Abstract
Purpose
The goals of this study were to optimize radiolabeling of renal lineages differentiated from human embryonic stem (hES) cells and use noninvasive imaging (positron emission tomography (PET) and bioluminescence imaging (BLI)) to detect the cells in fetal monkeys post-transplant.
Procedures
hES cells expressing firefly luciferase (5×106) were radiolabeled with the optimized concentration of 10 μCi/ml 64Cu-PTSM then transplanted under ultrasound guidance into early second trimester fetal monkey kidneys. Fetuses were imaged in utero with PET and tissues collected for analysis 3 days post-transplant. Fetal kidneys were imaged ex vivo (PET and BLI) post-tissue harvest, and serial kidney sections were assessed by PCR for human-specific DNA sequences, fluorescent in situ hybridization (FISH) for human-specific centromere probes, and immunohistochemistry (IHC) to assess engrafted cells.
Results
Transplanted cells were readily imaged in vivo and identified at the site of injection; tissue analyses confirmed the imaging findings. Using a semi-quantitative method, one in approximately 650 cells in the kidney was shown to be of human origin by PCR and FISH.
Conclusions
These studies suggest that hES cells differentiated toward renal lineages can be effectively radiolabeled, transplanted into fetal monkey kidneys under ultrasound guidance, monitored with PET post-transplant, and identified by PET, BLI, PCR, FISH, and IHC post-tissue harvest.
Keywords: PET, Bioluminescence imaging, Human embryonic stem cells, Fetal transplant, Rhesus monkey
Introduction
Congenital anomalies of the kidney are responsible for the majority of chronic renal failure and end-stage renal disease in young children, with congenital obstruction of the urinary tract the most common [1]. Data have shown little improvement in the 5-year survival among pediatric dialysis patients, with lowest survival in the youngest patients [2]. Congenital obstruction of the urinary tract during active nephrogenesis results in a well-described pattern of histopathological changes, with architectural disorganization and the development of immature glomeruli, primitive tubules surrounded by fibromuscular collars, interstitial fibrosis and mesenchymal expansion, and cystic transformation of tubules with injury to the developing collecting duct epithelium [3–5]. To better understand the pathogenesis of fetal obstructive nephropathy, we developed a rhesus monkey model of unilateral ureteric obstruction [6], have characterized the histopathological features of this model, and demonstrated that it very closely recapitulates obstructive uropathy in the human [4–8].
Investigations have focused on renal ontogeny in the monkey and the use of different stem and progenitor cell populations for novel renal transplant protocols [9, 10]. In vivo imaging technologies are important to develop for assessing the safety and efficiency of cell-based therapies for future human application and have included positron emission tomography (PET), bioluminescence imaging (BLI), and magnetic resonance imaging (MRI) [11–16]. Preclinical studies have advanced these applications to human clinical trials where in vivo imaging techniques have been effectively used including PET and MRI [17, 18]. Thus, one of the goals of this study was to initially assess the radiolabeling efficiency of differentiated human embryonic stem (hES) cells to identify the optimal dose of 64Cu-pyruvaldehyde-bis(N4-methylthiosemicarbazone) (PTSM) using our previously reported methods [19] and to show that cells differentiated toward renal lineages could be effectively radiolabeled without adverse effects. Studies then focused on determining if transplanted cells could be identified in vivo with BLI and PET and using established imaging techniques [20].
Materials and Methods
Animals
All animal procedures conformed to the requirements of the Animal Welfare Act, and protocols were approved prior to implementation by the Institutional Animal Care and Use Committee at the University of California, Davis. Normally cycling, adult female rhesus monkeys (Macaca mulatta) (N=3) with a history of prior pregnancy were bred and identified as pregnant using established methods [21]. Pregnancy in the rhesus monkey is divided into trimesters by 55-day increments with 0–55 days gestation representing the first trimester, 56–110 days gestation representing the second trimester, and 111–165 days gestation the third trimester (term 165±10 days). Activities related to animal care (diet, housing) were performed according to Primate Center standard operating procedures. Tissue harvests (N=3) were performed using established protocols (see below).
