Abstract
Cephalochordates, the sister group of tunicates plus vertebrates, have been called “living fossils” due to their resemblance to fossil chordates from Cambrian strata. The genome of the cephalochordate Branchiostoma floridae shares remarkable synteny with vertebrates and is free from whole-genome duplication. We performed RNA sequencing from larvae and adults of Asymmetron lucayanum, a cephalochordate distantly related to B. floridae. Comparisons of about 430 orthologous gene groups among both cephalochordates and 10 vertebrates using an echinoderm, a hemichordate, and a mollusk as outgroups showed that cephalochordates are evolving more slowly than the slowest evolving vertebrate known (the elephant shark), with A. lucayanum evolving even more slowly than B. floridae. Against this background of slow evolution, some genes, notably several involved in innate immunity, stand out as evolving relatively quickly. This may be due to the lack of an adaptive immune system and the relatively high levels of bacteria in the inshore waters cephalochordates inhabit. Molecular dating analysis including several time constraints revealed a divergence time of ∼120 Ma for A. lucayanum and B. floridae. The divisions between cephalochordates and vertebrates, and that between chordates and the hemichordate plus echinoderm clade likely occurred before the Cambrian.
Keywords: amphioxus, Branchiostoma, Asymmetron, chordate evolution, transcriptome, innate immunity
Introduction
Cephalochordates (amphioxus, lancelets) have long been prominent in the discussions of vertebrate evolution. Haeckel (1876), who regarded amphioxus as the simplest vertebrate, thought it key to understanding how the vertebrates evolved (Haeckel 1876). However, others thought amphioxus was possibly a degenerate form of the ancestral vertebrate (Lankester 1875) or a primitive offshoot from the vertebrate stem (MacBride 1898). On the basis of both morphology and 18s ribosomal DNA, cephalochordates had been placed as the sister group of vertebrates with tunicates basal in the chordates (Wada and Satoh 1994). However, molecular phylogenetic analyses with large sets of nuclear genes reversed the positions of tunicates and cephalochordates, placing cephalochordates basal within the chordates (Bourlat et al. 2006; Delsuc et al. 2008). Studies of genes and development of the cephalochordate genus Branchiostoma have confirmed that amphioxus is a close vertebrate relative that diverged before the evolution of jaws, neural crest, paired eyes, or vertebrae (Holland et al. 1992, 1996; Shimeld 1999; Meulemans and Bronner-Fraser 2002; Yu et al. 2007; Holland 2009).
There are three genera of cephalochordates: Branchiostoma with ∼28 species, Epigonichthys with 1 species, and Asymmetron with 2 recognized species, but probably additional cryptic ones. To date, the genomes of the three species of Branchiostoma have been or are in the process of being sequenced. The first was that of the Florida amphioxus (Branchiostoma floridae) (Holland et al. 2008; Putnam et al. 2008) (http://genome.jgi-psf.org/Brafl1/Brafl1.home.html, last accessed September 27, 2014). Analysis of the B. floridae genome confirmed that cephalochordates did not undergo the two rounds of whole-genome duplication that occurred in vertebrates and demonstrated that genomes of B. floridae and vertebrates share a surprisingly high degree of synteny. In fact, 17 linkage groups (chromosomes) that probably existed at the base of the chordates could be constructed (Holland et al. 2008; Putnam et al. 2008). The genome of one of the Asian species (B. belcheri) is available on a genome browser (http://mosas.sysu.edu.cn/genome/gbrowser_wel.php, last accessed September 27, 2014) (Huang et al. 2012), while sequencing of the genome of the European species (B. lanceolatum) is in progress (http://www.cns.fr/spip/-Branchiostoma-lanceolatum,585-.html, last accessed September 27, 2014). Given the conservation between the B. floridae and vertebrate genomes as well as the rather subtle morphological differences among the various species of Brachiostoma, it is perhaps not surprising that such genomic features as microRNAs (miRNAs) are highly conserved between Branchiostoma species (Dai et al. 2009; Yang et al. 2013).
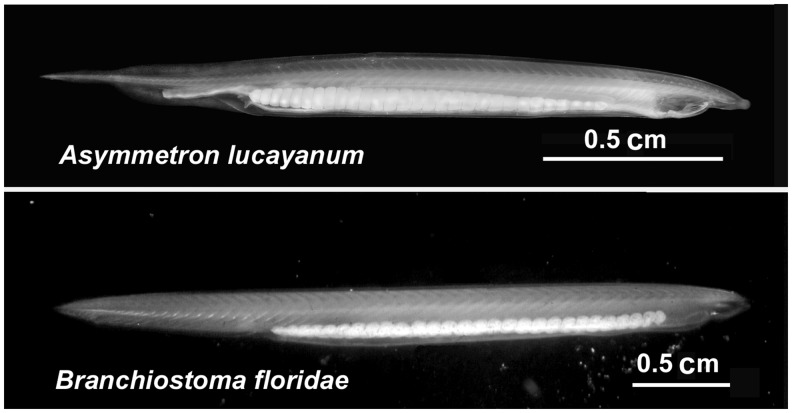
From mitochondrial DNA sequences, the various species of Branchiostoma have been calculated to have radiated between about 112 and 40 Ma (Nohara et al. 2004; Xiao et al. 2008; Zhao and Zhu 2011). In contrast, Epigonichthys and Branchiostoma split about 120 Ma (Kon et al. 2007), while the two species of Asymmetron identified to date, Asymmetron lucayanum and A. inferum, are calculated to have split from the Epigonichthys/Branchiostoma clade about 165 Ma. Morphologically, Asymmetron and Branchiostoma have several differences (fig. 1). Most striking is that Asymmetron has gonads only on the right side. In addition, the larvae of A. lucayanum have a pigmented tail fin, which B. floridae does not, although B. lanceolatum does. The first gill slits open more ventrally in A. lucayanum as does the anus, and the anterior coelom forms by schizocoely as opposed to enterocoely in Branchiostoma (Holland ND and Holland LZ 2010).
Fig. 1.—
Side views of living Asymmetron lucayanum (top) and Branchiostoma floridae (bottom). Asymmetron lucayanum has a single row of gonads on the right side only. Branchiostoma floridae has two rows of gonads—one on each side (anterior to the right).
Because Asymmetron has been placed as the sister group to the Epigonichthys/Branchiostoma clade (Kon et al. 2007), to gain insights into evolution within the chordates, we sequenced the A. lucayanum transcriptome from 20-h embryos (phylotypic stage) and adults. Together with the whole-genome gene set for B. floridae, this rich data set enabled us to perform large-scale comparative analyses to investigate the evolutionary rates of cephalochordate genes. We found that cephalochordates are evolving more slowly than the slowest evolving vertebrate known. We also investigated the evolutionary rates of cephalochordate genes across functional categories. Genes involved in innate immunity stood out as evolving relatively quickly. In addition, we estimated divergence times between protostomes and hemichordates/echinoderms, cephalochordates, and vertebrates among the deuterostomes. Our calculations place the separations between protostomes and deuterostomes, between hemichordates/echinoderms and chordates, and between cephalochordates and vertebrates in the Proterozoic, before their appearance in the fossil record, suggesting either that most animals failed to fossilize prior to the Cambrian, or, if they did, their fossils are yet to be found.
Materials and Methods
Animal Sampling and Sequencing
Adult specimens of A. lucayanum were collected from Bimini, Bahamas, and maintained in the laboratory on a diet of the haptophytes (Tisochrysis lutea [CCMP 463]) and (Isochrysis sp. [CCMP 1244] and the diatom Thalassiosira floridana [CCMP 985]). Animals were kept under a moonlight regime (Fishbowl Innovations, Spokane, WA). Animals spawned 2 days before the new moon. RNA was extracted in TRIzol (Invitrogen, Carlsbad, CA), from adult animals and 20-h larvae (phylotypic stage) pooled from several females. Two RNA sequencing (RNA-Seq) libraries, one from the adult RNA and one from larval RNA, were constructed with the TruSeq kit (Illumina Inc., San Diego, CA). Library construction and paired-end sequencing (2× 100 bp) in a single lane of a flow cell was performed by the BioGem facility at University of California, San Diego, La Jolla, CA.
Processing and Assembly of Reads
For each RNA-Seq library, raw reads were processed by Trimmomatic (v0.32) (Bolger et al. 2014), prinseq (v0.20.4) (Schmieder and Edwards 2011b), and Deconseq (v0.4.3) (Schmieder and Edwards 2011a), respectively. We first used Trimmomatic to remove the potential Illumina adapter contamination and conducted reads clipping and trimming. It was reported that the first 13 bp of Illumina RNA-Seq reads are likely to have guanine–cytosine content (GC%) bias due to random hexamer priming (Hansen et al. 2010), and such bias may affect the assembly quality, so we used Trimmomatic to clipp off the first 13 bp of all our reads. Quality trimming was performed using a 5-bp sliding window with a mean quality cutoff of 30. Prinseq was subsequently employed to remove poly-A/T tails as well as low-complexity reads. In addition, we used Deconseq to remove potential contaminated reads from sources like ribosomal RNA, human, bacteria, and virus. Finally, a minimum length cutoff of 36 bp was applied for the reads that passed all previous quality control steps. The transcriptome assembly was conducted by Trinity (r20131110) (Grabherr et al. 2011). To evaluate the quality of the assembly, we aligned the RNA-Seq reads back to the contigs from the assembly using Bowtie (v0.12.8) (Langmead et al. 2009) and examined summary statistics such as the percentage of properly aligned and paired reads. Trinity can detect potential isoforms from alternative splicing and label them with the same prefix. In this case, we selected the longest contig for each isoform group as a unique representation for that group. TransDecoder (http://transdecoder.sourceforge.net, last accessed September 27, 2014) was then used to extract likely coding sequences (CDSs) for A. lucayanum based on the Trinity assemblies. The CDSs and the translated protein sequences were used for our analyses.
Correction of B. floridae Gene Models
The B. floridae CDSs were retrieved based on the Gene Feature Format (GFF) file downloaded from http://genome.jgi-psf.org/Brafl1/Brafl1.home.html (last accessed September 27, 2014). Some contained internal stop codons, indicating that the original gene models were inaccurate. Therefore, to correct these erroneous annotations, we used Exonerate (Slater and Birney 2005) to align the corresponding protein sequences to the B. floridae reference genome (v2.0) (Putnam et al. 2008) and, based on the protein–genome alignments, updated our copy of the GFF file.
