Abstract
Recently, DNA has been evaluated as a chiral scaffold for metal complexes to construct so called ‘DNA-based hybrid catalysts’, a robust and inexpensive alternative to enzymes. The unique chiral structure of DNA allows the hybrid catalysts to catalyze various asymmetric synthesis reactions. However, most current studies used aqueous buffers as solvents for these asymmetric reactions, where substrates/products are typically suspended in the solutions. The mass transfer limitation usually requires a long reaction time. To overcome this hurdle and to advance DNA-based asymmetric catalysis, we evaluated a series of ionic liquids (ILs), inorganic salts, deep eutectic solvents (DES), glymes, glycols, acetonitrile and methanol as co-solvents/additives for the DNA-based asymmetric Michael addition. In general, these additives induce indistinguishable changes to the DNA B-form duplex conformation as suggested by circular dichroism (CD) spectroscopy, but impose a significant influence on the catalytic efficiency of the DNA-based hybrid catalyst. Conventional organic solvents (e.g. acetonitrile and methanol) led to poor product yields and/or low enantioselectivities. Most ILs and inorganic salts cause the deactivation of the hybrid catalyst except 0.2 M [BMIM][CF3COO] (95.4% ee and 93% yield) and 0.2 M [BMIM]Cl (93.7% ee and 89% yield). Several other additives have also been found to improve the catalytic efficiency of the DNA-based hybrid catalyst (control reaction without additive gives >99% ee and 87% yield): 0.4 M glycerol (>99% ee and 96% yield at 5 °C or 96.2% ee and 83% yield at room temperature), 0.2 M choline chloride/glycerol (1:2) (92.4% ee and 90% yield at 5 °C or 94.0% ee and 88% yield at room temperature), and 0.5 M dipropylene glycol dimethyl ether (>99% ee and 87% yield at room temperature). The use of some co-solvents/additives allows the Michael addition to be performed at a higher temperature (e.g. room temperature vs 5 °C) and a shorter reaction time (24 h vs 3 days). In addition, we found that a brief pre-sonication (5 min) of DNA in MOPS buffer prior to the reaction could improve the performance of the DNA-based hybrid catalyst. We have also shown that this DNA-based catalysis method is suitable for a variety of different substrates and relatively large-scale reactions. In conclusion, a judicious selection of benign co-solvents/additives could improve the catalytic efficiency of DNA-based hybrid catalyst.
Keywords: DNA, asymmetric synthesis, hybrid catalyst, ionic liquid, deep eutectic solvent, glyme
Introduction
In nature, DNA is the blueprint for living organisms and is typically associated with gene replication. Since the 1990s, certain DNA molecules have been identified to have catalytic capability through a systematic evolution of ligands by the exponential enrichment (SELEX) process.1, 2 These DNAs are known as DNAzymes (or deoxyribozymes or DNA enzymes). They usually serve as catalysts to cleave or ligate two RNA substrates, and also as photolyases (to photocleave thymine cyclobutane dimers), or peroxidases (for DNA detection), etc.2, 3 Therefore, these catalytic nucleic acids have gained fast growth in sensor applications, nanotechnology, and logic gate operations.4 In addition, the Li group5 found that double-stranded DNA from herring sperm is capable of catalyzing the dithioacetalization in water for a variety of aldehydes under mild reaction conditions. Recently, G-quadruplex DNA has been demonstrated as a promising catalyst for enantioselective Diels–Alder6–8 and Friedel–Crafts9 reactions.
More excitingly, the unique chiral structure of DNA has inspired the development of another new class of catalysts: DNA can be used as a chiral scaffold for metal complexes to construct so called ‘DNA-based hybrid catalysts’. These new hybrid catalysts are becoming very promising for asymmetric synthesis,10–12 and have shown high activities and enantioselectivities in several reactions including the Michael addition,13 Diels-Alder reaction,14–16 Friedel-Crafts alkylation,17 intramolecular cyclopropanation,18 electrophilic fluorination of β-keto esters,19 and asymmetric hydration,15 etc. The Roelfes group20 indicated that the addition of organic solvents (10–33%, v/v) including acetonitrile, THF and alcohols usually has no negative impact on the enantiomeric excess (ee) values of DNA-based catalytic Diels–Alder, Michael addition and Friedel–Crafts reactions, and could even accelerate the Michael addition and Friedel–Crafts alkylation reaction. The rate acceleration is likely due to a faster dissociation of the product but not the faster conjugate addition reaction. More interestingly, Wang et al.21 found that when the mirror image (L-DNA) of D-DNA is used as the chiral scaffold, a different enantioselectivity is achieved in DNA-based asymmetric Michael addition and Friedel–Crafts reaction. This enables a reliable and predictable access to both enantiomers for a particular reaction.
The only chirality in the DNA-based asymmetric catalysis is introduced through DNA helical structures. The likely catalytic mechanism has been proposed as the formation of supramolecular assembly of a DNA-based catalyst from DNA and a metal (such as Cu2+) complex of an achiral ligand.10, 15 The proximity of the catalytically active metal complex to the chiral DNA double helix leads to the preferential formation of one of the enantiomers of the product. There are several potential advantages of using DNA as biomolecular scaffold to develop hybrid catalysts for asymmetric catalysis: (1) DNA is readily available biomass and is inexhaustible in nature, (2) DNA is biodegradable, (3) DNA is inexpensive, (4) DNA is generally more stable than most enzymes, (5) DNA is rigid rod-like polymer and is ideal as catalyst scaffold, and (6) DNA can introduce chirality to the asymmetric reaction.
However, there are also a number of issues associated with current asymmetric catalysis using DNA-based hybrid catalysts. Since DNA is highly compatible with aqueous environment, most DNA-based asymmetric catalytic reactions have been carried out in aqueous buffer or aqueous organic solvents.20 In most cases, organic substrates are insoluble in aqueous solutions and thus reactions are typically very slow; in addition, these reactions are conducted at low temperature (< 5 °C),10 and thus it takes days to complete the reactions. Therefore, there is a strong need for developing a co-solvent system that can not only promote the substrate dissolution, but also produce a DNA-compatible environment.
Recently, several types of non-volatile solvents (e.g. ionic liquids, deep eutectic solvents, and glymes/glycols) have been actively explored in the field of biocatalysis, especially in enzyme-catalyzed reactions. One of these solvents is known as ionic liquids (ILs), which consist of ions and remain liquid at temperatures lower than 100 °C. ILs could dissolve many polar and non-polar substrates, and have tunable physical properties that make them highly compatible with various enzymes.22–24 Another new type of solvents, so called ‘deep eutectic solvents’ (DES), is formed by mixing a solid organic salt (such as choline chloride) and a suitable complexing agent (such as urea or glycerol). Recent studies have demonstrated the DES could be highly compatible with enzymes such as lipases, epoxide hydrolase, potato epoxide hydrolase StEH1, subtilisin, and α-chymotrypsin.25–30 In addition to their high compatibility with biomolecules, many DES are also inexpensive, highly biodegradable and non-toxic. On the other hand, two unique types of organic solvents, i.e. glycols and glymes, could also become benign solvents for biocatalysis. Glycols such as polyethylene glycols (PEGs)31–33 and PEG-based aqueous biphasic systems (ABS)34 are known media for enzymatic reactions. Glycol diethers, known as glymes, are not common solvents for biocatalysis.35 A few studies suggested the high enzyme activity and/or stability in aqueous glymes.36–39 Our group40 found that long-chain glymes are highly compatible with immobilized Candida antarctica lipase B.
