Abstract
The primitive red alga Cyanidioschyzon merolae inhabits acidic hot springs and shows robust resistance to heat shock treatments up to 63 °C. Microarray analysis was performed to identify the key genes underlying the high temperature tolerance of this organism. Among the upregulated genes that were identified, we focused on two small heat shock proteins (sHSPs) that belong to a unique class of HSP families. These two genes are located side by side in an inverted repeat orientation on the same chromosome and share a promoter. These two genes were simultaneously and rapidly upregulated in response to heat shock treatment (>1,000-fold more than the control). Interestingly, upregulation appeared to be triggered not by a difference in temperatures, but rather by the absolute temperature. Similar sHSP structural genes have been reported in the green alga Chlamydomonas reinhardtii, but the threshold temperature for the expression of these sHSP-encoding genes in Ch. reinhardtii was different from the threshold temperature for the expression of the sHSP genes from Cy. merolae. These results indicate the possible importance of an absolute temperature sensing system in the evolution and tolerance of high-temperature conditions among unicellular microalgae.
Keywords: unicellular algae, heat resistance, HSP, microarray, environmental adaptation
Introduction
High temperature is a major environmental stress factor that can cause life-threatening distress in organisms, including the denaturation of proteins and nucleic acids, the dissociation of protein complexes, and the destabilization of membrane structure. Despite this, many organisms thrive in extremely high temperature environments. Most studies have focused on the prokaryotes that inhabit extreme environments, often providing practical benefits, such as thermostable DNA polymerases (Innis et al. 1988). However, some eukaryotes can also exist in extremely hot environments.
The red algae in Cyanidiaceae inhabit sulfate-rich hot springs (pH 0–3.0, 30–60 °C) throughout the world, and are considered to be the most heat-tolerant photosynthetic eukaryotes (Ciniglia et al. 2004; Yoon et al. 2006). Cyanidioschyzon merolae, a Cyanidiaceae, is a eukaryote with a very simple cell architecture, namely a single nucleus, a single mitochondrion, and a single chloroplast (Merola et al. 1981; Kuroiwa et al. 1994). The Cy. merolae genome was the first alga genome to be sequenced (Matsuzaki et al. 2004), and one of the first eukaryotic genomes to be 100% sequenced (Nozaki et al. 2007). Furthermore, microarray techniques have revealed the organism’s transcription profiles under varied conditions (Fujiwara et al. 2009). In this study, we investigated the heat-resistance strategy of Cy. merolae by microarray analysis using cells with and without transient heat shock treatments. We focused especially on two small heat shock proteins (sHSPs) that belong to a unique class of HSPs, among many other candidates. These sHSPs were associated with gene regulatory systems that could sense species-specific absolute temperatures, and may be linked to fitness in extreme thermal environments.
Materials and Methods
Growth Conditions
Cyanidioschyzon merolae strain 10D was cultured in 2 × Allen’s medium (Allen 1959) at pH 2.3 under continuous light (40 W/m2) at 42 °C or at room temperature (28 °C). The green alga Ch. reinhardtii strain CC125 (in family Chlamydomonadaceae) was cultured in tris-acetate-phosphate (TAP) medium (Harris 1989) under continuous light (40 W/m2).
Heat Shock Treatment
To test the survival temperature limits of Cy. merolae and Ch. reinhardtii, 200 µl of cell cultures were placed in PCR (polymerase chain reaction) tubes and heated using a thermal cycler (Bio-Rad Laboratories, CA). Cell cultures were put in glass test tubes (φ 25 mm × 1.3 mm) and incubated in a constant temperature bath for 20 min in preparation for RNA extraction. The cell cultures were kept in the dark during the heat shock treatments. It took less than 5 min to heat samples up to the temperature of the bath using this method. Color tone changes were analyzed by quantifying the amount of chlorophyll a and phycocyanin, as described previously (Arnon et al. 1974).
RNA Extraction
Cells were collected by centrifugation at 6,000 × g for 2 min. Pellets were lysed with nucleic acid extraction buffer (300 mM NaCl, 2% Tris–HCl [pH 7.5], 100 mM ethylenediaminetetraacetic acid, 4% SDS) and vortexed well. Two-fold volumes of phenol–chloroform mixture were added to the samples and centrifuged at 15,000 × g for 10 min. Supernatants were removed to new tubes and the same volume of 2-propanol was added and centrifuged at 15,000 × g for 20 min. Pellets were rinsed with 70% ethyl alcohol and lysed with 180 µl of nuclease-free water. DNA was digested using DNase I (Takara Bio, Shiga, Japan). After DNA degradation, the same volume of phenol–chloroform mixture was added to the samples and centrifuged at 15,000 × g for 10 min. Supernatants were again removed to new tubes and the same volume of 2-propanol was added and centrifuged at 15,000 × g for 20 min to pellet DNA. Pellets were rinsed with 70% ethyl alcohol and lysed with 50 µl of nuclease-free water.
Microarray Analysis
All steps in the microarray analysis were performed essentially as described previously (Fujiwara et al. 2009). Five micrograms of total RNA samples were reverse transcribed in 20 µl reaction mixes containing 50 ng/μl of oligo (dT) primer, 2 U/μl of RNase inhibitor, 0.5 mM dNTP (deoxynucleotide triphosphate) mixture, and 1 µl of reverse transcriptase. Amino-allyl amplified RNA (aRNA) was synthesized using an Amino Allyl MessageAmp II aRNA Kit (Ambion, TX), according to the manufacturer’s instructions.
