Abstract
We report a preliminary functional and complete structural characterization of a highly unusual geldanamycin analog, natalamycin A, that was isolated from Streptomyces strain M56 recovered from a South African nest of Macrotermes natalensis termites. Bioassay-guided fractionation based on antifungal activity led to the isolation of natalamycin A, and a combination of NMR spectroscopy and X-ray crystallographic analysis, including highly-accurate quantum-chemical NMR calculations on the largest and most conformationally-flexible system to date, revealed natalamycin A’s three-dimensional solid- and solution-state structure. This structure along with the structures of related compounds isolated from the same source suggest a geldanamycin-like biosynthetic pathway with unusual post-PKS modifications.
Introduction
Bacterially-produced small molecules modulate interactions between the microbial producers and their relatives, allies, competitors, and hosts. These molecules have long interested chemists because of their potential therapeutic value, their remarkable molecular structures, and the challenges posed by their synthesis and biosynthesis. In a systematic effort to discover new members of this family, we have been exploring the bacterial symbionts of insect agricultural systems.
Agriculture in insects is ancient, originating millions of years ago when insect farmers – ants, beetles, and termites, – formed obligate mutualisms with specialized food fungi.1 The insect benefits by feeding on the fungus, and the fungus benefits by the insect providing plant material for the fungus to consume in protected gardens. A third party to this symbiosis, bacteria, fend off pathogens by producing antibacterial and antifungal agents that suppress specialist and generalist crop pathogens. The secondary metabolites produced by the bacteria in these agricultural systems are expected to undergo multiple rounds of alteration and selection, which likely makes them promising therapeutic candidates. 1,2
In an effort to extend earlier studies, we investigated termite-associated Actinobacteria and their potential for producing biologically active small molecules.3 Screening bacterial isolates against the cultivar (Termitomyces spp.) and pathogen (Pseudoxylaria spp.) revealed that while most of the Actinobacteria produced molecules with antifungal activity, Streptomyces strain M56, isolated from the fungal comb material of a Macrotermes natalensis Mn802 colony, showed exceptionally high and broad-range antifungal activity. Further investigation of Streptomyces sp. M56 led to the isolation of natalamycin A (1) and related ansa macrolides (Figure 1).
Fig. 1.
Natalamycin A, geldanamycin and other analogs isolated from Streptomyces sp. M56
Results and Discussion
Fungal comb material was collected from a South African M. natalensis Mn802 colony (Figure 2) and bacteria from the comb were grown and subcultured to yield a Streptomyces isolate called M56. M56 is most closely related to S. malaysiensis 1160 as judged by 100% 16S rRNA sequence identity. On a preparative scale, M56 was grown on ISP-2 agar for 14 d and was subsequently extracted using iPrOH. The concentrated crude extract was subjected to bioassay-guided fractionation against Pseudoxylaria X802 using a disk assay. Fractionation began with C18 solid phase extraction followed by MeOH/H2O elution. Two fractions (80% MeOH in H2O and 100% MeOH) exhibited antifungal activity, and they were combined and subjected to preparative-scale C18 reverse-phase HPLC. 1H NMR analysis of a contiguous set of bioactive fractions indicated that geldanamycin (2),4 a known antifungal agent, was present. However, additional compounds were observed in these fractions. Through a careful series of semi-preparative reverse-phase HPLC experiments with a phenyl-hexyl column, reblastatin (3),5 17-O-demethyl-geldanamycin (4),6 19-S-methylgeldanamycin (5),7 17-amino-17-demethoxy-geldanamycin (6),8 methyl geldanamycinate derivative 7,9 and recently reported geldanamycin analog 9 were resolved and isolated.10 There were also two previously unreported metabolites. The first was 17-amino-17-demethoxy-methyl geldanamycinate (8), which is a plausible biosynthetic precursor to 6,11 and it was characterized through MS and 2D NMR analysis. The second and more structurally challenging metabolite was a fused bicyclic [6.4.0] ansa macrolide, which we named natalamycin A (1) after its association with M. natalensis. The yield of 1 when grown on 50 × 14 cm diameter ISP-2 agar plates was 0.7 mg. We were unable to observe natalamycin A production if the bacteria were grown in ISP-2 liquid media.
Fig. 2.
South African Macrotermes natalensis colony and fungal comb. The white nodules are the Termitomyces fungal cultivar.
