Abstract
Under certain circumstances, X-linked loci are expected to experience more adaptive substitutions than similar autosomal loci. To look for evidence of faster-X evolution, we analyzed the evolutionary rates of coding sequences in two sets of Drosophila species, the melanogaster and pseudoobscura clades, using whole-genome sequences. One of these, the pseudoobscura clade, contains a centric fusion between the ancestral X chromosome and the autosomal arm homologous to 3L in D. melanogaster. This offers an opportunity to study the same loci in both an X-linked and an autosomal context, and to compare these loci with those that are only X-linked or only autosomal. We therefore investigated these clades for evidence of faster-X evolution with respect to nonsynonymous substitutions, finding mixed results. Overall, there was consistent evidence for a faster-X effect in the melanogaster clade, but not in the pseudoobscura clade, except for the comparison between D. pseudoobscura and its close relative, Drosophila persimilis. An analysis of polymorphism data on a set of genes from D. pseudoobscura that evolve rapidly with respect to their protein sequences revealed no evidence for a faster-X effect with respect to adaptive protein sequence evolution; their rapid evolution is instead largely attributable to lower selective constraints. Faster-X evolution in the melanogaster clade was not related to male-biased gene expression; surprisingly, however, female-biased genes showed evidence for faster-X effects, perhaps due to their sexually antagonistic effects in males.
Keywords: faster-X effect, Drosophila melanogaster, Drosophila pseudoobscura, positive selection, sex-biased gene expression
Introduction
Sex chromosomes have many properties that distinguish them from autosomes, allowing insights into evolutionary processes through comparisons between them (Meisel and Connallon 2013). When males are the heterogametic sex, for example, rare variants at loci on the hemizygous X chromosome that have recessive effects on fitness are exposed to natural selection, both positive and negative, whereas these effects would be masked on the autosomes in a randomly mating population (Haldane 1924). This unmasking of alleles in males has several evolutionary consequences. For instance, it may affect the relative values of neutral diversity on the X chromosome and the autosomes, due to different effects of selection at linked sites on the two types of chromosomes, involving either background selection caused by deleterious mutations (Aquadro et al. 1994; Charlesworth 2012) or selective sweeps of positively selected mutations (Betancourt et al. 2004).
Another consequence is that positively selected X-linked mutations can, under some conditions, be substituted more rapidly than those on the autosomes. In particular, with a 1:1 sex ratio among breeding individuals and equal variances of fitness in males and females, when beneficial mutations are recessive or partially recessive, genes on the X chromosome will experience a higher rate of substitution than genes with similar properties on autosomes, unless their fitness effects are limited to females (Charlesworth et al. 1987). Conversely, the rate of substitution of recessive or partially recessive deleterious mutations is expected to be lower for X-linked genes. The conditions for such faster-X evolution for beneficial mutations are somewhat more relaxed when the effective sex ratio is biased toward females, or there is a higher variance of fitness in males (Vicoso and Charlesworth 2009). Under other circumstances, however, faster-X evolution with respect to adaptive evolution is not expected to occur (Orr and Betancourt 2001), especially when adaptation proceeds mainly by fixing formerly deleterious alleles that were previously segregating at mutation–selection balance.
In view of this diverse set of predictions, it is worth establishing whether or not, or how often, faster-X evolution occurs, as its existence suggests that some modes of evolution are more common than others (Meisel and Connallon 2013). Tests for adaptive faster-X evolution have been carried out using data from Drosophila (reviewed in Presgraves 2008), birds (Mank et al. 2007; Ellegren 2009; Mank, Nam, et al. 2010), and mammals (Khaitovich et al. 2005; Torgerson and Singh 2006; Kousathanas et al. 2014). The results of these studies have been mixed, and somewhat taxon specific. Drosophila protein sequence divergence data show a general trend toward faster-X effects, with some exceptions (Presgraves 2008); studies of divergence in gene expression in Drosophila also show a faster-X effect (Kayserili et al. 2012; Meisel et al. 2012a). Although divergence data by themselves cannot distinguish between adaptive and other causes of rapid divergence, additional studies using polymorphism data suggest significantly more adaptive evolution of protein sequences of X-linked genes (Langley et al. 2012; Mackay et al. 2012; Campos et al. 2014). Similar results were obtained for mammals (e.g., Torgerson and Singh 2006), with polymorphism data from mice providing strong evidence for adaptive faster-X evolution (Kousathanas et al. 2014). Data from birds, which have a ZW sex-determination system, also show faster Z chromosome divergence, but gene expression patterns indicate that this may not be due to adaptive evolution (Mank, Nam, et al. 2010).
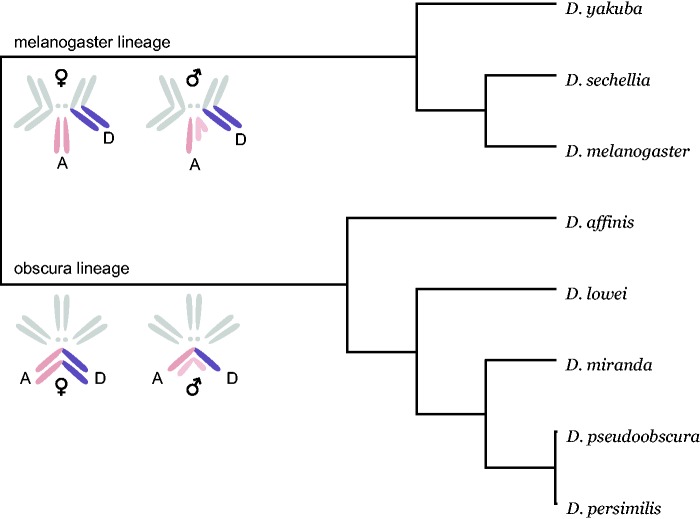
One possible confounding factor in these comparisons is that the X chromosomes and autosomes may contain loci that are inherently different in their rates of evolution (Hu et al. 2013); for example, the X chromosome contains a greater fraction of genes with narrow expression breadth (Meisel et al. 2012b), and different densities of sex-biased genes (reviewed in Vicoso and Charlesworth 2006), both of which may affect rates of protein sequence evolution. To partly circumvent this difficulty, several studies (Counterman and Noor 2004; Thornton et al. 2006; Vicoso et al. 2008) have taken advantage of an X–autosome fusion in the obscura subgroup of the genus Drosophila, where the 3L arm of the Drosophila melanogaster subgroup (Muller element D; (Muller 1940) has become X-linked in the clade containing Drosophila pseudoobscura and its relatives (fig. 1; Ashburner et al. 2005). A comparison of orthologous genes between the melanogaster and the pseudoobscura clades thus allows the separation of chromosome location from gene-specific attributes of chromosomes, when interpreting differences in rates of evolution.
Fig. 1.—
Phylogenetic tree and karyotypes of the eight species analyzed.
Here, we systematically investigate the melanogaster and pseudoobscura clades of Drosophila for evidence of higher X-linked rates of protein sequence divergence, using whole-genome coding sequence data and incorporating information about sex-biased expression. Like Counterman et al. (2004), we use the X–autosome fusion in the pseudoobscura clade to distinguish X-linkage from other factors affecting locus-specific rates of evolution. Faster protein sequence divergence could be due to either higher rates of adaptive evolution or relaxed purifying selection, but these factors can be teased apart using information from polymorphism data (Smith and Eyre-Walker 2002), so that we have combined sequence comparisons among species with analyses of polymorphism data. Overall, we find evidence for faster-X effects at nonsynonymous sites in the melanogaster comparisons. In the pseudoobscura clade however, only a comparison of a pair of very closely related species appears to show faster-X evolution, possibly reflecting changes in selection pressures around the time of speciation events.
Materials and Methods
Genome-Wide Coding Sequence Data
We downloaded coding sequences (CDS) of the following genome sequence releases from FlyBase (www.flybase.org): D. melanogaster 5.43, Drosophila sechellia 1.3, Drosophila yakuba 1.3, D. pseudoobscura 2.26, and Drosophila persimilis 1.3. In addition, sequences of 6,110 coding regions from Drosophila lowei were kindly provided by Noor et al., and sequences of 10,272 coding regions from Drosophila miranda by Bachtrog et al. (Zhou and Bachtrog 2012), and is available under the GenBank accession number AJMI00000000.2.
