Abstract
Purpose of review
The purpose of this review is to highlight recent progress in autoantibody detection technologies and describe how these methods are providing novel information and insights into autoimmune disorders.
Recent findings
In recent years, alternative methods such as comprehensive phage display, fluid-phase immunoassays, and antigen microarrays have been developed for autoantigen discovery and profiling autoantibody responses. Compared to classic approaches such as Western blot and ELISA, these methods show improved diagnostic performance, the ability to measure antibody responses to multiple targets, and/or allow for more quantitative analyses. Specific notable findings include uncovering previously unrecognized autoantigens, the improved classification of patient clinical phenotypes, and the discovery of pathogenic autoantibodies promoting disease.
Summary
Advances in immunoassay technologies offer many opportunities for understanding the relationship between autoantibody detection and the myriad complex, clinical phenotypes characteristic of most autoimmune diseases. Further simplification and standardization of these technologies may allow routine integration into clinical practice with improved diagnostic and therapeutic outcomes.
Keywords: Antigen arrays, autoantibodies, autoimmunity, immunoassay, fluid-phase immunoassays, LIPS, pathogenic autoantibodies, phage display
Introduction
The complete spectrum of autoimmune diseases is complex, and the diseases cause significant patient morbidity, mortality and commensurate societal costs. While the exact causes of most autoimmune diseases are not yet know, current evidence suggests that environmental factors trigger these conditions in genetically susceptible individuals. Autoimmune disorders can be difficult to diagnose properly, and the detection of autoantibodies is often a critical determinant of an accurate diagnosis. In fact, many autoimmune diseases, including systemic lupus erythematosus (SLE) and idiopathic inflammatory myopathies (IIM), present clinically as syndromes consisting of multiple, overlapping disease sub-phenotypes sharing certain clinical signs and symptoms. In these instances, the detection of specific autoantibodies may distinguish disease sub-phenotypes, and correlate with differences in therapeutic responses and long-term prognoses. Evidence also suggests that autoantibodies are sometimes produced in patients several years before clinical disease is evident [1–3] making such tests important as early predictors for risk and possible therapeutic intervention aimed at preventing or slowing disease progression.
Immunoassays based on immunofluorescence, Western blotting and ELISA are still commonly used to detect known autoantibody responses, but each of these technologies is limited in specificity, sensitivity and sample throughput. In this review, we describe several, relatively new immunoassay technologies used to investigate autoantibody production and highlight novel findings that influence our understanding and treatment of these complex diseases. Moreover, we review the advantages and disadvantages of these newer approaches and how future advances can promote these methods into routine clinical practice.
Molecular autoantibody discovery using peptides from the entire human proteome
The complete list of informative autoantibody targets associated with different human autoimmune disorders remains unknown. Autoantibody responses range from the production of low-affinity IgM subclasses to more robust production of class-switched, affinity-matured IgG subclasses associated with a strong, autoantigen-driven lymphocyte response. While a better understanding of autoantibody profiles may improve diagnostic accuracy and clinical sub-phenotyping, and may also yield possible insights into disease pathogenesis, there is no bioinformatic approach that reliably predicts associations between candidate autoantigens and autoimmune disorders. To date, most studies describing the identification of novel autoantigens have used numerous methods, including expression cloning, mass spectroscopy, proteomic methods, and the candidate protein approach [4].