While the fractions containing mixtures of natalamycin A, geldanamycin and its analogs exhibited antifungal activity against the Pseudoxylaria X802 fungus, activity was also observed against the Termitomyces T112 cultivar. This differs from the selectivities seen in beetle and ant systems, but it agrees with recent competition studies by Visser and co-workers in which Actinobacteria from termite nests were found to inhibit both cultivar and pathogenic fungus.12 They concluded that a mutualistic symbiosis would still be possible if the antibiotics were applied with spatial localization. It should also be noted that M56 produces not fully characterized metabolites with antifungal activity that are not geldanamycin analogs. Furthermore, natalamycin A exhibited weak or no activity against the fungal cultivar, S. cerevisiae, B. subtilis, and other isolated fungi. Studies are underway to further evaluate natalamycin A’s bioactivity in a larger set of assays.
In regards to the structural elucidation of natalamycin A (1), analysis through HR-ESI-MS provided the unfamiliar molecular formula of C35H50N2O11 (m/z: 697.3310 [M + Na]+), and initial further analysis relied heavily on NMR spectroscopy. NMR analysis revealed that it contains the macrolactam core of geldanamycin (2). HMBC data and comparison of 1 to the NMR data for 3 revealed that the six-membered carbocycle is a substituted phenol, not the substituted benzoquinone of 2. NMR analysis revealed an additional spin-system which appeared to form, in conjunction with the phenolic core, an unprecedented fused bicyclic [6.4.0] fragment that had not been found in any previously reported ansamycins. COSY and HMBC analysis allowed for the determination of bond connectivities for the 8-membered ring. It is interesting to note that while 1 was originally analyzed in CD3OD, the crucial HMBC correlation establishing the connectivity of the acetal moiety to the aromatic core was only observed by NMR analysis in CDCl3. This allowed for the assignment of C20 and C21 as the bridgehead carbons versus fusion through C19 and C20. Altogether, NMR analysis provided bonding arrangements for 1, but it was unable to clearly define the relative stereochemistries of the methoxy and methyl groups on the eight-membered heterocyclic ring or to connect those relative stereochemistries with those of the macrolactam.
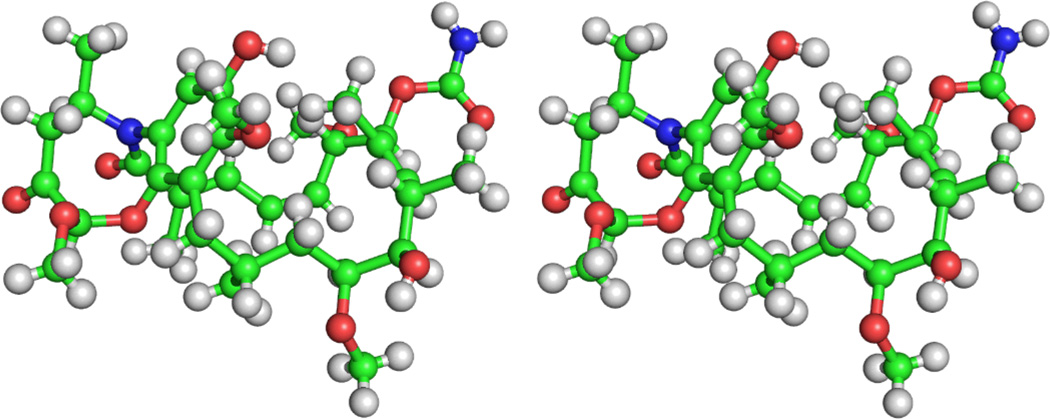
In order to fully elucidate the relative and absolute structure of 1, X-ray diffraction analysis was used. After multiple attempts, small crystals of 1 were grown through slow evaporation from CHCl3 with minimal MeOH. Achieving sufficient resolution and data quality for the measurement of these crystals virtually precluded the use of in-house diffractometers. Therefore, data collection was performed using high-flux synchrotron radiation with a helium cold stream (ChemMatCARS, Advanced Photon Source, Argonne National Laboratories), and the crystal structure was successfully solved and refined with sufficient resolution and quality (Figure 3).13 Fortuitous inclusion of three CHCl3 solvent molecules within the asymmetric unit cell allowed for the unambiguous determination of absolute stereochemistry at a shorter typical synchrotron wavelength (0.41 Å) affording a Hooft y parameter of 0.00(4).14 The methoxy and methyl groups on the eight-membered ring were both determined as 1′R and 4′R and the stereocenters on the macrolactam ring matched those of geldanamycin. Natalamycin A crystallizes in a closed C-clamp conformation in a similar fashion to 19-(2-furyl)geldanamycin synthesized by Moody and co-workers with its cis-configured macrolactam,15 and in a fashion different from the solid-state conformation of 17-azetidinyl-17-demethoxygeldanamycin, which has an open extended conformation with its trans-configured macrolactam.16
Fig. 3.
