Abstract
Transposable elements (TEs) are one of the most important features of genome architecture, so their evolution and relationship with host defense mechanisms have been topics of intense study, especially in model systems such as Drosophila melanogaster. Recently, a novel small RNA-based defense mechanism in animals called the Piwi-interacting RNA (piRNA) pathway was discovered to form an adaptive defense mechanism against TEs. To investigate the relationship between piRNA and TE content between strains of a species, we sequenced piRNAs from 16 inbred lines of D. melanogaster from the Drosophila Genetic Reference Panel. Instead of a global correlation of piRNA expression and TE content, we found evidence for a host response through de novo piRNA production from novel TE insertions. Although approximately 20% of novel TE insertions induced de novo piRNA production, the abundance of de novo piRNAs was low and did not markedly affect the global pool of ovarian piRNAs. Our results provide new insights into the evolution of TEs and the piRNA system in an important model organism.
Keywords: piRNA, transposable elements, Drosophila melanogaster, de novo piRNA production
Introduction
Transposable elements (TEs) are a ubiquitous component of virtually all genomes across the tree of life (Burt and Trivers 2008). TEs are generally thought to be deleterious and therefore need to be repressed in order to ensure the viability of the species. Drosophila melanogaster has served as a valuable model system for studying TE biology for many decades (Bergman et al. 2006). In D. melanogaster, the reference strain has 5,390 individual TE insertions comprising approximately 5.5% of the genome (Bergman et al. 2006). At a genome-wide scale, the pericentromeric regions of the chromosomes and the heterochromatic fourth chromosome have a high proportion of TE-derived sequences, which are mostly incomplete and nested. There are also euchromatic copies of TEs which are more often complete copies and potentially actively transposing.
Recently, a class of small RNAs called Piwi-interacting RNAs (piRNAs) that are found in animal germlines has been discovered to repress TEs (reviewed in [Siomi et al. 2011]). piRNAs are approximately 23–29 nt RNAs that are related to the more well-known microRNAs and small interfering RNAs. Biochemically, piRNAs are distinguished by binding to Piwi class Argonaute proteins, whereas miRNAs and siRNAs bind to Ago class Argonaute proteins (Siomi et al. 2011). piRNAs were initially discovered as repeat-associated small RNAs (Aravin et al. 2001) and later found to play an important role in TE repression (Brennecke et al. 2007). In Drosophila, piRNAs are generally derived from degraded copies of TEs and are deposited into the embryo maternally. The piRNA system helps explain classical evolutionary phenomena such as the existence of TE-repressing loci that encoded degraded copies of TEs but no protein-coding genes (Brennecke et al. 2007), as well as hybrid dysgenesis in which immunity to TEs is inherited only maternally (Brennecke et al. 2008). piRNAs are most prominently expressed in animal germlines and are often found in large genomic clusters, usually in heterochromatic regions. piRNA evolution is rapid such that adaptation to a novel TE can occur even within the lifetime of a single individual (Khurana et al. 2011).
Unlike small RNAs in other species that amplify the small RNA signal using an RNA-dependent RNA polymerase, piRNAs amplify their expression levels using a positive feed-back loop called the “ping pong cycle” in which primary and secondary piRNAs alternatively cleave sense and antisense copies of transposon transcripts (Brennecke et al. 2007; Gunawardane et al. 2007). Thus, the act of cleaving primary transposon transcripts both represses the transposon mRNA and also produces a novel secondary piRNA in the process. Similar to other Argonaute proteins, the cleavage event invariably occurs after the tenth base pair from the 5′-end of the piRNA (Brennecke et al. 2007; Gunawardane et al. 2007).
Although piRNAs are known to play a role in TE regulation in D. melanogaster, the overall relationship of piRNA and TE content between strains has not been studied systematically except in two recent studies which each investigated only two strains (Kelleher and Barbash 2013; Shpiz et al. 2014). Two simple hypotheses might be that Drosophila strains increase their piRNA content to repress more actively transposing TE families or that lower piRNA content leads to more active TE families. However, the relationship between piRNAs and TEs could also be a complex mix of these two scenarios. Recently, a community resource called the Drosophila Genetic Reference Panel (DGRP) was released, consisting of genome sequences and phenotypic data for over 100 inbred lines of D. melanogaster sampled from a North Carolina population (Mackay et al. 2012). Here, we used 16 strains from the DGRP panel to examine the relationship between TE and piRNA content and study the importance of piRNA-mediated regulation in D. melanogaster.
Materials and Methods
Drosophila Stocks and Fly Husbandry
DGRP inbred fly lines were obtained from the Bloomington Drosophila Stock Center. All flies were maintained on standard yeast-cornmeal-dextrose medium at 25 °C. Virgin flies were collected within 6 h of eclosion and maintained in single-sex groups for 2–3 days before mating. All virgin female flies were mated with males in groups of 20 or less for 5 h. Males were removed and females were incubated at 25 °C for 24 h from the end of the mating period. Ovaries were dissected from females with the presence of sperm in the seminal receptacle and used for piRNA library preparation.
Total RNA Preparation, Periodate Oxidation, and β-Elimination Treatment
Dissected ovaries from approximately 60 females in each line were placed in Trizol reagent (Invitrogen) and briefly homogenized and stored at −80 °C. Total RNA was extracted according to manufacturer’s instructions and then purified using PurelinkTM RNA Minikit (Ambion). Three micrograms of total RNA was processed following the modified Periodate Oxidation and β-elimination protocol described in Kirino and Mourelatos (2007): A 50 µl mixture consisting of 3 µg of total RNA and 20 mM NaIO4 was incubated at 0 °C for 40 min at dark, 5 µl of 2M rhamnose was added to quench unreacted NaIO4 and incubated at 0 °C for additional 30 min. Fifty-five microliters of 2M Lys-HCl (PH = 8.5) was then added and the solution was incubated at 45 °C for 90 min for β-elimination, followed by standard ethanol precipitation for RNA purification. After ethanol precipitation, periodate-treated RNA was dissolved in 8 µl RNase free H2O. The concentration of periodate-treated RNA was determined by Nanodrop.
piRNA Library Construction
The piRNA libraries were constructed using the NEBNext Multiplex Small RNA Library Prep for Illumina Set 1 (E7300S). The main difference between this protocol and that previously published by several of the authors for mammals (Ha et al. 2014) was the removal of the 2S rRNAs using the terminator oligo blocking, following Wickersheim and Blumenstiel (2013). Briefly, periodate-treated RNAs (100–1,000 ng) were ligated to a Multiplex 3′-SR adaptor. The ligation product was then hybridized to the multiplex SR Reverse Transcription Primer. The 2S rRNA depletion was performed during the hybridization step. Specifically, six pmoles 2S rRNA block oligo (for 3 µg total RNAs) were added in the hybridization reaction without changing the concentration of other reaction components and the total volume (25.5 µl) of the hybridization reaction. The sequence of the 2S rRNA block DNA oligo is: 5′-AGT CTT ACA ACC CTC AAC CAT ATG TAG TCC AAG CAG CAC T-3′, which is complementary to the Drosophila 2S rRNA (Wickersheim and Blumenstiel 2013). During the hybridization step, 2S rRNA hybridizes with the block DNA oligo and forms double-stranded 2S rRNA/DNA fragments. The hybridization products were then ligated to the 5′-SR Adaptors. The double-stranded 2S rRNA/DNA fragments are not substrates for the single-strand RNA ligation of the 5′-SR Adaptors and are therefore excluded from the next step (reverse transcription).