We obtained a genome sequence from a fourth species, Drosophila affinis, evolutionarily more distant from D. pseudoobscura than D. loweii or D. persimilis, as this comparison increases the power of tests for a faster-X effect in the obscura subgroup. Drosophila affinis Nebraska line no. 0141.2 (Drosophila Species Resource Center) was sequenced in collaboration with V. Nolte, N. Palmieri, and C. Schlötterer from the Institute of Population Genetics, Vetmeduni, Vienna, Austria (Palmieri et al. 2014). Genomic DNA was extracted from females, and libraries with insert sizes of 310 and 630 bp (including the sequenced ends) were prepared. These libraries were then sequenced on one lane each of an Illumina GAIIX to obtain 42,657,732 (for the short insert library) and 39,630,082 (for the long insert library)101-bp paired-end reads. The data were then processed using the standard Illumina pipeline v. 1.7.
To obtain a genome assembly, we first trimmed low quality sequence (using the trim_fastq.pl script from PoPoolation; Kofler et al. 2011), then obtained a de novo assembly using CLC Genomic Workbench version 4.6 (http://www.clcbio.com/products/clc-genomics-workbench/, last accessed October 22, 2014), and finally used nucmer (Delcher et al. 2002) with parameters -c 30 –g 1000 –l 15 to scaffold the assembled contigs against D. pseudoobscura. To annotate this genome, we masked interspersed repeats on our assembled D. affinis genome using RepeatMasker 3.2.9 (Smit et al. 1996); parameters: –q –gff -nolow –norna –species drosophila, and then annotated protein-coding genes based on the D. pseudoobscura genome annotation using Exonerate 2.2.0 (Slater and Birney 2005); parameters: -model protein2genome –bestn 1 –showtargetgff. This annotation was filtered to remove CDS containing frame shifts or premature stop codons. The raw reads are available on the EBI Short Read Archive under the study accession number ERP001460.
Polymorphism Data
We collected polymorphism data from one representative species from each group, D. pseudoobscura and D. melanogaster. The D. pseudoobscura data were collected by sequencing genes from 12 lines originally collected in July 2005 from Mesa Verde National Park, Mesa Verde, CO, and kindly provided by Stephen Schaeffer, as described in Haddrill et al. (2010). A data set of the orthologous genes was obtained from the DPGP resequencing project (http://www.dpgp.org/, last accessed October 22, 2014; Pool et al. 2012) from the Rwandan sample of 17 D. melanogaster haploid genomes, filtered for introgression from European populations based on the recommendations in Pool et al. (2012), as described in Campos et al. (2014).
We selected three sets of genes for use in the polymorphism analysis: 1) Fast-evolving XR genes, which are genes that are newly X-linked in D. pseudoobscura (i.e., on 3L in D. melanogaster and on XR in D. pseudoobscura); 2) fast-evolving autosomal genes, which are genes that are autosomal in both the D. melanogaster and D. pseudoobscura lineages; and 3) fast-evolving XR and autosomal female-biased genes, which are genes that are strongly female-biased in both lineages, and therefore not expected to experience faster-X evolution (Charlesworth et al. 1987; Meisel and Connallon 2013). For both the XR and strictly autosomal data set, we aimed to enrich our set for loci undergoing adaptive evolution, as a previous study suggested that a faster-X effect was marginally significant for the faster-evolving genes in the D. pseudoobscura–D. affinis comparison (Vicoso et al. 2008).
Accordingly, we chose for the polymorphism analyses genes with high rates of evolution in the D. yakuba lineage (as estimated by Clark et al. 2007) under the M0 model in PAML; note that the D. yakuba lineage was not further analyzed in the polymorphism analysis. We restricted the data set to those genes with rates of protein evolution corresponding to the 70–100% quantiles on 3L, that is, with ω > 0.096. We filtered out long and short genes, using only genes falling within two intermediate quantiles for length in D. melanogaster, between 1,279 and 4,571 bp, as gene length is correlated with the rate of nonsynonymous evolution (Comeron et al. 1999). We further excluded any genes showing strong sex-biased gene expression in either D. yakuba or D. pseudoobscura, as assessed by Sturgill et al. (2007).
This procedure resulted in a set of 75 XR genes, and 48 strictly autosomal genes, from which we obtained part of the coding sequence for 54 and 31 genes, respectively. For the female-biased expression control data set, we restricted the list of genes to those that showed significantly female-biased expression in both D. yakuba and D. pseudoobscura (again as assessed by Sturgill et al. 2007), and applied the same criteria for the rate of evolution as above. As this resulted in a candidate pool of only 17 XR genes, and 36 autosomal genes, we did not further restrict this data set by gene length. From these female-biased genes, we obtained sequence from 6 3L/XR genes and 17 strictly autosomal genes.
Sequencing Methods
We sequenced the above genes from the 12 D. pseudoobscura Mesa Verde lines using standard polymerase chain reaction (PCR) and Sanger sequencing methods (Haddrill et al. 2010). A complete list of the PCR primers as well as the cycling conditions used for each gene are available on request. PCR-amplified products were treated with ExoSAP-IT (USB, Cleveland, OH) and sequenced from both strands using BigDye chemistry and a 3730 automated sequencer (Applied Biosystems, Foster City, CA) at the University of Edinburgh GenePool sequencing service, with PCR primers used as sequencing primers. Not all genes were sequenced from all strains; the average number of strains sequenced per gene was 11 (see supplementary table S6, Supplementary Material online). Sequences have been submitted to the GenBank database under the accession numbers JX409935–JX411616.
Analyses of Genome-Wide Rates of Protein Sequence Evolution
For this analysis, we retained only orthologs whose location on the same Muller element (equivalent to a chromosome arm, Muller 1940) was conserved between D. melanogaster and D. pseudoobscura, resulting in a data set of 10,273 protein-coding sequences. Pairwise in-frame CDS alignments were performed for orthologous-coding sequences within the melanogaster (D. melanogaster, D. sechellia, and D. yakuba) and pseudoobscura (D. pseudoobscura, D. lowei, D. affinis, and D. persimilis) groups using MAFFT (Katoh and Standley 2013). Sequence alignments are posted in DRYAD (http://datadryad.org/, last accessed October 22, 2014) under doi:10.5061/dryad.3hh83.
Two sets of pairwise divergence estimates were obtained: One set (denoted by KA and KS) using the site-counting method of Comeron (1995) implemented in G-estimator (http://molpopgen.org/software/lseqsoftware.html), and the other (denoted by dN and dS) obtained using the maximum-likelihood method implemented in the PAML program codeml (Yang 2007). As estimates of divergence based on site-counting and maximum-likelihood methods gave qualitatively equivalent results, only counting estimates are shown here (for a discussion of the different methods, see Bierne and Eyre-Walker 2003). We then excluded from the analysis genes shorter than 100 amino acids, genes that had KS or dS estimates below 0.01 or above 3 (as recommended in the PAML manual; http://abacus.gene.ucl.ac.uk/software/, last accessed October 22, 2014), and genes for which KA/KS or dN/dS estimates could not be calculated (usually due to low synonymous divergence). The total number of genes analyzed for each pair of species is shown in supplementary table S1, Supplementary Material online. We also used PAML to estimate dN, dS, and dN/dS over the entire phylogeny for genes that occurred in all species, using the M0 model of codeml to estimate a single dN/dS for each gene, separately for the obscura and melanogaster clades; transition–transversion rates were estimated from the data, and codon frequencies from the nucleotide frequencies. For the melanogaster group, we used the single unrooted tree to relate the three species; for the obscura group, we estimated rates for all 15 unrooted trees, taking the values from the model yielding the highest likelihood. This procedure is equivalent to an exhaustive likelihood search, and has the advantage of estimating the phylogeny and the rates under the same model of sequence evolution.
Fixed inversions between D. pseudoobscura and D. persimilis were defined as in Machado et al. (2007), with 2,322 loci classed as inside an inversion, 801 loci within 2 MB of inversion breakpoints, and 7,139 outside inverted regions and mapped to D. pseudoobscura scaffolds based on the information in Schaeffer et al. (2008).
