Abstract
ZMM proteins have been defined in budding yeast as factors that are collectively involved in the formation of interfering crossovers (COs) and synaptonemal complexes (SCs), and they are a hallmark of the predominant meiotic recombination pathway of most organisms. In addition to this so-called class I CO pathway, a minority of crossovers are formed by a class II pathway, which involves the Mus81-Mms4 endonuclease complex. This is the only CO pathway in the SC-less meiosis of the fission yeast. ZMM proteins (including SC components) were always found to be co-occurring and hence have been regarded as functionally linked. Like the fission yeast, the protist Tetrahymena thermophila does not possess a SC, and its COs are dependent on Mus81-Mms4. Here we show that the ZMM proteins Msh4 and Msh5 are required for normal chiasma formation, and we propose that they have a pro-CO function outside a canonical class I pathway in Tetrahymena. Thus, the two-pathway model is not tenable as a general rule.
Keywords: meiosis, synaptonemal complex, DSB repair, crossover pathways, recombination
MEIOSIS is a specialized cell division by which the diploid somatic chromosome set is halved prior to the production of gametes. This reduction is achieved by the separation of homologous chromosomes. To ensure faithful segregation, homologous chromosomes must first identify each other and become stably linked. This is achieved by physical connections between homologs due to the invasion and exchange of homologous DNA strands (see Ehmsen and Heyer 2008). An invading single strand forms a heteroduplex with the complementary strand of its target double-stranded DNA (dsDNA). Subsequent strand ligation leads to a more stable intermediate, a Holliday junction (HJ). A possible outcome of HJ formation are crossovers (COs), which mature into chiasmata and hold homologs together and, at the same time, are the basis of genetic recombination. However, a subset of strand invasion events, ranging from less than half in budding yeast (Mancera et al. 2008; Qi et al. 2009) to a considerable excess in Drosophila (Comeron et al. 2012; Miller et al. 2012), Arabidopsis (Yang et al. 2012), and humans (Wang et al. 2012), result in noncrossovers (NCOs), which are tractable as genetic or sequence conversion events.
COs and NCOs are initiated by DNA double-strand breaks (DSBs) generated by the meiosis-specific nuclease Spo11 (see Keeney 2001; de Massy 2013), and the mechanisms of DSB end processing and homologous strand invasion are largely conserved. However, the downstream pathways that convert transiently connected (joint) DNA molecules (JMs) into COs are diverse (Kohl and Sekelsky 2013). The fact that in Caenorhabditis elegans, but not in the budding yeast, COs are entirely dependent on the MutS family protein HIM-14/Msh4 led Zalevsky et al. (1999) to propose that two CO pathways may operate in the yeast. Moreover, COs in C. elegans are all subject to interference (i.e., the suppression of neighboring COs), whereas the budding yeast features both interfering and noninterfering COs. Copenhaver et al. (2002) found the situation in Arabidopsis similar to that in yeast, and therefore they postulated two CO pathways also for this organism. The existence of two CO pathways in yeast was then clearly stated by de los Santos et al. (2003). They found that one (class I) is dependent on Msh4 and Msh5 and that it produces interfering COs, whereas noninterfering class II COs rely on the Mus81-Mms4(Eme1) protein complex. Further insight into the nature of the two pathways revealed that class I COs are dependent on a set of so-called ZMM proteins and accompanied by the formation of a synaptonemal complex (SC), the meiotic pairing structure (Bishop and Zickler 2004; Börner et al. 2004; Hollingsworth and Brill 2004). ZMM (an acronym for Zip1, Zip2, Zip3, Zip4, Msh4, Msh5, Mer3) proteins comprise components for the assembly of the SC as well as proteins used for processing COs in the context of the SC (see Lynn et al. 2007). In budding yeast, SC proteins Zip3, Zip2, and Zip1 assemble at DSB sites and Zip3 recruits the CO-promoting factors Msh4-Msh5 and Mer3. This ensemble then is believed to promote Zip1 polymerization into the central element of the SC with the help of Zip4 and Spo16 (Shinohara et al. 2008). SC polymerization, in turn, was identified as a factor contributing to CO interference in mammals and C. elegans (Lian et al. 2008; Libuda et al. 2013), with this role being controversial in the budding yeast (Börner et al. 2004; Fung et al. 2004; Shinohara et al. 2008; Klutstein et al. 2009; Zhang et al. 2014). Indeed, in yeast, interference may be the result of multiple layers of control (Berchowitz and Copenhaver 2010). In addition to transmitting an interference signal, completed synapsis could be a signal to discontinue DSB formation and homology search (Hayashi et al. 2010; Kauppi et al. 2013; Thacker et al. 2014).
The budding yeast class II pathway produces non-interfering COs without the help of ZMM proteins. The only known class II-specific factor is the Mus81-Mms4(Eme1) structure-selective endonuclease, which can resolve (nicked) HJs to produce COs (Boddy et al. 2001; Osman et al. 2003; Schwartz et al. 2012). The corresponding role in the class I pathway is probably played by the MutLγ complex (Mlh1-Mlh3) with the help of Exo1 (Zakharyevich et al. 2012; Ranjha et al. 2014). Most organisms seem to primarily use the class I pathway, with the class II pathway as a backup. However, the fission yeast, which does not form an SC (see Loidl 2006), depends exclusively on the class II pathway (Villeneuve and Hillers 2001; Smith et al. 2003). On the other hand, most COs in C. elegans (Zalevsky et al. 1999; Kelly et al. 2000) and Drosophila (Kohl et al. 2012) are through a class I pathway, but use different HJ resolvases (see Bellendir and Sekelsky 2013). The class II pathway is believed to be evolutionarily primordial because it is simpler as it relies primarily on proteins that have roles in somatic DNA repair and are not exclusive to meiosis (Kohl and Sekelsky 2013).
Tetrahymena thermophila is a member of the ciliates, which are single-celled protists. Tetrahymena possess a germline and a soma, each represented by a single nucleus within one cell. Tetrahymena can propagate by vegetative divisions, during which the diploid germline nucleus (also known as the micronucleus, or MIC) undergoes a mitotic cycle. The somatic nucleus (macronucleus, or MAC) is polyploid and divides by amitotic splitting (see Karrer 2012). Genes are expressed only from the MAC. Tetrahymena switch to sexual reproduction when starved. Under these conditions, cells of complementing mating types form pairs (conjugate) and their MICs perform synchronous meioses (see Cole and Sugai 2012). Meiosis is followed by mutual fertilization and a complex sequence of mitoses and nuclear degradation, leading to four sexual progeny per mating pair with new MICs and MACs developed from zygote nuclei (see Cole and Sugai 2012).