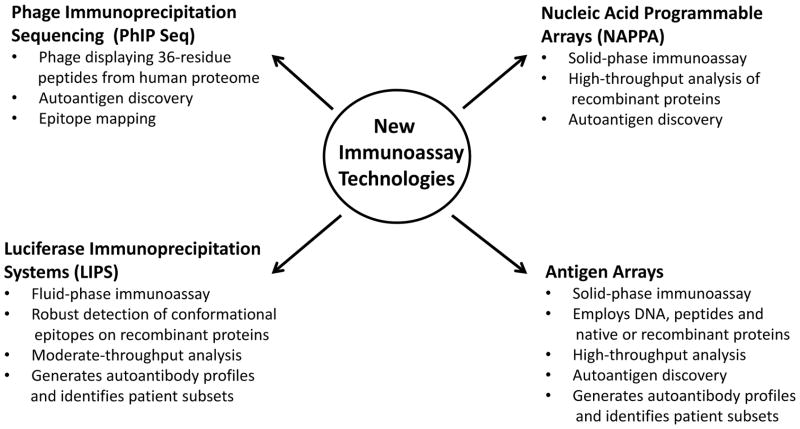
With the completion of the draft human proteome, one new strategy for discovering potential autoantibody targets involves phage immunoprecipitation sequencing (PhIP seq) [5]. This method is based on a T7 bacteriophage system displaying more than 413,000, 36-residue, overlapping peptides of all known, human protein sequences (Fig. 1). The resulting library of phage-expressing human peptides is applied to immobilized autoantibodies derived from patients’ sera. High affinity peptides bound by autoantibodies are subsequently purified by multiple rounds of stringent washing and immunoprecipitation. Bacteriophages encoding these high affinity antigens are then analyzed directly by high-throughput DNA sequencing. Larman et al have used PhIP seq to identify potentially novel autoantigens in patients with encephalitis [5], rheumatoid arthritis (RA) [6], and multiple sclerosis (MS) [6]. Compared to healthy control subjects, RA patients showed enriched autoantibodies against multiple novel target proteins including ADAM33, Honerin and HCN3. Despite these encouraging findings, other established disease autoantigens were not identified in these studies; perhaps consistent with the poor detection of conformational epitopes when using smaller peptide antigens (36 amino acids) [6; 5]. Nevertheless, this approach appears to be a valuable tool for validating potential antigenic epitopes identified in other studies. For example, PhIP seq screening of MS patients’ sera identified a short consensus peptide sequence in multiple human proteins that matched a previously described EBV protein epitope [6]. Conversely, a peptide derived from the proposed Kir4.1 MS autoantigen [7] was not detected [6]. The reason for this discrepancy remains unclear but Nerrant et al. [8] also failed to identify anti-Kir4.1 autoantibodies by ELISA in MS patients, highlighting the challenges of cross-validation of certain autoantigens.
Figure 1. New Autoantibody Detection Technologies.
Several new technologies have emerged over the last decade for identifying novel autoantigens and defining composite autoantibody profiles for autoimmune disease. Several advantages of each technology are listed.
Sporadic inclusion body myositis (IBM) is an increasingly recognized autoimmune condition among the idiopathic inflammatory myositis syndromes, including polymyositis and dermatomyositis, whose differential diagnosis is often difficult [9]. One exciting development using PhIP seq was the characterization of a previously described 43 kDa muscle autoantigen immunoreactive with IBM patient sera [10]. Using a combination of protein sequencing and PhIP seq screening, the cytosolic 5′-nucleosidase 1A (cN1A) was identified as a potential autoantigenic target. Autoantibodies recognizing cN1A were confirmed using recombinant full-length protein by Western blot analysis. Immunoreactive peptides identified by PhIP seq employed in a dot blot immunoassay also showed approximately 70% sensitivity and 92% specificity for the diagnosis of IBM [11]. Additional analysis of archived, pre-clinical serum samples revealed that two of the IBM patients had high titer autoantibodies with cN1A immunoreactivity five and eight years before disease onset. Using mass spectrometry, Pluk et al. also independently and contemporaneously identified cN1A/Mup44 as a diagnostic autoantigen for IBM [12]. Particularly encouraging is the possibility of using the anti-cN1A autoantibody as an alternative test to the otherwise invasive and expensive muscle biopsy generally used for the clinical diagnosis of IBM.
Fluid-phase Immunoassays for Detecting Autoantibodies
Many human autoantibodies are directed against conformational epitopes rather than linear peptide sequences. Solid phase immunoassays such as ELISA show poor diagnostic performance for detecting conformational epitopes from autoantigens [13]. In contrast, fluid- phase radiobinding assays (RBA), wherein the autoantigen is tested in solution rather than immobilized on a solid surface, remains the assay of choice for detecting linear and conformational autoantibodies for multiple autoimmune disorders including type I diabetes (T1D), celiac and thyroid disease [13]. These assays employ radiolabeled proteins that are typically generated by in vitro transcription/translation. One advantage of the RBA is that any protein can be produced in this format including large proteins and proteins that are not commercially available. For example, a recently described target of pathogenic autoantibodies in interstitial lung disease, BPIFB1, was radiolabeled by in vitro transcription/translation and used in immunoprecipitation assays for autoantibody detection [14]. BPIFB1 seropositivity was subsequently detected among 15% of patients with interstitial lung disease and 12% of patients with other idiopathic lung diseases. This study, as well as many others, utilized the RBA for achieving exceptionally high sensitivity and specificity, unlike that associated with conventional ELISA or other solid-phase immunoassays.