Next, RNAs that were ligated to Multiplex 3′ and 5′-SR Adaptors were reverse transcribed. The reverse transcribed cDNA products were PCR amplified using Index (X) Primer* (primer 1–12, using a different primer for each sample). The PCR conditions were: An initial step at 94 °C for 30 s, 15 cycles at 94 °C for 15 s, 62 °C for 30 s, and 70 °C for 15 s, followed by 75 °C for 5 mins for final extension. The amplified libraries were electrophoresed and size fractionated through a 3% NuSieve GTG (Lonza) 3:1 GenePure LE agarose gel (Bioexpress). A gel slice in the approximately 150-bp size range was excised and purified following standard gel extraction procedures (Wizard SV Gel and PCR Clean-up System, Promega). Libraries were eluted in 50 µl nuclease-free H2O.
Quantification of libraries was performed using KAPA Library Quantification Kits for Illumina sequencing platforms (KAPA) following the kit protocol. Absolute quantification method was applied to accurately quantify the number of amplifiable molecules in a library. Eight libraries with distinct indexes were pooled with same amount of molecules (0.02 pmole). Two libraries pooled from 16 samples were further validated for size, purity, and concentration before sequencing using Illumina HiSeq2500 with single-end 50-bp format.
Computational Pipeline to Analyze piRNA Sequences
We first removed 3′-adapter sequences from raw sequencing reads using the Cutadapt software (Martin 2014) and removed reads that were smaller than 5 or larger than 45 nt. Then, we removed reads that were aligned to all known small RNAs using Bowtie (Langmead et al. 2009) allowing up to one mismatch ([-k 1 -v 1]). We then mapped reads to the reference genome (dm3) using Bowtie allowing up to one mismatch and multiple best matches ([-a —best —strata -v 1]) and removed unmapped reads. Finally, we selected reads of size between 23 and 29 nt as piRNAs. We plotted the distribution of read sizes after removing reads mapping to (non-piRNA) small RNAs as well as unmapped reads. We confirmed that there was a peak in the size distribution in the range 23–29 nt for all 16 samples. For some samples, there was also a small peak at the 13 nt. We found that these were mostly fragments of (non-piRNA) small RNAs that were not removed at the step of removing small RNA-derived reads. When we allowed up to two mismatch to remove reads mapped to small RNAs, the peak at 13 nt completely disappeared while almost nothing changed in the 23–29 nt (<1% of reads were removed from the 23–29 nt peak). It is possible that some small RNA-derived reads have natural mutations, RNA-editing or base changes induced by the experimental protocol. Because there was very little change in the amount of putative piRNAs whether the one or two mismatch option was used, we used one mismatch for the results reported in this manuscript. We treated the 13 nt peak as degradation products so we did not consider it further.
After read mapping, the number of reads mapped to a particular genomic position was normalized by the number of the possible mapping positions, following Brennecke et al. (2007). For example, a read that mapped to ten positions in the genome was counted as 0.1 reads at each position. We allowed up to one mismatch due to possible genomic differences between the DGRP strains and the reference genome. For mapping to the reference genome, we used the following chromosomes, chr2L, chr2R, chr3L, chr3R, chrX, chr4, chr2LHet, chr2RHet, chr3LHet, chr3RHet, chrXHet, and chrU. We also mapped reads to each strain’s genome by introducing the alleles from the single nucleotide polymorphism (SNP) calls from DGRP into the reference genome. Overall, there was less than 1% of increase in the amount of putative piRNA reads if we mapped reads to each strain’s genome because most piRNAs are in heterochromatic regions but all available SNP calls are in euchromatic regions. Thus, we used the reference genome to map piRNA reads for all strains.
To compute the ping-pong signal, we counted the number of overlaps between all pairs of piRNA reads at all overlap sizes from 23 nt to 1 nt. We plotted the distribution of the overlaps between pairs of piRNA reads, where the copy number of pairs of piRNAs was defined as the product of the copy numbers of the two piRNAs.
DGRP and TE Annotation Data Sets
The DGRP is a collection of inbred D. melanogaster lines collected from Raleigh, NC (Mackay et al. 2012). Because the lines have been kept in the lab since 2003, there are novel TE insertions in the lines that are not in the reference genome and are unique to a single line. Currently, there are three published TE annotation methods (Linheiro and Bergman 2012; Cridland et al. 2013; Zhuang et al. 2014) and one unpublished TE annotation method (Fiston-Lavier et al. 2014) that have been run on the DGRP lines. We downloaded annotations of TE insertions for the first three methods because no annotations were available for the fourth method.
For each data set, we removed TE insertions that were either not in euchromatic regions (chr2L, chr2R, chr3L, chr3R, chr4, or chrX) or were found in more than one strain at the same position. We took 16 lines from DGRP (313, 358, 362, 375, 379, 380, 391, 399, 427, 437, 555, 705, 707, 712, 714, and 732) for which transcriptome data from Affymetrix arrays are available from Ayroles et al. (2009).
Variation of piRNA Cluster Expressions between Strains
The piRNA cluster expression level for each strain was computed from the expression level of piRNAs mapped to the piRNA cluster using the RPKM (Reads Per Kilobase per Million reads) measure, which was defined as the normalized number of piRNA reads mapped within the cluster divided by the cluster size in Kb divided by the total normalized number of piRNA reads. Note that we made an assumption that both efficiency of generating piRNAs from the precursor transcript and degradation rate of processed piRNAs are not highly variable among the clusters. Also, we used the genomic DNA length rather than the transcript length to normalize the RPKM.
To compute the variation of piRNA cluster expression between strains, we required that at least five samples should have expression level with RPKM ≥ 10. The coefficient of variation (CV) for each cluster is defined as the standard deviation of the expression levels of strains divided by its average.
We ran Bowtie (bowtie2-2.0.2) (Langmead et al. 2009) with default options for three sets of piRNA reads for the 16 strains—all mapped reads, uniquely mapped reads, and all mapped reads except reads sense to TEs. We acknowledge that the mapping of piRNAs to the genome is inherently difficult because of their repetitive nature, so we have chosen to present a range of possible solutions from underconservative to overconservative. Many piRNA clusters are bidirectional so the 141 known piRNA clusters were considered on each strand separately and there were a total of 282 cluster transcripts. Then, we ran Cufflinks (Cufflinks-2.2.0) (Trapnell et al. 2010) with the multiread-correct option and with the 282 cluster transcripts as the isoform transcript annotations. We followed the pipeline of Cufflinks by running Cuffmerge, Cuffquant, and Cuffdiff (Trapnell et al. 2013) consecutively.
Correlation Analysis
We downloaded Affymetrix array gene expression data set for the DGRP strains from Ayroles et al. (2009). Based on a literature search, we used the following list of 19 piRNA pathway genes for our analysis: AGO3, Gasz, Pimet, UAP56, armi, asterix, aub, cuff, krimp, kumo, mael, piwi, rhi, shu, spn-E, squ, vas, vret, and zuc.
In all of our correlation analyses, we controlled for the genome sequencing coverage which can affect the sensitivity of the TE calls (e.g., a strain with higher genome coverage would be expected to have a higher number of TEs called present).
De Novo piRNA Production Signature
We defined the de novo piRNA production signature to be the presence of piRNAs in the flanking regions of a TE insertion in an asymmetric way such that there was at least RPKM ≥ 0.5 in both 1-Kb flanking regions and at least 70% of piRNAs in each flanking region was skewed on the minus strand upstream or on the plus strand downstream in the genome. This definition is consistent with Shpiz et al. (2014). We also varied these thresholds to check that our conclusions are robust (table 4). For this analysis, we filtered out piRNAs that were TE-derived or within the annotated piRNA clusters, and also novel TE insertions that were inserted into known TE regions or the annotated piRNA clusters.