Cell Culture and Transduction
A karyotypically normal hES cell line (HSF-6, passage 37) was cultured on a monolayer of irradiated mouse embryonic fibroblasts in high-glucose Dulbecco’s modified Eagle medium (DMEM) containing 20% Knockout™ Serum Replacement, 2 mM L-glutamine, 0.1 mM MEM, nonessential amino acids solution, 4 ng/ml recombinant human fibroblast growth factor basic (Invitrogen Corp., Carlsbad, CA), and 0.1 mM β-mercaptoethanol (Sigma, St. Louis, MO) as previously described [9]. Cells were transduced with an HIV-1-derived lentiviral vector expressing firefly luciferase under the control of the MND promoter at 1×107 infectious units/ml overnight [22]. Cells were washed with phosphate-buffered saline (PBS), replenished with fresh medium, and expanded to establish a bank of hES cells expressing firefly luciferase for all experiments. A subset of the bank was assessed by real-time polymerase chain reaction (RT-PCR) (OCT-4) and immunofluorescence analysis (Tra-1-60, Tra-1-81, SSEA-3, SSEA-4; not shown) [9]. Cells were also evaluated for the ability to differentiate into the three primary germ layers by RT-PCR (endoderm, α-fetoprotein, and transthyretin; mesoderm, Brachyury, and Flk-1; and neural ectoderm, NCAM) as well as a normal karyotype. Expression of firefly luciferase was determined by adding 100 μg/ml of D-luciferin to cells and imaging with the IVIS200® Imaging System (Caliper, Hopkinton, MA). Once cells were shown with a normal karyotype and documented to express luciferase, hES cell colonies were removed from underlying feeder layers by collagenase treatment and cultured in high-glucose DMEM medium containing 10% fetal bovine serum (Hyclone, Logan, UT), 1% Penicillin–Streptomycin, 1% L-glutamine, 100 μM 1-thioglycerol (Sigma) supplemented with 0.1 μM trans-Retinoic Acid (EMD Biosciences, San Diego, CA), 10 ng/ml Activin-A (R&D Systems, Minneapolis, MN), and 50 ng/ml recombinant human BMP-7 (R&D Systems) as previously described [9].
64Cu-PTSM Radiolabeling
hES cells expressing firefly luciferase were dissociated using trypsin–EDTA (Invitrogen), counted, and plated in a 96-well plate in triplicate at 1.0×105, 5.0×104, 2.5×104, 1.3×104, 6.3×103, 3.1×103, and 1.6×103 cells per well. After adding 100 μg/ml of D-luciferin, the plate was imaged using BLI. Photon emission was quantified as described below. Experiments were conducted four times.
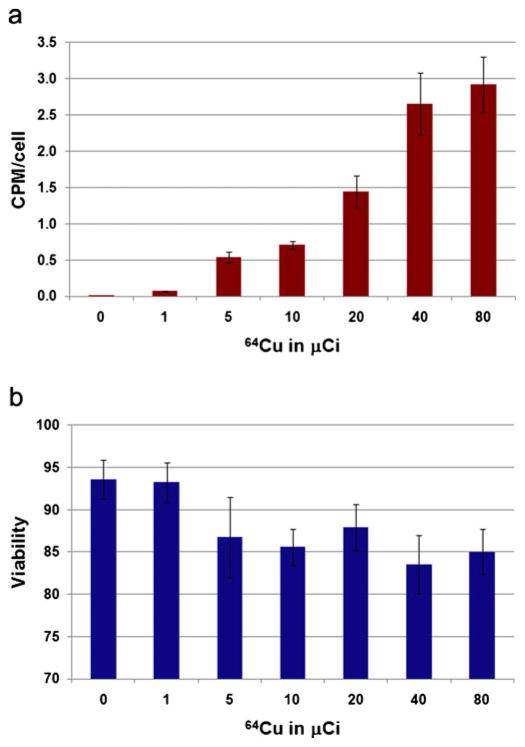
64Cu was produced by cyclotron irradiation of 64Ni at the Washington University at St. Louis Medical School, Department of Radiological Science using established methods, and the 64Cu-PTSM conjugation was performed as previously described [19]. These prior studies have also shown that nonradioactive copper does not negatively impact the cells, and that different cell populations may have a different 64Cu radiolabeling efficiency. Differentiated hES cells prepared as noted above were radiolabeled with 64Cu-PTSM at 0 to 80 μCi/ml in 1 ml of culture medium at 37°C, 5% CO2 conditions for 3 h. After incubation, all free 64Cu-PTSM was removed by washing three times with PBS. Cells radiolabeled with 64Cu were counted for viability, with the degree of radiolabeling assessed using the Wallac Wizard 1470 gamma counter (Perkin-Elmer, Shelton, CT; Fig. 1) and cultured further for 7 days to evaluate growth and gene expression (NCAM, Brachyury, FLK-1, AFP, TTN, PAX2, WT-1, CD24, OSR1) by RT-PCR (Fig. 2).
Fig. 1.
Labeling of hES cells with 64Cu-PTSM. Efficiency of radiolabeling of hES cells with 64Cu-PTSM was dependent on concentration (a), and viability was noted to decrease with concentrations greater than 10 μCi 64Cu-PTSM (b). Mean± standard deviation (N=4 experiments).
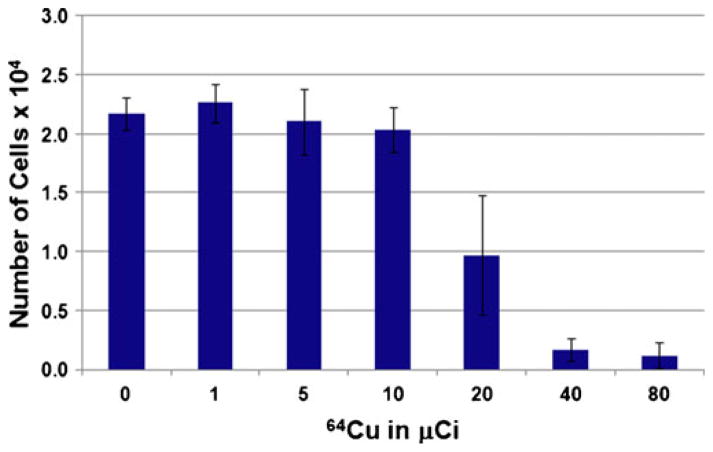
Fig. 2.