In addition, recent studies found that DNA molecules could maintain their secondary duplex structures in some ILs and DES. The Hud group41 indicated that the oligonucleotide d(CG)8 exhibits secondary-structure formation in [HMIM][BF4], however, the 32 bp DNA duplex seems partially denatured in this IL. On the other hand, the Prasad group42 dissolved up to 3.5 wt% DNA (from salmon testes) in two biomaterial-based ILs, namely choline indole-3-acetate and choline indole-3-butyrate. DNA maintains B-form structures and a long-term stability (up to 6 months) in the first IL but is denatured by the second one. The MacFarlane group43 observed the long-term (up to 6 months) stability of salmon testes DNA in choline lactate, choline dihydrogenphosphate (containing 20–50 wt% water) and choline nitrate (containing 20% water), as suggested by the circular dichroism (CD) and fluorescence spectra as well as gel electrophoresis. The Hud group41 investigated the secondary structures of DNA and RNA in a neat DES known as choline chloride/urea (1:2) using CD spectroscopy. They suggest that the 32 bp mixed-sequence DNA exhibits an A-form duplex in the neat DES but B-form helix in aqueous salt solutions (such as 3.7 m NaCl), and the oligonucleotide [d(AT)16]2 has a similar B-form helix in the neat DES as in aqueous solutions. However, the melting transition midpoints (TM) of these DNAs in the DES are lower than those in aqueous solutions, suggesting a lower duplex stability in dehydrating and high ionic strength conditions. The Prasad group44 has demonstrated that two DES (choline chloride/ethylene glycol 1:2 and choline chloride/glycerol 1:2) could dissolve up to 5.5 wt% and 2.5 wt% DNA from salmon testes. They observed that DNA maintains B-form helical structures in DES and the regenerated DNA shows a high thermal and pH stability.
Built upon these exciting developments, we plan to incorporate co-solvents (ILs, DES, glymes and glycols) in asymmetric synthesis catalyzed by DNA-based hybrid catalyst, aiming to provide an DNA-compatible environment, a better substrate dissolution, a higher DNA-metal ligand binding, as well as a more enantioselective and efficient catalytic system. To demonstrate the proof-of-concept, we chose an asymmetric Michael addition catalyzed by a DNA/Cu2+/ligand hybrid catalyst to evaluate the role of these co-solvents/additives.
Experimental section
Materials
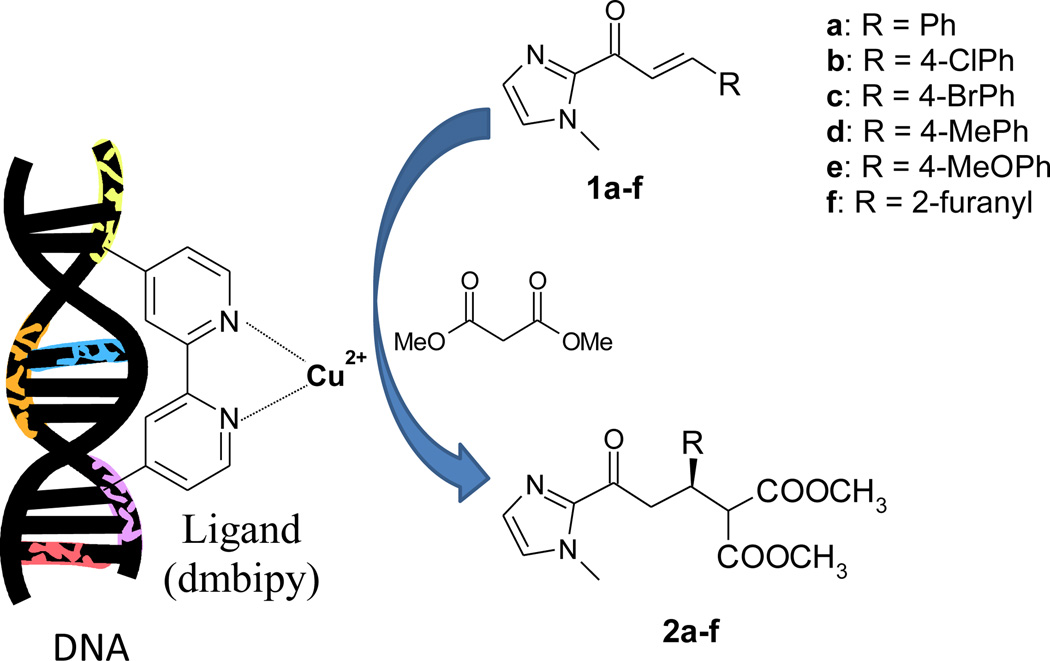
Deoxyribonucleic acid (DNA) sodium salt from salmon testes (catalog # D1626), N-Methylimiazole, n-butyllithium, trans-cinnamic acid, 4,4'-dimethyl-2,2'-dipyridyl (dmbipy), 3-(N-morpholino) propanesulfonic acid (MOPS), MOPS sodium salt, diethylene glycol dimethyl ether (G2), and tetraethylene glycol dimethyl ether (G4) were acquired from Sigma-Aldrich and used as received. 1,2-Dimethoxyethane (G1), triethylene glycol dimethyl ether (G3), and methanol-d4 were acquired from Alfa Aesar Company (Ward Hill, MA, USA). Dipropylene glycol dimethyl ether (P2) is a kind gift from Novolyte Technologies (Cleveland, Ohio). ILs were purchased or prepared as shown in our earlier paper.45 Deep eutectic solvents, choline chloride/glycerol (1:2, molar ratio) and choline acetate/glycerol (1:2) was prepared following our earlier study.28 The synthesis of 2-acyl imidazole substrates (1a-f) was a modification of literature methods46–48 and is described in details in Electronic supplementary information (ESI). The preparation of copper complex, Cu(dmbipy)(NO3)2, was based on a literature method14 and is also shown in ESI.
DNA-based catalytic enantioselective Michael reaction (small scale)
A solution of salmon testes DNA (5.0 mL of a 2.0 mg/mL solution in 30 mM pH 6.5 MOPS buffer) was sonicated in ice bath for 5 min. The DNA solution was added to a solution of 5.0 mL 0.45 mM [Cu(dmbipy)(NO3)2] in 30 mM pH 6.5 MOPS to prepare a buffered solution of DNA bound catalyst (1.0 mM salmon testes DNA and 0.15 mM [Cu(dmbipy)(NO3)2]). Additional co-solvent/additive in water was added to make a total volume of 15.0 mL. To this mixture, 15 µmol of substrate (1a-f) in 50 µL acetonitrile/2-propanol (3/2, v/v) was added and cooled to below 5 °C. The reaction was initiated by adding 100 eq. dimethylmalonate and stirred at 5 °C for 3 days. The product was isolated by extraction with diethyl ether followed by drying with Na2SO4 and removal of the solvent. The crude product (2a-f), was dissolved in 1.0 mL CD3OD for analysis by 1H-NMR and HPLC (Chiralpak AD 2.1×150 mm, 10 µm, n-heptane/i-PrOH 90/10, 0.2 mL/min, 210 nm).13
Large-scale Michael addition
A solution of salmon testes DNA (100 mL of a 2.0 mg/mL solution in 30 mM pH 6.5 MOPS buffer) was sonicated in ice bath for 5 min. The DNA solution was added to a solution of 100 mL 0.45 mM [Cu(dmbipy)(NO3)2] in 30 mM pH 6.5 MOPS to prepare a buffered solution of DNA bound catalyst (1.0 mM salmon testes DNA and 0.15 mM [Cu(dmbipy)(NO3)2]). Glycerol (final concentration 0.4 M) was dissolved in MOPS buffer to make a total volume of 100 mL. To this mixture, 0.064 g (0.3 mmol) of substrate (1a) was added and cooled to below 5 °C. The reaction was initiated by adding 100 eq. dimethylmalonate (5.65 g) and stirred at 5 °C for 7 days. The product was isolated by extraction with diethyl ether followed by drying with Na2SO4 and removal of the solvent. The crude product (2a) was then purified by column chromatography (Silica 60, ethyl acetate/hexane, 2:3 v/v) and analyzed by NMR and HPLC (Chiralpak AD 2.1×150 mm, 10 µm, n-heptane/i-PrOH 90/10, 0.2 mL/min, 210 nm). 1H NMR (300 MHz, CD3OD) δ/ppm = 3.40 (s, 3H), 3.36–3.48 (m, 1H), 3.68 (s, 3H), 3.83 (s, 3H), 3.69–3.87 (m, 2H), 3.93 (m, 1H), 7.05 (s, 1H), 7.10–7.27 (m, 6H). 13C NMR (CD3OD) δ/ppm = 35.1, 40.7, 42.4, 51.4, 51.7, 57.2, 126.8, 127.6, 128.0, 128.1, 128.2, 140.6, 142.7, 168.5, 168.8, 189.5.
