, and unequal segregation at anaphase II (Figure S1B).
In light of the genetic evidence for high rates of both meiosis I and meiosis II NDJ, the absence of obvious abnormalities in homolog pairing and segregation during meiosis I in the DAPI analysis would have been surprising had the same behavior not been previously observed in ord and solo mutants (Miyazaki and Orr-Weaver 1992; Bickel et al. 1997; Yan et al. 2010). As in those cases, abnormalities in prometaphase I and metaphase I bivalent morphology consistent with premature loss of sister chromatid cohesion—loose chromatid packing, extruded single kinetochore regions and, occasionally, fully separated chromatids—were seen in acetic–orcein preparations of mutant chromosomes, presumably because of harsher fixation procedures than are normally used in DAPI staining (Figure S1C). These observations suggested that although homologs remain paired throughout meiosis I in sunn mutants, defects in sister chromatid cohesion might underlie the abnormal segregation patterns.
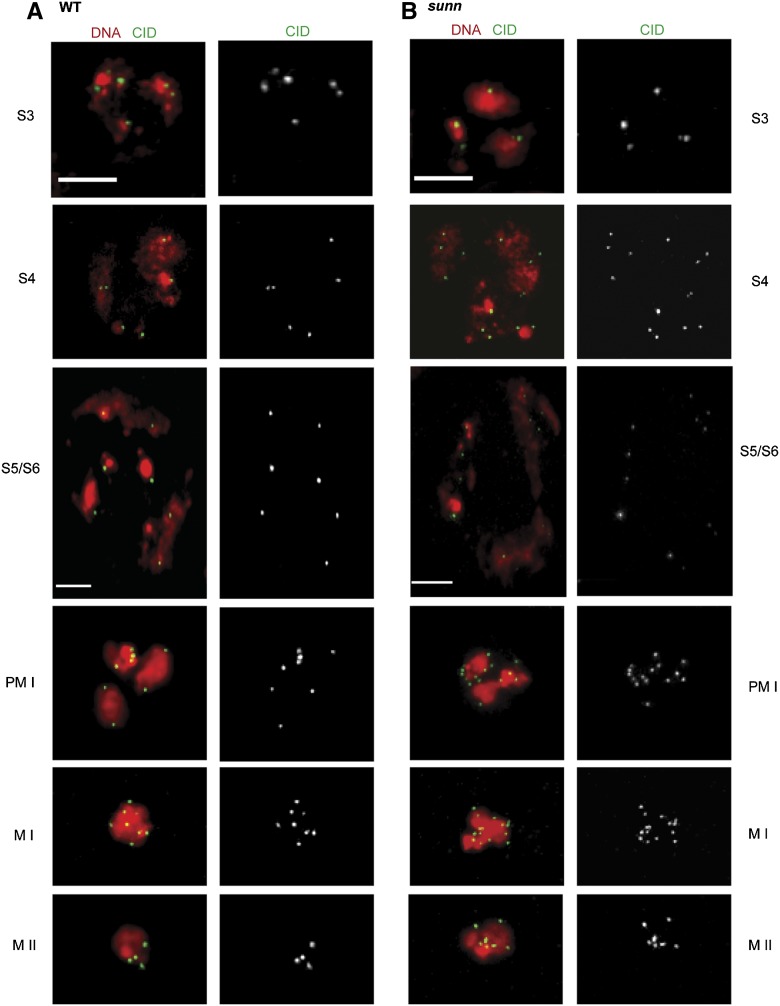
To examine sister chromatid cohesion directly, we immunostained sunn and WT spermatocytes with an antibody against the centromere marker CID (Blower and Karpen 2001) (Figure 1). The cohesion status of sister centromeres was determined by counting the number of discrete CID spots per nucleus (Table 4). When homologous centromeres are unpaired but sister centromere cohesion is intact, as is generally the case after stage S3, spermatocytes are expected to show maxima of 8 CID spots during meiosis I and 4 CID spots during meiosis II (Vazquez et al. 2002; Yan et al. 2010). As expected WT spermatocytes rarely exhibited >8 CID spots per nucleus during meiosis I (mean CID spot numbers of 6.1–7.2 from stage S4 through metaphase I) or >4 CID spots during meiosis II. sunn mutants did not differ from WT in early prophase I but began to diverge from WT by stage S4 when 34% of spermatocytes showed >8 CID spots. By late prophase I (stages S5 and S6) and throughout the division stages, >90% of sunn spermatocytes showed >8 spots, with a mean of ∼11–12 spots per spermatocyte (Table 4). A total of 14–16 CID spots were seen in a substantial fraction of sunn spermatocytes at prometaphase I and metaphase I, indicating that sunn function is required for cohesion of all eight Drosophila chromosomes. In meiosis II, 82% of sunn spermatocytes showed >4 CID spots. Thus the data show that sunn mutants begin losing centromere cohesion by stage S4 and exhibit extensive cohesion loss by stage S5, long before chromosomes begin orienting on the meiosis I spindle.
Figure 1.
Sister centromere cohesion is lost during prophase I in sunn spermatocytes. Immunostaining was performed using anti-CID antibody, which marks centromeres (green). DNA was stained with DAPI (red). PM I, prometaphase I; M I, metaphase I; M II, metaphase II. (A) In WT (Df/+) spermatocytes, spot numbers never exceeded eight during meiosis I or four during meiosis II. Representative images of S3, S4, S5/S6, PM I, M I, and M II show 6, 6, 7, 8, 8, and 4 CID spots, respectively. (B) In sunn (sunnZ3-5839/Df) spermatocytes, CID spot numbers exceeded 8 in most meiosis I spermatocytes from stage S4 onwards and exceeded 4 in most meiosis II spermatocytes. Representative images of S4, S5/S6, PM I, M I, and M II stages show 14, 12, 15, 14, and 7 spots, respectively. S3 bars apply to S4, PM I, M I, and M II. Bars, 5 μM. See Table 4 for quantification. Df = Df(3L)ED4470.
Table 4. Quantification of CID spots in sunn mutant spermatocytes.
| sunna | WTb | |||||
|---|---|---|---|---|---|---|
| ≤8 spots | >8 spots | Mean spot no. | ≤8 spots | >8 spots | Mean spot no. | |
| Meiosis I stages | ||||||
| S1 | 75 (100) | 0 | 3.96 | 71 (100) | 0 | 3.31 |
| S2 | 55 (100) | 0 | 3.82 | 60 (100) | 0 | 3.14 |
| S3 | 96 (98.97) | 1 (1.03) | 4.12 | 53 (100) | 0 | 3.96 |
| S4 | 60 (65.9) | 31 (34.1) | 7.62 | 58 (98.25) | 1 (1.75) | 6.08 |
| S5, S6 | 9 (6.2) | 138 (93.8) | 10.41 | 134 (99.3) | 1 (0.7) | 6.92 |
| PM I | 5 (3.9) | 122 (96.1) | 10.75 | 68 (100) | 0 | 6.89 |
| Metaphase I | 1 (9) | 10 (91) | 12.20 | 11 (100) | 0 | 7.23 |
| sunna | WTb | |||||
| ≤4 spots | >4 spots | Mean spot no. | ≤4 spots | >4 spots | Mean spot no. | |
| Meiosis II stage | ||||||
| Metaphase II | 8 (17.4) | 38 (82.6) | 6.22 | 79 (93) | 6 (7) | 3.80 |
Number in parentheses indicates percentage values calculated from the total number of nuclei scored at each spermatocyte stage. PM I, prometaphase I.
sunnZ3-5839/Df.
