Abstract
Increasing evidence suggests that inflammation is one pathophysio-logical mechanism in Alzheimer's disease (AD). Recent studies have identifiedan association between the poly (ADP-ribose) polymerase 1 (PARP1) gene and AD. This gene encodes a protein that is involved in many biological functions, including DNA repair and chromatin remodeling, and is a mediator of inflammation. Therefore, we performed a targeted genetic association analysis to investigate the relationship between the PARP1 polymorphisms and brain micro-glial activity as indexed by [11C]PBR28 positron emission tomography (PET). Participants were 26 non-Hispanic Caucasians in the Indiana Memory and Aging Study (IMAS). PET data were intensity-normalized by injected dose/total body weight. Average PBR standardized uptake values (SUV) from 6 bilateral regions of interest (thalamus, frontal, parietal, temporal, and cingulate cortices, and whole brain gray matter) were used as endophenotypes. Single nucleotide polymorphisms (SNPs) with 20% minor allele frequency that were within +/− 20 kb of the PARP1 gene were included in the analyses. Gene-level association analyses were performed using a dominant genetic model with translocator protein (18-kDa) (TSPO) genotype, age at PET scan, and gender as covariates. Analyses were performed with and without APOE ε4 status as a covariate. Associations with PBR SUVs from thalamus and cingulate were significant at corrected p<0.014 and <0.065, respectively. Subsequent multi-marker analysis with cingulate PBR SUV showed that individuals with the “C” allele at rs6677172 and “A” allele at rs61835377 had higher PBR SUV than individuals without these alleles (corrected P<0.03), and individuals with the “G” allele at rs6677172 and “G” allele at rs61835377 displayed the opposite trend (corrected P<0.065). A previous study with the same cohort showed an inverse relationship between PBR SUV and brain atrophy at a follow-up visit, suggesting possible protective effect of microglial activity against cortical atrophy. Interestingly, all 6 AD and 2 of 3 LMCI participants in the current analysis had one or more copies of the “GG” allele combination, associated with lower cingulate PBR SUV, suggesting that this gene variant warrants further investigation.
1 Introduction
Alzheimer's disease (AD) is the most common form of dementia and a progressive, degenerative disorder resulting in loss of memory at first, and eventually affecting all cognition and behavior. Increasing evidence suggests that failed or dysregulated immune response is one candidate mechanism contributing to the pathogenesis of AD [1-4]. Recent large-scale genome-wide association studies (GWAS) have identified several candidate genetic variants in CLU, CR1, ABCA7, BIN1, PICALM, CD33, CD2AP, EPHA1 and MS4A6A/MS4A6E in addition to the most robust candidate gene, APOE [5-8]. Several of these genes are known to be involved in immune system functioning [2, 3].
The poly (ADP-ribose) polymerase 1 (PARP1) gene plays roles in many biological functions including chromatin remodeling, DNA repair, telomere maintenance and others and is known to be a mediator of inflammation via regulation of NF-κB and other transcription factors [9]. Several studies have investigated the PARP1 gene in relation to AD [9-12] reporting risk and protective haplotypes [10], enhanced activity of PARP1 in AD brain [12], and association with rate of hippocampal atrophy [11].
The peripheral benzodiazepine receptor (PBR; official name – translocator protein (18kDa), TSPO) is expressed at low levels in relatively inactive microglia. Microglia play an early critical role in activation of the immune response in the central nervous system [13]. Because activated microglia express higher levels of PBR than inactive microglia, PBR has been considered a useful marker to detect neuroinflammation. Positron emission tomography (PET) imaging of the N-acetyl-N-(2-methoxybenzyl)-2-phenoxy-5-pyridinamine ([11C]PBR28) radioligand has shown high specific signal for microglial activity in vivo [14]. The goal of this study was to investigate the relationship between PARP1 gene variation and microglial activity indexed by [11C]PBR28 PET.
