Abstract
Monotremes (platypus and echidna) are the descendants of the oldest ancestor of all extant mammals distinguished from other mammals by mode of reproduction. Monotremes lay eggs following a short gestation period and after an even briefer incubation period, altricial hatchlings are nourished over a long lactation period with milk secreted by nipple-less mammary patches located on the female’s abdomen. Milk is the sole source of nutrition and immune protection for the developing young until weaning. Using transcriptome and mass spectrometry analysis of milk cells and milk proteins, respectively, a novel Monotreme Lactation Protein (MLP) was identified as a major secreted protein in milk. We show that platypus and short-beaked echidna MLP genes show significant homology and are unique to monotremes. The MLP transcript was shown to be expressed in a variety of tissues; however, highest expression was observed in milk cells and was expressed constitutively from early to late lactation. Analysis of recombinant MLP showed that it is an N-linked glycosylated protein and biophysical studies predicted that MLP is an amphipathic, α-helical protein, a typical feature of antimicrobial proteins. Functional analysis revealed MLP antibacterial activity against both opportunistic pathogenic Staphylococcus aureus and commensal Enterococcus faecalis bacteria but showed no effect on Escherichia coli, Pseudomonas aeruginosa, Staphylococcus epidermidis, and Salmonella enterica. Our data suggest that MLP is an evolutionarily ancient component of milk-mediated innate immunity absent in other mammals. We propose that MLP evolved specifically in the monotreme lineage supporting the evolution of lactation in these species to provide bacterial protection, at a time when mammals lacked nipples.
Keywords: mammals, monotreme, platypus, echidna, lactation, antibacterial protein
Introduction
Lactation evolved in the late Triassic period, and although it is a defining feature of mammals it predated their origin (Oftedal 2002a, 2013; Lefèvre, Sharp, et al. 2010). Monotremes are the mammals most distant phylogenetically from the placental mammals. They probably retain ancestral traits that have been lost/changed in therians (e.g., lay eggs, lack nipples) (Oftedal 2002a, 2013; Lefèvre, Sharp, et al. 2010). The Monotremata lineage comprises platypus (Ornithorhynchus anatinus) and two genera of echidnas, short-beaked echidna (Tachyglossus aculeatus) and the long-beaked echidnas (Zaglossus spp.) (Springer and Krajewski 2009). Monotremes exhibit a fascinating combination of reptilian, avian, and mammalian features (Warren et al. 2008). Monotremes are oviparous (egg-laying) mammals and eggs are incubated exterior to the mother’s body until hatching (Musser 2005; Springer and Krajewski 2009). Monotreme reproductive systems are highly specialized to facilitate both the production of eggs and milk (Grützner et al. 2008).
After a short gestation period of 15–21 days (platypus) (Holland and Jackson 2002; Hawkins and Battaglia 2009) and 21 days (short-beaked echidna), leathery-shelled eggs are laid (Morrow et al. 2009). The short-beaked echidna incubates its egg in a functional incubatorium pouch on the abdomen (Griffiths et al. 1969), whereas those of the platypus are probably held between the curled tail and the abdomen of the female in the nesting burrow (Burrell 1927; Griffiths 1978). The egg is incubated externally for 10–12 days in the platypus (Griffiths 1978; Hawkins and Battaglia 2009), and 10–10.5 days in the short-beaked echidna, until hatching (Griffiths et al. 1969). The monotreme young hatch at a very early stage of development resembling an early fetal stage of development in eutherians (Grant 1995; Hughes and Hall 1998). Young rely on milk for the remainder of development during a long lactation period extending to 114–145 days in platypus (Grant and Griffiths 1992; Holland and Jackson 2002; Hawkins and Battaglia 2009) and 160–210 days in echidna (Morrow et al. 2009; Morrow and Nicol 2013), depending on geographical location. In comparison, eutherian mammals have evolved with a longer gestation allowing for prolonged development in utero, and the neonate is more developed at birth (Hamilton et al. 2011; Power and Schulkin 2013). During this period, the young are housed in a burrow and do not leave until weaning (Grant 2007). This implies that young excrete within the burrow environment. The difficulties of gaining access to breeding burrows during the 4-month period of lactation from hatching until emergence have prevented any direct evidence for management of excretory waste products.
Much of the development of monotreme hatchlings, including their immune system occurs before weaning (Whittington et al. 2009; Wong et al. 2009; Wang et al. 2011). During this time, the young are situated within the pouch environment in short-beaked echidnas and later move to a potentially pathogen-laden burrow (Wong et al. 2009; Wang et al. 2011). The platypus lacks a pouch, so from an early age platypus nestlings are deposited in the burrow which is likely to be pathogen-laden (Hawkins and Battaglia 2009). Interestingly, monotreme mammary gland structure is similar in both the short-beaked echidna and the platypus despite the short-beaked echidna being a terrestrial mammal and platypus being an amphibious mammal (Griffiths et al. 1969, 1973; Musser 2005). The alveoli and duct system is similar to that of placental mammals, but the areolae where the milk emerges are hidden by fur in platypus, covered by hairs in echidna and there are no nipples (Griffiths et al. 1969, 1973). Monotreme mammary glands therefore represent the first known attempt to produce milk by a sophisticated mammary gland structure (Griffiths et al. 1969, 1973; Oftedal 2002a).
It has been proposed that monotreme lactation originally evolved to prevent desiccation of the eggs or for protection against microbes, and subsequently evolved a nutritional role (Warren et al. 2008; Oftedal 2012, 2002b; Urashima et al. 2012). Several studies have shown that a variety of antimicrobial proteins are expressed in either mammary glands or milk in marsupials, humans, cow and mouse, and these factors are believed to contribute to a lower incidence of infections (Aniansson et al. 1990; Strömqvist et al. 1995; Murakami et al. 2005; Hettinga et al. 2011; Nicholas et al. 2012; Dallas et al. 2013). Historically it was generally thought that monotreme milk contained antimicrobial proteins (Blackburn et al. 1989), and identification of various immunological factors such as lysozyme and transferrin in monotreme milk supports the idea (Hopper and McKenzie 1974; Teahan and McKenzie 1990). Recently, our laboratory reported a monotreme protein EchAMP (Echidna Antimicrobial Protein) secreted in milk with antibacterial activity which was shown to be specific to the monotreme lineage (Bisana et al. 2013). These antimicrobial milk proteins may have evolved as a consequence of the hatchlings needing to survive without an adaptive immune system in pathogen-laden environments. This interest extends to revealing further antimicrobial proteins in monotreme milk, which could uncover more of the diversity of antibacterial proteins.
In order to study the genes expressed in mammary glands during lactation in monotremes, we previously used cDNA sequencing to elucidate the monotreme milk cell transcriptome. A novel cDNA that was common to both platypus and short-beaked echidna that had apparent similarity to C6orf58 was identified (Lefèvre et al. 2009; Lefèvre, Menzies, et al. 2010; Lefèvre, Sharp, et al. 2010). Here, we determine that the gene producing this cDNA is not C6orf58 and is renamed Monotreme Lactation Protein (MLP). Our data suggest that MLP has evolved as a gene which is highly expressed in milk cells and the protein comprises a significant fraction of monotreme milk proteins. MLP is present only in the monotreme lineage and appears to protect the young from microbial infections during lactation. This novel protein extends the existence of anti-infectious molecules in monotreme milk and adds to the complexity of species-specific antibacterial activity to protect immunocompromised hatchlings.
Materials and Methods
Ethics Statement
This work was carried out under permit from the Tasmanian Department of Primary Industries, Water & Environment, and the South Australian Department of Environment and Heritage, and the University of Tasmania and University of Adelaide Animal Ethics Committees. This work complies with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (2004), Scientific Licence (SL100489) NSW Office of Environment and Heritage, and NSW Department of Primary Industries Animal Research Authority (Trim 09/3535 May 2012).
Monotreme Milk and Tissue Collection
Short-beaked echidna (T. aculeatus) milk samples were collected during the reproductive season near Hobart, Tasmania, Australia (supplementary table S1, Supplementary Material online). Milk was collected at different times of lactation from platypuses (O. anatinus) in rivers on the southern tableland of New South Wales, Australia (supplementary table S2, Supplementary Material online). Anaesthetized short-beaked echidnas and conscious platypuses were injected intramuscularly with 0.2 ml of synthetic oxytocin (2 IU, Syntocin; Sandoz-Pharma, Basel, Switzerland) and mammary glands gently massaged to collect milk. All milk samples were transferred to −80 °C storage.
Tissue from an adult male echidna was collected from one animal (heart, intestine, kidney, liver, penis, submandibular glands, spleen, stomach, testis, and thyroid glands) and frozen at −80 °C.
Protein Concentration Determination
Protein concentration of monotreme skim milk samples was determined using a BCA (bicinchoninic acid) protein assay kit (Thermo Fisher Scientific, Australia), according to the manufacturer’s instructions.
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis and Mass Spectrometry
Milk samples were thawed at 4 °C and diluted with 1:2 1× PBS (pH 7.5). The samples were centrifuged at 16,000 × g for 20 min at 4 °C to remove the fat and cells. The skim milk fraction was carefully transferred to a fresh Eppendorf tube and centrifuged at 16,000 × g at 4 °C for 15 min, and any residual fat removed. The platypus and short-beaked echidna skim milk fractions (30 µg) were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) alongside molecular weight markers (Precision Plus Protein Kaleidoscope standards, BioRad) using 12.5% criterion Tris–HCl polyacrylamide gels (BioRad). Proteins were electrophoresed at a constant current of 200 V for 1 h at room temperature and stained using Coomassie Brilliant Blue R-250 overnight. The stained gel spot of interest was excised and cut into smaller pieces. MLP bands were transferred to microcentrifuge tubes, washed twice with 50% acetonitrile and 50 mM triethyl ammonium bicarbonate (TEAB) buffer. Reduction was performed with reducing buffer (10 mM tris (2-carboxyethyl) phosphine in 50 mM TEAB), followed by alkylation 100 mM iodoacetamide in 50 mM TEAB and incubation in the dark for 1 h. Samples were then dried, rehydrated with trypsin (1 µg) in 50 mM TEAB, and digested at 37 °C for 16 h. The reaction was stopped by the addition of 10% formic acid. The supernatant containing the digested peptides was collected and analyzed by ESI-TOF MS/MS using an Agilent 1100 Series HPLC coupled to an Agilent LC/MSD Trap XCT plus Mass Spectrometer (Bio21, Melbourne, Australia). The data were analyzed by correlating the peptide masses obtained to the predicted peptide masses from MLP in the monotreme milk cell transcriptome data and Ensembl platypus genome database, and identified using search algorithms ProteinPilot and Mascot.
SDS-PAGE Analysis of MLP Expression in Milk Collected across Lactation
Samples containing 50 μg of total protein were mixed with SDS sample buffer. Samples were heated at 100 °C for 5 min and loaded onto SDS-PAGE gels. The short-beaked echidna skim fractions were prepared as previously described and analyzed on a 12% precast criterion TGX SDS-PAGE (BioRad, Australia) and platypus milk samples were analyzed on a 12.5% precast criterion Tris–HCl SDS-PAGE (BioRad, Australia). The gels were stained with Coomassie Brilliant Blue R-250 dye for overnight and destained with a methanol/acetic acid/water solution (50/40/10). Protein bands were visualized in the gel and analyzed on a BioRad ChemiDoc XRS + system (BioRad).
RNA Preparation and Reverse Transcriptase Polymerase Chain Reaction Analysis
RNA was extracted from echidna thyroid gland, submandibular glands, liver, spleen, stomach, kidney, ileum, jejunum, duodenum, heart, testis, and penis tissue using Tripure reagent (Roche Applied Science) according to the manufacturer’s instructions. RNA was resuspended in nuclease free water and stored at −80 °C.
Total RNA (1 µg) was used as a template to synthesize cDNA using Super Script III Reverse Transcriptase (Invitrogen). Reverse transcriptase polymerase chain reaction (RT-PCR) was performed using gene-specific primers, echidna MLP (For: 5′-ACCATGGCGTTCTCCCTC-3′, Rev: 5′-GGAGGATGTGAAGTTT CGACG-3′). Platypus-specific primers were made and used to amplify short-beaked echidna glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (For: 5′-AAGGCTGTGGGCAAGG TCAT-3′, Rev: 5′-CTGTTGAAGTCACAGG AGAC-3′) as previously reported (Ordoñez et al. 2008). cDNA was used with Gotaq green master mix (Promega) with initial denaturation at 94 °C for 3 min followed by 30 cycles of 94 °C for 30 s, 60 °C for 1 min, 72 °C for 1 min, and a final extension at 72 °C for 10 min. PCR products were resolved on a 1% agarose gel using electrophoresis and visualized with SYBR Safe DNA gel stain (Invitrogen) using a BioRad ChemiDoc XRS + system (BioRad).