Functional Annotation
The translated protein sequences of both the corrected B. floridae CDSs and the assembled A. lucayanum CDSs were annotated with the Blast2GO pipeline (Conesa et al. 2005). Two rounds of homology searches were performed by BLASTP (E-value cutoff = 1 × 10−3): all queries were first blasted against the SwissProt database. Those queries that failed to find any hit were then blasted against the National Center for Biotechnology Information (NCBI) nonredundant (nr) protein database. The gene ontology (GO) terms associated with each BLAST hit were mapped and assigned to each query (E-value cutoff = 1 × 10−6, annotation cutoff = 55, GO weight = 5). Finally, InterProScan was performed to identify more associated GO terms based on domain/motif conservation.
Construction of Orthologous Alignments
To identify the orthologous gene pairs between A. lucayanum and B. floridae, we employed Proteinortho (v5.05) (Lechner et al. 2011), a reciprocal best hits based method that is suitable for large-scale analysis. In the case where multiple genes from a species are identified as a co-orthologous group (which means gene duplication occurred after the speciation event), we selected the first member as a unique representative for that co-orthologous group. PhyloTreePruner pipeline was used to refine the orthology identification and to pick the unique representative sequence for each species based on the gene tree (Kocot et al. 2013). Muscle (v3.8) (Edgar 2004) and RevTrans (v1.4) (Wernersson and Pedersen 2003) were used to generate protein/CDS alignment for each gene pair between A. lucayanum and B. floridae. These alignments were further trimmed by Gblock (v0.91b) (Castresana 2000) to remove poorly aligned regions. Trimmed alignments that contained fewer than 20 amino acids/codons were eliminated.
Corresponding to our two RNA-Seq libraries, two sets of A. lucayanum–B. floridae orthologous gene alignments were constructed following the method described above. In addition, we constructed a set of mouse (Mus musculus)-opossum (Monodelphis domestica) orthologous gene alignments for the parallel GO term enrichment analysis. Finally, 15-way orthologous gene alignments were constructed with a high conservation stringency setting (connectivity cutoff = 0.5) in Proteinortho for the following phylogenetic analysis. For this construction, we sampled human (Homo sapiens), mouse (M. musculus), opossum (Mo. domestica), medaka fish (Oryzias latipes), zebrafish (Danio rerio), fugu fish (Takifugu rubripes), frog (Xenopus tropicalis), elephant shark (Callorhinchus milii), coelacanth (Latimeria chalumnae), lamprey (Petromyzon marinus), acorn worm (Saccoglossus kowalevskii), sea urchin (Strongylocentrotus purpuratus), and limpet (Lottia gigantea) in addition to the two cephalochordates that we studied, A. lucayanum and B. floridae. The proteome sequences of these sampled species were retrieved from online databases (supplementary table S1, Supplementary Material online). For each of these additional species, in the case of alternative splicing where multiple protein products correspond to a single gene in that species, the longest protein product was selected from the downloaded proteome as the unique representative for the corresponding gene. The 15-way orthologous protein alignments were further constructed by Muscle (Edgar 2004) and trimmed by Gblock (Castresana 2000). Trimmed alignments that contained fewer than 20 amino acids were eliminated.
Analysis of Molecular Evolution
For each A. lucayanum–B. floridae orthologous gene pair, CDS alignments with fewer than three nucleotide substitutions were eliminated. Sequence divergence for each gene was calculated by applying the Jukes–Cantor correction for the pairwise nucleotide differences. The synonymous substitution rate (Ks) and nonsynonymous substitution rate (Ka) values were calculated by KaKs_calculator (v1.2) (Zhang et al. 2006) based on a maximum-likelihood (ML) method (Goldman and Yang 1994). We defined genes with the top 5% Ka and Ka/Ks (alternative cutoffs such as top 10% and top 15% were also examined) as fast-evolving genes in cephalochordates (i.e., CDS alignments showed that these genes are much more divergent between Asymmetron and Branchiostoma than the genome-wide average). The enrichment of functional classes (GO terms) for these fast-evolving genes was examined by Fisher’s exact test (Fisher 1922) implemented in Blast2GO pipeline (Conesa et al. 2005). False discover rate was employed for multiple test correction (Benjamini and Hochberg 1995). The list of significantly enriched GO terms was further pared by GO trimming (v2.0) (Jantzen et al. 2011) to remove those redundant terms. In addition, a parallel analysis for mouse (M. musculus) and opossum (Mo. domestica) to compare with the results from the two cephalochordates was done, because placental-marsupial has a comparable divergence scale. This parallel analysis was performed with the same definition and rate cutoffs for fast-evolving genes in mouse–opossum orthologous gene pairs.
Phylogenetic Analysis
We employed ProtTest (v.3.4) (Darriba et al. 2011) to identify the most appropriate amino acid substitution model for our phylogenetic analysis based on both Akaike information criterion (AIC) and Bayesian information criterion (BIC). The LG+I+G+F model (Le and Gascuel 2008) was recommended by ProtTest as the best model based on both AIC and BIC criteria. Where LG model was not available (e.g., LG model was not implemented in MrBayes), the JTT+I+G+F model (Jones et al. 1992) was recommended as the secondary best choice. We constructed the ML trees using PhyML (v3.1) (Guindon et al. 2010) with the LG+I+G+F model. An approximate likelihood-ratio test was used to access the stability of internal nodes. We also constructed the Bayesian trees using MrBayes (v3.2.2) (Ronquist et al. 2012) with the JTT+I+G+F model. An automatic stopping rule setting and 25% burn-in were used. The branch lengths of these trees were summarized by Newick utilities (v1.6) (Junier and Zdobnov 2010) and visualized in FigTree (v1.4) (http://tree.bio.ed.ac.uk/software/figtree/, last accessed September 27, 2014).
Estimation of Divergence Time
We performed Bayesian estimation for the divergence time of the sampled 15 species to construct the time frame of cephalochordate evolution. The divergence nodes of human–mouse (61.50–100.50 Ma), human/mouse–opossum (124.50–138.40 Ma), human/mouse/opossum–frog (330.40–350.10 Ma), medaka–fugu (96.90–150.90 Ma), medaka/fugu–zebrafish (149.85–165.20 Ma), human/mouse/opossum/frog–medaka/fugu/zebrafish (416.00–421.75 Ma), human/mouse/opossum/frog/coelacanth/medaka/fugu/zebrafish–elephant shark (421.75–462.50 Ma), human/mouse/opossum/frog/coelacanth/medaka/fugu/zebrafish/elephant shark–lamprey (460.60–? Ma), human/mouse/opossum/frog/coelacanth/medaka/fugu/zebrafish/elephant shark/lamprey/asymmetron/branchistoma–sea urchin/acorn worm (518.50–? Ma), and human/mouse/opossum/frog/coelacanth/medaka/fugu/zebrafish/elephant shark/lamprey/asymmetron/branchistoma/sea urchin/acorn worm–limpet (531.50–? Ma) were used as calibration points (Benton et al. 2009). Markov chain Monte Carlo Tree (MCMCTree) requires a safe upper bound for the root age in its molecular dating analysis. For this upper bound, Benton et al. suggested that the maximum constraint for the crown bilaterian divergence time could be placed at 581.5 Ma and also used this date as the maximum constraint for the last three calibration points that we used. However, some other references gave the support for older divergence times for the crown bilaterian. For example, Peterson et al. (2008) estimated this age as approximately 640–730 Ma, while Blair (2009) proposed even older age as approximately 700–900 Ma. Therefore, we explored different maximum constraints (600, 700, 800, and 900 Ma) for the crown bilaterian divergence time in our MCMCTree analysis.
The molecular dating analysis was carried out using MCMCTree from the PAML package (v4.8) (Reis and Yang 2011) with the concatenated gene matrix used for the phylogenetic analysis. Based on the fixed topology revealed by our previous phylogenetic analysis, we first used the codeml from the PAML package (v4.8) (Reis and Yang 2011) to obtain the ML estimates of the branch lengths, the gradient vector, and Hessian matrix. These estimates together with the PAML’s LG substitution matrix (Le and Gascuel 2008) were fed into MCMCTree. Under a time unit of 1 million years, we used the same gamma prior G(1, 100) to specify the substitution rate and rate drift parameters. MCMCTree used uniform distribution with a power decay on the left side tail and an exponential decay on the right side tail to model the time span of fossil calibrations when both lower and upper bounds were specified (Yang and Rannala 2006). When only the lower bounds were specified, a truncated Cauchy distribution was implemented in MCMCTree to model the probability distribution (Inoue et al. 2010). Both the lower and upper bounds were “soft” so that there is a small probability (2.5% at both tails) that the age is beyond the bound. An autocorrelated lognormal relaxed clock model (Rannala and Yang 2007) was used to estimate the posterior distribution of divergence time, given these priors. The burn-in for the MCMC chain was set to be 10,000,000 generations. Sampled parameters were recorded every 1,000 steps and a total of 100,000 samples were collected. For each data set (asymAD and asym20h), at least two independent runs were performed to ensure the convergence of MCMC. The mean and 95% confidence interval for each Bayesian MCMC analysis were summarized by MCMCTree and visualized in FigTree (v1.4) (http://tree.bio.ed.ac.uk/software/figtree/, last accessed September 27, 2014). We also ran MCMCTree without using the sequence data but with specified calibration priors to make sure that the effective priors are sensible when compared with the specified priors.