Wall-eye stereoview of the crystal structure of 1 exhibiting a C-clamp conformation, with CHCl3 solvent hidden for clarity. Ball-and-stick representation displayed; see Supporting Information for ORTEP plots.
Next, we wanted to predict 1H and 13C NMR chemical shifts for 1 with sufficient accuracy to understand its predominant solution conformation, to verify the assignment of experimental chemical shifts, and to establish reliable procedures for structural elucidation of other natural products in the laboratory. The conformation of geldanamycin analogs also has special significance as it affects binding to Hsp90, geldanamycin’s therapeutic target. Previous conformational studies of 2,17 17-azetidinyl-17-demethoxygeldanamycin,16 and 19-(2-furyl)geldanamycin15 used NMR and/or in silico analysis, and Moody and co-workers outlined the importance of geldanamycin analogs adopting a C-clamp conformation for target binding.15
We used a recent quantum-chemical NMR calculation method developed by Tantillo and co-workers18 that successfully revised the structures of aquatolide18a and nobilistine A18b as it promised the sensitivity we needed. The method depends on finding relevant conformers, which is not trivial for 1 due to its size and flexibility. Natalamycin A contains 98 atoms and has 294 degrees of freedom. This poses a particular challenge in regards to finding conformers, energetically ranking conformers and eliminating irrelevant ones through very computationally intensive DFT optimization and frequency calculations, and performing high-level NMR calculations, all in a high-throughput fashion. Through the use of multiple conformational search tools along with the aid of the X-ray crystal structure and subsequent DFT geometry optimizations using cluster hardware, two relevant conformers of 1 – C-clamp and extended – were afforded, and the C-clamp is the global minimum (ΔG298.15 K= 1.08 kcal/mol, 86% population). NMR calculations with implicit CHCl3 solvation, empirical scaling of the isotropic shifts, averaging of equivalent proton shifts, and Boltzmann averaging afforded the calculated 1H and 13C shifts relative to TMS (Figure 4).
Fig. 4.
Comparison of Boltzmann-averaged computed NMR chemical shifts (bolded) and experimental values (underlined and italicized) for 1 shown on the major C-clamp conformer with error statistics for a) 1H and b) 13C. Computed values were obtained using the SCRF(chloroform)-mPW1PW91/6-311+G(2d,p)//B3LYP/6–31+G(d,p) level of theory, and experimental values were obtained in CDCl3.
The calculated 1H and 13C NMR chemical shifts correspond very well to experimental values. The respective corrected mean absolute deviation (CMAD) for 1H and 13C shifts are 0.13 ppm and 2.1 ppm, and the respective maximum shift deviation for each is 0.57 ppm and 4.2 ppm. These error statistics values are low and closely correspond to those obtained by Tantillo and co-workers in their NMR calculation studies.18 The data strongly suggest that 1 adopts a C-clamp solution conformation due to significant differences in the error statistics for each individual conformer arising from 1H and 13C shift discrepancies. For the C-clamp conformer, the CMAD values for 1H and 13C shifts are 0.12 ppm and 2.2 ppm, respectively, whereas for the extended conformer it is 0.38 ppm and 3.2 ppm, respectively. The extended conformer has larger shift discrepancies overall. The C-clamp conformer geometry also aligns with the experimental ROESY correlations. These results show that given properly identified relevant conformers, highly accurate chemical shifts can be computed for large and flexible secondary metabolites, which further validates the power and versatility of the method by Tantillo and co-workers for natural product structural studies. To the best of our knowledge, this is the largest and most conformationally flexible molecule in which this quantum-mechanical NMR calculation method has been generally used, and is the first time that it has been employed for the purposes of resolving conformational versus bond connectivities.