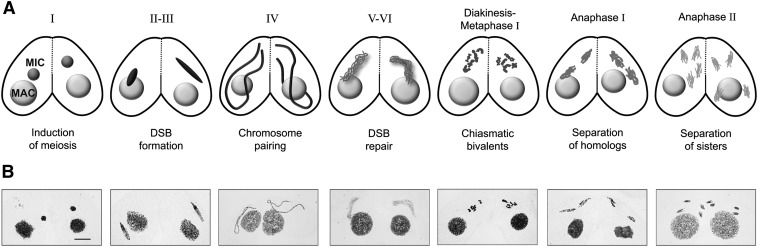
Previous studies of Tetrahymena meiosis have revealed some remarkable features, the most striking being the extreme elongation of MICs during prophase to about twice the length of the cell. Nuclear elongation is triggered by DSB formation (Mochizuki et al. 2008; Loidl and Mochizuki 2009), and it begins ∼2 hr after induction of meiosis (Figure 1). Within an elongated nucleus, chromosomes are arranged in a stretched bouquet-like manner, with centromeres and telomeres attached to opposite ends of the nucleus. This ultimate bouquet arrangement is believed to promote the juxtapositioning of homologous regions and, thereby, homologous pairing and CO formation (Loidl and Scherthan 2004; Loidl et al. 2012).
Figure 1.
Meiosis in Tetrahymena. (A) Meiosis is initiated by the mating of two starving cells (I). Each contains a diploid generative MIC and a somatic MAC. The MICs undergo synchronous meiosis in the two partners whereas the MACs do not participate. The MIC elongates during meiotic prophase (II–III) in response to DSB formation. During the stage of maximal elongation (IV), homologous chromosomes pair. When DSBs are repaired by homologous recombination, the MIC shortens again (V–VI), and distinct chromatin threads become visible. Five condensed bivalents appear from diakinesis to metaphase I. Chromosomes and chromatids separate during first and second meiotic division, respectively. Classification of early meiotic stages (I–VI) according to Sugai and Hiwatashi (1974). (B) Microscopical images (Giemsa staining) of corresponding stages. Bar, 10 µm.
At the molecular level, the early steps in the meiotic recombination pathway are similar to those in canonical model organisms. Meiotic DSBs are induced by Spo11, and initial DSB end processing is promoted by Sae2 (Com1) and Mre11 (Lukaszewicz et al. 2010). Both Rad51 and Dmc1 are required for CO formation, but primarily Dmc1 can be seen localizing to chromatin (Howard-Till et al. 2011). Hop2 recombinase (and presumably its partner Mnd1) are required for interhomolog crossing over (Mochizuki et al. 2008). However, in contrast to budding yeast and multicellular organisms, Mus81 and its partner Mms4 (Eme1) have a prominent role in CO formation, as they were found to be required for the resolution of JMs (Lukaszewicz et al. 2013). Similarly, homologous chromosomes failed to separate in the absence of Sgs1 (Lukaszewicz et al. 2013). This is consistent with Sgs1’s various contributions to prevent deleterious excessive CO formation by channeling intermediates into a NCO pathway or in undoing aberrant JMs (Oh et al. 2008; de Muyt et al. 2012). Notably, structural proteins of the SC were neither detected by bioinformatic searches, nor were these structures observed cytologically (Wolfe et al. 1976; Mochizuki et al. 2008; Chi et al. 2014).
Because of the dependence of COs on Mus81-Mms4 and the lack of SCs (which are considered a characteristic of the class I pathway; see above), we posited that the class II pathway is the predominant, if not the only, CO pathway in Tetrahymena (Lukaszewicz et al. 2013). However, recently, homologs of the ZMM proteins Msh4 and Msh5 were reported for Tetrahymena (Chi et al. 2014). Here, we investigate if these proteins have a function in meiosis, which may challenge the concept of a conserved set of collaborating ZMM proteins.
Materials and Methods
Cell growth and induction of meiosis
T. thermophila wild-type (WT) strains B2086 and Cu428, being of complementing mating types, served as controls and as the source material for the construction of knockout strains. Strains were obtained from the Tetrahymena Stock Center at Cornell University (http://tetrahymena.vet.cornell.edu/). Cells were cultured in Neff’s medium at 30° according to standard methods (see Orias et al. 2000), and they were made competent for sexual reproduction by starvation in 10 mM Tris–Cl (pH 7.4) for 12–16 hr. Meiosis was induced by mixing starved cultures of cells of complementing mating types at equal densities (∼2 × 105 cells/ml). For the investigation of recombination-related DNA synthesis, cells were fed with BrdU at a final concentration of 2 × 10−4 M 2.5 hr after mixing. Cells were harvested for analysis of meiotic stages 3–4.5 hr after mixing.
Macronuclear gene knockout
For the construction of msh4 knockout (KO) strains, ∼500-bp fragments of genomic Tetrahymena DNA upstream and downstream of the ORF were amplified using primers MSH4KO 5′FW (gaa act gat aac att tag caa gac), MSH4KO5′RV (GTC TAT CGA ATT CCT GCA GCC C tgg ttt tgg agc tat atc ag), MSH4KO 3′FW (CTG GAA AAA TGC AGC CC gag cga tat ata tcg taa gg), and MSH4KO 3′RV (ctt cat aat cta aca act cta cc). The underlined portions are homologous to the flanking regions of the knockout cassette (Mochizuki 2008). The cassette consists of the NEO4 resistance gene under the Cd2+-inducible MTT1 metallothionein promoter (Shang et al. 2002). These flanking fragments were then joined to the ends of the cassette using overlapping PCR (Mochizuki 2008). The knockout construct was introduced into B2086 and Cu428 cells by biolistic transformation as described previously (Bruns and Cassidy-Hanley 2000). The transformants were selected in media containing CdCl2 (1 µg/ml at the start of selection and 0.05 µg/ml at the end of selection) and increasing concentrations of paromomycin (from 120 µg to 40 mg/ml) until the WT chromosomes were completely replaced by the knockout chromosomes in the somatic nucleus.
msh5 knockout strains were constructed in the same way. The primers were MSH5KO 5′FW (tct tgt caa ttg agc ttc atc), MSH5KO5′ RV (GTC TAT CGA ATT CCT GCA GCC C ctc gcc ata ata aaa tgt tg), MSH5KO 3′FW (CTG GAA AAA TGC AGC CC att taa tgg tat tat tag agc), and MSH5KO 3′RV (gtt ttt agg ttc aat cag tag g).
Gene knockdown by RNA interference
To elicit a double msh4 sgs1 phenotype, Sgs1 was depleted by RNA interference (RNAi) in msh4 KO strains. msh4 strains were transformed with a construct carrying two copies of an ∼500-bp fragment of the SGS1 ORF in reverse tandem arrangement under a Cd2+-inducible MTT1 metallothionein promoter and a cycloheximide resistance cassette (Lukaszewicz et al. 2013). High-copy transformants were selected under increasing cycloheximide concentrations. Transcription of hairpin dsRNA was activated by the addition of CdCl2 to premeiotic starving cells (see Lukaszewicz et al. 2013).
Cytological methods
For subsequent immunostaining, cells were prepared by a formaldehyde fixation method (Mochizuki et al. 2008). In short, 250 µl 10% Triton X-100 and 500 µl 37% formaldehyde were added to 5 ml of cell suspension. After 30 min, the fixed cell suspension was centrifuged and the pellet was resuspended in 500 µl of 4% paraformaldehyde + 3.4% sucrose solution. A drop of this mixture was spread onto a slide and air-dried. Immunostaining of Dmc1 associated with DNA required a modified fixation protocol, using a higher concentration of Triton X-100 detergent, which removes free protein from the nucleus (Howard-Till et al. 2011).
