Abstract
A significant limitation when testing the putative single origin of primary plastids and the monophyly of the Archaeplastida supergroup, comprised of the red algae, viridiplants, and glaucophytes, is the scarce nuclear and organellar genome data available from the latter lineage. The Glaucophyta are a key algal group when investigating the origin and early diversification of photosynthetic eukaryotes. However, so far only the plastid and mitochondrial genomes of the glaucophytes Cyanophora paradoxa (strain CCMP 329) and Glaucocystis nostochinearum (strain UTEX 64) have been completely sequenced. Here, we present the complete mitochondrial genomes of Gloeochaete wittrockiana SAG 46.84 (36.05 kb; 33 protein-coding genes, 6 unidentified open reading frames [ORFs], and 28 transfer RNAs [tRNAs]) and Cyanoptyche gloeocystis SAG 4.97 (33.24 kb; 33 protein-coding genes, 6 unidentified ORFs, and 26 tRNAs), which represent two genera distantly related to the “well-known” Cyanophora and Glaucocystis. The mitochondrial gene repertoire of the four glaucophyte species is highly conserved, whereas the gene order shows considerable variation. Phylogenetic analyses of 14 mitochondrial genes from representative taxa from the major eukaryotic supergroups, here including novel sequences from the glaucophytes Cyanophora tetracyanea (strain NIES-764) and Cyanophora biloba (strain UTEX LB 2766), recover a clade uniting the three Archaeplastida lineages; this recovery is dependent on our novel glaucophyte data, demonstrating the importance of greater taxon sampling within the glaucophytes.
Keywords: glaucophyta, phylogenetics, Cyanophora, Plantae
Introduction
It is a widely accepted hypothesis that red algae, viridiplants (green algae and land plants), and glaucophytes, collectively termed the Plantae sensu lato or Archaeplastida (Adl et al. 2005), are monophyletic and descended from a common photosynthetic ancestor (Palmer 2003; Rodríguez-Ezpeleta et al. 2005; Price et al. 2012). Further, the primary plastid common to each of the three Archaeplastida groups is thought to have originated from a single endosymbiotic partnership between a cyanobacterium and a eukaryotic cell approximately 1.5 Ga (Hedges et al. 2004; Yoon et al. 2004). However, the monophyly of the Archaeplastida remains uncertain, as the three groups are not recovered as a monophyletic clade in some nuclear multigene phylogenies (Parfrey et al. 2006; Stiller 2007; Kim and Graham 2008; Hampl et al. 2009; Nozaki et al. 2009; Burki et al. 2012; Yabuki et al. 2014). Out of the red algae, viridiplants, and glaucophytes the latter are by far the least studied group, and this is reflected in the paucity of available organelle genome data. There are currently 118 sequenced mitochondrial genomes (mtDNAs) from viridiplants and 22 from red algae. In contrast, only two glaucophyte mtDNAs have been sequenced: Cyanophora paradoxa and Glaucocystis nostochinearum (Price et al. 2012). The scarcity of glaucophyte genomic data has been a major limitation when testing the monophyly of the Archaeplastida. Resolving this uncertainty is essential to our understanding of the origin and early evolution of all photosynthetic eukaryotes (Bhattacharya et al. 2004; Reyes-Prieto et al. 2007; Keeling 2010).
Glaucophytes are a rare group of algae apparently limited to freshwater environments. Only 4 genera and 13 species have been described (Kies and Kremer 1986), but recent molecular analyses suggest cases of hidden diversity, and possible cryptic species, in the genera Cyanophora and Glaucocystis (Chong et al. 2014). Unlike red algal and viridiplant plastids, glaucophyte photosynthetic organelles exhibit the remains of a peptidoglycan wall between the two organelle membranes (Pfanzagl et al. 1996), and carboxysome-like structures likely associated with carbon concentrating mechanisms localized in the stroma (Mangeney et al. 1987; Burey et al. 2005). These characteristics are thought to be a remnant of the plastid’s cyanobacterial ancestor. The recent sequencing of the C. paradoxa and G. nostochinearum mitochondrial genomes indicates that at least some glaucophyte species contain a large mtDNA gene complement, comparable to the most gene-rich mtDNAs known in green and red algae (Price et al. 2012). Both C. paradoxa and G. nostochinearum mtDNAs contain over 30 protein-coding genes, a large and small subunit ribosomal RNA (rRNA) gene together with a rrn5 rRNA gene, and approximately 25–30 transfer RNA (tRNA) genes that are capable of servicing all the codons in the mitochondrial genes, with the exception of ACN codons (see below, and table 1). A number of unidentified open reading frames (ORFs) are also present in both species. Overall these mtDNAs have characteristics similar to those of many Archaeplastida: They are circular-mapping with no apparent unusual gene or genome structures. However, the diversity of mtDNA gene complements and genome architectures throughout other glaucophyte genera is unknown. Here, we present the first complete organellar genome data from the glaucophytes Cyanoptyche gloeocystis and Gloeochaete wittrockiana, which represent taxa distantly related to the “well-known” Cyanophora and Glaucocystis (Chong et al. 2014), and use mitochondrial sequences from diverse eukaryote groups to assess the presumed monophyly of Archaeplastida.
Table 1.
Gene Repertoires of Glaucophyte Mitochondrial Genomes
 |
Note.—Cp, C. paradoxa; Gn, Glaucocystis nostochinearum; Cg, Cyanoptyche gloeocystis; Gw, Gloeochaete wittrockiana; gray boxes, not present.
aTwo copies, one unique gene sequence.
bThree unique genes.
cFour copies, three unique gene sequences.
dTwo unique genes.