Circular dichroism (CD) spectra of DNA in aqueous solutions
DNA from salmon testes was dissolve in distilled water to make a nominal concentration of 2.0 mg/mL. The actual DNA concentration was determined by NanoDrop 2000 as 1.8 mg/mL. The DNA stock solution (300 µL) was diluted 5 times by an aqueous solution of co-solvents/additives. Co-solvents/additives were weighted to meet the desired final concentrations. An aliquot of the mixture was scanned in the range of 220–320 nm by a JASCO J-825 CD Spectrometer. The instrument parameters were set as data pitch 0.1 nm, scanning speed 100 nm/min, band width 1.00 nm, slit width 100 um, DIT 16 sec, standard sensitivity, 10 accumulations, and the cell temperature of 5 °C.
DNA binding to Cu2+ ligand in aqueous solutions of organic solvents or ionic liquids
DNA from salmon testes was dissolve in distilled water to make a nominal concentration of 2.0 mg/mL. The actual DNA concentration was determined by NanoDrop 2000 as 1.8 mg/mL. The concentration of base pairs was calculated from spectrophometrical absorbance at 260 nm using ε260 =12,800 M−1 cm−1. An aliquot of stock DNA solution (0, 30, 60 and 90 µL) was added into 100 µL of 0.45 mM Cu(dmbipy)(NO3)2 in MOPS (30 mM, pH 6.5), followed by the addition of MOPS buffer to make the total volume of 1.5 mL. The copper complex was maintained at 30 mM. The mixture was then analyzed by NanoDrop 2000 for its absorbance at 260 nm. The binding constant (Kb) can be determined by the following equation,20, 49
where ∆εap = |εa - εf|, ∆ε = |εb - εf|, εa, εf and εb the apparent, free and bound extinction coefficients for the complex respectively, and D is the DNA concentration in base pairs. D/∆εap was plotted against D, and the Kb value was calculated from the ratio of the slope to the y-intercept.
Results and discussion
The DNA-based hybrid catalyst was constructed by mixing salmon testes DNA in MOPS buffer (30 mM, pH 6.5) with Cu(dmbipy)(NO3)2 in MOPS (dmbipy = 4,4'-dimethyl-2,2'-dipyridyl), as shown in Scheme 1.13, 20 To evaluate the catalytic capability of the hybrid catalyst, a Michael addition reaction was chosen as our primary model system first: the conversion of (E)-(1-methyl-1H-imidazole-2-yl)-3-phenylprop-2-en-1-one (1a) to (R)-dimethyl 2-(3-(1-methyl-1H-imidazol-2-yl)-3-oxo-1-phenylpropyl)malonate (2a) (see Scheme 1). Without the presence of salmon testes DNA, a control reaction (trial 1 in Table 1) produced a very low yield (7%) and a poor enantioselectivity (10.3% ee); this signifies the important role of DNA in the hybrid catalyst.
Scheme 1.
DNA-based asymmetric Michael addition (dmbipy = 4,4′-dimethyl-2,2′-bipyridine).
Table 1.
Effect of co-solvent/additive on the enantioselectivity and yield of Michael addition a
| Trial | Co-solvent/additive | ee (%) | HPLC yield (%) |
Binding constant Kb/104 (M−1) |
|---|---|---|---|---|
| 1 | MOPS buffer, no DNA | 10.3 (−) | 7 | |
| 2 | MOPS buffer, DNA not sonicated | 99.8 (−) | 74 | 2.02 |
| 3 | MOPS buffer, DNA sonicated 5 min | >99 (−) | 87 | 7.15 |
| 4 | MOPS, r.t., 24 h | 95.9 (−) | 77 | |
| 5 | MOPS, r.t., 48 h | 96.5 (−) | 99 | |
| 6 | MOPS, r.t., 72 h | 93.8 (−) | 96 | |
| 7 | 0.2 M acetonitrile | 98.0 (−) | 69 | 7.91 |
| 8 | 0.5 M acetonitrile | >99 (−) | 50 | 1.23 |
| 9 | 10% acetonitrile (1.9 M) | 98.3 (−) | 58 | 0.42 |
| 10 | 25% acetonitrile (4.7 M) | 93.0 (−) | 36 | 0.69 |
| 11 | 10% methanol (2.3 M) | 71.5 (−) | 61 | 1.13 |
| Ionic liquids as co-solvents | ||||
| 12 | 0.2 M [BMIM]Cl | 93.7 (−) | 89 | 0.64 |
| 13 | 0.2 M [BMIM][CF3COO] | 95.4 (−) | 93 | 3.87 |
| 14 | 0.2 M [BMIM][CF3COO], r.t., 24 h | 85.7 (−) | 85 | |
| 15 | 0.5 M [BMIM][CF3COO] | 53.1 (−) | 36 | 0.28 |
| 16 | 0.2 M [EMIM][OAc] | 25.2 (−) | 1.2 | 1.57 |
| 17 | 0.2 M [BMIM][OAc] | 75.5 (−) | 3.3 | 0.39 |
| 18 | 0.2 M [HMIM][OAc] | 91.0 (−) | 1.9 | 2.20 |
| 19 | 0.2 M [CH3(OCH2CH2)3-2-Me-Et-Im] [OAc] |
67.7 (−) | 4.6 | 1.46 |
| 20 | 0.2 M [Choline][OAc] | 61.5 (−) | 15 | 1.90 |