Results
Sequencing, Assembly, and Annotation
As shown in table 1, ∼146 million and ∼177 million raw reads (paired end 100 bp) were obtained for the adult and pooled 20-h larvae libraries, respectively. For simplicity, we designate these two libraries as asymAD and asym20h. For the asymAD library, the raw Trinity assembly generated 107,972 contigs with lengths of 201–17,353 bp and the weighted median (N50) of 1,886 bp (table 1). For the asym20h library, we obtained 157,696 contigs with lengths of 201–20,851 bp and an N50 of 1,635 bp (table 1). When aligning the RNA-Seq paired-end reads back to these contigs, 89.28% (asymAD) and 83.27% (asym20h) of the reads properly aligned, suggesting that the quality of these two assemblies is reasonably good. After removing detectable redundancy due to alternative splicing, the contig counts for our two libraries were reduced to 63,243 (asymAD) and 82,723 (asym20h), respectively. Based on these contigs, we extracted 42,148 (asymAD) and 43,479 (asym20h) likely CDSs for our downstream analyses. These likely CDSs correspond to 23,245 (asymAD) and 22,941 (asym20h) original contigs because many contigs may correspond to two CDSs (both strands were considered in the CDS inference by TransDecoder). The GC% of the A. lucayanum coding regions was estimated as ∼53.38% (53.11% for asymAD and 52.64% for asym20h), which is close to that of B. floridae (53.00%). Functional annotation via the Blast2GO pipeline gave matches for 21,550 (asymAD) and 20,649 (asym20h) contigs in the SwissProt + NCBI nr database based on the likely CDSs inferred above (table 1).
Table 1.
Summary Statistics for Sequencing, Assembly, and Annotation
| asymAD | asym20h | |
|---|---|---|
| Raw reads | ∼146 million | ∼177 million |
| Raw contigs | 107,972 | 157,696 |
| Raw contig N50 | 1,886 bp | 1,635 bp |
| Reduced contigs | 63,243 | 82,723 |
| Reduced contigs with likely CDSs | 23,245 | 22,941 |
| Reduced contigs with BLAST2GO annotations | 21,550 | 20,649 |
Cephalochordates Are Evolving Particularly Slowly
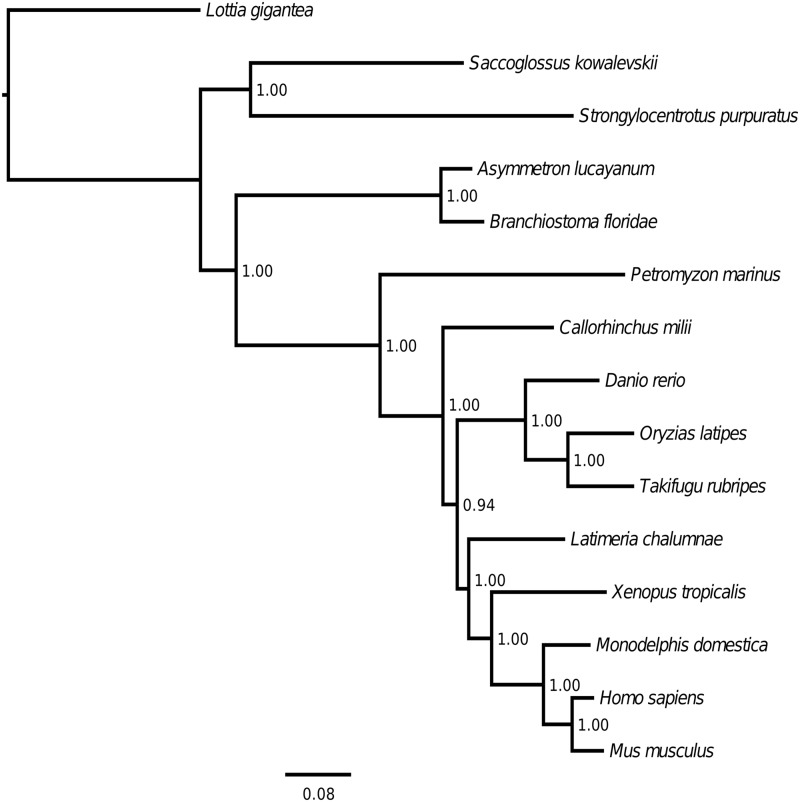
Cephalochordates have been referred to as “living fossils” due to their resemblance to fossil chordates found in Cambrian strata. To test whether such morphological conservation also holds at the molecular level, we first performed a phylogenetic analysis with two cephalochordates (A. lucayanum and B. floridae), ten vertebrates (lamprey, elephant shark, coelacanth, zebrafish, medaka fish, fugu, frog, opossum, mouse, and human), and three outgroups (acorn worm, sea urchin, and limpet). We constructed a 15-way concatenated gene matrix based on ∼430 conserved orthologous gene groups (427 for asymAD and 437 for asym20h) and examined the branch length (i.e., the expected amino acid substitution rate) of each evolutionary lineage based on ML and Bayesian analyses. Our analysis recovered phylogenetic relationships previously calculated from other data sets (Dunn et al. 2008) (see also http://metazome.org, last accessed September 27, 2014). Regardless of which RNA-Seq library (asymAD or asym20h) or which phylogenetic method (ML or Bayesian) we used, the branch lengths of the two cephalochordates were consistently shorter than those of their vertebrates, even when compared with the previously reported slowest evolving vertebrate, the elephant shark (C. millii) (Venkatesh et al. 2014) (fig. 2 and supplementary figs. S1–S3, Supplementary Material online). In addition, the clear difference in branch lengths since their last common ancestor indicated that A. lucayanum is evolving even more slowly than B. floridae (fig. 2 and supplementary figs. S1–S3, Supplementary Material online). From the difference in branch lengths, we estimate that the amino acid substitution rate of B. floridae is ∼1.5 times higher than that of A. lucayanum. In addition, we performed Tajima’s relative rate test (Tajima 1993) based directly on the concatenated gene alignment. All pairwise tests between cephalochordates and vertebrates and between the two cephalochordates reveal significant statistical supports (P value < 2.2 × 10−16) for both observed evolutionary rate differences, no matter which outgroup we used. In addition, the hemichordate, S. kowalevskii, also appears to be evolving slowly, as indicated by the short branch length in both ML and Bayesian trees.
Fig. 2.—
Fifteen-way maximum-likelihood phylogenetic tree inferred from a concatenated orthologous gene matrix (427 orthologous gene groups). The branch length is proportional to the expected amino acid substitution rate and the scale bar represents 0.08 expected amino acid substitutions per site. The numbers at the internal nodes show the statistical support for the topology of the tree based on the approximate logarithm-likelihood test. The result shown here is based on the analysis for the asymAD library. The Bayesian tree for the asymAD library and parallel analyses for the asym20h library are shown in supplementary figs. S1–S3, Supplementary Material online.
Genes Involved in Innate Immunity Are Fast Evolving in Cephalochordates
To identify the types of genes that are evolving most rapidly, we calculated the mean nucleotide divergence (D), nonsynonymous substitution rate (Ka), synonymous substitution rate (Ks), and nonsynonymous to synonymous substitution rate ratio (Ka/Ks) for orthologous gene pairs from B. floridae and A. lucayanum. First we repaired the annotated gene models that mapped to the B. floridae v2.0 genome assembly (http://genome.jgi-psf.org/Brafl1/Brafl1.download.ftp.html, last accessed September 27, 2014). Among the 28,667 B. floridae genes in this assembly, 793 contained stop codons within CDS regions, suggesting spurious gene models. We corrected 719 of them based on their protein sequences and excluded the rest because protein sequences (downloaded from http://genome.jgi-psf.org/Brafl1/Brafl1.download.ftp.html, last accessed September 27, 2014) could not be aligned with the presumed CDS. This left 28,593 genes for subsequent analyses. Searches with the Blast2GO pipeline against the SwissProt + NCBI nr database successfully annotated 28,586 of these genes.
When A. lucayanum CDSs were matched to these 28,586 B. floridae genes, more than 8,000 orthologous gene pairs were recovered (8,333 for asymAD and 8,715 for asym20h). Approximately 72% of these Asymmetron–Branchiostoma orthologous gene pairs (6,016 for asymAD and 6,305 for asym20h) had nearly full-length CDS alignments. After trimming and gap removal, the alignment coverage was ≥80%, and the length of the alignments ranged from 138 to 13,160 bp. The means of nucleotide divergence (D), nonsynonymous substitution rate (Ka), synonymous substitution rate (Ks), and nonsynonymous to synonymous substitution rate ratio (Ka/Ks) across these ∼8,500 genes are shown in table 2. Regardless of which library we used, more than 40% of the genes had Ks > 1, suggesting considerable saturation at the synonymous sites—that is, that the forward and backward mutations are in steady state. Therefore, in addition to using Ka/Ks, we also employed Ka as the measurement for molecular evolutionary rate in defining the fast-evolving genes. Ka has been recommended as a superior proxy in sorting genes by their evolutionary rate especially when the divergence scale is large (Wang et al. 2011). These analyses measure the degree of divergence between the A. lucayanum and B. floridae genes and do not distinguish whether a given gene is evolving faster in one species compared with the other. Two sets of particularly fast-evolving genes were identified for each library by using top 5% Ka and Ka/Ks criteria, respectively. No matter which proxy we used (Ka/Ks or Ka) when defining the fast-evolving gene set, we found a 3- to 4-fold difference in evolutionary rates between the fast-evolving set and the genome-wide average (table 2).
Table 2.
Genome-Wide Nucleotide Divergence (D), Nonsynonymous Substitution Rate (Ka), Synonymous Substitution Rate (Ks), and Nonsynonymous to Synonymous Substitution Rate Ratio (Ka/Ks) in the Asymmetron–Branchiostoma comparison
| Mean of All Genes |
Mean of Fast-Evolving Genes Sorted by Ka |
Mean of Fast-Evolving Genes Sorted by Ka/Ks |
||||
|---|---|---|---|---|---|---|
| asymAD | asym20h | asymAD | asym20h | asymAD | asym20h | |
| D | 0.2630 | 0.2549 | 0.8823 | 0.8674 | 0.6413 | 0.6530 |
| Ka | 0.1448 | 0.1379 | 0.6735 | 0.6692 | 0.5152 | 0.5246 |
| Ks | 1.1520 | 1.1170 | 2.0680 | 2.0880 | 1.1930 | 1.2560 |
| Ka/Ks | 0.1211 | 0.1162 | 0.3656 | 0.3568 | 0.4668 | 0.4365 |
To examine whether genes involved in certain biological functions are overrepresented in our fast-evolving list, we examined the enrichment of GO terms based on the contrast between our fast-evolving gene list and all those ∼8,500 tested genes. To ensure the robustness of this analysis of fast-evolving genes, we examined alternative Ka and Ka/Ks cutoffs (top 10% and top 15%) and only considered those enriched GO terms that are insensitive to the choice of cutoff. We found 76 and 19 GO terms that are overrepresented in our fast-evolving gene sets based on Ka and Ka/Ks, respectively (tables 3 and 4). Most GO terms identified by Ka/Ks were included in the terms identified by Ka.