Mitochondrial Genomes of Cyanoptyche gloeocystis and Gloeochaete wittrockiana
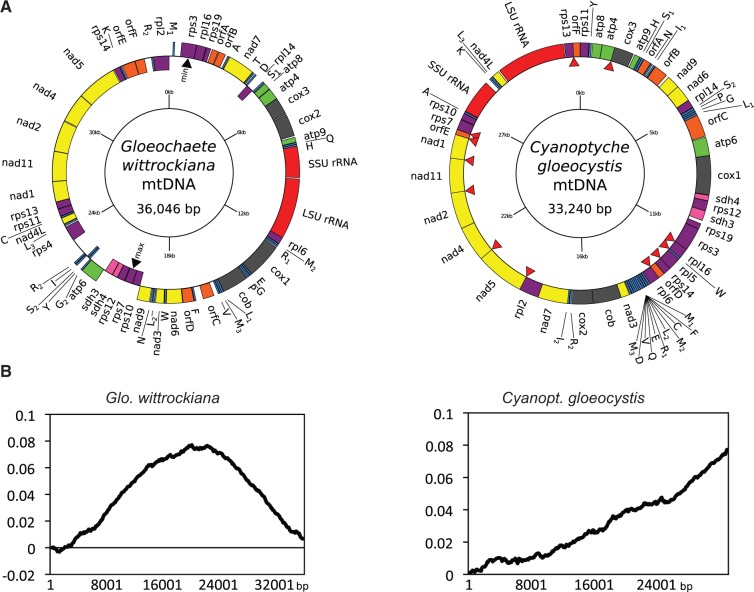
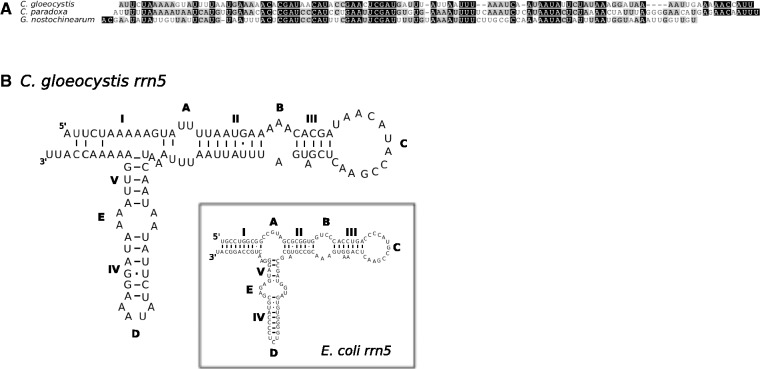
We Illumina-sequenced the complete mtDNAs from Cyanopt. gloeocystis (strain SAG 4.97) and Glo. wittrockiana (strain SAG 48.84). Both assembled mtDNAs consist of a single circular-mapping molecule, 33.24 kb (Cyanopt. gloeocystis) and 36.05 kb (Glo. wittrockiana) in length (fig. 1A and table 2). The Cyanopt. gloeocystis mtDNA contains 33 protein-coding genes, a large and small subunit rRNA gene (LSU and SSU rRNA), and 26 tRNAs corresponding to 23 unique anticodons, whereas the Glo. wittrockiana mtDNA contains 33 protein-coding genes, an LSU and SSU rRNA, and 28 tRNAs corresponding to 26 unique anticodons (table 1, also showing a comparison with C. paradoxa and G. nostochinearum mtDNAs). The gene complement is largely identical between the two mtDNAs, with identified protein products consisting exclusively of respiratory-chain complex subunits and ribosomal proteins. In comparison to Cyanopt. gloeocystis we could not identify rpl5 in Glo. wittrockiana, whereas Cyanopt. gloeocystis apparently lacks rps4. Using hidden Markov model (HMM) profiles of rrn5 sequences from C. paradoxa, G. nostochinearum, and nonglaucophyte taxa (see Materials and Methods), we detected a putative rrn5 gene in Cyanopt. gloeocystis which could be modeled to form a secondary structure consistent with known rrn5 RNAs (fig. 2). However, we could not detect an rrn5 gene in Glo. wittrockiana, perhaps due to sequence divergence in this short gene (∼110 nucleotides). A canonical start codon is used for all protein-coding genes in Cyanopt. gloeocystis and Glo. wittrockiana except for Cyanopt. gloeocystis nad3, which appears to use either the alternative initiation codon UUG or GUG. With this exception the genetic code appears to be standard in both taxa (although see discussion of putative unidentified ORFs below). Both Cyanopt. gloeocystis and Glo. wittrockiana use all three standard stop codons, with UAA the most common, whereas only UAA and UAG are employed in C. paradoxa and G. nostochinearum. As in C. paradoxa and G. nostochinearum, no introns were found in Glo. wittrockiana mtDNA genes, whereas a single 212-nt intron occurs in Cyanopt. gloeocystis LSU rRNA. This intron is identified as group I (derived from subgroup A) by RNAweasel (Lang et al. 2007), and is positioned between nucleotides 2500 and 2501 relative to the Escherichia coli LSU rRNA; this corresponds to a single-stranded region of the peptidyl-transferase center, within domain V.
Fig. 1.—
Gloeochaete wittrockiana and Cyanoptyche gloeocystis mtDNAs and cumulative GC-skew plots. (A) Circular gene maps of Glo. wittrockiana (strain SAG 46.84) and Cyanopt. gloeocystis (strain SAG 4.97) mitochondrial genomes. Colored bars identify types of genes/ORFs: rRNAs (red), tRNA (blue), ribosomal proteins (purple), complex I (yellow), complex II (pink), complex III and IV (dark gray), complex V (green), uORFs (orange). Red arrowheads indicate overlaps between genes. (B) Cumulative GC-skew plots for Glo. wittrockiana and Cyanopt. gloeocystis mtDNAs. y axis: cumulative GC skew, x axis: position in the genome.
Table 2.
Characteristics of Glaucophyte Mitochondrial Genomes
| Strain | Size (kb) | Shape | Total A+T Content (%) | A+T Content Proteinsa (%) | A+T Content tRNAs/ rRNAs (%) | A+T Content Noncodinga (%) | % Intergenic Sequencea (%) | Average Intergenic Size (bp)a,b | Intron Type | Genetic Code |
|---|---|---|---|---|---|---|---|---|---|---|
| Cg | 33.2 | Circular-mapping | 71.5 | 72.8/72.3 | 60.3/65.3 | 83.8/79.1 | 3.9/12.1 | 23.2/81.6 (1–202/1–956) | Group I | Standard (UUG/GUG start in nad3) |
| Gw | 36.1 | Circular-mapping | 69.5 | 70.0/69.7 | 58.3/62.6 | 79.3/76.5 | 11.3/18.3 | 59.9/108.1 (1–833/1–1,535) | / | Standard (UUG start in uORF) |
| Cp | 51.6 | Circular-mapping | 74.0 | 75.2/74.4 | 57.7/64.1 | 80.8/78.7 | 15.3/34.8 | 102.6/256.7 (1–665/1–5,026) | / | Standard |
| Gn | 34.1 | Circular-mapping | 74.3 | 75.5/75.1 | 59.8/66.8 | 85.6/85.0 | 8.0/11.2 | 47.9/68.4 (5–504/5–820) | / | Standard |
Note.—Cp, Cyanophora paradoxa; Gn, Glaucocystis nostochinearum; Cg, Cyanoptyche gloeocystis; Gw, Gloeochaete wittrockiana.
aValues including uORFs as bona fide genes/values not including uORFs as bona fide genes.
bValues in parentheses are the minimum and maximum intergenic sizes.
Fig. 2.—
Putative rrn5 sequence from Cyanoptyche gloeocystis mtDNA, aligned with rrn5 sequences from C. paradoxa and G. nostochinearum (A); putative secondary structure of Cyanopt. gloeocystis rrn5 (B). The inset shows the proposed structure of the E. coli rrn5 for reference. Helices are shown by Roman numerals (I–V), and single-stranded regions are shown by upper case letters (A–D).