2 Materials and Methods
2.1 Participants
In order to reduce the potential bias of population stratification, analyses were restricted to 26 non-Hispanic Caucasian participants from the Indiana Memory and Aging Study (IMAS) cohort. IMAS is an ongoing longitudinal study, including euthymic older adults with subjective cognitive decline (SCD) including memory concerns (e.g., self-perceived decline) in the context of cognitive test performance that is within the normal range, patients with early and late amnestic mild cognitive impairment (EMCI and LMCI) or mild AD, and age-matched cognitively normal controls (NC) without significant cognitive complaints or concerns. Details regarding participant selection criteria and characterization have been described previously [15, 16]. This study was approved by the institutional review board and written informed consent was obtained from all participants. The 26 participants in the study included 7 NC, 6 CC, 4 EMCI, 3 LMCI, and 6 AD. Table 1 shows the sample characteristics. APOE ε2/ε3/ε4 genotype, genome-wide genotyping data, and [11C]PBR28 PET scans were available for all participants. It has been shown that the rs6971 variant in the TSPO gene affects in vivo binding affinity of the [11C]PBR28 ligand [17, 18]. Samples with mixed or high affinity at the TSPO were included in the study; one non-binder was excluded.
Table 1.
Sample Characteristics
| Characteristics | All | NC | SCD | EMCI | LMCI | AD |
|---|---|---|---|---|---|---|
| Number of Samples | 26 | 7 | 6 | 4 | 3 | 6 |
| Age at PET scan (years; mean±SD) | 71.3±7.49 | 68.4±2.64 | 70.3±9.81 | 74.5±6.95 | 72.7±5.69 | 72.7±10.48 |
| Education (years; mean±SD) | 16.4±2.78 | 16.3±1.70 | 17.3±1.21 | 15.5±4.12 | 15.3±3.06 | 16.5±4.18 |
| Gender (M/F) | 9/17 | 1/6 | 2/4 | 2/2 | 2/1 | 2/4 |
| APOE (ε4−/ε4+) | 15/11 | 3/4 | 4/2 | 3/1 | 2/1 | 3/3 |
| TSPO binding affinity (Mixed/High) | 9/17 | 2/5 | 1/5 | 3/1 | 2/1 | 1/5 |
2.2 Data and quality control procedure
Genetic data
Genotyping was performed on genomic DNA from blood using the Illumina HumanOmniExpress BeadChip (Illumina, Inc., San Diego, CA), which contains over 700,000 SNP (single nucleotide polymorphism) markers, according to the manufacturer's protocols (Infinium HD Assay; Super Protocol Guide; Rev. A, May 2008). APOE ε2/ε3/ε4 genotyping was separately performed. All genotype data, including two APOE SNPs (rs429358 and rs7412), underwent standard quality control (QC) assessment using PLINK v1.07 [19] as described previously [20]. SNPs were imputed using the 1000 Genomes reference panel (http://www.1000genomes.org/) following the Enhancing Neuroimaging Genetics through Meta-Analysis 2 (ENIGMA 2) imputation protocol (http://enigma.loni.ucla.edu/wp-content/uploads/2012/07/ENIGMA2_1KGP_v3.pdf [27 July 2012]). Some imputed SNPs were removed based on the following criteria: r2 < 0.5 between imputed and the nearest genotyped SNPs. After all QC steps, 96 SNPs with 20% minor allele frequency that were within +/− 20 kb of the PARP1 gene were included in the analyses.
Imaging data
Dynamic PET scans, acquired on a Siemens HR+, were initiated with injection of ~555 MBq of [11C]PBR28. Data were acquired for 90 min (10x30s, 9x60s, 2x180s, 8x300s, 3x600s). PET data were processed as described previously [18]. In brief, PET data were motion-corrected and normalized to MNI space. Static images were created from data between 40-90 min, and were normalized by injected dose/total body weight to produce standardized uptake value (SUV) images. Regions of interest (ROIs) were generated from each subject's anatomic MRI, which was concurrently acquired on a Siemens Tim Trio using an MPRAGE sequence and post processed using Freesurfer v4.0.1 (http://surfer.nmr.mgh.harvard.edu/). Average [11C]PBR28 SUV values were extracted from 6 bilateral ROIs (thalamus, frontal, parietal, temporal, and cingulate cortices, and whole brain gray matter including cingulate and sensory motor areas) and used as endophenotypes.