Autoantibodies against Znt8, a major autoantigen for type I diabetes (T1D), were studied extensively by RBA [15]; they are poorly detected by solid-phase immunoassay [16]. Interestingly, the detection of Znt8 autoantibodies is largely dependent on specific amino acid polymorphisms present in T1D patients [17]. More specifically, Znt8 protein variants have one of three amino acid differences, arginine (R325), glutamine (Q325), and tryptophan (W325), that are differentially responsive in the RBA and consistent with the corresponding genotype of the patient. For example, T1D patients harboring R325 produce strong signals in the RBA, while Q325 or W325 variants react poorly [17]. Based on these findings, it is possible that the detection of some autoantibodies may be missed because the peptide used in the immunoassay did not contain a necessary structural polymorphism or mutation present in some patients.
Particularly intriguing is a recent report describing the presence of anti-RPC1 autoantibodies in a subset of scleroderma patients with cancer that had somatically acquired mutations within the RPC1-encoding gene, POLR3A [18]. In this study, point mutations or other genetic variations of the POLR3A gene were detected in six of eight patients with anti-RPC1 autoantibodies, but not from RPC1 seronegative patients, suggesting that aberrant structural variations within the protein triggered the humoral immune response. However, patient autoantibodies examined by RBA were found to react equally well with both wild type and variant RPC1 mutant proteins, suggesting that patient autoantibodies are not necessarily specific for the variant region [18]. The full implications of these findings are not yet known, but they imply that structural variations in target antigens may promote autoantibody production in certain autoimmune disorders, and that fluid-phase immunoassays may be required for their for optimum detection.
Despite the high sensitivity of RBA, the requirement for radioactivity is a major barrier to its widespread use. One alternative fluid-phase immunoassay that does not require radioactivity is Luciferase Immunoprecipitation Systems (LIPS) [19]. LIPS is based on generating light-emitting recombinant antigens which are then used in a high-throughput immunoprecipitation format to generate high quality autoantibody binding data (Fig. 1). LIPS has been used to study autoantibodies in a number of autoimmune rheumatologic disorders including Sjögren’s syndrome [20], SLE [21] and the IIM [22]. One distinct advantage of the LIPS technology is the ability to quantitate autoantibody levels over a large dynamic range. The ability of LIPS to yield highly quantitative results against multiple antigen targets allows the potential to differentiate subsets of patients based on autoantibody levels. Using LIPS technology, Ching et al. profiled sixteen autoantigens in SLE patients and found two major autoantibody clusters [21]. In addition, a subgroup of SLE patients with elevated levels of autoantibodies targeting interferon-α (IFN-α), a proinflammatory cytokine, was associated with a more favorable clinical outcome, consistent with a previous report [23]. The exact mechanism regulating the production of these autoantibodies in some subsets of SLE patients is not known, but it is consistent with increased levels of IFN-α production and the corresponding IFN-α gene signature characteristic of some SLE patients [24].
LIPS was also used to study the effect of genetic and environmental factors in disease-discordant twins with SLE and IIM [22]. Autoantibody profiling of eighteen distinct candidate autoantigens revealed that 42% of the disease-affected twins showed significant seropositivity while unrelated, healthy controls in the study appeared seronegative. Eleven of the thirteen affected twins produced elevated levels of autoantibodies directed against two or more autoantigens commonly associated with systemic autoimmune diseases (e.g., Ro52, Ro60, and RNP-70). In contrast, only 10% of the unaffected twins showed seropositivity and were not against known rheumatological target antigens. In the unaffected twins, autoantibodies recognized only a single antigen per subject, and immunoreactivity was directed against autoantigens not typically associated with systemic rheumatic disease. These findings may be attributable in part to non-genetic, environmental influences.