Table 3.
Enriched GO Terms Identified based on Ka Sorting in Both RNA-Seq Libraries for Fast-Evolving Genes in Cephalochordate Evolution
| GO ID | Term | Category | FDR |
|
|---|---|---|---|---|
| asymAD | asym20h | |||
| GO:0004888 | Transmembrane signaling receptor activity | F | 4.69 × 10−11 | 4.20 × 10−6 |
| GO:0042330 | Taxis | P | 2.84 × 10−7 | 2.46 × 10−4 |
| GO:0044459 | Plasma membrane part | C | 2.84 × 10−7 | 2.91 × 10−6 |
| GO:0005886 | Plasma membrane | C | 3.75 × 10−7 | 2.28 × 10−5 |
| GO:0007155 | Cell adhesion | P | 3.93 × 10−7 | 2.32 × 10−9 |
| GO:0031012 | Extracellular matrix | C | 4.55 × 10−7 | 5.09 × 10−16 |
| GO:0004930 | G-protein coupled receptor activity | F | 6.29 × 10−7 | 3.42 × 10−4 |
| GO:0006935 | Chemotaxis | P | 7.08 × 10−7 | 1.34 × 10−4 |
| GO:0044707 | Single–multicellular organism process | P | 2.29 × 10−6 | 4.75 × 10−5 |
| GO:0051239 | Regulation of multicellular organismal process | P | 4.70 × 10−6 | 8.81 × 10−7 |
| GO:0044421 | Extracellular region part | C | 5.88 × 10−6 | 4.51 × 10−14 |
| GO:0007411 | Axon guidance | P | 1.42 × 10−5 | 1.93 × 10−3 |
| GO:0005216 | Ion channel activity | F | 1.43 × 10−5 | 3.62 × 10−3 |
| GO:0022838 | Substrate-specific channel activity | F | 1.73 × 10−5 | 4.44 × 10−3 |
| GO:0005262 | Calcium channel activity | F | 2.30 × 10−5 | 6.49 × 10−4 |
| GO:0015267 | Channel activity | F | 2.33 × 10−5 | 2.41 × 10−3 |
| GO:0022803 | Passive transmembrane transporter activity | F | 2.33 × 10−5 | 2.41 × 10−3 |
| GO:0048666 | Neuron development | P | 2.33 × 10−5 | 4.44 × 10−3 |
| GO:0044425 | Membrane part | C | 3.06 × 10−5 | 3.66 × 10−4 |
| GO:0032501 | Multicellular organismal process | P | 3.54 × 10−5 | 1.62 × 10−4 |
| GO:0005578 | Proteinaceous extracellular matrix | C | 5.06 × 10−5 | 6.53 × 10−8 |
| GO:0002689 | Negative regulation of leukocyte chemotaxis | P | 5.15 × 10−5 | 2.05 × 10−5 |
| GO:0031175 | Neuron projection development | P | 7.86 × 10−5 | 2.41 × 10−3 |
| GO:0007166 | Cell surface receptor signaling pathway | P | 9.19 × 10−5 | 2.53 × 10−4 |
| GO:0048699 | Generation of neurons | P | 1.39 × 10−4 | 4.09 × 10−4 |
| GO:0065008 | Regulation of biological quality | P | 1.41 × 10−4 | 1.10 × 10−2 |
| GO:0005261 | Cation channel activity | F | 1.44 × 10−4 | 2.86 × 10−2 |
| GO:0022836 | Gated channel activity | F | 1.44 × 10−4 | 7.27 × 10−3 |
| GO:0022839 | Ion-gated channel activity | F | 1.44 × 10−4 | 7.27 × 10−3 |
| GO:0007186 | G-protein coupled receptor signaling pathway | P | 2.00 × 10−4 | 3.72 × 10−2 |
| GO:0021834 | Chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration | P | 2.00 × 10−4 | 9.32 × 10−4 |
| GO:0035385 | Roundabout signaling pathway | P | 2.00 × 10−4 | 1.43 × 10−3 |
| GO:0051414 | Response to cortisol stimulus | P | 2.00 × 10−4 | 9.32 × 10−4 |
| GO:0061364 | Apoptotic process involved in luteolysis | P | 2.00 × 10−4 | 1.43 × 10−3 |
| GO:0070100 | Negative regulation of chemokine-mediated signaling pathway | P | 2.00 × 10−4 | 1.43 × 10−3 |
| GO:0001960 | Negative regulation of cytokine-mediated signaling pathway | P | 2.73 × 10−4 | 1.23 × 10−2 |
| GO:0006928 | Cellular component movement | P | 2.73 × 10−4 | 4.59 × 10−6 |
| GO:0048495 | Roundabout binding | F | 3.44 × 10−4 | 9.32 × 10−4 |
| GO:0009605 | Response to external stimulus | P | 4.37 × 10−4 | 2.28 × 10−5 |
| GO:0048731 | System development | P | 4.86 × 10−4 | 3.65 × 10−4 |
| GO:0009888 | Tissue development | P | 6.42 × 10−4 | 4.71 × 10−5 |
| GO:0040011 | Locomotion | P | 9.94 × 10−4 | 1.82 × 10−4 |
| GO:0015276 | Ligand-gated ion channel activity | F | 1.04 × 10−3 | 1.81 × 10−2 |
| GO:0030198 | Extracellular matrix organization | P | 1.04 × 10−3 | 3.65 × 10−4 |
| GO:0001948 | Glycoprotein binding | F | 1.51 × 10−3 | 7.65 × 10−8 |
| GO:0030154 | Cell differentiation | P | 1.79 × 10−3 | 8.44 × 10−4 |
| GO:0021772 | Olfactory bulb development | P | 1.80 × 10−3 | 7.69 × 10−4 |
| GO:0009653 | Anatomical structure morphogenesis | P | 2.03 × 10−3 | 3.76 × 10−6 |
| GO:0048846 | Axon extension involved in axon guidance | P | 2.20 × 10−3 | 1.71 × 10−3 |
| GO:0007610 | Behavior | P | 2.51 × 10−3 | 3.60 × 10−3 |
| GO:0072358 | Cardiovascular system development | P | 2.74 × 10−3 | 1.01 × 10−6 |
| GO:0032101 | Regulation of response to external stimulus | P | 2.84 × 10−3 | 1.65 × 10−3 |
| GO:0050920 | Regulation of chemotaxis | P | 2.84 × 10−3 | 4.59 × 10−4 |
| GO:0051606 | Detection of stimulus | P | 2.84 × 10−3 | 2.85 × 10−2 |
| GO:0009986 | Cell surface | C | 3.19 × 10−3 | 2.55 × 10−3 |
| GO:0050900 | Leukocyte migration | P | 3.31 × 10−3 | 5.90 × 10−5 |
| GO:0032879 | Regulation of localization | P | 4.65 × 10−3 | 2.32 × 10−4 |
| GO:0044420 | Extracellular matrix part | C | 5.20 × 10−3 | 8.87 × 10−6 |
| GO:0031224 | Intrinsic to membrane | C | 6.41 × 10−3 | 3.33 × 10−2 |
| GO:0048513 | Organ development | P | 6.56 × 10−3 | 1.13 × 10−3 |
| GO:0007275 | Multicellular organismal development | P | 7.76 × 10−3 | 3.51 × 10−4 |
| GO:0043394 | Proteoglycan binding | F | 8.24 × 10−3 | 9.79 × 10−4 |
| GO:0006816 | Calcium ion transport | P | 8.72 × 10−3 | 2.41 × 10−3 |
| GO:0007417 | Central nervous system development | P | 9.18 × 10−3 | 2.41 × 10−3 |
| GO:0021537 | Telencephalon development | P | 1.02 × 10−2 | 1.40 × 10−3 |
| GO:0042221 | Response to chemical stimulus | P | 1.11 × 10−2 | 4.51 × 10−3 |
| GO:0060326 | Cell chemotaxis | P | 1.41 × 10−2 | 1.43 × 10−3 |
| GO:0014912 | Negative regulation of smooth muscle cell migration | P | 1.57 × 10−2 | 6.12 × 10−5 |
| GO:0032102 | Negative regulation of response to external stimulus | P | 1.74 × 10−2 | 1.55 × 10−2 |
| GO:0016477 | Cell migration | P | 1.90 × 10−2 | 4.09 × 10−4 |
| GO:0051674 | Localization of cell | P | 2.56 × 10−2 | 1.42 × 10−3 |
| GO:0006929 | Substrate-dependent cell migration | P | 3.40 × 10−2 | 2.86 × 10−2 |
| GO:0035295 | Tube development | P | 3.73 × 10−2 | 1.23 × 10−3 |
| GO:0001568 | Blood vessel development | P | 3.80 × 10−2 | 8.29 × 10−6 |
| GO:0034220 | Ion transmembrane transport | P | 4.57 × 10−2 | 1.20 × 10−2 |
| GO:0051240 | Positive regulation of multicellular organismal process | P | 4.81 × 10−2 | 3.51 × 10−3 |
Note.—For the GO term category column, “C” stands for cellular component, “F” for biological function, and “P” biological process. The statistical significance was assessed by Fisher’s exact test with FDR correction.
FDR = false discover rate.
Table 4.