| 21 | 0.2 M [BMIM][OTf] | 35.3 (−) | 0.55 | 2.12 |
| 22 | 0.2 M [BMIM][BF4] | 90.0 (−) | 74 | 8.88 |
| 23 | 0.2 M [BMIM][NO3] | 10.0 (−) | 25 | 1.42 |
| 24 | 0.2 M [BMIM][SCN] | 69.9 (−) | 0.18 | 0.46 |
| 25 | 0.2 M [BMIM][dca] | 90.7 (−) | 0.55 | 0.33 |
| 26 | 0.2 M [Choline][H2PO4] | 18.4 (−) | 1.6 | 4.13 |
| 27 | 0.2 M [Choline][bitartrate] | 25.2 (−) | 0.16 | 1.11 |
| 28 | 0.5 M [Choline][Tf2N] | 8.7 (−) | 56 | |
| 29 | 0.2 M [BMIM][Tf2N] | 23.4 (−) | 85 | |
| 30 | 0.2 M [CH3(OCH2CH2)3-Et-Im][Tf2N] | 2.1 (+) | 58 | 0.95 |
| 31 | 0.5 M [CH3(OCH2CH2)3-Et3N][Tf2N] | 11.0 (+) | 30 | 0.71 |
| Inorganic salts as additives | ||||
| 32 | 0.2 M LiCl | 81.5 (−) | 6 | 0.39 |
| 32 | 0.2 M NaCl | 63.6 (−) | 43 | 0.73 |
| 33 | 0.2 M KCl | 62.4 (−) | 52 | 1.26 |
| 34 | 0.2 M Na[CF3COO] | 55.9 (−) | 73 | 2.43 |
| 35 | 0.2 M NaOAc | 26.2 (−) | 8 | 4.32 |
| 36 | 0.2 M Na[OTf] | 47.2 (−) | 59 | 1.64 |
| 37 | 0.2 M BaBF4 | 62.6 (−) | 14 | 2.19 |
| 38 | 0.2 M NaNO3 | 87.6 (−) | 51 | 0.73 |
| 39 | 0.2 M NaSCN | 26.5 (−) | 2 | 0.44 |
| 40 | 0.2 M Na[dca] | 68.1 (−) | 1 | 1.06 |
| 41 | 0.2 M Li[Tf2N] | 4.9 (−) | 28 | 1.95 |
| 42 | 0.2 M urea | 98.1 (−) | 71 | 4.24 |
| DES and related compounds as co-solvents | ||||
| 43 | 0.2 M Choline chloride/glycerol (1:2) | 92.4 (−) | 90 | 1.14 |
| 44 | 0.5 M Choline chloride/glycerol (1:2) | 58.6 (−) | 53 | 0.39 |
| 45 | 0.2 M Choline chloride/glycerol (1:2), r.t., 24 h | 94.0 (−) | 88 | |
| 46 | 0.2 M Choline chloride/glycerol (1:2), r. t., 48 h | 86.9 (−) | 95 | |
| 47 | 0.2 M Choline chloride/glycerol (1:2), r.t., 3 days | 87.1 (−) | 97 | |
| 48 | 0.2 M Choline acetate/glycerol (1:1.5) | 71.8 (−) | 16 | 4.10 |
| 49 | 0.5 M Choline acetate/glycerol (1:1.5) | 16.1 (−) | 3.5 | 4.01 |
| 50 | 0.2 M Choline chloride | 92.8 (−) | 80 | 1.90 |
| 51 | 0.5 M Choline chloride | 59.6 (−) | 38 | 1.27 |
| 52 | 0.2 M Choline chloride, r.t., 24 h | 71.1 (−) | 29 | |
| 53 | 0.2 M Glycerol | 98.4 (−) | 65 | 0.80 |
| 54 | 0.4 M Glycerol | >99 (−) | 96 | 8.15 |
| 55 | 0.8 M Glycerol | >99 (−) | 63 | 0.67 |
| 56 | 0.4 M Glycerol, r.t, 24 h | 96.2 (−) | 83 | |
| Glymes and glycols as co-solvents | ||||
| 57 | 0.5 M monoglyme | >99 (−) | 73 | 13.06 |
| 58 | 0.5 M diglyme | >99 (−) | 56 | 1.91 |
| 59 | 0.5 M triglyme | >99 (−) | 75 | 3.45 |
| 60 | 0.2 M tetraglyme | 99.8 (−) | 87 | 12.08 |
| 61 | 0.5 M tetraglyme | >99 (−) | 76 | 3.87 |
| 62 | 0.2 M tetraglyme, r.t., 24 h | 96.3 (−) | 78 | |
| 63 | 0.5 M dipropylene glycol dimethyl ether | >99 (−) | 49 | 2.77 |
| 64 | 0.5 M dipropylene glycol dimethyl ether, r.t., 24 h | >99 (−) | 87 | |
| 65 | 0.2 M ethylene glycol | 98.3 (−) | 79 | 0.94 |
| 66 | 0.2 M diethylene glycol | 99.3 (−) | 59 | 2.36 |
| 67 | 0.2 M triethylene glycol | 97.8 (−) | 61 | 8.59 |
| 68 | 0.4 M tetraethylene glycol | 99.8 (−) | 71 | 20.51 |
Note
reaction conditions (except noted otherwise): 10 mg DNA (Sigma D1626) in 5.0 mL MOPS buffer sonicated for 5 min in ice bath, 15 µmol acyl donor (1a), 100 eq. dimethylmalonate, 0.15 mM [Cu(dmbipy)(NO3)2], 30 mM pH 6.5 MOPS, reaction volume 15.0 mL, 5 °C for 3 days (except noted otherwise); bold values of yields and ees indicate they are relatively high.
Effect of pre-sonication and organic solvents on the DNA’s catalytic efficiency
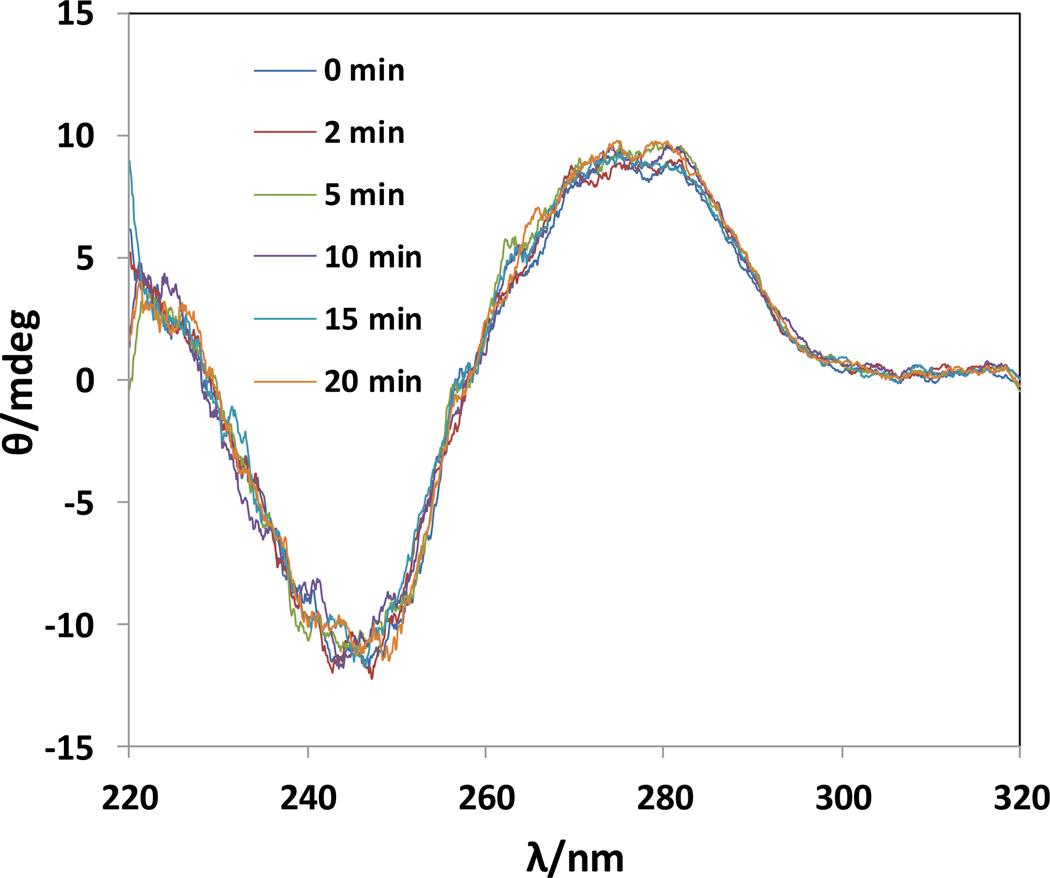
When salmon testes DNA was fully dissolved in MOPS buffer without sonication, 74% yield and >99% ee were achieved for the Michael reaction in 3 days at 5 °C. When the DNA was pre-sonicated in ice bath prior to the reaction, we found that 5 min of sonication gave the best results: 87% yield and >99% ee (see ESI, Fig. S1). Gel electrophoresis of salmon testes DNA (see Fig. S2 in ESI) suggested that 5 min of sonication led to the lowest base pairs compared with those without sonication or sonicated for other times. It seems that a short sonication time (such as 5 min) enables a better dispersion and less aggregation of DNA molecules in aqueous solutions.