Defining new autoimmune disease phenotypes will require highly specific and sensitive autoantibody detection methods. Particularly relevant is the identification of human diseases where autoantibodies might directly promote disease by binding cell-surface or secreted molecules. Browne et al. used LIPS to study a cohort of patients with disseminated non-tuberculosis mycobacterial infection (dNTM) to understand the role that anti-cytokine autoantibodies might contribute to disease pathogenesis [25]. Patients with dNTM present clinically with numerous infections of both rapid- and slow-growing mycobacteria. Analysis of autoantibodies against over 40 cytokines and other immune targets revealed that 88% of the dNTM patients produced interferon-γ (IFN-γ) autoantibodies that were routinely 1000-fold higher than the levels found in disease controls and healthy blood donors. Moreover, circulating IFN-γ autoantibody levels measured by LIPS in the dNTM patients’ sera correlated well with their ability to neutralize downstream IFN-γ signaling activity by in vitro assays. The cohort of dNTM patients showed little significant autoantibody seropositivity against many of the other cytokines in the LIPS panel. Besides the discovery of autoantibodies to IFN-γ promoting opportunistic infections with mycobacteria, autoantibodies against another cytokine, GMCSF, normally associated with the autoimmune condition alveolar proteinosis [26], have also been associated with opportunistic infections of the central nervous system with Cryptococcus [27; 28]. These recent findings suggest that there are pathogenic anti-cytokine or even anti-cytokine receptor autoantibodies yet to be discovered by LIPS or other immunoassays.
Detecting Autoantibodies by Antigen Microarray Technologies
Antigen microarrays represent another powerful approach for autoantibody detection. Because most patients with autoimmune disorders have heterogeneous phenotypes and divergent autoantibody responses, assays targeting multiple potential autoantigens produce greater disease detection sensitivity. For these arrays, antigenic targets are spotted on a membrane or other solid support, incubated with sera, washed to remove unbound antibody and subsequently imaged using a secondary anti-IgG antibody conjugated to reporter molecules often with a fluorescence-based readout [29]. An important advantage of the antigen microarray approach is the potential to simultaneously measure autoantibody responses to large numbers of potential targets (e.g. 50 to 5000 targets) on a single platform matrix thus allowing for the identification of autoantibody response profiles not generally observable using smaller scale formats.
While it would be theoretically ideal to evaluate autoantibodies against recombinant proteins representing a partial or complete human proteome, arrays produced with several thousand recombinant proteins, generated from either bacterial expression or in vitro transcription/translation systems, have proven less useful than expected in studies of autoimmune disease. Such mass proteomic arrays generally suffer from high non-specific background, multiple false positive signals, and failure to detect known conformational epitopes [30]. For example, nucleic acid programmable protein arrays (NAPPA) can produce large numbers of recombinant proteins using in vitro transcription/translation wherein the corresponding epitope-tagged proteins are directly immobilized onto arrays (Fig. 1). Using NAPPA, novel autoantibodies have been identified in juvenile arthritis [31], ankylosing spondylitis [32] and T1D [16]. Despite the identification of a number of new potential autoantigens, the validation and diagnostic importance of many of these protein reactivities remains to be confirmed by other immunoassay methods. Particularly troubling is the inability of NAPPA to detect several well-established T1D autoantigens including GAD65, IA2 and Znt8, highlighting the difficulty of producing recombinant proteins with conformational epitopes suitable for diagnosis [16].
In contrast to antigen arrays composed entirely of recombinant proteins, arrays utilizing diverse targets such as DNA, peptides and recombinant or native proteins have been more informative [33–36] (Fig. 1). Due to difficulties in obtaining relatively pure proteins, these studies often focused on a defined spectrum of candidate antigens that were obtained from diverse sources, including commercial suppliers, customized recombinant proteins produced in the laboratory and synthetic peptides. One of the first large-scale antigen arrays of this type analyzed autoantibody responses against 196 distinct molecules and identified autoantibody responses to several known autoantigenic targets among eight different autoimmune diseases [36]. Other customized antigen arrays were used to analyze prospective autoantigens in RA [33], explore autoantibodies as potential biomarkers for renal involvement in lupus [34], and examine subsets of patients with lupus or lupus-like signs and symptoms [37]. More recently, Balboni et al. used protein arrays to screen for autoantibodies to a customized set of candidate autoantigens in juvenile dermatomyositis (JDM), a systemic autoimmune disorder of skeletal muscle and skin [38]. Autoantibody responses were evaluated using 80 distinct protein targets derived from 41 different candidate antigens including multiple known rheumatologic markers and antigenic peptides from common human infectious agents such as EBV. Twenty-seven of JDM serum samples (n=36) were immunoreactive with at least one of the 41 candidate autoantigens on the microarray. Although the cutoff value for the different antigens on the array was based on the means plus three standard deviations of the ten controls, no value for specificity was presented for the controls. The autoantibody responses were also correlated with the ability of patients’ sera to induce IFN-α expression using a novel, cell-based reporter system. Interestingly, there was a significant association of seropositive autoantibody responses against several ribonucleoprotein complexes including Ro60, La, and Sm with the induction of IFN-α activity (p=0.03). These findings required large-scale and simultaneous analysis of many different target antigens; an experimental design generally infeasible using traditional ELISA or Western blot approaches.