Geldanamycin was isolated from S. hygroscopicus in 1970, and since then the ansamycin family of structurally complex and therapeutically promising molecules has grown considerably,4,19 serving as an impetus for biosynthetic studies. These studies have revealed that ansamycins are assembled from a 3-amino-5-hydroxybenzoic acid (AHBA) starter unit followed by the addition of extender units – malonate, methylmalonate, and methoxymalonate – to form their polyketide backbones.20 After chain extension, the resulting seco-progeldanamycin is released with concomitant macrolactam formation to release progeldanamycin.21 Progeldanamycin undergoes a variety of post-PKS modification – C-17 hydroxylation, C-17 O-methylation, C-21 oxidation, C-7 carbamoylation, and a late stage C-4/C-5 dehydrogenation – to form family members. The recent isolation of several new metabolites functionalized at C-19 highlight the possibilities of other family members from alternative post-PKS modifications.7,9,22 Another approach to ansamycin structural diversification using chain extender units has recently been reported.23
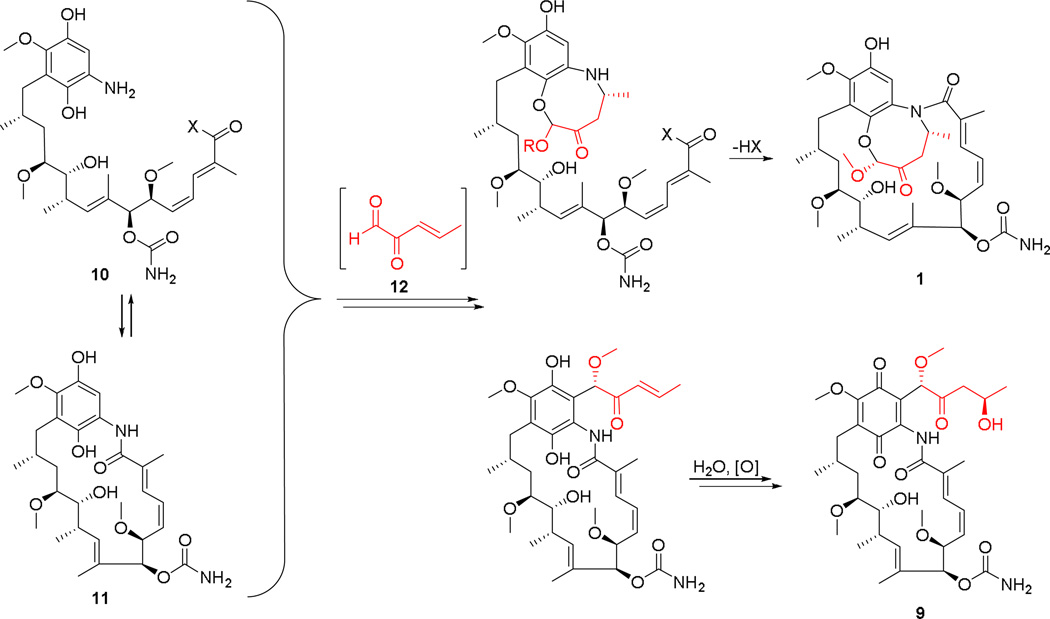
The pathway for ansamycin biosynthesis in S. hygroscopicus coupled with the identification of compounds 1–9 in this study on M56 allows a plausible biosynthetic hypothesis for 1 and 9 to be drawn (Figure 5). In this formulation the PKS ansamycin pathway could lead to 10 through early release of seco-progeldanamycin followed by the known modifications listed above. Alternatively progeldanamycin could undergo the same modifications to give 11, the macrolactam version of 10. A five-carbon fragment with the sort of functionality shown in 12 could react with either 10 or 11 to eventually give 1 or 9. It is hypothetically possible that a five-carbon fragment of this nature can be derived from 4-hydroxy-2-oxopentanoate, which is an intermediate in 3-phenylpropionate catabolism. This catabolic pathway is known in Escherichia coli and other microbes.24 For actinomycetes, an enzyme within this pathway that forms 4-hydroxy-2-oxopentanoate from 2-oxopenta-4-enoate (2-oxopent-4-enoate hydratase) is present in an annotated gene sequence belonging to Streptomyces sp. SirexAA-E (GenBank accession number for hydratase: AEN11956.1), which is a member of a bacterial/fungal symbiotic community associated with Sirex noctilio, an invasive pinewood-boring wasp.25
Fig. 5.