Cy3-conjugated aRNA in hybridization solution (5 × SSC [saline sodium citrate], 0.5% SDS, 4 × Denhalt’s solution, 10% formamide, 100 ng/ml salmon sperm DNA) was hybridized to spotted microarray slides and covered with a cover glass (Matsunami Glass Ind., Ltd., Osaka, Japan) in a slide hybridization chamber (Sigma-Aldrich, MO) for 18 h at 45 °C. Hybridized slides were washed in 1 × SSC/0.03% SDS for 6 min at 45 °C, followed by 0.2 × SSC for 5 min and 0.05 × SSC for 4 min, and then spin-dried before scanning. Microarray slides were scanned using a FLA-8000 scanner (Fujifilm Corp., Tokyo, Japan) at a wavelength of 532 and 635 nm at 5-mm resolution. Microarray image gene spot signal strength was measured using the microarray analyzing software ArrayGauge version 2 (Fujifilm Corp.). Every gene was spotted at two locations on the microarray slides to confirm reproducibility. Genes with signal strength ratios between both spots from 0.5 to 2.0 were extracted and included in the data.
RNA Gel Blot Analysis
Fifteen micrograms of total RNA was electrophoresed on a 1.2% agarose gel and blotted onto Biodyne Nylon Transfer Membranes (Pall Corp., NY). RNA was cross-linked using ultraviolet radiation (1,200 × 100 μJ/cm2; Spectrolinker XL-1500, Spectronics Corp., NY). Ten micrograms of DNA were labeled using Amersham Gene Images AlkPhos Direct Labeling and Detection System (GE Healthcare, Little Chalfont, United Kingdom) and added to hybridization buffer for 20 h at 55 °C. Chemiluminescence was detected for 2,000 s with a lumino-image analyzer LAS3000 (Fujifilm Corp.) after washing the membranes, and analyzed with Multi Gauge version 3.0 software (Fujifilm Corp.). As a loading control, the gels were also stained by ethidium bromide.
Quantitative Real-Time PCR
Quantitative real-time PCR (qRT-PCR) was performed using the FastStart SYBR Green Master (ROX, Roche Diagnostics, Basel, Switzerland) and a Mx3000P system, following the manufacturer’s instructions. The quantitative estimations were calculated with MxPro software using the ΔΔCt (cycle threshold) method (Stratagene, Agilent Technologies, CA).
Antibody Preparation
Full-length HSP1 (CmsHSP1) and HSP22F (CreHSP22F) genes were amplified and cloned in pQE-80 L (Qiagen, Venlo, The Netherlands). The vectors were transformed into Escherichia coli BL21 and selected on Luria–Bertani (LB) agar medium containing 50 µg/ml Carbenicillin (Nacalai Tesque, Kyoto, Japan). The cultures were grown in LB medium at 37 °C for several hours and isopropyl β-D-1-thiogalactopyranoside (IPTG) was added at OD 600 0.7–1 to a final concentration of 1 mM. Proteins were purified using Ni-NTA agarose (Qiagen), following the manufacturer’s instructions. The isolated CmsHSP1 and CreHSP22F fusion proteins were injected into mice.
Immunoblot
Proteins were separated by 12.5% (w/v) SDS-PAGE and electrotransferred onto polyvinylidene fluoride membranes. A 1:5,000 antibody dilution was added, and the protein-antibody complexes were labeled using an ECL Plus Western blot detection kit (GE Healthcare). Chemiluminescence was detected with a lumino-image analyzer LAS3000 (Fujifilm Corp.) and analyzed using Multi Gauge version 3.0 software (Fujifilm Corp.). As a loading control, the gels were also stained by Coomassie Brilliant Blue.
Phylogenic Analysis
Small HSP homologs were collected using BLAST searches against public databases. The sequences were aligned using ClustalW in MEGA 5.0 (Tamura et al. 2011). Eighty-one amino acid residues corresponding to the α-crystallin (αC) domain that is conserved in sHSPs were used for the phylogenetic analyses. Bayesian inference was performed using MrBayes version 3.2 (Ronquist et al. 2012). Eleven million generations were completed, and trees were collected every 5,000 generations, after discarding trees corresponding to the first 25% (burn-in), to generate a consensus phylogenetic tree. Bayesian posterior probabilities were estimated as the proportion of trees sampled after burn-in.
Results
Cyanidioschyzon merolae Survive a Heat Shock of 63 °C for 20 min
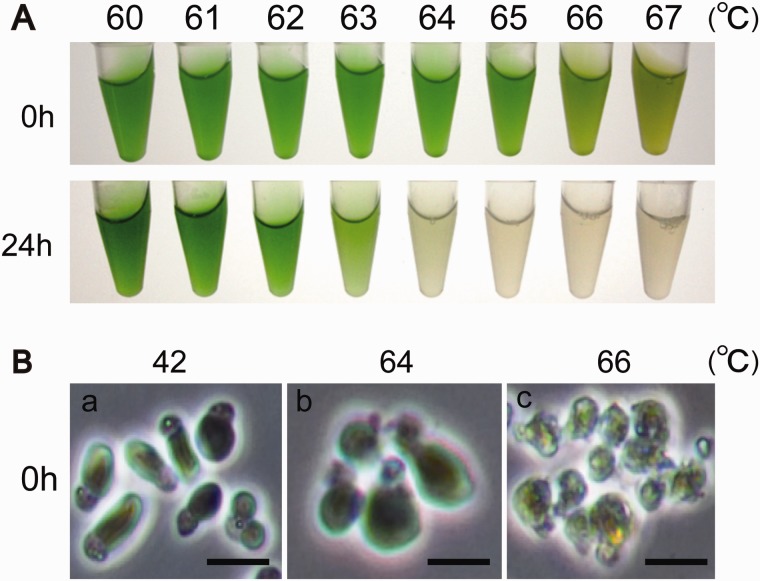
The optimal growth temperature for Cy. merolae in a laboratory is 42 °C. We first investigated Cy. merolae’s upper temperature heat shock limit of survival. Cell cultures were divided and treated with high temperatures (60–67 °C) for 20 min. Cells were discolored immediately after the heat shock treatment when exposed to temperatures higher than 66 °C (fig. 1A). Photosynthetic pigments were quantified colorimetrically. Chlorophyll a content began to drop above 66 °C, while the content of phycocyanin declined more gradually beginning at temperatures above 50 °C (supplementary fig. S1, Supplementary Material online). Intactness of cells was studied by phase-contrast microscopy. We found that heat shock treatments at or above 66 °C caused severe cell structural damage, including a loss of membrane integrity and cell shrinkage (fig. 1B). After the heat shock treatment, cells were incubated at 42 °C to see whether they could recover from the damage. One day after heat shock treatment, cell cultures exposed to temperatures higher than 64 °C bleached out and could not recover, whereas cells treated with temperatures lower than 63 °C recovered successfully and stayed green (fig. 1A). These results suggest that Cy. merolae could survive a transient heat elevation up to 63 °C, which is ∼20 °C higher than its optimal growth temperature.