Using protein arrays, SLE patients were screened for the presence of novel autoantibodies against cytokines, chemokines, and other circulating proteins [35]. For the array, 59 different cytokines and other immunoregulatory molecules combined with 101 known, tissue-derived autoantigen targets were examined. Significant immunoreactivity was detected against a variety of cytokine targets including IL-2, IL-15, TGF-β, TNF, and BAFF. The identification of BAFF autoantibodies in the lupus patients was of particular interest because a previous study had shown that mice overexpressing BAFF developed a SLE-like phenotype and produced autoantibodies [39]. Additional in vitro experiments confirmed that sera from SLE patients with BAFF autoantibodies exhibited neutralizing activity in blocking BAFF-signaling pathways [35]. Further analysis of BAFF seropositive patients from whom adequate clinical information was available, revealed that these SLE patients had a more severe disease course and elevated interferon signatures. Although the mechanistic significance of BAFF autoantibodies in SLE is not known, co-production of anti-BAFF and anti-IFN-α autoantibodies in certain subsets of SLE patients may have important clinical implications. To this end, new clinical trials in SLE patients utilizing neutralizing monoclonal antibodies directed against IFN-α [40] and BAFF [41] have been undertaken. It is likely that the future success of such therapeutic agents will rely upon the accurate, predetermination of autoantibody profiles using antigen arrays or similar technologies.
Conclusion
We have reviewed several new discovery technologies for detecting autoantibodies associated with systemic rheumatic disease. Each of these technologies has distinct advantages and disadvantages compared to traditional detection methods such as ELISA and Western blotting. Phage display technology, while potentially useful for autoantigen discovery, may not be adaptable to routine, clinical testing. Liquid phase immunoassays are limited in size and are not well-suited to an array format required for simultaneously testing large numbers of candidate autoantigens. Clearly, antigen arrays are preferable for screening larger numbers of potential autoantibodies or autoantigens, including those representing DNA, modified peptides or native proteins; however, uncertainty persists about their capacity to detect conformational epitopes for many recombinant proteins. It is also important to emphasize that the development of improved immunoassays remains focused on the discovery of novel autoantibody targets and the relationships between autoantibody profiles and various clinical phenotypes and proposed mechanisms of disease pathogenesis. Since no standardized autoantibody/antigen profiling format has been adopted, the technologies employed remain dependent on the nature of the molecules being studied and the laboratory performing the analysis.
At present, cross-validation of newer versus traditional immunoassay methods is required to assess their reproducibility, specificity and sensitivity. Based upon published findings, it is conceivable that a highly sensitive technology, such as a fluid-phase immunoassay, used to profile less than 100 established autoantigens could prove sufficient for the broad diagnosis of many common autoimmune disorders. Ultimately, such broad autoantibody profiles or signatures might be integrated with other data sets describing, for example, gene expression patterns, SNP variants, and DNA sequences, to provide important mechanistic insights into disease development and progression. The recent identification of specific genetic variations in scleroderma patients in association with anti-RPC1 autoantibodies represents one example of such integrative approaches [18]. With more routine application, it is envisioned that autoantibody profiles will be better utilized in guiding personalized therapy. Autoantibody response profiles have already been used to monitor therapeutic outcomes for several autoimmune disorders including for treating anti-cytokine autoantibodies [42–44]. An extension of this idea would be the use of autoantibody profiles to predict the most efficacious therapy a priori. For example, it is conceivable that patient subsets will be predefined by clinical, genetic and autoantibody characteristics which correlate with disease severity and prove useful in the prescription of individually tailored therapies. Such broad quantitative autoantibody profiling is still in its infancy, but offers the possibility of improving patient outcomes.