In both Cyanopt. gloeocystis and Glo. wittrockiana, mtDNA-encoded tRNAs are sufficient to decode all codons used in the mtDNA-encoded proteins (table 1), with the exception of ACN codons (Threonine). Gloeochaete wittrockiana mtDNA encodes a trnT(ggu) gene which decodes ACC and ACU codons, but we could not detect a tRNA to decode ACA and ACG using a variety of search strategies (tRNAscan-SE, ARWEN, hidden Markov and covariance model profiles built from trnT-gene alignments; see Materials and Methods). Cyanoptyche gloeocystis mtDNA appears to lack trnT genes entirely. This is similar to the situation in the C. paradoxa and G. nostochinearum mtDNAs, where C. paradoxa lacks any trnT gene whereas G. nostochinearum encodes trnT(ggu) (Price et al. 2012). As for these latter two species it is possible that Cyanopt. gloeocystis relies on import of cytoplasmic trnT, or that differential RNA editing of the anticodon might produce two rRNAs from a single mitochondrial gene as reported in opossum (Borner et al. 1996).
In addition to known genes, both mtDNAs also contain a number of ORFs of unknown function (unidentified ORFs [uORFS]), here defined as an ORF greater than 200 bases with either canonical start and stop codons, or an alternative start codon that has been previously identified in other organisms (see https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi?mode=c, last accessed October 9, 2014). Both Glo. wittrockiana and Cyanopt. gloeocystis contains six such uORFs with an average size of 421 and 431 bases, respectively. In comparison, C. paradoxa and G. nostochinearum mtDNAs contain ten and two uORFs, respectively (table 1). To determine whether these uORFs have characteristics similar to canonical genes, and hence might indeed represent transcribed protein-coding regions, we compared their A+T content, amino acid composition, and start and stop codon usage (supplementary tables S1–S3, Supplementary Material online). With regard to A+T content and amino acid composition, we could not distinguish a pattern that clearly delineated between canonical genes and uORFs in any glaucophyte species. In Cyanopt. gloeocystis uORFs use standard start codons and all three stop codons, consistent with canonical genes in this mtDNA. In comparison, canonical genes in C. paradoxa and G. nostochinearum use only UAA and UAG stop codons, whereas one uORF in each species terminates with an UGA stop codon, perhaps indicating that these latter uORFs are not genuine genes. Similarly, one uORF in Glo. wittrockiana appears to use the alternative start codon UUG, whereas canonical genes exclusively use AUG start codons. To further investigate the uORFs, each sequence was scanned for conserved protein domains by comparison to the Pfam database using HMMscan. Using manually curated cutoff scores (gathering thresholds) provided in Pfam domain HMM files, no uORFs returned significant hits to any domain. Finally, pairwise tBLASTx comparisons using an e value cutoff of 10−3 did not reveal any similarity between uORFs among the four glaucophyte taxa, although within-species BLAST (Basic Local Alignment Search Tool) comparisons revealed some similarity between C. paradoxa orf166 and orf229 #2 (e value 1.21×10−24; orf533, orf544, and orf427 also had regions of similarity with each other, but this is due to repeats in the C. paradoxa mtDNA). BLAST searching all uORF sequences against the National Center for Biotechnology Information expressed sequence tag (EST) database did recover several ESTs matching one uORF, orf535 in C. paradoxa (1,608 nucleotides). The longest of these ESTs (GenBank accession number ES236117) is 5′ truncated by 1,079 bases in comparison to orf535 sequence, and the 3′-end is truncated by 138 bp; this latter truncation occurs at a region of 16 consecutive adenosine nucleotides in orf535, and likely corresponds to mispriming of the oligo-dT primer used to prime first-strand cDNA synthesis. The presence of an EST for orf535 suggests that this putative gene does code for a protein; another possibility is that the EST represents a fragment of a larger polycistronic transcript, and may not be coding sequence. Overall, it is unclear from the available evidence whether the majority of uORFs in each glaucophyte species are in fact protein-coding genes, or ORFs arising by chance.
When considering uORFs as bona fide genes, gene density is high in both Cyanopt. gloeocystis and Glo. wittrockiana mtDNAs with intergenic sequence comprising only 3.9% and 11.3% of total sequence, respectively (12.1% and 18.3% when uORF sequence is considered intergenic; see table 2). Although genes occur on both mtDNA strands in Glo. wittrockiana, C. paradoxa, and G. nostochinearum, in Cyanopt. gloeocystis all genes are encoded on the same strand (fig. 1A). This latter gene arrangement is not common but is known in mtDNAs from diverse eukaryotes including some amoebozoans, choanoflagellates, green algae, fungi, haptophytes, cryptophytes, bivalves, annelid worms, brachiopods, and sponges. (Burger et al. 1995, 2003; Denovan-Wright et al. 1998; Turmel et al. 1999; Ogawa et al. 2000; Helfenbein et al. 2001; Sánchez Puerta et al. 2004; Dreyer and Steiner 2006; Kim et al. 2008; Rosengarten et al. 2008; Wu et al. 2009). Such an organization raises the possibility that all genes are transcribed as a single polycistronic transcript from a single promoter, with subsequent endonucleolytic processing producing single-gene transcripts. However, the Cyanopt. gloeocystis mtDNA is more highly compacted than the other three glaucophyte species, with 12 cases of overlapping genes (fig. 1A, red arrows; overlaps range from 4 to 32 nucleotides, and 4 of 12 involve uORFs). Hence, if polycistronic transcription occurs in Cyanopt. gloeocystis, subsequent processing must be regulated in a manner that produces full-length transcripts for each overlapping gene. No overlapping genes were found in the Glo. wittrockiana mtDNA, whereas C. paradoxa and G. nostochinearum contain three (7–32 nucleotides, two involving an uORF) and four cases (10–31 nucleotides, one involving an uORF), respectively. An overlap between the 3′-end of rps3 and the 5′-end of rpl16 is shared between Cyanopt. gloeocystis, C. paradoxa, and G. nostochinearum, whereas in Glo. wittrockiana rps3 terminates nine nucleotides upstream of rpl16.