Biosynthetic hypothesis for natalamycin A (1) and 9 from Streptomyces sp. M56
Conclusions
This study explored the biosynthetic potential of Streptomyces sp. M56 isolated from the fungal comb of a Macrotermes natalensis Mn802 nest with an initial focus on natalamycin A (1). Natalamycin A was structurally characterized with NMR spectroscopy and X-ray crystallography. Additionally, quantum-chemical NMR calculations were successfully used on the largest and most conformationally-flexible system to date to predict its solution conformation, as opposed to previous applications of this technique to solely determine bond connectivities. The identification of structurally related metabolites that are likely biosynthetic precursors coupled with the known geldanamycin biosynthetic pathway allowed a plausible biosynthetic pathway to compounds 1 and 9 to be constructed as a basis for further studies. Efforts to sequence and assemble the G+C rich / highly redundant Streptomyces sp. M56 genome and efforts to maximize production of 1 for biological testing are currently underway in our laboratory.
Supplementary Material
Acknowledgements
We are grateful for financial support through the NIH (R01-GM086258 to J.C., U19-AI109673 and RC4-GM096347 to J.C. and C.R.C, and F32-GM108415 to T.R.R.) and through the German National Academy of Sciences Leopoldina for a postdoctoral fellowship to C.B. We thank Dr. Yu-Sheng Chen (ChemMatCARS, Advanced Photon Source, Argonne National Laboratory), for assistance with single-crystal data collection. ChemMatCARS Sector 15 is principally supported by the National Science Foundation/Department of Energy under grant number NSF/CHE-0822838. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. We also thank Dr. Shao-Liang Zheng (Department of Chemistry and Chemical Biology, Harvard University) for assistance with single-crystal data collection and helpful discussions on X-ray crystallography. Quantum-chemical calculations were run on the Odyssey cluster supported by the FAS Division of Science, Research Computing Group at Harvard University. We thank Dr. Duur K. Aanen for providing T112 and Mr. Saria Otani for providing the termite/fungal comb images for Figure 2 and the graphical abstract.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental and computational procedures, bioassay data, characterization data, NMR spectra, X-ray crystal data, Cartesian coordinates, and calculated NMR shifts. See DOI: 10.1039/b000000x/
Notes and references
- 1.For a review, see: Ramadhar TR, Beemelmanns C, Currie CR, Clardy J. J. Antibiot. 2014;67:53. doi: 10.1038/ja.2013.77. and references therein.
- 2.(a) Bode HB. Angew. Chem. Int. Ed. 2009;48:6394. doi: 10.1002/anie.200902152. [DOI] [PubMed] [Google Scholar]; (b) Kaltenpoth M. Trends Microbiol. 2009;17:529. doi: 10.1016/j.tim.2009.09.006. [DOI] [PubMed] [Google Scholar]; (c) Brownlie JC, Johnson KN. Trends Microbiol. 2009;17:348. doi: 10.1016/j.tim.2009.05.005. [DOI] [PubMed] [Google Scholar]; (d) Crawford JM, Clardy J. Chem. Commun. 2011;47:7559. doi: 10.1039/c1cc11574j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Seipke RF, Kaltenpoth M, Hutchings MI. FEMS Microbiol. Rev. 2012;36:862. doi: 10.1111/j.1574-6976.2011.00313.x. [DOI] [PubMed] [Google Scholar]