Fig. 1.—
Effect of heat shock for 20 min on the red alga Cyanidioschyzon merolae. (A) Macroscopic appearance of cultures incubated for 0 or 24 h at 42 °C and exposed to a heat shock for 20 min. (B) Microscopic appearance of the cells exposed to a heat shock for 20 min. The cells at 66 °C for 20 min were corrupted. Bar is 1 µm.
Heat Stress Induced Two Small HSPs
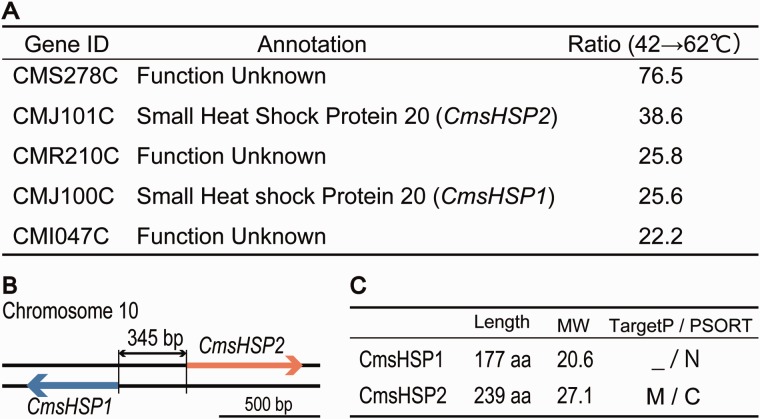
We compared the gene expression profiles of cells cultured at 42 °C and cells transferred to 62 °C, using a microarray system that covered >96% of the total predicted gene models (4,586 genes out of 4,775 predicted genes) (Matsuzaki et al. 2004; Fujiwara et al. 2009), to characterize the molecular basis for the Cy. merolae heat response. Cells cultured at 28 °C and transferred to 50 °C were analyzed as a reference. We found 57 genes with messenger RNA (mRNA) accumulation levels that were elevated more than 3-fold in cells that were transferred from 42 to 62 °C, and 44 genes that were upregulated in cells that were transferred from 28 to 50 °C (supplementary table S1, Supplementary Material online). Most of these upregulated genes had unknown functions, and may be specific to C. melorae. However, among them, we identified two genes (CMJ100C and CMJ101C: accession numbers AB979124 and AB979125) that were dramatically upregulated after the heat elevation (fig. 2A and supplementary table S2, Supplementary Material online). These two genes were homologous to two sHSPs identified in the Cy. merolae Genome Project (http://merolae.biol.s.u-tokyo.ac.jp/, last accessed October 10, 2014).
Fig. 2.—
Transcriptome analysis of Cyanidioschyzon merolae cells exposed to heat shock for 20 min at 62 °C. (A) The representative genes that were upregulated by the heat shock. Ratio indicates the increase in signal strength ratios between the spots in the microarray analysis. (B) Schematic representation of CMJ100C and CMJ101C on chromosome 10. (C) Nomenclature and molecular properties of the small heat shock protein CmsHSP1 and CmsHSP2. Length is peptide length (aa, amino acids); MW is molecular weight; and TargetP/PSORT indicates the localizations predicted by the corresponding programs where M is mitochondria, C is chloroplast, and N is nucleus.
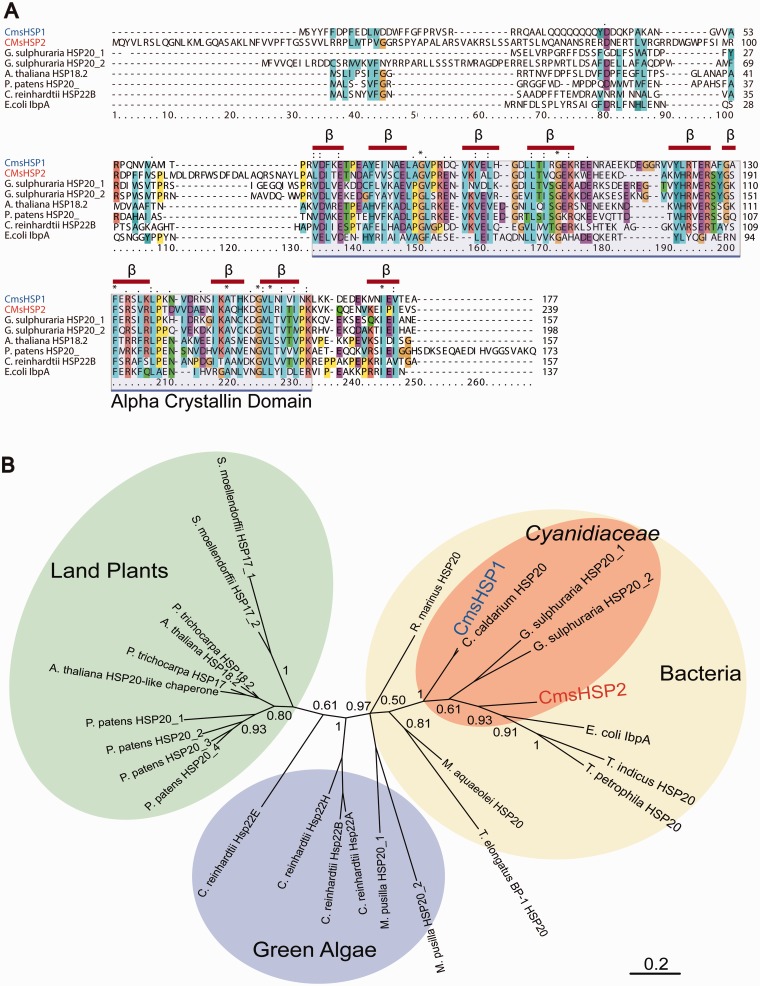
The amino acid sequences encoded by CMJ100C (deduced molecular weight: 20.6 kDa) and CMJ101C (27.1 kDa) were aligned to homologous sHSPs from other species (fig. 3A). This alignment showed that the C-termini of the CMJ100C and CMJ101C protein products possessed a well-conserved αC domain, composed of eight to nine regions likely to form β sheet structures, whereas the N-termini were less conserved. It has been assumed that the N-termini of sHSPs play important roles in substrate recognition (Ahrman et al. 2007; Jaya et al. 2009; McHaourab et al. 2009). These are defining features of sHSP/αC proteins; therefore, we tentatively designated CMJ100C and CMJ101C as CmsHSP1 and CmsHSP2, respectively.