MLP cDNA Cloning
RNA from monotreme milk cells was extracted as previously described (Lefèvre et al. 2009), followed by cDNA preparation using SuperScript III Reverse Transcriptase kit (Invitrogen). RT-PCR was performed using primers designed specifically for the platypus MLP gene (For: 5′-CACCATGGCGCTCTCCCTCT-3′, Rev: 5′-GAGACTGAGCTGGACGATGTT-3′). PCR amplification was conducted in 25 μl of reaction mixture containing GoTaq Green master mix (Promega). PCR reaction was performed as follows: 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 60°C for 1 min, 72°C for 1 min, and final extension 72°C for 5 min. PCR products were resolved on agarose gel and then cleaned using the Qiagen gel extraction kit (Qiagen). The DNA was ligated into pGEM-T easy vector at 4 °C over night according to manufacturer’s instructions (pGEM-T easy vector system, Promega). The products were transformed into JM109 competent cells, then plated on LB-ampicillin blue/white selection plates and incubated overnight at 37 °C. Positive colonies from each MLP clone were selected for DNA extraction and grown overnight in LB-ampicillin 37 °C. Plasmid DNA was prepared using the Qiagen Miniprep Kit (Qiagen), and the presence of the fragment of interest was verified by NOT1 restriction enzyme digestion. The DNA insert was sequenced before the plasmid was used as the template for the following PCRs.
Construction of MLP Expression Vector
A construct containing a cDNA for platypus MLP with a FLAG tag (a single Flag epitope DYKDDDDK) fused to its C-terminus was ligated into the pTarget expression vector (Promega).
To generate insert for subcloning, MLP cDNA was amplified using the forward primer pMLP (For2: 5′-CACCATGGCGCTCTCCCTCT-3′, Rev2: 5′-TCACTTGTCATCG TCGTCCTTGTAATCGACGGGAGTGGGGATCC-3′) under the following PCR conditions; 30 cycles of 94 °C for 3 min, 94 °C for 30 s, 60 °C for 1 min, 72 °C for 1 min followed by 72 °C final extension for 5 min. The amplified DNA was examined on a 0.8% agarose gel with SYBR-safe staining under ultraviolet (UV) light, excised and subsequently purified using an agarose gel extraction kit (Qiagen) following the instructions provided by the manufacturer. The DNA was ligated into pTARGET expression vector (Promega) using T4 DNA ligase from Promega, following the manufacturer’s instructions to generate pTarget-MLP. All plasmids were validated using DNA sequencing.
Cell Culture and Transfection
Human Embryonic Kidney cell line (HEK293T) was maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% l-glutamine, and 1% penicillin/streptomycin. All cells were grown in a humidified atmosphere containing 5% CO2 at 37 °C. HEK293T cells (106 cells/6-well plates) were seeded in 2% FBS/serum free Opti-MEM media into individual wells of 6-well plates. After 24 h cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 6 h the transfection media was removed and cells were incubated with 2 ml of fresh serum free Opti-MEM media. pGFP (pmaxGFP expressing green fluorescent protein) was used as a fluorescent marker for transfection efficiency. Conditioned media was collected 48 h after transfection and centrifuged at 5,000 × g for 10 min to remove cell debris and stored at −80 °C until needed.
Protein Purification
Recombinant Flag-tagged MLP was purified by affinity column chromatography using anti-Flag M2 affinity gel (Sigma, USA) according to manufacturer’s standard protocol. The conditioned media was loaded onto a 10-ml column of anti-Flag M2 affinity gel and washed with 100 ml of TBS buffer (10 mM Tris–HCl, 150 mM NaCl, pH 7.4) to remove the unbound proteins. After washing twice, bound protein was eluted with 100 μg/ml Flag peptide (Sigma, USA) in TBS buffer.
SDS-PAGE and Western Blotting Analysis
The pTarget (empty vector control) conditioned media, pTarget-MLP conditioned media, and affinity purified MLP (30 µg) were analyzed on a 15% SDS-PAGE (Tris–HCl SDS gels; Biorad, Australia) and visualized with Coomassie Brilliant Blue R-250 dye. For western blot analysis, 50 μg of each sample was electrophoresed in 10% SDS-polyacrylamide gels (Tris–HCl SDS gels; BioRad, Australia) and transferred onto a PVDF membrane using a semidry blotter (BioRad). Immunoblotting was conducted using a horseradish peroxidase-conjugated anti-FLAG M2 mouse monoclonal antibody (Origene) and visualized using ECL chemiluminescence (GE Healthcare, USA). The reactive protein was visualized by enhanced chemiluminescence under a BioRad ChemiDoc XRS + system (BioRad).
Antibacterial Assay
Bacterial strains, Staphylococcus aureus ATCC 25923, Enterococcus faecalis ATCC 10100, Escherichia coli ATCC 2348/69, Pseudomonas aeruginosa ATCC 27853, Staphylococcus epidermidis ATCC 12228, Es. coli ATCC O157:H7, and Salmonella enterica ATCC 43971 were streaked on Iso-Sensitest agar (ISA; Oxoid) plates and incubated overnight at 37 °C. Isolated colonies of each strain were inoculated into 3 ml of Iso-Sensitest broth and grown overnight at 37 °C with agitation in a shaker incubator. The overnight cultures were inoculated into fresh ISA broth at a ratio of 1:100 and grown at 37 °C with agitation until an optical density at 600 nm (OD600) of 0.6. An empirically determined count of 100 cell suspension in 50 µl of ISA broth was added to each well of a 96-well black, clear-bottom plate (Costar; Corning Incorporated, Corning, NY). MLP purified protein (20 μg/ml) and Alamar Blue (10 µl; Invitrogen, #152763SA) were added to each well. Bacitracin (100 μg/ml) was used as a positive control in the assay and it is active against only Gram-positive bacteria (Sigma, #11702). Bovine serum albumin (BSA; 20 μg/ml) and protein elution TBS buffer were used as negative controls. Plates were incubated at 37 °C with shaking and fluorescence was measured every hour at an excitation of 544 nm wavelength whereas emission was measured at 590 nm using the Glomax Multi Detection System (Promega). All treatments were performed in triplicate and experiments were repeated at least thrice.
N-Linked Glycans Release by PNGase F
Affinity-purified MLP (20 µg) was denatured in denaturing buffer (5% SDS, 0.4 M DTT) at 100 °C for 5 min. After samples were cooled to room temperature, they were incubated with 5 units of PNGase F enzyme (New England Biolabs, glycerol free) at 37 °C for 16 h. An accurate molecular mass of the purified, glycosylated MLP was determined by directly injecting the protein (100 µg) onto an Accurate TOF LC/MS 6220 (Agilent Technologies) through an ESI source, followed by deconvolution of the primary mass-to-charge data using the program MassHunter (Agilent Technologies).
Circular Dichroism Spectroscopy
Circular dichroism (CD) spectra analysis of MLP (protein 6.25–12.5 μg/ml) was carried out under a nitrogen atmosphere on a Jasco J-815 CD spectrophotometer (ATA Scientific, NSW, Australia) and analyzed at room temperature using a quartz cell of 0.1 cm path length (Starna Pty. Ltd., Atascadero, CA) using purified recombinant MLP. Wavelength scans were collected using a 1.0 nm bandwidth, 1.0 s averaging time, and an average of five scans for each sample. Spectra were smoothed but not baseline corrected and signals were recorded in ellipticity (θ) given in millidegrees (mdeg).
Calculations of the MLP grand average hydropathy (GRAVY) index and positive charge were performed using the ProtParam tool of the Expert Protein Analysis System (ExPASy) Proteomics server.
Phylogenetic Tree Construction
To search for the possible MLP sequences in other species, we performed a BLASTP search against the nonredundant archive database at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/, last accessed April 25, 2014) and Ensembl database (version 74) protein annotations of vertebrate genomes (http://www.ensembl.org, last accessed April 25, 2014). Sequences of 49 different species showing similarity to MLP were selected and aligned using ClustalX2 with default parameters (Larkin et al. 2007). To account for uncertainty in the phylogeny, the protein sequence alignment was bootstrap sampled to generate 100 data sets using the SEQBOOT program from the PHYLIP software package (Felsenstein 1989). The pairwise genetic distances between MLP and related sequences in each of the 100 data sets were measured with the PROTDIST program utilizing the Jones–Taylor–Thornton matrix in the PHYLIP software. The protein evolutionary phylogenetic tree constructed from the program PHYLIP was viewed with Tree View (Page 1996). (See supplementary table S3, Supplementary Material online, for sources of protein sequences.) Sequences with C6orf58 annotations were investigated where possible genomic locations were examined to confirm gene identity. Duplicated C6orf58 sequences were designated C6orf58-like and appeared to be cis duplications of the C6orf58 locus.
Comparison of MLP and C6orf58 Proteins in Vertebrate Species
A standard TBLASTN was used to search against the nonredundant archive database at the NCBI website (http://www.ncbi.nlm.nih.gov/, last accessed April 25, 2014) and Ensembl database (version 74) protein annotations of vertebrate genomes (http://www.ensembl.org, last accessed April 25, 2014). All protein sequences were aligned using ClustalW2 with default parameters and manual adjustments.
Gene Structure Analysis of Platypus MLP and Related Genes in Vertebrates
tBLASTn searches were used using MLP and the nonredundant archive database at the NCBI website (http://www.ncbi.nlm.nih.gov/) and Ensembl database (version 74) nucleotide annotations of complete vertebrate genomes to identify MLP orthologs and related genes (genes information given in supplementary table S4, Supplementary Material online).
Synteny Analysis
To compare the flanking loci genes of platypus MLP and C6orf58 from other vertebrates, the genes were identified in the zebrafish, frog, chicken, platypus, opossum, cow, sheep, dog, mouse, and human genomes and retrieved from the latest Ensembl database (Ensembl genes 74). The accession numbers of the gene sequence names for gene structure comparison analyses are given in supplementary table S4, Supplementary Material online.
In Silico Analysis of MLP
The putative signal peptide sequence prediction was performed using the HMM (hidden Markov models) method in Signal 4.0 server (http://www.cbs.dtu.dk/services/SignalP/, last accessed April 25, 2014 ). The probability of a signal peptidase cleavage site, located between positions 17 and 18, is 0.909. The protein molecular mass and pI (isoelectric point) was analyzed using web-based tools hosted on the ExPASy proteomics server portal (http://web.expasy.org/compute_pi/, last accessed April 25, 2014). The prediction of N-glycosylation sites was confirmed at the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/, last accessed April 25, 2014) (Blom et al. 2004). The PSIPRED server graphical viewer from the Brunel Bioinformatics group of Brunel University was used for the secondary structure prediction (McGuffin et al. 2000). Multiple sequence alignments were obtained using the ClustalW2 on the EMBL-EBI web site.
Statistical Analysis
Data were analyzed for statistical significance by a two-tailed t-test with significance accepted at P < 0.05.
Results
Identification of MLP in Monotreme Milk Cells
Previous studies in our laboratory have examined the monotreme milk cell transcriptome using a noninvasive approach involving cDNA analysis of cells collected from mammary gland secretions (Lefèvre et al. 2009; Lefèvre, Menzies, et al. 2010). In our analysis of both platypus and short-beaked echidna peak lactation milk cells, a number of expressed whey and casein protein genes were identified, including a highly expressed novel transcript with similarity to C6orf58 (Lefèvre et al. 2009; Lefèvre, Menzies, et al. 2010). Our previous study identified this cDNA as similar to human C6orf58, encoding a novel protein with unknown function. However, we have since determined that this annotation is incorrect and this novel sequence represents a novel gene renamed MLP. Transcriptome analysis revealed that MLP was highly expressed representing 4.1% of total transcripts in platypus milk cells and 6.8% in short-beaked echidna milk cells (Lefèvre et al. 2009; Lefèvre, Menzies, et al. 2010) (see supplementary table S5, Supplementary Material online).