2.3 Statistical analyses
In order to investigate the overall influence of PARP1 variants on microglial activity indexed by average [11C]PBR28 SUV values in 6 bilateral ROIs, a set-based analysis method in PLINK was adopted. In brief, this method evaluates the association of individual SNPs in a given set with a given phenotype and selects a set of independent (based on r2 threshold) and significant (based on p threshold) SNPs to represent the overall effect of the set. Then, the significance of the overall set effect is assessed using permutation to correct for multiple SNPs within a set while taking into account the linkage disequilibrium (LD) structure among SNPs. In this study, the analysis was performed using the following settings: (1) r2 threshold: 0.3, (2) p threshold: 0.05, (3) maximum number of independent and significant SNPs: 99999 in order to use all independent and significant SNPs, and (4) number of permutation: 50,000. Due to the limited number of samples, only a dominant genetic model was assessed. Age at PET scan, gender and TSPO binding affinity based on rs6971 genotype were added to the model as covariates. Analysis was performed with and without APOE ε4 status as a covariate.
When more than one independent and significant SNP were identified from significant associations, a subsequent multi-marker analysis was performed using a haplo-type analysis method in PLINK with the same set of covariates in the model. The association p-value was corrected for multiple testing (the number of SNP combinations) using 50,000 permutations. Although the PLINK set-based approach provides the significance of the PARP1 gene and the list of independent and significant SNPs in PARP1, it does not show the joint influence of multiple SNPs on average [11C]PBR28 SUV values. This multi-marker method allowed us to further study the combinatorial effect of multiple SNPs on average [11C]PBR28 SUV values.
3 Results
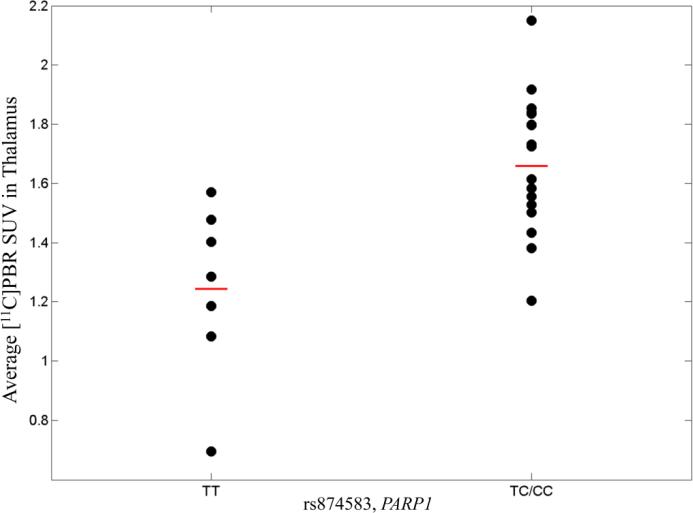
PARP1 variation was associated with average PBR SUV from thalamus at p<0.014 after adjusting for APOE ε4 status. This association was driven by rs874583, located in the intergenic area downstream of the gene. Samples with one or more minor allele (“C”) of rs874583 showed higher SUV in thalamus (Fig.1). Another association with average PBR SUV in cingulate showed marginal significance at p<0.065 after APOE ε4 adjustment and was driven by two SNPs (rs6677172 and rs61835377). Both SNPs are intergenic and downstream of the gene. Minor alleles of these two SNPs (rs6677172: “G”, rs61835377: “A”) showed an inverse relationship with average PBR SUV in cingulate.
Fig. 1.
Average [11C]PBR SUV in thalamus in PARP1 variant, rs874583 (minor allele: C). SUV was adjusted for age at PET scan, gender, TSPO binding affinity, and APOE ε4 status. The horizontal bars represent the mean PBR SUV for each genotype group.
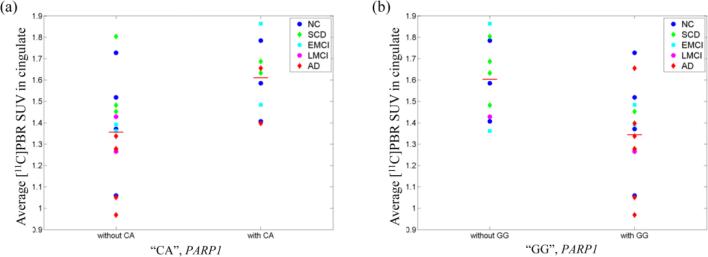
Two SNPs (rs6677172 and rs61835377) ere jointly associated with average PBR SUV in cingulate. Therefore, a subsequent multi-marker analysis was performed to investigate influence of the allele combination of the SNPs on the same phenotype. The analysis identified three different combinations of alleles (“CA”,”CG”, and “GG”), of which two were significantly associated with average PBR SUV in cingulate at uncorrected p<0.05. One (“CA”) was significant after correction for multiple testing at corrected p<0.05. Table 2 summarizes the multi-marker analysis results. “CA” allele combination was positively correlated with the PBR SUV and “GG” allele combination was negatively correlated with the phenotype. Average PBR SUVs in cingulate are displayed in Fig.2 for samples with and without “CA” allele combination (Fig.2 (a)) and with and without “GG” allele combination (Fig.2 (b)). Interestingly, all 6 AD and 2 out of 3 LMCI participants in the current analysis had one or two copies of the “GG” allele combination, associated with lower average cingulate PBR SUV.