Df/+.
sunn mutants disrupt sister centromere mono-orientation
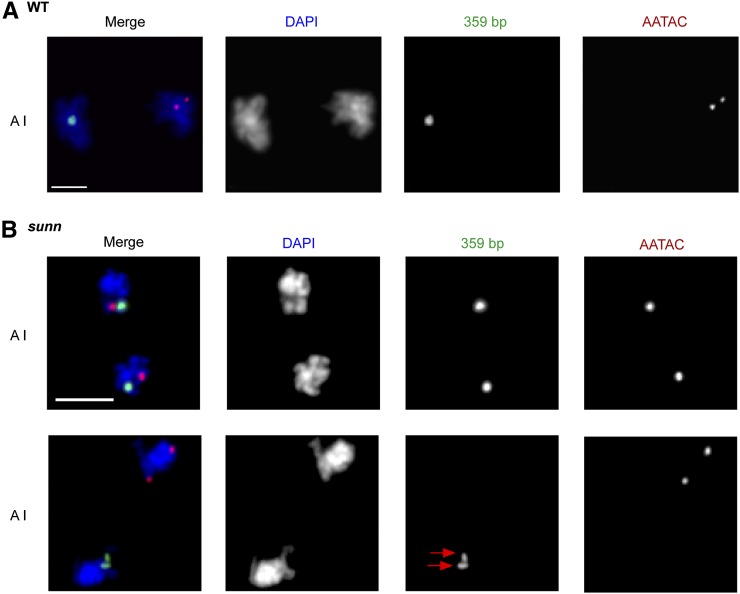
The absence of cohesion between most sister centromere pairs during prometaphase I might impair sister centromere mono-orientation and thereby disrupt reductional segregation. To track the segregation of the X and Y chromatids at anaphase I, we performed FISH using probes that bind to the 359 bp satellite repeats in the pericentromeric region of the X chromosome and to a block of AATAC satellite repeats in the long arm of the Y chromosome. Signals were scored both during anaphase I and prometaphase II/metaphase II. In WT, as expected, only reductional segregations (XX-YY) were observed, as shown by a complete absence of anaphase I poles or prometaphase II/metaphase II nuclei with both X and Y signals or with no signals (Figure 2A and Table 5). The segregation pattern in sunn mutants was completely different. Only 31% of the 553 sunn poles/nuclei scored exhibited the reductional segregation pattern. Most (60%) of the sunn poles/nuclei exhibited one X signal and one Y signal, reflecting an XY-XY equational segregation pattern (Figure 2B; Table 5). The remaining 9% of sunn nuclei exhibited either three signals (2 X and 1 Y or vice versa) or one signal (either X or Y, reflecting unbalanced XXY-Y or XYY-X segregations (Table 5). No completely unbalanced (XXYY-O) segregations were observed.
Figure 2.
X and Y chromatids segregate both equationally and reductionally at meiosis I in sunn spermatocytes. FISH was performed using probes for the 359-bp repeats on the X chromosome (green) and a block of AATAC repeats on the Y chromosome (red). DNA was stained with DAPI (blue). A I, anaphase I. (A) Meiosis I segregation is exclusively reductional in WT (Df/+) spermatocytes. Representative image of a reductional anaphase I segregation. The single 359 bp spot reflects maintenance of cohesion at and near the X centromere. Bar, 3 μM. (B) Both reductional and equational segregation occur at meiosis I in sunn (sunnZ3-5839/Df) spermatocytes. Representative images at anaphase I showing normal reductional segregation (bottom) and abnormal equational segregation (top). The two pericentromerically located 359 bp signal spots are separated in sunn mutants (red arrows), reflecting premature loss of X centromere cohesion. Bar, 5 μM. See Table 5 for quantification.
Table 5. Quantification of X-Y chromatid segregation patterns in sunn mutant spermatocytes.
| Chromatid pattern | sunna | WTb |
|---|---|---|
| Anaphase I | ||
| XX/YY | 47 (30%) | 62 (100%) |
| XY/XY | 95 (61%) | 0 |
| XXY/Y | 9 (6%) | 0 |
| XYY/X | 5 (3%) | 0 |
| Total | 156 (100%) | 62 (100%) |
| Prometaphase II or metaphase II | ||
| XX or YY | 78 (32%) | 121 (100%) |
| XY | 143 (59%) | 0 |
| XXY or Y | 11 (5%) | 0 |
| XYY or X | 9 (4%) | 0 |
| XXYY or O | 0 | 0 |
| Total | 241 (100%) | 121 (100%) |
X and Y chromatids were identified by FISH using probes for the 359 bp and AATAC loci as described in legend of Fig. 2 and Materials and Methods.
sunnZ3-5839/Df.
Df/+.
Absence of sister centromere cohesion in sunn mutants was also evident in the FISH data. Unlike in WT in which the two X signals were usually fused or overlapping (due to cohesion of pericentric regions), sister X signals in sunn nuclei were usually separate even when they co-segregated (Figure 2B). The presence of two separate AATAC (Y chromosome arm) spots in most WT spermatocytes reflects the fact that arm cohesion is lost early in male meiosis—by stage S3 (Vazquez et al. 2002; Yan et al. 2010). The complete absence of sister chromatid cohesion in sunn mutants was also apparent in many prometaphase II and metaphase II nuclei in which sister 359 bp or AATAC signals were present in separate DAPI-stained masses (Figure 3B). In conclusion, sunn mutations perturb the segregation pattern of the X and Y chromosomes at anaphase I by prematurely eliminating sister centromere cohesion, thereby disrupting sister centromere mono-orientation.
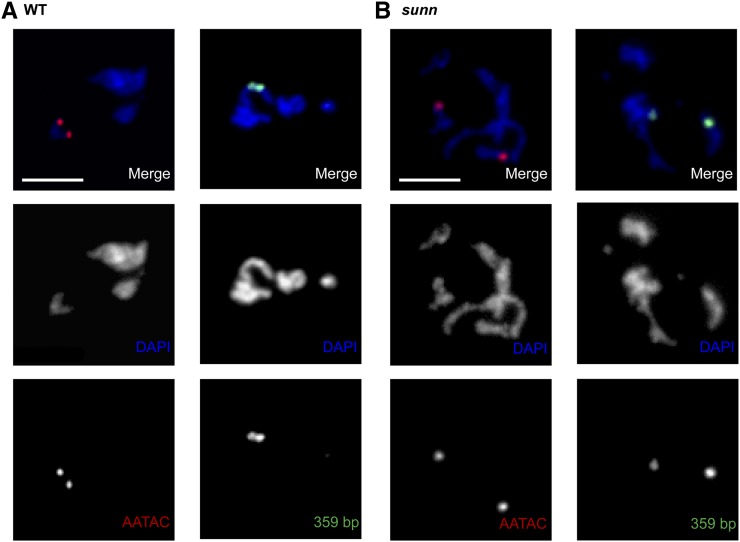
Figure 3.
Sister chromatid separation in prometaphase II spermatocytes of sunn mutants. X and Y chromatids were identified by the presence of 359 bp (green) and AATAC (red) probes, respectively. (A) Sister chromatid cohesion is maintained in WT (Df/+). In two prometaphase II spermatocytes bearing a Y chromosome (left) or an X chromosome (right), both AATAC signals and both 359 bp signals are situated in the same chromosome territory. (B) Premature sister chromatid separation in sunn (sunnZ3-5839/Df) spermatocytes. In two prometaphase II spermatocytes bearing two separated Y chromatids (left) or two separated X chromatids (right), the two AATAC signals and the two 359 bp signals are located in completely separate chromosome territories. See Table 5 for quantification. Bars, 5 μM.
sunn is not required for mitotic segregation or for arm cohesion
In WT male meiosis, arm cohesion is established in meiotic S phase but, unlike centromere cohesion, it is lost during midprophase I (late S2/S3). Consequently, when a single chromosome arm site is labeled by FISH or the GFP–LacI/lacO assay, only one spot is observed during early prophase I but two separate (sister) spots are generally observed at later stages of meiosis I (Vazquez et al. 2002; Yan et al. 2010). Thus the effect of a mutation on arm cohesion can be assayed by counting spots (one vs. two) during early prophase I. We examined arm cohesion in sunn spermatocytes by labeling a heterozygous lacO array inserted in the euchromatin of chromosome 2 with GFP–LacI expressed under control of the hsp83 promoter. In both WT and sunn mutants, a single spot was observed in the great majority of stage S1 and S2 spermatocytes, indicating that sunn is not required for arm cohesion in early prophase I (Figure S2). Similar results were reported for solo mutants (Yan et al. 2010).