Enriched GO Terms Identified based on Ka/Ks Sorting in both RNA-Seq Libraries for Fast-Evolving Genes in Cephalochordate Evolution
| GO ID | Term | Category | FDR |
|
|---|---|---|---|---|
| asymAD | asym20h | |||
| GO:0031012 | Extracellular matrix | C | 4.38 × 10−5 | 2.45 × 10−5 |
| GO:0004872 | Receptor activity | F | 1.02 × 10−4 | 2.45 × 10−5 |
| GO:0048495 | Roundabout binding | F | 1.02 × 10−4 | 1.10 × 10−3 |
| GO:0050919 | Negative chemotaxis | P | 1.02 × 10−4 | 1.18 × 10−4 |
| GO:0021834 | Chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration | P | 1.53 × 10−4 | 1.10 × 10−3 |
| GO:0035385 | Roundabout signaling pathway | P | 1.53 × 10−4 | 1.99 × 10−3 |
| GO:0051414 | Response to cortisol stimulus | P | 1.53 × 10−4 | 1.10 × 10−3 |
| GO:0061364 | Apoptotic process involved in luteolysis | P | 1.53 × 10−4 | 1.99 × 10−3 |
| GO:0070100 | Negative regulation of chemokine-mediated signaling pathway | P | 1.53 × 10−4 | 1.99 × 10−3 |
| GO:0007155 | Cell adhesion | P | 3.84 × 10−4 | 8.51 × 10−4 |
| GO:0005576 | Extracellular region | C | 5.91 × 10−4 | 1.13 × 10−10 |
| GO:0002689 | Negative regulation of leukocyte chemotaxis | P | 6.73 × 10−4 | 1.00 × 10−4 |
| GO:0005578 | Proteinaceous extracellular matrix | C | 1.07 × 10−3 | 7.80 × 10−3 |
| GO:0050922 | Negative regulation of chemotaxis | P | 1.15 × 10−3 | 3.58 × 10−3 |
| GO:0048846 | Axon extension involved in axon guidance | P | 1.72 × 10−3 | 3.22 × 10−2 |
| GO:0044421 | Extracellular region part | C | 4.46 × 10−3 | 2.44 × 10−8 |
| GO:0021772 | Olfactory bulb development | P | 6.56 × 10−3 | 8.90 × 10−3 |
| GO:0004888 | Transmembrane signaling receptor activity | F | 1.16 × 10−2 | 1.98 × 10−2 |
| GO:0086070 | SA node cell to atrial cardiac muscle cell communication | P | 4.02 × 10−2 | 2.55 × 10−2 |
Note.—For the GO term category column, “C” stands for cellular component, “F” for biological function, and “P” for biological process. The statistical significance was assessed by Fisher’s exact test with FDR correction.
FDR = false discover rate.
Among the enriched GO terms, those related to the innate immune system stood out. For example, 56 of the 76 GO terms identified by Ka are included in a curated collection of neural/immune-related GO terms (Geifman et al. 2010). The nervous and immune systems (both innate and adaptive) are known to interact with each other; the nervous system modulates the function of the immune system, which in turn influences the nervous system (Sternberg 2006; Ziv et al. 2006; Ransohoff and Brown 2012; Wraith and Nicholson 2012). This strong connection between the fast-evolving genes and the neural/immune systems was also observed in our parallel analysis on the placental–marsupial (mouse–opossum) comparison (supplementary tables S2 and S3, Supplementary Material online). A relatively rapid evolutionary rate of immune genes also occurs in other lineages, such as Drosophila (Schlenke and Begun 2003), Daphnia (McTaggart et al. 2012), chickens (Downing et al. 2009), rodents (Hurst and Smith 1999), and primates (Nielsen et al. 2005).
Many of the fast-evolving genes identified by the Asymmetron–Branchiostoma comparison of GO terms are transmembrane cellular receptors involved in the response to external stimuli, suggesting a collective role in innate immunity. The most notable GO terms are those related to “chemotaxis”—that is, the unidirectional movement of cells in response to chemical stimuli. The chemotactic movement of immune cells such as leukocytes (neutrophils in particular) is an important feature of innate immune systems (Mantovani et al. 2011). Several GO terms in table 3 echo this feature, such as chemotaxis (GO:0006935), “negative regulation of leukocyte chemotaxis” (GO:0002689), “regulation of chemotaxis” (GO:0050920), and “leukocyte migration” (GO:0050900). Other enriched GO terms like “axon guidance” (GO:0007411), “roundabout signaling pathway” (GO:0035385), and “roundabout binding” (GO:0048495) are known to be associated with the Robo/Slit signaling pathway, which not only can regulate nervous system development (Kidd et al. 1998; Borrell et al. 2012) but also down-regulates neutrophil migration (Wu et al. 2001). This highlights the functional links among these enriched GO terms in the context of innate immunity. In addition, some well-known innate immune components of amphioxus, such as the NOD-like receptor (NLR) genes, scavenger receptor cysteine-rich (SRCR) receptor genes, and C1q-like genes, are also included in the fast-evolving genes we identified. In contrast, the enriched GO terms identified in the fast-evolving genes in the placental–marsupial comparison are highly associated with adaptive immunity as suggested by multiple terms associated with T-cells (supplementary tables S2 and S3, Supplementary Material online).
Reconstructing the Time Frame of Cephalochordate Evolution
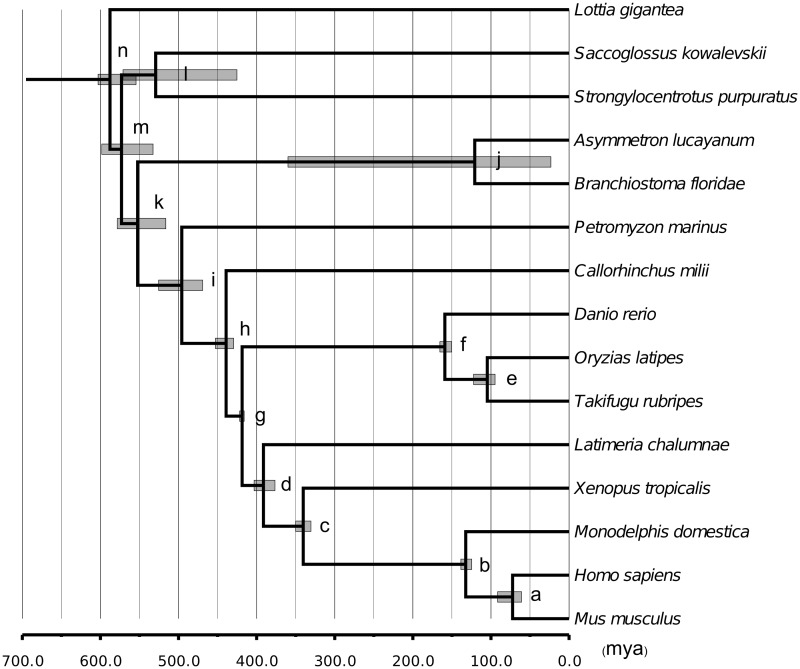
In addition to allowing comparisons of evolutionary rates between cephalochordates and vertebrates, the 15-way alignments also permitted reconstruction of the time frame of cephalochordate evolution. For each RNA-Seq library, we performed Bayesian MCMC estimations of the divergence times among these 15 species. We used 11 time constraints across the phylogenetic tree shown in figure 2 (see Materials and Methods). Different maximum constraints for the crown bilaterian divergence time were explored ranging from 600 to 900 Ma based on previous reports (Peterson et al. 2008; Benton et al. 2009; Blair 2009). According to the Bayesian MCMC estimation, the split between Asymmetron and Branchiostoma occurred ∼120 Ma, regardless of how we capped the crown bilaterian divergence time (tables 5 and 6, supplementary tables S4 and S5, Supplementary Material online, and fig. 3). The divergence time between cephalochordates and vertebrates was estimated as ∼552, 620, 680, or 730 Ma depending on the specific assumption for the upper bound of the crown bilaterian divergence time (600, 700, 800, or 900 Ma, respectively) (tables 5 and 6, supplementary tables S4 and S5, Supplementary Material online, and fig. 3).
Table 5.
Bayesian MCMC Estimations for the Divergence Time of Each Internal Node Shown in Figure 3, Assuming the Crown Bilaterian Divergence Occurred No Earlier than 600 Ma
| Node Index | Node Name | Calibration Constraints | asymAD |
asym20h |
||
|---|---|---|---|---|---|---|
| [Min, Max] (Ma) | Mean (Ma) | 95% CI (Ma) | Mean (Ma) | 95% CI (Ma) | ||
| a | Eutheria | [61.50, 100.50] | 72.30 | [60.80–91.48] | 72.16 | [60.81–91.49] |
| b | Mammalia | [124.50, 138.40] | 132.30 | [124.69–138.52] | 132.30 | [124.67–138.51] |
| c | Tetrapoda | [330.40, 350.10] | 340.34 | [330.44–350.09] | 340.31 | [330.41–350.11] |
| d | Sarcopterygii | — | 391.29 | [376.53–403.28] | 391.51 | [376.65–403.36] |
| e | Acanthopterygii | [96.90, 150.90] | 104.69 | [94.77–122.23] | 104.74 | [94.79–122.43] |
| f | Actinopterygii | [149.85, 165.20] | 159.05 | [150.24–165.40] | 159.05 | [150.25–165.40] |
| g | Osteichthyes | [416.00, 421.75] | 418.53 | [415.95–421.69] | 418.58 | [415.96–421.70] |
| h | Gnathostomata | [421.75, 462.50] | 437.60 | [428.82–450.21] | 438.61 | [429.15–452.18] |
| i | Vertebreta | [460.60, —] | 495.86 | [469.11–525.45] | 488.56 | [468.91–525.34] |
| j | Cephalochordata | — | 120.77 | [23.25–359.86] | 120.66 | [23.13–357.97] |
| k | Chordata | — | 552.38 | [516.31–578.43] | 552.85 | [516.89–578.81] |
| l | Ambulacraria | — | 529.27 | [425.21–571.10] | 529.29 | [425.02–571.03] |
| m | Deuterostomia | [518.50, —] | 573.10 | [532.67–598.27] | 573.07 | [533.08–598.29] |
| n | Bilateria | [531.50, —] | 587.78 | [554.43–602.99] | 587.64 | [554.05–602.95] |
Note.—The nodes are labeled in figure 2. The calibration constraints were based on fossil records summarized by Benton et al. (2009).
CI = confidence interval.
Table 6.