Table 2.
Multi-marker analysis results. Allele, F, BETA, P, and Corrected P represent allele combination, frequency of allele combination, regression coefficient, uncorrected p, and corrected p for the number of allele combination, respectively.
| PHENOTYPE | Allele | F | BETA | P | Corrected P |
|---|---|---|---|---|---|
| Average PBR SUV Cingulate | CA | 0.212 | 0.204 | 0.0105 | 0.02962 |
| GG | 0.385 | −0.164 | 0.0241 | 0.06426 |
Fig. 2.
Scatter plots of average PBR SUV in cingulate for (a) “CA” allele group and (b) “GG” allele group. Average PBR SUV was adjusted for age at PET scan, gender, TSPO binding affinity, and APOE ε4 status. The horizontal bars represent the mean PBR SUV for each allele group.
4 Conclusions
This preliminary study investigated the relationship between variation in PARP1 and microglial activity indexed by [11C]PBR PET and identified significant associations of the gene with average PBR SUVs in thalamus and cingulate. The subsequent multi-marker analysis also identified two allele combinations from the gene-based analysis associated with average PBR SUV in cingulate. These identified associations confirmed the role of PARP1 in immune activation. Microglia can perform different functions [1, 13] and the specific role in the current sample of older adults at risk for AD is not known and may include both adaptive and adverse aspects. However, one interesting observation in the current study is that 8 out of 9 participants with AD or LMCI had one or two copies of the “GG” allele combination, which was associated with lower average PBR SUV in cingulate compared to non-“GG” carriers. A previous study with the same cohort showed an inverse relationship between PBR SUV and brain atrophy at a follow-up visit, suggesting a possible protective effect of microglial activity against cortical atrophy [21], which warrants further investigation. A major limitation of this preliminary study is the modest sample size which attenuates power and the findings require replication in larger, independent samples as a future direction. The relationship between PARP1 and microglial activity also warrants experimental molecular validation. Despite the limited sample size, this preliminary study identified interesting significant in vivo associations in an important pathway related to AD pathobiology. This approach combining neuroimaging and genetics data appears promising and can be applied to many related fields of research.
Acknowledgment
This study was supported in part by the National Institutes of Health, National Institute on Aging (R01 AG19771, P30AG10133), National Library of Medicine (R01 LM011360), National Science Foundation (IIS-1117335), NIH Clinical and Translational Sciences Institute Pre-doctoral Training Fellowship (Training Grant TL1 RR025759), and National Library of Medicine (K99 LM011384).
References
- 1.Hickman SE, Allison EK, El Khoury J. Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer's disease mice. J Neurosci. 2008;28:8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones L, Holmans PA, Hamshere ML, Harold D, Moskvina V, Ivanov D, Pocklington A, Abraham R, Hollingworth P, Sims R, Gerrish A, et al. Genetic evidence implicates the immune system and cholesterol metabolism in the aetiology of Alzheimer's disease. PLoS One. 2010;5:e13950. doi: 10.1371/journal.pone.0013950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert JC, Grenier-Boley B, Chouraki V, Heath S, Zelenika D, Fievet N, Hannequin D, Pasquier F, Hanon O, Brice A, Epelbaum J, Berr C, Dartigues JF, Tzourio C, Campion D, Lathrop M, Amouyel P. Implication of the immune system in Alzheimer's disease: evidence from genome-wide pathway analysis. J Alzheimers Dis. 2010;20:1107–1118. doi: 10.3233/JAD-2010-100018. [DOI] [PubMed] [Google Scholar]