In later stages of meiosis I (after arm cohesion is lost), the number of spots in single-locus arm labeling experiments provides a reliable measure of chromatid copy number, useful to diagnose aneuploidy due to mitotic NDJ. The Y chromatids are particularly useful for such studies because they are normally present in two copies but can be absent altogether (XO) or present in four copies (XYY) without blocking spermatocyte development. Thus, in the FISH analysis reported above (Figure 2 and Table 5), mitotic NDJ of the Y chromosome in sunn spermatogonia would be expected to generate XYY or XO spermatocytes exhibiting four or no AATAC signals, respectively, by late prophase I. However, 100% of the anaphase I sunn spermatocytes (N = 156) reported in Table 5 showed two AATAC spots (sometimes at opposite poles, sometimes at the same pole) as did 100% of prometaphase I and metaphase I sunn spermatocytes (N = 68, data not shown). These results strongly suggest that there is no significant mitotic NDJ in sunn mutants.
sunn mutations disrupt centromere clustering, pairing, and cohesion in female meiosis
To determine if centromere cohesion is also lost prematurely in female meiosis, CID spot numbers were scored in pro-oocytes and oocytes from sunnZ3-5839/Df females and WT sibling controls. Analysis of CID spot numbers in sunn oocytes was also of interest because of recent evidence for clustering of centromeres throughout prophase I in WT female meiosis and for the dependence of that clustering on ord and solo as well as on the genes encoding SC components, c(3)G and cona and kinetochore components, cenp-c and cal-1 (Khetani and Bickel 2007; Takeo et al. 2011; Tanneti et al. 2011; Unhavaithaya and Orr-Weaver 2013; Yan and McKee 2013).
In female Drosophila, meiosis occurs in ovaries, which contain 10–30 ovarioles, each consisting of linear arrays of oocytes of increasing developmental age from stem cells to metaphase I arrested oocytes. Meiosis initiates in the germarium, the anterior-most compartment of each ovariole. Region 1, at the anterior end of the germarium, contains stem cells and pre-meiotic cysts undergoing mitotic amplification. Regions 2A, 2B, and 3 of the germarium contain 16-cell cysts in the zygotene (region 2A) or pachytene (regions 2A, 2B, and 3) stages of meiosis. Meiosis initiates and SCs begin forming in up to four pro-oocytes in each cyst in region 2A but by region 3, only a single oocyte retains SC. The other 15 germ cells in each cyst develop as polyploid nurse cells that support the oocyte during its development. The maturing cysts leave the germarium and continue developing in the vitellarium. SC is disassembled in vitellarial stages 5–7, marking the end of pachytene (McKim et al. 2002; Lake and Hawley 2012). The oocyte subsequently enters an arrested late prophase I state termed the karyosome, in which the chromosomes are highly compact. Nuclear envelope breakdown in stage 12 is followed by prometaphase I and metaphase I in stages 13 and 14.
In agreement with the reports cited above, we found 1–3 CID foci in nearly all (∼90–97%) nuclei in WT pro-oocytes/oocytes in germarial regions 2A, 2B, and 3 nuclei (average of 2.1–2.6 CID foci/nucleus), indicative of pairing and clustering of centromeres. Clustering was also present in stage 2 of WT vitellaria (Figure 4A, Table 6). However, in sunn mutants, 1–3 CID foci were observed in only 11.5, 0, 5, and 0% of nuclei in regions 2A, 2B, 3, and stage 2, respectively, indicating an absence of centromere clustering during prophase I. Most sunn pro-oocytes in regions 2A (89%) and 2B (85%) exhibited 4–8 CID spots, with means of 5.1 and 7 spots per nucleus, respectively, indicating that pairing of homologous centromeres was also compromised, somewhat more completely in region 2B than in region 2A.
Figure 4.
Centromeric clustering is disrupted in sunn mutant females. Whole-mount ovaries were immunostained with anti-CID and anti-C(3)G, which serve as markers for centromeres and SCs, respectively (Blower and Karpen 2001; Page and Hawley 2001). (A) Centromeres are paired and clustered in WT (Df /TM3) oocytes. Pro-oocytes/oocytes showed one to three CID foci throughout the germarium in regions 2A, 2B, and 3 and in the vitellarium at stage 2. Representative oocyte/pro-oocyte images all show 2 large CID foci in region 2A, region 2B, region 3, and stage 2. (B) Loss of centromere clustering, pairing, and cohesion in sunn (sunnZ3-5839/Df) oocytes. Pro-oocytes/oocytes averaged >4 CID spots throughout pachytene. Representative images show 5, 8, 10, and 13 CID foci in region 2A, region 2B, region 3, and stage 2, respectively. Bars, 5 μM. See Table 6 for quantification.
Table 6. Quantification of CID spots in WT and sunn mutant oocytes/pro-oocytes.
| Oocyte stages | ||||
|---|---|---|---|---|
| Genotype | Region 2A | Region 2B | Region 3 | Stage 2 |
| WTa | 2.14 ± 0.91 (N = 21) | 2.19 ± 0.79 (N = 36) | 2.6 ± 1.07 (N = 10) | 3 ± 1.12 (N = 12) |
| <4 CID (%) | 95.2 | 97.2 | 90 | 75 |
| ≥4, ≤ 8 CID (%) | 4.8 | 2.8 | 10 | 25 |
| >8 CID (%) | 0 | 0 | 0 | 0 |
| sunnb | 5.15 ± 1.43 (N = 26) | 6.96 ± 1.96 (N = 32) | 8.5 ± 2.8 (N = 20) | 8.8 ± 2.6 (N = 9) |
| <4 CID (%) | 11.5 | 0 | 5 | 0 |
| ≥4, ≤ 8 CID (%) | 88.5 | 84.4 | 45 | 44.4 |
| >8 CID (%) | 0 | 15.6 | 50 | 55.6 |
Table entries in the “WT” and “sunn” lines are mean CID spots per oocyte with standard deviations. N indicates the total number of oocytes counted in the indicated region of the ovariole.
Df/TM3.
sunnZ3-5839/Df.
Cohesion was intact in region 2A since no pro-oocytes with >8 CID spots were observed. However, more than 8 CID spots were observed in 16% of region 2B pro-oocytes, 50% of region 3 oocytes, and 56% of stage 2 oocytes in sunn mutants (Figure 4B, Table 6). Thus, cohesion begins deteriorating by region 2B and is extensively compromised by region 3 in sunn mutants. As in male meiosis, this cohesion loss occurs long before centromeres must orient on the meiosis I spindle and thus provides a likely explanation for the high levels of meiosis I NDJ.
SUNN is a novel protein produced from the CG32088 locus
Using deficiency complementation and candidate gene sequence analysis, sunn was mapped to the CG32088 locus in region 68D3 of chromosome arm 3L. We note that a gene named mei(3)M20 that exhibited mutant phenotypes similar to those of sunn was previously reported and mapped to the 68C8–11;69A4–5 interval (Hirai et al. 2004). Complementation analysis will be required to determine whether sunn and mei(3)M20 are allelic. Genomic DNA sequencing of CG32088 exons from the three alleles of sunn revealed single mutations in each line: a nonsense mutation predicted to truncate the protein 132 amino acids from the C terminus (Z3-1956); a missense mutation predicted to substitute arginine for a conserved glycine (G170) (Z3-4085); and a 8-bp deletion (Z3-5839) which creates a frameshift that leads to a predicted in-frame stop codon near the middle of the coding sequence (Figure 5). As no full-length cDNAs for sunn were available, a cDNA was obtained by reverse transcription and PCR from ovary RNA using primers designed on the basis of genomic sequence. The resulting sunn cDNA consists of 10 exons and contains a predicted coding sequence of 2856 bp corresponding to a protein 952 amino acids in length (Figure 5). Rapid amplification of cDNA ends (RACE) revealed short 5′ and 3′ UTRs, 69 bp and 75 bp in length, respectively, that contained no potential alternative start or stop codons.
Figure 5.
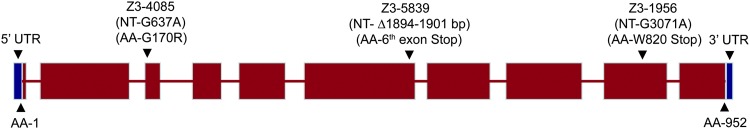
Structure of sunn gene. Gene structure of sunn and location and identity of sunn mutations. Sequencing of ovary cDNAs showed that there are 10 exons in sunn. The red boxes show the predicted coding sequence of sunn and the blue boxes show the UTRs. The length of the predicted coding sequence of sunn is 2856 bp and the lengths of the 5′ and 3′ UTRs are 69 bp and 75 bp, respectively. The locations and identities of the sequenced sunn mutations discussed in the text are shown above. NT denotes genomic nucleotide position (introns included) from the start of the translation unit (position 1 is the first nucleotide of the predicted initiator AUG) and AA depicts the respective predicted amino acid numbers.