Growth of hES cells labeled with 64Cu-PTSM. Cells radioloabeled with 64Cu-PTSM were cultured for 7 days to evaluate growth and expression of genes important in kidney development. At radiolabeling concentrations greater than 10 μCi, cell proliferation was noted to decline. No differences in gene expression were noted across the range of 64Cu-PTSM concentrations tested (not shown). Mean±standard deviation (N=4 experiments).
Transplantation and Tissue Harvest
All fetuses were sonographically assessed to confirm normal growth and development prior to cell transplantation under ultrasound guidance [21]. The dams were administered ketamine hydrochloride (10 mg/kg, intramuscular (IM)) for these and subsequent ultrasound examinations. On the day of cell transplantation, the dams (N=3) were administered Telazol (5–8 mg/kg, IM) and were aseptically prepared for transabdominal ultrasound-guided fetal cell transplantation. A total volume of approximately 50 μl with 5×106 differentiated hES cells expressing firefly luciferase and radiolabeled with 10 μCi/ml of 64Cu-PTSM in suspension was injected under aseptic conditions into fetal kidneys using established techniques at 85±5 days gestation (mid-second trimester) and using a 25-gauge spinal needle attached to a 1-ml syringe [21]. Imaging was performed as described below and hysterotomies scheduled for fetal tissue harvests 3 days post-transplant using established methods [22]. D-luciferin was injected into the fetal circulation (100 mg/kg) for BLI. Kidney length, width, and weight were assessed post-imaging, and each kidney was divided in half and snap frozen after embedding in OCT embedding compound (Sakura, Japan) for immunohistochemistry (IHC) and fluorescent in situ hybridization (FISH).
microPET and BLI
The microPET P4 imaging system (Siemens Preclinical Solutions, Inc., Malvern, PA) has a 22-cm bore, 20-cm transaxial field of view, and 8-cm axial field of view, and sensitivity of the unit is 2.25% at the center of the field of view with an energy window of 250–750 keV and a timing window of 10 ns (default values) [20]. With maximum a posteriori reconstruction incorporating an accurate system model (standard reconstruction algorithm used), image resolution is ~1.8 mm isotropically (6-μl volumetric resolution). Levels of PET signal were analyzed with AMIDE software. Average counts were obtained from each slice and converted into radioactivity (with 6,448.48 applied as a constant to calculate microcurie. Immediately post-transplant fetuses were imaged in vivo with PET. The anatomical position of the fetus in relation to the dam and the fetal kidneys was determined by ultrasound prior to PET imaging to provide the anatomical information needed to determine the PET scanning bed position. PET imaging was repeated 24 and 48 h post-transplant in Telazol-sedated animals (15 min per scan). Three days post-transplantation, fetal tissue harvests were performed using established methods [22], and kidneys and other organs were imaged with BLI followed by PET. Bioluminescence and photographic images were superimposed using Living Image 2.50 software. Regions of interest (ROI) were defined by selecting areas with bioluminescence. Numbers of total photons per second per centimeter detected in ROI were recorded. Kidneys were embedded in OCT embedding compound and serial sectioned for IHC and FISH to detect human cells.
Real-Time PCR
Serial sections (6 μm) of kidneys embedded in OCT were obtained using a cryostat (Leica CM 3050S, Leica Microsystems Inc., Bannockburn, IL). Genomic DNA was extracted from every fifth section after washing with PBS and using the Gentra Puregene Tissue kit (Qiagen, Valencia, CA) as recommended by the manufacturer to identify the sections for IHC and FISH. Other sections were mounted on poly-L-lysine-coated (0.01%, Sigma) Superfrost®/Plus slides (ThermoFisher Scientific, Pittsburgh, PA) and stored at ≤−20°C. For quantitative PCR analysis, primers were designed for the human MAS-related GPR, member X4 (MRGPRX4) gene with Primer Express software (Perkin-Elmer, Foster City, CA). The primers for the human MRGPRX4 sequence were as follows: forward 5′-CCGTCCCAGTCTTCGGTACA-3′, reverse 5′-CAGTCCGACAAGGGAAATGATG-3′, and probe 5′-FAM-CAACGGACGTGAGGAGACTCCTTGCTA-TAMSp-3′. The ε-globin system was utilized as an internal control (housekeeping gene) for DNA isolation and PCR reactions [22]. Primer sequences for ε-globin were as follows: forward 5′-TGGCAAG GAGTTCACCCCT-3′, reverse 5′-AATGGCGACAGCAGA CACC-3′, and probe 5′-FAM-TGCAGGCTGCCTGGCAGAAGC-TAMSp-3′. Quantitative RT-PCR analysis was carried out in 96-well optical plates using the 7900 ABI Sequence Detection System (Applied Biosystems, Foster City, CA) and the TaqMan Universal PCR Master Mix (Applied Biosystems) according to the manufacturer’s protocols [22]. PCR reactions were run in duplicate in separate wells and contained 1× TaqMan universal master mix with 400 nM of forward and reverse primers and 100 nM probe in a 25-μl reaction volume. The PCR protocol consisted of one cycle of 2 min at 50°C, 15 min at 95°C, followed by 40 cycles at 15 s at 95°C, and 60 s at 60°C.