- 4.Zhang R, Miller RG, Madison C, Jin X, Honrada R, Harris W, Katz J, Forshew DA, McGrath MS. Systemic immune system alterations in early stages of Alzheimer's disease. J Neuroimmunol. 2013;256:38–42. doi: 10.1016/j.jneuroim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 8.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kauppinen TM, Suh SW, Higashi Y, Berman AE, Escartin C, Won SJ, Wang C, Cho SH, Gan L, Swanson RA. Poly(ADP-ribose)polymerase-1 modulates microglial responses to amyloid beta. J Neuroinflammation. 2011;8:152. doi: 10.1186/1742-2094-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu HP, Lin WY, Wu BT, Liu SH, Wang WF, Tsai CH, Lee CC, Tsai FJ. Evaluation of the poly(ADP-ribose) polymerase-1 gene variants in Alzheimer's disease. J Clin Lab Anal. 2010;24:182–186. doi: 10.1002/jcla.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nho K, Corneveaux JJ, Kim S, Lin H, Risacher SL, Shen L, Swaminathan S, Ramanan VK, Liu Y, Foroud T, Inlow MH, Siniard AL, et al. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.24. doi:10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strosznajder JB, Czapski GA, Adamczyk A, Strosznajder RP. Poly(ADP-ribose) polymerase-1 in amyloid beta toxicity and Alzheimer's disease. Mol Neurobiol. 2012;46:78–84. doi: 10.1007/s12035-012-8258-9. [DOI] [PubMed] [Google Scholar]
- 13.Gehrmann J, Matsumoto Y, Kreutzberg GW. Microglia: intrinsic immuneffector cell of the brain. Brain Res Brain Res Rev. 1995;20:269–287. doi: 10.1016/0165-0173(94)00015-h. [DOI] [PubMed] [Google Scholar]
- 14.Kreisl WC, Fujita M, Fujimura Y, Kimura N, Jenko KJ, Kannan P, Hong J, Morse CL, Zoghbi SS, Gladding RL, Jacobson S, Oh U, Pike VW, Innis RB. Comparison of [(11)C]-(R)-PK 11195 and [(11)C]PBR28, two radioligands for translocator protein (18 kDa) in human and monkey: Implications for positron emission tomographic imaging of this inflammation biomarker. Neuroimage. 2010;49:2924–2932. doi: 10.1016/j.neuroimage.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Risacher SL, Wudunn D, Pepin SM, MaGee TR, McDonald BC, Flashman LA, Wishart HA, Pixley HS, Rabin LA, Pare N, Englert JJ, Schwartz E, Curtain JR, West JD, O'Neill DP, Santulli RB, Newman RW, Saykin AJ. Visual contrast sensitivity in Alzheimer's disease, mild cognitive impairment, and older adults with cognitive complaints. Neurobiol Aging. 2013;34:1133–1144. doi: 10.1016/j.neurobiolaging.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saykin AJ, Wishart HA, Rabin LA, Santulli RB, Flashman LA, West JD, McHugh TL, Mamourian AC. Older adults with cognitive complaints show brain atrophy similar to that of amnestic MCI. Neurology. 2006;67:834–842. doi: 10.1212/01.wnl.0000234032.77541.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owen DR, Yeo AJ, Gunn RN, Song K, Wadsworth G, Lewis A, Rhodes C, Pulford DJ, Bennacef I, Parker CA, StJean PL, Cardon LR, Mooser VE, Matthews PM, Rabiner EA, Rubio JP. An 18-kDa translocator protein (TSPO) polymorphism explains differences in binding affinity of the PET radioligand PBR28. J Cereb Blood Flow Metab. 2012;32:1–5. doi: 10.1038/jcbfm.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoder KK, Nho K, Risacher SL, Kim S, Shen L, Saykin AJ. Influence of TSPO genotype on [11C]PBR28 standardized uptake values. Journal of nuclear medicine. 2013 doi: 10.2967/jnumed.112.118885. doi:10.2967/jnumed.112.118885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, Foroud T, Pankratz N, Moore JH, Sloan CD, Huentelman MJ, Craig DW, Dechairo BM, Potkin SG, Jack CR, Jr., Weiner MW, Saykin AJ. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. Neuroimage. 2010;53:1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risacher SL, Kim S, Yoder KK, Shen L, West JD, McDonald BC, Wang Y, Nho K, Tallman E, Hutchins GD, Fletcher JW, Ghetti B, Gao S, Farlow MR, Saykin AJ. Relationship of microglial activation measured by [11C]PBR28 PET, atrophy on MRI, and plasma biomarkers in individuals with and at-risk for Alzheimer's disease. Alzheimer's Association International Conference 2013. 2013 (Abstract number: 39417) [Google Scholar]