Gene Expression Data
We extracted the ratio of male to female expression level from the Sebida database v. 3.0 (Jiang and Machado 2009; www.sebida.de), with the classification of genes as male, female, or unbiased taken from this database, expected to yield a 20% false-positive rate (Gnad and Parsch 2006). For the pseudoobscura clade species, genes with an M/F expression ratio lower than 0.9 or greater than 1.1 were classified as female- and male-biased, respectively, and genes with an M/F expression ratio between 0.9 and 1.1 were classified as unbiased. Values used for the melanogaster group were measured in D. melanogaster, whereas those used for the obscura group were measured in D. pseudoobscura.
Statistical Analyses
To compare rates of sequence evolution, we used two-tailed nonparametric Kruskal–Wallis or Mann–Whitney U tests. For the Mann–Whitney U tests, multitest corrections were applied using the false discovery rate method by Benjamini and Hochberg (1995). All statistical analyses were performed using R version 2.14.0 or later.
To analyze the polymorphism data sets, we calculated polymorphism and divergence summary statistics for all genes using custom Python scripts. To perform McDonald–Kreitman tests, we used the method of Welch (2006). To estimate the distribution of fitness effects of deleterious nonsynonymous mutations and the proportion of sites under positive selection, we used the DFE-α method of Eyre-Walker and Keightley (2009), which uses data on interspecies divergence and the folded site frequency spectra of variants at synonymous and nonsynonymous sites.
Recombination Rate Bins for D. melanogaster
For the purpose of examining the possible effects of recombination rates on sequence evolution in the melanogaster clade, we divided genes up into low, medium, and high recombination rate categories based on rates from Fiston-Lavier et al. (2010), according to the criteria described in Campos et al. (2012).
Results
Faster-X Evolution in the melanogaster Clade
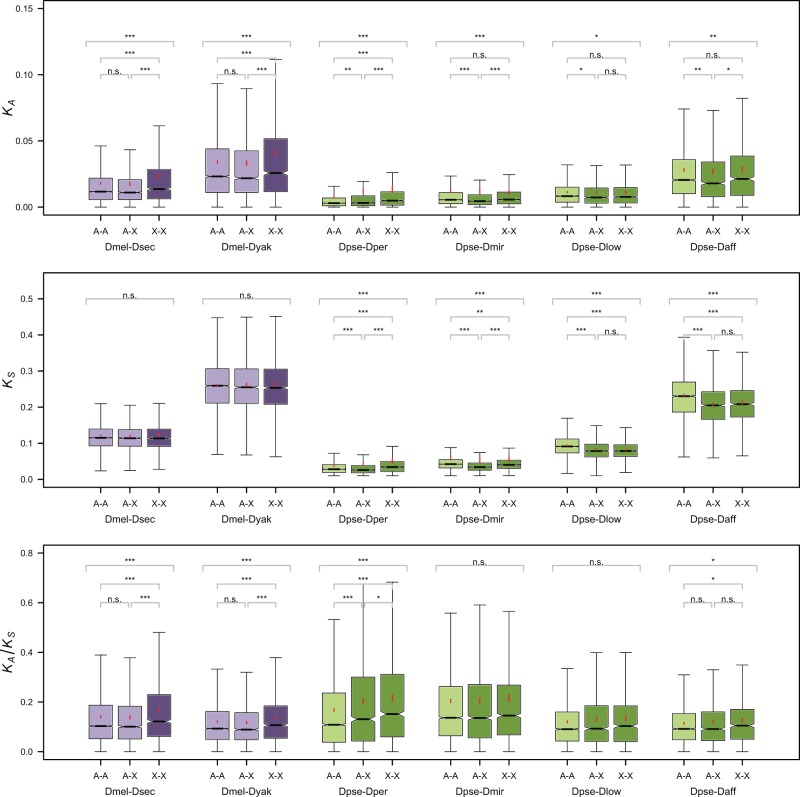
Summary results on nonsynonymous and synonymous divergence between D. melanogaster and its relatives, and for D. pseudoobscura and its relatives, using a counting measure of divergence (see Materials and Methods) are shown in figure 2 (see also supplementary table S1, Supplementary Material online; results from maximum-likelihood estimates are shown in supplementary table S2 and fig. S1, Supplementary Material online). We compared rates of nonsynonymous and synonymous sequence evolution among three classes of genes: XX genes (X-linked in both the melanogaster and pseudoobscura clades), AA genes (autosomal in both clades), and AX genes (autosomal in the melanogaster, but linked to XR in the pseudoobscura clade). To ensure that any differences among comparisons do not reflect differences in the sets of genes that were analyzed, we carried out many of the analyses described below for the orthologous genes present in all species (the “common” genes in supplementary table S1, Supplementary Material online), as well as for all genes that could be analyzed for a given pair (“all genes” in supplementary table S1, Supplementary Material online), after the filtering described in Materials and Methods. The general patterns found for all genes also hold for the common genes subset, so we focus on results from the larger data set.
Fig. 2.—
Notched boxplots of KA (upper panel), KS (middle panel), and KA/KS (lower panel) for six pairs of species analyzed and the three categories of genes (AA, XX, and AX). The boundary of the box closest to zero indicates the 25th percentile and that farthest from zero the 75th percentile. The whiskers indicate 1.5 times the interquartile range. A line within a box marks the median and the notches represent 95% CIs for the medians. A red point marks the mean and the red lines the 95% CIs for the mean (which are usually too narrow to be visible). Outliers not shown. Stars above the boxplot indicate statistical significance levels (***P < 0.001, **P < 0.01, *P < 0.05, and ns, not significant). Stars above all three boxplots for a species pair indicate significant heterogeneity among chromosome types (determined through a Kruskal–Wallis test). For species with heterogeneity among chromosome types, the significance of pairwise comparisons between A–A, A–X, and X–X loci is shown (determined with a Mann–Whitney U test).
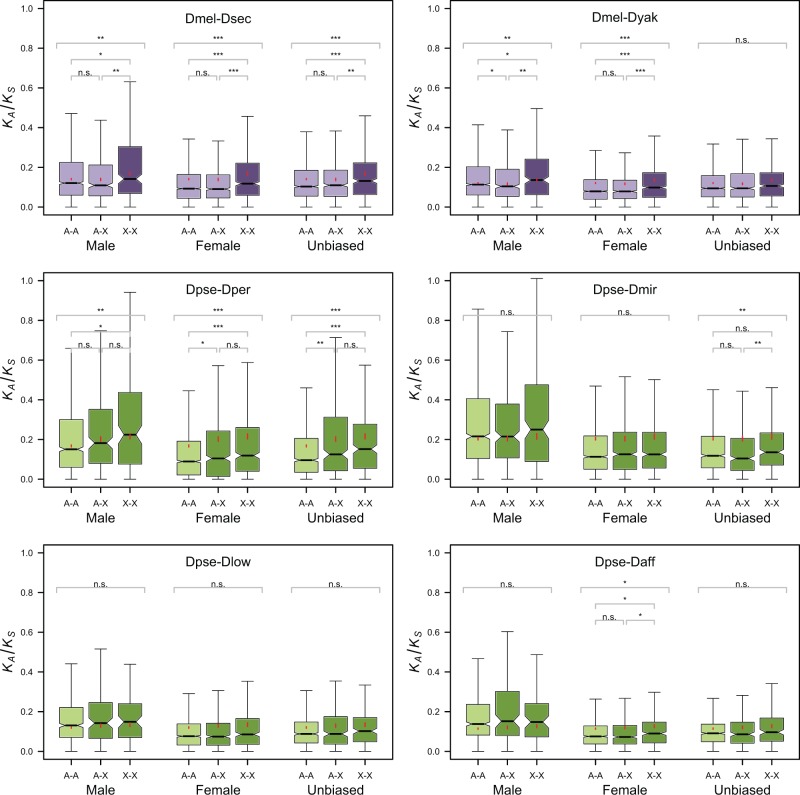
In the melanogaster clade, nonsynonymous divergence was significantly higher for X-linked than for autosomal genes (XX vs. AA, AX), whereas synonymous divergence was not significantly different (fig. 2 and supplementary fig. S1 and tables S1 and S2, Supplementary Material online). These results are consistent with those for the maximum-likelihood estimates using PAML (Yang 2007), except for the D. yakuba–D. melanogaster comparison, where both dN and dS for X-linked loci were elevated relative to the autosomes, yielding an overall nonsignificantly higher value of dN/dS for the X chromosome compared with the autosomes. Furthermore, division of the genes into classes based on their sex-specific levels of expression shows that the faster-X effect is more marked for sex-biased genes, particularly those with female-biased expression (see fig. 3 and supplementary fig. S2 and tables S3 and S4, Supplementary Material online).