To verify that CG32088 corresponds to sunn, we cloned the full-length cDNA-derived coding sequence of sunn (without its 5′ and 3′ UTRs) into an upstream activator sequence (UAS) vector in frame with C-terminal Venus, (enhanced yellow fluorescent protein, eYFP). Transgenic insertions of UAS–SUNN::Venus were generated and found to complement the meiotic NDJ phenotypes of sunn mutants in both sexes when expressed under control of the germline specific driver nos-GAL4::VP16 (Table S2 and Table S3). In addition to verifying the identity of sunn and CG32088, these results show that SUNN::Venus functions similarly to wild-type SUNN protein and that the sunn cDNA sequence used for the construct likely represents the true sunn coding region.
SUNN exhibits structural similarity to the cohesin protein SA
Predicted secondary structure for SUNN indicated a primarily α-helical protein (Figure S3). To identify homologs of SUNN, the complete amino acid sequence of SUNN was used to search the protein sequence database using the BLASTp tool offered by FlyBase (http://flybase.org/blast/). We found single SUNN homologs in all of the sequenced species of Drosophila (see File S1 for details). No homologs of SUNN were found in other eukaryotes and no conserved domains were identified by searching a conserved domain database at National Center for Biotechnology Information (NCBI). However, structure-based searches, described in detail in File S1, proved more informative. In particular, the fold-recognition/threading programs MUSTER and I-TASSER revealed statistically significant similarity of SUNN to multiple templates, all of which belong to the HEAT repeat protein family (Table 7 and Table S4) (Wu and Zhang 2008; Roy et al. 2010). HEAT repeats are conserved domains that form all-α superhelices and are involved in protein–protein interactions. They are particularly abundant in chromosomal proteins involved in cohesion and condensation, including the cohesin cofactors Nipped-B and Pds5 and the condensin subunits Cap-G and Cap-D2. The cohesin subunit SA also exhibits weak similarity to HEAT repeats (Neuwald and Hirano 2000; Andrade et al. 2001; Nasmyth and Haering 2009; Hirano 2012).
Table 7. Threading/fold recognition (Z scores) results of hits generated by SUNN, stromalin (SA), and some Drosophila chromosomal HEAT repeat proteins using MUSTER.
| PDB ID | Protein | SUNN | SA | Pds5 | Nipped-B | CapG | CapD2 |
|---|---|---|---|---|---|---|---|
| 1wa5C | Exportin CSE1P | 8.333 | 8.711 | 10.255 | — | 10.343 | 10.313 |
| 3m1iC | Exportin-1 | 8.116 | 8.839 | 11.174 | 7.733 | 9.980 | 11.181 |
| 1qgkA | Importin-β | 7.995 | 10.060 | 10.933 | 7.637 | 11.106 | 11.310 |
| 3ea5B | Importin-β1 | 7.969 | 9.981 | 11.422 | — | 11.083 | 11.386 |
| 4fgvA | Exportin 1 | 7.918 | — | 10.350 | 7.416 | 9.901 | 10.350 |
| 2x1gF | Importin-13 | 7.914 | 9.278 | 10.838 | — | 9.904 | 10.727 |
| 1u6gC | Cand1/TIP120 | — | 8.912 | 12.815 | 7.945 | 11.406 | 12.807 |
| 3icqT | Exportin-T | 7.480 | 8.494 | — | — | — | — |
| 2x19B | Importin-13 | 7.389 | 8.596 | 10.140 | — | — | 10.092 |
| 3a6pA | Exportin-5 | 7.386 | — | — | — | — | — |
| 3gjxA | Exportin-1 | 7.156 | — | — | — | 9.648 | — |
| 1b3uA | PP2A | — | 8.258 | — | — | — | — |
| 3nowA | UNC45 | — | 8.221 | — | — | — | — |
| 3w3tA | Importin-β3 | — | — | 12.645 | 8.222 | 12.213 | 12.645 |
| 1qbkB | Karyopherin-β2 | — | — | — | 7.759 | — | — |
| 4c0oA | Transportin 3 | — | — | 11.585 | 8.295 | 10.977 | 11.596 |
| 1vw1A | TcdA1 | — | — | — | 8.405 | — | — |
| 4acqA | α-2-Macroglobulin | — | — | — | 7.984 | — | — |
| 4jspB | mTOR | — | — | — | 7.580 | — | — |
Bold Z scores represent the top six PDB templates matched by the proteins tested. Above depicted proteins were derived from the following organisms: 1wa5C: Exportin CSE1P (Saccharomyces cerevisiae). 3m1iC: Exportin-1 (S. cerevisiae). 1qgkA: Importin-β (human). 3ea5B: Importin-β1 subunit (S. cerevisiae). 4fgvA: Exportin 1 (Chaetomium thermophilum). 2x1gF: Importin-13 (Drosophila melanogaster). 1u6gC: Cand1/TIP120 (human). 3icqT: Exportin-T (S. pombe). 2x19B: Importin-13 (human). 3a6pA: Exportin-5 (human). 3gjxA: exportin-1 (mouse). 1b3uA: PP2A (human). 3nowA: UNC45 (D. melanogaster). 3w3tA: importin subunit β3 (S. cerevisiae) (Kap121p). 1qbkB: karyopherin-β2 (human). 4c0oA: transportin 3 (human). 1vw1A: TcdA1 (Photorhabdus luminescens). 4acqA: α-2-macroglobulin (human). 4jspB: (human serine/threonine-protein kinase mTOR).
Detailed comparisons of the MUSTER and I-TASSER analyses revealed a stronger similarity of SUNN to SA than to the other Drosophila chromosomal HEAT repeat proteins. In the MUSTER analysis 5 of the top 6 matches for SA were also among the top 6 matches for SUNN (Table 7). Similarly, 8 of the top 10 matches for SUNN and SA in the I-TASSER analysis overlapped, and 6 of these common templates overlapped with the top common templates hit by SUNN and SA in MUSTER (Table S4). Although some of the top-matching templates for the other Drosophila chromosomal HEAT repeat proteins, Nipped-B, Pds5, Cap-G, and Cap-D2, overlapped with the templates matched by SUNN, the overlap was much less extensive than for SA. For example, in the MUSTER analysis, the highest-scoring and second-highest scoring templates for Cap-G, Cap-D2, Nipped-B, and Pds5 were not among the 10 best matches for SUNN (Table 7). Taken together, these results suggest that SUNN is a distant member of the HEAT-repeat family and exhibits stronger structural similarity to the cohesin subunit SA than to other Drosophila chromosomal HEAT-repeat proteins. Structure-based pairwise alignments of SUNN and SA with the most similar template protein (1qgkA) from the MUSTER search are shown in Figure S4 and a graphical alignment of the α-helices is shown in Figure S5.