Table 4.
The Percentage of Novel TE Insertions with the De Novo piRNA Signature As the RPKM and Skewness Cutoffs Are Varied (Materials and Methods)
| RPKM Cutoffa | Skewness Cutoffb(%) | Median of the Percentages of Novel TE Insertions with De Novo piRNA Signature over Drosophila Strains |
||
|---|---|---|---|---|
| Cridland (%) | Linheiro (%) | TEMP (%) | ||
| 0.5 | 70 | 19.6 | 21.6 | 5.6 |
| 1 | 70 | 11.2 | 13.7 | 3.6 |
| 3 | 70 | 3.8 | 3.8 | 1.6 |
| 0.5 | 90 | 9.2 | 9.6 | 3.7 |
| 1 | 90 | 6.7 | 5.9 | 2.5 |
| 3 | 90 | 2.4 | 0 | 1.2 |
aRPKM cutoff x means the number of piRNAs is at least RPKM ≥x on each 1-Kb flanking region
bSkewness cutoff y means at least y% of piRNAs in each flanking region are skewed on the minus strand upstream or on the plus strand downstream in the genome.
For the sensitivity analysis, we slid a 1-Kb window by 100 bp steps along each chromosome. We defined a window to be a piRNA dense region if the RPKM measure was ≥1. The two ends of each window were cut at the last piRNA. If we found that two consecutive piRNA dense regions within 100 bp satisfied the following criteria, we called them as de novo piRNA signals—at least 70% of piRNAs of the left (and right) region were skewed on the minus (and plus) strand and at least 70% of piRNAs on the minus strand of the left region (and the plus strand of the right region) had a 5′-uridine. If two de novo piRNA signals overlapped, we kept the one with a higher number of piRNAs.
Gene Expression Data Sets and Analysis
We downloaded an RNA-tiling array data set from Oregon R adult female mated ovaries (4 days after eclosion) from modEncode (modEncode_2340) (modENCODE Consortium et al. 2010). If transcripts from the array covered at least half of all exons for a gene, we called the gene ovary-expressed.
We call a protein-coding gene “piRNA producing” if the number of piRNAs in the gene divided by the total number of piRNAs per million reads >15.
Results
Sequencing of piRNAs from 16 Inbred Strains of D. melanogaster
We used a published piRNA sequencing protocol and computational pipeline to annotate piRNA loci, clusters, and expression levels in ovaries from 16 inbred D. melanogaster lines from the DGRP (Ha et al. 2014) (Materials and Methods). The main modification we made to our previous sequencing protocol was to deplete 2S rRNAs from the samples, which is a necessary step in Drosophila but not mammals (Wickersheim and Blumenstiel 2013). We sequenced 6.0–23.0 million piRNA reads for each of the 16 strains (the analysis of the pooled sample of all 16 strains is shown in table 1 and fig. 1). We verified that the piRNAs we sequenced have the previously described characteristics of piRNAs. Specifically, 74.6–78.7% of the sequenced piRNAs had a uridine in the first position from the 5′-end (referred to as “1U” from now on) and the piRNAs showed evidence of a ping-pong signature (Brennecke et al. 2007; Gunawardane et al. 2007) (fig. 2, Materials and Methods). We found that 80.7–83.6% of the sequenced piRNAs were derived from TEs, which is comparable to the percentage of TE-derived piRNAs in published data from immunoprecipitation of three piRNA-binding proteins (Piwi, Aub or Ago3) (68–78%; Brennecke et al. 2007). The slightly higher percentage of TE-derived piRNAs that we observed in our data may correspond to TE-derived piRNAs in the cell that are not bound by these proteins. We also found that the percentage of genic piRNAs in our sample was 2.9–4.5%.
Table 1.
The Number of Sequencing Reads at Each Step of Our Computational Pipeline for the Pooled Sample of All 16 Strains
| Step | Number of Reads in the Pool (Million) |
|---|---|
| Raw | 558.4 |
| 5 ≤ size ≤ 45 | 546.3 |
| Removed small RNA-derived reads | 334.0 |
| Removed unmapped reads | 269.8 |
| 23 ≤ size ≤ 29 | 226.1 |
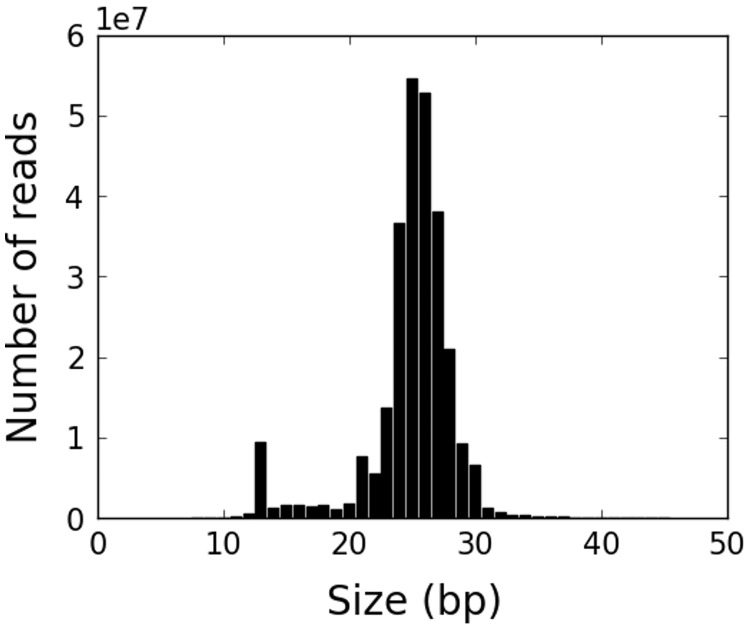
Fig. 1.—
The distribution of read sizes for all mapped putative piRNA reads in the pooled sample. There is a clear peak in the range 23–29 nt.
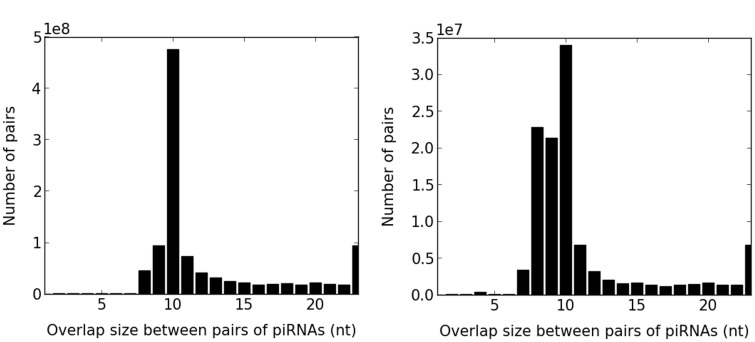
Fig. 2.—
Distribution of overlap sizes between all pairs of overlapping piRNAs for strain 705. There is a clear ping-pong signal with a peak at 10 nt. The peak of the distribution for uniquely mapped reads is not as sharp as for all mapped reads, presumably due to missing reads from bona fide ping-pong pairs.