For Giemsa staining, fluorescence in situ hybridization (FISH) and BrdU staining, cells were prepared by the method of Bruns and Brussard (1981). In short, 495 µl of Schaudinn’s fixative (saturated HgCl2 + absolute ethanol 2:1) was supplemented with 5 µl of acetic acid and squirted into a cell pellet prepared from 5 ml of culture. The suspension was kept for 1 hr at room temperature and centrifuged, and the pellet was resuspended in 1 ml of 70% EtOH. One hundred µl of the suspension was pelleted, and the pellet was resuspended in 300 µl of methanol/acetic acid 3:1. Several drops were applied to a clean slide and air-dried. For Giemsa staining, preparations were hydrolyzed with 5 N HCl (100 µl under a coverslip) for 2 min at room temperature, rinsed in distilled water, and air-dried. Slides were incubated in 4% Giemsa solution in 10 mM sodium phosphate buffer (pH 6.8) for 10 min, rinsed with distilled water, air-dried, and mounted with Euparal.
For immunostaining, slides were washed with 1×PBS and 1×PBS + 0.05% Triton X-100. Primary and fluorochrome-labeled secondary antibodies were applied as described previously, and the preparations were mounted under a coverslip in Vectashield antifading agent (Vector Laboratories; Burlingame, CA) supplemented with 0.05 mg/ml 40,60-diamidino-2-phenylindole (DAPI) (Howard-Till et al. 2011). Dmc1 was detected with an antibody that reacts with both Dmc1 and Rad51 (1:50 mouse monoclonal, Clone 51RAD01, NeoMarkers, Fremont, CA). Incorporated BrdU was detected by denaturing slides in 70% formamide for 2 min at 65° and immunostaining with rat anti-BrdU antibody (1:40; Abcam, Cambridge, UK) (Loidl et al. 2012).
For FISH, a compound probe was produced by amplifying a 22.1-kb intercalary chromosomal locus by PCR (Loidl and Mochizuki 2009). The purified PCR products were Cy3-labeled by nick translation. The probe and chromosomal DNA were denatured by hot formamide and hybridized for 36–48 hr at 37° (Loidl and Scherthan 2004). Slides were washed in PBS and mounted in Vectashield + DAPI as above.
Microscopic images were recorded using the MetaVue software (Molecular Devices, Sunnyvale, CA), and distances between FISH signals were measured using the program’s measuring tool and converted to micrometers. Dmc1 and BrdU foci were counted on enlarged and contrast-enhanced prints of immunostained spread nuclei as described in Howard-Till et al. (2011).
Results
Tetrahymena possesses homologs of MSH4 and MSH5
It was shown previously that Tetrahymena possesses a Msh4 homolog (Malik et al. 2008; Chi et al. 2014). It is encoded by ORF TTHERM_00857890 [Tetrahymena Genome Database (TGD) http://www.ciliate.org/; Stover et al. (2006)]. A Msh5 homolog also was detected (Chi et al. 2014). Its transcript is denoted as gene_000007168 in the Tetrahymena Functional Genomic Database (TFGD, http://tfgd.ihb.ac.cn/; Miao et al. 2009). It should be noted that the corresponding ORF TTHERM_00763040 is incorrectly annotated in the current version of the TGD. Tetrahymena TTHERM_00857890 predicted protein produces the best reciprocal BLAST hits for both human and Arabidopsis Msh4. Similarly, the Tetrahymena protein sequence derived from transcript gene_000007168 gives the best reciprocal BLAST hits for human and Arabidopsis Msh5 homologs (Supporting Information, Figure S1). Their relationships to the budding yeast Msh4 and Msh5, respectively, are more distant (Figure 2A). Warren et al. (2007) identified the five conserved domains of bacterial MutS in its eukaryotic homologs Msh2 and Msh6, which have a role in DNA mismatch repair. The first (mismatch binding) domain is specific to Msh2 and Msh6, whereas domains II-V are present in Msh4 and Msh5 as well. As shown in Figure S2, domains II-V are present in TTHERM_00857890p and the predicted protein of transcript gene_000007168, which suggests functional conservation. Moreover, TTHERM_00857890 and gene_000007168 sequences are exclusively transcribed early in conjugation, i.e., the time when meiotic recombination genes are expressed according to TFGD. Together, this suggests that they are the Msh4 and Msh5 orthologs. In the following, the Tetrahymena genes and corresponding proteins will be referred to as MSH4, MSH5 and Msh4, Msh5, respectively.
Figure 2.
Tetrahymena Msh4 and Msh5 homologs and msh4 and msh5 knockout strains. (A) Unrooted phylogenetic tree of MutS1 homologs from T. thermophila, human, and budding and fission yeast reconstructed from a protein sequence alignment of the MutS_II-MutS_V domain regions. Tetrahymena MutS homologs were collected by searching the Tetrahymena proteome derived from the TetraFGD RNAseq transcriptome with the Pfam MutS_V domain model. Sequences for all other species are based on Lin et al. (2007). The MrBayes consensus tree is shown with Bayesian posterior probabilities in percent illustrating the degree of support for each node on the tree (Ronquist et al. 2012). The scale bar indicates the branch length representing the expected number of substitutions per site. (B) Full replacement of wild-type MSH4 copies by KO cassettes is shown by Southern hybridization with a sequence flanking the 3′ end of MSH4 as a probe to BglII fragments of wild type (WT) and msh4 knockout (mating type B and C clones) genomic DNA. The msh5 knockout strains (mating type B and C) show extensive but not complete replacement of wild-type MSH5 copies by knockout cassettes. Also for MSH5 KO, the 3′ flanking sequence was used as a Southern probe to BglII fragments of genomic DNA.
To study the roles of the two proteins, we knocked out the ∼45 macronuclear (i.e., transcribed) gene copies by gene replacement. Southern hybridization with a probe against the wild-type MSH4 gene demonstrated that the knockout was complete in both mating types (Figure 2B). Thus, the observed defects may be considered as the msh4 null phenotype. The knockout of MSH5 was only partial (Figure 2B). For both knockout strains sexual progeny production was reduced. At least 100 conjugating cells were isolated and tested for progeny viability (for viability testing, see Karrer 2000). The viability of sexual progeny of msh4 and msh5 matings was 48.1 and 52.8%, respectively, compared to 68% viable progeny of wild-type matings.
Neither an antibody against Msh4 nor HA-tagged Msh4 allowed us to detect the protein cytologically (data not shown), which could be due to the low abundance of the protein.