SUNN co-localizes with CID during meiosis
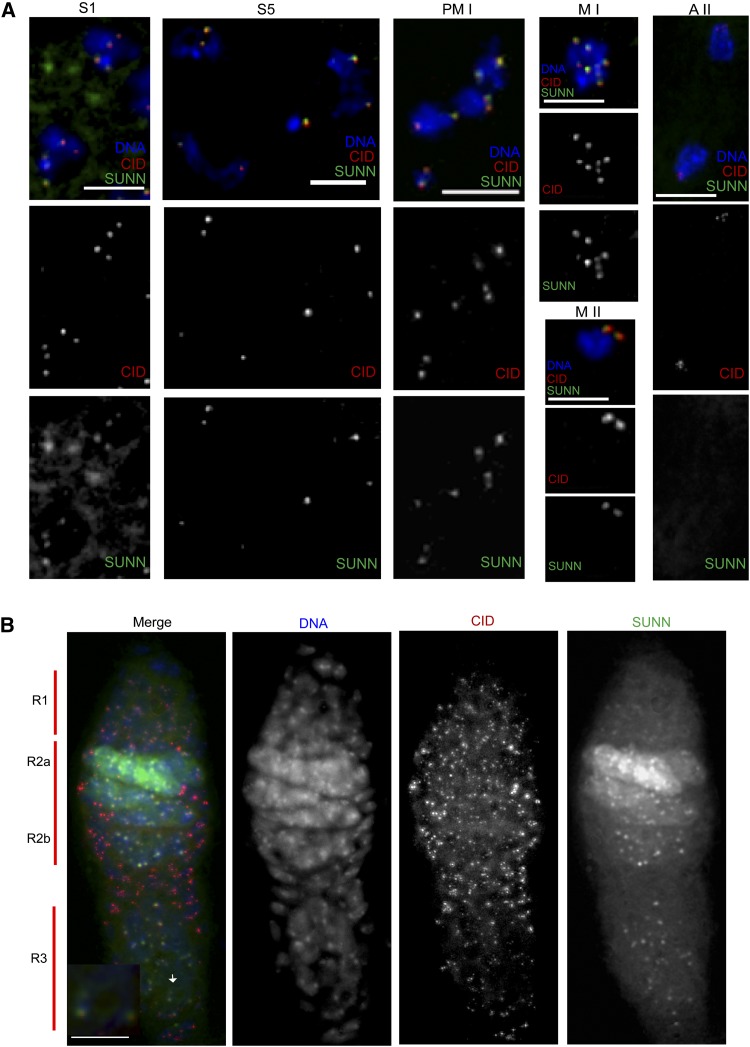
Immunostaining of spermatocytes expressing SUNN::Venus with anti-CID antibody revealed bright Venus foci that co-localized with CID spots (Figure 6A and Figure S6B). These centromeric SUNN::Venus foci were present at all stages of meiosis through metaphase II but were absent at later stages. The SUNN::Venus foci sometimes showed small extensions, suggesting that SUNN localizes to heterochromatic domains that extend beyond the centromeres. However, no localization of SUNN::Venus to chromosome arms in male germ cells was detected at any stage. SUNN::Venus also co-localized with CID in premeiotic 8-cell cysts, but these centromeric signals were weaker than in spermatocytes. In addition, SUNN::Venus formed large foci and bright smears outside the DNA in premeiotic 8-cell cysts (Figure S6A). These non-chromosomal staining patterns were still present but greatly attenuated in the earliest 16-cell cysts, and were not seen in later stages (Figure 6A and Figure S6). The significance of this non-chromosomal staining is unknown; the possibility that it is an artifact of ectopic expression cannot be excluded. We conclude that SUNN localizes to centromeric regions prior to the beginning of male meiosis and persists at centromeres through metaphase II but is removed by anaphase II.
Figure 6.
Co-localization of SUNN::Venus with CID. (A) SUNN and CID co-localize in male meiosis. Spermatocytes from sunnZ3-5839/Df, nos-GAL4::VP16 males carrying UASp-SUNN::Venus were immunostained with anti-CID antibody. SUNN::Venus forms bright spots, which co-localize with CID spots. At S1 stage, SUNN::Venus also shows diffuse signals and large foci, which do not localize with CID and are present predominantly on the nuclear membrane and outside the nucleus in the cytoplasm. In representative images, SUNN::Venus and CID both form three to four CID spots, eight spots and four spots at S1, PM I, and M II, respectively. SUNN::Venus signals are absent at A II (anaphase II). (B) Co-localization of SUNN::Venus and CID in female germ cells. Immunostaining was performed using anti-CID antibody on whole-mount ovaries from UASp-SUNN::Venus; sunnZ3-5839/Df, nos-GAL4::VP16 females. SUNN::Venus foci were observed in germ cells in all regions of the germarium including region 1, and they co-localized with CID spots. SUNN::Venus expression was absent from the follicle cells due to germ cell-specific expression directed by the nos-GAL4 driver. Arrow shows enlarged germ cell used in the inset. Bars, 5 μM.
To evaluate the localization pattern of SUNN in female meiosis, we immunostained ovaries expressing SUNN::Venus with anti-CID. Discrete SUNN::Venus foci that co-localized with CID foci were observed in the germ cells, but not the somatic follicle cells, in the proximal half of region 1, where four- and eight-cell premeiotic gonial cysts reside, throughout regions 2A, 2B, and 3 of the germarium (Figure 6B) and in the vitellarium at least up to stage 3 (not shown). In addition to the centromeric foci, SUNN::Venus also exhibited diffuse localization in female germ cells, particularly in regions 2A and 2B (Figure 6B). The nature of this non-centromeric localization could not be ascertained from the whole-mount squash preparations and remains under investigation. SUNN’s localization to the centromeres in both sexes could explain the phenotypes of NDJ, centromeric cohesion, and centromeric clustering defects associated with sunn mutants
Mutual co-dependence of SUNN, SOLO, and SMC1 centromere foci
The cohesion proteins SMC1, ORD, and SOLO also localize to centromeric regions in spermatocytes and SMC1 centromeric foci have been shown to depend on both ord and solo (Balicky et al. 2002; Thomas et al. 2005; Khetani and Bickel 2007; Yan et al. 2010). To test for dependence of SMC1 centromeric foci on sunn, we expressed Venus::SMC1 using the nos-GAL4::VP16 driver in both WT and sunn mutant backgrounds. Although bright Venus::SMC1 foci were observed throughout WT meiosis, no Venus::SMC1 foci were seen at any stage of meiosis in sunn spermatocytes (Figure 7A). In WT females, SMC1 forms bright foci at the centromeres and it localizes to the chromosome arms in oocytes/pro-oocytes. However, in sunn mutants, SMC1 is absent from the centromeres but is still present (although weakly) on the chromosome arms (Figure 7B). Thus stable centromere localization of SMC1 requires wild-type function of sunn.
Figure 7.
Interactions among SUNN, SMC1, and SOLO. (A) SMC1 foci in spermatocyte nuclei require sunn function. Venus::SMC1 expressed under the control of nos-GAL4::VP16 formed DNA-associated foci in WT but not sunn (sunnZ3-5839/Df) spermatocytes. (B) SMC1 localization to centromeres in pro-oocytes/oocytes requires sunn. Whole-mount ovaries where immunostained with anti-SMC1 and anti-CID antibodies. In WT, SMC1 forms centromeric foci (white arrows) and localizes to chromosome arms, whereas in sunn (sunnZ3-5839/Df), SMC1 is lost from the centromeres (red arrows) observed as gaps in C(3)G staining but is weakly present on chromosome arms. R2b, region 2b; R3, region 3. (C) Centromeric SUNN::Venus foci in spermatocyte nuclei require solo function. Spermatocytes expressing SUNN::Venus under control of nos-GAL4::VP16 were immunostained with anti-CID antibody. DNA-associated SUNN::Venus foci co-localized with CID in WT spermatogonia (eight-cell stage) and spermatocytes (S1 and S4 stages) but were absent at the same stages in solo (Df(2L)A267/soloZ2-0198) spermatogonia and spermatocytes. Bars, 5 μM.
To investigate whether SUNN localization depends on solo, SUNN::Venus was expressed using the nos-GAL4::VP16 driver in both WT and solo mutant backgrounds. Although SUNN::Venus foci that co-localized with CID were readily observed throughout meiosis in WT spermatocytes, no SUNN::Venus foci were observed at any stage of meiosis in solo spermatocytes. Premeiotic SUNN–Venus foci were also absent in solo spermatocytes (Figure 7C). We conclude that localization of SUNN to centromere regions requires wild-type solo function.
To determine whether SOLO localization requires sunn function, we expressed Venus::SOLO (Yan et al. 2010) in WT and sunn mutant backgrounds in male meiosis. Venus::SOLO foci are visible on chromosomes in WT males throughout meiosis, but no Venus::SOLO foci were detected at any stage of meiosis in sunn mutants (Figure S7). Thus, SOLO and SUNN foci are reciprocally codependent. This pattern is consistent with SUNN and SOLO participating in the same cohesion complex.