Fig. 3.—
Multiple sequence alignment and phylogenetic tree of CmsHSP1 and CmsHSP2 and other sHSPs. (A) Multiple sequence alignment of CmsHSP1 and CmsHSP2 protein sequences with their closest homologs Red bars indicate regions predicted to form beta-sheet structures. (B) Unrooted phylogenetic tree of sHSPs based on Bayesian inference methods.
A phylogenetic analysis was performed using the amino acid sequences of sHSPs from bacteria, red and green algae, and land plants. The results showed that the sHSPs from the Cyanidiaceae (Cy. merolae, Cyanidium caldarium, and Galdieria sulphuraria) were more closely related to bacterial genes than to the sHSPs from the other algae or land plants (fig. 3B).
The sHSPs can act as adenosine triphosphate (ATP)-independent molecular chaperones. They bind denaturing proteins and thereby protect cells from damage caused by the formation of irreversible protein aggregates. Plants encode sHSPs that are targeted to specific cellular compartments (Scharf et al. 2001). Organelle-targeted sHSPs are unique to plants, the only exception being a mitochondrion-targeted sHSP in Drosophila melanogaster (Wadhwa et al. 2010; Basha et al. 2012). The proteins encoded by CmsHSP1 and CmsHSP2 were predicted by TargetP and PSORT (Nakai and Horton 1999; Emanuelsson et al. 2000) to be targeted to different intracellular compartments: CmsHSP1 to the cytosol or nucleus, and CmsHSP2 to chloroplast or mitochondria (fig. 2C). Consistent with this prediction, the amino acid sequence alignment showed that CmsHSP2 possessed an ∼30 amino acid extension at the N-terminus that was absent in other sHSPs that may function as a transit peptide (fig. 3A).
sHSP Genes Are Regulated by a Survival Threshold Temperature
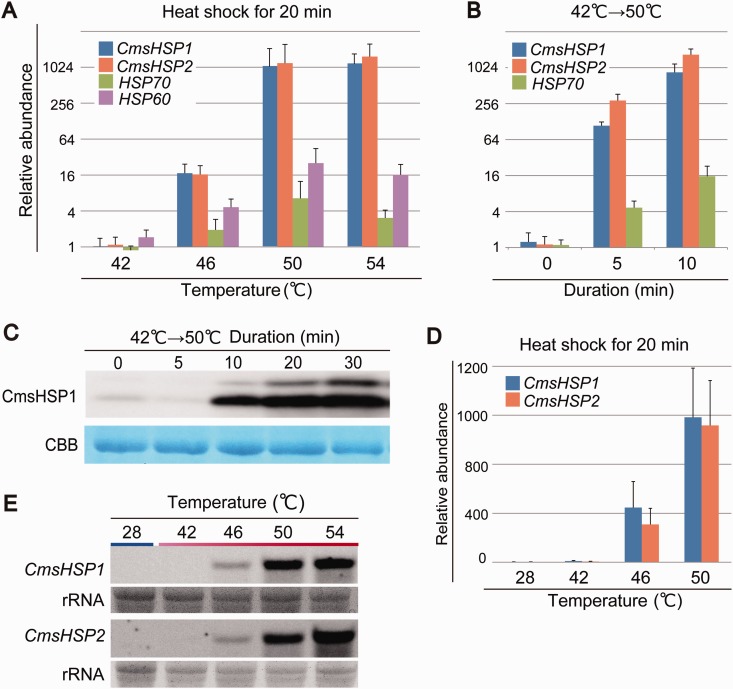
The CmsHSP1 and CmsHSP2 proteins are encoded by genes on chromosome 10 that are side by side in an inverted repeat orientation, which was likely a common promoter for the two genes (fig. 2B). We performed a qRT-PCR analysis to compare the expression profiles of these two genes with two other HSP genes, HSP70 and HSP60, that were predicted to encode proteins that localized to cytosol, and to mitochondria and chloroplast by TargetP and PSORT (Nakai and Horton 1999; Emanuelsson et al. 2000). This analysis showed that the expressions of both the CmsHSP1 and CmsHSP2 genes were enhanced by ∼1,000-fold on exposure to 50 °C, while HSP70 and HSP60 were more moderately upregulated (∼16-fold) (fig. 4A). In the microarray analysis, other HSP-encoding genes such as HSP60, HSP70, and HSP100 were enhanced by only 1.17- to 1.5-fold (supplementary table S1, Supplementary Material online). It has been reported previously that these three HSP genes were highly expressed under normal growth conditions (42 °C, pH 2.3) (Matsuzaki et al. 2004). For HSP60, HSP70, and HSP100, the expression levels were already high and the microarray signals were almost saturated even under the normal growth conditions (42 °C, pH 2.3). Therefore, further elevation was difficult to detect using microarray.