- 3.(a) Carr G, Poulsen M, Klassen JL, Hou Y, Wyche TP, Bugni TS, Currie CR, Clardy J. Org. Lett. 2012;14:2822. doi: 10.1021/ol301043p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Um S, Fraimout A, Sapountzis P, Oh D-C, Poulsen M. Sci. Rep. 2014;3 doi: 10.1038/srep03250. 3250:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBoer C, Meulman PA, Wnuk RJ, Peterson DH. J. Antibiot. 1970;23:442. doi: 10.7164/antibiotics.23.442. [DOI] [PubMed] [Google Scholar]
- 5.Takatsu T, Ohtsuki M, Muramatsu A, Enokita R, Kurakata SI. J. Antibiot. 2000;53:1310. doi: 10.7164/antibiotics.53.1310. [DOI] [PubMed] [Google Scholar]
- 6.Barziley E, Ben-Califa N, Supino-Rosin L, Kashman Y, Hirschberg K, Elazar Z, Neumann D. J. Biol. Chem. 2004;279:6847. doi: 10.1074/jbc.M312799200. [DOI] [PubMed] [Google Scholar]
- 7.(a) Liu X, Li J, Ni S, Wu L, Wang H, Lin L, He W, Wang Y. J. Antibiot. 2011;64:519. doi: 10.1038/ja.2011.39. [DOI] [PubMed] [Google Scholar]; (b) Li S, Cui J, Lu X, Zheng Z, Liu X, Ni S, Wang Y, Wu L. J. Antibiot. 2013;66:499. doi: 10.1038/ja.2013.31. [DOI] [PubMed] [Google Scholar]
- 8.(a) Yin M, Lu T, Zhao LX, Chen Y, Huang SX, Lohman JR, Xu LH, Jiang CL, Shen B. Org. Lett. 2011;13:3726. doi: 10.1021/ol201383w. [DOI] [PubMed] [Google Scholar]; (b) Tadtong S, Meksuriyen D, Tanasupawat S, Isobe M, Suwanborirux K. Bioorg. Med. Chem. Lett. 2007;17:2939. doi: 10.1016/j.bmcl.2006.12.041. [DOI] [PubMed] [Google Scholar]
- 9.Hu Z, Liu Y, Tian ZQ, Ma W, Starks CM, Regentin R, Licari P, Myles DC, Hutchinson CR. J. Antibiot. 2004;57:421. doi: 10.7164/antibiotics.57.421. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Ni S, Wu L, Li L, Jiang B, Wang H, Sun G, Gan M, Li J, He W, Lin L, Wang Y, Bai S, Si S. J. Na. Prod. 2013;76:969. doi: 10.1021/np4000679. [DOI] [PubMed] [Google Scholar]
- 11.Though iPrOH was initially used for extraction, there is a possibility that 8 could instead result from methanolysis of 6 during C18 fractionation.
- 12.Visser AA, Nobre T, Currie CR, Aanen DK, Poulsen M. Microb. Ecol. 2012;63:975. doi: 10.1007/s00248-011-9987-4. [DOI] [PubMed] [Google Scholar]
- 13.CCDC 994285 contains the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif
- 14.Hooft RW, Straver LH, Spek AL. J. Appl. Cryst. 2008;41:96. doi: 10.1107/S0021889807059870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitson RRA, Chang C-H, Xiong R, William HEL, Davis AL, Lewis W, Dehn DL, Siegel D, Roe SM, Prodromou P, Ross D, Moody CJ. Nat. Chem. 2013;5:307. doi: 10.1038/nchem.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnur RC, Corman ML. J. Org. Chem. 1994;59:2581. [Google Scholar]
- 17.Thepchatri P, Eliseo T, Cicero DO, Myles D, Snyder JP. J. Am. Chem. Soc. 2007;129:3127. doi: 10.1021/ja064863p. [DOI] [PubMed] [Google Scholar]
- 18.(a) Lodewyk MW, Soldi C, Jones PB, Olmstead MM, Rita J, Shaw JT, Tantillo DJ. J. Am. Chem. Soc. 2012;134:18550. doi: 10.1021/ja3089394. [DOI] [PubMed] [Google Scholar]; (b) Lodewyk MW, Tantillo DJ. J. Nat. Prod. 2011;74:1339. doi: 10.1021/np2000446. [DOI] [PubMed] [Google Scholar]; (c) Lodewyk MW, Siebert MR, Tantillo DJ. Chem. Rev. 2012;112:1839. doi: 10.1021/cr200106v. [DOI] [PubMed] [Google Scholar]; (d) [accessed November 22, 2013]; CHESHIRE Chemical Shift Repository. http://cheshirenmr.info. [Google Scholar]
- 19.For selected reviews, see: Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Proc. Natl. Acad. Sci. USA. 1994;91:8324. doi: 10.1073/pnas.91.18.8324. Floss HG, Yu TW. Chem. Rev. 2005;105:621. doi: 10.1021/cr030112j. Fukuyo Y, Hunt CR, Horikoshi N. Cancer Lett. 2010;290:24. doi: 10.1016/j.canlet.2009.07.010. Franke J, Eichner S, Zeilinger C, Kirschning A. Nat. Prod. Rep. 2013;30:1299. doi: 10.1039/c3np70012g.