Bivalent formation is reduced in the msh4Δ mutant
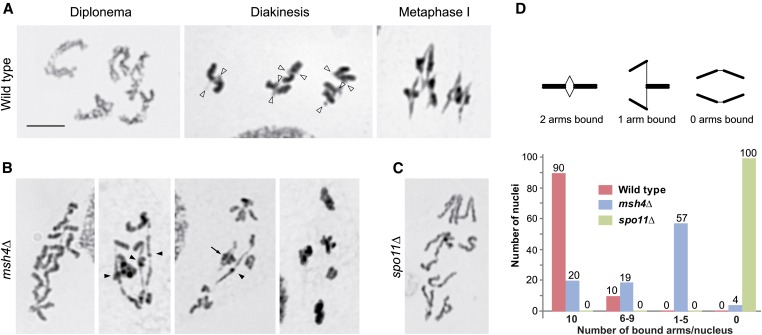
The cytological phenotype of msh4Δ early meiotic stages was inconspicuous. However, there was a striking reduction in bivalent formation at diakinesis-metaphase I (Figure 3). In the wild type, five well-condensed bivalents can be observed in well-preserved diplotene-diakinesis nuclei, and mostly ring-shaped bivalents are arranged in a usually dense metaphase I plate (Figure 3A). However, in msh4Δ, a variable number of chromatin bodies are present in the nucleus (Figure 3B). This suggests that the mutant forms a mixture of bivalents and univalents, where bivalents have a reduced number of chiasmata, since they are mostly rod-, rather than ring-, shaped.
Figure 3.
Reduced chiasma formation in the msh4Δ mutant. (A) Five bivalents in the wild type. In diplonema, homologous chromosomes twist around each other. In diakinesis, homologous arms are tightly associated, and only the centromeric regions (arrowheads) are separated. In metaphase I, tension at the kinetochores separates the proximal regions of homologous arms. Bar, 5 µm. (B) In the mutant, diakinesis-metaphase I nuclei show only univalents, mixtures of univalents and bivalents, or only bivalents (from the left). Arrowheads denote rod bivalents; the arrow indicates a ring bivalent. (C) Univalents in a spo11Δ control. Chromosomes are stained with Giemsa in A–C. (D, Top) Since bound arms accomodate an unknown number of chiasmata, chiasma reduction was indirectly estimated by scoring diakinesis-metaphase I chromosomes as having two arms bound (ring bivalents), one arm bound (rod bivalents), and no arms bound (pairs of univalents). (Bottom) Numbers of bound arms/nucleus scored for the wild type, the msh4Δ mutant, and the spo11Δ control (n = 100 nuclei for each).
In Tetrahymena, it is not possible to count COs or chiasmata. During diplonema, bivalents show numerous junctions (Figure 3A), but these may be chiasmata or twisted chromatids. At diakinesis and metaphase I, bivalent arms are closely bound, again concealing the number of chiasmata. Since bound arms accommodate an unknown number of COs, we counted ring and rod bivalents vs. univalents in diakinesis-metaphase I cells to roughly quantify the mutant defect (Figure 3D). In the wild type, 90% of cells (n = 100) formed only ring bivalents; in the spo11Δ control, 100% (n = 100) showed only univalents. In the mutant, 20% showed the full set of ring bivalents (n = 100 cells), 4% showed only univalents, and the rest showed mixed sets of ring and rod bivalents and univalents (Figure 3D). The mutant rarely showed typical diakineses or metaphase I plates with little or no stretched bivalents, like the ones shown in Figure 3A for the wild type. Due to attenuated cohesiveness of bivalents, this arrangement may be unstable and arms may separate even upon slight tension at the centromeres.
Reduced bivalent formation is not due to reduced DSB or JM formation
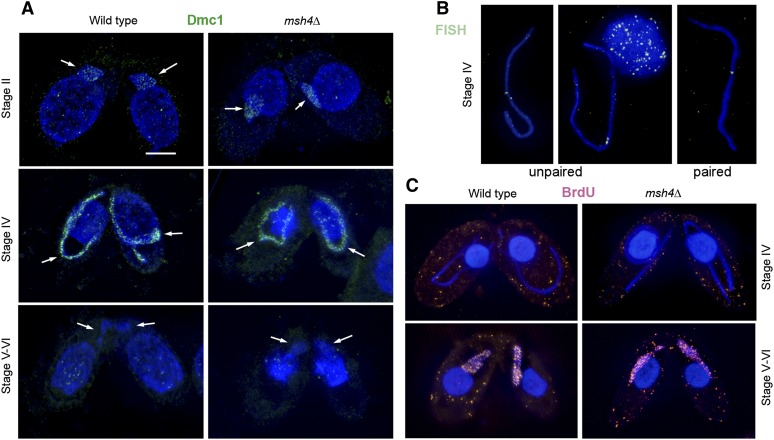
Reduced bivalent formation could be a consequence of impaired DSB formation or processing. Elongation of the meiotic nucleus in the mutant indicated that at least some DSBs were formed because DSBs are known to trigger nuclear elongation (Mochizuki et al. 2008). We therefore tested whether their number was reduced. In Tetrahymena, the number of Dmc1 foci is a reliable indication of the number of DSBs because they are long lived. They appear at the beginning of the elongation stage, and their number remains stable until the stage after maximal elongation when DSBs become repaired (Loidl et al. 2012) (Figure 4A). We compared the number of Dmc1 foci in elongated nuclei of the wild type and the msh4Δ mutant. The average number of Dmc1 foci determined in 10 wild-type nuclei was 165.7 ± 15.3 (SD), which conforms with previous observations (Howard-Till et al. 2011). In the mutant, the number was 183.4 ± 15.8 (SD; n = 10). Thus, DSBs are not reduced in the mutant.
Figure 4.
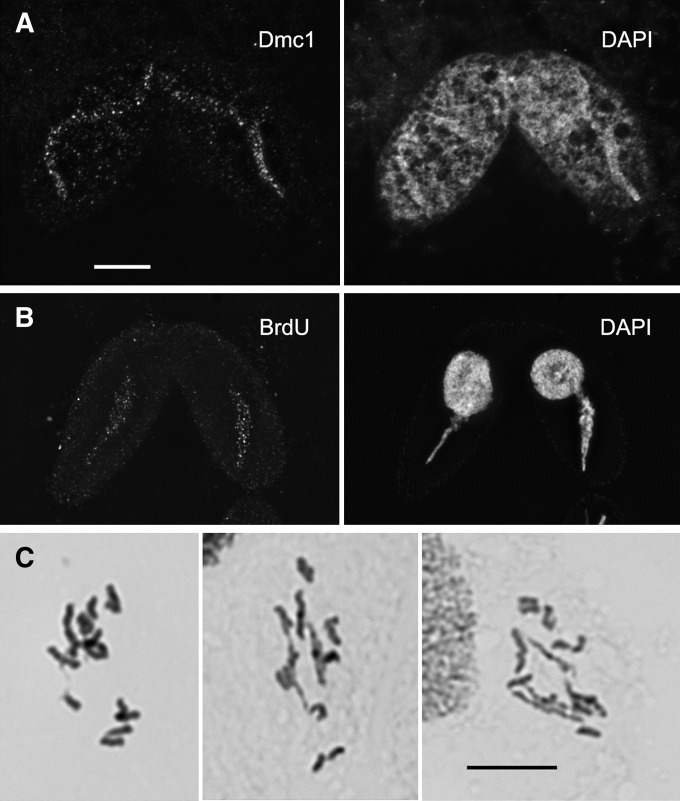
DSB and JM formation appear normal in the msh4Δ mutant. (A) Dmc1 foci, cytological markers of DSBs, appear similar in meiotic nuclei (arrows) of the wild type and the mutant. Foci appear from stage II onward, reach a maximum at stage IV, and disappear by stage V. Cells were subjected to a spreading technique, which removes free protein from nuclei. The antibody used detects chromatin-bound Dmc1 in the MICs but also Rad51 in the MACs (Howard-Till et al. 2011). (B) FISH to elongated nuclei was used to assess pairing of homologous loci (see text). Examples of unpaired and paired loci are shown. (C) BrdU administered to cells early in meiosis is not incorporated during stage IV, but only from stage V onward. Stage numbers correspond to the categories shown in Figure 1A. Bar, 10 µm.