Key Points.
Although the full spectrum of autoantigens associated with different human autoimmune disorders remains unknown, new technologies such as PhIP seq and antigen arrays show promise for discovering novel autoantigens.
Fluid-phase immunoassays, such as RBA and LIPS, remain the immunoassay of choice for measuring many autoantibody responses because of their ability to efficiently detect conformational epitopes that are often missed by solid-phase formats.
Antigen arrays testing immunoreactivities against fifty or more targets simultaneously are ideal for generating personalized autoantibody profiles.
Broad quantitative autoantibody profiling technologies are still in development, but will lead to new tools for diagnosis, monitoring, predicting therapeutic responses and understanding disease pathogenesis.
Footnotes
Conflicts of Interest
This research was supported by the Intramural Research Program of the NIH, Division of Intramural Research, National Institute of Dental and Craniofacial Research and National Institute of Environmental Health Sciences. The authors have no conflicts of interest to declare.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of the review, have been highlighted as:
* of special interest
** of outstanding interest.
- 1.Arbuckle MR, McClain MT, Rubertone MV, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349:1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 2.Eriksson C, Kokkonen H, Johansson M, et al. Autoantibodies predate the onset of systemic lupus erythematosus in northern Sweden. Arthritis Res Ther. 2011;13:R30. doi: 10.1186/ar3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonsson R, Theander E, Sjostrom B, et al. Autoantibodies present before symptom onset in primary Sjogren syndrome. JAMA. 2013;310:1854–1855. doi: 10.1001/jama.2013.278448. [DOI] [PubMed] [Google Scholar]
- 4.Robinson WH, Steinman L. Human peptidome display. Nat Biotechnol. 2011;29:500–502. doi: 10.1038/nbt.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larman HB, Zhao Z, Laserson U, et al. Autoantigen discovery with a synthetic human peptidome. Nat Biotechnol. 2011;29:535–541. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larman HB, Laserson U, Querol L, et al. PhIP-Seq characterization of autoantibodies from patients with multiple sclerosis, type 1 diabetes and rheumatoid arthritis. J Autoimmun. 2013;43:1–9. doi: 10.1016/j.jaut.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srivastava R, Aslam M, Kalluri SR, et al. Potassium channel KIR4. 1 as an immune target in multiple sclerosis. N Engl J Med. 2012;367:115–123. doi: 10.1056/NEJMoa1110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nerrant E, Salsac C, Charif M, et al. Lack of confirmation of anti-inward rectifying potassium channel 4.1 antibodies as reliable markers of multiple sclerosis. Mult Scler. 2014 doi: 10.1177/1352458514531086. [DOI] [PubMed] [Google Scholar]
- 9.Hohlfeld R, Engel AG, Goebels N, Behrens L. Cellular immune mechanisms in inflammatory myopathies. Curr Opin Rheumatol. 1997;9:520–526. doi: 10.1097/00002281-199711000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Salajegheh M, Lam T, Greenberg SA. Autoantibodies against a 43 KDa muscle protein in inclusion body myositis. PLoS One. 2011;6:e20266. doi: 10.1371/journal.pone.0020266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11**.Larman HB, Salajegheh M, Nazareno R, et al. Cytosolic 5′-nucleotidase 1A autoimmunity in sporadic inclusion body myositis. Ann Neurol. 2013;73:408–418. doi: 10.1002/ana.23840. Important publication describing the use of Phip Seq for identifying a new major autoantigen (cN1A) in sporadic inclusion myositis. The study also describes diagnostic performance of cN1A autoantibodies and their presence before clinical diagnosis. [DOI] [PubMed] [Google Scholar]
- 12.Pluk H, van Hoeve BJ, van Dooren SH, et al. Autoantibodies to cytosolic 5′-nucleotidase 1A in inclusion body myositis. Ann Neurol. 2013;73:397–407. doi: 10.1002/ana.23822. [DOI] [PubMed] [Google Scholar]