- 20.(a) Rascher A, Hu Z, Viswanathan N, Schirmer A, Reid R, Nierman WC, Lewis M, Hutchinson CR. FEMS Microbiol. Lett. 2003;218:223. doi: 10.1016/S0378-1097(02)01148-5. [DOI] [PubMed] [Google Scholar]; (b) Hong Y-S, Lee D, Kim W, Jeong J-K, Kim C-G, Sohng JK, Lee J-H, Paik S-G, Lee JJ. J. Am. Chem. Soc. 2004;126:11142. doi: 10.1021/ja047769m. [DOI] [PubMed] [Google Scholar]; (c) Rascher A, Hu Z, Buchanan GO, Reid R, Hutchinson CR. Appl. Environ. Microbiol. 2005;71:4862. doi: 10.1128/AEM.71.8.4862-4871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Lee D, Lee K, Cai XF, Dat NT, Boovanahalli SK, Lee M, Shin JC, Kim W, Jeong JK, Lee JS, Lee C-H, Lee J-H, Hong Y-S, Lee JJ. ChemBioChem. 2006;7:246. doi: 10.1002/cbic.200500441. [DOI] [PubMed] [Google Scholar]; (e) Shin J-C, Na Z, Lee D-H, Kim W-C, Lee K, Shen Y-M, Paik S-G, Hong Y-S, Lee J-J. J. Microbiol. Biotechnol. 2008;18:1101. [PubMed] [Google Scholar]; (f) Kim W, Lee D, Hong SS, Na Z, Shin JC, Roh SH, Wu C-Z, Choi O, Lee K, Shen Y-M, Paik S-G, Lee JJ, Hong Y-S. ChemBioChem. 2009;10:1243. doi: 10.1002/cbic.200800763. [DOI] [PubMed] [Google Scholar]; (g) Ni S, Jiang B, Wu L, Wang Y, Zhou H, He W, Wang H, Zhu J, Li S, Li T, Zhang K. J. Antibiot. 2014;67:183. doi: 10.1038/ja.2013.94. [DOI] [PubMed] [Google Scholar]; (h) Li T, Ni S, Jia C, Wang H, Sun G, Wu L, Gan M, Shan G, He W, Lin L, Zhou H, Wang Y. J. Nat. Prod. 2012;75:1480. doi: 10.1021/np3001738. [DOI] [PubMed] [Google Scholar]; (i) Cheng-Zhu W, Jang J-H, Ahn JS, Hong Y-S. J. Microbiol. Biotechnol. 2012;22:1478. doi: 10.4014/jmb.1206.06026. [DOI] [PubMed] [Google Scholar]
- 21.Eichner S, Eichner T, Floss HG, Fohrer J, Hofer E, Sasse F, Zeilinger C, Kirschning A. J. Am. Chem. Soc. 2012;134:1673. doi: 10.1021/ja2087147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin L, Ni S, Wu L, Wang Y, Wang Y, Tao P, He W, Wang X. Biosci. Biotechnol. Biochem. 2011;75:2042. doi: 10.1271/bbb.110361. [DOI] [PubMed] [Google Scholar]
- 23.Ding L, Maier A, Fiebig H-H, Görls H, Lin W-H, Peschel G, Hertweck C. Angew. Chem. Int. Ed. 2011;50:1630. doi: 10.1002/anie.201006165. [DOI] [PubMed] [Google Scholar]
- 24.Turlin E, Perrotte-piquemal M, Danchin A, Biville F. J. Mol. Microbiol. Biotechnol. 2001;3:127. [PubMed] [Google Scholar]
- 25.(a) Adams AS, Jordan MS, Adams SM, Suen G, Goodwin LA, Davenport KW, Currie CR, Raffa KF. ISME J. 2011;5:1323. doi: 10.1038/ismej.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Takasuka TE, Book AJ, Lewin GR, Currie CR, Fox BG. Sci. Rep. 2013;3:1030. doi: 10.1038/srep01030. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Takasuka TE, Acheson JF, Bianchetti CM, Prom BM, Bergeman LF, Book AJ, Currie CR, Fox BG. PLoS One. 2014;9:e94116. doi: 10.1371/journal.pone.0094166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.