Fig. 3.—
Notched boxplots of KA/KS for the six pairs of species, the three categories of genes, and the three levels of sex bias analyzed. Boxplots, means, and statistical significance levels are as in figure 2.
The effect of sex-bias on faster-X evolution may be a consequence of its effect on rates of protein evolution (table 1); all else being equal, a high rate of substitution, particularly of adaptive substitutions, will yield more power to detect faster-X evolution. But if positive selection is the basis of faster-X evolution, the robustness of the faster-X effect for female-biased genes is surprising, as no faster-X evolution should occur for genes experiencing selection only in females (Charlesworth et al. 1987). It could be the case, however, that sex-biased expression is not an adequate measure of sex-specific selection. One reason for this might be that the definition of sex-bias we have used is too liberal and includes too many genes experiencing selection in both sexes; in fact, the criterion for female-biased expression that we used does not preclude a reasonable level of expression in males. Using more stringent criteria, however, does not appear to change the results: Genes with the strongest female-bias in expression show a faster-X effect roughly equivalent to that of the half with the weakest female-bias. For the D. melanogaster–D. yakuba comparison, for example, the half of the female-biased genes with the strongest bias have median autosomal KA/KS = 0.0822 versus X-linked KA/KS = 0.100, P = 0.00015, which is similar to the pattern for the half with the weakest bias, KA/KS = 0. 077 (A) versus 0.102 (X), P < 1.5 × 10−6; comparisons based on dN/dS and on other species pairs in the melanogaster clade show similar results (results not shown). If we use a 2-fold expression difference between males and females as the cutoff for male- and female-biased expression instead of the cutoffs provided by the Sebida database (see Materials and Methods), the faster-X effect for female-biased genes remains significant (median autosomal KA/KS = 0.0965 vs. X-linked KA/KS = 0.123, P = 3.4 × 10−5). We also checked for a quantitative weakening of the faster-X effect with the level of female-bias among female-biased genes, as might be expected if these genes are merely enriched for those experiencing selection in females only, but do not exclusively consist of such genes. We found no evidence of such an effect (fitting a linear model with X-linkage and sex-bias as factors to the log-transformed data shows a significant interaction between these factors, but in the wrong direction; see supplementary table S5, Supplementary Material online).
Table 1.
Spearman Correlation Coefficients (ρ) between Gene Expression Bias (the Ratio of Male to Female Mean Expression Levels) and KA/KS
| All | Male | Unbiased | Female | |||||
|---|---|---|---|---|---|---|---|---|
| Dmel–Dsec | 0.098 | *** | 0.181 | *** | −0.010 | ns | −0.034 | * |
| Dmel–Dyak | 0.146 | *** | 0.269 | *** | −0.020 | ns | −0.039 | * |
| Dpse–Dper | 0.147 | *** | 0.076 | ** | 0.017 | ns | 0.036 | ns |
| Dpse–Dmir | 0.187 | *** | 0.167 | *** | −0.010 | ns | −0.033 | ns |
| Dpse–Dlow | 0.179 | *** | 0.109 | ** | 0.013 | ns | 0.026 | ns |
| Dpse–Daff | 0.226 | *** | 0.229 | *** | 0.019 | ns | 0.026 | ns |
Note.—Dmel, Drosophila melanogaster; Dsec, Drosophila sechellia; Dyak, Drosophila yakuba; Dpse, Drosophila pseudoobscura; Dper, Drosophila persimilis; Dmir, Drosophila miranda; Dlow, Drosophila lowei; Daff, Drosophila affinis; *** P < 0.001, ** P < 0.01, * P < 0.05, and ns, not significant.
How, then, can we explain a faster-X effect that occurs regardless of sex-specific selection? One possibility is a difference in mutation rate between X and autosomes; if adaptive evolution is affected by the mutation rate, as is assumed in models based on the approach of Charlesworth et al. (1987) that assumes fixation of unique, new mutations, X-linked loci could evolve faster if they experience a higher mutation rate (Kirkpatrick and Hall 2004). Assuming that KS reflects the mutation rate, the data are not consistent with this scenario, as KS is usually somewhat lower, not higher, for X-linked loci (supplementary table S1, Supplementary Material online). Furthermore, the faster-X effect does not appear to be an artifact of lower KS for the X chromosome in the melanogaster clade, as the higher KA/KS for X-linked than for autosomal loci appears to be largely due to their higher KA (supplementary table S1, Supplementary Material online and fig. 2).
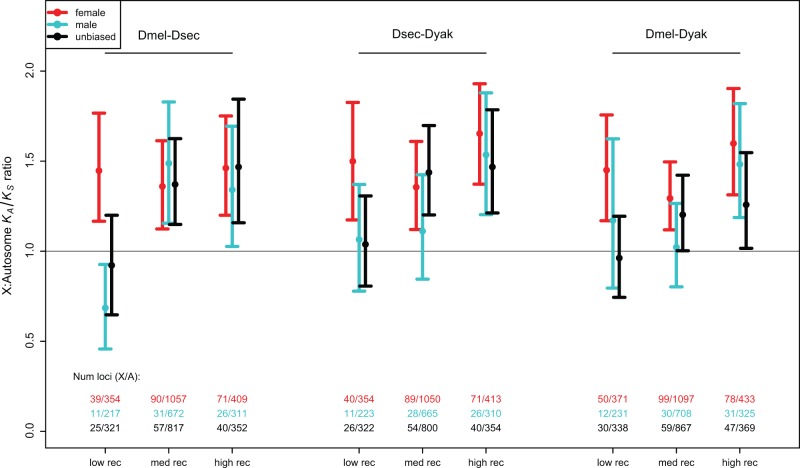
Another possible cause of the faster-X effect is a difference in the population effective recombination rate between X chromosomes and autosomes: In Drosophila, the lack of recombination in males implies a higher rate of recombination for X-linked genes than for autosomes, for a given rate of recombination in females, due to the fact that an X chromosome spends only one-third of its time in males, whereas an autosome spends half of its time in males (Langley et al. 1988; Charlesworth 2012). Thus, an adaptive faster-X effect might occur due to this higher effective recombination rate, which may alleviate the effects of Hill–Robertson interference among sites subject to selection, and thus yield a higher rate of fixation of adaptive alleles at X-linked loci (Connallon 2007). We tested for this by looking at regions of the genome for which X-linked and autosomal loci have roughly equivalent effective recombination rates as far as population genetic processes are concerned, following the procedure of Campos et al. (2013). We again find a faster-X effect for these genes, suggesting that it is not a simple consequence of the high X chromosome recombination rate (grouping genes by X- or autosomal linkage in the melanogaster group, KA/KS comparisons give P < 1.68 × 10−6 for all three species pairs; for female-biased genes, P < 8.09 × 10−7; for dN/dS comparisons, P < 0.030).
A second possibility is that the faster-X effect is not due to adaptive evolution, but is instead caused by the fixation of slightly deleterious mutations by genetic drift. This could occur if the X experiences an even lower effective population size relative to A than the “null” value of 75% expected with a 1:1 adult sex ratio and equal variances in reproductive success in the two sexes (Mank, Vicoso, et al. 2010). However, current East African populations of D. melanogaster, which inhabit the putatively ancestral range of this species, have an overall Ne for the X that is similar to that for the autosomes (Andolfatto 2001; Singh et al. 2007; Campos et al. 2013). It is possible that this does not reflect the long-term situation, but the fact that codon usage is generally higher on the X than A in several species of Drosophila is inconsistent with a lower than expected X/A ratio of Ne (Singh et al. 2005, 2008). In addition, if there were a faster rate of fixation of deleterious mutations on the X relative to A, we would expect the effect to be most extreme for genes in low recombination regions, due to the greater intensity of Hill–Robertson interference effects in these regions (Campos et al. 2014).