Discussion
SUNN is a Drosophila-specific meiotic cohesion protein
Several components and/or regulators of the meiotic cohesion machinery in Drosophila have been identified, but critical questions about meiotic cohesion remain unanswered. Chief among these are the composition of meiotic cohesin(s) and the role(s) of cohesion factors in pairing, synapsis, and recombination. The core cohesin subunits SMC1 and SMC3 are required for SC formation and have been implicated in centromere cohesion (Khetani and Bickel 2007; Yan et al. 2010; Tanneti et al. 2011; Yan and McKee 2013). The non-SMC components of meiotic cohesin remain uncharacterized. No meiotic role of SA has been reported, and although RAD21 has recently been shown to localize to SCs and to play a role in SC stability, it appears to be dispensable for meiotic cohesion and for proper chromosome segregation at both meiotic divisions (Urban et al. 2014). The Drosophila genome encodes meiosis-specific paralogs of both SA and RAD21, SNM, and C(2)M, respectively, but neither protein is required for meiotic cohesion in either sex (Manheim and McKim 2003; Heidmann et al. 2004; Thomas et al. 2005).
Heretofore, the best-characterized meiotic cohesion factors are two meiosis-specific proteins, ORD and SOLO, not found outside of the genus Drosophila but required for all aspects of meiotic cohesion. This report adds a third protein, SUNN, to this group of Drosophila-specific meiotic cohesion factors. Like fluorescently tagged versions of ORD and SOLO, SUNN::Venus localizes to centromeres of pre-meiotic gonial chromosomes (most clearly in eight-cell cysts) and meiotic chromosomes in both sexes. The disappearance of SUNN from spermatocyte centromeres at anaphase II is similar to the timing of ORD and SOLO removal and coincident with the disappearance of SMC1. Like solo and ord, mutations in sunn abolish SMC1 centromere foci and disrupt centromere cohesion during prophase I, well in advance of prometaphase I when sister centromeres would normally mono-orient. The result is high frequencies of meiosis I and meiosis II NDJ in both sexes, as previously described for ord and solo mutants (Miyazaki and Orr-Weaver 1992; Bickel et al. 1997; Yan et al. 2010; Yan and McKee 2013). The similarities among the phenotypes and localization patterns are striking and suggest that ORD, SOLO, and SUNN play closely related roles in meiotic cohesion and cohesion-related processes.
SUNN is required for centromere clustering and pairing
Both homologous and non-homologous centromere pairing have been described during meiosis in yeast, plants, and Drosophila (Stewart and Dawson 2008). In Drosophila female meiosis, centromeres typically aggregate into one to three clusters throughout prophase I from zygotene until prometaphase I. This phenomenon is meiosis specific since it is not observed in pre-meiotic gonia or in nurse cells (Takeo et al. 2011). Centromere clustering requires the SC proteins C(3)G and CONA, the cohesion proteins ORD and SOLO (Khetani and Bickel 2007; Takeo et al. 2011; Tanneti et al. 2011; Yan and McKee 2013), and the centromere proteins CENP-C and CAL-1 (Unhavaithaya and Orr-Weaver 2013). Our data show that SUNN is also required for centromere clustering. The frequency of sunn oocytes with fewer than four CID spots (the hallmark of clustering) was <12% in region 2A and ≤5% in all later stages. Moreover, the great majority of oocytes in all stages exhibited more than four CID spots, indicating a substantial disruption of homologous centromere pairing as well (Table 6). It remains to be determined whether pairing and clustering of centromeres are somehow consequences of centromere cohesion or whether they reflect separate functions of SUNN.
SUNN is required for centromere cohesion in both male and female meiosis
Sister centromeres normally remain tightly cohesive throughout meiosis until anaphase II, when the release of centromere cohesion triggers sister chromatid segregation. Sister centromere cohesion underlies not only the proper bipolar orientation of sister kinetochores during meiosis II but also their mono-orientation during meiosis I. Mutations in ord and solo were previously shown to disrupt centromere cohesion prior to prometaphase I in both sexes (Balicky et al. 2002; Bickel et al. 2002; Yan et al. 2010; Takeo et al. 2011; Tanneti et al. 2011; Yan and McKee 2013). In this article, we have shown that sunn mutations have similar effects. In spermatocytes, FISH analysis revealed substantial loss of cohesion at the pericentromeric X chromosome 359 bp repeat locus by prometaphase l, consistent with similar observations in ord and solo mutants (Balicky et al. 2002; Yan et al. 2010). CID spot counts showed that although centromere cohesion remains intact throughout stages S1–S3 of prophase I in sunn mutants, it begins deteriorating by stage S4 and is largely absent by stage S5–S6. Results of CID spot counts for solo mutants were similar except that no cohesion loss was detected until stage S5.
Results of CID spot counts in sunn oocytes were similar. No region 2A prooocytes with more than eight CID spots were seen, indicating that sister centromere cohesion remained intact in early pachytene. However, by region 2B, 16% of prooocytes exhibited more than eight CID spots and by region 3 and stage 2, at least half of oocytes did. Again, the progressive loss of cohesion during pachytene broadly parallels results of similar studies in ord and solo mutants. However, no pro-oocytes/oocytes with more than eight spots were observed in any region of the germarium in those mutants. Oocytes with more than eight spots were present in vitellarial stages 5–7 in ord and solo mutants (Takeo et al. 2011; Yan and McKee 2013). Thus cohesion is compromised at an earlier stage of pachytene in sunn oocytes than in ord or solo oocytes, paralleling the difference in timing of cohesion loss between sunn and solo spermatocytes.
Three conclusions seem warranted. First, SUNN, SOLO, and ORD work together during prophase I to maintain centromere cohesion in both sexes. In the absence of any of the three proteins, centromere cohesion is completely lost by the onset of prometaphase I. In light of the shared phenotype of loss of centromeric SMC1 foci in ord, solo, and sunn mutants, it seems likely that this cohesion pathway is mediated by a cohesin complex, although this inference remains to be verified. Second, since centromere cohesion is intact during early prophase in both spermatocytes and oocytes in all three groups of mutants, there must be at least one additional centromere cohesion mechanism that does not require ORD, SOLO, or SUNN. This mechanism may be independent of cohesin as well although the possibility that below-detection levels of SMC cohesins remain at centromeres in the absence of these proteins cannot be excluded. The nature of this alternative mechanism (or mechanisms) remains to be elucidated. Third, since there is an earlier onset of centromeric cohesion loss in sunn mutants than in ord or solo mutants in both male and female meiosis, SUNN may play a minor role in SOLO/ORD-independent early-prophase cohesion. Although we cannot completely exclude the possibility that the earlier onset of cohesion loss in sunn mutants resulted from a background genotype effect, we think this explanation is unlikely as the sunn and solo alleles used in these studies were derived on the same strain background.
SUNN is required for sister centromere mono-orientation
Reductional segregation during meiosis I requires sister centromeres to mono-orient so that homologous centromeres can reliably biorient. Mono-orientation requires that sister centromeres form a functionally single kinetochore by adopting a side-by-side configuration instead of a back-to-back configuration, which is characteristic of meiosis II and mitosis (Hauf and Watanabe 2004; Yokobayashi and Watanabe 2005). How sister centromeres achieve this unique orientation is poorly understood but genetic studies in several model eukaryotes have pinpointed sister centromere cohesion as a necessary prerequisite for mono-orientation. Mutation of cohesin genes including smc3, rec8, and scc3/sa in budding yeast, fission yeast, Arabidopsis, and Caenorhabditis elegans have been found to disrupt mono-orientation and cause chaotic and/or equational meiosis I segregation (Klein et al. 1999; Watanabe and Nurse 1999; Pasierbek et al. 2001; Wang et al. 2003; Chelysheva et al. 2005; Golubovskaya et al. 2006; Severson et al. 2009).
In Drosophila, no cohesins have been shown directly to be required for mono-orientation, but the detailed FISH analyses of meiosis I segregation reported previously for solo mutants (Yan et al. 2010) and herein for sunn mutants show that the products of these essential centromere cohesion genes are also essential for mono-orientation, at least in male meiosis. The simplest interpretation is that the mono-orientation defect is a consequence of the cohesion defect, although the possibility that SUNN or SOLO has an independent role in mono-orientation cannot be excluded. The fact that SMC1 centromere foci are absent in sunn mutants, as in ord and solo mutants, is consistent with the idea that mono-orientation in Drosophila requires cohesin (again presumably derivative of its role in cohesion), but direct proof of this inference is lacking as yet. The data available so far also do not address the question of whether the known cohesion factors are sufficient for mono-orientation. It would not be surprising if additional factors were needed since the same proteins mediate cohesion during both meiosis I and II but mono-orientation is restricted to meiosis I. Specific mono-orientation factors have been identified in both S. cerevisiae and S. pombe but no such factors have been identified as yet in higher eukaryotes (Toth et al. 2000; Yokobayashi and Watanabe 2005).