Cyanoptyche gloeocystis mtDNA is also unique among the four glaucophyte mtDNAs examined in having a linear cumulative GC-skew (fig. 1B). This skew pattern is typical of circular plasmids which replicate through rolling circle replication (RCR) (Arakawa et al. 2009), and is consistent with the linear skew observed for the mtDNA of the yeast Candida glabrata, which also likely replicates through RCR (Maleszka et al. 1991; Burger et al. 2013). In contrast, cumulative GC-skew plots for each of the other three glaucophytes reveal clear bimodal skew curves with distinct global minima and maxima (fig. 1B and supplementary fig. S1, Supplementary Material online). This latter pattern is typical of bidirectional theta genome replication in bacteria, where the global minimum of the curve usually represents the replication origin, and the global maximum represents the replication termination point (Grigoriev 1998). In the Glo. wittrockiana mtDNA, genes encoded from nucleotides 1 to approximately 19500 (relative to fig. 1A) are transcribed largely on one strand, whereas for the remainder of the genome they are transcribed mostly on the opposite strand. These switch points approximately coincide with the global minimum and maximum values of the cumulative GC-skew (fig. 1A, black arrows). A similar pattern is evident for C. paradoxa and G. nostochinearum (data not shown), perhaps indicating selective pressure to organize genes such that their replication direction is the same as their transcription orientation, as observed in some groups of bacteria (Rocha 2004); this is consistent with the single-strand gene organization in the Cyanopt. gloeocystis mtDNA, where putative RCR would occur in a single direction.
With the exception of a cluster of five consecutive nad genes (nad1, nad11, nad2, nad4, nad5), there is no obvious conservation of gene order between Cyanopt. gloeocystis and Glo. wittrockiana. This is true for all four glaucophyte mtDNAs, which, despite having very similar gene complements, are highly rearranged in comparison to one other (fig. 3). A distinctive feature of the Cyanopt. gloeocystis mtDNA is a bank of 11 consecutive tRNAs. In C. paradoxa, the mtDNA contains a number of repeat regions not present in the other three taxa, and these account for the larger size of this mtDNA. Finally, like C. paradoxa and G. nostochinearum, both Cyanopt. gloeocystis and Glo. wittrockiana mtDNAs are A+T rich, with an average A+T content of 71.5% and 69.5%, respectively (table 2). Intergenic regions have the highest A+T content at 83.8% and 79.3%, respectively (values calculated considering uORFS as bona fide genes), whereas protein-coding sequences are somewhat lower (72.8% and 70.0%, respectively) with tRNAs and rRNAs lower still (60.3% and 65.3%, and 58.3% and 62.6%, respectively).
Fig. 3.—
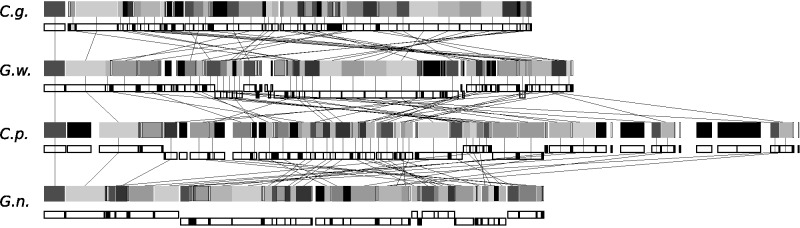
Gene rearrangements in glaucophyte mtDNAs. Individual genes are shown as grayscale rectangles, with genome organization and gene strand location shown below. Black lines connect genes found in each mtDNA (tRNA genes excluded). Rectangles with no lines represent unique ORFs, or genes not found in all taxa. C.g., Cyanopt. gloeocystis; G.w., Glo. wittrockiania; C.p., C. paradoxa; G.n., G. nostochinearum.
Testing the Archaeplastida Monophyly Using mtDNA-Encoded Genes
The Archaeplastida monophyly has been recovered in several phylogenetic studies of plastid genes (Rodríguez-Ezpeleta et al. 2005; Qiu et al. 2012), nuclear genes (Rodríguez-Ezpeleta et al. 2005; Hackett et al. 2007; Burki et al. 2009; Zhao et al. 2012), and genome-scale analyses (Price et al. 2012). Other studies have not recovered the Archaeplastida groups united in a single clade (Parfrey et al. 2006; Nozaki et al. 2009; Burki et al. 2012; Yabuki et al. 2014), and so the monophyly is still contentious (Stiller 2007; Howe et al. 2008). Mitochondrial sequences are, in principle, an additional source of phylogenetic information to investigate Archaeplastida monophyly. In contrast to plastid data, the use of mtDNA genes allows the inclusion of a broader sample of eukaryotes (i.e., not just plastid-bearing organisms). Further, mtDNA data are likely less affected than nuclear data by events of horizontal or endosymbiotic gene transfer. A significant weakness of all analyses to date has been the lack of comparative and comprehensive genomic data from diverse glaucophyte taxa. In this study, we tested whether mtDNA nucleotide and amino acid data support Archaeplastida monophyly using mitochondrial genes from our additional glaucophyte taxa, Cyanopt. gloeocystis and Glo. wittrockiana, together with novel sequences from three Cyanophora species: C. paradoxa (strain NEIS 763), Cyanophora tetracyanea (NEIS 764), and Cyanophora biloba (UTEX 2766), all available from ongoing sequencing projects in our laboratory. We used conceptual protein translations of 14 glaucophyte mitochondrial genes (atp6, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7) as queries to collate homologous sequences from representative taxa of the major eukaryote “supergroups” (opisthokonts, amoebozoans, haptophytes, cryptophytes, SAR, Archaeplastida and excavates; supplementary table S4, Supplementary Material online), and generated a concatenated multiple-gene alignment (see supplementary table S5, Supplementary Material online, for missing data percentages). Our analysis is the first to include mitochondrial data from a broad taxon sampling of the Archaeplastida, including multiple glaucophyte genera and species, together with viridiplantae representatives from Chlorophyta, Streptophyta, and “Prasinophyta,” and red algae from Florideophyceae, Bangiophyceae, and Cyanidiales. Using our expanded data set, we performed both maximum likelihood (ML) and Bayesian phylogenetic analyses.