Fig. 4.—
Expression patterns of CmsHSP1 and CmsHSP2. (A) Expression profiles of CmsHSP1, CmsHSP2, HSP60, and HSP70 in cells cultured at 42 °C. The cDNAs were prepared from cells cultured at 42 °C and exposed to a heat shock at 46, 50, or 54 °C for 20 min. Relative accumulation levels were normalized to the levels for the cells at 42 °C. (B) Changes in the mRNA levels of CmsHSP1, CmsHSP2, and HSP70 during heat shock treatment obtained by qRT–PCR. The cDNAs were prepared from cells cultured at 42 °C and exposed to heat shock at 50 °C for 5 and 10 min. (C) Western blot showing the accumulation of CmsHSP1 protein from 5 to 30 min. Coomassie Brilliant Blue is a loading control. (D) Expression profiles of CmsHSP1 and CmsHSP2 in cells cultured at 28 °C. The cDNAs were prepared from cells cultured at 28 °C and exposed to heat shock at 42, 46, or 50 °C for 20 min. Relative accumulation levels were normalized to the levels for the cells at 28 °C. (E) Northern blot showing the expression profiles of CmsHSP1 and CmsHSP2. rRNA is a loading control. Total RNA was prepared from cells cultured at 28 °C and exposed to heat shock at 42, 46, 50, or 54 °C for 20 min.
We also monitored the accumulation levels of CmsHSP1 mRNA and CmsHSP1 protein after the temperature shift (42–50 °C). The qRT-PCR results indicated that the CmsHSP1 mRNA levels were elevated already after 5 min of the heat shock (fig. 4B and C), and the accumulation of the CmsHSP1 protein also began within 10 min (fig. 4C), confirming the rapid response of the CmsHSP1 gene triggered by the temperature shift (42–50 °C). We detected an extra band above the protein band being investigated in our immunoblots (fig. 4C). This upper band was considered to be a cross-reaction of the antibody with CmsHSP2.
We then questioned whether the sHSP genes were regulated by the difference in temperatures or by the absolute temperature. To address this question, we compared cells shifted from 28 to 50 °C and from 42 to 62 °C. Microarray analysis indicated the expression level of CmsHSP1 and CmsHSP2 were upregulated drastically under both conditions (supplementary table S1, Supplementary Material online). This response was confirmed by qRT-PCR, RNA gel blot, and immunoblot analyses (fig. 4). When cells cultured at 28 or 42 °C were exposed to increasing temperatures (42, 46, 50, and 54 °C for 20 min), CmsHSP1 and CmsHSP2 were upregulated drastically by a threshold temperature of 46–50 °C in both initial growth conditions (fig. 4A and D). When cells cultured at 28 °C were incubated at 42 °C, neither CmsHSP1 nor CmsHSP2 were induced, despite a temperature difference of 14 °C. These results imply that the two sHSPs were regulated not by the relative difference in temperatures, but rather by the absolute temperature (46–50 °C) (fig. 4A, D, and E).
sHSP Genes in the Green Alga Ch. reinhardtii Are Regulated by a Threshold Temperature
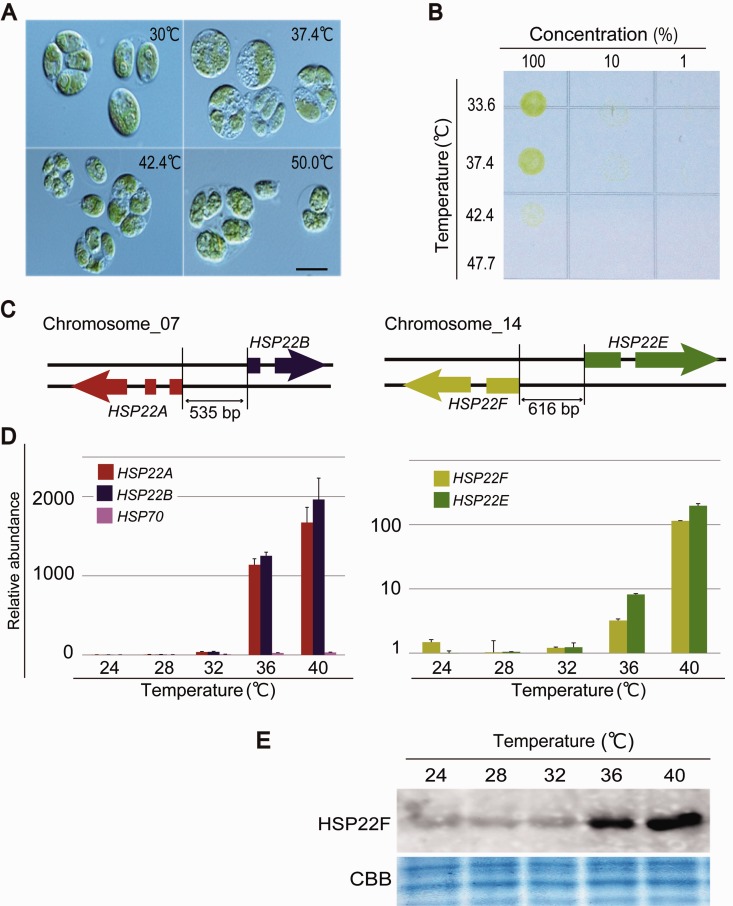
The gene pairs Cre07.g318800 (HSP22A) and Cre07.g318850 (HSP22B), and Cre14.g617400 (HSP22F) and Cre14.g617450 (HSP22E) are homologous to CmsHSP1 and CmsHSP2 and have also been reported to be located side by side in inverted repeat orientations, sharing promoter regions (fig. 5C: Phytozome version 9.1 [http://www.phytozome.net/, last accessed October 10, 2014]; Schroda 2004; Schroda and Vallon 2008). Therefore, we tested whether a threshold temperature might also regulate the expression of these Ch. reinhardtii genes.
Fig. 5.—
Effect of heat shock on the green alga Ch. reinhardtii. (A) Microscopic images of the cells exposed to a heat shock for 20 min. Bar is 5 µm. (B) Survival rate of cells exposed to a heat shock for 20 min. Numbers across the top indicate the relative quantity of cells originally plated on TAP medium. (C) Schematic representation of HSP22F and HSP22F located in tandem on the chromosome. (D) Expression profiles of HSP22A, HSP22B, and HSP70, and of HSP22F and HSP22E. (E) Western blot showing the accumulation of the HSP22F protein in cells exposed to a heat shock for 20 min.