Monotreme MLP cDNA and Gene Structure
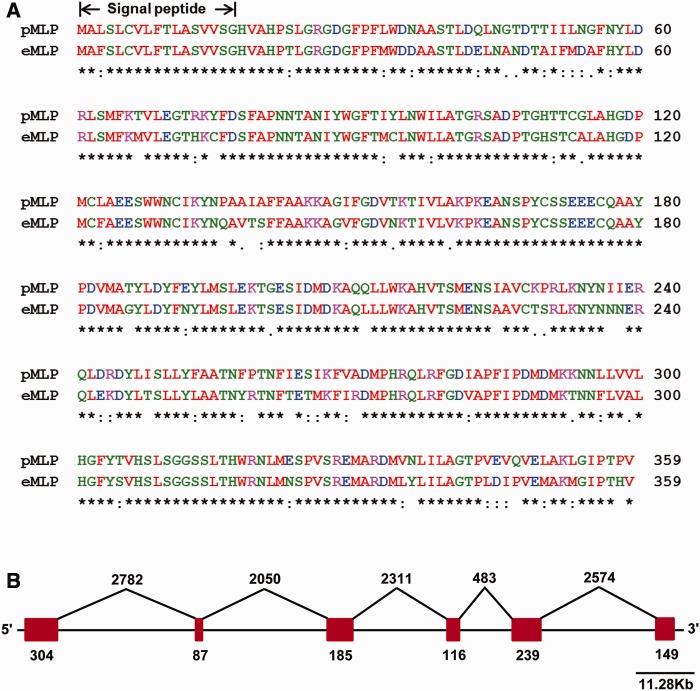
The complete nucleotide sequence of platypus MLP and short-beaked echidna MLP cDNA was determined and both were shown to comprise an open reading frame of 1,080 bp encoding a protein of 359 amino acids with a molecular mass of 38.4 kDa (excluding the signal peptide) and an estimated pI value of 5.60. Based on signal peptide analysis (Bendtsen et al. 2004), the first 17 amino acid residues encode a signal peptide in the N-terminus for both species. Protein sequence conservation between the two monotreme MLPs showed 82% identity and 95% similarity (fig. 1A). In order to assign putative function to MLP, a search for evolutionarily conserved domains was performed using the conserved domain database search options of the NCBI (Marchler-Bauer et al. 2011). Both MLP sequences revealed a conserved uncharacterized “Domains of Unknown Function 781” (DUF781) motif spanning the region from 1 to 350 amino acids. Alignment of platypus MLP cDNA nucleotide sequence to the platypus Ensembl genome assembly revealed MLP localized to supercontig 5 and contig 11343:6533–17761. Alignment of MLP cDNA sequence with its respective genomic DNA sequence indicated the gene was divided into six exons and five introns, spanning 11.28 kb of sequence (fig. 1B).
Fig. 1.—
Multiple sequence alignment of platypus and short-beaked echidna MLPs. (A) Alignment of the amino acid sequence of the platypus MLP (pMLP) and short-beaked echidna MLP (eMLP) shows 82% sequence identity and 95% similarity. Sites that are conserved in both species are indicated, respectively, by an asterisk (*), a colon (:) and dot (.). (B) MLP gene organization. Schematic representation of platypus MLP encoded by the 1,080 bp derived from six exons. Exons are shown with vertical red color boxes and introns as lines connecting exons. The length of exons and introns is shown.
Identification of MLP in Monotreme Milk
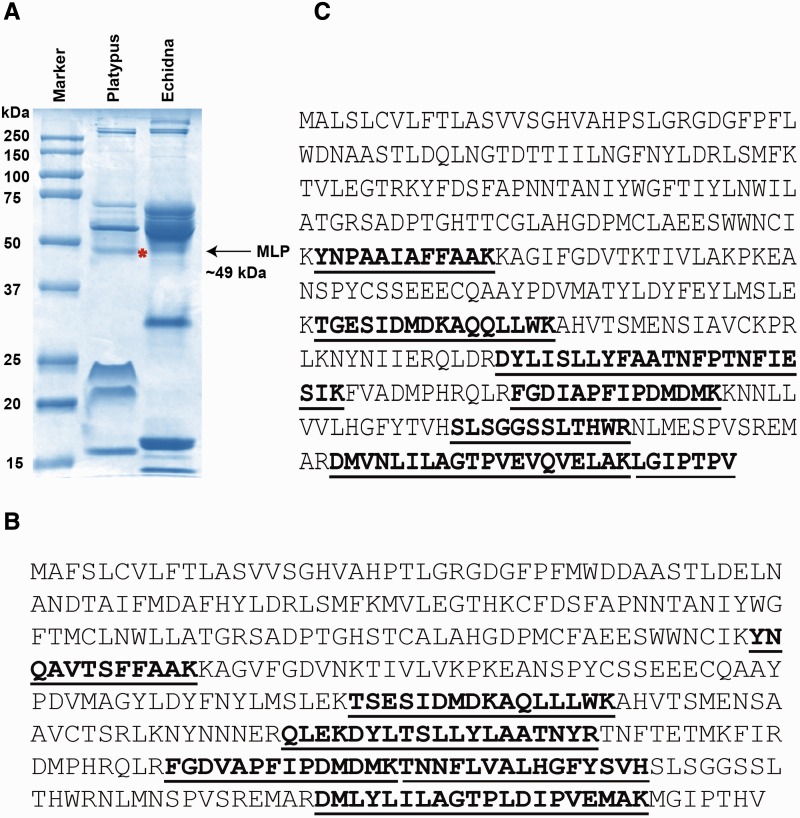
In order to confirm that MLP was secreted into milk, samples from platypus and short-beaked echidna milk were analyzed by SDS-PAGE and mass spectrometry (fig. 2A). A total of seven peptides at a confidence level of 99% in platypus MLP and six peptides in short-beaked echidna MLP were identified within the protein band migrating at approximately 49 kDa (fig. 2B and C; see supplementary table S6, Supplementary Material online, for detailed information regarding peptides identified by mass spectrometry). The spectral counting of peptides was achieved with high accuracy mass measurement. One of the most abundant fragment peaks at [M + Z]+ = 2,680.72 had the sequence of the tryptic peptide 244–267 in platypus MLP. The identified peptide sequences covered 29% and 27% of platypus and short-beaked echidna MLP, respectively.
Fig. 2.—
Identification of monotreme MLP by SDS-PAGE and mass spectrometry. (A) Coomassie stained SDS-PAGE gel and ESI-TOF mass spectrometry identifying MLP of platypus (asterisk) and short-beaked echidna (arrow) in milk. Lane 1: molecular weight marker, lane 2: platypus skim fraction, and lane 3: echidna skim fraction. (B) The complete amino acid sequence of platypus MLP and (C) short-beaked echidna MLP are shown overlaid with identified peptides (bold and underlined) showing sequence coverage of 29% and 27%, respectively.
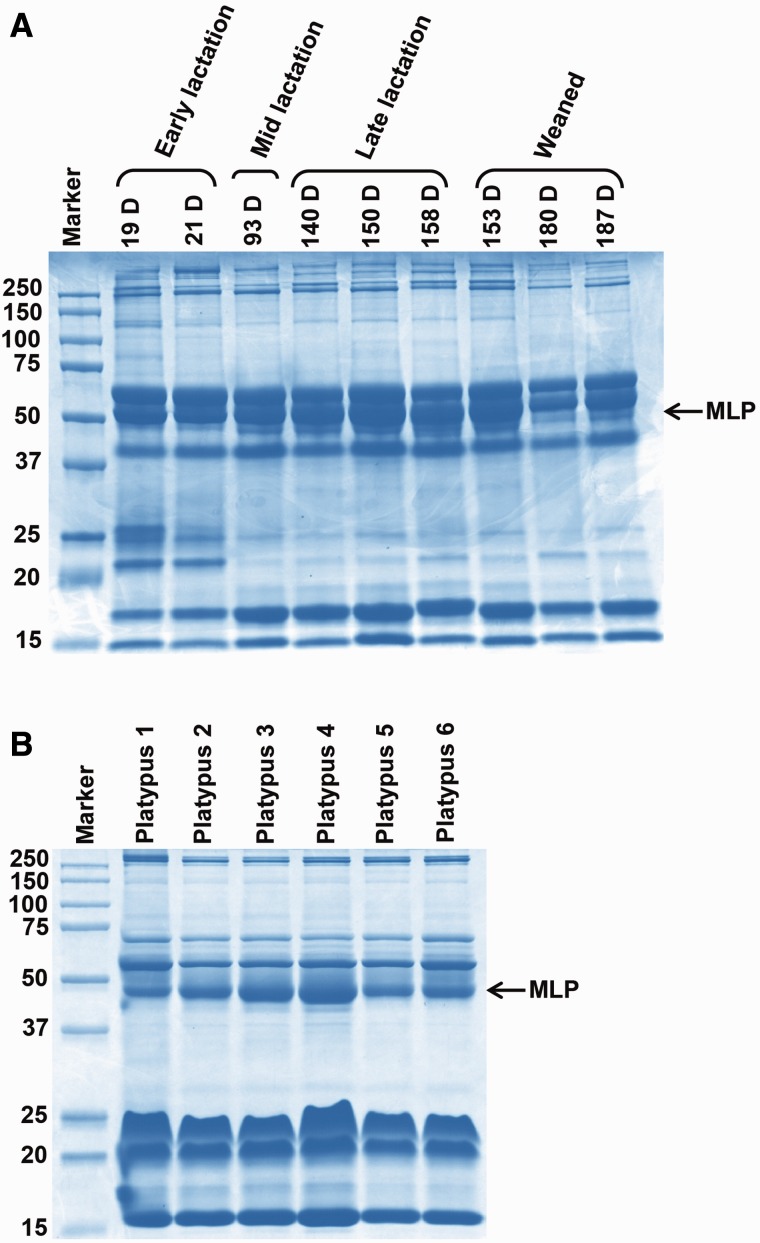
To test whether the pattern of MLP secretion varies over the lactation period in short-beaked echidna and platypus milk, a range of milk samples were collected during different stages of lactation from animals caught in the wild. Electrophoresis of the skim fraction showed that MLP is expressed throughout lactation in short-beaked echidna milk collected during the period of early lactation (days 19, 21), mid lactation (day 93), late lactation (days 140, 150, 158) and weaning (days 153, 180, 187), and platypus milk collected from six individuals during peak lactation (fig. 3A and B).
Fig. 3.—
Detection of MLP in short-beaked echidna and platypus milk during lactation. Coomassie stained SDS-PAGE electrophoretograms of (A) short-beaked echidna and (B) platypus milk samples during lactation showing MLP detection in all echidna and platypus milk samples. The sizes of the protein molecular weight markers (Precision Plus Protein Kaleidoscope standards, Biorad) are indicated. D: Lactation day.
Expression of MLP in Short-Beaked Echidna Tissues
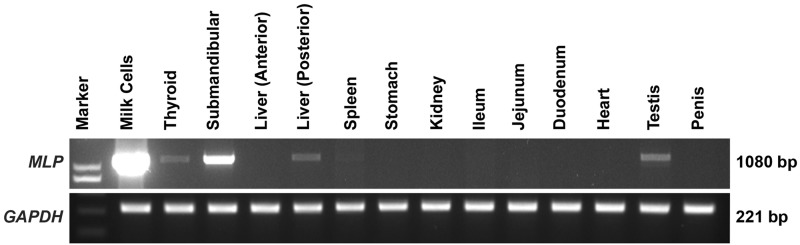
RNA extracted from thyroid gland, submandibular glands, liver, spleen, stomach, kidney, ileum, jejunum, duodenum, heart, testis, and penis from short-beaked echidna was analyzed for MLP gene expression. The MLP gene was expressed at higher levels in short-beaked echidna milk cells but lower levels of expression detected in submandibular glands, thyroid gland, liver, spleen, and testis, but was not detected in other tissues (fig. 4). GAPDH has been used as a house keeping gene to study the relative expression of many genes in the mammary gland during pregnancy and lactation (Miller et al. 2006; Li et al. 2012). Based on this evidence the differences in template concentration between tissues in this study were taken into consideration by normalization to the GAPDH housekeeping gene. However, it has been reported that housekeeping genes can have variable expression under different systems; hence, the stability of housekeeping gene expression should be validated for each experimental system (Bas et al. 2004; Huggett et al. 2005). GAPDH has been used as a house keeping gene to normalize gene expression profiles across tissues for other monotreme studies (Ordoñez et al. 2008; Hrdličková et al. 2012).
Fig. 4.—
Analysis of echidna MLP expression in short-beaked echidna tissues. MLP transcript was detected by RT-PCR in various short-beaked echidna tissues. The upper panel shows MLP mRNA is highly expressed in milk cells and submandibular glands whereas low level of expression was observed in thyroid gland, liver, spleen, and testis. The expression was normalized to the internal control GAPDH gene (lower panel).