To confidently and conservatively identify primary piRNA clusters, we first restricted our data to only the uniquely mapped reads, which left 20.5–23.9% of the reads in each strain. The choice to remove multiply mapped reads is important and commonly made in the literature to remove secondary piRNAs produced from TE transcripts. We note that this is a conservative approach because it may remove some bona fide primary piRNA clusters. We verified that the uniquely mapped piRNAs still had a high percentage of piRNAs with a 5′-uridine (1U) (72.5–76.9%). Among the uniquely mapped reads, we observed a slightly lower percentage of TE-derived piRNAs (56.5–65.5%), which includes both primary and secondary piRNAs. In this set of reads, we also observed a higher percentage of genic piRNAs (12.7–18.9%), most of which were not TE derived, as well as a ping-pong signal that was less apparent compared with all mapped reads (figs. 2 and 3). Taken together, these descriptive statistics such as the ping-pong signal, percentage of 1U nucleotides and percentage of TE-derived and genic piRNAs were broadly concordant with previous immunoprecipitation-based studies and thus validated the accuracy of our sequencing protocol and computational pipeline.
Fig. 3.—
Distribution of piRNA-generating loci across different classes of genomic elements. In order not to double-count overlapping regions, TE-derived piRNAs are defined as TE-derived but not within annotated piRNA clusters and genic piRNAs are defined as genic but neither TE-derived nor within annotated clusters.
Most piRNAs Map to Known piRNA Clusters, TE Fragments, or Genes
Overall, 226.1 million putative piRNA reads mapped to the reference genome (which we refer to as “all mapped reads”), of which 50.1 million mapped uniquely to the genome. We confirmed that the major loci that produce piRNAs in our sample correspond to the piRNA clusters previously annotated by Brennecke et al. (2007). These loci produced 36.4–40.1% of all mapped piRNAs and 52.7–63.6% of the uniquely mapped piRNAs. To examine the loci from which the other piRNAs were generated, we combined the piRNAs from all samples and examined their genomic origin (fig. 3). A large fraction of piRNAs that were not from the previously annotated piRNA clusters came from other TE fragments annotated in the reference genome. Excluding the annotated piRNA clusters, 51.0% of all mapped and 15.3% of the uniquely mapped reads were derived from other TE fragments. The third major source of piRNAs was protein-coding genes, particularly, 3′-untranslated regions (3′-UTRs) (Robine et al. 2009). Most genic piRNAs were euchromatic, consistent with the fact that most known piRNA clusters are located in heterochromatic and peri-centrometric regions where few genes exist. Genic piRNAs accounted for 14.1% of the uniquely mapped reads and 3.3% of all mapped reads. Thus taken together, we could explain approximately 90% of the mapped reads and uniquely mapped reads as being produced from annotated piRNA clusters, other TE fragments found in the reference genome or genic piRNAs (fig. 3). It is also possible that some of the remaining reads come from novel piRNA clusters.
It is interesting to consider the piRNAs that map to TE fragments. The Oregon R strain used by Brennecke et al. (2007) for their analysis differs from the DGRP strains and the reference y; cn bw sp strain. Many of these piRNAs might be produced from TE insertions in the DGRP strains that do not appear in the Oregon R genome or are not expressed in the Oregon R strain, but whose sequence exists in the reference genome. We cannot distinguish from our data whether the additional TE-derived piRNAs come from primary piRNA clusters or if they are secondary piRNAs processed from TE transcripts.
Relatively Low Variation of piRNA Cluster Expression between Strains
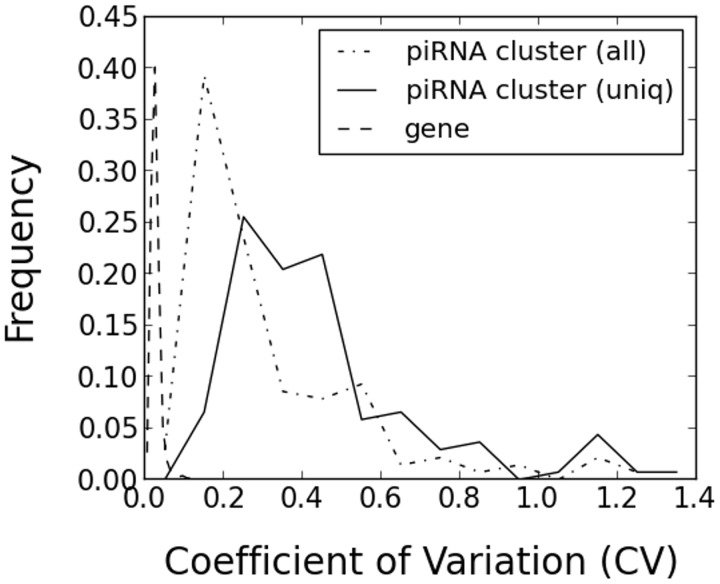
Because piRNAs are thought to be processed from long precursor transcripts produced from piRNA clusters, we computationally inferred the expression level of each piRNA cluster from the expression levels of piRNA reads that mapped to the cluster. We used established RNA-seq computational methods to infer the expression levels. Our procedure is slightly different from most current RNA-seq protocols which often use longer reads and paired-end reads but is consistent with older RNA-seq protocols which typically used shorter, single-end reads. In principle, it is possible that the processing of the primary piRNA cluster transcripts into piRNAs or the stability of individual piRNAs might vary between clusters. However, assuming that these effects do not produce strong biases, the piRNA read counts should give a reasonable estimate of the piRNA cluster expression levels. For each annotated piRNA cluster, we computed the number of piRNAs within the cluster in each strain and compared the piRNA cluster expression levels between the strains using the commonly used “RPKM” measure (Materials and Methods). For all mapped reads, we considered the 140 out of 141 clusters that had at least five strains with RPKM ≥ 10 and found that only 4 out of the 140 clusters had a CV > 1. We used the CV instead of the variance because in general there is a positive correlation between the mean and variance of gene expression levels. For uniquely mapped reads, we considered the 137 out of 141 clusters that had at least five strains with RPKM ≥ 10 and found that only 9 out of the 137 clusters had CV > 1 (fig. 4). Thus, using the commonly used CV threshold of one, few of the known piRNA clusters had high expression variation between the strains.
Fig. 4.—
The distribution of CVs of piRNA cluster expression levels between strains computed from all mapped and uniquely mapped piRNA reads. The distribution of CVs of protein-coding gene expressions between strains is plotted as a reference.
To compare the piRNA cluster expression variation against a baseline, we compared it with the variation in protein-coding gene expression taken from the same strains (fig. 4). Caution should be used when interpreting this data because the protein-coding gene expression was measured by tiling arrays and the piRNA cluster expression by RNA-seq. Nonetheless, taking this caveat into account, we observed higher variation in the piRNA cluster expression than protein-coding gene expression, suggesting the possibility of relaxed selective constraint on piRNA cluster expression compared with protein-coding genes.
Although the RPKM measure is commonly used in the literature, it is susceptible to inflation of the apparent variability of genes between samples. This is because a single highly expressed and highly variable gene across samples will increase the apparent variability of the other genes, even if they are in fact expressed at constant levels, because RNA-seq can give only information about the relative abundances of genes. To address this issue, we examined alternative ways to normalize the RNA-seq data (reviewed in [Rapaport et al. 2013]). In general, these normalization methods make the assumption that most genes do not change in expression between samples and so they remove outliers from the distribution of read counts (e.g., by taking medians and geometric means instead of arithmetic means).