It has been shown previously that, while nuclear elongation is sufficient to roughly align homologs, strand exchange at multiple sites along chromosomes is required for precise pairing in Tetrahymena (Lukaszewicz et al. 2010; Howard-Till et al. 2011) as in other organisms (Peoples-Holst and Burgess 2005). Thus, reduced JM formation would cause a reduction in homologous pairing. To determine whether this was the case in the msh4 mutant, we performed FISH of the elongated nucleus (Figure 4B). It was found that in 56% of nuclei (n = 50) homologous loci were merged into a single signal and the average distance of signal twins was 1.6 ± 0.8 µm. This was similar to the wild type with 54% (n = 50 nuclei) of the FISH signals merged and an average distance of 1.7 ± 0.8 µm of the remaining ones.
If DSB formation and strand exchange take place normally, DNA repair synthesis completes the DSB repair process. If BrdU is provided at the beginning of meiosis, it is incorporated only during stage V–VI (see Figure 1), i.e., after Dmc1 is removed from the chromatin (Loidl et al. 2012). We performed this experiment to see if recombination-related DNA synthesis is normal in the mutant. Similar numbers of BrdU foci were counted in the wild type (107.0 ± 7.2 SD; n = 10 nuclei) and in the mutant (104.7 ± 17.3 SD; n = 10 nuclei), indicating that strand invasion and DSB repair synthesis was taking place normally (Figure 4C).
Together, these observations indicate that neither reduced DSB formation nor reduced JM formation is responsible for bivalent reduction. Therefore, the reduction must be due to either preferential intersister COs or reduced conversion of JMs into COs.
Bivalent loss in msh4Δ is rescued by the depletion of Sgs1 helicase
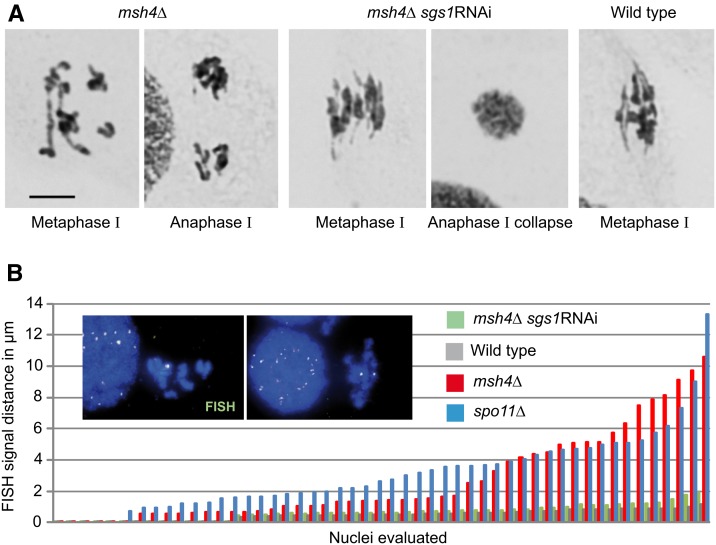
The helicase Sgs1 has a complex role in meiosis, with both anti-CO activity due to its involvement in synthesis-dependent strand annealing (SDSA) and in the dissolution of double HJs, and pro-CO activity in collaboration with ZMM proteins (see Klein and Symington 2012). In budding yeast, it was proposed that Msh4-Msh5 stabilize recombination intermediates against the anti-CO activity of Sgs1 in SDSA (Jessop et al. 2006; Oh et al. 2007; De Muyt et al. 2012; see Kohl and Sekelsky 2013). To determine whether Msh4-Msh5 and Sgs1 may also act as antagonists in Tetrahymena, we studied a double mutant (a sgs1 RNAi-mediated knockdown in a msh4 deletion background; for RNAi efficiency, see Figure S3). Chromatin at diakinesis-metaphase I looked fuzzy in the double mutant (Figure 5A), which resembles the aspect described previously for the sgs1 RNAi single mutant (Lukaszewicz et al. 2013). Chromatin fuzziness precluded the direct visualization of whether bivalent formation was restored in the double mutant. Therefore, we determined the distances of homologous chromosomes at diakinesis-metaphase I. For this, we measured the distance between the the two copies of an intercalary locus highlighted by FISH (Figure 5B). If two homologs are unpaired or form a (rod) bivalent with a low number of chiasmata, the average distance is expected to be greater than in a bivalent with numerous chiasmata. It was found that in the msh4Δ mutant homologous distances were similar to those in a spo11Δ achiasmatic control, whereas in the msh4 sgs1 double mutant the separation of homologous loci was reduced to almost the level of the wild type (Figure 5B). Also, the double mutant failed to separate chromosomes at anaphase I. All 200 post-metaphase I cells analyzed collapsed back into a single nucleus (Figure 5A). This anaphase collapse mirrors the behavior of the sgs1 (RNAi) single mutant, which was shown to be due to the requirement of Sgs1 for the resolution of HJs (Lukaszewicz et al. 2013). Anaphase collapse is additional evidence for the formation of CO intermediates or other JMs in the double mutant, since these are expected to remain unresolved due to the lack of Sgs1’s HJ resolution activity.
Figure 5.
Lack of Sgs1 restores bivalent formation in the msh4Δ mutant. (A) The msh4Δ mutant shows a mixture of bivalents and univalents, whereas bivalents in the msh4Δ sgs1 RNAi double mutant resemble those of the wild type except for a slight condensation defect. Giemsa staining. Bar, 5 µm. (B) Comparison of distances between homologous loci in diakinesis-metaphase I nuclei of the three genotypes and of spo11Δ as a nonbivalent forming control. Fifty nuclei of each genotype are plotted in the order of increasing distances. (Inset) Examples of a wild-type diakinesis-metaphase I nucleus with a single FISH signal where the corresponding chromosome arms are connected (left), and of a msh4Δ nucleus with two signals, indicating separated arms (right). Chromatin was stained with DAPI.
Bivalent formation is reduced in the msh5Δ mutant
Human and probably yeast Msh4 and Msh5 work as a dimer in meiosis (Snowden et al. 2004; Clark et al. 2013; Rakshambikai et al. 2013). Since in Tetrahymena the expression profiles of the two genes are slightly divergent (http://tfgd.ihb.ac.cn/), we wanted to see if the msh5 mutant also shows a meiotic defect. We did not achieve a complete macronuclear deletion of MSH5 (Figure 2B). Nevertheless, msh5 meiosis presented a cytological phenotype very similar to that of msh4Δ. While DSB and JM formation was normal, bivalent formation was reduced and most of the remaining bivalents were rod-shaped (Figure 6). This suggests that Msh4 and Msh5 work together in Tetrahymena and that their cooperation in meiosis is conserved across eukaryotes.