Recombinant Expression of Platypus MLP
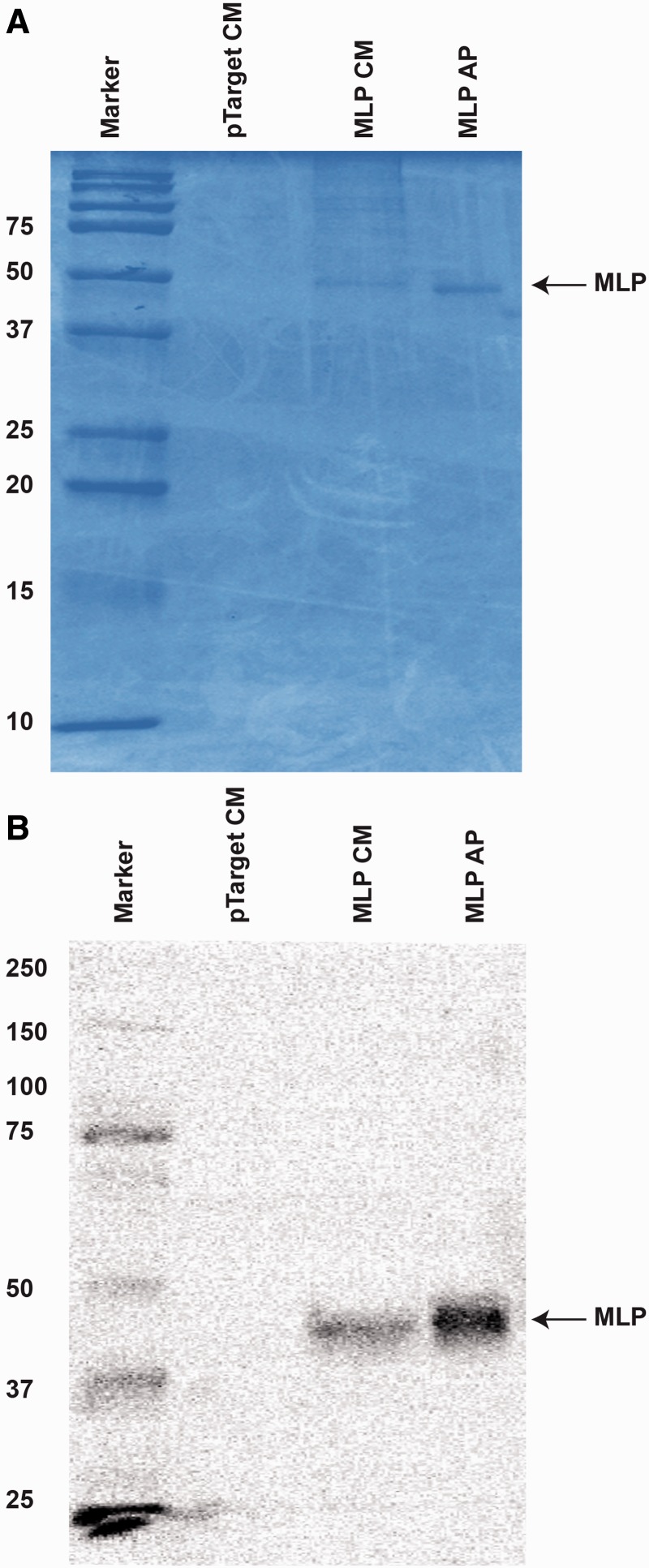
In order to study the structure and function of MLP, recombinant MLP was produced. Full-length platypus MLP cDNA was cloned into a mammalian expression vector which included an in-frame carboxy-terminal FLAG tag. The recombinant construct was expressed transiently in HEK293T cells. Conditioned media was collected 48 h after transfection and FLAG-tagged MLP was affinity purified using an anti-FLAG M2 affinity gel. Platypus recombinant MLP was detected by Coomassie staining as a single band corresponding to the predicted size on SDS-PAGE in both conditioned media and affinity purified samples. It was not detected in cells transfected with empty vector control (fig. 5A). The identity of the platypus recombinant MLP was further confirmed by western blot analysis using a mouse anti-DDK to recognize the FLAG-tag fusion MLP with an apparent molecular mass of approximately 49 kDa in the conditioned media and affinity purified samples (fig. 5B).
Fig. 5.—
Expression and detection of platypus recombinant MLP in HEK293T cells. (A) SDS-PAGE gel showing the expression and affinity purification of platypus MLP from conditioned medium collected from pMLP-FLAG transfected HEK293T cells. Lane 1, molecular weight marker; lane 2, pTARGET (empty vector) conditioned media; lane 3, MLP with pTARGET conditioned media; and lane 4, affinity purified MLP samples were run through a 15% SDS-PAGE and the gel was stained with Coomassie Brilliant Blue. (B) Western blot analysis of conditioned media collected from HEK293T cells transfected with FLAG tagged MLP plasmid. MLP-FLAG protein was detected using HRP-conjugated anti-DDK monoclonal antibody (Origene). Lane 1, molecular weight marker; lane 2, pTARGET (empty vector) conditioned media; lane 3, MLP with pTARGET conditioned media, and lane 4, affinity purified MLP samples. The antibody recognizes specific MLP-FLAG protein in conditioned media and affinity purified samples but not in an empty vector conditioned media control.
Antibacterial Activity of Platypus Recombinant MLP
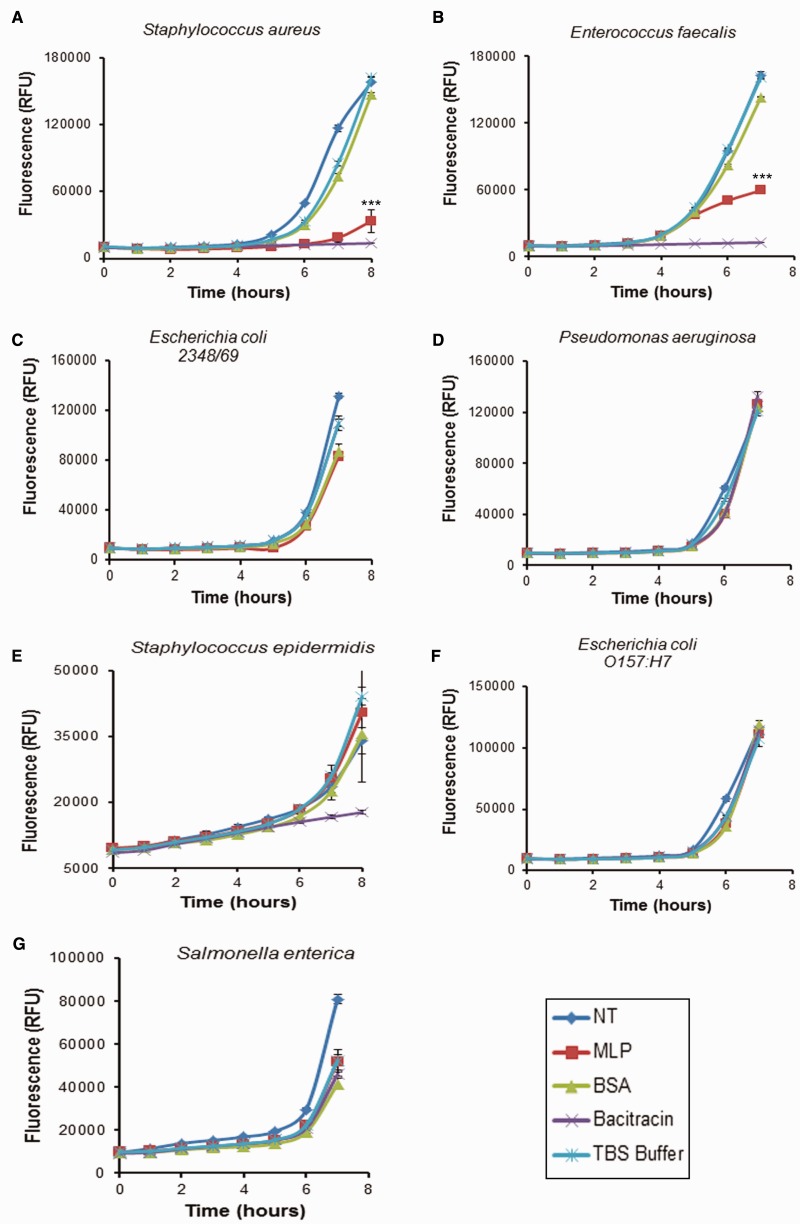
The antibacterial activity of the purified MLP was assayed using common Gram-positive and Gram-negative bacterial species (Gram-positive bacteria: St. aureus, St. epidermidis, En. faecalis; and Gram-negative bacteria: Es. coli, Sa. enterica, P. aeruginosa) and growth inhibition was measured by Alamar blue screening. Bacitracin activity showed antibacterial activity against all Gram-positive bacteria, whereas MLP showed significant antibacterial activity against two Gram-positive bacterial species St. aureus and En. faecalis (P < 0.05) but had no effect on the third Gram-positive species, St. epidermidis, nor the Gram-negative Es. coli 2348/69, P. aeruginosa, Es. coli O157:H7 and Sa. enterica (fig. 6). Antimicrobial activity was observed but levels of activity varied between St. aureus and En. faecalis. A BSA control was used for these analyses and no significant inhibition of bacterial growth was observed. The results demonstrate that the ancient MLP milk protein exhibits potent antibacterial activity not only against commensal but also against pathogenic bacteria of clinical importance.
Fig. 6.—
Antibacterial activity of platypus recombinant MLP. Bacteriostatic assay using purified MLP against (A) St. aureus, (B) En. faecalis, (C) Es. coli 2348/69, (D) P. aeruginosa, (E) St. epidermidis, (F) Es. coli O157:H7, and (G) Sa. enterica determined by using alamar blue reagent assay. MLP showed significant inhibition of growth on St. aureus and En. faecalis when compared with the TBS buffer control and BSA treatment (*statistically significant result P < 0.05). MLP showed no inhibition of growth of Es. coli 2348/69, P. aeruginosa, St. epidermidis, Es. coli O157:H7, and Sa. enterica (P > 0.05). *P < 0.05, **P < 0.01, and ***P < 0.001. Experiments were repeated three times and standard error bars are shown. NT, no treatment; MLP, purified MLP; Bacitracin: bacitracin antibiotic; TBS, Tris-buffered saline buffer.
MLP Posttranslational N-Linked Glycosylation
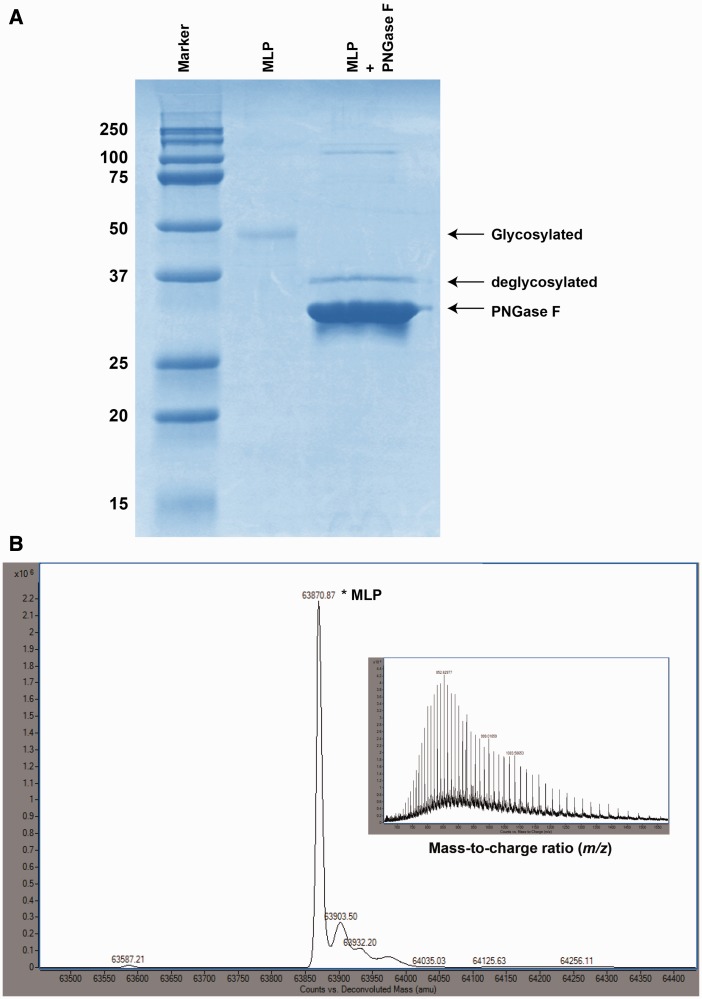
Platypus MLP was analyzed for the presence of N-glycosylation consensus sites using the N-glycosylation prediction tool NetNGlyc 1.0 server. MLP showed two putative N-glycosylation sites with a threshold glycosylation potential ≥0.5 in its N-terminal region at positions 45–48 and 82–85. Comparison of SDS-PAGE analyses of recombinant MLP and MLP derived from milk demonstrated that both proteins migrated at a similar molecular weight (49 kDa) and were therefore glycosylated to a similar degree (figs. 3 and 5A). To establish the nature of possible N-linked glycosylation modification, the purified recombinant MLP was treated with endoglycosidase PNGase F enzyme to remove N-glycans, followed by SDS-PAGE analysis. MLP deglycosylation revealed a clear reduction in the apparent molecular mass of the protein (∼49 kDa–38.4 kDa) (fig. 7A). This observation is in agreement with the theoretical average molecular mass of 38.4 kDa for MLP calculated from amino acid sequence. Analysis of glycosylated MLP using ESI-TOF LC/MS detected a main peak with a more precise molecular weight of 63,870.8 Da (fig. 7B). The observed difference in the mass between calculated molecular mass of the MLP and the experimental values suggests that there is approximately 25.4 kDa of carbohydrate linked to the protein.