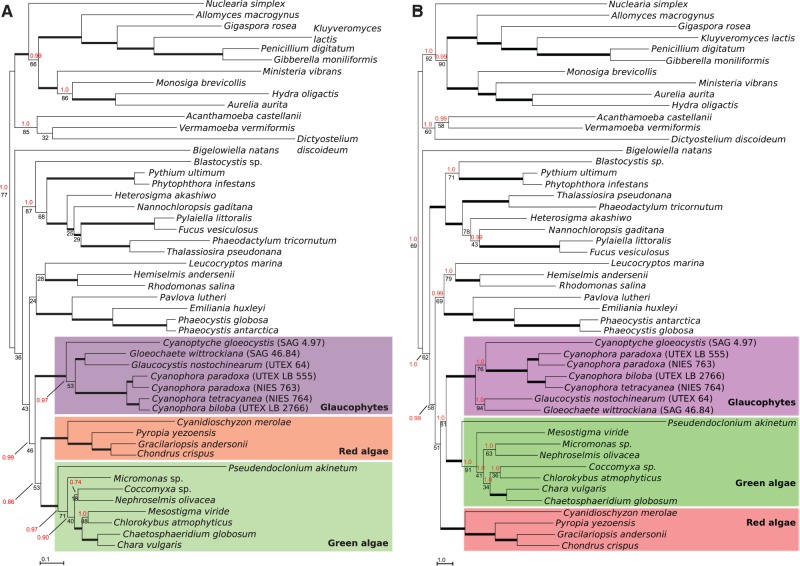
In our initial analyses of amino acid data excavate taxa were polyphyletic, most likely due to phylogenetic artifacts (see supplementary fig. S2, Supplementary Material online, and Hampl et al. 2009 for details), and so we removed all excavate sequences from our alignment. We also excluded alveolate sequences (ciliates, dinoflagellates, and apicomplexans) due to extremely long branches observed in single-gene ML analyses (trees not shown). The ML tree of the resulting alignment is shown in figure 4A. A “unikont” clade is recovered (77% bootstrap support [BS] and 1.0 Bayesian posterior probability [PP]), distinguishing Amoebozoa and opisthokonts with moderate to strong branch support (≥85% BS, 1.0 PP for each clade). A stramenopile clade is moderately supported (87% BS, 1.0 PP), and the only rhizarian (Bigelowiella natans) in our data set branches separately. Cryptophytes, katablepharids, and haptophytes (members of the “Hacrobia,” although the monophyly of this group has recently been questioned [Burki et al. 2012]) are resolved in a single clade with no branch support. A clade uniting the three Archaeplastida groups is recovered with no BS but a high posterior probability (46% BS, 0.99 PP). Hence, despite the low BS, phylogenetic conclusions with our mitochondrial data are consistent with the monophyly of the Archaeplastida and a single origin of their primary plastids, in tentative agreement with previous analyses using plastid-encoded genes (Rodríguez-Ezpeleta et al. 2005; Price et al. 2012; Qiu et al. 2012) and some analyses using nuclear-encoded genes (Rodríguez-Ezpeleta et al. 2005; Hackett et al. 2007; Burki et al. 2009; Zhao et al. 2012), but in contrast with other nuclear gene-based analyses (Parfrey et al. 2006; Nozaki et al. 2009; Burki et al. 2012; Yabuki et al. 2014). It is important to note that Archaeplastida monophyly is only recovered in our analyses when we include our additional glaucophyte data; ML and Bayesian analyses performed after removal of all glaucophyte taxa other than the previously published C. paradoxa and G. nostochinearum failed to recover the Archaeplastida as a single clade (supplementary figs. S3 and S4, Supplementary Material online), demonstrating the importance of greater taxon sampling within the glaucophytes. In previous analyses using mitochondrial genes from diverse eukaryotes, glaucophyte sequences were either unavailable (Oudot-Le Secq et al. 2006) or included only C. paradoxa and G. nostochinearum (Burger et al. 2013). In the Bayesian tree presented in this latter study all short basal branches with less than 60% support were collapsed, including those for the Archaeplastida groups, and hence it is unclear whether the Archaeplastida formed a clade.
Fig. 4.—
Phylogenetic analysis of 14 mitochondrial genes. ML phylogenetic tree estimated from conceptual translations (A) and nucleotide sequences (B) of atp6, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, and nad7. Numbers above and below branches represent Bayesian PP and RAxML BS proportions, respectively. Thick branches are supported by PP and BS values greater than 95. Branches without PP values were only recovered in the ML analysis. Branch lengths are proportional to the number of substitutions per site.
To further test the monophyly of the Archaeplastida using our amino acid data set, we manually defined tree topologies that disrupted the Archaeplastida monophyly and performed approximately unbiased (AU) topology tests. Most of these alternative topologies were statistically worse than the best estimated ML tree (P values < 0.05; supplementary table S6, Supplementary Material online). Exceptions to this were a topology placing red algae as a sister branch to haptophytes outside the Archaeplastida, and a topology uniting red algae and haptophytes within the Archaeplastida (supplementary fig. S5A and B, Supplementary Material online), which were not rejected by the AU test (P value 0.116 and 0.062, respectively). Interestingly, in phylogenetic analyses using nuclear genes haptophytes have also been observed branching within the Archaeplastida with robust BS, as sister to either viridiplants or red algae, and this relationship does not appear to be due to long-branch attraction artifacts (Hampl et al. 2009). As discussed by Hampl et al. (2009), the position of haptophytes in their analyses might be due to phylogenetic signal in genes endosymbiotically or horizontally transferred from primary algal lineages. However, although mtDNA to mtDNA horizontal gene transfer is known to occur in land plants (Bergthorsson et al. 2004; Archibald and Richards 2010; Xi et al. 2013), to our knowledge there is no conclusive evidence for this process occurring in algae or microbial eukaryotes. Further, alternative tree topologies grouping glaucophytes with stramenopiles or Bigelowiella (supplementary fig. S5C and D, Supplementary Material online, respectively) were not rejected by the AU test (supplementary table S4, Supplementary Material online). The inability to reject these alternative topologies is likely due to the relatively weak phylogenetic signal of this mitochondrial gene set.
To evaluate possible systematic biases caused by fast-evolving sites in our alignment, we calculated site-specific substitution rates for our concatenated data set and progressively removed the fastest-evolving sites to retain a minimum percentage length (95%, 80%, and 70%) of our 14-protein original alignment. ML analyses of these alignments recovered tree topologies very similar to figure 3, with comparable bootstrap values for the major groups (supplementary table S7A, Supplementary Material online); for the 80% and 70% alignments the branching position of rhizarians and katablepharids did differ, but this is difficult to interpret given the severely limited taxon sampling for these groups in our data set (one taxon each). For the Archaeplastida, removal of fast-evolving sites caused a marginal decrease in BS (46%, 47%, 35%, and 36% BS for the 100%, 95%, 80%, and 70%-length alignments, respectively). These results suggest that the branch stability (i.e., support value) for the opisthokont, amoebozoan, stramenopile, and Archaeplastida clades is not strongly affected by systematic biases arising from fast-evolving sites in our mitochondrial-gene data set.
Finally, to determine the effect of using mtDNA nucleotide data rather than derived protein sequences on support for Archaeplastida monophyly, we generated a DNA alignment using the same taxa and gene positions as our amino acid alignment and performed Bayesian and ML analyses. The topology of the resulting ML tree is very similar (fig. 4B), with recovery of the amoebozoa, opisthokonts, stramenopiles, Hacrobia, and Archaeplastida. Support for Archaeplastida clade is comparably low (51% BS) to the BS obtained with the amino acid data set, and systematic removal of fast-evolving sites from the nucleotide alignment did not increase the branch support (51%, 52% and 47% BS for the 100%, 95% and 80%-length alignment, respectively, supplementary table S7B, Supplementary Material online; in the 70% alignment, the Hacrobia clade branched within the Archaeplastida as sister to the red algae with 44% BS). To test for sequence composition biases specific to Archaeplastida taxa that might account for the poor resolution of this clade, we examined both the amino acid and nucleotide data sets using the compositional homogeneity test implemented in PhyloBayes. Results show that although the majority of sequences from the Archaeplastida taxa do indeed have compositional biases that violate assumptions of the models used, this is also true for 42 and 43 of 49 taxa (amino acid and nucleotide alignments, respectively; supplementary table S8, Supplementary Material online). Given that we recovered good support for many major eukaryotic groups despite this issue, it seems unlikely that compositional biases are a major contributor toward the low support values for the Archaeplastida clade. Overall, ML phylogenetic analyses of both the amino acid and nucleotide sequences present a similar picture, with weak support for Archaeplastida monophyly.