Is SUNN also required for mono-orientation during meiosis I in female Drosophila? Since we did not conduct cytological analysis of meiosis I segregation in females, our data do not provide direct evidence on this point. In principle, the observed combination of homolog and sister chromatid NDJ in the cross experiments could be explained without invoking any mono-orientation defects. The homolog NDJ products could result solely from dyad–dyad NDJ due to a failure of arm cohesion during meiosis I, and the sister chromatid NDJ products could result solely from meiosis II NDJ. However, we think this explanation is unlikely, mainly because of our data showing that centromere cohesion is extensively compromised during prophase I. It is difficult to see how prematurely separated sister centromeres could mono-orient on the meiosis I spindle. Although we cannot rule out the possibility that the dissociated sister centromeres somehow reassociate by prometaphase I, no such reassociation was seen in a FISH-based analysis of X chromatid segregation in ord females, which exhibit very similar centromere cohesion loss and chromatid missegregation phenotypes as sunn females. Instead, X chromatids often appeared fully separate after the onset of prometaphase I and segregated chaotically (Bickel et al. 2002). Thus, we suggest that meiosis I missegregation in both sexes in sunn mutants is likely due mainly to the premature loss of centromere cohesion and the resulting failure of sister centromeres to mono-orient.
Balanced vs. unbalanced segregations in the absence of cohesion
Our FISH analyses showed that in >90% of sex chromosome bivalents in both solo (Yan et al. 2010) and sunn (this article, Figure 2, and Table 5) males, the chromatids segregate in numerical balance (two toward each pole) at anaphase I even though the cross data indicate very high rates of meiosis I NDJ. Our FISH data show that the explanation for the high meiosis I NDJ is random sister chromatid partner choice, which results in a 1:2 ratio of reductional to equational sister chromatid segregation. This “random 2←→2” segregation pattern requires the homolog conjunction complex, since snm mutations in a solo mutant background result in completely random segregation (Yan et al. 2010).
Might the random 2←→2 mechanism also apply to meiosis I segregation in sunn females? Unfortunately, cross data are not informative on this point because the predicted ratio of sister vs. homolog NDJ products among XX eggs is identical (1:2) whether segregation is completely random or random 2←→2, and the unbalanced segregation products (XXX and XXXX) which are critical to distinguishing which mechanism is operative cannot be recovered. Nevertheless, we favor the fully random model in females for two reasons. First, the homolog conjunction complex that is essential for the random 2←→2 mechanism in male meiosis is absent in female meiosis (Thomas et al. 2005). Although females also have a robust achiasmate segregation mechanism (Hawley et al. 1992), it bears little mechanistic resemblance to that in males. Second, the FISH analysis of meiotic segregation of X chromatids in ord females mentioned above found no indication of orderly segregation (Bickel et al. 2002). It seems likely, then, that the random 2←→2 mechanism is male specific and that the uncohesive chromatids in sunn females segregate fully randomly during both meiotic divisions. It will be important to test this prediction experimentally. The mechanism underlying random 2←→2 segregation in male meiosis also remains to be investigated.
What role does SUNN play in cohesion?
Several of the findings summarized above are consistent with SUNN functioning as a component of a cohesion-providing complex along with SOLO and ORD. One possibility is that all three proteins are subunits, along with SMC1 and SMC3, of a specialized meiotic cohesin complex, perhaps replacing either or both of the mitotic non-SMC subunits RAD21 and SA, neither of which has been shown as yet to have a role in meiosis. This idea is supported by several lines of evidence. First, mutations in ord, solo, and sunn abolish detectable centromeric foci of SMC1 at all stages in both male and female meiosis (Khetani and Bickel 2007; Yan et al. 2010; Tanneti et al. 2011; Yan and McKee 2013). Second, centromeric foci of SOLO require both ord and sunn function and centromeric foci of SUNN require solo function, suggesting reciprocal co-dependence of the three proteins (Yan et al. 2010). Third, survival of centromeric foci of SMC1, ORD, and SOLO beyond metaphase I in male meiosis depends on mei-S332 (Balicky 2005; Yan et al. 2010), the Drosophila Shugoshin homolog. We expect that SUNN centromere foci will prove to be similarly dependent on mei-S332, although this remains to be shown. Fourth, SOLO interacts physically with SMC1 in coimmunoprecipation assays from ovarian extracts (Yan and McKee 2013) and with both SMC1 and SMC3 in yeast two-hybrid analysis (Q. Ma and B.D. McKee, unpublished data). At minimum, these data indicate very strong interactions of ORD, SOLO, and SUNN with each other and with the SMC cohesins and are consistent with roles as cohesin components. This idea does not exclude the possibility of other meiotic cohesin complexes, perhaps involving the mitotic non-SMC subunits and/or C(2)M. Multiple meiosis-specific cohesin complexes have been demonstrated in several higher eukaryotic systems (Nasmyth and Haering 2009; Severson et al. 2009; Llano et al. 2012).
The bioinformatic analysis of SUNN presented above is of interest in light of these considerations. Although no homologs of SUNN were identified in sequence- or profile-based searches, fold recognition and structural analysis indicated that SUNN may be a homolog of the cohesin protein SA. Although this line of reasoning is inconclusive, it suggests that the shared roles of SA and SUNN in sister chromatid cohesion may have a basis in a shared overall structure and raises the possibility that SUNN might serve as a meiosis-specific substitute for SA in some meiotic cohesin complexes. It will clearly be important to establish the role, if any, of SA in meiotic cohesion. More detailed biochemical and genetic studies of SUNN and its partners will be required to resolve the precise functions of these intriguing proteins in cohesion.
Supplementary Material
Acknowledgments
We thank B. Wakimoto, C. Zuker, Terry Orr-Weaver, Kim McKim, and the Bloomington Stock Center, Indiana University, for providing Drosophila stocks; R. Scott Hawley, Mary Lily, and Sharon Bickel for providing antibodies; Drosophila Genomics Resource Center (DGRC) and Roger Tsien for providing the pPVW (1093) and pPWV (1094) Venus tag containing P-element vector; and Joseph A. May at the Molecular Biology Resource facility, University of Tennessee, Knoxville for performing DNA sequencing. Funding for this work was provided by the National Institute of General Medical Sciences, grant no. R01 GM040489, and by a grant from the University of Tennessee jointly funded by the Office of Research Administration, College of Arts and Sciences and Department of Biochemistry and Cellular and Molecular Biology.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.166009/-/DC1.