Fig. 7.—
Analysis of the N-linked glycosylation of MLP. (A) The SDS-PAGE shows the difference in protein migration due to N-linked glycosylation of recombinant MLP (∼49 kDa), as compared with the deglycosylated MLP (38.4 kDa). Lane 1, molecular weight marker; lane 2, purified MLP (untreated); lane 3, purified MLP treated with PNGase F (deglycosylated). Enzymatic deglycosylation converted MLP into a single faster migrating band. (B) Deconvoluted ESI-TOF mass spectrum of glycosylated MLP, with the mass of the highest peak (63,870.87 Da) corresponding to the relative molecular weight of MLP, indicating that heavy glycosylation of protein. Inset: Raw mass-to-charge spectrum of MLP. Raw MS data plotted as intensity versus mass-to-charge ratio.
MLP Secondary Structure
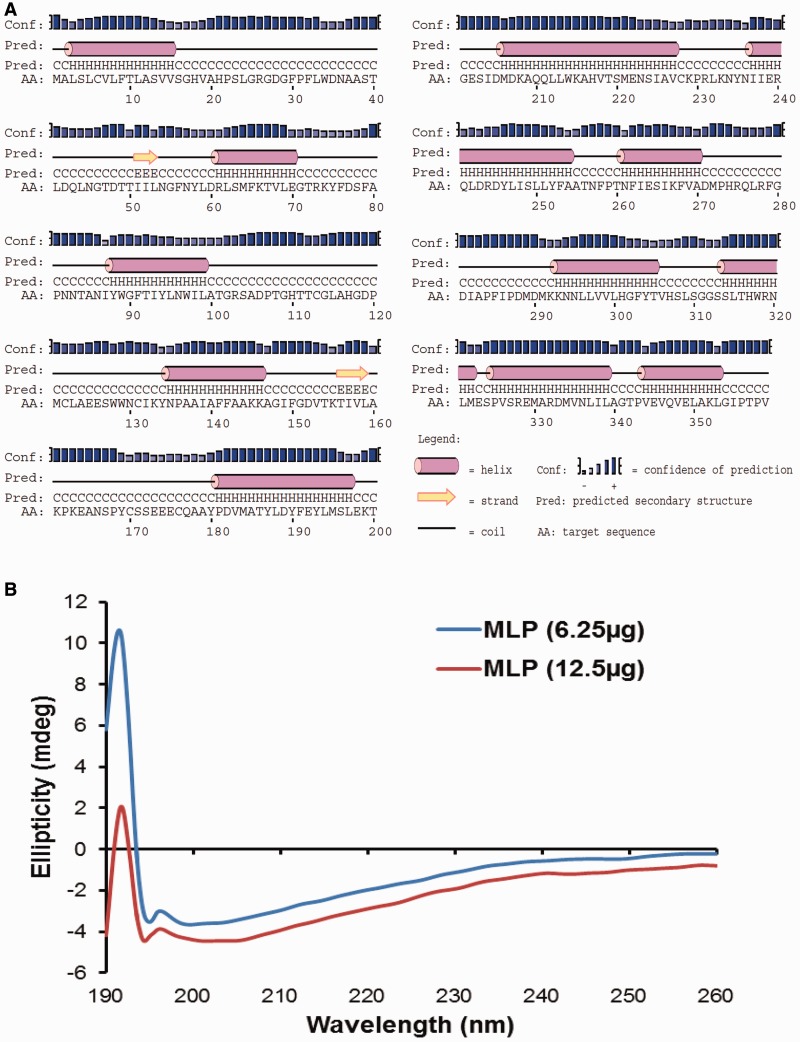
For several antibacterial proteins, a close correlation between α-helical secondary structure and antibacterial activity against Gram-positive and Gram-negative bacteria has been described (Brogden 2005; Wiesner and Vilcinskas 2010). To elucidate the secondary structural features of the platypus MLP, a bioinformatics analysis of amino acid sequence using protein secondary structure prediction (PSIPRED) server estimated a high percentage of α-helical elements (fig. 8A). The predicted secondary structure of platypus MLP was studied qualitatively using purified MLP by CD spectroscopy. CD spectra were collected between 190 and 260 nm at room temperature (fig. 8B). The MLP CD profile exhibited a positive dichroic band with a maximum at 190 nm characteristic of a protein with high α-helix content.
Fig. 8.—
MLP secondary structure analysis. MLP is an α-helical protein determined by PSIPRED prediction followed by CD signal in the far-UV region. (A) Platypus MLP secondary structure predicted by PSIPRED server shows high proportion of alpha helix content. Helical and β-sheet secondary structure elements are depicted by pink barrels and yellow arrows, respectively. Prediction confidence is represented by blue bars. (B) Platypus MLP secondary structure analysis determined by CD. The CD spectra acquired in the range of 190–260 nm for recombinant MLP. Analysis of CD spectrum shows a positive dichroic band with a maximum at 190 nm that highlights the content of α-helices in the protein.
In addition, sequence properties of GRAVY analysis suggest that MLP has low GRAVY indices −0.101, the negative GRAVY score indicates the possibility of better interaction with water. This was in agreement with the respective percentages of cationic amino acid residues. It is noteworthy that MLP is rich in negatively charged amino acids (37 residues: 20 Asp and 17 Glu), as compared with positively charged amino acids (29 residues: 17 Lys, 12 Arg) (supplementary table S7, Supplementary Material online). Positively charged amino acids are present in the alpha-helices region (see supplementary fig. S1A, Supplementary Material online). Kyte–Doolittle hydrophobicity plot analysis using window size n = 7 shows hydrophobic and hydrophilic regions appeared alternatively with random tendency at 1–359 amino acids of MLP (supplementary fig. S1B, Supplementary Material online). Based on a hydropathy plot, MLP is primarily an amphipathic protein.
MLP Homologous Genes
To elucidate the apparent evolution of the platypus MLP gene, we used multiple approaches. We first searched for orthologs of platypus and short-beaked echidna MLP in NCBI and Ensembl protein databases. The search resulted in the identification of a large number of secreted protein coding genes possess a conserved DUF781 domain. We found that the closest relative of MLP appears to be paralog C6orf58 (chromosome 6 open reading frame 58) which is present in the platypus genome (Ensembl Gene ID: ENSOANG00000008387). We searched for MLP closely related sequences in 49 vertebrate genomes and constructed a phylogenetic tree based on amino acid sequences (see supplementary fig. S2, Supplementary Material online). Phylogenetic analysis confirmed that platypus and short-beaked echidna MLPs are closely related homologs and grouped together in the same clade. Interestingly, Tasmanian devil, guinea pig, cow, and sheep C6orf58 proteins were positioned as a sister group to monotreme MLP. Together this clade does not represent the normal pattern of evolution for these species (i.e., monotreme, marsupials, and eutherian lineages), suggesting that these genes are not homologs but share sequence similarity. Platypus C6orf58 formed a group with other well-conserved C6orf58 sequences in fish, reptiles, amphibians, avian, and mammals and formed a separate clade and was representative of the expected pattern of evolution for these species.
In order to examine the degree of similarity and validate the relationships between monotreme MLP and C6orf58, and monotreme MLP and Tasmanian devil, guinea pig, cow and sheep C6orf58 proteins, multiple sequence similarities, identities, and alignments were performed. We compared the C6orf58 amino acid sequences from four species representative of the large clade comprising different eutherian orders (human, mouse, dog, and cow), marsupial (opossum), and monotreme (platypus) with monotreme MLP (supplementary fig. S3, Supplementary Material online). Comparison of the monotreme MLP and mammalian C6orf58 proteins revealed significant divergence and identity variation from 24% to 82% depending on the species. Platypus MLP deduced amino acid sequence shares 27.58%, 30.27%, 26.60%, 28.57%, 24.13%, 28.36% identity with C6orf58 sequences from human, mouse, dog, cow, opossum and platypus, respectively (supplementary table S8, Supplementary Material online). In contrast, platypus MLP shares a high level of amino acid identity (82.17%) with short-beaked echidna MLP sequence.
Conserved amino acid residues within a protein provide an indication of sites essential for structure and biological function. Several key amino acid residues were conserved between MLP and C6orf58 proteins. Conserved cysteine residues were observed at four positions, which are predicted to be essential for disulphide bonding. Both platypus and short-beaked echidna MLP contains two and four potential N-glycosylation sites, respectively. These occur at positions 45 (NGTD) and 82 (NNTA) in platypus MLP and positions 47 (NDTA), 82 (NNTA), 154 (NKTI), and 261 (NFTE) in short-beaked echidna MLP. The N-glycosylation site reported for human C6orf58 protein at 69 (NQT) (Ramachandran et al. 2008) was retained for each of the vertebrate C6orf58 sequences examined, with exception of mouse, opossum, and platypus. Predicted N-glycosylation sites were observed at other positions, 117 (NESL), 171 (NVSD), 196 (NSSD), 257 (NRTY) for mouse, 260 (NKTY) for opossum, and 105 (NVTT), 168 (NVTD) for platypus.
Multiple sequence similarities, identities, and alignments were also performed with species from the C6orf58-like sequences (Tasmanian devil, guinea pig, cow, and sheep) that made up the sister group to MLP. Amino acid sequence alignment showed low similarity between MLP and the sister group (supplementary fig. S4, Supplementary Material online), which was further supported by amino acid similarity and identity (supplementary table S9, Supplementary Material online). C6orf58-like sequences from eutherian species within the sister group shared between 53–90% identity and 80–96% similarity, 53% identity and 70% similarity with the marsupial (Tasmanian devil), and 21–25% identity and 62–67% similarity with monotreme MLP. The marsupial (Tasmanian devil) showed 25% identity and 65–67% similarity with monotreme MLP.
MLP Genomic Organization
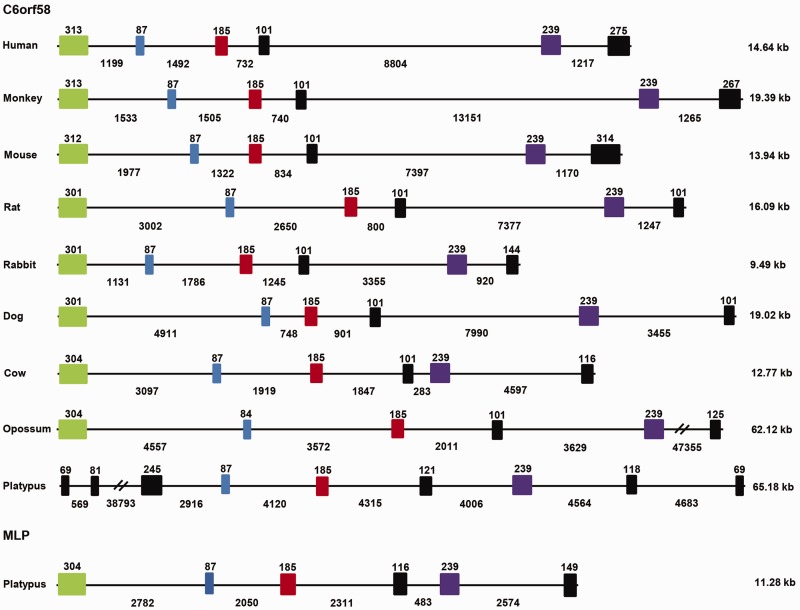
To understand the evolutionary relationship between MLP and C6orf58 genes, we compared the genomic structures of these genes from various species including platypus, opossum, cow, dog, rabbit, rat, mouse, monkey, and human. Analysis of gene structure revealed that platypus MLP comprised six exons and five introns which appeared to be highly similar to the gene structure of C6orf58 from other mammals (fig. 9). Interestingly, C6orf58 gene structure appeared more divergent in platypus with additional exon/intron boundaries at both the 5′- and 3′-end of the gene.