In both ML and Bayesian analyses of the full amino acid data set glaucophytes appear as the earliest diverging lineage within the Archaeplastida clade, and this is consistent with plastid-gene phylogenies (Rodríguez-Ezpeleta et al. 2005; Qiu et al. 2012). However, the early-diverging position of glaucophytes is ambiguous in our analyses given that the branch uniting red algae and viridiplants is only weakly supported (53% BS, 0.86 PP). Manually defined tree topologies that alternatively positioned red algae or viridiplants as the earliest branch within Archaeplastida were not rejected by AU tests (P values > 0.05; supplementary table S6, Supplementary Material online). Moreover, ML analyses using nucleotide rather than amino acid sequences show red algae branch earliest, with 81% BS for the branch uniting glaucophytes and green algae (fig. 4B), whereas in the Bayesian analysis the Hacrobia branch within the Archaeplastida as sister to the red algae (0.99 PP for both the Archaeplastida + Hacrobia and Hacrobia + red algae clades, tree not shown). The position of Cyanopt. gloeocystis within the glaucophyte clade is also equivocal. In ML analyses of amino acid sequences Cyanopt. gloeocystis branches first, but the branch uniting the strongly supported Cyanophora clade with the Glo. wittrockiana/G. nostochinearum clade receives only 53% BS (0.97 PP). In contrast, ML analysis of nucleotide data shows Cyanopt. gloeocystis branching as sister to the Cyanophora clade with 76% BS (1.0 PP).
To further examine intergenus relationships within the glaucophyte clade we compared amino acid and stop codon usage between the taxa (supplementary tables S3 and S9, Supplementary Material online), but could not detect a pattern clarifying the position of Cyanopt. gloeocystis; Cyanopt. gloeocystis and G. nostochinearum uniquely share an UAG stop codon in rps7, but G. nostochinearum and Glo. wittrockiana uniquely share an UAG stop in rps14. In terms of gene synteny, Glo. wittrockiana and Cyanopt. gloeocystis uniquely share a large syntenic region encompassing nad5, nad4, nad2, nad11, and nad1. Although this might suggest that these two taxa share a more recent common ancestor with each other than with the other glaucophytes, this inference is not supported by phylogenetic analyses of both amino acid and nucleotide data which group Glo. wittrockiana and G. nostochinearum together with strong ML and Bayesian support (fig. 4). The synteny observed in Glo. wittrockiana and Cyanopt. gloeocystis might reflect a conserved gene order that was inherited from a distant common ancestor and subsequently rearranged in other glaucophytes, or it could reflect convergence.
Conclusions
Sequencing of the complete mtDNAs of Cyanopt. gloeocystis and Glo. wittrockiana shows that, despite a highly conserved gene repertoire in glaucophytes, the order of mitochondrial genes is highly rearranged in different genera. Future studies of mtDNAs from more closely related species will be required to determine the degree of gene rearrangement at the intragenus level; such rearrangements could help clarify cases of putative cryptic diversity within the genera Cyanophora and Glaucocystis, as noted recently by Chong et al. (2014).
Our phylogenetic analyses are the first to use mtDNA data from a broad taxon sampling across eukaryotes, including data from all available glaucophyte genera. We provide a novel line of genomic evidence consistent with the monophyly of the Archaeplastida, and with a single origin of primary plastids within this supergroup. The branching history of the Archaeplastida lineages is a question that still needs a clear answer. Studies of plastid-encoded genes and plastid-targeted proteins have alternatively suggested that glaucophytes (Rodríguez-Ezpeleta et al. 2005; Reyes-Prieto and Bhattacharya 2007) or viridiplants (Deschamps and Moreira 2009) are the earliest-diverging group, whereas genome-scale analyses indicate a sister relationship between viridiplantae and glaucophytes (Price et al. 2012). Our mitochondrial data do not provide a conclusive result. However, although mtDNA data have been largely overlooked for resolving deep nodes of eukaryotic evolution (due to typically accelerated substitution rates in mtDNA genes), our analyses do recover most of the major supergroups with moderate to strong branch support. The current inability to resolve the relationships among the Archaeplastida lineages with molecular phylogenetics using multilocus data sets suggests that, after the origin of primary plastids, early Archaeplastida diversification followed accelerated evolutionary paths (e.g., rapid radiations or different rates of gene duplications/divergence/losses/replacements that result in potential unidentified paralogies) that cannot be reconstructed from the limited and conflicting phylogenetic signals present in the genomes of highly derived extant lineages. Additional studies of genomes across diverse glaucophyte taxa could help resolve this problem with increased scope for phylogenetic analyses and the potential identification of diagnostic genome characteristics, including gene content and gene and genome architectures.
Materials and Methods
DNA Extraction, Sequencing, mtDNA Assembly, and Gene Annotation
Total DNA was extracted from Cyanopt. gloeocystis (strain SAG 4.97) and Glo. wittrockiana (strain SAG 48.84) using standard phenol/chloroform-based methods. Total DNA was sequenced using Illumina sequencing technology (HiSeq2000 at the Roy J. Carver Center for Comparative Genomics, University of Iowa) producing approximately 60 × 106 and approximately 82 × 106 100-base reads from approximately 450-nt-insert paired-end libraries, for Cyanopt. gloeocystis and Glo. wittrockiana, respectively. Reads were assembled using the parallel MPI-based assembler Ray v2.2.0 (Boisvert et al. 2010) considering alternative k-mer lengths (21, 27, 31, 37). Contigs containing mitochondrial genes were identified by BLAST similarity searches using the reported mitochondrial gene sequences of Cyanophora and Glaucocystis (Price et al. 2012) as queries, and were assembled into scaffolds comprising complete genomes using read-mapping approaches with Geneious v6 (Biomatters). The average Illumina read coverage for the complete mtDNAs was 82 read/site for Cyanopt. gloeocystis (min 3, max 349), and 760 for Glo. wittrockiana (min 555, max 1,029). tRNA genes were predicted with the tRNAscan-SE Search Server (http://lowelab.ucsc.edu/tRNAscan-SE/, last accessed October 9, 2014), and additional tRNA searches were conducted using ARWEN (http://mbio-serv2.mbioekol.lu.se/ARWEN/, last accessed October 9, 2014). To search for trnT genes we used WAR (webserver for aligning structural RNAs: http://genome.ku.dk/resources/war/, last accessed October 9, 2014) to produce an alignment of available trnT sequences from glaucophytes and green algae, annotated with the consensus secondary structure. This alignment was used to generate a covariance model (CM) using the cmbuild and cmcalibrate tools from the Infernal software package v1.1.1 (Nawrocki and Eddy 2013), and the trnT CM profile was used to search the glaucophyte mtDNAs using the cmsearch tool. mtDNAs were also searched using an HMM profile generated from the trnT WAR alignment with the HMMER software package v3.1b1 (http://hmmer.janelia.org/, last accessed October 9, 2014). To search for rrn5 genes we generated an HMM profile from an alignment of available glaucophyte, brown algal, and Jakobid sequences (latter two taxa chosen based on sequence similarity and ease of alignment). Modeling of the Cyanopt. gloeocystis rrn5 secondary structure was guided by a minimum free energy structure calculated using RNAshapes (http://bibiserv.techfak.uni-bielefeld.de/rnashapes/submission.html, last accessed October 9, 2014). Mitochondrial genome figures were initially generated using GenomeVx (http://wolfe.ucd.ie/GenomeVx/, last accessed October 9, 2014). Cumulative GC-skew plots were generated with the GenSkew Java application (http://genskew.csb.univie.ac.at/, last accessed October 9, 2014), using a window and step size of 100 nucleotides.