We used the Cufflinks and Cuffdiff pipeline (Trapnell et al. 2013) to search for significant differences in piRNA cluster expression between pairs of strains (Materials and Methods). This pipeline is a state-of-the-art method that in principle addresses many of the shortcomings of the RPKM normalization approach, while also resolving the expression of repetitive transcripts using a statistical method (Expectation–Maximization) that takes into account both the uniquely and multiply mapped reads. When restricting to the uniquely mapped reads, there were 35 nominally significant pairs of strains that were differentially expressed (DE) among 32,525 total tests, and there were seven piRNA clusters with at least one DE pair. For all mapped reads, there were 69 significantly DE pairs of strains among 33,479 tests, and there were 11 piRNA clusters with at least one DE pair. To further filter out possible contamination from putative secondary piRNAs, we removed all reads mapping in a sense orientation to TEs. After this procedure, we found that there were 69 significantly DE pairs of strains among 30,590 tests, and there were 10 clusters with at least one DE pair. We examined the most variable piRNA clusters and found that they were enriched for the telomeric TE, TART, which plays a unique role in telomere formation in D. melanogaster (Pardue and DeBaryshe 2008). Considering just the uniquely mapped reads, five out of seven variable piRNA clusters contained only TART elements (supplementary table S1, Supplementary Material online). The relationship between piRNAs and telomeric TEs in Drosophila has been studied and found to differ from the transposon-silencing function of piRNAs (Khurana et al. 2010; Shpiz and Kalmykova 2011). Our data does not allow us to speculate on possible functional consequences of the variability of these telomeric piRNA clusters but they may be interesting clusters for future studies.
Taken together, these results suggested that piRNA cluster expression was relatively stable between strains, with the exception of the telomeric piRNA clusters. Nonetheless, piRNA expression overall appeared to be more variable than protein-coding gene expression, which might indicate a relaxation of selective constraint at piRNA loci than genic loci. We also cannot rule out the possibility of diversifying selection, which would be consistent with the genome defense mechanism of the piRNA pathway. Although we did not include biological replicates in our experiment, the main quantity of interest is the variability of piRNA cluster expression levels across strains, not the mean expression level for each strain. The variability of expression levels within a strain appears to be quite low because our comparison of piRNA populations from mated and unmated females from the same strain using the same Cuffdiff pipeline described above found no significantly DE piRNA clusters at all.
No Simple Linear Correlation between Global piRNA Expression and Novel TE Abundance
A simple hypothesis is that the number of piRNAs related to each TE family is positively correlated with the number of novel TE insertions in each strain because the host genome might respond to the increased number of TE copies by increasing the amount of piRNA expression. On the other hand, an equally plausible hypothesis is that increased piRNA expression would decrease the number of TE insertions, causing a negative correlation. In actual fact the relationship between these two quantities could also be more complex than a simple linear correlation because of the presence of other evolutionary forces, such as natural selection, or other molecular mechanisms of host defense against TEs.
To test these two hypotheses, we examined the relationship between TE and piRNA abundance. There were 25 TE families with both novel TE insertions in the Cridland TE annotation data set (Materials and Methods) and piRNA expression data for at least five strains. Among the 25 TE families, 13 and 12 TE families showed a positive and negative correlation, respectively, between the number of piRNAs and the number of novel TE insertions (Materials and Methods, table 2). However, all of the correlations were not statistically significant after Bonferroni correction for multiple testing. We also checked if gene expression of the piRNA pathway genes was correlated with the number of novel TE insertions (Materials and Methods). We found that none of the 19 piRNA pathway genes showed a significant correlation with novel TE abundance across the strains. Taken together, we did not find any statistically significant linear correlations of piRNA cluster expression or piRNA pathway gene expression with TE abundance, as would be predicted by simple models of piRNA-mediated TE repression.
Table 2.
Pearson Correlation between the Normalized Number of All Mapped piRNAs and the Number of Novel TE Insertions for Each TE Family
| TE Family | Pearson’s R | P value | TE Family | Pearson’s R | P value |
|---|---|---|---|---|---|
| Stalker2 | −0.70 | 0.02 | roo | 0.49 | 0.13 |
| G2 | −0.69 | 0.02 | pogo | 0.48 | 0.14 |
| Rt1b | −0.59 | 0.05 | Max | 0.48 | 0.14 |
| gypsy12 | −0.30 | 0.37 | mdg1 | 0.46 | 0.15 |
| I | −0.24 | 0.49 | Cr1a | 0.44 | 0.18 |
| transib3 | −0.17 | 0.61 | 412 | 0.35 | 0.29 |
| 3S18 | −0.15 | 0.66 | Quasimodo | 0.32 | 0.34 |
| Doc | −0.14 | 0.67 | FB | 0.29 | 0.39 |
| Transpac | −0.14 | 0.68 | flea | 0.26 | 0.44 |
| INE-1 | −0.08 | 0.80 | Tirant | 0.24 | 0.49 |
| Tabor | −0.05 | 0.87 | Juan | 0.21 | 0.54 |
| Burdock | −0.02 | 0.95 | F | 0.12 | 0.73 |
| S | 0.09 | 0.79 |
De Novo piRNA Production at Novel TE Sites May Explain Part of the Host Response
The lack of correlative results raises the question of how the piRNA pathway responds to de novo TE insertions. Recently, Shpiz et al. (2014) and Mohn et al. (2014) independently found that novel TE insertions can produce piRNAs from the insertion sites and their flanking regions. This de novo piRNA production produces a characteristic signature on the left and right flanking regions of novel TE insertions, consistent with bidirectional transcription from the novel TE insertion. Similar results on de novo piRNA production were published for Drosophila transgene insertions (Olovnikov et al. 2013). Another study suggested a role for transcription from an endogenous piRNA cluster (Kawaoka et al. 2012) which may also play a role in the de novo piRNA response.
To test if the left–right signature described by Shpiz et al. (2014) and Mohn et al. (2014) is present in our data, we examined all novel TE insertions unique to individual DGRP strains using three different TE annotation data sets for the DGRP strains that were recently published (Linheiro and Bergman 2012; Cridland et al. 2013; Zhuang et al. 2014 [TEMP]). These novel TE insertions are presumed to have occurred during the inbreeding phase of strain construction in the DGRP (Cridland et al. 2013). Because the three TE data sets gave annotated TE insertions mostly in euchromatic regions, we observed very few novel TE insertions in the annotated piRNA clusters, most of which were heterochromatic. For example, we found that only 21 (1.7%) out of 1,239 combined TE insertions in strain 391 were inserted in the annotated piRNA clusters.
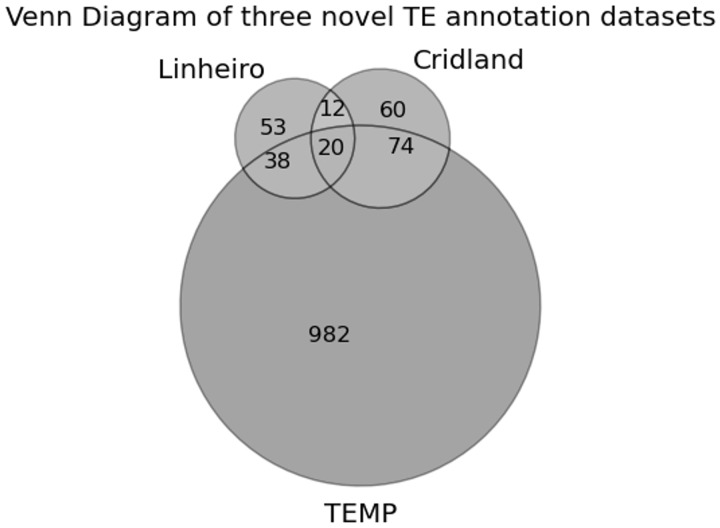
There was only one strain (strain 391) in which TEs were annotated by all three methods among the DGRP strains we sequenced. For this strain, we found that the overlap between the different annotation methods was low. Of the 1,239 total predicted TE insertions, only 20 (1.6%) overlapped between all three methods (fig. 5). Because there is no gold standard TE annotation data set available for these strains, it is not clear whether the small overlap between the methods is due to false positives or false negatives. We thus analyzed all three annotations using the de novo piRNA expression signature. Note that the small overlap between the data sets does not necessarily mean that all the three data sets have low accuracy. For example, it is possible that one data set is very accurate but overlaps poorly with the other two data sets.