Fig. 9.—
Comparison of gene structure between the platypus MLP and C6orf58 genes. Gene structure schematic diagrams of the platypus MLP and C6orf58 genes in other vertebrates showing conservation and divergence of gene structure between species. Conserved exons are illustrated in green, blue, red and purple colors, and nonconserved exons are in black. Exons (shaded boxes) and introns (lines) are shown, based on Ensembl gene prediction. The length (base pairs) of exons and introns is shown above and below the boxes and lines, respectively. The total length of exons and introns for each gene is represented in kilobase. Genes are drawn approximately to scale.
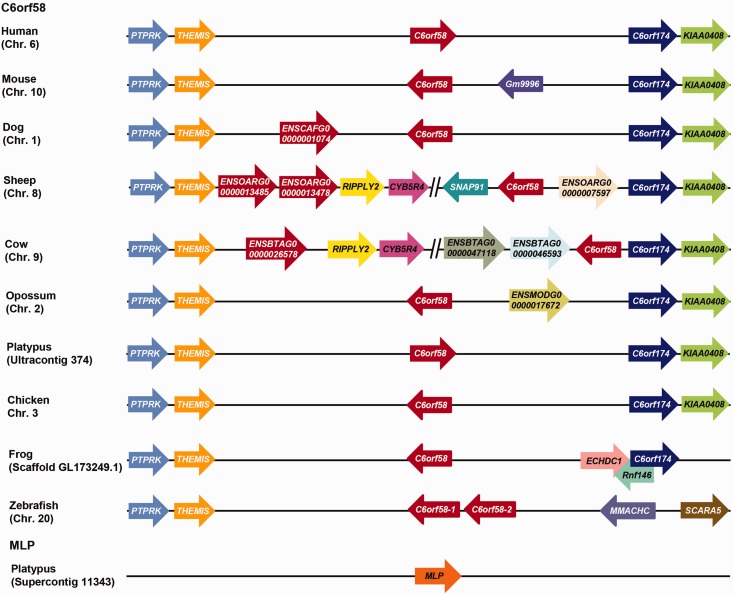
To gain further insight into the evolutionary relationship of MLP and C6orf58, and in particular the genes encoding the proteins that appeared to form a sister clade to MLP (i.e., the C6orf56-like genes), we investigated the location and arrangement of MLP, C6orf58, and C6orf56-like flanking genes. Synteny analysis of the region containing C6orf58 showed strong conservation among vertebrate species. The genomic region surrounding the C6orf58 region in monotreme, marsupial, and therians contains four protein coding single copy genes, protein tyrosine phosphatase receptor type K (PTPRK), thymocyte-expressed molecule involved in selection (THEMIS), chromosome 6 open reading frame 174 (C6orf174), uncharacterized protein KIAA0408 (KIAA0408), and defines the boundaries of this locus (fig. 10). This analysis revealed that the multiple C6orf58-like copies found in cow, sheep and dog all reside within this locus. In zebrafish, two C6orf58-like homologs are at an adjacent locus and C6orf174 and KIAA0408 are replaced by methylmalonic aciduria and homocystinuria cblC protein (MMACHC) and scavenger receptor class A member 5 (SCARA5). In the frog genome, there is an insertion of enoyl CoA hydratase domain containing 1 (ECHDC1) and ring finger protein 146 (rnf146); however, it retains the C6orf174 gene boundary similar to the marsupials. In opossum, there was an insertion of an uncharacterized gene between C6orf58 and C6orf174. In mouse, there was an insertion of an uncharacterized gene (Gm9996) in between C6orf58 and C6orf174. Unfortunately, the available platypus genome assembly has not yet been fully annotated and assembled in the region corresponding to the MLP flanking locus to allow systematic analysis in the MLP region. However, analysis of the C6orf58 locus does show that it is absent within this region.
Fig. 10.—
Comparison of platypus MLP and C6orf58 gene synteny from various vertebrate species. Schematic representation of chromosomal regions containing platypus and other vertebrate C6orf58 flanking genes. Arrows indicate the arrangement and orientation of genes. Gene sizes and intergenic distances are not drawn to scale.
Discussion
Monotreme hatchlings exit the egg in an altricial state with an underdeveloped immune system and enter a nonsterile environment (Wong et al. 2009; Bisana et al. 2013). They suck milk directly from the areolae on the abdomen but are not attached to them (Griffiths et al. 1969; Musser 2005). The nipple-less areolae of monotreme species are central interfaces between animals and their hatchlings in an external environment, and possibly contain their own microbiota due to potential pathogenic burrow or pouch environments. It is quite possible to speculate the mother-to-hatchling transfer of pathogens. Milk, as the first and only source of nutrition, may play a key role in transfer of antimicrobial proteins in protecting the immunologically vulnerable young during early monotreme development (Renfree and Shaw 2001; Wong et al. 2009; Kuruppath et al. 2012; Bisana et al. 2013). Recently, the echidna milk protein gene, EchAMP which is highly expressed in both short-beaked echidna and platypus mammary cells during lactation has been identified as an antibacterial protein which is implicated in milk protein-mediated innate immunity (Bisana et al. 2013). The marsupials do not lay eggs but like the monotremes have a very short gestation (Renfree and Shaw 2001), and give birth to a highly altricial young without functional immunological tissues and the young is exposed to nonsterile environment (Edwards and Deakin 2012; Edwards et al. 2012). Their growth and immune system develops through an elaborate and sophisticated lactation (Renfree and Shaw 2001). Therefore, new born are heavily reliant on passive immunity through lactation by permanent attachment to one of the mother’s teats and in many marsupials within a pouch (Edwards and Deakin 2012; Edwards et al. 2012). The marsupial mammary gland transfers immunoglobulins and other immune components in milk, and antimicrobial compounds secreted in the pouch for immunologically naive young to survive pathogen challenge (Edwards et al. 2012; Nicholas et al. 2012). Several immune protection compounds, notably antimicrobial proteins with diverse functions have been identified in marsupial milk such as cathelicidin, whey acidic protein (WAP) four-disulfide core domain 2, β-lactoglobulin, and transferrin (Edwards et al. 2012; Nicholas et al. 2012). The newly hatched monotreme very much resembles that of new born marsupials (Renfree and Shaw 2001). In monotreme milk secretion β-lactoglobulin (Lefèvre et al. 2009), transferrin (Teahan and McKenzie 1990), lysozyme (Teahan, McKenzie, Shaw, et al. 1991), and WAP four-disulfide core domain 2 (Sharp et al. 2007) have been identified, but not the cathelicidin protein (Whittington et al. 2009; Wang et al. 2011), suggesting that similar immune antimicrobial component secretion in monotreme milk could act to protect the immunocompromised hatchlings (Wong et al. 2009; Kuruppath et al. 2012).
Expression of MLP was first identified in cells collected from monotreme milk (Lefèvre et al. 2009). Cells in milk were harvested from monotreme milk during the lactation period and are likely an ambiguous mixture of cells that could include skin cells, immune cells, exfoliated epithelial cells from ducts and mammary or sebaceous glands. However, cDNA sequencing of this cell population showed that several casein and whey protein gene cDNAs were detected at high levels, indicating that monotreme milk cells harvested during lactation are enriched in exfoliated mammary epithelial cells (Lefèvre et al. 2009). The unavailability of cells or RNA from the mammary gland of nonlactating monotremes constrained us from determining the endogenous expression of MLP during the nonlactating period.
In this study we have identified a novel antibacterial protein which is highly expressed and found exclusively in monotremes, including monotreme milk. There are detailed reports that monotremes, both in the wild and captivity are susceptible to common pathogenic Gram-positive and Gram-negative bacterial infections including Aeromonas hydrophila, Salmonella typhimurium, Proteus, Streptococcus spp., Staphylococcus, Es. coli, Streptococcus zooepidemicus, Leptospira interogans Serovar hardjo, Edwardsiella sp., Mycobacterium, and Anaplasma (McColl 1983; Butler 1986; Whittington 1993; Munday et al. 1998; Jackson 2003; Macgregor et al. 2012). Interestingly, A. hydrophila and Es. coli are part of the normal gut flora of the platypus but sometimes become pathogenic (e.g., causing pneumonia) (Whittington 1993). In support of this, a dead platypus was found in the wild due to aspiration pneumonia, resulted from the lung infection. The infected lungs contained commensal A. hydrophila and Es. coli isolate (Whittington and McColl 1983).
Our experiments demonstrated that the recombinant MLP selectively exhibited significant bacteriostatic activity against two Gram-positive bacteria, St. aureus and En. faecalis, but not St. epidermidis and it was not active against all Gram-negative bacteria examined; Es. coli, P. aeruginosa, and Sa. enterica. We suggest that the high level of secretion of MLP in milk acts to provide immunological protection to the young against either potentially pathogenic bacteria or commensals. MLP demonstrated potent antistaphylococcal activity and may also protect the mother’s mammary gland from infection caused by St. aureus (Delgado et al. 2011). Enterococcus faecalis is an ubiquitous commensal of mammalian gastrointestinal flora (McBride et al. 2007) and although it is a major commensal there are some reports that En. faecalis causes infections in immune deficient mice (Ruiz et al. 2005; Hooper 2009; Hooper and Macpherson 2010). Recent studies have shown that En. faecalis is causal in a majority of human enterococcal infections (McBride et al. 2007). More recent studies have shown that the immune system may not only protect from invasive pathogens but also promote the optimal configuration of commensal bacterial flora to the gut (Hepworth et al. 2013). Group 3 type innate lymphoid cells (ILCs) maintain intestinal homeostasis through CD4+ T-cell immune response, which limits pathological, adaptive immune cell responses to pathogenic and commensal bacteria in the intestine (Hepworth et al. 2013). It has been reported that monotremes do not possess CD4 T-cell memory immune and lymph nodes (LNs) responses (Withers et al. 2012), which are normally associated with mammalian immunity (Withers et al. 2012). In the platypus, the precise timing of immune maturity and subsequent immune system development is unclear but immunohistological studies have revealed that when the young emerge as adults, they possess lymphoid tissue which is similar to those of therian mammals (Connolly et al. 1999; Whittington et al. 2009; Wang et al. 2011). During the weaning period monotreme young undergo a transition from milk to solid food which is presumably associated with dramatic changes in intestinal microbial invasion and composition, as well as withdrawal of passive immune protection afforded by the mother’s milk (Bisana et al. 2013; Morrow and Nicol 2012). The transition to primary colonization of the intestinal tract presents a crucial challenge for the immune system of the young. The ability of commensal bacteria to produce infection is a consequence of immature host immunity. In the case of En. faecalis, we propose that MLP secretion is elicited as part of a compensatory response to maintain gut homeostasis. These findings are consistent with our observation that MLP is expressed throughout the entire lactation cycle and therefore may be critical for limiting gut microbe infections to maintain homeostasis during the vulnerable periods of organ differentiation during early development.
Recently, the glycosylation of human milk proteins has been gaining increasing attention as a way of producing a major class of anti-infective agents in milk (Dallas et al. 2012), and glycosylated milk proteins have also been observed in monotreme milk (Teahan, McKenzie, Griffiths 1991; Bisana et al. 2013). N-linked glycosylation is a key posttranslational modification for proteins that are essential mediators in bacterial recognition, proteolytic resistance, intracellular signaling, and antigen binding and presentation (Marth and Grewal 2008; Dallas et al. 2012). For example, lactoferrin, one of the most abundant proteins in human milk, has an N-linked glycan essential in mediating interactions between specific enteropathogenic bacteria (Barboza et al. 2012). It has been assumed that monotreme mammary glands produce abundant oligosaccharides mainly as antimicrobial defense factors due to the presence of most of the glycosyltransferases along with antibodies, transferrin and lysozyme in milk (Urashima et al. 2012). Glycans on the MLP may have differential effects on each bacterial species that could modulate species-specific antibacterial activity, although there is currently no evidence to support this in MLP.