Division of genes into low, medium, and high recombination categories, and by sex-biased expression category, shows that this is not the case. Instead, the faster-X effect appears to be stronger for the high and medium recombination rate regions than for the low recombination rate regions, as would be expected under adaptive evolution (fig. 4). The partitioning by recombination rate also shows that unbiased, female-biased, and male-biased genes all have similar X:A ratios of KA/KS. Further, this effect of recombination suggests that the faster-X effect we observe is not an artifact of lower quality sequence for the X chromosome (and thus a higher contribution to KA/KS from sequencing errors), as might occur due to lower coverage when males (or a mixture of males and females) are sequenced. Finally, estimates of the extent of adaptive evolution of nonsynonymous mutations from combinations of polymorphism and divergence data suggest very strongly that the faster-X effect in the melanogaster clade is due to positive selection (Langley et al. 2012; Mackay et al. 2012; Campos et al. 2014). Campos et al. (2014) also found no evidence for adaptive evolution of nonsynonymous mutations in the very low recombination regions of autosomes, in contrast to significant adaptive evolution in the low recombination X chromosome regions.
Fig. 4.—
Mean and 95% bootstrap confidence intervals from 1,000 bootstraps for KA/KS for the three pairs of melanogaster group species, divided according to sex bias and recombination rate category. Recombination rates are based on recombination maps for D. melanogaster (Fiston-Lavier et al. 2010), with rates for X-linked loci adjusted by 4/3 for to correct for the lack of recombination in males. The set of genes is restricted to those in the range where recombination rates for the X chromosomes and autosomes overlap, and divided into bins corresponding to low ([1.00–1.4 cM/MB)), medium ([1.4–1.75)), and high ([1.75–2.1]) recombination rate regions.
Faster-X Evolution in the pseudoobscura Clade
In the pseudoobscura clade, on the other hand, different pairwise comparisons produced contrasting results (fig. 2 and supplementary fig. S1 and tables S1 and S2, Supplementary Material online). The D. pseudoobscura–D. persimilis pair, as was seen previously (Grath and Parsch 2012), shows evidence of faster-X evolution, with higher KA/KS for X-linked genes (pooling A–X genes with the X–X genes, median X-linked KA/KS = 0.144 vs. median autosomal KA/KS = 0.111, P = 3.87; for KA the medians were X-linked = 0.00385 vs. autosomal = 0.00300, P = 1.21 × 10−13). This elevation was seen for both the ancestral X chromosome (XL) and for the derived XR chromosome; furthermore, the median KA/KS for XR (0.131) was substantially higher than that for the equivalent AX comparisons in the melanogaster clade. It should be noted, however, that the proportion of filtered genes for this species pair (see Materials and Methods) was 2 orders of magnitude higher than for the rest of the clade (14.2% vs. <0.5%), mainly due to genes with low synonymous divergence (KS < 0.01).
In contrast, the other pairwise comparisons D. pseudoobscura–D. miranda, D. pseudoobscura–D. lowei, and D. pseudoobscura–D. affinis showed no evidence of faster-X evolution. There was no significant difference for nonsynonymous divergence between AA and XX genes, whereas AX genes showed significantly lower values than AA and XX genes. Synonymous divergence was significantly lower for X-linked genes (XX and AX) for the comparisons of D. pseudoobscura with D. miranda, D. lowei, and D. affinis. In the case of D. miranda we ignored the fact that the Muller element C has become a neo-X chromosome since its split with D. pseudoobscura (Ashburner et al. 2005), because these loci were autosomal for at least half of the divergence time for this species pair, and faster evolution of the loci on the neo-X may reflect a short-term response to their new genomic environment rather than the faster-X effect as usually understood (Bachtrog et al. 2009). Treating these loci as autosomal is thus conservative. Faster-X evolution of this chromosome may have contributed to the higher KA/KS that is seen for the autosomes in the D. pseudoobscura–D. miranda comparison (supplementary table S1, Supplementary Material online) relative to the other comparisons, especially as there is evidence for a higher rate of adaptive protein sequence evolution on this chromosome in the miranda lineage (Bachtrog et al. 2009).
Comparisons of Rates of Evolution Using a Phylogenetic Approach
We also used a maximum-likelihood-based approach, which allows estimation of ω = dN /dS along different branches of the phylogenetic tree connecting all the species (see Materials and Methods). This allows us to compare rates of nonsynonymous evolution at the same loci in an X-linked and in an autosomal context, controlling for locus-specific rates of evolution, for the subset of the data for which we have gene sequences for all species. As expected, there appear to be locus-specific rates of evolution, with a strong correlation between rates of evolution in the two clades (rS = 0.562, P < 2.2 × 10−16). There is also an overall faster-X effect (median autosomal ω = 0.0587, median X-linked ω = 0.0673, Wilcoxon rank sum test with continuity correction P = 0.000029). Overall, therefore, this analysis confirms the conclusions based on the pairwise species comparisons.
Polymorphism and Divergence Analyses
We have attempted to use polymorphism and divergence data to distinguish the contributions of adaptive and slightly deleterious mutations to nonsynonymous divergence in the pseudoobscura clade (Fay et al. 2002). We collected polymorphism data from a population of D. pseudoobscura, focusing on genes with high rates of nonsynonymous sequence evolution, as these are likely to show either the most adaptive evolution or the highest number of fixations due to slightly deleterious mutations (to avoid confounding our results, we chose these genes based on their rates of evolution in the melanogaster clade, not in the pseudoobscura clade), without reference to their patterns of sex-biased gene expression (see Materials and Methods). As a control, we also selected a set of fast-evolving genes with female-biased expression. We then compared autosomal and XR genes in order to determine whether the latter showed evidence of faster-X effects.
Table 2 shows summary divergence and polymorphism statistics for the genes that we studied, using divergence from D. affinis for the KA and KS estimates (see supplementary table S6, Supplementary Material online, for results for individual genes). As might be expected, mean KA and KA/KS for the fast-evolving genes were high when compared with those for a D. pseudoobscura polymorphism data set of slow-evolving genes, where the mean KA values were 1.5% for both X and A, and the ratios of mean KA to mean KS were 5% and 6%, respectively (Haddrill et al. 2010). In this data set, however, mean KA and the ratio of mean KA to mean KS were much higher for the autosomes than for the XR genes in the unbiased set of genes. This was also observed in the D. melanogaster clade data set, indicating that gene-specific selective constraints drive this pattern (for further evidence on this point, see the Discussion). The change from an autosomal context to an X-linked context has not reversed or decreased this difference, as we would expect on the hypothesis of faster-X evolution, consistent with the lack of evidence for faster-X effects described above.
Table 2.
Summary of Polymorphism and Divergence Statistics
| πA (%) | πS (%) | πA/πS (%) | KA (%) | KS (%) | KA/KS (%) | Tajima’s D (Nonsynonymous) | Tajima’s D (Synonymous) | |
|---|---|---|---|---|---|---|---|---|
| Unbiased | ||||||||
| XR (n = 54) | 0.136 | 1.52 | 8.88 | 2.06 | 22.4 | 9.18 | −0.763 | −0.799 |
| (0.0264) | (0.158) | (1.96) | (0.315) | (1.33) | (1.46) | (0.123) | (0.183) | |
| A (n = 31) | 0.356 | 2.16 | 16.5 | 6.23 | 28.8 | 21.6 | −0.966 | −0.881 |
| (0.0606) | (0.267) | (3.48) | (0.815) | (3.32) | (2.95) | (0.120) | (0.106) | |
| Female-biased | ||||||||
| X (n = 8) | 0.359 | 1.82 | 19.8 | 8.42 | 27.4 | 30.8 | −0.937 | −1.03 |
| (0.220) | (0.663) | (23.1) | (2.24) | (8.42) | (12.5) | (0.269) | (0.268) | |
| A (n = 17) | 0.197 | 1.17 | 16.8 | 7.55 | 31.9 | 26.4 | −1.41 | −1.07 |
| (0.0552) | (0.262) | (6.06) | (1.32) | (0.755) | (6.96) | (0.0612) | (0.117) | |
Note.—Standard errors are in parentheses; these were calculated directly from the individual gene values, except for the ratios πA/πS and KA/KS, which were estimated using the delta method (Bulmer 1980). Divergence is measured from D. affinis.