- 13.Liu E, Eisenbarth GS. Accepting clocks that tell time poorly: fluid-phase versus standard ELISA autoantibody assays. Clin Immunol. 2007;125:120–126. doi: 10.1016/j.clim.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Shum AK, Alimohammadi M, Tan CL, et al. BPIFB1 is a lung-specific autoantigen associated with interstitial lung disease. Sci Transl Med. 2013;5:206ra139. doi: 10.1126/scitranslmed.3006998. This study describes the identification a potential new pathogenic lung autoantigen in interstitial lung disease and uses the fluid-phase RBA for analyzing autoantibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wenzlau JM, Hutton JC. Novel diabetes autoantibodies and prediction of type 1 diabetes. Curr Diab Rep. 2013;13:608–615. doi: 10.1007/s11892-013-0405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miersch S, Bian X, Wallstrom G, et al. Serological autoantibody profiling of type 1 diabetes by protein arrays. J Proteomics. 2013;94:486–496. doi: 10.1016/j.jprot.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Wenzlau JM, Liu Y, Yu L, et al. A common nonsynonymous single nucleotide polymorphism in the SLC30A8 gene determines ZnT8 autoantibody specificity in type 1 diabetes. Diabetes. 2008;57:2693–2697. doi: 10.2337/db08-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Joseph CG, Darrah E, Shah AA, et al. Association of the autoimmune disease scleroderma with an immunologic response to cancer. Science. 2014;343:152–157. doi: 10.1126/science.1246886. Outstanding publication identifying genetic mutations as the cause of autoantibodies against RPC1 in scleroderma patients. Studies described combining autoantibodies evaluation with high-throughput sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burbelo PD, Ching KH, Bren KE, Iadarola MJ. Searching for biomarkers: humoral response profiling with luciferase immunoprecipitation systems. Expert Rev Proteomics. 2011;8:309–316. doi: 10.1586/epr.11.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burbelo PD, Leahy HP, Issa AT, et al. Sensitive and robust luminescent profiling of anti-La and other autoantibodies in Sjogren’s syndrome. Autoimmunity. 2009;42:515–524. doi: 10.1080/08916930902911738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ching KH, Burbelo PD, Tipton C, et al. Two major autoantibody clusters in systemic lupus erythematosus. PLoS One. 2012;7:e32001. doi: 10.1371/journal.pone.0032001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gan L, O’Hanlon TP, Gordon AS, et al. Twins discordant for myositis and systemic lupus erythematosus show markedly enriched autoantibodies in the affected twin supporting environmental influences in pathogenesis. BMC Musculoskelet Disord. 2014;15:67. doi: 10.1186/1471-2474-15-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morimoto AM, Flesher DT, Yang J, et al. Association of endogenous anti-interferon-alpha autoantibodies with decreased interferon-pathway and disease activity in patients with systemic lupus erythematosus. Arthritis Rheum. 2011;63:2407–2415. doi: 10.1002/art.30399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wahren-Herlenius M, Dorner T. Immunopathogenic mechanisms of systemic autoimmune disease. Lancet. 2013;382:819–831. doi: 10.1016/S0140-6736(13)60954-X. [DOI] [PubMed] [Google Scholar]
- 25.Browne SK, Burbelo PD, Chetchotisakd P, et al. Adult-onset immunodeficiency in Thailand and Taiwan. N Engl J Med. 2012;367:725–734. doi: 10.1056/NEJMoa1111160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-Dov I, Segel MJ. Autoimmune pulmonary alveolar proteinosis: clinical course and diagnostic criteria. Autoimmun Rev. 2014;13:513–517. doi: 10.1016/j.autrev.2014.01.046. [DOI] [PubMed] [Google Scholar]
- 27.Rosen LB, Freeman AF, Yang LM, et al. Anti-GM-CSF autoantibodies in patients with cryptococcal meningitis. J Immunol. 2013;190:3959–3966. doi: 10.4049/jimmunol.1202526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saijo T, Chen J, Chen SC, et al. Anti-granulocyte-macrophage colony-stimulating factor autoantibodies are a risk factor for central nervous system infection by Cryptococcus gattii in otherwise immunocompetent patients. MBio. 2014;5:e00912–00914. doi: 10.1128/mBio.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharp V, Utz PJ. Technology insight: can autoantibody profiling improve clinical practice? Nat Clin Pract Rheumatol. 2007;3:96–103. doi: 10.1038/ncprheum0404. [DOI] [PubMed] [Google Scholar]