Fig. 5.—
The overlap of TE predictions between the three novel TE annotation data sets for one strain that was annotated by all three methods.
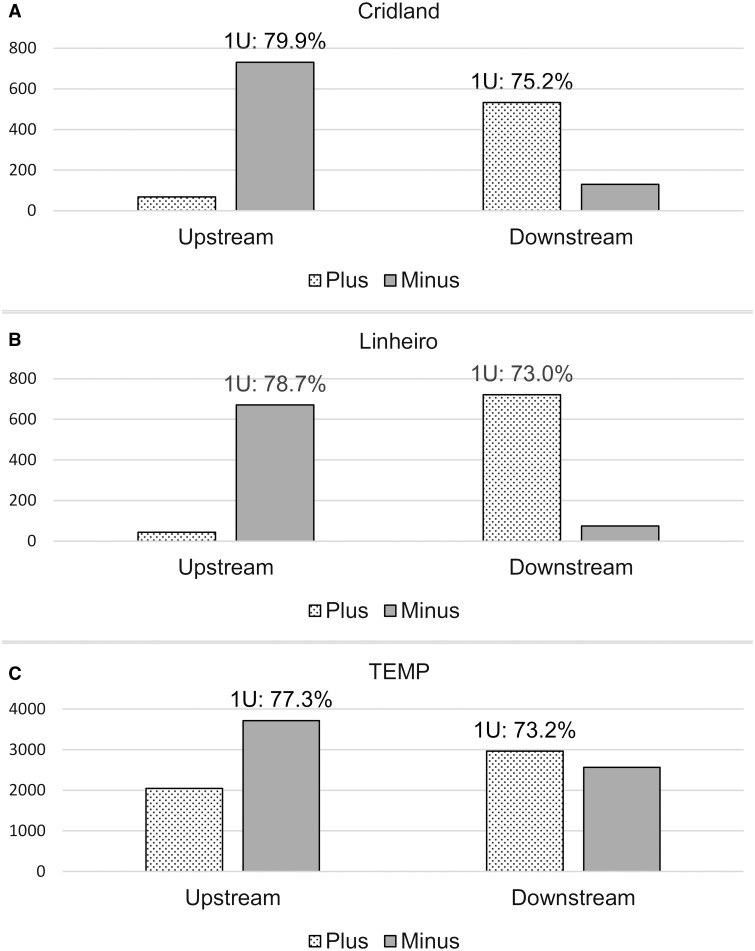
After excluding novel TE insertions within other existing TEs in the reference genome or the annotated piRNA clusters, we found that 7.3–27.5% (median: 19.6%), 4.2–31.6% (median: 21.6%), 1.2–6.2% (median: 5.6%) of novel TE insertions from the Cridland, Linheiro, and TEMP annotations, respectively, showed evidence of the de novo signature in each sample (Materials and Methods). We note that 16.1% (56 out of 348) of the novel TE insertions showed evidence for the de novo piRNA signature in Shpiz et al. (2014). This percentage is in a similar range to our results above for the Cridland (median: 19.6%) and Linheiro (median: 21.6%) data sets. Because Shpiz et al. (2014) used the reference genome which has a different TE annotation method from the Cridland and Linheiro data sets and the latter two data sets have similar numbers of called TEs, the concordance between these percentages suggests that the Cridland and Linheiro TE annotation methods may have similar error rates despite a relatively small overlap. When combining all of the novel TE insertions in the genome, we observed a clear strand bias among the upstream and downstream piRNAs for all three TE annotations for strain 391 (fig. 6). The signature was present but not as strong for the TEMP data set. If we interpret the signature as a validation of TE insertions, this indicates that TEMP includes a higher number of false positive predictions than the other two methods. We also give a more detailed breakdown for all strains when combining only the TE insertions that show evidence of the de novo signature above our threshold (Materials and Methods, table 3). Taken together, our data supports the de novo piRNA production model in a larger data set than previous studies.
Fig. 6.—
Number of piRNA reads found in the flanking regions (±1 kb) of novel TE insertions in DGRP strain 391. PiRNA reads are mostly found on the minus strand upstream (i.e., to the left) of novel TE insertions and on the plus strand downstream (i.e., to the right) of novel TE insertions. The percentage of piRNAs with a 5′-uridine (1U) confirms that the reads are likely to be piRNAs. The y axis shows the actual number of sequencing reads.
Table 3.
When Considering All Novel TE Insertions with De Novo piRNA Signatures, Most piRNAs in the Left Flanking Region Are on the Minus Strand with High 1U Percentage and Most piRNAs in the Right Flanking Region Are on the Plus Strand with High 1U Percentage
| TE Annotation Data Set | Strain | % of piRNAs on Minus Strand Upstream | % 1U of piRNAs on Minus Strand Upstream | % of piRNAs on Plus Strand Downstream | % 1U of piRNAs on Plus Strand Downstream |
|---|---|---|---|---|---|
| Cridland | 375 | 91.4 | 74.1 | 94.5 | 73.8 |
| 380 | 92.3 | 74.8 | 92.5 | 74.3 | |
| 391 | 95.6 | 79.1 | 95.0 | 77.6 | |
| 399 | 96.8 | 66.0 | 98.7 | 69.5 | |
| 427 | 94.7 | 70.3 | 96.8 | 64.4 | |
| 437 | 95.8 | 75.2 | 96.8 | 76.5 | |
| 555 | 97.4 | 74.3 | 93.6 | 81.8 | |
| 705 | 94.4 | 81.8 | 97.8 | 70.8 | |
| 707 | 95.4 | 74.5 | 96.2 | 73.9 | |
| 714 | 90.5 | 100.0 | 95.4 | 68.6 | |
| Linheiro | 313 | 100.0 | 100.0 | 100.0 | 73.3 |
| 358 | 100.0 | 66.7 | 86.2 | 66.7 | |
| 362 | 94.6 | 63.3 | 83.8 | 75.5 | |
| 375 | 93.5 | 75.0 | 93.2 | 73.1 | |
| 379 | 90.9 | 75.2 | 97.8 | 68.8 | |
| 380 | 90.3 | 76.2 | 95.2 | 79.9 | |
| 391 | 96.6 | 78.5 | 96.5 | 73.6 | |
| 399 | 88.6 | 68.0 | 79.6 | 79.5 | |
| 555 | 94.1 | 79.7 | 89.3 | 76.0 | |
| 705 | 99.0 | 78.3 | 86.8 | 72.3 | |
| 707 | 90.8 | 70.8 | 98.3 | 79.7 | |
| 712 | 94.9 | 77.1 | 87.9 | 79.8 | |
| 714 | 92.0 | 70.4 | 96.9 | 75.3 | |
| 732 | 94.8 | 78.3 | 96.5 | 77.5 | |
| TEMP | 362 | 93.9 | 71.6 | 94.6 | 70.0 |
| 391 | 95.1 | 77.4 | 96.2 | 77.5 | |
| 437 | 92.2 | 74.9 | 95.9 | 72.6 |
To find further evidence for the existence of the de novo piRNA signature, we applied more stringent cutoffs to define the de novo piRNA signature and found that the signature was robust to our parameter settings because the number of novel TEs satisfying the cutoffs was almost always higher than 0 (table 4). We further controlled for other factors that might make some parts of the genome more likely to have de novo piRNA production (e.g., base composition or chromatin structure). To do so, for each strain we computed the fraction of novel TE insertions that have de novo piRNA signatures at the same genomic position in the other strains. In this test, we found that a negligible percentage of novel TE insertions (<1%) had de novo piRNA signature in any of the other strains. These controls give additional statistical confidence to the robustness of the de novo piRNA signatures we observed.