Phylogenetic Analyses
Individual gene alignments for each of the 14 genes were generated using conceptual protein translations in Geneious v6.0, and alignments were manually edited to discard ambiguous and poorly aligned regions. For each alignment the best-fit protein substitution model was selected with ProtTest (http://darwin.uvigo.es/software/prottest.html, last accessed October 9, 2014), using the AIC model selection criterion. Ten individual ML trees were estimated with RAxML v7.2.6 (Stamatakis 2006) using the corresponding best-fit model, and the tree with the highest likelihood value was selected. Branch support was assessed with 500 nonparametric bootstrap replicates, and single-protein trees were assessed, as described above. Subsequently, all alignments were concatenated to produce a multigene supermatrix 3,267 amino acids long, and the best-fit model was determined (LGF). ML trees were estimated with RAxML as described for single-gene alignments.
Bayesian analyses of the concatenated alignment were performed with PhyloBayes MPI v1.4 (Lartillot et al. 2013) using the LG model with four gamma categories modeling the relative substitution rates across sites. Two independent Markov chain Monte Carlo runs were performed for a total of 13,000 cycles. Convergence was assessed by comparing the difference in frequency for all tree bipartitions (<0.1), with the exception of the first 2,600 cycles which were discarded as burn-in. Trees from both chains were sampled every ten cycles after burn-in, and a majority-rule posterior consensus tree was generated.
A nucleotide alignment was generated using the concatenated protein alignment as a guide, using the same taxa, genes, and gene positions. Partitions and the best-fit model (GTRI) were selected using Partition Finder (Lanfear et al. 2012), and ML and Bayesian trees were estimated with RAxML and PhyloBayes as above (10,500 cycles, 2,100 burn-in for Phylobayes analysis). Phylogenetic trees were visualized and figures prepared using Archaeopteryx beta v0.997 (Han and Zmasek 2009). Compositional homogeneity tests were performed using PhyloBayes v3.3 with the ppred –comp option.
Estimation of Site-By-Site Substitution Rate Variability
Per-site substitution rates were calculated for our alignments using HyPhy (Pond et al. 2005), and the fastest evolving sites were removed using SiteStripper v.1.01 (http://www.phycoweb.net/software/SiteStripper/index.html, last accessed October 9, 2014), leaving a percentage length of the original alignment (95%, 80%, and 70%).
Hypothesis Testing
To test alternative hypotheses to the Archaeplastida monophyly, we manually generated 31 competing phylogenetic trees (supplementary fig. S4, Supplementary Material online) with MacClade v4.08a (http://macclade.org/index.html, last accessed October 9, 2014), using the ML tree generated from the protein alignment as a reference. Sitewise log-likelihoods of the 31 alternative topologies were calculated with RAxML. AU test P values (Shimodaira 2002) were computed with Consel v0.20 (Shimodaira and Hasegawa 2001).
Supplementary Material
Supplementary figures S1–S5 and tables S1–S9 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This research work was supported by the Natural Sciences and Engineering Research Council of Canada (grant number: 402421-2011), the Canada Foundation for Innovation (grant number: 28276), and the New Brunswick Innovation Foundation (grant number: RIF2012-006) awarded to A.R.P.
Literature Cited
- Adl SM, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Arakawa K, Suzuki H, Tomita M. Quantitative analysis of replication-related mutation and selection pressures in bacterial chromosomes and plasmids using generalised GC skew index. BMC Genomics. 2009;10:640. doi: 10.1186/1471-2164-10-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM, Richards TA. Gene transfer: anything goes in plant mitochondria. BMC Biol. 2010;8:147. doi: 10.1186/1741-7007-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, Palmer JD. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc Natl Acad Sci U S A. 2004;101:17747–17752. doi: 10.1073/pnas.0408336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Yoon HS, Hackett JD. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays. 2004;26:50–60. doi: 10.1002/bies.10376. [DOI] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 2010;17:1519–1533. doi: 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GV, Morl M, Janke A, Paabo S. RNA editing changes the identity of a mitochondrial tRNA in marsupials. EMBO J. 1996 15:5949–5957. [PMC free article] [PubMed] [Google Scholar]
- Burey SC, et al. The central body of the cyanelles of Cyanophora paradoxa: a eukaryotic carboxysome? Can J Bot. 2005;83:758–764. [Google Scholar]
- Burger G, Forget L, Zhu Y, Gray MW, Lang BF. Unique mitochondrial genome architecture in unicellular relatives of animals. Proc Natl Acad Sci U S A. 2003;100:892–897. doi: 10.1073/pnas.0336115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray MW, Forget L, Lang BF. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 2013;5:418–438. doi: 10.1093/gbe/evt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Plante I, Lonergan KM, Gray MW. The mitochondrial DNA of the amoeboid protozoon, Acanthamoeba castellanii: complete sequence, gene content and genome organization. J Mol Biol. 1995;245:522–537. doi: 10.1006/jmbi.1994.0043. [DOI] [PubMed] [Google Scholar]
- Burki F, et al. Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, telonemia and centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol Evol. 2009;1:231–238. doi: 10.1093/gbe/evp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Okamoto N, Pombert J-F, Keeling PJ. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc Biol Sci. 2012;279:2246–2254. doi: 10.1098/rspb.2011.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Jackson C, Kim JI, Yoon HS, Reyes-Prieto A. Molecular markers from different genomic compartments reveal cryptic diversity within glaucophyte species. Mol Phylogenet Evol. 2014;76:181–188. doi: 10.1016/j.ympev.2014.03.019. [DOI] [PubMed] [Google Scholar]
- Denovan-Wright EM, Nedelcu AM, Lee RW. Complete sequence of the mitochondrial DNA of Chlamydomonas eugametos. Plant Mol Biol. 1998;36:285–295. doi: 10.1023/a:1005995718091. [DOI] [PubMed] [Google Scholar]
- Deschamps P, Moreira D. Signal conflicts in the phylogeny of the primary photosynthetic eukaryotes. Mol Biol Evol. 2009;26:2745–2753. doi: 10.1093/molbev/msp189. [DOI] [PubMed] [Google Scholar]