To estimate the fraction of nonsynonymous differences between D. pseudoobscura and D. affinis or D. lowei that were caused by positive selection (α), we used both the MacDonald–Kreitman test approach implemented in Welch (2006), and the DFE-α method of Eyre-Walker and Keightley (2009) (table 3). As a basis for comparison, we also applied these methods to polymorphism data on the Rwandan population of D. melanogaster from the DPGP (Pool et al. 2012) with D. yakuba as the outgroup, following the methods of Campos et al. (2014).
Table 3.
Estimates of α and ωα for the X-Linked and Autosomal Loci Drosophila pseudoobscura and Drosophila melanogaster Polymorphism Data Sets, Using the DFE-α Method
| Group | Sites | Chromosome | α | ωα |
|---|---|---|---|---|
| melanogaster | 0 and 4-fold | X | 0.733 (0.536, 0.833) | 0.080 (0.050, 0.106) |
| Synonymous and nonsynonymous | X | 0.721 (0.539, 0.824) | 0.079 (0.052, 0.103) | |
| 0 and 4-fold | Autosomal | 0.417 (0.049, 0.677) | 0.099 (0.011, 0.167) | |
| Synonymous and nonsynonymous | Autosomal | 0.414 (0.086, 0.694) | 0.094 (0.020, 0.169) | |
| pseudo obscura | 0 and 4-fold | XR | 0.390 (0.142, 0.731) | 0.051 (0.014, 0.104) |
| Synonymous and nonsynonymous | XR | 0.328 (0.131, 0.680) | 0.036 (0.012, 0.081) | |
| 0 and 4-fold | Autosomal | 0.668 (0.188, 0.880) | 0.142 (0.035, 0.238) | |
| Synonymous and nonsynonymous | Autosomal | 0.624 (0.289, 0.866) | 0.125 (0.051, 0.201) |
The analyses using the method of Welch (2006) showed no evidence for a faster rate of adaptive amino acid fixations for the D. pseudoobscura–D. affinis or D. lowei comparisons on XR compared with the autosomes, with statistically significant α values for the autosomes for the fast-evolving genes in both comparisons, but not for XR. Curiously, female-biased genes show significant evidence for positive selection in the comparison with D. lowei, with an α value very similar to that for the autosomes. The results for the same set of genes in D. melanogaster suggest that the fast-evolving genes that are autosomal in the pseudoobscura clade have a lower α value than the genes that are on XR in this clade, but the estimates are too noisy to be interpreted with confidence. The female-biased genes give results that are broadly similar to those for the pseudoobscura clade.
Estimating α by the DFE-α method gives slightly different results for the D. pseudoobscura clade, but in the same direction as those obtained by the Welch (2006) method. XR-linked genes show consistently less adaptive evolution than autosomal genes in the unbiased gene expression data set. The ωα estimate gives the rate of adaptive nonsynonymous substitutions relative to synonymous substitutions (Gossmann et al. 2010): These estimates are close to 0 for unbiased XR-linked genes and around 15% for the unbiased autosomal genes (table 3).
Discussion
The Existence and Causes of Faster-X Effects
In this study, we have evidence for faster-X evolution at nonsynonymous sites in the melanogaster clade, in agreement with findings from previous studies (Grath and Parsch 2012; Hu et al. 2013). Evidence that the faster-X signal reflects a higher rate of fixation of advantageous mutations on the X chromosome rather than of slightly deleterious mutations has come from analyses of genome-wide polymorphism data and between-species divergence estimates (Mackay et al. 2012; Campos et al. 2014; this study). Surprisingly, however, we find a faster-X effect in the melanogaster clade that is as strong for female-biased genes as for other genes, whereas the standard theory predicts a lack of a faster-X effect for genes with female-specific fitness effects (Charlesworth et al. 1987). Some of this may be due to misclassification of sex-bias genes: Female-biased genes can be difficult to identify (Assis et al. 2012), and imperfect dosage compensation may skew X-linked genes toward female-biased expression regardless of their sex-specific fitness effects (Meiklejohn and Presgraves 2012).
Further, genes that are female-biased in expression may not experience selection exclusively in females. Many are expressed in both sexes at some point in development (Perry et al. 2014), and many are expressed in somatic tissues present in both males and females (Meisel 2011). Studies of deleterious mutations indicate that the effects of mutations in sex-biased genes are often not sex-limited (Connallon and Clark 2011), and our criteria for female-biased expression do not preclude substantial expression in males. Furthermore, in spite of the apparent general enrichment of female-biased genes on the X chromosome (Vicoso and Charlesworth 2006), X-linked mutations may have particularly strong effects on males (Mallet et al. 2011).
It is likely that the surprisingly robust faster-X effect seen for female-biased genes is partly due their selective effects in males. One way in which an association with female-bias and faster-X could arise is these genes have a prior history of selection to minimize negative fitness effects on males, where they are still expressed. For genes with a pattern of sexually antagonistic fitness effects, nonsynonymous mutations that reduce the functionality of the protein might be beneficial to males but harmful to females. If this reduction is partially recessive, as is plausible, then its beneficial effect in hemizygous males could outweigh the deleterious effects on females for mutations on the X chromosome, but not the autosomes, leading to a faster-X effect (see fig. 6 of Vicoso and Charlesworth 2009). Consistent with this idea, the faster-X effect found for gene expression divergence (Meisel et al. 2012a; Kayserili et al. 2012), while generally found for female-biased genes, is not found for genes primarily expressed in female reproductive tissues, though this may be partially due to a lack of power (Meisel et al. 2012a). This effect might be particularly strong for low recombination regions, where the female-biased genes, unlike other genes, still show faster-X effects (fig. 4 of Campos et al. 2014). In these regions, the effective size of the X appears to be greater than that of the autosomes, probably because of smaller effects of background selection (Campos et al. 2014); other things being equal, a higher X:A ratio of Ne favors adaptive faster-X effects (Vicoso and Charlesworth 2009). In addition, if the female faster-X effect is driven by mutations that reduce function, it may be less mutation limited than other kinds of faster-X evolution, as these mutations are likely to be more common than other kinds of beneficial mutations. When the supply of beneficial mutations is abundant, a reduced effective population size due to low recombination rates may have little impact on the rate of adaptive evolution (Maynard Smith 1968; Orr 2000).
Differences among Different Species Comparisons
The results for the pseudoobscura clade are substantially different; we found no convincing evidence for a faster-X effect, with the exception of the D. pseudoobscura–D. persimilis comparison (see also Grath and Parsch 2012). One possible explanation for these conflicting results is there are fewer genes analyzed for D. lowei and D. affinis than for the other species (supplementary table S1, Supplementary Material online), reducing our power to detect a faster-X effect. As the numbers of genes involved are still very large and the confidence intervals for these species are nearly as narrow as in the other cases, however, this factor does not seem likely to be important. Furthermore, the numbers of genes analyzed for D. miranda are comparable to those of the other species, yet this species also yielded a negative result. It therefore seems likely that the contrast between the melanogaster clade comparisons and most of the pseudoobscura clade comparisons is a real one. This result is also consistent with the lack of evidence for a higher α value for the XR genes, compared with the autosomal genes, in the polymorphism-divergence study (table 3).
These results raise several questions. The first is why there is no faster-X effect in most of the pseudoobscura clade comparisons, in contrast to the melanogaster clade. The answer is unclear. One possibility is a difference in the X/A ratio of effective population sizes (Ne) between the two clades. As discussed by Charlesworth (2012), synonymous diversity values suggest that this ratio is close to 1 for the chromosomes as a whole in D. melanogaster, whereas in D. pseudoobscura and D. miranda it is not significantly different from the null value of 0.75 expected with an equal sex ratio, as would occur if there are no sex differences in the variance in reproductive success. Deviations from an X/A Ne of 0.75 in the direction of higher X-linked Ne as seen in D. melanogaster are expected to result in faster-X effects for a broader range of dominance parameters (Vicoso and Charlesworth 2009; Connallon et al. 2012).
The contrasting X/A ratios are consistent with the considerably higher rates of recombination per base pair in the D. pseudoobscura group (McGaugh et al. 2012), as argued in Charlesworth (2012). In D. melanogaster, the lack of recombination in males reduces the effective population rate of recombination on the autosomes relative to the X, so that they suffer more from the reduction in the rate of adaptive evolution due to Hill–Robertson effects. This possibility is supported by the fact that the analysis of Campos et al. (2014) shows that the α and ωα values for the autosomes in D. melanogaster are both positively correlated with the rate of recombination experienced by a gene, and approach those for the X chromosome in regions with very high rates of recombination (see their table 4). In D. pseudoobscura, in contrast, the overall higher rate of recombination on both X chromosomes and autosomes is likely to mitigate this X/A difference in the intensity of interference.