Earlier studies50, 51 have suggested that sonication of low concentrations (such as 7.5–70 µg/mL) of DNA solutions for 30 sec to a few minutes could lead to considerable DNA fragmentation; the reason is that vibrations of ultrasonic waves create gaseous cavitations in the liquid, which fragments DNA molecules through resonance vibration. However, our electrophoresis data in Fig. S2 appear not supporting any substantial DNA fragmentation under a short sonication in ice bath. To further confirm the double helical structures of DNA upon sonication, we compared the CD spectra of DNA samples under different sonication times (Fig. 1). All the CD spectra showed characteristics of B-form DNA with a long wave positive band at 278 nm corresponding to p–p base packing and a shorter wave negative band at 243 nm corresponding to helicity.52 Therefore, sonication of DNA in ice bath for up to 20 min has no impact on the helical structures of DNA. As shown in Table 1 (trials 2 and 3), the binding constant (Kb) of DNA with Cu2+ ligand was determined to be 2.02 × 104 M−1 for DNA without pre-sonication, while a higher Kb value of 7.15 × 104 M−1 was found for the DNA sonicated for 5 min. Therefore, the improved reaction yield for DNA sonicated for 5 min is likely due to a better dispersion and less aggregation of DNA in aqueous media, further resulting in a strong binding between the DNA molecule and Cu2+ ligand. When the same reaction was performed at room temperature (trials 4–6), a nearly quantitative yield (up to 99%) was obtained in 48 h; however, the product ee decreased gradually with the reaction time to 93.8% at 72 h.
Fig. 1.
Effect of sonication time on the CD spectra of salmon testes DNA in 30 mM pH 6.5 MOPS.
Megens and Roelfes20 suggested that the use of water-miscible organic co-solvents (10–33%, v/v) has no appreciable effect on the enantioselectivity of DNA-based asymmetric reaction, although the rate of Diels–Alder reaction became slower while the Michael addition and Friedel–Crafts alkylation reaction were accelerated. We examined the addition of 0.2–4.7 M acetonitrile to the Michael addition (trials 7–10 in Table 1), and found a slight decrease in ee value at higher acetonitrile contents but a dramatic decrease in product yields (from 69% to 36%). The addition of 10% methanol (2.3 M) resulted in a low ee (71.5%) (trial 11). Therefore, the use of organic co-solvents results in no improvement in both the enantioselectivity and yield.
Effect of IL, inorganic salts and DES as co-solvents/additives
We systematically evaluated a series of ILs as co-solvents for the Michael addition (trials 12–31), and noticed that the IL structures have a considerable influence on the catalytic efficiency of DNA-based hybrid catalyst. Among 18 ILs/organic salts studied, only three of them afforded relatively high ees and yields: 0.2 M [BMIM][CF3COO] (95.4% ee and 93% yield; trial 13), 0.2 M [BMIM]Cl (93.7% ee and 89% yield; trial 12) and 0.2 M [BMIM][BF4] (90.0% ee and 74% yield; trial 22). Other anions-based ILs (such as OAc−, OTf−, NO3−, SCN−, dca− and Tf2N−) tend to significantly reduce the yield and/or ee. In particular, Tf2N−-containing ILs are more hydrophobic and typically compatible with enzymes;22–24 however, none of them (trials 28–31) seems to be compatible with the DNA-based hybrid catalyst. Through inspecting the effect of cations (trials 16–18), we found that a more hydrophobic IL leads to a higher enantioselectivity but no improvement in product yield. Ether-functionalized ILs (trials 19, 30 and 31) are compatible with lipases and proteases;53, 54 however, poor yield and/or ee values were observed in the presence of these ionic solvents. The addition of most choline-based salts (trials 20, 26–28) also led to poor catalytic performance of the DNA-based hybrid catalyst; only 0.2 M choline chloride (trial 50) afforded a relatively high yield (80%) and enantioselectivity (92.8%). A higher IL concentration (such as 0.5 M [BMIM][CF3COO], trial 15) often results in a low yield (36%) and ee (53.1%). A higher reaction temperature is more detrimental to the Michael reaction in IL solutions than in MOPS buffer; for example, in 0.2 M [BMIM][CF3COO] at room temperature (trial 14), 85% yield and 85.7% ee were obtained while the same reaction at 5 °C produced 95.4% ee and 93% yield (trial 13).
To further understand the effect of individual ions on the catalysis, we also evaluated a variety of inorganic salts as additives (0.2 M) (trials 32–41). The addition of these inorganic salts induced a drastic decrease in both the yield and enantioselectivity. Anions of CF3COO−, NO3−, Cl− and OTf− cause less catalyst deactivation than other anions (OAc−, BF4−, SCN−, dca− and Tf2N−). The product yield increased in the order of Li+ < Na+ < K+. This is in agreement with the case of ILs to some extent that the addition of [BMIM][CF3COO] and [BMIM]Cl results in relatively high yields and ees. The enzyme-denaturing urea actually imposes a minimum impact on the DNA-based catalysis (71% yield and 98.1% ee in 0.2 M urea, trial 42).
We also examined the use of DES as co-solvents in the DNA-based asymmetric catalysis of Michael addition (trials 43–49). In general, choline-based DES are less catalyst-deactivating than most ILs and inorganic salts. 0.2 M Choline chloride/glycerol (1:2) produced 90% yield with 92.4% ee at 5 °C for 3 days (trial 43) and 88% yield with 94.0% ee at room temperature for 24 h (trial 45), while the same reaction in MOPS buffer gave 87% yield with > 99% ee at 5 °C for 3 days (trial 3) and 77% yield with 95.9% ee at room temperature for 24 h (trial 4). Therefore, the use of this DES as co-solvent could accelerate the reaction rate. Choline acetate-based DES is less effective in promoting the DNA-based catalysis (trials 48 and 49). Furthermore, we evaluated the effect of individual components of DES on the catalysis reaction. As mentioned earlier, choline chloride causes less deactivation than other choline salts (trials 50–52). Interestingly, glycerol has been shown quite compatible with the DNA-based hybrid catalyst: 0.4 M glycerol affords 96% yield with >99% ee at 5 °C for 3 days (trial 54) and 83% yield with 96.2% ee at room temperature for 24 h (trial 56). These reaction rates are much faster than those without glycerol.