Bayesian MCMC Estimations for the Divergence Time of Each Internal Node Shown in Figure 3, Assuming the Crown Bilaterian Divergence Occurred No Earlier than 700 Ma
| Node Index | Node Name | Calibration Constraints | asymAD |
asym20h |
||
|---|---|---|---|---|---|---|
| [Min, Max] (Ma) | Mean (Ma) | 95% CI (Ma) | Mean (Ma) | 95% CI (Ma) | ||
| a | Eutheria | [61.50, 100.50] | 70.20 | [60.62–86.77] | 70.11 | [60.64–86.84] |
| b | Mammalia | [124.50, 138.40] | 132.57 | [124.77–138.55] | 132.57 | [124.74–138.55] |
| c | Tetrapoda | [330.40, 350.10] | 339.11 | [330.27–349.83] | 339.12 | [330.26–349.85] |
| d | Sarcopterygii | — | 388.93 | [376.60–399.61] | 389.21 | [376.74–399.90] |
| e | Acanthopterygii | [96.90, 150.90] | 102.63 | [94.27–117.61] | 102.70 | [94.33–117.79] |
| f | Actinopterygii | [149.85, 165.20] | 159.63 | [150.49–165.47] | 159.61 | [150.49–165.45] |
| g | Osteichthyes | [416.00, 421.75] | 418.72 | [415.98–421.72] | 418.73 | [415.98–421.72] |
| h | Gnathostomata | [421.75, 462.50] | 444.75 | [433.64–458.91] | 444.03 | [432.96–458.34] |
| I | Vertebreta | [460.60, —] | 529.34 | [490.15–574.11] | 528.40 | [488.22–573.34] |
| J | Cephalochordata | — | 119.34 | [25.02–379.87] | 116.83 | [24.74–343.19] |
| K | Chordata | — | 620.47 | [560.67–665.04] | 619.55 | [557.46–665.14] |
| L | Ambulacraria | — | 589.71 | [463.44–652.45] | 589.15 | [473.08–652.14] |
| M | Deuterostomia | [518.50, —] | 653.52 | [585.50–698.50] | 651.58 | [580.92–698.00] |
| N | Bilateria | [531.50, —] | 679.01 | [618.90–708.77] | 678.07 | [615.82–708.83] |
Note.—The nodes are labeled in figure 2. The calibration constraints were based on fossil records summarized by Benton et al. (2009).
CI = confidence interval.
Fig. 3.—
The time frame of cephalochordate evolution inferred by MCMCTree based on the asymAD library. The Bayesian estimations for all internal nodes (nodes a–n) are tabulated in table 5. Each grey bar represents the 95% confidence interval for the corresponding estimate.
Discussion
Divergence Time Estimation
Divergence times calculated from fossil records and molecular data are often disparate. In our analysis with ∼430 orthologous gene groups, the two cephalochordates split from one another about 120 Ma, cephalochordates split from vertebrates more than 550 Ma, while hemichordates and echinoderms split with chordates more than 570 Ma. The specific dates of the last two divergences depend on the divergence time chosen for the crown bilaterian (tables 5 and 6 and supplementary tables S4 and S5, Supplementary Material online). Unfortunately, this divergence time is highly uncertain. The earliest known bilaterians such as Spriggina, a possible arthropod from the Ediacaran (635–542 Ma), are Precambrian (Glaessner 1958). Different maximal bounds for timing of this divergence point have been proposed, ranging from ∼600 to ∼900 Ma or even earlier (Peterson et al. 2008; Benton et al. 2009; Blair 2009). Benton et al. (2009) proposed using 581 Ma as the soft maximal bound for the crown bilaterian divergence based on the most conservative fossil evidence. Using different methods and parameter settings, Peterson et al. (2008) obtained an estimate 643–733 Ma for the timing of the crown bilaterian divergence, which corresponded to a maximal bound of 671–870 Ma. However, Blair (2009) argued that the method of Peterson et al. (2008) may suffer from underestimation and proposed an even older time for the crown bilaterian divergence. Because this is still an open question, and the current fossil record cannot illuminate this point clearly, we explored different divergence times for the crown bilaterian.
When using 600 Ma to cap the crown bilaterian divergence time, we obtained an estimate of 550 Ma for the cephalochordate–vertebrate divergence and 570 Ma for the split between chordates and the hemichordate + echinoderm clade. These two dates should be taken as the most conservative estimates for these two divergence points. An alternative assumption of 700 Ma for the maximal crown bilaterian divergence time led to the estimate of 620 and 650 Ma for those two divergence points respectively. These estimates are consistent with recent studies with much larger taxonomic samplings. For example, Edgecombe et al. (2011) obtained an estimate of 584 and 617 Ma for the cephalochordate–vertebrate and the chordate to hemichordate + echinoderm divergences, respectively, by using 635 Ma as the maximal constraints for the bilaterian divergence. Using 800 or 900 Ma for the maximal bound of the crown bilaterian divergence further pushes back the estimated time of these two divergence points by another 60–100 Ma.
If the chordates radiated before the Cambrian as molecular analyses suggest, there are only rather dubious chordate fossils in earlier strata. Fossils from the Ediacaran about 555 Ma have been interpreted as possible tunicates (Fedonkin et al. 2012), but this identification is far from certain. Fedonkin and Waggoner (1997) have also described mollusk-like animals from the same period. There are few fossils from earlier in the Proterozoic, and most bear little or no resemblance to animals in the Cambrian. Sponges from the Cryogenian 760 Ma have been described (Brain et al. 2012) as well as presumed sponge microfossils from the Riphean (800–1,400 Ma) (German and Podkovyrov 2012), and it has been suggested that the lack of siliceous spicules in the Precambrian is due to lack of their preservation and not to the absence of sponges (Sperling et al. 2010). A divergence time of 800–900 Ma for early bilaterians is prior to the “snowball earth” when it was theorized that the earth was completely covered in ice with the underlying ocean anoxic (Hoffman and Schrag 2002). However, the idea of a stagnant, relatively lifeless ocean beneath the ice has been disputed by Ashkenazy et al. (2013), who calculated that near the continents, coastal upwelling and strong ocean circulation combined to create high melting rates and may have created conditions facilitating photosynthesis.
Divergence of Asymmetron and Branchiostoma
On the basis of nuclear genes, we estimated the divergence time between A. lucayanum and B. floridae as about 120 Ma, which is somewhat less than ∼162 Ma estimated from mitochondrial gene sequences (Nohara et al. 2005; Kon et al. 2007). A divergence time of 120 Ma is comparable with that of the marsupial/placental split (Benton et al. 2009). Our estimate of the A. lucayanum–B. floridae divergence is insensitive to the choice of the maximal bound for the crown bilaterian divergence, suggesting that it is quite robust.
Given this long evolutionary time, it is surprising that B. floridae and A. lucayanum are morphologically far more alike than marsupials and placentals, which have a comparable divergence scale (Benton et al. 2009). The major differences between these two amphioxus genera are in size (Branchiostoma is up to 5 cm long, while Asymmetron does not exceed 1.5 cm) and in the presence of gonads only on the right side of A. lucayanum and on both sides in B. floridae. The second group of invertebrate chordates, the tunicates, provides a sharp contrast. The five groups of tunicates ascidians, salps, doliolids, pyrosomes, and appendicularians are extremely diverse in lifestyle, reproductive modes, development, and morphology. The genomes of Oikopleura, an appendicularian, and Ciona, an ascidian, have been sequenced. Their genomes are very small, ∼70 (Seo et al. 2001) and 160 mb (Dehal et al. 2002), respectively. In addition, they have lost many genes but not necessarily the same ones in both species. For example, while both have lost several Hox genes, they have lost different sets of Hox genes. Not surprisingly, it has been calculated that C. intestinalis is evolving about 50% faster than vertebrates. Taking this into consideration, a divergence time of ∼180 Ma was calculated for two species of Ciona (Berná et al. 2009). This is greater than the divergence times between the three genera of cephalochordates. Based on a relaxed molecular clock method, a phylogenetic analysis using 129 nuclear genes calculated the divergence time between C. intestinalis and vertebrates at about 620 Ma (Douzery et al. 2004). Presumably, the divergence times of the five major groups of tunicates would also be quite old.
Asymmetron Is Evolving More Slowly than Branchiostoma
Because the two amphioxus genera are morphologically very much alike in spite of their diverging ∼120 Ma (fig. 1), we asked whether this slow evolution in morphology holds at the molecular level. Our results indicate that it does; both Asymmetron and Branchiostoma are evolving significantly more slowly than vertebrates, even compared with the elephant shark, which to date is the slowest evolving vertebrate known (Venkatesh et al. 2014). Nothing is known of the molecular evolutionary rate of the third genus of cephalochordates, Epigonichthys. However, as an analysis based on mitochondrial DNA sequences placed Branchiostoma and Epigonichthys as sister groups, with Asymmetron diverging earlier from these two in the cephalochordates, it would be surprising if Epigonichthys were not also evolving slowly.
Given the slow evolutionary rate of cephalochordates as a whole, we found that Asymmetron is evolving even more slowly than Branchiostoma. Molecular evolution rates are a combined product of mutation, random genetic drift, and natural selection. Several factors have been identified that can affect these parameters, including efficiency of DNA replication and DNA repair, effective population size, metabolic rate, speciation rate, and generation time (Ohta 1972; Bousquet et al. 1992; Martin and Palumbi 1993). However, aside from molecular phylogenetic studies based on mitochondrial DNA sequences (Nishikawa 2004; Kon et al. 2006), two studies on reproduction and development (Holland ND and Holland LZ 2010; Holland 2011), a few reports on its occurrence in various localities, and our current study, nothing is known about any species of Asymmetron.
In contrast, there are several studies on population structure of Asian species of Branchiostoma (Chen et al. 2007; Li et al. 2013). These studies have demonstrated that both B. japonicum and B. belcheri have very high genetic diversity of both mitochondrial DNA and microsatellites (Li et al. 2013). However, mitochondrial DNA failed to show any difference among populations, although microsatellite data showed there were some differences among local populations. These results indicated high gene flow along the east coast of China with evidently some discontinuity of habitats (Li et al. 2013). Similarly, there was no genetic diversity detected by analysis of 12S ribosomal RNA sequences in populations of B. japonicum or B. belcheri between Hong Kong in the south, Xiamen in the center, and Qingdao in the north, a distance of over 1,700 km, indicating very high gene flow among populations (Chen et al. 2007). However, some differentiation among the populations of B. japonicum was shown by AFLP analysis, although it was quite low (<10%; Chen et al. 2007). A. lucayanum and B. floridae also have broad distributions—A. lucayanum over at least 2,500 km (Bimini, Bahamas to Bermuda) and B. floridae in the Gulf of Mexico from at least Alabama to the Dry Tortugas (∼1,100 km) and possibly further into the Caribbean. Because the species within a genus are morphologically nearly identical, it is uncertain to what extent B. floridae and B. caribaeum, known from Jamaica to Brazil, overlap (Poss and Boschung 1996). Certainly, the geographic distribution and population structure of amphioxus species in the Atlantic Ocean should be studied by molecular methods.