The upper survival temperature limit of Ch. reinhardtii was determined to be 42 °C or higher (fig. 5A and B). Next, cells cultured at 24 °C were transferred to various temperatures (24–40 °C) for 20 min and the expression profiles of the sHSP genes were compared (fig. 5D). The qRT-PCR analysis indicated that the sHSP transcripts were 10- to 1,000-fold upregulated at temperatures higher than 36 °C. This protein level upregulation at 36 °C was also observed in the immunoblot analysis using an antibody against HSP22F (fig. 5E). These results suggest that the threshold temperature for activating sHSP expression in Ch. reinhardtii is ∼36 °C, which is significantly different from our findings for Cy. merolae (46–50 °C).
Discussion
High temperatures create a series of challenges for eukaryotes. One of the early events caused by high temperatures is the simultaneous denaturation and/or destabilization of proteins, lipids, nucleic acids, and other biomolecules in multiple cellular compartments. One possible outcome is impaired electron transport efficiency within mitochondria and chloroplasts related to an elevated production of reactive oxygen species that can result in power and energy production deficiencies (Yamamoto et al. 2008; Foyer and Noctor 2009).
The optimal growth temperature for Cy. merolae in a laboratory is 42 °C. When exposed to temperatures higher than 66 °C, the cells discolored immediately (fig. 1). Photosynthetic pigments were quantified colorimetrically, which confirmed the breakdown of chlorophyll a (supplementary fig. S1, Supplementary Material online). Interestingly, although chlorophyll a content began to drop only above 66 °C, phycocyanin content declined gradually after 50 °C. It is not immediately clear what caused these two different behaviors, but one explanation may be that the stability of chlorophyll a is likely determined by a noncovalent association with photosynthetic proteins and this association may break down at ∼66 °C, leading to the observed rapid degradation of chlorophyll a. However, phycocyanin is covalently associated with phycobiliprotein (Brown et al. 1979; Grantt 1981) and so gradual structural changes in phycobilprotein in response to the heat stress may have influenced the absorption spectrum, making phycocyanin appear more sensitive.
Small HSPs are considered to be “first responders” capable of immediately binding unfolding proteins. Indeed, sHSPs/αC can suppress protein aggregation in an ATP-independent manner (Horwitz 1992). Many sHSP/αC proteins have been shown to bind up to an equal weight of nonnative proteins, keeping these proteins accessible to other ATP-dependent components of the protein quality control network for further processing (Mogk, Deuerling, et al. 2003; Mogk, Schlieker, et al. 2003; McHaourab et al. 2009; Mymrikov et al. 2011).
Furthermore, sHSPs are targeted to various intercellular components including the cytoplasm, nucleus, endoplasmic reticulum (ER), mitochondria, and chloroplasts. In chloroplasts, for example, sHSPs participate in the protection of photosystem II (PSII) (specifically, the H2O-oxidizing, quinone-reducing complex PSII), which is usually the most heat sensitive component of the chloroplast thylakoid membrane protein complex involved in photosynthesis (Heckathorn et al. 1998). In mitochondria, sHSPs protect respiratory complexes (especially complex I, which is the most heat sensitive component of the respiratory chain) from degradation and proteolysis (Lenne and Douce 1994; Downs and Heckathorn 1998).
In this study, we found that CmsHSP1 and CmsHSP2 are located side by side in the chromosome and share a promoter region that probably functions as the common promoter for the two sHSP genes (fig. 2B). CmsHSP1 and CmsHSP2 most likely target different cellular compartments in Cy. merolae: CmsHSP1 to the nucleus or cytoplasm and CmsHSP2 to chloroplasts and/or mitochondria (fig. 2C). This system may be advantageous for achieving the emergent and rapid activation/delivery of sHSPs to multiple cellular compartments, with minimum time and cost.
Expression of CmsHSPs Is Regulated in Response to Absolute Temperatures
Living organisms adapt to various environments. Therefore, it is likely that emergency response systems will be different in different species, depending on the specific environmental niche that each organism occupies.
The green alga Ch. reinhardtii inhabits soil and water and has an optimal growth temperature of ∼22 °C. It is reasonable, therefore, for this alga to activate heat response sHSP genes at ∼36 °C (fig. 5). However, Cy. merolae inhabits acidic hot springs where 36 °C should not be the temperature at which heat response genes are activated. It is crucial, therefore, that Cy. merolae possess a mechanism that can activate the sHSP genes at temperatures of ∼50 °C (fig. 5).
We found that the threshold temperature for sHSP expression in Ch. reinhardtii was 36 °C, while in a previous study, a threshold temperature of 39 °C was reported (Tanaka et al. 2000). One reason for the difference may be that different strains of Ch. reinhardtii were used; that is, in this study, the CC125 strain was used, while in the previous study, the CC2986 strain was used (Tanaka et al. 2000). Ch. reinhardtii was reported to accumulate spontaneous mutations even under laboratory conditions (Morgan et al. 2014), which might have led to their subtly different responses to the heat stress.
Absolute temperature sensing mechanisms are still largely unknown; however, heat shock factors (HSFs) are possible candidates. HSP expression is regulated by HSFs acting as transcription factors. Heat shock transcription factor 1 (HSF1) is the major and most well-studied HSF in eukaryotes. HSF1 is inhibited by associating with HSPs. When cells are stressed by, for example, a rapid rise in temperature, HSPs bind to the denatured proteins and dissociate from HSF1. This frees HSF1, which then forms trimers, which translocate to the cell nucleus and activate transcription (Prahlad and Morimoto 2009). In yeast and Ch. reinhardtii, HSF1s form constitutive trimers and their activation appears to occur exclusively by hyperphosphorylation (Rabindran et al. 1993; Lee et al. 1995; Schulz-Raffelt et al. 2007). Three HSFs are encoded in the Cy. merolae genome (Matsuzaki et al. 2004). It is possible that some of the HSF activation steps, such as HSF–HSP complex formation or HSF trimerization, may be sensitive to the absolute temperature, or that HSF1s also form constitutive trimers in Cy. merolae and that hyperphosphorylation of HSP1 is induced at the survival threshold temperature.