There is, however, a problem with postulating that differences in X/A ratios of Ne as an explanation for the faster-X effect differences between the clades. It is not clear that the situation in D. melanogaster is representative of the melanogaster group: In Drosophila simulans, the current evidence suggests that the X/A ratio of silent site diversity in East African and Madagascan populations is substantially less than 0.75 (Obbard D, Campos J, personal communication), perhaps reflecting the fact that D. simulans also has substantially higher rates of recombination than D. melanogaster (Sturtevant 1929; True et al. 1996). Nonetheless, the rate of recombination measured in D. melanogaster appears to be correlated with the X/A ratio of KA/KS even in the D. sechellia–D. yakuba comparison (fig. 4). If fine-scaled genetic maps, together with genome-wide surveys of polymorphism levels, become available for all the species in the melanogaster clade, it may be possible to rigorously test for the role of recombination. In the absence of such information, we cannot exclude the possibility that the difference between the two clades reflects some biological differences between them that we have not taken into account. Given the fact that the faster-X effect is observed even with female-biased genes, it seems unlikely that this is related to potential differences in the intensity of sexual selection. A difference between the two groups in the relative contribution of standing variants versus new mutations to adaptation is a potential cause: No faster-X effect is expected when adaptation uses standing variation, at least with an X/A ratio for Ne of 0.75, as appears to be roughly true for D. pseudoobscura (Charlesworth et al. 1987; Orr and Betancourt 2001; Connallon et al. 2012).
The next question is whether the faster-X effect for the D. pseudoobscura–D. persimilis comparison is genuine, or is an artifact of their close phylogenetic relatedness. It is well-known that ancestral shared polymorphism may be misinferred as divergence when closely related species are studied, and that this can cause biases in inferences concerning the action of selection. Grath and Parsch (2012) were aware of this concern, and stated that their divergence estimates were “likely to be inflated by the presence of ancestral polymorphism.” Nevertheless, the authors dismissed the possibility that their inference of a faster-X effect was affected, as they claimed that such inflation is expected to be a general pattern across the genome, and would affect synonymous and nonsynonymous divergence equally.
We have investigated this possibility in more detail, as described at length in the supplementary text, Supplementary Material online. Briefly, we first confirm that ancestral polymorphism is likely to be a major component of neutral divergence between these species, by showing that the divergence times estimated from sequence data are sufficiently small (shorter than 4Ne generations [Charlesworth et al. 2005, eqs. 14 and 15]). To show this, we use KS, corrected for within species diversity (Haddrill et al. 2010), as an estimate of 2u times the divergence time, and πS, an estimate of 4Neu. The ratio of these two quantities thus constitutes a rough estimate of the time separating the species in units of 2Ne generations. The divergence time estimates obtained for X-linked and autosomal loci are 0.88 and 1.80, respectively, well within the range for which ancestral polymorphisms are expected to have a large contribution to neutral fixations.
Next, we ask whether the higher KA values for the X-linked versus autosomal loci can be explained solely by the fixation by genetic drift of ancestral polymorphisms, which might occur more rapidly on the X chromosome than the autosomes, given that its Ne is smaller. In general, the contribution of ancestral polymorphisms to the expected neutral divergence between two independently evolving lineages is equal to the pairwise neutral diversity in the ancestor, π-anc (Charlesworth et al. 2005). If we assume that nonsynonymous variants are neutral, and that the current πA values for D. pseudoobscura represent the ancestral values (this is likely to be conservative, given the lower diversity values in D. persimilis, as described in the supplementary text, Supplementary Material online), we can estimate the expected contribution to the KA values from ancestral polymorphisms. In reality, the assumption of neutrality provides an upper limit, as πA values must include a contribution from deleterious mutations, whose fixation is resisted by selection and hence do not contribute to KA. Using the highest estimate of πA in table 2, we obtain a maximum contribution of ancestral polymorphism to KA of 0.00136, only about 10% of the observed KA value for the X-linked loci (with values for XL and XR combined). Because ancestral polymorphism contributes only a tiny amount to X-linked divergence, it seems impossible to account for the faster-X effect in these species by fixations of ancestral polymorphisms. The magnitude of this discrepancy is so large that it has a very low probability of arising by chance: Even when not adjusting for the contribution of within-species polymorphism to KS, the KA values adjusted for within-species polymorphism still result in a significantly higher KA/KS for X-linked loci (mean adjusted KA to unadjusted KS values for X-linked loci is 0.168, compared with an autosomal value of 0.121; Mann–Whitney U test, P = 2 × 10−13).
This analysis ignores, however, the possible effects of ongoing gene flow between the two species, for which there is statistical support from the use of the IM algorithm (Hey and Nielsen 2004). To yield an apparent faster-X effect for nonsynonymous mutations, however, there would have to be a difference among X and A genes in the extent of introgression, with lower rates of introgression for X genes, for which there was no evidence in the (admittedly very limited) data set analyzed by Hey and Nielsen (2004). Furthermore, the theory of drift, mutation, and selection in subdivided populations implies that purifying selection against deleterious mutations leads to lower divergence among populations connected by migration than for neutral sites (Charlesworth B and Charlesworth D 2010, p. 355). If a lower rate of introgression for X-linked genes were the only factor involved, nonsynonymous sites would be less diverged than the more weakly selected synonymous sites, which is the opposite of what we observed.
These arguments seem to leave only the possibility that these patterns are caused by higher rates of fixation of nonsynonymous mutations on the X chromosome arms in either D. pseudoobscura or D. persimilis. This could be due either to a higher mutation rate or to a higher rate of adaptive evolution on the X. Given that KA/KS is significantly elevated in the pse–per comparison (even using the estimates of KS that are uncorrected for within-species polymorphism), the latter seems to be the only viable explanation. This then raises the question of why a faster-X effect is detected for pse–per but not for the other pseudoobscura clade comparisons.
One possibility is that there is increased accumulation of species-specific differences in divergent chromosomal arrangements, as these are associated with hybrid sterility (Noor et al. 2000, 2007; McGaugh and Noor 2012). That is, because these constitute large blocks of loci that cannot introgress between species, they are free to accumulate species-specific adaptations. As roughly a third of each arm of the X is associated with inversion differences between the species, X-linked loci may be disproportionately affected. But analyzing loci in noninverted regions separately shows that the faster-X effect occurs in these regions as well (median KA/KS values for A–A = 0.106, A–X = 0.138, and X–X = 0.155, Kruskal–Wallis test P = 6 × 10−11 and Mann–Whitney U comparisons between A–A and A–X P = 0.0001, between A–A and X–X, P = 1 × 10−9). Comparisons between X-linked and autosomal loci inside and near inversions are also consistent with a faster-X effect, but nonsignificant, which is probably due to the smaller number of loci in these regions.
The Relationship between Diversity and Rate of Protein Sequence Evolution
The estimates of synonymous nucleotide site diversity for our data set of fast-evolving genes appear to be similar to those for a data set of more highly conserved genes (Haddrill et al. 2010, table 1), as noted above. In contrast, the mean nonsynonymous site diversity values are substantially lower for the more highly conserved set (conserved gene set πA = 0.00066 for both A and X vs. fast-evolving genes πA = 0.0036 and 0.0014 for A and X, respectively). This suggests that differences in levels of selective constraint play a major role in causing the differences between the two sets of genes, with the fast-evolving genes being under weaker constraints with respect to purifying selection. This in turn implies that the more rapid protein sequence evolution of these genes mainly reflects weaker purifying selection, not more intense positive selection, consistent with the fact that the α values in table 3 are not exceptionally large in comparison to those from other studies of Drosophila species (Sella et al. 2009; Campos et al. 2014). Further, despite the large differences in KA between our set of fast-evolving genes and the set of conserved genes from Haddrill et al. (2010), πS is barely different between the two data sets. There is also no evidence for a negative correlation between KA and πS for these genes (supplementary table S7, Supplementary Material online), contrary to what was found for fast-evolving genes in D. melanogaster in a previous study (Haddrill et al. 2011). This is consistent with the interpretation that the difference in KA is largely due to relaxed selective constraints on the fast-evolving genes, so that πS is not being reduced by the localized effects of selective sweeps in genes as appears to be the case for fast-evolving genes in D. melanogaster (Andolfatto 2007; Sella et al. 2009; Jensen and Bachtrog 2010; Haddrill et al. 2011). In addition, πA is significantly positively correlated with KA (supplementary table S7, Supplementary Material online), as was also found for the more highly conserved D. pseudoobscura set of genes (Haddrill et al. 2011); this is also hard to reconcile with major effects of selective sweeps on variability within the genes affected.