DNA structural changes and binding characteristics in the presence of ionic additives
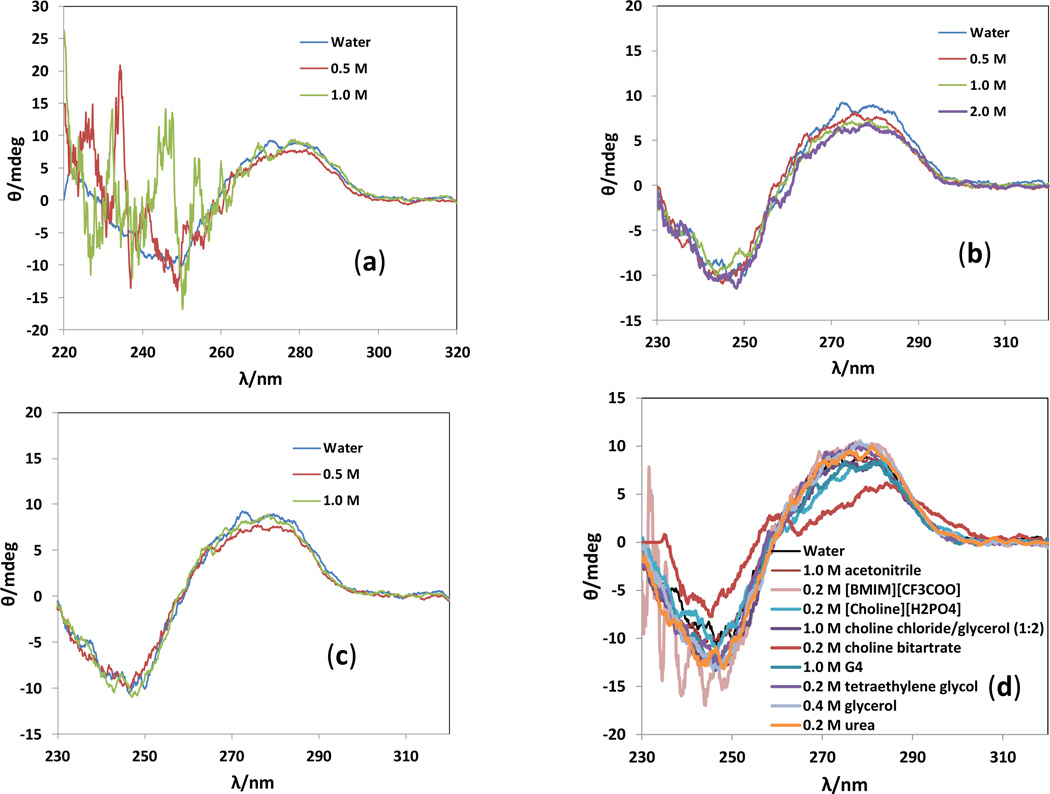
In an attempt to explain how ionic additives (ILs, inorganic salts and DES) could affect the DNA-based asymmetric catalysis, we firstly examined the possible DNA structural changes by CD spectroscopy. Our results suggested that DNA molecules maintain their characteristic B-form helical duplex structures in up to 2.0 M ILs (Fig. 2b, 2d, and Fig. S3 in ESI), 0.2 M inorganic salts (Fig. S4), and up to 1.0 M DES (Fig. 2d and Fig. S3). Since imidazolium-based ILs (such as [BMIM][OAc] in Fig. 2a and [BMIM][CF3COO] in Fig. 2d) typically have a strong absorption at the far-UV wavelength range (≤ 200–250 nm), they tend to provide a messy interference to the CD spectra of biomolecules;55 therefore, the perturbance to the CD spectra in Figs. 2a and 2d may not be caused by the DNA conformational changes. The only notable shift in CD spectrum was observed in 0.2 M choline bitartrate (Fig. 2d). In conclusion, most ionic salts in low concentrations have no significant impact on the DNA duplex structures. This conclusion is also in agreement with a recent study by Chandran et al.,56 who performed the molecular dynamics simulations and spectroscopic studies (CD, UV-Vis and fluorescence) to examine the DNA stability in hydrated ILs. This group suggested the model DNA (Dickerson−Drew dodecamer) preserved the B-conformation in hydrated (up to 80 wt%) ILs ([BMIM]Cl, [BMIM][NO3], [BMIM][lactate], [choline][NO3] and [choline][lactate]).
Fig. 2.
CD spectra of salmon testes DNA in (a) aqueous [BMIM][OAc], (b) aqueous [Choline][OAc], (c) aqueous dipropylene glycol dimethyl ether, and (d) aqueous solutions of organic solutes.
However, a number of studies have indicated that there are strong interactions between DNA and ionic solvents.57 The major interaction between ILs (or DES) and DNA is the electrostatic attraction of organic cations and DNA phosphate backbone, followed by the hydrophobic and polar interactions between ILs and DNA major and minor grooves. 44, 56, 58–60 Past studies have also discussed how di- and monovalent metal cations preferentially interact with the major and minor grooves of DNA duplexes.61–63 Those DNA-ionic solvent interactions are not necessarily strong enough to modify the DNA duplex conformation, but could considerably affect the ligand binding to DNA molecules (Scheme 1). When ionic species bind to DNA phosphates, the ligand binding to DNA becomes a competitive process and is likely to be reduced; the catalytic capability of DNA-based hybrid catalyst is also affected by this binding competition. As shown in Table 1, in some cases (such as trials 13 and 22), high binding constants were observed for ionic additives that afford high yields and ee values; however, there are several exceptions to this trend (such as trials 12, 26, 35, 43 and 48). Therefore, this binding constant may not actually reflect the competitive bindings between DNA and ionic solvents, as well as between DNA and Cu2+ ligand. The other possibility is that ionic species compete with Cu2+ ions in binding with the ligand, lowering the catalytic ability of the catalyst.
Effect of glymes and glycols as co-solvents
We also evaluated the possibility of using glycol-based media (PEGs and glymes) as co-solvents in our DNA-based asymmetric Michael addition. As shown in Table 1 (trials 57–68), high enantioselectivities were also obtained in 0.2–0.5 M glymes and glycols whilst the yields varied in the range of 49–87%. In particular, two very encouraging results were achieved in 0.2 M tetraglyme at 5 °C (trial 60: 99.8% ee and 87% yield), and 0.5 M dipropylene glycol dimethyl ether at room temperature for 24 h (trial 64: >99% ee and 87% yield). As illustrated in Fig. 2c and 2d, salmon testes DNA maintains its B-form conformation in up to 1.0 M glycols and glymes. The binding constants in Table 1 suggest that high binding constants are generally associated with high yields for the same type of solvents. Therefore, glymes and glycols can affect the binding of ligand with DNA, which becomes favorable to the reaction in some cases.
Other substrates and large-scale synthesis
To evaluate the adaptability of this DNA-based catalysis method to other substrates, we prepared substrates 1b-f (see Scheme 1 and ESI) and used them in the small-scale Michael addition in 0.4 M glycerol or 0.2 M [BMIM][CF3COO] as co-solvent. As shown in Table 2 (trials 54, 69–74), high ee values (≥ 97%) were obtained for all substrates 1a-f; however, product yields varied with substrates: 1b, 1c and 1f led to yields below 50%, while 1a, 1d and 1e produced yields in the range of 77–96%. In particular, when the reaction of 1d was carried out in 0.2 M [BMIM][CF3COO], a high yield of 95% and a high ee of >99% were achieved.