- Dreyer H, Steiner G. The complete sequences and gene organisation of the mitochondrial genomes of the heterodont bivalves Acanthocardia tuberculata and Hiatella arctica—and the first record for a putative Atpase subunit 8 gene in marine bivalves. Front Zool. 2006;3:13. doi: 10.1186/1742-9994-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JD, et al. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol Biol Evol. 2007;24:1702–1713. doi: 10.1093/molbev/msm089. [DOI] [PubMed] [Google Scholar]
- Hampl V, Hug L, Leigh J. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups.”. Proc Natl Acad Sci U S A. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfenbein KG, Brown WM, Boore JL. The complete mitochondrial genome of the articulate brachiopod Terebratalia transversa. Mol Biol Evol. 2001;18:1734–1744. doi: 10.1093/oxfordjournals.molbev.a003961. [DOI] [PubMed] [Google Scholar]
- Howe CJ, Barbrook AC, Nisbet RER, Lockhart PJ, Larkum AWD. The origin of plastids. Philos Trans R Soc Lond B Biol Sci. 2008;363:2675–2685. doi: 10.1098/rstb.2008.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365:729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kies L, Kremer BP. Typification of the Glaucocystophyta. Taxon. 1986;35:128–133. [Google Scholar]
- Kim E, et al. Complete sequence and analysis of the mitochondrial genome of Hemiselmis andersenii CCMP644 (Cryptophyceae) BMC Genomics. 2008;9:215. [Google Scholar]
- Kim E, Graham LE. EEF2 analysis challenges the monophyly of Archaeplastida and Chromalveolata. PLoS One. 2008;3:e2621. doi: 10.1371/journal.pone.0002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- Lang BF, Laforest M-J, Burger G. Mitochondrial introns: a critical view. Trends Genet. 2007;23:119–125. doi: 10.1016/j.tig.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 2013;62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- Maleszka R, Skelly PJ, Clark-Walker GD. Rolling circle replication of DNA in yeast mitochondria. EMBO J. 1991;10:3923–3929. doi: 10.1002/j.1460-2075.1991.tb04962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeney E, Hawthornthwaite AM, Codd GA, Gibbs SP. Immunocytochemical localization of phosphoribulose kinase in the cyanelles of Cyanophora paradoxa and Glaucocystis nostochinearum. Plant Physiol. 1987;84:1028–1032. doi: 10.1104/pp.84.4.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki H, et al. Phylogenetic positions of Glaucophyta, green plants (Archaeplastida) and Haptophyta (Chromalveolata) as deduced from slowly evolving nuclear genes. Mol Phylogenet Evol. 2009;53:872–880. doi: 10.1016/j.ympev.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Ogawa S, et al. The mitochondrial DNA of Dictyostelium discoideum: complete sequence, gene content and genome organization. Mol Gen Genet. 2000;263:514–519. doi: 10.1007/pl00008685. [DOI] [PubMed] [Google Scholar]
- Oudot-Le Secq M-P, Loiseaux-de Goër S, Stam WT, Olsen JL. Complete mitochondrial genomes of the three brown algae (Heterokonta: Phaeophyceae) Dictyota dichotoma, Fucus vesiculosus and Desmarestia viridis. Curr Genet. 2006;49:47–58. doi: 10.1007/s00294-005-0031-4. [DOI] [PubMed] [Google Scholar]
- Palmer JD. The symbiotic birth and spread of plastids: how many times and whodunit? J Phycol. 2003;39:4–12. [Google Scholar]
- Parfrey LW, et al. Evaluating support for the current classification of eukaryotic diversity. PLoS Genet. 2006;2:e220. doi: 10.1371/journal.pgen.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanzagl B, et al. Primary structure of cyanelle peptidoglycan of Cyanophora paradoxa: a prokaryotic cell wall as part of an organelle envelope. J Bacteriol. 1996;178:332–339. doi: 10.1128/jb.178.2.332-339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Price DC, et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012;335:843–847. doi: 10.1126/science.1213561. [DOI] [PubMed] [Google Scholar]
- Qiu H, Yang EC, Bhattacharya D, Yoon HS. Ancient gene paralogy may mislead inference of plastid phylogeny. Mol Biol Evol. 2012;29:3333–3343. doi: 10.1093/molbev/mss137. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Bhattacharya D. Phylogeny of nuclear-encoded plastid-targeted proteins supports an early divergence of glaucophytes within Plantae. Mol Biol Evol. 2007;24:2358–2361. doi: 10.1093/molbev/msm186. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Weber APM, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- Rocha EPC. The replication-related organization of bacterial genomes. Microbiology. 2004;150:1609–1627. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, et al. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Rosengarten RD, Sperling EA, Moreno MA, Leys SP, Dellaporta SL. The mitochondrial genome of the hexactinellid sponge Aphrocallistes vastus: evidence for programmed translational frameshifting. BMC Genomics. 2008;9:33. doi: 10.1186/1471-2164-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Puerta MV, Bachvaroff TR, Delwiche CF. The complete mitochondrial genome sequence of the haptophyte Emiliania huxleyi and its relation to heterokonts (supplement) DNA Res. 2004;11:67–68. doi: 10.1093/dnares/11.1.67. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stiller JW. Plastid endosymbiosis, genome evolution and the origin of green plants. Trends Plant Sci. 2007;12:391–396. doi: 10.1016/j.tplants.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Turmel M, et al. The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor. Two radically different evolutionary patterns within green algae. Plant Cell. 1999;11:1717–1730. doi: 10.1105/tpc.11.9.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, et al. Phylogenetic analyses of complete mitochondrial genome of Urechis unicinctus (Echiura) support that echiurans are derived annelids. Mol Phylogenet Evol. 2009;52:558–562. doi: 10.1016/j.ympev.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Xi Z, et al. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet. 2013;9:e1003265. doi: 10.1371/journal.pgen.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki A, et al. Palpitomonas bilix represents a basal cryptist lineage:insight into the character evolution in Cryptista. Sci Rep. 2014;4:4641. doi: 10.1038/srep04641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- Zhao S, et al. Collodictyon—an ancient lineage in the tree of eukaryotes. Mol Biol Evol. 2012;29:1557–1568. doi: 10.1093/molbev/mss001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.