IHC and FISH Double Staining
Kidney sections adjacent to those identified positive for human-specific DNA sequences by PCR were rinsed in PBS for 5 min, fixed in a 3:1 solution of ethanol/acetic acid (ThermoFisher Scientific) for 10 min at ≤−20°C, air dried, then washed in PBS for 5 min. Sections were then incubated in 2× SSC (ThermoFisher Scientific) for 10 min, treated with RNase (100 μg/ml, Sigma) for 30 min at 37°C, washed for 5 min each in 2× SSC and PBS, dehydrated through a graded series of ethanol, and air dried. Sections were then denatured in 70% formamide (Sigma)/2× SSC for 5 min at 65°C, and then dehydrated through a graded series of ethanol. Pan Centromeric Chromosome Paint probe was denatured at 85°C for 10 min and immediately chilled on ice, and then 12 μl of the denatured probe was placed directly on the sections. Sections were covered with hybridization coverslips (Research Products International Corp., Mt. Prospect, IL) and hybridized in a humidified chamber at 37°C for 16–18 h. After incubation, the sections were washed one time in 2× SSC (at 37°C), twice in 50% formamide/2× SSC for 10 min each at 37°C, and rinsed in 2× SSC at room temperature. Sections were blocked in a solution of 10% goat serum (Sigma-Aldrich) in 1% BSA (Sigma) for 1 h at room temperature and then incubated overnight at 4°C with the following primary antibodies in blocking solution: monoclonal mouse anti-human vimentin (clone RV202, 1:500 dilution, BD, San Jose, CA), polyclonal rabbit anti-human PAX2 (1:250 dilution, Invitrogen), monoclonal mouse anti-human WT1 (clone 6F-H2, prediluted, Invitrogen), and polyclonal rabbit pan-cytokeratin (1:500 dilution, Abcam, Cambridge, MA), all previously tested and known to only cross-react with human cells, with no cross-reactivity with monkey cells. Sections were washed with PBS for 5 min two times and incubated with goat anti-rabbit or mouse Alexa Fluor® 594 secondary antibodies (1:1,000, Invitrogen) for 2 h at room temperature. After two washes with PBS for 5 min, a coverslip was placed on the slides with ProLong Gold® antifade reagent with DAPI (Invitrogen), and sections were examined with an Olympus BX61 fluorescent microscope (Olympus America Inc., Melville, NY).
Results
Radiolabeling of Differentiated hES Cells Expressing Firefly Luciferase with 64Cu-PTSM
Our prior studies have shown the efficiency of retinoic acid, Activin-A, and BMP-7 (termed “RA7”) for differentiating hES cells towards renal lineages [9]. For radiolabeling studies (N=4), established banks of hES cells transduced with a lentiviral vector expressing firefly luciferase were used. Cells were cultured overnight and incubated with 1 ml of medium containing various concentrations (0, 1, 5, 10, 20, 40, or 80 μCi) of 64Cu-PTSM for 3 h. Cells were washed with PBS, counted for radioactivity, and then viability and proliferation were assessed. Uptake of 64Cu-PTSM (counts per minute per cell (cpm/cell)) was found to increase with increasing concentration; 0, 1, 5, 10, 20, 40, and 80 μCi incubations resulted in 0.0049, 0.099, 0.72, 1.1, 3.6, 7.2, and 7.6 cpm/cell, respectively, and a dose-dependent labeling efficiency was observed (Fig. 1a). A decrease in cell viability was noted with increasing 64Cu-PTSM concentrations (Fig. 1b). The modest decline in cell viability observed with 5–80 μCi was likely due to the increase in copper concentration since trypan blue stains cells based on the integrity of the cell membrane, supporting that the findings were not likely related to radiotoxicity. An increase in radiolabeling efficiency was observed between 10 and 40 μCi, which paralleled a significant decline in cell proliferation. These findings suggested that radiotoxicity occurred at doses >10 μCi.
Cells radiolabeled with 64Cu-PTSM were also evaluated for growth and gene expression (NCAM, Brachyury, FLK-1, AFP, TTN, PAX2, WT1, CD24, OSR1, and housekeeping gene). Cell growth declined at radiolabeling concentrations >10 μCi 64Cu-PTSM (Fig. 2). No differences in expression of genes associated with differentiation were noted across the range of concentrations of 64Cu-PTSM tested. Based on these findings, 10 μCi was determined to be the optimal dose for the labeling and imaging studies. These findings are important because they show for the first time successful methods for radiolabeling hES cells for PET imaging in vitro without effects on growth, proliferation, and key genes.