Table 2.
Michael addition with different substrates and at large scales
| Trial | Substrate | Conditions | ee (%) | HPLC yield (%) |
|---|---|---|---|---|
| 54 | 1a | Small scale, 15 µmol 1a, 0.4 M glycerol | >99 (−) | 96 |
| 69 | 1b | Small scale, 15 µmol 1b, 0.4 M glycerol | 98.4 | 40 |
| 70 | 1c | Small scale, 15 µmol 1c, 0.4 M glycerol | 96.8 | 27 |
| 71 | 1d | Small scale, 15 µmol 1d, 0.4 M glycerol | >99 | 85 |
| 72 | 1d | Small scale, 15 µmol 1d, 0.2 M [BMIM][CF3COO] | >99 | 95 |
| 73 | 1e | Small scale, 15 µmol 1e, 0.4 M glycerol | 97.0 | 77 |
| 74 | 1f | Small scale, 15 µmol 1f, 0.4 M glycerol | >99 | 49 |
| 75 | 1a | Large scale, 0.3 mmol 1a (× 20 times), 0.4 M glycerol | >99 (−) | 93 |
| 76 | 1a | Large scale, 0.94 mmol 1a (× 63 times), 0.4 M glycerol | >99 (−) | 60 |
| 77 | 1a | Large scale, 2.4 mmol 1a (× 160 times), 0.4 M glycerol | >99 (−) | 20 |
Furthermore, we scaled up the reaction of 1a from a small scale (15 µmol 1a in 15 mL reaction mixture) to 20–160 times (trials 75–77 in Table 2), and found a scale-up of 1a to 20 times resulting in 93% yield and >99% ee (trial 75). However, a further scale-up of the substrate to 63 and 160 times resulted in lower product yields (60% and 20% respectively) although high ee values (>99%) were not affected.
Conclusions
The addition of ILs, inorganic salts, DES, glymes and glycols as co-solvents (or additives) in the DNA-based asymmetric Michael addition typically causes no considerable changes to the DNA B-form conformation; however, these additives interact with DNA molecules at various extents, which in turn affects the DNA-ligand binding and the catalytic efficiency. In particular, several additives could improve the efficiency of DNA-based hybrid catalyst: 0.4 M glycerol (5 °C for 3 days or room temperature for 24 h), 0.2 M [BMIM][CF3COO] (5 °C for 3 days), 0.2 M choline chloride/glycerol (1:2) (5 °C for 3 days or room temperature for 24 h), and 0.5 M dipropylene glycol dimethyl ether (room temperature for 24 h). The use of some co-solvents/additives allows us to carry out the asymmetric reaction at a higher temperature (e.g. room temperature vs 5 °C) and a shorter reaction time (24 h vs 3 days). In addition, we found that a brief pre-sonication (5 min) of DNA in MOPS buffer prior to the reaction could improve the performance of DNA-based hybrid catalyst. This DNA-based catalysis method could be applied to a variety of different substrates and relatively large-scale reactions. Future studies will explore the mechanisms of how these additives interact with the DNA-based hybrid catalyst.
Supplementary Material
Acknowledgements
HZ acknowledges the supports by the Henry Dreyfus Teacher-Scholar Award (2012–2017), NIH MBRS-RISE grant (1R25GM096956), NIH NIBIB contract award (HHSN268201200011C), and the National Natural Science Foundation of China (21328601).
Notes and references
- 1.Breaker RR, Joyce GF. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 2.Sen D, Geyer CR. Current Opinion in Chemical Biology. 1998;2:680–687. doi: 10.1016/s1367-5931(98)80103-8. [DOI] [PubMed] [Google Scholar]
- 3.Höbartner C, Silverman SK. Biopolymers. 2007;87:279–292. doi: 10.1002/bip.20813. [DOI] [PubMed] [Google Scholar]
- 4.Willner I, Shlyahovsky B, Zayats M, Willner B. Chem. Soc. Rev. 2008;37:1153–1165. doi: 10.1039/b718428j. [DOI] [PubMed] [Google Scholar]
- 5.Wang C, Li Y, Jia G, Lu S, Liu Y, Li C. Acta Chim. Sinica. 2013;71:36–39. [Google Scholar]
- 6.Roe S, Ritson DJ, Garner T, Searle M, Moses JE. Chem. Commun. 2010;46:4309–4311. doi: 10.1039/c0cc00194e. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Jia G, Zhou J, Li Y, Liu Y, Lu S, Li C. Angewandte Chemie International Edition. 2012;51:9352–9355. doi: 10.1002/anie.201204850. [DOI] [PubMed] [Google Scholar]
- 8.Wilking M, Hennecke U. Org. Biomol. Chem. 2013;11:6940–6945. doi: 10.1039/c3ob41366g. [DOI] [PubMed] [Google Scholar]
- 9.Wang C, Li Y, Jia G, Liu Y, Lu S, Li C. Chem. Commun. 2012;48:6232–6234. doi: 10.1039/c2cc31320k. [DOI] [PubMed] [Google Scholar]
- 10.Boersma AJ, Megens RP, Feringa BL, Roelfes G. Chem. Soc. Rev. 2010;39:2083–2092. doi: 10.1039/b811349c. [DOI] [PubMed] [Google Scholar]
- 11.Park S, Sugiyama H. Molecules. 2012;17:12792–12803. doi: 10.3390/molecules171112792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park S, Sugiyama H. Angewandte Chemie International Edition. 2010;49:3870–3878. doi: 10.1002/anie.200905382. [DOI] [PubMed] [Google Scholar]
- 13.Coquière D, Feringa BL, Roelfes G. Angewandte Chemie International Edition. 2007;46:9308–9311. doi: 10.1002/anie.200703459. [DOI] [PubMed] [Google Scholar]
- 14.Roelfes G, Boersma AJ, Feringa BL. Chem. Commun. 2006:635–637. doi: 10.1039/b516552k. [DOI] [PubMed] [Google Scholar]
- 15.Rosati F, Roelfes G. ChemCatChem. 2011;3:973–977. [Google Scholar]
- 16.Boersma AJ, Feringa BL, Roelfes G. Org. Lett. 2007;9:3647–3650. doi: 10.1021/ol7015274. [DOI] [PubMed] [Google Scholar]
- 17.Boersma AJ, Feringa BL, Roelfes G. Angewandte Chemie International Edition. 2009;48:3346–3348. doi: 10.1002/anie.200900371. [DOI] [PubMed] [Google Scholar]
- 18.Oelerich J, Roelfes G. Chem. Sci. 2013;4:2013–2017. [Google Scholar]
- 19.Shibata N, Yasui H, Nakamura S, Toru T. Synlett. 2007;7:1153–1157. [Google Scholar]
- 20.Megens RP, Roelfes G. Org. Biomol. Chem. 2010;8:1387–1393. doi: 10.1039/b921385f. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Benedetti E, Bethge L, Vonhoff S, Klussmann S, Vasseur J-J, Cossy J, Michael Smietana M, Arseniyadis S. Angewandte Chemie International Edition. 2013;52:11546–11549. doi: 10.1002/anie.201306232. [DOI] [PubMed] [Google Scholar]