Due to the protected nature of monotremes within Australia, we were limited in the availability of echidna and platypus tissues. The MLP tissue expression profile examined in this study employed only male tissues, and as such we have assumed that male and female expression of MLP in other tissues is similar between the sexes. This remains to be verified. The pattern of MLP expression corresponding to male submandibular gland revealed similarity to C6orf58 expression in human salivary fluids (Ramachandran et al. 2006, 2008); however, the function of C6orf58 protein is currently unknown. Dog and cow C6orf58 proteins share a putative conserved N-glycosylation site at position 68–72 amino acid, consistent with the site identified for human C6orf58 (IILN*QTAR). The authors suggested that N-linked glycosylated salivary proteins may protect against infections by binding to oral pathogens and eliminating them from the oral cavity (Ramachandran et al. 2008), but this remains to be determined experimentally. These findings support the notion that MLP may play similar protective functions in milk. Most of the antibacterial proteins maintain certain common characteristic features that are necessary for the activity, such as α-helical structures, and carry a positive net charge that is thought to allow them to interact with negatively charged bacterial cell membranes which contain a huge proportion of phospholipids and hydrophobic fatty acid chains (Brogden 2005; Wiesner and Vilcinskas 2010). This unique arrangement confers a negative charge to the surface (Brogden 2005; Wiesner and Vilcinskas 2010). Several studies have shown that effective antimicrobial milk proteins contain α-helical content such as lactoferrin and lysozyme (Benkerroum 2008; Wiesner and Vilcinskas 2010). Computational analysis suggested that MLP is an amphipathic protein and the positive charge in the α-helix region may be required for MLP interactions with bacterial membranes. Therefore, α-helices most likely play a key role in MLP antibacterial activity. The MLP CD spectra were confirmed with the solved crystal structure of platypus MLP (manuscript in preparation). Furthermore, antimicrobial proteins with α-helical secondary structures are often more active (Brogden 2005).
Phylogenetic analysis suggests that MLP and C6orf58 have evolved from a common ancestral gene by an ancient duplication event. This is supported by comparative and genomic evidence for monotreme MLP, C6orf58 and vertebrate C6orf58 genes and encoding proteins, which share some key features of protein and gene structure, including similar exon structure.
Although our phylogenetic analysis suggested that there was a sister group of MLP which comprised C6orf58-like genes from other species, this relationship is likely due to convergent evolution of these proteins as the phylogenetic tree did not support a direct evolutionary relationship of homologous genes. Further, gene loci suggested that the C6orf58-like genes were recent duplications occurring within the C6orf58 locus. We therefore concluded that we failed to find any true orthologs of MLP and that MLP is specific to monotremes. We suggest that monotremes may have gained MLP by duplication of C6orf58, a gene known to be expressed in salivary secretions (Karn et al. 2013) and that is considered diagnostic of saliva (Yang et al. 2013). C6orf58 has been associated with liver development in zebrafish (Chang et al. 2011) and pancreatic cancer survival prognosis (Wu et al. 2011); however, its function is unknown. Its presence in other tissues suggests that MLP evolved from C6orf58 as a secretory antimicrobial protein similar to other secretory proteins that are also found in multiple types of secretions, such as transferrin and lysozyme. This evolutionary process need not to have been directly associated with lactation, even though MLP was subsequently incorporated into milk, and perhaps assumed an important role (whether solely antimicrobial or also nutritional) in lactation. MLP incorporation into milk may have evolved in order to succeed in nipple-less milk delivery, which was overcome in the therian lineage by the evolution of nipples as a more sophisticated mode of milk delivery during the passage to viviparity, which is estimated to have occurred about 166–220 Ma (Lefèvre, Sharp, et al. 2010). Alternatively MLP may have been lost in the therian lineage. To identify the possible pseudogenes of MLP in placental mammals, platypus MLP gene flanking genomic region needs to be sequenced. The linkage between platypus MLP flanking genes conservation in marsupial and eutherian species provides information for the monotreme MLP evolution and pseudogenes in placental mammals. The loss of some milk protein genes in some mammalian lineages is supported by cases of more recent losses, for example, WAP is absent from the milk of cows, sheep and humans (Hajjoubi et al. 2006), β-lactoglobulin is also absent from the milk of many eutherian mammals, including guinea pig, domestic rabbits, camels, lamas, and humans (Oftedal 2012). The loss of specific milk protein genes from various species, such as β-lactoglobulin and whey acidic protein, which are retained in other species reflect a unique pattern of evolution of milk protein genes which may be associated with such activities as domestication.
The protective, prebiotic, or innate immune role in monotremes would be further supported by additional studies. The most valuable proteins in milk, that is, lactoferrin, immune globulins, lactoperoxidase and lysozyme, together with bioactive peptides, possess active domains that are resistant to pepsin and trypsin activity (Florisa et al. 2003). Proteins such as lactoferrin are susceptible to digestion by gastrointestinal proteases and give rise to highly active peptides (lactoferricin) with exceptional bioactivity (Furlund et al. 2013). The resistance of these peptides to intestinal conditions ensures that the peptides survive long enough to arrive at the right place to exert their beneficial function. It would be of interest to study the effects of different protease activities on MLP to ascertain if it will survive digestion in the gut and to decipher the MLP cryptic peptidome. It is important to note that monotreme intestinal development is still unknown, but if similar to marsupials the intestine of the young will first comprise a glandular type and later develop gastric secretion (Kwek, De Iongh, Nicholas, et al. 2009). In marsupials, milk has been shown to be involved in this process (Kwek, De Iongh, Digby, et al. 2009). Milk peptide function is not only limited to antimicrobial activity, several milk peptides, such as lactoferricin, have also been shown to possess antifungal and antiviral activities (Garcia-Montoya et al. 2012). These properties should also be assessed in MLP.
In summary, we have reported that MLP represents a novel secreted protein with selective bacteriostatic activity and is highly expressed in monotreme milk. We suggest that MLP is expressed in milk and represents a more evolutionarily primitive mechanism of milk protein-mediated innate immunity to protect the young. The purported function in the living animal is, at present, a viable hypothesis but not yet proven. The molecular basis of the MLP antibacterial mechanism remains to be established and these findings may provide a new mechanism to target St. aureus and En. faecalis infections. The increasing resistance to antibiotics in pathogenic bacteria over the past 30 years to established antibacterial drugs possesses a serious threat to human health and has increased the demand for new antimicrobial compounds (Simmons et al. 2010). A greater understanding of functional properties, structural basis of the interaction and mechanism of action of monotreme antimicrobial milk proteins may reveal new inhibitors that could provide therapies as “natural host antibiotics” against invading resistant pathogenic bacteria in humans.
Supplementary Material
Supplementary figures S1–S4 and tables S1–S9 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Ms. Swathi Bisana (CCMB—India) for sharing valuable mass spectrometry analysis and Paul O’Donnell (Bio21, Melbourne, Australia) for support and assistance in mass spectrometry analysis. This work was supported by the in-house funds of the Deakin University, Australia and grants to S.C.N. from the National Geographic Committee for Research and Exploration (grant number 9171-12) and also supported by a Deakin University International Research Scholarship (DUIRS), Deakin University, Australia to A.K.E. The authors declare that they have no competing financial interests. A.K.E., J.A.S., C.M.L., and K.R.N. conceived and designed the experiments; A.K.E. performed the experiments; A.K.E., J.A.S., C.M.L., and K.R.N. analyzed and interpreted the results; T.R.G. and S.C.N. were responsible for monotreme milk sample collection; J.A.S., C.M.L., and K.R.N. supervised the study; all authors were involved in data discussions and the final version of the manuscript; and A.K.E. wrote the manuscript.
Literature Cited
- Aniansson G, Andersson B, Lindstedt R, Svanborg C. Anti-adhesive activity of human casein against Streptococcus pneumoniae and Haemophilus influenzae. Microb Pathog. 1990;8:315–323. doi: 10.1016/0882-4010(90)90090-d. [DOI] [PubMed] [Google Scholar]
- Barboza M, et al. Glycosylation of human milk lactoferrin exhibits dynamic changes during early lactation enhancing its role in pathogenic bacteria-host interactions. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.015248. M111.015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bas A, Forsberg G, Hammarström S, Hammarström ML. Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand J Immunol. 2004;59:566–573. doi: 10.1111/j.0300-9475.2004.01440.x. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, Heijne GV, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Benkerroum N. Antimicrobial activity of lysozyme with special relevance to milk. Afr J Biotechnol. 2008;7:4856–4867. [Google Scholar]
- Bisana S, et al. Identification and functional characterization of a novel monotreme-specific antibacterial protein expressed during lactation. PLoS One. 2013;8:e53686. doi: 10.1371/journal.pone.0053686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn DG, Hayssen V, Murphy CJ. The origins of lactation and the evolution of milk: a review with new hypotheses. Mamm Rev. 1989;19:1–26. [Google Scholar]
- Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Brogden K. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Burrell H. Sydney (NSW): Angus and Robertson; 1927. The platypus. [Google Scholar]
- Butler R. Bacterial diseases. Monotremes and marsupials. In: Fowler ME, editor. Zoo and wild animal medicine. 2nd, ed. Philadelphia (PA): W. B. Saunders Co.; 1986. pp. 572–576. [Google Scholar]
- Chang C, et al. Liver-enriched gene 1a and 1b encode novel secretory proteins essential for normal liver development in zebrafish. PLoS One. 2011;6:e22910. doi: 10.1371/journal.pone.0022910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly JH, Canfield PJ, McClure SJ, Whittington RJ. Histological and immunohistological investigation of lymphoid tissue in the platypus (Ornithorhynchus anatinus) J Anat. 1999;195:161–171. doi: 10.1046/j.1469-7580.1999.19520161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas DC, et al. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. J Proteome Res. 2013;12:2295–2304. doi: 10.1021/pr400212z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas DC, Sela D, Underwood MA, German JB, Lebrilla CB. Protein-linked glycan degradation in infants fed human milk. J Glycomics Lipidomics. 2012 doi: 10.4172/2153-0637.S1-002. Special issue S1-002:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S, et al. Characterization of Staphylococcus aureus strains involved in human and bovine mastitis. FEMS Immunol Med Microbiol. 2011;62:225–253. doi: 10.1111/j.1574-695X.2011.00806.x. [DOI] [PubMed] [Google Scholar]
- Edwards MJ, Deakin JE. The marsupial pouch: implications for reproductive success and mammalian evolution. Aust J Zool. 2012;61:41–47. [Google Scholar]
- Edwards MJ, Hinds LA, Deane EM, Deakin JE. A review of complementary mechanisms which protect the developing marsupial pouch young. Dev Comp Immunol. 2012;37:213–220. doi: 10.1016/j.dci.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Florisa R, Recio I, Berkhout B, Visser S. Antibacterial and antiviral effects of milk proteins and derivatives thereof. Curr Pharm Des. 2003;9:1257–1275. doi: 10.2174/1381612033454810. [DOI] [PubMed] [Google Scholar]
- Furlund CB, et al. Identification of lactoferrin peptides generated by digestion with human gastrointestinal enzymes. J Dairy Sci. 2013;96:75–88. doi: 10.3168/jds.2012-5946. [DOI] [PubMed] [Google Scholar]