Figure 6.
Meiosis in msh5Δ. (A) Dmc1 localization to elongated prophase nuclei (left) is normal. (B) Incorporation of BrdU (left) indicates normal recombination-related DNA synthesis and JM formation. (C) Three examples of abnormal diakinesis-metaphase I stages with univalents and bivalents. Giemsa staining. Bar, 10 µm.
Discussion
Msh4 and Msh5 stabilize chiasmata in Tetrahymena
Msh4 and Msh5 are meiosis-specific homologs of the bacterial DNA mismatch-repair protein MutS (Kolas and Cohen 2004). They form a heterodimer that binds and stabilizes DNA strand-exchange intermediates to promote crossing over (Snowden et al. 2004). Together with Mer3 helicase, which stimulates DNA heteroduplex extension in the 3′–5′ direction (Mazina et al. 2004), they may stabilize JMs en route to interference-sensitive COs (Lynn et al. 2007). Msh4-Msh5 and Mer3 partner with other ZMM proteins at future CO sites, whereby Zip3/RNF212 may be an initializing or stabilizing factor (Shinohara et al. 2008; Reynolds et al. 2013; Serrentino et al. 2013). In budding yeast, SC formation by Zip1 polymerization may start preferentially from these sites.
Here, gene knockouts revealed that the lack of Msh4 or Msh5 causes a notable reduction in bivalent formation in Tetrahymena. This phenotype resembles that of Arabidopsis msh4 mutants with an increased abundance of rod bivalents and univalents at the cost of ring bivalents (Higgins et al. 2004). While it has not yet been possible to determine the number of COs or chiasmata in Tetrahymena, the presence of univalents and rod bivalents instead of ring bivalents in the msh4 and msh5 mutants indicates that the number of chiasmata is notably reduced. This could be explained by the destabilization of chiasmata causing the precocious separation of homologs. The alternative is a smaller number of JMs being transformed into COs. Currently, these two possibilities cannot be discriminated. However, given the conserved pro-CO role of Msh4 and Msh5, we consider it likely that a Msh4-Msh5 dimer may promote the maturation of JMs into COs.
The observation that bivalent pairing is closer in a msh4 sgs1 double mutant than in a msh4 single mutant could suggest that Msh4 (together with Msh5) acts as a stabilizing factor of recombination intermediates against the anti-CO activity of Sgs1. Thus, if Sgs1’s anti-CO activity were missing, COs could be formed without the help of Msh4. However, it is also conceivable that Sgs1 dissolves HJs into NCOs and/or undoes aberrant JMs that cannot be resolved by Mus81-Mms4 (or by any other resolvase). In the absence of Sgs1, these unresolved NCO precursors or abnormal intermediates would persist. In the absence of both Sgs1 and Msh4, these intermediates would contribute to the connection of bivalents. Both possibilities are consistent with proposed roles of Sgs1 in budding yeast (Jessop et al. 2006; Oh et al. 2007); however, in the light of the inability of homologous chromosomes to separate in the double mutant, the latter is more likely.
Tetrahymena possesses a partial set of ZMM proteins
The meiotic function of Msh4 and Msh5 homologs in Tetrahymena is unexpected because they are ZMM proteins considered to be part of the class I CO pathway, which is accompanied by SC formation. No homologs to any of the Zip proteins or to Red1 or Hop1, which all contribute to SC formation, have been detected in the Tetrahymena genome (Chi et al. 2014). Also, the ZMM protein Mer3 is missing. Thus, Msh4-Msh5 must work outside of its usual ZMM context. It was suggested that SC components Zip1, -2, and -3 recruit the other ZMM proteins to recombination sites (Shinohara et al. 2008), and it is unclear how a partial ZMM ensemble would be formed in Tetrahymena. However, since Zip1 polymerization, i.e., genuine synapsis, takes place downstream of of the other ZMM-dependent events, it is easy to imagine the origin of CO by a subset of ZMM proteins without SC formation. Moreover, some Msh4 functions need not necessarily depend on other ZMM proteins, since in the fungus Sordaria Msh4 was shown to act independently in prealignment during leptonema (Storlazzi et al. 2010).
Notably, TTHERM_01044360p was recently identified as the best Mlh3 homolog in Tetrahymena (Chi et al. 2014), and its expression pattern (TFGD, http://tfgd.ihb.ac.cn/) is consistent with a function in meiosis. The Mlh1-Mlh3 (Mutγ) complex with the help of Exo1 is believed to resolve JMs as COs in the class I pathway (Zakharyevich et al. 2012). We produced mlh3 knockout strains, but, except for a reduction in viable sexual progeny to 76% compared to the wild type, we did not observe a meiotic defect (File S1). Similarly, we previously found that the formation of bivalents and the segregation of chromosomes and chromatids at anaphase I and II were not affected in a mlh1 mutant (Lukaszewicz et al. 2013). Thus, either the two proteins may not represent functional homologs of Mlh3 and Mlh1, respectively, or they have lost their function as part of a conserved class I pathway.
Information on whether COs are interfering or noninterfering would be crucial for the further elucidation of Tetrahymena CO pathway(s). While the presence of a limited number of COs per bivalent would suggest the action of interference to ensure the obligatory CO, the lack of a marker for COs and uncertainty in the counting of chiasmata have so far precluded the solution of this question. Altogether, despite the presence and the meiotic function of some ZMM protein homologs, we conclude that a canonical class I CO pathway is missing in Tetrahymena.
Noncanonical meiotic models are questioning the two-pathway paradigm
Meiotic COs are believed to be formed via two main pathways, with interfering class I COs being dependent on ZMM proteins and accompanied by SC formation, and noninterfering class II COs involving the endonuclease Mus81-Mms4 in the absence of a SC (see Kohl and Sekelsky 2013 and literature cited therein). The class II pathway is believed to be evolutionarily older; however, most organisms have evolved the more sophisticated class I pathway, which allows a more efficient control of COs, and use both pathways in parallel (Kohl and Sekelsky 2013). There is the possibility of yet another CO pathway because, upon elimination of both the class I and the class II pathway in budding yeast (Abdullah et al. 2004; Argueso et al. 2004) or Arabidopsis (Higgins et al. 2008), residual COs still remained (see also Schwartz and Heyer 2011). The resolution of JMs generated in the class I pathway results in different variants. Budding yeast probably uses MutLγ+Exo1, whereas in Drosophila, class I COs depend on the Rad1/Xpf1/MEI-9 endonuclease (see Kohl and Sekelsky 2013). In C. elegans, a large proportion of class I COs relies on the cooperation of Mus81 and Slx1 (Agostinho et al. 2013; O’Neil et al. 2013; Saito et al. 2013), where Slx1 was proposed to produce nicks in HJs, which makes them a better substrate for cleavage by Mus81 (Agostinho et al. 2013). The use of Mus81 in the generation of interfering COs undermines the paradigm of two separate CO pathways. Also, the two-pathway distinction is blurred by the following recent observation in the mouse: In the absence of Msh4/5-containing pre-CO complexes almost all chiasmata are eliminated, whereas 10% or more are maintained when the downstream Mlh1/3 CO pathway is blocked. This correlation raises the possibility that Msh4/5 is responsible for promoting all meiotic COs, including those derived from recombination intermediates processed by Mus81 (Holloway et al. 2014).