64Cu-PTSM and Firefly Luciferase for PET Short-Term Tracking
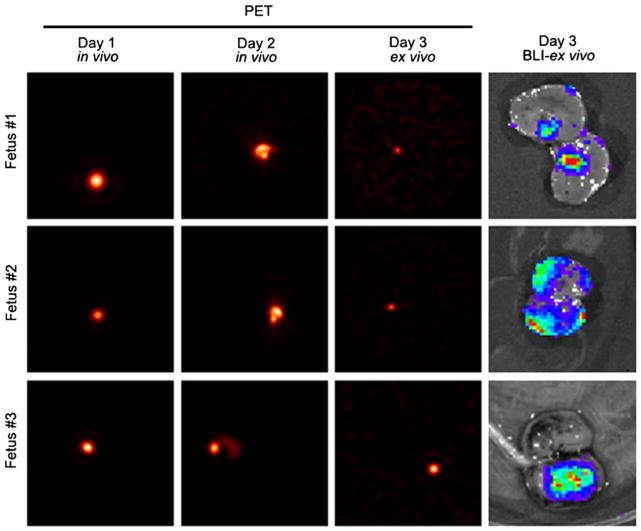
hES cells (5×106 differentiated cells radiolabeled with 10 μCi 64Cu-PTSM) were transplanted into second trimester fetal rhesus monkey kidneys under ultrasound guidance using established methods [6, 21]. For each of the fetuses, a specific location in the kidney was chosen for injection of the cells (e.g., cortex or medulla). The fetal position in relation to the dam was documented for anatomical detail by ultrasound, and then immediately post-transplant the dam was placed on the PET scanning bed for fetal imaging. The ultrasound analysis of fetal position was repeated at 24 and 48 h post-transplant in Telazol-sedated dams for subsequent fetal PET imaging. Fetal kidneys were readily identified by PET in vivo in transverse, coronal, and sagittal planes at all scanning sessions of the dam (Fig. 3, first two panels (days 1 and 2]). On day 3 post-transplant, fetal tissue harvests were performed for ex vivo imaging; the timing of tissue harvest was based on the half-life of 64Cu-PTSM (12.7 days) [19]. Kidneys and other tissues were imaged ex vivo post-collection with the IVIS200® Imaging System first to confirm that only the kidneys showed a positive imaging outcome. Kidneys were then imaged ex vivo with PET using the same orientation and parallel images obtained. Three examples of in vivo (first two panels—days 1 and 2) and ex vivo imaging outcomes (third panel, PET—day 3; fourth panel, BLI—day 3) are shown (Fig. 3). These studies clearly indicated that we can effectively transplant radiolabeled differentiated hES cells expressing firefly luciferase under ultrasound guidance into defined anatomical locations of the fetal kidney in utero, monitor the fetuses in vivo with PET, identify the cells within the transplanted fetal kidney when scanning the dam, and obtain correlative data with BLI and PET ex vivo on the day of tissue harvest. The in vivo imaging also confirmed that the cells were localized within the kidney precisely where they had been placed (e.g., cortex, medulla) and with no evidence of trafficking to other anatomical sites.
Fig. 3.
PET in vivo imaging. PET (in vivo images of the dam; transverse plane, days 1 and 2; ex vivo day 3) and BLI of transplanted differentiated hES cells radiolabeled with 64Cu-PTSM and expressing firefly luciferase (ex vivo day 3). PET images of the gravid dam (fetus in utero #1, #2, #3) showed a focal density indicating the cells within the fetal kidney (PET; left panels day 1, day 2, and day 3 ex vivo). BLI ex vivo clearly showed the transplanted cells expressing firefly luciferase in defined areas of the fetal kidney (right, day 3 BLI). Fetus #1 (cells transplanted near the hilum), Fetus #2 (cells transplanted within the cortex), and Fetus #3 (cells transplanted within the medulla) were all clearly shown with BLI ex vivo on day 3.
Molecular and Histological Analysis of Transplanted Cells
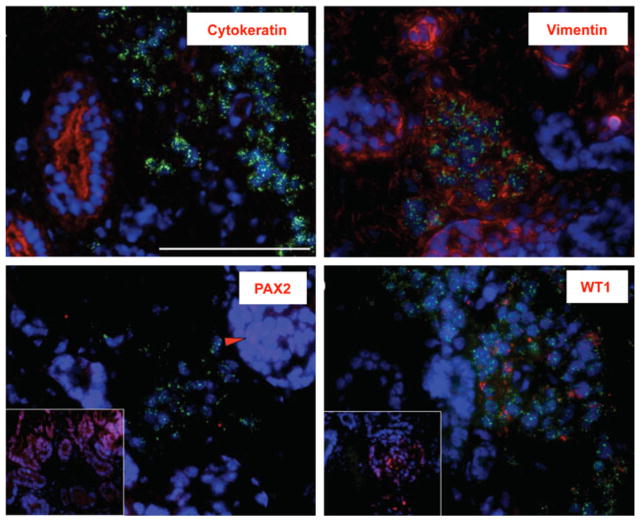
Transplanted and contralateral (control) kidneys were collected at tissue harvest, embedded in OCT, sectioned serially, and evaluated for the presence of the human MRGPRX4 gene by RT-PCR. Transplanted kidney sections were found to contain 76.7 copies of MRGPRX4 per 50,000 copies ε-globin (housekeeping gene), equivalent to approximately one human cell per 650 monkey kidney cells. To further characterize transplanted cells, sections adjacent to those found to be positive for human genes by PCR were analyzed by FISH using a human-specific centromere probe (does not cross-react with monkey) followed by IHC for antigens consistent with kidney differentiation including cytokeratin, vimentin, PAX2, and WT1. Transplanted human cells identified by green nuclear fluorescence (FISH) were negative for cytokeratin and PAX2 and shown to be positive for vimentin and WT1 (Fig. 4) in a staining pattern consistent with metanephric mesenchyme.