- Garcia-Montoya IA, Cendon TS, Arevalo-Gallegos S, Rascon-Cruz Q. Lactoferrin a multiple bioactive protein: an overview. Biochim Biophys Acta. 2012;1820:226–236. doi: 10.1016/j.bbagen.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant TR. Kensington: University of New South Wales Press; 1995. The Platypus: A Unique Mammal. [Google Scholar]
- Grant TR, Griffiths M. Aspects of lactation and determination of sex ratios and longevity in a free-ranging population of platypuses, Ornithorhynchus anatinus, in the Shoalhaven River, NSW. In: Augee ML, editor. Platypus and echidnas. Royal Zoological Society of NSW (Sydney) 1992. pp. 80–89. [Google Scholar]
- Grant TR. Melbourne (Vic): CSIRO Publishing; 2007. Platypus; p. 168. [Google Scholar]
- Griffiths M. New York: Academic Press; 1978. The biology of the monotremes. [Google Scholar]
- Griffiths M, Elliott MA, Leckie RMC, Schoefl GI. Observations of the comparative anatomy and ultrastructure of mammary glands and on the fatty acids of the triglycerides in platypus and echidna milk fats. J Zool Lond. 1973;169:255–279. [Google Scholar]
- Griffiths M, McIntosh DL, Coles REA. The mammary gland of the echidna, Tachyglossus aculeatus with observations on the incubation of the egg and on the newly-hatched young. J Zool Lond. 1969;158:371–386. [Google Scholar]
- Grützner F, Nixon B, Jones RC. Reproductive biology in egg-laying mammals. Sex Dev. 2008;2:115–127. doi: 10.1159/000143429. [DOI] [PubMed] [Google Scholar]
- Hajjoubi S, et al. Ruminants genome no longer contains Whey Acidic Protein gene but only a pseudogene. Gene. 2006;370:104–112. doi: 10.1016/j.gene.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Hamilton MJ, Davidson AD, Sibly RM, Brown JH. Universal scaling of production rates across mammalian lineages. Proc Biol Sci. 2011;278:560–566. doi: 10.1098/rspb.2010.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins M, Battaglia A. Breeding behaviour of the platypus(Ornithorhynchus anatinus) in captivity. Aust J Zool. 2009;57:283–293. [Google Scholar]
- Hepworth MR, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettinga K, et al. The host defense proteome of human and bovine milk. PLoS One. 2011;6:e19433. doi: 10.1371/journal.pone.0019433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland N, Jackson SM. Reproductive behaviour and food consumption associated with the captive breeding of platypus (Ornithorhynchus anatinus) J Zool. 2002;256:279–288. [Google Scholar]
- Hooper LV. Do symbiotic bacteria subvert host immunity? Nat Rev Microbiol. 2009;7:367–374. doi: 10.1038/nrmicro2114. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Hopper KE, McKenzie HA. Comparative studies of alpha-lactalbumin and lysozyme: echidna lysozyme. Mol Cell Biochem. 1974;3:93–108. doi: 10.1007/BF01659181. [DOI] [PubMed] [Google Scholar]
- Hrdličková R, Nehyba J, Lim SL, Grützner F, Bose HR. Insights into the evolution of mammalian telomerase: platypus TERT shares similarities with genes of birds and other reptiles and localizes on sex chromosomes. BMC Genomics. 2012;13:216. doi: 10.1186/1471-2164-13-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Hughes RL, Hall LS. Early development and embryology of the platypus. Philos Trans R Soc Lond B Biol Sci. 1998;353:1101–1114. doi: 10.1098/rstb.1998.0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. Melbourne (Vic.): CSIRO Publishing; 2003. Australian mammals: biology and captive management. [Google Scholar]
- Karn RC, Chung AG, Laukaitia CM. Shared and unique proteins in human, mouse and rat saliva proteomes: footprints of functional adaptation. Proteomes. 2013;1:275–289. doi: 10.3390/proteomes1030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuruppath S, et al. Monotremes and marsupials: comparative models to better understand the function of milk. J Biosci. 2012;37:581–588. doi: 10.1007/s12038-012-9247-x. [DOI] [PubMed] [Google Scholar]
- Kwek J, De Iongh R, Nicholas K, Familari M. Molecular insights into evolution of the vertebrate gut: focus on stomach and parietal cells in the marsupial, Macropus eugenii. J Exp Zool B Mol Dev Evol. 2009;312:613–624. doi: 10.1002/jez.b.21227. [DOI] [PubMed] [Google Scholar]
- Kwek JH, De Iongh R, Digby MR, et al. Cross-fostering of the tammar wallaby (Macropus eugenii) pouch young accelerates fore-stomach maturation. Mech Dev. 2009;126:449–463. doi: 10.1016/j.mod.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lefèvre CM, Menzies K, Sharp JA, Nicholas KR. Comparative genomics and transcriptomics of lactation. In: Pontarotti P, editor. Evolutionary biology—concepts, molecular and morphological evolution. Berlin (Germany): Springer; 2010. pp. 115–132. [Google Scholar]
- Lefèvre CM, Sharp JA, Nicholas KR. Characterisation of monotreme caseins reveals lineage-specific expansion of an ancestral casein locus in mammals. Reprod Fertil Dev. 2009;21:1015–1027. doi: 10.1071/RD09083. [DOI] [PubMed] [Google Scholar]
- Lefèvre CM, Sharp JA, Nicholas KR. Evolution of lactation: ancient origin and extreme adaptations of the lactation system. Annu Rev Genomics Hum Genet. 2010;11:219–238. doi: 10.1146/annurev-genom-082509-141806. [DOI] [PubMed] [Google Scholar]
- Li HP, et al. IL-8 mRNA expression in the mouse mammary glands during pregnancy and lactation. Genet Mol Res. 2012;11:4746–4753. doi: 10.4238/2012.October.9.10. [DOI] [PubMed] [Google Scholar]
- Macgregor JW, Holyoake CS, Munks SA, Robertson ID, Warren KS. Preliminary investigation into the prevalence of mucormycosis in the platypus (Ornithorhynchus anatinus) in three catchments in north-west Tasmania. Aust Vet J. 2012;88:190–196. doi: 10.1111/j.1751-0813.2010.00568.x. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth JD, Grewal PK. Mammalian glycosylation in immunity. Nat Rev Immunol. 2008;8:874–887. doi: 10.1038/nri2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, Fischetti VA, Leblanc DJ, Moellering RC, Jr, Gilmore MS. Genetic diversity among Enterococcus faecalis. PLoS One. 2007;2:e582. doi: 10.1371/journal.pone.0000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl KA. Pathology in captive platypus (Ornithorhynchus anatinus) in Victoria, Australia. J Wildl Dis. 1983;19:118–122. doi: 10.7589/0090-3558-19.2.118. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Miller N, et al. Effect of stage of lactation and parity on mammary gland cell renewal. J Dairy Sci. 2006;89:4669–4677. doi: 10.3168/jds.S0022-0302(06)72517-6. [DOI] [PubMed] [Google Scholar]
- Morrow G, Andersen NA, Nicol SC. Reproductive strategies of the short-beaked echidna—a review with new data from a long-term study on the Tasmanian subspecies (Tachyglossus aculeatus setosus) Aust J Zool. 2009;57:275–282. [Google Scholar]
- Morrow GE, Nicol SC. Maternal care in the Tasmanian echidna (Tachyglossus aculeatus setosus) Aust J Zool. 2012;60:289–298. [Google Scholar]
- Munday BL, Whittington RJ, Stewart NJ. Disease conditions and subclinical infections of the platypus (Ornithorhynchus anatinus) Philos Trans R Soc Lond Biol Sci. 1998;353:1093–1099. doi: 10.1098/rstb.1998.0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Dorschner RA, Stern LJ, Lin KH, Gallo RL. Expression and secretion of cathelicidin antimicrobial peptides in murine mammary glands and human milk. Pediatr Res. 2005;57:10–15. doi: 10.1203/01.PDR.0000148068.32201.50. [DOI] [PubMed] [Google Scholar]
- Musser AM. 2005. Monotremata. In. eLS: John Wiley & Sons, Ltd. New York. [Google Scholar]
- Nicholas K, et al. The tammar wallaby: a model system to examine domain-specific delivery of milk protein bioactives. Semin Cell Dev Biol. 2012;23:547–556. doi: 10.1016/j.semcdb.2012.03.016. [DOI] [PubMed] [Google Scholar]
- Oftedal OT. The mammary gland and its origin during synapsid evolution. J Mammary Gland Biol Neoplasia. 2002a;7:225–252. doi: 10.1023/a:1022896515287. [DOI] [PubMed] [Google Scholar]
- Oftedal OT. The origin of lactation as a water source for parchment-shelled eggs. J Mammary Gland Biol Neoplasia. 2002b;7:253–266. doi: 10.1023/a:1022848632125. [DOI] [PubMed] [Google Scholar]
- Oftedal OT. The evolution of milk secretion and its ancient origins. Animal. 2012;6:355–368. doi: 10.1017/S1751731111001935. [DOI] [PubMed] [Google Scholar]
- Oftedal OT. Origin and evolution of the major constituents of milk. In: McSweeney PLH, Fox PF, editors. Advanced dairy chemistry: Volume 1A proteins: basic apsects. 4th ed. New York: Springer; 2013. pp. 1–42. [Google Scholar]
- Ordoñez GR, et al. Loss of genes implicated in gastric function during platypus evolution. Genome Biol. 2008;9:R81. doi: 10.1186/gb-2008-9-5-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. Tree View: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Power ML, Schulkin J. Maternal regulation of offspring development in mammals is an ancient adaptation tied to lactation. Appl Transl Genomics. 2013;130:55–63. doi: 10.1016/j.atg.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, et al. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J Proteome Res. 2006;5:1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- Ramachandran P, et al. Comparison of N-linked glycoproteins in human whole saliva, parotid, submandibular, and sublingual glandular secretions identified using hydrazide chemistry and mass spectrometry. Clin Proteomics. 2008;4:80–104. doi: 10.1007/s12014-008-9005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree M, Shaw G. 2001. Reproduction in monotremes and marsupials. . In: eLS. John Wiley & Sons, Ltd. [Google Scholar]
- Ruiz PA, Shkoda A, Kim SC, Sartor RB, Haller D. IL-10 gene-deficient mice lack TGF-β/Smad signaling and fail to inhibit proinflammatory gene expression in intestinal epithelial cells after the colonization with colitogenic Enterococcus faecalis. J Immunol. 2005;174:2990–2999. doi: 10.4049/jimmunol.174.5.2990. [DOI] [PubMed] [Google Scholar]
- Sharp JA, Lefèvre C, Nicholas KR. Molecular evolution of monotreme and marsupial whey acidic protein genes. Evol Dev. 2007;9:378–392. doi: 10.1111/j.1525-142X.2007.00175.x. [DOI] [PubMed] [Google Scholar]
- Simmons KJ, Chopra I, Fishwick CWG. Structure-based discovery of antibacterial drugs. Nat Rev Microbiol. 2010;8:501–510. doi: 10.1038/nrmicro2349. [DOI] [PubMed] [Google Scholar]
- Springer MS, Krajewski CW. Monotremes (Prototheria) In: Hedges SB, Kumar S, editors. The timetree of life. Oxford University Press; 2009. pp. 462–465. [Google Scholar]
- Strömqvist M, et al. Human milk K-casein and inhibition of Helicobacter pylori adhesion to human gastric mucosa. J Pediatr Gastroenterol Nutr. 1995;21:288–296. doi: 10.1097/00005176-199510000-00006. [DOI] [PubMed] [Google Scholar]
- Teahan CG, McKenzie HA. Iron(III) binding proteins of echidna (Tachyglossus aculeatus) and platypus (Ornithorhynchus anatinus) Biochem Int. 1990;22:321–328. [PubMed] [Google Scholar]
- Teahan CG, McKenzie HA, Griffiths M. Some monotreme milk “whey” and blood proteins. Comp Biochem Physiol B. 1991;99:99–118. doi: 10.1016/0305-0491(91)90014-5. [DOI] [PubMed] [Google Scholar]
- Teahan CG, McKenzie HA, Shaw DC, Griffiths M. The isolation and amino acid sequences of echidna (Tachyglossus aculeatus) milk lysozyme I and II. Biochem Int. 1991;24:85–95. [PubMed] [Google Scholar]
- Urashima T, Fukuda K, Messer M. Evolution of milk oligosaccharides and lactose: a hypothesis. Animal. 2012;6:369–374. doi: 10.1017/S1751731111001248. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. Ancient antimicrobial peptides kill antibiotic-resistant pathogens: Australian mammals provide new options. PLoS One. 2011;6:e24030. doi: 10.1371/journal.pone.0024030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington CM, Sharp JA, Papenfuss A, Belov K. No evidence of expression of two classes of natural antibiotics (cathelicidins and defensins) in a sample of platypus milk. Aust J Zool. 2009;57:211–217. [Google Scholar]
- Whittington RJ. Diseases of monotremes. In: Fowler ME, editor. Zoo and wild animal medicine. 1993. . Current therapy 3. Philadelphia: W.B. Saunders Company. p. 269–276. [Google Scholar]
- Whittington RJ, McColl KA. Aspiration pneumonia in a wild platypus Ornithorhynchus anatinus. Aust Vet J. 1983;60:277. doi: 10.1111/j.1751-0813.1983.tb07107.x. [DOI] [PubMed] [Google Scholar]
- Wiesner J, Vilcinskas A. Antimicrobial peptides: the ancient arm of the human immune system. Virulence. 2010;1:440–464. doi: 10.4161/viru.1.5.12983. [DOI] [PubMed] [Google Scholar]
- Withers DR, et al. Cutting edge: lymphoid tissue inducer cells maintain memory CD4 T cells within secondary lymphoid tissue. J Immunol. 2012;189:2094–2098. doi: 10.4049/jimmunol.1201639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ESW, Papenfuss AT, Miller RD, Belov K. Hatching time for monotreme immunology. Aust J Zool. 2009;57:185–198. [Google Scholar]
- Wu TT, Gong H, Clarke EM. A transcriptome analysis by lasso penalized Cox regression for pancreatic cancer survival. J Bioinform Comput Biol. 2011;9:63–73. doi: 10.1142/s0219720011005744. [DOI] [PubMed] [Google Scholar]
- Yang H, Zhou B, Deng H, Prinz M, Siegel D. Body fluid identification by mass spectrometry. Int J Legal Med. 2013;127:1065–1077. doi: 10.1007/s00414-013-0848-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.