We found that CmsHSP1 and CmsHSP2 were upregulated simultaneously; therefore, we speculate that they may be regulated by a single heat shock element (HSE) and consist of at least three contiguous of the pentameric sequence nGAAn (Amin et al. 1988; Xiao and Lis 1988; Perisic et al. 1989). To identify the HSE, the promoter sequences of CmsHSP1 and CmsHSP2 were investigated using the TFBIND program (Tsunoda and Takagi 1999) to predict TATA boxes and HSEs. In the CmsHSP1 promoter, a TATA box and two putative HSEs were found, while in the CmsHSP2 promoter, no distinct TATA box was found and information about the 5′-untranslated region (UTR) was not available because the expressed sequence tag for CmsHSP2 was not cloned in the Cy. merolae Genome Project, probably because of its low expression level (Matsuzaki et al. 2004). We found putative HSEs in both the forward and reverse directions at the center of the CmsHSP1 and CmsHSP2 promoters (supplementary fig. S2B, Supplementary Material online). This was actually the candidate HSE that was closest to the TATA box of CmsHSP1. We think that this putative HSE sequence may be the HSE that is critical for the absolute temperature-dependent regulation of the two Cy. merolae HSPs (supplementary fig S2A, Supplementary Material online).
Small HSPs are known to evolve at a relatively rapid rate compared with other HSPs such as HSP70 (Basha et al. 2012). Previous phylogenetic analyses have found that algal sHSPs are not members of the land plant sHSP families (Waters and Rioflorido 2007). We performed a phylogenetic analysis of the sHSPs encoded in the Cyanidiaceae: Cy. merolae, Cy. caldarium, and G. sulphuraria. All these red algae can grow at pHs from 0 to 4 and at temperatures of up to ∼60 °C. The phylogenetic analysis revealed that these three sHSPs were more closely related to bacterial sHSPs than to green algal sHSPs (fig. 3B).
Recently, Schonknecht et al. (2013) reported that at least 75 genes may have been horizontally transferred from archaea or bacteria to G. sulphuraria. Some of these genes may be linked to fitness-related traits such as archaeal ATPases, which may contribute to heat tolerance and bacterial monovalent cation/proton antiporters that may confer tolerance to high salinity. Other speculations are that the sHSPs in Cyanidiaceae may have been acquired from archaea or bacteria by horizontal gene transfer, or that primitive forms of sHSPs have been conserved in Cyanidiaceae, despite the relatively fast evolutionary rate of most sHSPs. Red algae are generally considered the most basal lineage within the bikonts sensu (Cavalier-Smith 2003) or the Plantae sensu (Nozaki et al. 2003). Cyanidiaceae inhabit an extreme environment, speculated to be similar to the environment when primitive life emerged. This environment may have prevented the Cyanidiaceae sHSPs from evolving at relatively rapid evolutionary rates.
Supplementary Material
Supplementary figures S1 and S2 and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Dr. Toshiharu Shikanai (Kyoto University), Dr. Sota Fujii (Kyoto University), Akiko Fujita (Nara Institute of Science and Technology), and Dr. Haruko Kuroiwa (Rikkyo University) for their support. This work was supported, in part, by the Ministry of Education, Culture, Sports, Science, and Technology of Japan grants to T.K. and O.M.; Core Research for Evolutional Science and Technology (CREST) of Program of Japan Science and Technology Agency (JST) (to O.M.); Grant-in-Aid for JSPS Fellows (grant 26·786 to Y.K.); and the Funding Program for Next Generation World-Leading Researchers (NEXT Program Grant GS015).
Literature Cited
- Ahrman E, Lambert W, Aquilina JA, Robinson CV, Emanuelsson CS. Chemical cross-linking of the chloroplast localized small heat-shock protein, Hsp21, and the model substrate citrate synthase. Protein Sci. 2007;16:1464–1478. doi: 10.1110/ps.072831607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MB. Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Arch Microbiol. 1959;32:270–277. doi: 10.1007/BF00409348. [DOI] [PubMed] [Google Scholar]
- Amin J, Ananthan J, Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. 1988;8:3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI, McSwain BD, Tsujimoto HY, Wada K. Photochemical activity and components of membrane preparations from blue-green algae. I. Coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochim Biophys Acta. 1974;357:231–245. doi: 10.1016/0005-2728(74)90063-2. [DOI] [PubMed] [Google Scholar]
- Basha E, O’Neill H, Vierling E. Small heat shock proteins and alpha-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AS, Offner GD, Ehrhardt MM, Troxler RF. Phycobilin-apoprotein linkages in the alpha and beta subunits of phycocyanin from the unicellular rhodophyte, Cyanidium caldarium. Amino acid sequences of 35S-labelled chromopeptides. J Biol Chem. 1979;254:7803–7811. [PubMed] [Google Scholar]
- Cavalier-Smith T. The excavate protozoan phyla Metamonada Grasse emend. (Anaeromonadea, Parabasalia, Carpediemonas, Eopharyngia) and Loukozoa emend. (Jakobea, Malawimonas): their evolutionary affinities and new higher taxa. Int J Syst Evol Microbiol. 2003;53:1741–1758. doi: 10.1099/ijs.0.02548-0. [DOI] [PubMed] [Google Scholar]
- Ciniglia C, Yoon HS, Pollio A, Pinto G, Bhattacharya D. Hidden biodiversity of the extremophilic Cyanidiales red algae. Mol Ecol. 2004;13:1827–1838. doi: 10.1111/j.1365-294X.2004.02180.x. [DOI] [PubMed] [Google Scholar]
- Downs CA, Heckathorn SA. The mitochondrial small heat-shock protein protects NADH:ubiquinone oxidoreductase of the electron transport chain during heat stress in plants. FEBS Lett. 1998;430:246–250. doi: 10.1016/s0014-5793(98)00669-3. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid Redox Signal. 2009;11:861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, et al. Periodic gene expression patterns during the highly synchronized cell nucleus and organelle division cycles in the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2009;16:59–72. doi: 10.1093/dnares/dsn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantt E. Phycobilisomes. Annu. Rev. Plant Physiol. 1981;32:327–347. [Google Scholar]