- 30.Burbelo PD, Ching KH, Bush ER, et al. Antibody-profiling technologies for studying humoral responses to infectious agents. Expert Rev Vaccines. 2010;9:567–578. doi: 10.1586/erv.10.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibson DS, Qiu J, Mendoza EA, et al. Circulating and synovial antibody profiling of juvenile arthritis patients by nucleic acid programmable protein arrays. Arthritis Res Ther. 2012;14:R77. doi: 10.1186/ar3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright C, Sibani S, Trudgian D, et al. Detection of multiple autoantibodies in patients with ankylosing spondylitis using nucleic acid programmable protein arrays. Mol Cell Proteomics. 2012;11:M9 00384. doi: 10.1074/mcp.M9.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hueber W, Kidd BA, Tomooka BH, et al. Antigen microarray profiling of autoantibodies in rheumatoid arthritis. Arthritis Rheum. 2005;52:2645–2655. doi: 10.1002/art.21269. [DOI] [PubMed] [Google Scholar]
- 34.Li QZ, Xie C, Wu T, et al. Identification of autoantibody clusters that best predict lupus disease activity using glomerular proteome arrays. J Clin Invest. 2005;115:3428–3439. doi: 10.1172/JCI23587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Price JV, Haddon DJ, Kemmer D, et al. Protein microarray analysis reveals BAFF-binding autoantibodies in systemic lupus erythematosus. J Clin Invest. 2013;123:5135–5145. doi: 10.1172/JCI70231. Important publication describing the use of antigen arrays for identifying anti-BAFF and potentially other cytokine autoantibodies in patients with autoimmune disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson WH, DiGennaro C, Hueber W, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 37.Li QZ, Zhou J, Wandstrat AE, et al. Protein array autoantibody profiles for insights into systemic lupus erythematosus and incomplete lupus syndromes. Clin Exp Immunol. 2007;147:60–70. doi: 10.1111/j.1365-2249.2006.03251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Balboni I, Niewold TB, Morgan G, et al. Interferon-alpha induction and detection of anti-ro, anti-la, anti-sm, and anti-rnp autoantibodies by autoantigen microarray analysis in juvenile dermatomyositis. Arthritis Rheum. 2013;65:2424–2429. doi: 10.1002/art.38038. Study describes the use of antigen arrays for identifying patient subgroups with juvenile dermatomyositis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackay F, Woodcock SA, Lawton P, et al. Mice transgenic for BAFF develop lymphocytic disorders along with autoimmune manifestations. J Exp Med. 1999;190:1697–1710. doi: 10.1084/jem.190.11.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petri M, Wallace DJ, Spindler A, et al. Sifalimumab, a human anti-interferon-alpha monoclonal antibody, in systemic lupus erythematosus: a phase I randomized, controlled, dose-escalation study. Arthritis Rheum. 2013;65:1011–1021. doi: 10.1002/art.37824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace DJ, Stohl W, Furie RA, et al. A phase II, randomized, double-blind, placebo-controlled, dose-ranging study of belimumab in patients with active systemic lupus erythematosus. Arthritis Rheum. 2009;61:1168–1178. doi: 10.1002/art.24699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baerlecken N, Jacobs R, Stoll M, et al. Recurrent, multifocal Mycobacterium avium-intercellulare infection in a patient with interferon-gamma autoantibody. Clin Infect Dis. 2009;49:e76–78. doi: 10.1086/605581. [DOI] [PubMed] [Google Scholar]
- 43.Browne SK, Zaman R, Sampaio EP, et al. Anti-CD20 (rituximab) therapy for anti-IFN-gamma autoantibody-associated nontuberculous mycobacterial infection. Blood. 2012;119:3933–3939. doi: 10.1182/blood-2011-12-395707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macdougall IC, Rossert J, Casadevall N, et al. A peptide-based erythropoietin-receptor agonist for pure red-cell aplasia. N Engl J Med. 2009;361:1848–1855. doi: 10.1056/NEJMoa074037. [DOI] [PubMed] [Google Scholar]