Fig. 4.
Analysis of hES cell localization and phenotype. All images were captured at 40× magnification and oriented with the renal cortex at the top left and medulla at bottom right. Blue DAPI nuclear localization, green human centromere detection, red antibodies including cytokeratin, vimentin, PAX2 (inlay shows cortical PAX2 as light red/pink staining within the blue nuclear staining; arrowhead) supporting a typical expression pattern of nuclear localization, and WT1 (inlay shows cortical staining with typical expression pattern of combined nuclear and cytosolic localizations). Scale bar=100 μm.
Discussion
The development of new stem and progenitor cell therapies requires noninvasive methods to assess the dynamic survival, fate, and engraftment of transplanted cells. Although cell fate can be determined using histopathologic techniques, methods are needed to monitor the cells in real time, particularly when considering applications in humans. Ideal noninvasive imaging technologies should be nontoxic, have no effect on gene expression or the functional capabilities of transplanted cells, be highly sensitive, and permit in vivo imaging over extended periods of time. Our prior studies in monkeys have demonstrated the feasibility of utilizing BLI and PET to monitor transgene expression longitudinally and for many years after fetal gene transfer [20, 23, 24]. In the studies described herein, we have identified an optimized 64Cu-PTSM radiolabeling concentration for a sensitive hES cell population and have utilized PET and BLI for in vivo short-term tracking of transplanted cells in fetal kidneys. Our results demonstrate that differentiated hES cells can be transplanted and readily monitored in vivo, establishing a valuable method for monitoring survival and fate immediately post-transplantation.
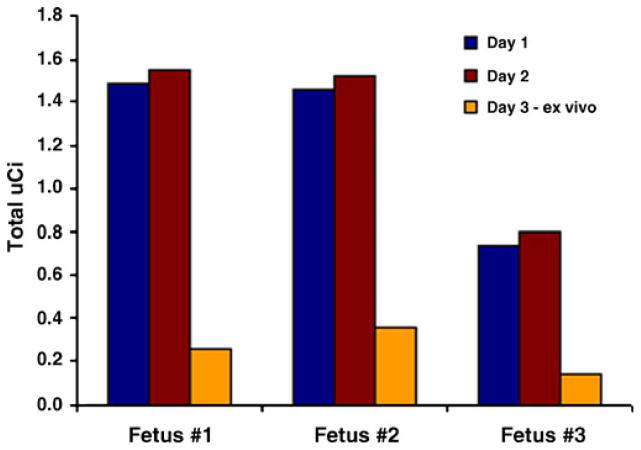
PET signals were detected in these studies over three sequential days and indicated that the majority of cells remained within the kidney for at least 24 h. A decline in signal was observed on day 3 post-transplant and within approximately 20% of the signal found on day 1 (Fig. 5), suggesting a loss of cell viability over time. Transduced hES cells in these studies showed light emissions of approximately 75 photons per second per 100 cells, and based on the standard curve generated for semi-quantitation (Fig. 6), roughly 5×105 cells remained in the kidney on day 3 post-transplant. Taken together, approximately 20% of transplanted cells were found at the time of tissue harvest and may also indicate cellular apoptosis. Further studies are in progress to determine the outcome of cells post-transplant for extended periods of time and whether a more defined environment is needed to support transplanted cells to ensure long-term viability and proliferation.
Fig. 5.
Quantitation of PET imaging. Results for individual fetuses are shown. A decline in the level of radioactivity was observed in all three fetuses over time likely due to cell loss. Decay-corrected values indicate that cells radiolabeled with 64Cu-PTSM persisted in the kidney for 24 h post-transplantation, followed by a decline in radioactivity in all three fetuses.
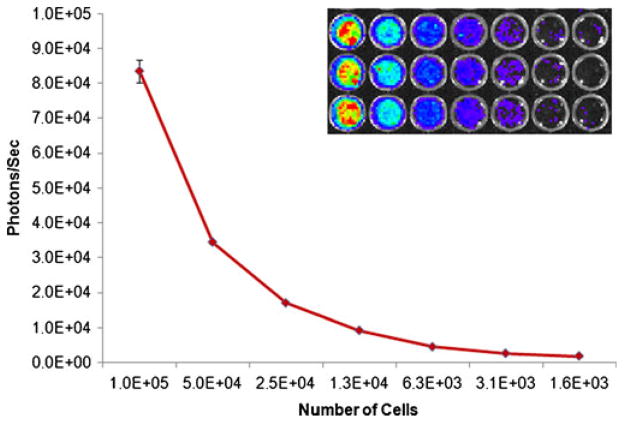
Fig. 6.