What our data underscore is the very slow rate of evolution of cephalochordates in general. Not surprisingly, modern cephalochordates are morphologically very much like the fossil cephalochordate from the mid-Cambrian, Pikaia, and, except for the absence of eyes and a smaller brain, very much like Haikouella, from the early Cambrian, which has been placed as the sister group of vertebrates (craniates) (Mallatt and Chen 2003). This low level of morphological evolution is congruent with the low rate of evolution at the genome level. Even so, there have been wide swings in atmospheric CO2 and air/ocean temperatures over the last 500 million years. Over much of this time, CO2 level and temperatures were higher than today. Exceptions occurred about 340 Ma in the Carboniferous when average surface temperatures were lower than today and during the glacial cycles in the Cenozoic (Rothman 2002). The ability of animals to cope with environmental change is some combination of the ability of individuals to acclimate and of the species to adapt (Whitehead 2012). Modern amphioxus, which typically live in shallow waters protected from wave action, have a remarkable ability to acclimate to short-term environmental change. Populations of amphioxus in Old Tampa Bay can experience wide fluctuations of both temperature and salinity. Over a period of 5 years, salinity at one location in the northern part of the bay varied from 19 to 28.5 parts per thousand during summer, while water temperatures ranged from 15 °C in winter to 32 °C in summer (Stokes and Holland 1996). This ability to acclimate is clearly advantageous to amphioxus. Presumably, natural selection occurred for certain gene regulatory networks that regulate the physiological parameters that enable amphioxus to acclimate. It is tempting to speculate that the ability to acclimate may help amphioxus to maintain their ability to accommodate environmental changes, and therefore allow their very slow background evolutionary rates and morphological stasis through time. The physiology, ecology, and population dynamics of amphioxus species have received relatively little attention. In depth studies of these parameters may help to clarify the reasons why amphioxus is evolving so very slowly.
Many Fast Evolving Genes in Cephalochordates Are Involved in Innate Immunity
In the absence of an adaptive immune system, cephalochordates must rely on their innate immune system to fight against various pathogens in the environment. Indeed, many gene families involved in innate immunity, including those with toll interleukin-1 receptor domains and those in a nucleotide triphosphatase family (NACHT domains), have undergone considerable expansion in amphioxus (Zhang et al. 2008). Therefore, it is not altogether surprising that the fast evolving genes include many of those involved in innate immunity. Particularly noteworthy are those related to chemotaxis of important immune cells like neutrophils and several well-known innate immune receptor genes such as NLRs, SRCR receptor genes, and the C1q-like genes in the complement system. Pattern recognition receptors like NLRs and SRCRs can sense the intruding signal of a wide range of bacterial and fungal pathogens and trigger the downstream innate immune response. Previous studies have found significant expansion of these gene families in animals that lack the adaptive immune system, including amphioxus and sea urchins (Huang et al. 2008; Dishaw et al. 2012). Finally, the complement system consists of some small proteins in the circulation system and functions in proteolysis to clear pathogens from the host. It has been shown that the amphioxus C1q-like genes may function similarly as the lamprey C1q genes via the lectin pathway (Yu et al. 2008).
Populations of both Branchiostoma and Asymmetron burrow in the sand in shallow in-shore waters, which would be expected to be impacted by runoff of nutrient-rich water, which could in turn fuel growth of microorganisms, creating an incentive for adaptive evolution of the innate immune system. High levels of nitrogen and phosphate as well as bacteria have been measured during summer in Old Tampa Bay, Florida, where the B. floridae specimen for genome sequencing was obtained, and indeed a bacterium causing the “red disease”—most likely a Rhodobacter (Bartlett D, personal communication)—is endogenous in amphioxus populations and will kill them if they are stressed (Bert 1867; Arey 1915; Ravitch-Stcherbo 1936; Silva et al. 1995). Given that bacterial populations can evolve rapidly as environmental conditions change (Boto and Martínez 2011), a relatively fast-evolving innate immune system would be advantageous for amphioxus.
Supplementary Material
Supplementary figures S1–S4 and tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Dr Tereza Manousaki for providing the script for conducting Tajima’s relative rate test. We thank the three anonymous reviewers for their valuable comments that helped us to improve the manuscript significantly. This study used the Shared University Grid at Rice funded by NSF grant EIA-0216467, and a partnership among Rice University, Sun Microsystems, and Sigma Solutions, Inc. and the Extreme Science and Engineering Discovery Environment (XSEDE), supported by the National Science Foundation OCI-1053575 and computing resources of the National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. This work was supported by an University of California, San Diego academic senate grant (MBR099S to L.Z.H.) and the National Science Council Taiwan grants (NSC101-2923-B-001-004-MY2 and NSC102-2311-B-001-011-MY3 to J.K.Y.) and a Career Development Award from Academia Sinica Taiwan (AS-98-CDA-L06 to J.K.Y.). We thank N.D. Holland for critically reading the manuscript and collecting Asymmetron lucayanum adults.
Literature Cited
- Arey LB. The orientation of amphioxus during locomotion. J Exp Zool. 1915;19:37–44. [Google Scholar]
- Ashkenazy Y, et al. Dynamics of a snowball earth ocean. Nature. 2013;495:90–93. doi: 10.1038/nature11894. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Benton MJ, Donoghue PC, Asher RJ. Calibrating and constraining molecular clocks. In: Hedges SB, Kumar S, editors. The timetree of life. Oxford (United Kingdom): Oxford University Press; 2009. pp. 35–86. [Google Scholar]
- Berná L, Alvarez-Valin F, D’Onofrio G. How fast is the sessile ciona? Comp Funct Genomics. 2009:875901. doi: 10.1155/2009/875901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bert MP. Sur l'amphioxus. Comptes Rend Acad Sci Paris. 1867;65:364–367. [Google Scholar]
- Blair JE. Animals (Metazoa) In: Hedges SB, Kumar S, editors. The timetree of life. Oxford (United Kingdom): Oxford University Press; 2009. pp. 223–230. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell V, et al. Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron. 2012;76:338–352. doi: 10.1016/j.neuron.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boto L, Martínez JL. Ecological and temporal constraints in the evolution of bacterial genomes. Genes. 2011;2:804–828. doi: 10.3390/genes2040804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourlat SJ, et al. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- Bousquet J, Strauss SH, Doerksen AH, Price RA. Extensive variation in evolutionary rate of rbcL gene sequences among seed plants. Proc Natl Acad Sci U S A. 1992;89:7844–7848. doi: 10.1073/pnas.89.16.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brain CK, et al. The first animals: ca. 760-million-year-old sponge-like fossils from Namibia. S Afr J Sci. 2012;108:83–90. [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chen Y, Cheung SG, Kong RYC, Shin PKS. Morphological and molecular comparisons of dominant amphioxus populations in the China Seas. Mar Biol. 2007;153:189–198. [Google Scholar]
- Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Dai Z, et al. Characterization of microRNAs in cephalochordates reveals a correlation between microRNA repertoire homology and morphological similarity in chordate evolution. Evol Dev. 2009;11:41–49. doi: 10.1111/j.1525-142X.2008.00301.x. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Tsagkogeorga G, Lartillot N, Philippe H. Additional molecular support for the new chordate phylogeny. Genesis. 2008;46:592–604. doi: 10.1002/dvg.20450. [DOI] [PubMed] [Google Scholar]
- Dishaw LJ, Haire RN, Litman GW. The amphioxus genome provides unique insight into the evolution of immunity. Brief Funct Genomics. 2012;11:167–176. doi: 10.1093/bfgp/els007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzery EJ, Snell EA, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci U S A. 2004;101:15386–15391. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing T, Cormican P, O'Farrelly C, Bradley DG, Lloyd AT. Evidence of the adaptive evolution of immune genes in chicken. BMC Res Notes. 2009;2:254. doi: 10.1186/1756-0500-2-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgecombe GD, et al. Higher-level metazoan relationships: recent progress and remaining questions. Org Divers Evol. 2011;11:151–172. [Google Scholar]
- Fedonkin MA, Vickers-Rich P, Swalla BJ, Trusler P, Hall M. A new metazoan from the Vendian of the White Sea, Russia, with possible affinities to the ascidians. Paleontol J. 2012;46:1–11. [Google Scholar]
- Fedonkin MA, Waggoner BM. The late Precambrian fossil Kimberella is a mollusc-like bilaterian organism. Nature. 1997;388:868–871. [Google Scholar]
- Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. J R Stat Soc. 1922;1:87–94. [Google Scholar]
- Geifman N, Monsonego A, Rubin E. The Neural/Immune Gene Ontology: clipping the Gene Ontology for neurological and immunological systems. BMC Bioinformatics. 2010;11:458. doi: 10.1186/1471-2105-11-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German TN, Podkovyrov VN. Records of a new spongelike group in the Riphean biota. Paleontol J. 2012;46:219–227. [Google Scholar]
- Glaessner MF. New fossils from the base of the Cambrian in South Australia. Trans R Soc S Austr. 1958;81:185–188. [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Haeckel E. New York: Fowler; 1876. The evolution of man [English translation of third edition of Anthropogenie] [Google Scholar]
- Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010;38:e131. doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman PF, Schrag DP. The snowball Earth hypothesis: testing the limits of global change. Terra Nova. 2002;14:129–155. [Google Scholar]
- Holland LZ. Chordate roots of the vertebrate nervous system: expanding the molecular toolkit. Nat Rev Nerurosci. 2009;10:736–746. doi: 10.1038/nrn2703. [DOI] [PubMed] [Google Scholar]
- Holland LZ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland ND. Spawning periodicity of the lancelet, Asymmetron lucayanum (Cephalochordata), in Bimini, Bahamas. Ital J Zool. 2011;78:478–486. [Google Scholar]