Some organisms may have fully or partially abandoned elements of one pathway during evolution. The fission yeast produces all its COs in the class II pathway, yet its linear elements represent evolutionary relics of a SC (Loidl 2006) and, likely, a once-existing class I pathway. Similarly, the absence of a SC in Tetrahymena is believed to be a derived condition. Since (residual) SCs are present in other representatives of the ciliates, meiotic embellishments may have been reduced during the evolution of alveolates or ciliates (Chi et al. 2014). Yet other class I pathway components, such as MSH4, MSH5, and MLH3, are expressed in meiosis. Hence it is conceivable that an ancient class I CO pathway did not completely disappear and that functions of it have been integrated with the extant (class II?) pathway in Tetrahymena.
Supplementary Material
Acknowledgments
We thank Alexander Lorenz and Rachel Howard-Till for discussion and insightful comments. This work was supported by grant W1238-B20 from the Austrian Science Fund (FWF).
Footnotes
Available freely online through the author-supported open access option.
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.169698/-/DC1.
Communicating editor: M. P. Colaiácovo
Literature Cited
- Abdullah M. F. F., Hoffmann E. R., Cotton V. E., Borts R. H., 2004. A role for the MutL homologue MLH2 in controlling heteroduplex formation and in regulating between two different crossover pathways in budding yeast. Cytogenet. Genome Res. 107: 180–190. [DOI] [PubMed] [Google Scholar]
- Agostinho A., Meier B., Sonneville R., Jagut M., Woglar A., et al. , 2013. Combinatorial regulation of meiotic Holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 9: e1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso J. L., Wanat J., Gemici Z., Alani E., 2004. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168: 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellendir S. P., Sekelsky J., 2013. An elegans solution for crossover formation. PLoS Genet. 9: e1003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L. E., Copenhaver G. P., 2010. Genetic interference: don’t stand so close to me. Curr. Genomics 11: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. K., Zickler D., 2004. Early decision: meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117: 9–15. [DOI] [PubMed] [Google Scholar]
- Boddy M. N., Gaillard P.-H. L., McDonald W. H., Shanahan P., Yates J. R. , et al. , 2001. Mus81-Eme1 are essential components of a Holliday junction. Cell 107: 537–548. [DOI] [PubMed] [Google Scholar]
- Börner G. V., Kleckner N., Hunter N., 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Bruns P. J., Brussard T. E. B., 1981. Nullisomic Tetrahymena: eliminating germinal chromosomes. Science 213: 549–551. [DOI] [PubMed] [Google Scholar]
- Bruns P., Cassidy-Hanley D., 2000. Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 62: 501–512. [DOI] [PubMed] [Google Scholar]
- Chi J., Mahé F., Loidl J., Logsdon J., Dunthorn M., 2014. Meiosis gene inventory of four ciliates reveals the prevalence of a synaptonemal complex-independent crossover pathway. Mol. Biol. Evol. 31: 660–672. [DOI] [PubMed] [Google Scholar]
- Clark N., Wu X., Her C., 2013. MutS homologues hMSH4 and hMSH5: genetic variations, functions, and implications in human diseases. Curr. Genomics 14: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole E., Sugai T., 2012. Developmental progression of Tetrahymena through the cell cycle and conjugation, pp. 177–236 in Tetrahymena thermophila, edited by Collins K. Academic Press, San Diego. [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan, R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver G. P., Housworth E. A., Stahl F. W., 2002. Crossover interference in Arabidopsis. Genetics 160: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T., Hunter N., Lee C., Larkin B., Loidl J., et al. , 2003. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., 2013. Initiation of meiotic recombination: How and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 47: 563–599. [DOI] [PubMed] [Google Scholar]
- De Muyt A., Jessop L., Kolar E., Sourirajan A., Chen J., et al. , 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmsen K. T., Heyer W.-D., 2008. Biochemistry of meiotic recombination: formation, processing, and resolution of recombination intermediates, pp. 91–164 in Recombination and Meiosis. Models, Means, and Evolution, edited by Egel R., Lankenau D.-H. Springer-Verlag, Berlin; Heidelberg, Germany; New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung J. C., Rockmill B., Odell M., Roeder G. S., 2004. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116: 795–802. [DOI] [PubMed] [Google Scholar]
- Hayashi M., Mlynarczyk-Evans S., Villeneuve A. M., 2010. The synaptonemal complex shapes the crossover landscape through cooperative assembly, crossover promotion and crossover inhibition during Caenorhabditis elegans meiosis. Genetics 186: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. D., Armstrong S. J., Franklin F. C. H., Jones G. H., 2004. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 18: 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. D., Buckling E. F., Franklin F. C. H., Jones G. H., 2008. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54: 152–162. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N. M., Brill S. J., 2004. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 18: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway J. K., Sun X., Yokoo R., Villeneuve A. M., Cohen P. E., 2014. Mammalian CNTD1 is critical for meiotic crossover maturation and deselection of excess precrossover sites. J. Cell Biol. 205: 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Till R. A., Lukaszewicz A., Loidl J., 2011. The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. PLoS Genet. 7: e1001359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Rockmill B., Roeder G. S., Lichten M., 2006. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet. 2: 1402–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer K. M., 2000. Tetrahymena genetics: two nuclei are better than one, pp. 127–186 in Tetrahymena thermophila, edited by Asai D. J., Forney J. D. Academic Press, San Diego. [Google Scholar]
- Karrer K. M., 2012. Nuclear dualism, pp. 29–52 in Tetrahymena thermophila, edited by Collins K. Academic Press, San Diego. [Google Scholar]
- Kauppi L., Barchi M., Lange J., Baudat F., Jasin M., et al. , 2013. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 27: 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53. [DOI] [PubMed] [Google Scholar]
- Kelly K. O., Dernburg A. F., Stanfield G. M., Villeneuve A. M., 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H. L., Symington L. S., 2012. Sgs1: the maestro of recombination. Cell 149: 257–259. [DOI] [PubMed] [Google Scholar]
- Klutstein M., Xaver M., Shemesh R., Zenvirth D., Klein F., et al. , 2009. Separation of roles of Zip1 in meiosis revealed in heterozygous mutants of Saccharomyces cerevisiae. Mol. Genet. Genomics 282: 453–462. [DOI] [PubMed] [Google Scholar]
- Kohl K. P., Sekelsky J., 2013. Meiotic and mitotic recombination in meiosis. Genetics 194: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K. P., Jones C. D., Sekelsky J., 2012. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338: 1363–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas N. K., Cohen P. E., 2004. Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenet. Genome Res. 107: 216–231. [DOI] [PubMed] [Google Scholar]
- Lian J., Yin Y., Oliver-Bonet M., Liehr T., Ko E., et al. , 2008. Variation in crossover interference levels on individual chromosomes from human males. Hum. Mol. Genet. 17: 2583–2594. [DOI] [PubMed] [Google Scholar]
- Libuda D. E., Uzawa S., Meyer B. J., Villeneuve A. M., 2013. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 502: 703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Nei M., Ma H., 2007. The origins and early evolution of DNA mismatch repair genes: multiple horizontal gene transfers and co-evolution. Nucleic Acids Res. 35: 7591–7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J., 2006. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma 115: 260–271. [DOI] [PubMed] [Google Scholar]