In vitro BLI. A subset of hES cells expressing firefly luciferase used for transplantation was plated in a 96-well plate and imaged using the IVIS200® Imaging System. These cells emitted 75 photons per second per 100 cells. This level of photon emission was consistent between different numbers of cells plated in each well and was used as a semi-quantitative method for in vivo images.
Noninvasive imaging systems include a wide range of technologies that make use of different contrast mechanisms to provide images that can reflect anatomy, physiology, metabolism, and to track transplanted stem and progenitor cells. A range of technologies that are available include those adapted from the radiologic setting and others based on optical techniques that build on the methods of optical microscopy and diffuse optical tomography [25]. A powerful approach is to use imaging reporter genes or direct radio-labeling in conjunction with these imaging technologies [11, 12, 14]. The most common reporter system used has been the gene for firefly luciferase and its substrate luciferin for BLI for experimental studies. The main advantage of transducing cells with reporter genes such as luciferase is that the reporters will be found in all cells after proliferation and are not diluted over time. Reporter gene approaches have also been developed for PET imaging including those that encode for a phosphorylating enzyme (e.g., mutant herpes simplex virus thymidine kinase) such that a radio-labeled substrate for the enzyme is trapped only in cells that express the reporter gene [18]. Superparamagnetic iron oxide nanoparticles have also been used with MRI to label cells for imaging in experimental and clinical studies [17]. The advantages of these in vivo imaging techniques include the relatively noninvasive nature, the fact that each individual can be followed longitudinally, and individual variability is reduced as a confounding variable. In vivo imaging is also of great utility in situations where it is unclear where one should focus the anatomical analyses—cell transplantation is a prime example since cells can traffic to different anatomical sites. While in vivo imaging presents clear advantages, there are challenges specifically for stem and progenitor cell trafficking. For example, several factors limit the detection sensitivity of PET scanners for cell tracking such as background noise that does not emanate from the cells and can sometimes make interpretation difficult and image reconstruction and processing methods employed which have not been optimized for cell trafficking applications. Our prior in vivo imaging studies have used semi-quantitative methods to determine imaging outcomes, although some limitations of these modalities have been addressed including tissue attenuation of source signal and generation of surface-weighted images [23]. Prior studies in monkeys have focused on correlating in vivo findings at the tissue level and determining whether there is loss of bioluminescent signals through attenuation from the body surface and overlying tissues [23, 24]. These studies have shown very good correlations when comparing the imaging outcomes in vivo with findings at the tissue level.
In summary, results of this study have shown that transplanted differentiated hES cells expressing firefly luciferase can be radiolabeled for PET imaging with 64Cu-PTSM and monitored short-term post-transplant using PET and BLI. Further studies in progress in the rhesus monkey are exploring the fate of transplanted renal precursors and progenitors and the use of crucial in vivo imaging technologies to monitor transplant outcomes.
Acknowledgments
The authors wish to thank the animal care staff at the California National Primate Research Center and the members of the Center for Molecular and Genomic Imaging in the College of Engineering for their expert technical assistance. These studies were supported by the California Institute for Regenerative Medicine (CIRM) Comprehensive grant no. RC1-00144-1 and the Primate Center base operating grant no. RR00169.
Footnotes
Disclosure. The authors have nothing to disclose.