Finally, we compared the sensitivity of the three TE annotation data sets under the assumption that the asymmetric piRNA signals are a genuine indicator of novel TE insertions (Materials and Methods). For strain 391 where all three methods annotate novel TE insertions, we found 80 de novo piRNA signals using stringent cutoffs (Materials and Methods). We found that 9 out of the 80 (11.3%) piRNA signatures were annotated by at least one of the three methods. The relatively low sensitivity could indicate that the TE annotation data sets are incomplete or it might simply indicate that there are additional sites of de novo piRNA production in the genome that do not correspond to novel TE insertions. For example, we considered only unique insertions in our analysis and it is possible that other TE insertions also produce de novo piRNAs. The total number of de novo signatures in the genome is not high so the overlap between de novo piRNA producing regions and novel TE insertions is clearly nonrandom. Thus, taken together, our data provide support for the de novo piRNA production model of TE response.
Novel TE Insertions Into Genes May Affect Gene Expression
Next, we analyzed the genic piRNAs in the different strains. Almost all (>99%) of the genic piRNAs were not derived from TE sequences and 41.5–50.4% of genic piRNAs from uniquely mapped reads were 3′-UTR derived, consistent with previous results (Robine et al. 2009). We called a gene “3′-UTR enriched” if there were more piRNAs in the 3′-UTR than the 5′-UTR or coding regions compared with the expected proportion by their length in the gene. Among the piRNA-producing genes (Materials and Methods), 59.5–74.3% were 3′-UTR enriched genes, consistent with previous results. The set of 3′-UTR enriched genes in each strain from ovaries was highly overlapping with those in Drosophila ovary somatic sheet cells (Robine et al. 2009) (Hypergeometric P value < 2.4e-46 to 7.4e-60).
One model for the role of piRNAs in the evolution of gene regulation is that TE insertions in or near genes might affect gene expression in a piRNA-dependent manner, for example, by causing the transcript to be cleaved or causing local heterochromatin formation. We note that many other molecular mechanisms are possible, such as causing a change in mRNA splicing or introducing a microRNA-binding site. We found that 4.4–10.9%, 4.6–18.4%, and 6.9–9.2% of novel TE insertions from the Cridland, Linheiro, and TEMP data sets, respectively, were genic. These numbers are much lower than by chance (about 25.5% of the bases in the genome are genic), presumably because of purifying selection for TE insertions in genes (Cridland et al. 2013).
We tested if the genes that had novel TE insertions in their coding region, UTRs or promoter (up to 1 kb upstream of transcription start site) showed a significant difference in gene expression between strains with and without novel TE insertions. We found between 41 and 98 genes with novel TE insertions to be nominally significantly DE (table 5). Of these genes, we found that roughly half of them were expressed in ovaries, which is significant in all three TE data sets (table 5). There were 2,895 ovary-expressed genes among the 9,327 genes for which gene expression data for the DGRP strains was available (∼31%). When we restricted to DE genes that were significant after multiple testing correction, the statistical significance of the overlap with ovary-expressed genes dropped, presumably due in part to a loss of statistical power. Although this analysis does not prove that the change in gene expression is piRNA-dependent, it is consistent with the idea that novel TE insertion into genes might affect gene expression in a piRNA-dependent manner (Sienski et al 2012), and confirmation of this model could be interesting for further experimental investigations.
Table 5.
Ovary-Expressed Genes Tend to Show a Significant Difference in Gene Expression between Strains with and without Novel TE Insertions in the Gene
| TE Data Set | Number of Genes with Novel TE Insertions | Showing Significant Gene Expression Difference by Nominal T-Test (P < 0.05) |
Showing Significant Gene Expression Difference by T-Test (Bonferroni Corrected P < 0.05) |
||||
|---|---|---|---|---|---|---|---|
| Number of Genes | Number of Ovary-Expressed Genes | P valuea | Number of Genes | Number of Ovary-Expressed Genes | P valuea | ||
| Linheiro | 248 | 41 | 22 (53.7%) | 2.1e-3 | 9 | 7 (77.8%) | 5.3e-3 |
| Cridland | 344 | 73 | 40 (54.8%) | 2.0e-5 | 20 | 10 (50.0%) | 0.06 |
| TEMP | 3,211 | 98 | 43 (43.9%) | 4.8e-3 | 0 | 0 | N/A |
aP values are from Fisher exact tests.
Discussion
Here, we performed piRNA sequencing in ovaries from 16 strains of D. melanogaster taken from an important community resource—the DGRP. Our study is larger than two previous studies (Kelleher and Barbash 2013; Shpiz et al. 2014) which each looked at two Drosophila strains. These data allowed us to perform correlation analysis between piRNA abundance and TE copy number across a larger number of strains, to perform all of our de novo piRNA analyses using the same TE annotation methods for all the strains and to use D. melanogaster strains with similar genetic backgrounds. This differs from Shpiz et al. (2014) who used two strains that were more diverged from each other and had TE annotations from different methods. In addition, the larger number of strains allowed us to better estimate the variability of piRNA cluster expression and compare it with the variability of protein-coding gene expression.
Overall, we found that there was no statistically significant linear correlation of global piRNA abundance or piRNA pathway gene expression with the number of novel TE insertions, even when subdividing the TEs according to TE families. One simple evolutionary model might be that higher piRNA expression leads to lower rates of TE insertion, which would produce a negative correlation between the number of TE insertions and piRNA abundance. On the other hand, an equally plausible evolutionary model might be that piRNA abundance would increase in response to a higher number of TE insertions, an effect that would produce a positive correlation. The fact that we observe no consistent pattern of positive or negative correlations across TE families is consistent with a more complex scenario that is a mix of these two simple scenarios. We also cannot exclude that some part of this pattern is due to the relatively low statistical power in the tests with the number of strains available in our study. Our study would be powered to detect a Pearson correlation of 0.5 (a value reached by a number of our tests) with significance level and power both set to 0.22. Our results are consistent with that of Kelleher and Barbash (2013) who performed linear regression analysis for a number of different genomic features, including the transposition rate of active TEs, and concluded that there are limits to the optimization of host defense mechanisms. Because the piRNA pathway has been molecularly shown to be important for defense against TEs, our results indicate that the piRNA-pathway response to TE invasion is complex or possibly smaller than other mechanisms of TE control. On the other hand, we observed the existence of de novo piRNA production at novel TE loci which suggests that this may be one way the genome responds to novel TE insertions, consistent with the recent results of Mohn et al. (2014) and Shpiz et al. (2014).
A number of other groups have recently studied the determinants of TE abundance in D. melanogaster and some groups also considered their relationship with piRNAs. Petrov et al. (2011) studied euchromatic, nonnested TEs found in the reference D. melanogaster strain using a pooled-PCR approach. They concluded that the primary determinant of TE abundance is the strength of negative selection due to ectopic recombination, but they did not examine piRNAs closely. Kofler et al. (2012) used a Pool-Seq approach to study TE insertions in D. melanogaster. They found that different TE families have very different insertion rates and they suggested that the history of TE activity influences this pattern but they did not study piRNAs. Lee and Langley (2012) studied the determinants of TE abundance in the DPGP, a different community resource from the one studied here, and found that piRNA pathway genes often are adaptively evolving at the sequence level. Finally, a simulation study by Lu and Clark (2010) showed that piRNAs can allow TEs to increase in frequency in the population because they attenuate the deleterious effects of TE insertion. In addition, one of the authors previously performed an analysis of piRNAs and TE in humans and found that piRNA targeting is strongly correlated with the age of TE families (Lukic and Chen 2011). Finally, a study on inter-specific hybrids between D. melanogaster and Drosophila simulans attributed changes in TE abundance between species to mutations in piRNA pathway genes (Kelleher et al. 2012), consistent with the observation that there is adaptive evolution of many piRNA pathway genes between species (e.g., Simkin et al. 2013; Yi et al. 2014). We do not observe this pattern among the DGRP strains, at least at the level of gene expression variation.