Communicating editor: J. Sekelsky
Literature Cited
- Andrade M. A., Petosa C., O’Donoghue S. I., Muller C. W., Bork P., 2001. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309: 1–18. [DOI] [PubMed] [Google Scholar]
- Balicky, E. M., 2005 Regulation of Chromosome Segregation by ORD and dRING During Drosophila Meiosis. Ph.D. Dissertation. Department of Biological Sciences, Dartmouth College, Hanover, New Hampshire. [Google Scholar]
- Balicky E. M., Endres M. W., Lai C., Bickel S. E., 2002. Meiotic cohesion requires accumulation of ORD on chromosomes before condensation. Mol. Biol. Cell 13: 3890–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel S. E., Wyman D. W., Miyazaki W. Y., Moore D. P., Orr-Weaver T. L., 1996. Identification of ORD, a Drosophila protein essential for sister chromatid cohesion. EMBO J. 15: 1451–1459. [PMC free article] [PubMed] [Google Scholar]
- Bickel S. E., Wyman D. W., Orr-Weaver T. L., 1997. Mutational analysis of the Drosophila sister-chromatid cohesion protein ORD and its role in the maintenance of centromeric cohesion. Genetics 146: 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel S. E., Orr-Weaver T. L., Balicky E. M., 2002. The sister-chromatid cohesion protein ORD is required for chiasma maintenance in Drosophila oocytes. Curr. Biol. 12: 925–929. [DOI] [PubMed] [Google Scholar]
- Blower M. D., Karpen G. H., 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3: 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi, S. M. G. Giansanti, G. Cenci, and M. Gatti, 2000 Cytological Analysis of Spermatocyte Growth and Male Meiosis in Drosophila melanogaster. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Brar G. A., Hochwagen A., Ee L. S., Amon A., 2009. The multiple roles of cohesin in meiotic chromosome morphogenesis and pairing. Mol. Biol. Cell 20: 1030–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Dong F., Edelmann R. E., Makaroff C. A., 2003. The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J. Cell Sci. 116: 2999–3007. [DOI] [PubMed] [Google Scholar]
- Cenci G., Bonaccorsi S., Pisano C., Verni F., Gatti M., 1994. Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J. Cell Sci. 107(Pt 12): 3521–3534. [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Diallo S., Vezon D., Gendrot G., Vrielynck N., et al. , 2005. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J. Cell Sci. 118: 4621–4632. [DOI] [PubMed] [Google Scholar]
- Clift D., Marston A. L., 2011. The role of shugoshin in meiotic chromosome segregation. Cytogenet. Genome Res. 133: 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya I. N., Hamant O., Timofejeva L., Wang C. J., Braun D., et al. , 2006. Alleles of afd1 dissect REC8 functions during meiotic prophase I. J. Cell Sci. 119: 3306–3315. [DOI] [PubMed] [Google Scholar]
- Hauf S., Watanabe Y., 2004. Kinetochore orientation in mitosis and meiosis. Cell 119: 317–327. [DOI] [PubMed] [Google Scholar]
- Hawley R. S., Irick H., Zitron A. E., Haddox D. A., Lohe A., et al. , 1992. There are two mechanisms of achiasmate segregation in Drosophila females, one of which requires heterochromatic homology. Dev. Genet. 13: 440–467. [DOI] [PubMed] [Google Scholar]
- Heidmann D., Horn S., Heidmann S., Schleiffer A., Nasmyth K., et al. , 2004. The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma 113: 177–187. [DOI] [PubMed] [Google Scholar]
- Hirai K., Toyohira S., Ohsako T., Yamamoto M. T., 2004. Isolation and cytogenetic characterization of male meiotic mutants of Drosophila melanogaster. Genetics 166: 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., 2012. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 26: 1659–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetani R. S., Bickel S. E., 2007. Regulation of meiotic cohesion and chromosome core morphogenesis during pachytene in Drosophila oocytes. J. Cell Sci. 120: 3123–3137. [DOI] [PubMed] [Google Scholar]
- Kleckner N., 2006. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115: 175–194. [DOI] [PubMed] [Google Scholar]
- Klein F., Mahr P., Galova M., Buonomo S. B., Michaelis C., et al. , 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91–103. [DOI] [PubMed] [Google Scholar]
- Koundakjian E. J., Cowan D. M., Hardy R. W., Becker A. H., 2004. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics 167: 203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake C. M., Hawley R. S., 2012. The molecular control of meiotic chromosomal behavior: events in early meiotic prophase in Drosophila oocytes. Annu. Rev. Physiol. 74: 425–451. [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Orr-Weaver T. L., 2001. The molecular basis of sister-chromatid cohesion. Annu. Rev. Cell Dev. Biol. 17: 753–777. [DOI] [PubMed] [Google Scholar]
- Llano E., Herran Y., Garcia-Tunon I., Gutierrez-Caballero C., de Alava E., et al. , 2012. Meiotic cohesin complexes are essential for the formation of the axial element in mice. J. Cell Biol. 197: 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manheim E. A., McKim K. S., 2003. The Synaptonemal complex component C(2)M regulates meiotic crossing over in Drosophila. Curr. Biol. 13: 276–285. [DOI] [PubMed] [Google Scholar]
- Matthies, H. J. G., M. Clarkson, R. B. Saint, R. Namba, and R. S. Hawley, 2000 Analysis of meiosis of fixed and live oocytes by light microscopy, pp. 67–85 in Drosophila Protocols, edited by W. Sullivan, M. Ashburner, and R. S. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- McKim K. S., Jang J. K., Manheim E. A., 2002. Meiotic recombination and chromosome segregation in Drosophila females. Annu. Rev. Genet. 36: 205–232. [DOI] [PubMed] [Google Scholar]
- Miyazaki W. Y., Orr-Weaver T. L., 1992. Sister-chromatid misbehavior in Drosophila ord mutants. Genetics 132: 1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35: 673–745. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C. H., 2009. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43: 525–558. [DOI] [PubMed] [Google Scholar]
- Neuwald A. F., Hirano T., 2000. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 10: 1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., Hawley R. S., 2001. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev 15: 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., Hawley R. S., 2003. Chromosome choreography: the meiotic ballet. Science 301: 785–789. [DOI] [PubMed] [Google Scholar]
- Page S. L., Hawley R. S., 2004. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20: 525–558. [DOI] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., et al. , 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15: 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Siomos M. F., Nasmyth K., 2003. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112: 423–440. [DOI] [PubMed] [Google Scholar]
- Retelska D., Iseli C., Bucher P., Jongeneel C. V., Naef F., 2006. Similarities and differences of polyadenylation signals in human and fly. BMC Genomics 7: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y., 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A. F., Ling L., van Zuylen V., Meyer B. J., 2009. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 23: 1763–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. N., Dawson D. S., 2008. Changing partners: moving from non-homologous to homologous centromere pairing in meiosis. Trends Genet. 24: 564–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo S., Lake C. M., Morais-de-Sa E., Sunkel C. E., Hawley R. S., 2011. Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr. Biol. 21: 1845–1851. [DOI] [PubMed] [Google Scholar]
- Tanneti N. S., Landy K., Joyce E. F., McKim K. S., 2011. A pathway for synapsis initiation during zygotene in Drosophila oocytes. Curr. Biol. 21: 1852–1857. [DOI] [PubMed] [Google Scholar]
- Thomas S. E., Soltani-Bejnood M., Roth P., Dorn R., Logsdon J. M., Jr, et al. , 2005. Identification of two proteins required for conjunction and regular segregation of achiasmate homologs in Drosophila male meiosis. Cell 123: 555–568. [DOI] [PubMed] [Google Scholar]
- Toth A., Rabitsch K. P., Galova M., Schleiffer A., Buonomo S. B., et al. , 2000. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell 103: 1155–1168. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y., Orr-Weaver T. L., 2013. Centromere proteins CENP-C and CAL1 functionally interact in meiosis for centromere clustering, pairing, and chromosome segregation. Proc. Natl. Acad. Sci. USA 110: 19878–19883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban E., Nagarkar-Jaiswal S., Lehner C. F., Heidmann S. K., 2014. The cohesin subunit Rad21 is required for synaptonemal complex maintenance, but not sister chromatid cohesion, during Drosophila female meiosis. PLoS Genet. 10: e1004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J., Belmont A. S., Sedat J. W., 2002. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12: 1473–1483. [DOI] [PubMed] [Google Scholar]
- Wakimoto B. T., Lindsley D. L., Herrera C., 2004. Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics 167: 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Yoder J., Antoshechkin I., Han M., 2003. Caenorhabditis elegans EVL-14/PDS-5 and SCC-3 are essential for sister chromatid cohesion in meiosis and mitosis. Mol. Cell. Biol. 23: 7698–7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., 2005. Shugoshin: guardian spirit at the centromere. Curr. Opin. Cell Biol. 17: 590–595. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Nurse P., 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400: 461–464. [DOI] [PubMed] [Google Scholar]
- Wu S., Zhang Y., 2008. MUSTER: improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins 72: 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., McKee B. D., 2013. The cohesion protein SOLO associates with SMC1 and is required for synapsis, recombination, homolog bias and cohesion and pairing of centromeres in Drosophila meiosis. PLoS Genet. 9: e1003637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Thomas S. E., Tsai J. H., Yamada Y., McKee B. D., 2010. SOLO: a meiotic protein required for centromere cohesion, coorientation, and SMC1 localization in Drosophila melanogaster. J. Cell Biol. 188: 335–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S., Watanabe Y., 2005. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 123: 803–817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.