References
- 1.Gulati A, Sarwal MM. Pediatric renal transplantation: an overview and update. Curr Opin Pediatr. 2010;22:189–196. doi: 10.1097/MOP.0b013e32833683fd. [DOI] [PubMed] [Google Scholar]
- 2.U.S. Renal Data System. Annual Data Report. 2009 ( http://www.usrds.org/)
- 3.Chevalier RL, Thornhill BA, Forbes MS, Kiley SC. Mechanisms of renal injury and progression of renal disease in congenital obstructive nephropathy. Pediatr Nephrol. 2010;25:687–697. doi: 10.1007/s00467-009-1316-5. [DOI] [PubMed] [Google Scholar]
- 4.Hiatt MJ, Ivanova L, Toran N, Tarantal AF, Matsell DG. Remodeling of the fetal collecting duct epithelium. Am J Pathol. 2010;176:630–637. doi: 10.2353/ajpath.2010.090389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanova L, Hiatt MJ, Yoder MC, et al. Ontogeny of CD24 in the human kidney. Kidney Int. 2010;77:1123–1131. doi: 10.1038/ki.2010.39. [DOI] [PubMed] [Google Scholar]
- 6.Tarantal AF, Han VKM, Cochrum KC, et al. Fetal rhesus monkey model of obstructive renal dysplasia. Kidney Int. 2001;59:446–456. doi: 10.1046/j.1523-1755.2001.059002446.x. [DOI] [PubMed] [Google Scholar]
- 7.Butt MJ, Tarantal AF, Jimenez DF, Matsell DG. Collecting duct epithelial-mesenchymal transition in fetal urinary tract obstruction. Kidney Int. 2007;72:936–944. doi: 10.1038/sj.ki.5002457. [DOI] [PubMed] [Google Scholar]
- 8.Matsell DG, Mok A, Tarantal AF. Altered primate glomerular development due to in utero urinary tract obstruction. Kidney Int. 2002;61:1263–1269. doi: 10.1046/j.1523-1755.2002.00274.x. [DOI] [PubMed] [Google Scholar]
- 9.Batchelder CA, Lee CC, Matsell DG, et al. Renal ontogeny in the rhesus monkey (Macaca mulatta) and directed differentiation of human embryonic stem cells towards kidney precursors. Differentiation. 2009;78:45–56. doi: 10.1016/j.diff.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batchelder CA, Lee CC, Martinez ML, Tarantal AF. Ontogeny of the kidney and renal developmental markers in the rhesus monkey (Macaca mulatta) Anat Rec. 2010;293:1971–1983. doi: 10.1002/ar.21242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budde MD, Frank JA. Magnetic tagging of therapeutic cells for MRI. J Nucl Med. 2009;50:171–174. doi: 10.2967/jnumed.108.053546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau JF, Anderson SA, Adler E, Frank JA. Imaging approaches for the study of cell-based cardiac therapies. Nat Rev Cardiol. 2010;7:97–105. doi: 10.1038/nrcardio.2009.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Libani IV, Lucignani G, Gianelli U, et al. Labeling protocols for in vivo tracking of human skeletal muscle cells (HSkMCs) by magnetic resonance and bioluminescence imaging. Mol Imaging Biol. 2011 doi: 10.1007/s11307-011-0474-6. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen PK, Nag D, Wu JC. Methods to assess stem cell lineage, fate and function. Adv Drug Deliv Rev. 2010;62:1175–1186. doi: 10.1016/j.addr.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Buul GM, Kotek G, Wielopolski PA, et al. Clinically translatable cell tracking and quantification by MRI in cartilage repair using superparamagnetic iron oxides. PLoS ONE. 2011;6:e17001. doi: 10.1371/journal.pone.0017001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welling MM, Duijvestein M, Signore A, Weerd LV. In vivo biodistribution of stem cells using molecular nuclear medicine imaging. J Cell Physiol. 2010;226:1444–1452. doi: 10.1002/jcp.22539. [DOI] [PubMed] [Google Scholar]
- 17.Bulte JWM. In vivo MRI cell tracking: clinical studies. Am J Roentgenol. 2009;193:314–325. doi: 10.2214/AJR.09.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaghoubi SS, Jensen MC, Satyamurthy N, et al. Noninvasive detection of therapeutic cytolytic T cells with 18F-FHBG PET in a patient with glioma. Nat Clin Pract Oncol. 2009;6:53–58. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Lee CCI, Sutcliffe JL, et al. Radiolabeling rhesus monkey CD34+ hematopoietic and mesenchymal stem cells with 64Cu-pyruvaldehyde-bis(N4-methylthiosemicarbazone) for microPET imaging. Mol Imaging. 2008;7:1–11. [PubMed] [Google Scholar]
- 20.Tarantal AF, Lee CC, Jimenez DF, et al. Fetal gene transfer using lentiviral vectors: in vivo detection of gene expression by microPET and optical imaging in fetal and infant monkeys. Hum Gene Ther. 2006;17:1254–1261. doi: 10.1089/hum.2006.17.1254. [DOI] [PubMed] [Google Scholar]
- 21.Tarantal AF. Ultrasound imaging in rhesus (Macaca mulatta) and long-tailed (Macaca fascicularis) macaques: reproductive and research applications. In: Wolfe-Coote S, editor. The Laboratory Primate. Elsevier; Amsterdam: 2005. pp. 317–352. [Google Scholar]
- 22.Jimenez DF, Lee CI, O’Shea CE, et al. HIV-1-derived lentiviral vectors and fetal route of administration on transgene biodistribution and expression in rhesus monkeys. Gene Ther. 2005;12:821–830. doi: 10.1038/sj.gt.3302464. [DOI] [PubMed] [Google Scholar]
- 23.Tarantal AF, Lee CCI, Itkin-Ansari P. Real-time bioluminescence imaging of macroencapsulated fibroblasts reveals allograft protection in rhesus monkeys (Macaca mulatta) Transplantation. 2009;88:38–41. doi: 10.1097/TP.0b013e3181a9ee6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarantal AF, Lee CCI. Long-term luciferase expression monitored by bioluminescence imaging after adeno-associated virus-mediated fetal gene delivery in rhesus monkeys (Macaca mulatta) Hum Gene Ther. 2010;21:143–148. doi: 10.1089/hum.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gambhir SS, Herschman HR, Cherry SR, et al. Imaging transgene expression with radionuclide imaging technologies. Neoplasia. 2000;2:118–138. doi: 10.1038/sj.neo.7900083. [DOI] [PMC free article] [PubMed] [Google Scholar]