- Loidl J., Mochizuki K., 2009. Tetrahymena meiotic nuclear reorganization is induced by a checkpoint kinase-dependent response to DNA damage. Mol. Biol. Cell 20: 2428–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J., Scherthan H., 2004. Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J. Cell Sci. 117: 5791–5801. [DOI] [PubMed] [Google Scholar]
- Loidl J., Lukaszewicz A., Howard-Till R. A., Koestler T., 2012. The Tetrahymena meiotic chromosome bouquet is organized by centromeres and promotes interhomolog recombination. J. Cell Sci. 125: 5873–5880. [DOI] [PubMed] [Google Scholar]
- Lukaszewicz A., Howard-Till R. A., Novatchkova M., Mochizuki K., Loidl J., 2010. MRE11 and COM1/SAE2 are required for double-strand break repair and efficient chromosome pairing during meiosis of the protist Tetrahymena. Chromosoma 119: 505–518. [DOI] [PubMed] [Google Scholar]
- Lukaszewicz A., Howard-Till R. A., Loidl J., 2013. Mus81 nuclease and Sgs1 helicase are essential for meiotic recombination in a protist lacking a synaptonemal complex. Nucleic Acids Res. 41: 9296–9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn A., Soucek R., Börner V., 2007. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 15: 591–605. [DOI] [PubMed] [Google Scholar]
- Malik S. B., Pightling A. W., Stefaniak L. M., Schurko A. M., Logsdon J. M., 2008. An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS ONE 3: e2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina O. M., Mazin A. V., Nakagawa T., Kolodner R. D., Kowalczykowski S. C., 2004. Saccharomyces cerevisiae Mer3 helicase stimulates 3′- 5′heteroduplex extension by Rad51: implications for crossover control in meiotic recombination. Cell 117: 47–56. [DOI] [PubMed] [Google Scholar]
- Miao W., Xiong J., Bowen J., Wang W., Liu Y., et al. , 2009. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS ONE 4: e4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. E., Takeo S., Nandanan K., Paulson A., Gogol M. M., et al. , 2012. A whole-chromosome analysis of meiotic recombination in Drosophila melanogaster. G3 (Bethesda) 2: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., 2008. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene 425: 79–83. [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Novatchkova M., Loidl J., 2008. DNA double-strand breaks, but not crossovers, are required for the reorganization of meiotic nuclei in Tetrahymena. J. Cell Sci. 121: 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil N. J., Martin J. S., Youds J. L., Ward J. D., Petalcorin M. I. R., et al. , 2013. Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during Caenorhabditis elegans meiosis. PLoS Genet. 9: e1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. D., Lao J. P., Hwang P. Y.-H., Taylor A. F., Smith G. R., et al. , 2007. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. D., Lao J. P., Taylor A. F., Smith G. R., Hunter N., 2008. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell 31: 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias E., Hamilton E. P., Orias J. D., 2000. Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance, pp. 189–211 in Tetrahymena thermophila, edited by Asai D. J., Forney J. D. Academic Press, San Diego. [DOI] [PubMed] [Google Scholar]
- Osman F., Dixon J., Doe C. L., Whitby M. C., 2003. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12: 761–774. [DOI] [PubMed] [Google Scholar]
- Peoples-Holst T. L., Burgess S. M., 2005. Multiple branches of the meiotic recombination pathway contribute independently to homolog pairing and stable juxtaposition during meiosis in budding yeast. Genes Dev. 19: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Wijeratne A. J., Tomsho L. P., Hu Y., Schuster S. C., et al. , 2009. Characterization of meiotic crossovers and gene conversion by whole-genome sequencing in Saccharomyces cerevisiae. BMC Genomics 10.: 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakshambikai R., Srinivasan N., Nishant K. T., 2013. Structural insights into Saccharomyces cerevisiae Msh4-Msh5 complex function using homology modeling. PLoS ONE 8: e78753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjha L., Anand R., Cejka P., 2014. The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions. J. Biol. Chem. 289: 5674–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A., Qiao H., Yang Y., Chen J. K., Jackson N., et al. , 2013. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., et al. , 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. T., Lui D. Y., Kim H.-M., Colaiácovo M. P., 2013. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet. 9: e1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E. K., Heyer W. D., 2011. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120: 109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E. K., Wright W. D., Ehmsen K. T., Evans J. E., Stahlberg H., et al. , 2012. Mus81-Mms4 functions as a single heterodimer to cleave nicked intermediates in recombinational DNA repair. Mol. Cell. Biol. 32: 3065–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrentino M. E., Chaplais E., Sommermeyer V., Borde V., 2013. Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet. 9: e1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Song X., Bowen J., Corstanje R., Gao Y., et al. , 2002. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 99: 3734–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M., Oh S. D., Hunter N., Shinohara A., 2008. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 40: 299–309. [DOI] [PubMed] [Google Scholar]
- Smith G. R., Boddy M. N., Shanahan P., Russell P., 2003. Fission yeast Mus81·Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics 165: 2289–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T., Acharya S., Butz C., Berardini M., Fishel R., 2004. hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15: 437–451. [DOI] [PubMed] [Google Scholar]
- Storlazzi A., Gargano S., Ruprich-Robert G., Falque M., David M., et al. , 2010. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141: 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover N. A., Krieger C. J., Binkley G., Dong Q., Fisk D. G., et al. , 2006. Tetrahymena Genome Database (TGD): a new resource for Tetrahymena thermophila research. Nucleic Acids Res. 34(Database issue): D500–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai T., Hiwatashi K., 1974. Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in Tetrahymena pyriformis. J. Protozool. 21: 542–548. [DOI] [PubMed] [Google Scholar]
- Thacker D., Mohibullah N., Zhu X., Keeney S., 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature 510: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve A. M., Hillers K. J., 2001. Whence meiosis? Cell 106: 647–650. [DOI] [PubMed] [Google Scholar]
- Wang J., Fan H. C., Behr B., Quake S. R., 2012. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150: 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren J. J., Pohlhaus T. J., Changela A., Iyer R. R., Modrich P. L., et al. , 2007. Structure of the human MutSα DNA lesion recognition complex. Mol. Cell 26: 579–592. [DOI] [PubMed] [Google Scholar]
- Wolfe J., Hunter B., Adair W. S., 1976. A cytological study of micronuclear elongation during conjugation in Tetrahymena. Chromosoma 55: 289–308. [DOI] [PubMed] [Google Scholar]
- Yang S. H., Yuan Y., Wang L., Li J., Wang W., et al. , 2012. Great majority of recombination events in Arabidopsis are gene conversion events. Proc. Natl. Acad. Sci. USA 109: 20992–20997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Tang S., Ma Y., Hunter N., 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J., Duffy J. B., Kemphues K. J., Villeneuve A. M., 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wang S., Yin S., Hong S., Kim K. P., et al. , 2014. Topoisomerase II mediates meiotic crossover interference. Nature 511: 551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.