Overall, our study gives new insights into the response of a host genome to novel insertions of TEs. In addition, we also found several other results from our data that may be interesting for future studies. First, we found that piRNA clusters are generally not highly variable between strains, although based on current data sets they appear to be more variable than protein-coding genes. The most variable piRNA clusters tended to be located in telomeric regions and contain piRNAs derived from the telomeric TART TE. We cannot distinguish from our data whether the high variability of these clusters is due to their location in telomeric regions, which are regions of generally high variability at many levels, including sequence and expression variation, or whether there is any selective advantage for high diversity among this particular class of piRNAs. Second, we found patterns which are consistent with the idea that one-way TEs can affect gene expression variation is by inserting into genes and changing their expression patterns in a way that depends on their ovarian expression. Finally, because we performed our analyses in an important community resource, the DGRP, it is possible that our piRNA sequence data will also be useful for other groups as well, not only for TE biology but also for studies of biological processes other than TE defense such as regulation of protein-coding genes (Peng and Lin 2013).
Supplementary Material
Supplementary table S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The piRNA sequence data are available at the NIH SRA (SRP019948). The authors thank Mark Siegal for assistance with Drosophila husbandry, Sergio Lukic for comments on the manuscript, and Daniel Schrider and the Xing and Chen labs for helpful discussions. This work was partially supported by the National Institutes of Health (grant number R00HG005846 to J.X., R00HG004515 to K.C.C.).
Literature Cited
- Aravin A, et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Ayroles J, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman C, et al. Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol. 2006;7:R112. doi: 10.1186/gb-2006-7-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone C, Aravin A, Sachidanandam R. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A, Trivers R. 2008 Genes in conflict: the biology of selfish genetic elements. Cambridge (MA): Harvard University Press. [Google Scholar]
- Cridland J, Macdonald S, Long A, Thornton K. Abundance and distribution of transposable elements in two Drosophila QTL mapping resources. Mol Biol Evol. 2013;30:2311–2327. doi: 10.1093/molbev/mst129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiston-Lavier A, Barron M, Petrov D, Gonzalez J. T-lex2: genotyping, frequency estimation and re-annotation of transposable elements using single or pooled next-generation sequencing data. 2014 doi: 10.1093/nar/gku1250. In: bioRxiv. Available from: http://dx.doi.org/10.1101/002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane L, et al. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Ha H, et al. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics. 2014;15:545. doi: 10.1186/1471-2164-15-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, et al. A role for transcription from a piRNA cluster in de novo piRNA production. RNA. 2012;18:265–273. doi: 10.1261/rna.029777.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher E, Barbash D. Analysis of piRNA-mediated silencing of active TEs in Drosophila melanogaster suggests limits on the evolution of host genome defense. Mol Biol Evol. 2013;30:1816–1829. doi: 10.1093/molbev/mst081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher E, Edelman N, Barbash D. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol. 2012;10:e1001428. doi: 10.1371/journal.pbio.1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana J, et al. Adaptation to P Element Transposon Invasion in Drosophila melanogaster. Cell. 2011;147:1551–1563. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana J, Xu J, Weng Z, Theurkauf W. Distinct functions for the Drosophila piRNA pathway in genome maintenance and telomere protection. PLoS Genet. 2010;6:e1001246. doi: 10.1371/journal.pgen.1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2'-O-methylated at their 3' termini. Nat Struct Mol Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- Kofler R, Betancourt A, Schloetterer C. Sequencing of pooled DNA samples (Pool-Seq) uncovers complex dynamics of transposable element insertions in Drosophila melanogaster. PLoS Genet. 2012;8:e1002487. doi: 10.1371/journal.pgen.1002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Langley C. Long-term and short-term evolutionary impacts of transposable elements on Drosophila. Genetics. 2012;192:1411–1432. doi: 10.1534/genetics.112.145714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linheiro R, Bergman C. Whole genome resequencing reveals natural target site preferences of transposable elements in Drosophila melanogaster. PLoS One. 2012;7:e30008. doi: 10.1371/journal.pone.0030008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Clark A. Population dynamics of PIWI-interacting RNAs (piRNAs) and their targets in Drosophila. Genome Res. 2010;20:212–227. doi: 10.1101/gr.095406.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukic S, Chen K. Human piRNAs are under selection in Africans and Repress transposable elements. Mol Biol Evol. 2011;28:3061–3067. doi: 10.1093/molbev/msr141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T, et al. The Drosophila melanogaster genetic reference panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBNet J. 2014;17:10–12. [Google Scholar]
- modENCODE Consortium, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J. The rhino-deadlock- cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157:1364–1379. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- Olovnikov I, et al. De novo piRNA cluster formation in the Drosophila germ line triggered by transgenes containing a transcribed transposon fragment. Nucleic Acids Res. 2013;41:5757–5768. doi: 10.1093/nar/gkt310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M, DeBaryshe P. Drosophila telomeres: a variation on the telomerase theme. Fly. 2008;2(3):101–110. doi: 10.4161/fly.6393. [DOI] [PubMed] [Google Scholar]
- Peng J, Lin H. Beyond transposons: the epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr Opin Cell Biol. 2013;25:190–194. doi: 10.1016/j.ceb.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D, Fiston-Lavier A, Lipatov M, Lenkov K, Gonzalez J. Population genomics of transposable elements in Drosophila melanogaster. Mol Biol Evol. 2011;28:1633–1644. doi: 10.1093/molbev/msq337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport F, et al. Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol. 2013;14:R95. doi: 10.1186/gb-2013-14-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N, et al. A broadly conserved pathway generates 3'UTR-directed primary piRNAs. Curr Biol. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Kalmykova A. Role of piRNAs in the Drosophila telomere homeostasis. Mob Genet Elements. 2011;1:274–278. doi: 10.4161/mge.18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Ryazansky S, Olovnikov I, Abramov Y, Kalmykova A. Euchromatic transposon insertions trigger production of novel Pi- and endo-siRNAs at the target sites in the Drosophila germline. PLoS Genet. 2014;10:e1004138. doi: 10.1371/journal.pgen.1004138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A, Wong A, Poh Y, Theurkauf W, Jensen J. Recurrent and recent selective sweeps in the piRNA pathway. Evolution. 2013;67:1081–1090. doi: 10.1111/evo.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M, Sato K, Pezic D, Aravin A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersheim M, Blumenstiel J. Terminator oligo blocking efficiently eliminates rRNA from Drosophila small RNA sequencing libraries. Biotechniques. 2013;55:269–272. doi: 10.2144/000114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, et al. Rapid Evolution of piRNA pathway in the teleost fish: implication for an adaptation to transposon diversity. Genome Biol Evol. 2014;6:1393–1407. doi: 10.1093/gbe/evu105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, Wang J, Theurkauf W, Weng Z. TEMP: a computational method for analyzing transposable element polymorphism in populations. Nucleic Acids Res. 2014;42:6826–6838. doi: 10.1093/nar/gku323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.