- Harris EH. San Diego (CA): Academic Press; 1989. The Chlamydomonas sourcebook: a comprehensive guide to biology and laboratory use. [DOI] [PubMed] [Google Scholar]
- Heckathorn SA, Downs CA, Sharkey TD, Coleman JS. The small, methionine-rich chloroplast heat-shock protein protects photosystem II electron transport during heat stress. Plant Physiol. 1998;116:439–444. doi: 10.1104/pp.116.1.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci U S A. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis MA, Myambo KB, Gelfand DH, Brow MA. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci U S A. 1988;85:9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc Natl Acad Sci U S A. 2009;106:15604–15609. doi: 10.1073/pnas.0902177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T, et al. Comparison of ultrastructures between the ultrasmall eukaryote Cyanidioschyzon merolae and Cyanidium caldarium. Cytologia. 1994;59:149–158. [Google Scholar]
- Lee JH, Hubel A, Schoffl F. Derepression of the activity of genetically engineered heat shock factor causes constitutive synthesis of heat shock proteins and increased thermotolerance in transgenic Arabidopsis. Plant J. 1995;8:603–612. doi: 10.1046/j.1365-313x.1995.8040603.x. [DOI] [PubMed] [Google Scholar]
- Lenne C, Douce R. A low molecular mass heat-shock protein is localized to higher plant mitochondria. Plant Physiol. 1994;105:1255–1261. doi: 10.1104/pp.105.4.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- McHaourab HS, Godar JA, Stewart PL. Structure and mechanism of protein stability sensors: chaperone activity of small heat shock proteins. Biochemistry. 2009;48:3828–3837. doi: 10.1021/bi900212j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merola A, et al. Revision of Cyanidium caldarium. Three species of acidophilic algae. Giorn Bot Ital. 1981;115:189–195. [Google Scholar]
- Mogk A, Deuerling E, Vorderwülbecke S, Vierling E, Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50:585–595. doi: 10.1046/j.1365-2958.2003.03710.x. [DOI] [PubMed] [Google Scholar]
- Mogk A, Schlieker C, et al. Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J Biol Chem. 2003;278:31033–31042. doi: 10.1074/jbc.M303587200. [DOI] [PubMed] [Google Scholar]
- Morgan AD, Ness RW, Keightley PD, Colegrave N. Spontaneous mutation accumulation in multiple strains of the green alga, Chlamydomonas reinhardtii. Evolution. 2014;68:2589–2602. doi: 10.1111/evo.12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mymrikov EV, Seit-Nebi AS, Gusev NB. Large potentials of small heat shock proteins. Physiol Rev. 2011;91:1123–1159. doi: 10.1152/physrev.00023.2010. [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nozaki H, et al. The phylogenetic position of red algae revealed by multiple nuclear genes from mitochondria-containing eukaryotes and an alternative hypothesis on the origin of plastids. J Mol Evol. 2003;56:485–497. doi: 10.1007/s00239-002-2419-9. [DOI] [PubMed] [Google Scholar]
- Nozaki H, et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007;5:28. doi: 10.1186/1741-7007-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisic O, Xiao H, Lis JT. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell. 1989;59:797–806. doi: 10.1016/0092-8674(89)90603-x. [DOI] [PubMed] [Google Scholar]
- Prahlad V, Morimoto RI. Integrating the stress response: lessons for neurodegenerative diseases from C. elegans. Trends Cell Biol. 2009;19:52–61. doi: 10.1016/j.tcb.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science. 1993;259:230–234. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins) Cell Stress Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:tefoat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonknecht G, et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science. 2013;339:1207–1210. doi: 10.1126/science.1231707. [DOI] [PubMed] [Google Scholar]
- Schroda M. The Chlamydomonas genome reveals its secrets: chaperone genes and the potential roles of their gene products in the chloroplast. Photosynth Res. 2004;82:221–240. doi: 10.1007/s11120-004-2216-y. [DOI] [PubMed] [Google Scholar]
- Schroda M, Vallon O. Chaperones and proteases. In: Harris EH, Stern DB, Witman GB, editors. The Chlamydomonas sourcebook. 2nd ed. Vol. 2. San Diego (CA): Academic Press; 2008. [Google Scholar]
- Schulz-Raffelt M, Lodha M, Schroda M. Heat shock factor 1 is a key regulator of the stress response in Chlamydomonas. Plant J. 2007;52:286–295. doi: 10.1111/j.1365-313X.2007.03228.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Nishiyama Y, Murata N. Acclimation of the photosynthetic machinery to high temperature in Chlamydomonas reinhardtii requires synthesis de novo of proteins encoded by the nuclear and chloroplast genomes. Plant Physiol. 2000;124:441–449. doi: 10.1104/pp.124.1.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, et al. Proproliferative functions of Drosophila small mitochondrial heat shock protein 22 in human cells. J Biol Chem. 2010;285:3833–3839. doi: 10.1074/jbc.M109.080424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters ER, Rioflorido I. Evolutionary analysis of the small heat shock proteins in five complete algal genomes. J Mol Evol. 2007;65:162–174. doi: 10.1007/s00239-006-0223-7. [DOI] [PubMed] [Google Scholar]
- Xiao H, Lis JT. Germline transformation used to define key features of heat-shock response elements. Science. 1988;239:1139–1142. doi: 10.1126/science.3125608. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, et al. Quality control of photosystem II: impact of light and heat stresses. Photosynth Res. 2008;98:589–608. doi: 10.1007/s11120-008-9372-4. [DOI] [PubMed] [Google Scholar]
- Yoon HS, et al. Establishment of endolithic populations of extremophilic Cyanidiales (Rhodophyta) BMC Evol Biol. 2006;6:78. doi: 10.1186/1471-2148-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.