- Holland ND, Holland LZ. Laboratory spawning and development of the Bahama lancelet, Asymmetron lucayanum (Cephalochordata): fertilization through feeding larvae. Biol Bull. 2010;219:132–141. doi: 10.1086/BBLv219n2p132. [DOI] [PubMed] [Google Scholar]
- Holland ND, Panganiban G, Henyey EL, Holland LZ. Sequence and developmental expression of AmphiDll, an amphioxus Distal-less gene transcribed in the ectoderm, epidermis and nervous system: insights into evolution of craniate forebrain and neural crest. Development. 1996;122:2911–2920. doi: 10.1242/dev.122.9.2911. [DOI] [PubMed] [Google Scholar]
- Holland PWH, Holland LZ, Williams NA, Holland ND. An amphioxus homeobox gene: sequence conservation, spatial expression during development and insights into vertebrate evolution. Development. 1992;116:653–661. doi: 10.1242/dev.116.3.653. [DOI] [PubMed] [Google Scholar]
- Huang S, et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008;18:1112–1126. doi: 10.1101/gr.069674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, et al. HaploMerger: reconstructing allelic relationships for polymorphic diploid genome assemblies. Genome Res. 2012;22:1581–1588. doi: 10.1101/gr.133652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Smith NG. Do essential genes evolve slowly? Curr Biol. 1999;9:747–750. doi: 10.1016/s0960-9822(99)80334-0. [DOI] [PubMed] [Google Scholar]
- Inoue J, Donoghue PCJ, Yang Z. The impact of the representation of fossil calibrations on Bayesian estimation of species divergence times. Syst Biol. 2010;59:74–89. doi: 10.1093/sysbio/syp078. [DOI] [PubMed] [Google Scholar]
- Jantzen SG, Sutherland BJ, Minkley DR, Koop BF. GO trimming: systematically reducing redundancy in large Gene Ontology datasets. BMC Res Notes. 2011;4:267. doi: 10.1186/1756-0500-4-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Junier T, Zdobnov EM. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics. 2010;26:1669–1670. doi: 10.1093/bioinformatics/btq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd T, et al. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell. 1998;92:205–215. doi: 10.1016/s0092-8674(00)80915-0. [DOI] [PubMed] [Google Scholar]
- Kocot KM, Citarella MR, Moroz LL, Halanych KM. PhyloTreePruner: A phylogenetic tree-based approach for selection of orthologous sequences for phylogenomics. Evol Bioinform Online. 2013;9:429–435. doi: 10.4137/EBO.S12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Nohara M, Nishida M, Sterrer W, Nishikawa T. Hidden ancient diversification in the circumtropical lancelet Asymmetron lucayanum complex. Mar Biol. 2006;149:875–883. [Google Scholar]
- Kon T, et al. Phylogenetic position of a whale-fall lancelet (Cephalochordata) inferred from whole mitochondrial genome sequences. BMC Evol Biol. 2007;7:1–12. doi: 10.1186/1471-2148-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankester ER. On some new points in the structure of amphioxus, and their bearing on the morphology of vertebrata. Q J Microsc Sci. 1875;15:257–267. [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Lechner M, et al. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhong J, Wang Y. Genetic diversity and population structure of two lancelets along the coast of China. Zool Sci. 2013;30:83–91. doi: 10.2108/zsj.30.83. [DOI] [PubMed] [Google Scholar]
- MacBride EW. The early development of amphioxus. Q J Microsc Sci. 1898;40:589–612. [Google Scholar]
- Mallatt J, Chen JY. Fossil sister group of craniates: predicted and found. J Morphol. 2003;258:1–31. doi: 10.1002/jmor.10081. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- Martin AP, Palumbi SR. Body size, metabolic rate, generation time, and the molecular clock. Proc Natl Acad Sci U S A. 1993;90:4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart SJ, Obbard DJ, Conlon C, Little TJ. Immune genes undergo more adaptive evolution than non-immune system genes in Daphnia pulex. BMC Evol Biol. 2012;12:63. doi: 10.1186/1471-2148-12-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Amphioxus and lamprey AP-2 genes: implications for neural crest evolution and migration patterns. Development. 2002;129:4953–4962. doi: 10.1242/dev.129.21.4953. [DOI] [PubMed] [Google Scholar]
- Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa T. A new deep-water lancelet (Cephalochordata) from off Cape Nomamisaki, SW Japan, with a proposal of the revised system recovering the genus Asymmetron. Zool Sci. 2004;21:1131–1136. doi: 10.2108/zsj.21.1131. [DOI] [PubMed] [Google Scholar]
- Nohara M, Nishida H, Miya M, Nishikawa T. Evolution of the mitochondrial genome in Cephalochordata as inferred from complete nucleotide sequences from two Epigonichthys species. J Mol Evol. 2005;60:526–537. doi: 10.1007/s00239-004-0238-x. [DOI] [PubMed] [Google Scholar]
- Nohara M, Nishida M, Manthacitra V, Nishikawa T. Ancient phylogenetic separation between Pacific and Atlantic cephalochordates as revealed by mitochondrial genome analysis. Zool Sci. 2004;21:203–210. doi: 10.2108/zsj.21.203. [DOI] [PubMed] [Google Scholar]
- Ohta T. Population size and rate of evolution. J Mol Evol. 1972;1:305–314. [PubMed] [Google Scholar]
- Peterson KJ, Cotton JA, Gehling JG, Pisani D. The Ediacaran emergence of bilaterians: congruence between the genetic and the geological fossil records. Philos Trans R Soc Lond B Biol Sci. 2008;363:1435–1443. doi: 10.1098/rstb.2007.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss SG, Boschung HT. Lancelets (Cephalochordata: Branchiostomatidae): how many species are valid? Israel J Zool. 1996;42:S13–S66. [Google Scholar]
- Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Rannala B, Yang Z. Inferring speciation times under an episodic molecular clock. Syst Biol. 2007;56:453–466. doi: 10.1080/10635150701420643. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM, Brown MA. Innate immunity in the central nervous system. J Clin Invest. 2012;122:1164–1171. doi: 10.1172/JCI58644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravitch-Stcherbo J. De l'origine bactérienne du pigment rouge de l'Amphioxus lanceolatum, cause de sa mort et destruction. Trudy Sevastopol Biol Stan Akad Nauk SSSR. 1936;5:287–296. [Google Scholar]
- Reis M, Yang Z. Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol Biol Evol. 2011;28:2161–2172. doi: 10.1093/molbev/msr045. [DOI] [PubMed] [Google Scholar]
- Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman DH. Atmospheric carbon dioxide levels for the last 500 million years. Proc Natl Acad Sci U S A. 2002;99:4167–4171. doi: 10.1073/pnas.022055499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke TA, Begun DJ. Natural selection drives Drosophila immune system evolution. Genetics. 2003;164:1471–1480. doi: 10.1093/genetics/164.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One. 2011a;6:e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011b;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HC, et al. Miniature genome in the marine chordate Oikopleura dioica. Science. 2001;294:2506. doi: 10.1126/science.294.5551.2506. [DOI] [PubMed] [Google Scholar]
- Shimeld SM. The evolution of the hedgehog gene family in chordates: insights from amphioxus hedgehog. Dev Genes Evol. 1999;209:40–47. doi: 10.1007/s004270050225. [DOI] [PubMed] [Google Scholar]
- Silva JRMC, Mendes EG, Mariano M. Wound repair in the amphioxus (Branchiostoma platae), an animal deprived of inflammatory phagocytes. J Invertebr Pathol. 1995;65:147–151. doi: 10.1006/jipa.1995.1022. [DOI] [PubMed] [Google Scholar]
- Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling EA, Robinson JM, Pisani D, Peterson KJ. Where's the glass? Biomarkers, molecular clocks, and microRNAs suggest a 200-Myr missing Precambrian fossil record of siliceous sponge spicules. Geobiology. 2010;8:24–36. doi: 10.1111/j.1472-4669.2009.00225.x. [DOI] [PubMed] [Google Scholar]
- Sternberg EM. Neural regulation of innate immunity: a coordinated nonspecific host response to pathogens. Nat Rev Immunol. 2006;6:318–328. doi: 10.1038/nri1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MD, Holland ND. Reproduction of the Florida lancelet (Branchiostoma floridae): spawning patterns and fluctuations in gonad indexes and nutritional reserves. Invert Biol. 1996;115:349–359. [Google Scholar]
- Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Satoh N. Details of the evolutionary history from invertebrates to vertebrates, as deduced from the sequences of 18S rDNA. Proc Nat Acad Sci U S A. 1994;91:1801–1804. doi: 10.1073/pnas.91.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu F, Wang L, Huang S, Yu J. Nonsynonymous substitution rate (Ka) is a relatively consistent parameter for defining fast-evolving and slow-evolving protein-coding genes. Biol Direct. 2011;6:13. doi: 10.1186/1745-6150-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A. Comparative genomics in ecological physiology: toward a more nuanced understanding of acclimation and adaptation. J Exp Biol. 2012;215:884–891. doi: 10.1242/jeb.058735. [DOI] [PubMed] [Google Scholar]
- Wraith DC, Nicholson LB. The adaptive immune system in diseases of the central nervous system. J Clin Invest. 2012;122:1172–1179. doi: 10.1172/JCI58648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, et al. The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature. 2001;410:948–952. doi: 10.1038/35073616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YS, Zhang Y, Gao TX, Yabe M, Sakurai Y. Phylogenetic relationships of the lancelets of the genus Branchiostoma in China inferred from mitochondrial genome analysis. Afr J Biotechnol. 2008;7:3845–3852. [Google Scholar]
- Yang R, et al. Genome-wide analyses of amphioxus microRNAs reveal an immune regulation via miR-92d targeting C3. J Immunol. 2013;190:1491–1500. doi: 10.4049/jimmunol.1200801. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol Biol Evol. 2006;23:212–226. doi: 10.1093/molbev/msj024. [DOI] [PubMed] [Google Scholar]
- Yu JK, et al. Axial patterning in cephalochordates and the evolution of the organizer. Nature. 2007;445:613–617. doi: 10.1038/nature05472. [DOI] [PubMed] [Google Scholar]
- Yu Y, et al. A novel C1q family member of amphioxus was revealed to have a partial function of vertebrate C1q molecule. J Immunol. 2008;181:7024–7032. doi: 10.4049/jimmunol.181.10.7024. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, et al. Novel genes dramatically alter regulatory network topology in amphioxus. Genome Biol. 2008;9(8):R123. doi: 10.1186/gb-2008-9-8-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Zhu Q. Taxonomic and genetic status of lancelet in Weihai coastal waters based on mitochondrial DNA sequence. Chin J Oceanol Limnol. 2011;29:623–632. [Google Scholar]
- Ziv Y, et al. Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat Neurosci. 2006;9:268–275. doi: 10.1038/nn1629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.