- 22.van Rantwijk F, Sheldon RA. Chem. Rev. 2007;107:2757–2785. doi: 10.1021/cr050946x. [DOI] [PubMed] [Google Scholar]
- 23.Moniruzzaman M, Nakashima K, Kamiya N, Goto M. Biochem. Eng. J. 2010;48:295–314. [Google Scholar]
- 24.Zhao H. J. Chem. Tech. Biotechnol. 2010;85:891–907. [Google Scholar]
- 25.Gorke JT, Srienc F, Kazlauskas RJ. Chem. Commun. 2008:1235–1237. doi: 10.1039/b716317g. [DOI] [PubMed] [Google Scholar]
- 26.Lindberg D, de la Fuente Revenga M, Widersten M. J. Biotechnol. 2010;147:169–171. doi: 10.1016/j.jbiotec.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Attri P, Venkatesu P, Kumar A, Byrne N. Phys. Chem. Chem. Phys. 2011;13:17023–17026. doi: 10.1039/c1cp22195g. [DOI] [PubMed] [Google Scholar]
- 28.Zhao H, Baker GA, Holmes S. Org. Biomol. Chem. 2011;9:1908–1916. doi: 10.1039/c0ob01011a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao H, Zhang C, Crittle TD. J. Mol. Catal. B: Enzym. 2013;85–86:243–247. [Google Scholar]
- 30.Zhao H, Baker GA, Holmes S. J. Mol. Catal. B: Enzym. 2011;72:163–167. doi: 10.1016/j.molcatb.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohno H, Yamaguchi N. Bioconjugate Chem. 1994;5:379–381. doi: 10.1021/bc00029a001. [DOI] [PubMed] [Google Scholar]
- 32.Kawahara NY, Ohno H. Bioconjugate Chem. 1997;8:643–648. doi: 10.1021/bc9701196. [DOI] [PubMed] [Google Scholar]
- 33.Kurusu F, Ohno H. Electrochimica Acta. 2000;45:2911–2915. [Google Scholar]
- 34.Chen J, Spear SK, Huddleston JG, Rogers RD. Green Chem. 2005;7:64–82. [Google Scholar]
- 35.Tang S, Zhao H. RSC Adv. 2014;4:11251–11287. doi: 10.1039/C3RA47191H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshpe-Besancon I, Auriol D, Paul F, Monsan P, Gripon JC, Ribadeau-Dumas B. Biotechnol. Appl. Biochem. 1993;18:93–102. [PubMed] [Google Scholar]
- 37.Rosell CM, Terreni M, Fernandez-Lafuente R, Guisan JM. Enzyme Microb. Technol. 1998;23:64–69. [Google Scholar]
- 38.Berkowitz DB, Hartung RE, Choi S. Tetrahedron: Asymmetry. 1999;10:4513–4520. [Google Scholar]
- 39.Schroën CGPH, Nierstrasz VA, Kroon PJ, Bosma R, Janssen AEM, Beeftink HH, Tramper J. Enzyme Microb. Technol. 1999;24:498–506. [Google Scholar]
- 40.Tang S, Jones CL, Zhao H. Bioresour. Technol. 2013;129:667–671. doi: 10.1016/j.biortech.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 41.Mamajanov I, Engelhart AE, Bean HD, Hud NV. Angewandte Chemie International Edition. 2010;49:6310–6314. doi: 10.1002/anie.201001561. [DOI] [PubMed] [Google Scholar]
- 42.Mukesh C, Mondal D, Sharma M, Prasad K. Chem. Commun. 2013;49:6849–6851. doi: 10.1039/c3cc42829j. [DOI] [PubMed] [Google Scholar]
- 43.Vijayaraghavan R, Izgorodin A, Ganesh V, Surianarayanan M, MacFarlane DR. Angewandte Chemie International Edition. 2010;49:1631–1633. doi: 10.1002/anie.200906610. [DOI] [PubMed] [Google Scholar]
- 44.Mondal D, Sharma M, Mukesh C, Gupta V, Prasad K. Chem. Commun. 2013;49:9606–9608. doi: 10.1039/c3cc45849k. [DOI] [PubMed] [Google Scholar]
- 45.Xia S, Baker GA, Li H, Ravula S, Zhao H. RSC Adv. 2014;4:10586–10596. doi: 10.1039/C3RA46149A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans DA, Fandrick KR, Song H-J. J. Am. Chem. Soc. 2005;127:8942–8943. doi: 10.1021/ja052433d. [DOI] [PubMed] [Google Scholar]
- 47.Hayakawa S, Michiue T, Okamoto M, Hatakeyama S, Ohta S. Heterocycles. 1988;27:457–473. [Google Scholar]
- 48.Myers MC, Bharadwaj AR, Milgram BC, Scheidt KA. J. Am. Chem. Soc. 2005;127:14675–14680. doi: 10.1021/ja0520161. [DOI] [PubMed] [Google Scholar]
- 49.Wolfe A, Shimer GHJ, Meehan T. Biochemistry. 1987;26:6392–6396. doi: 10.1021/bi00394a013. [DOI] [PubMed] [Google Scholar]
- 50.Atienzar FA, Venier P, Jha AN, Depledge MH. Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 2002;521:151–163. doi: 10.1016/s1383-5718(02)00216-4. [DOI] [PubMed] [Google Scholar]
- 51.Knierim E, Lucke B, Schwarz JM, Schuelke M, Seelow D. PLoS One. 2011;6:e28240. doi: 10.1371/journal.pone.0028240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morris ER, Rees DA, Thom D. Carbohydrate Research. 1980;81:305–314. [Google Scholar]
- 53.Zhao H, Jones CL, Cowins JV. Green Chem. 2009;11:1128–1138. [Google Scholar]
- 54.Zhao H, Song Z, Olubajo O. Biotechnol. Lett. 2010;32:1109–1116. doi: 10.1007/s10529-010-0262-4. [DOI] [PubMed] [Google Scholar]
- 55.Wehofsky N, Wespe C, Cerovsky V, Pech A, Hoess E, Rudolph R, Bordusa F. ChemBioChem. 2008;9:1493–1499. doi: 10.1002/cbic.200800025. [DOI] [PubMed] [Google Scholar]
- 56.Chandran A, Ghoshdastidar D, Senapati S. J. Am. Chem. Soc. 2012;134:20330–20339. doi: 10.1021/ja304519d. [DOI] [PubMed] [Google Scholar]
- 57.Zhao H. J. Chem. Technol. Biotechnol. 2015 [Google Scholar]
- 58.Cheng D-H, Chen X-W, Wang J-H, Fang Z-L. Chem. Eur. J. 2007;13:4833–4839. doi: 10.1002/chem.200601544. [DOI] [PubMed] [Google Scholar]
- 59.Ding Y, Zhang L, Xie J, Guo R. J. Phys. Chem. B. 2010;114:2033–2043. doi: 10.1021/jp9104757. [DOI] [PubMed] [Google Scholar]
- 60.Singh PK, Sujana J, Mora AK, Nath S. J. Photochem. Photobiol. A: Chem. 2012;246:16–22. [Google Scholar]
- 61.Egli M. Chemistry & biology. 2002;9:277–286. doi: 10.1016/s1074-5521(02)00116-3. [DOI] [PubMed] [Google Scholar]
- 62.Hud NV, Polak M. Current Opinion in Structural Biology. 2001;11:293–301. doi: 10.1016/s0959-440x(00)00205-0. [DOI] [PubMed] [Google Scholar]
- 63.McFail-Isom L, Sines CC, Williams LD. Current Opinion in Structural Biology. 1999;9:298–304. doi: 10.1016/S0959-440X(99)80040-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.