Supplementary Material
Supplementary text, figures S1 and S2, and tables S1–S7 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by research grants BB/G003076/1 and BB/H006029/1 to B.C. from the Biotechnology and Biological Sciences Research Council of the United Kingdom, and by a Spanish Government fellowship to V.A. The authors thank the editor and three anonymous reviewers, whose comments greatly improved this article. They also thank H. Cowan for technical assistance, and J. Parsch and V. Nolte for helpful discussion, and P. Keightley for access to computational facilities.
Literature Cited
- Andolfatto P. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol Biol Evol. 2001;18:279–290. doi: 10.1093/oxfordjournals.molbev.a003804. [DOI] [PubMed] [Google Scholar]
- Andolfatto P. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 2007;17:1755–1762. doi: 10.1101/gr.6691007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquadro CF, Begun DJ, Kindahl EC. Selection, recombination, and DNA polymorphism in Drosophila. In: Brian Golding., editor. Non-neutral evolution. New York: Springer; 1994. pp. 46–56. [Google Scholar]
- Ashburner M, Golic KG, Hawley RS. Drosophila. A laboratory handbook. Cold Spring Harbor (NY): Cold Spring Harbor Press; 2005. [Google Scholar]
- Assis R, Zhou Q, Bachtrog D. Sex-biased transcriptome evolution in Drosophila. Genome Biol Evol. 2012;4:1189–1200. doi: 10.1093/gbe/evs093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Jensen JD, Zhang Z. Accelerated adaptive evolution on a newly formed X chromosome. PLoS Biol. 2009;7:e82. doi: 10.1371/journal.pbio.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Betancourt AJ, Kim Y, Orr HA. A pseudohitchhiking model of X vs. autosomal diversity. Genetics. 2004;168:2261–2269. doi: 10.1534/genetics.104.030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Eyre-Walker A. The problem of counting sites in the estimation of the synonymous and nonsynonymous substitution rates: implications for the correlation between the synonymous substitution rate and codon usage bias. Genetics. 2003;165:1587–1597. doi: 10.1093/genetics/165.3.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer MG. The mathematical theory of quantitative genetics. Oxford (United Kingdom): Clarendon Press; 1980. [Google Scholar]
- Campos JL, Charlesworth B, Haddrill PR. Molecular evolution in nonrecombining regions of the Drosophila melanogaster genome. Genome Biol Evol. 2012;4:278–288. doi: 10.1093/gbe/evs010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos JL, Halligan DL, Haddrill PR, Charlesworth B. The relation between recombination rate and patterns of molecular evolution and variation in Drosophila melanogaster. Mol Biol Evol. 2014;31:1010–1028. doi: 10.1093/molbev/msu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos JL, Zeng K, Parker DJ, Charlesworth B, Haddrill PR. Codon usage bias and effective population sizes on the X chromosome versus the autosomes in Drosophila melanogaster. Mol Biol Evol. 2013;30:811–823. doi: 10.1093/molbev/mss222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. The role of background selection in shaping patterns of molecular evolution and variation: evidence from variability on the Drosophila X chromosome. Genetics. 2012;191:233–246. doi: 10.1534/genetics.111.138073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Bartolomé C, Noël V. Estimating the incidence of ancestral polymorphisms. Genet Res. 2005;86:149–157. doi: 10.1017/S0016672305007743. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. Elements of evolutionary genetics. Greenwood Village (CO): Roberts & Company; 2010. [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987;130:113–146. [Google Scholar]
- Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Comeron JM. A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J Mol Evol. 1995;41:1152–1159. doi: 10.1007/BF00173196. [DOI] [PubMed] [Google Scholar]
- Comeron JM, Kreitman M, Aguade M. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics. 1999;151:239–249. doi: 10.1093/genetics/151.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T. Adaptive protein evolution of X-linked and autosomal genes in Drosophila: implications for faster-X hypotheses. Mol Biol Evol. 2007;24:2566–2572. doi: 10.1093/molbev/msm199. [DOI] [PubMed] [Google Scholar]
- Connallon T, Clark AG. Association between sex-biased gene expression and mutations with sex-specific phenotypic consequences in Drosophila. Genome Biol Evol. 2011;3:151–155. doi: 10.1093/gbe/evr004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Singh ND, Clark AG. Impact of genetic architecture on the relative rates of X versus autosomal adaptive substitution. Mol Biol Evol. 2012;29:1933–1942. doi: 10.1093/molbev/mss057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counterman BA, Noor MA. Using comparative genomic data to test for fast-X evolution. Evolution. 2004;58:565–660. [PubMed] [Google Scholar]
- Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Genomic evidence for a large-Z effect. Proc Biol Sci. 2009;276:361–366. doi: 10.1098/rspb.2008.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 2009;26:2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wyckoff GJ, Wu CI. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- Fiston-Lavier AS, Singh ND, Lipatov M, Petrov DA. Drosophila melanogaster recombination rate calculator. Gene. 2010;463:18–20. doi: 10.1016/j.gene.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Gnad F, Parsch J. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics. 2006;22:2577–2579. doi: 10.1093/bioinformatics/btl422. [DOI] [PubMed] [Google Scholar]
- Gossmann TI, et al. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol Biol Evol. 2010;27:1822–1832. doi: 10.1093/molbev/msq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grath S, Parsch J. Rate of amino acid substitution is influenced by the degree and conservation of male-biased transcription over 50 myr of Drosophila evolution. Genome Biol Evol. 2012;4:346–359. doi: 10.1093/gbe/evs012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Loewe L, Charlesworth B. Estimating the parameters of selection on nonsynonymous mutations in Drosophila pseudoobscura and D. miranda. Genetics. 2010;185:1381–1396. doi: 10.1534/genetics.110.117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Zeng K, Charlesworth B. Determinants of synonymous and nonsynonymous variability in three species of Drosophila. Mol Biol Evol. 2011;28:1731–1743. doi: 10.1093/molbev/msq354. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. A mathematical theory of natural and artificial selection. Trans Camb Philos Soc. 1924;23:19–41. [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Eisen MB, Thornton KR, Andolfatto P. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 2013;23:89–98. doi: 10.1101/gr.141689.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JD, Bachtrog D. Characterizing recurrent positive selection at fast-evolving genes in Drosophila miranda and Drosophila pseudoobscura. Genome Biol Evol. 2010;2:371–378. doi: 10.1093/gbe/evq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang ZF, Machado CA. Evolution of sex-dependent gene expression in three recently diverged species of Drosophila. Genetics. 2009;183:1175–1185. doi: 10.1534/genetics.109.105775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayserili MA, Gerrard DT, Tomancak P, Kalinka AT. An excess of gene expression divergence on the X chromosome in Drosophila embryos: implications for the faster-X hypothesis. PLoS Genet. 2012;8:e1003200. doi: 10.1371/journal.pgen.1003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Hall DW. Male-biased mutation, sex linkage, and the rate of adaptive evolution. Evolution. 2004;58:437–440. [PubMed] [Google Scholar]
- Kofler R, et al. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One. 2011;6:e15925. doi: 10.1371/journal.pone.0015925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A, Halligan DL, Keightley PD. Faster-X adaptive protein evolution in house mice. Genetics. 2014;196:1131–1143. doi: 10.1534/genetics.113.158246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, et al. Genomic variation in natural populations of Drosophila melanogaster. Genetics. 2012;192:533–598. doi: 10.1534/genetics.112.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, Montgomery E, Hudson R, Kaplan N, Charlesworth B. On the role of unequal exchange in the containment of transposable element copy number. Genet Res. 1988;52:223–235. doi: 10.1017/s0016672300027695. [DOI] [PubMed] [Google Scholar]
- Machado CA, Haselkorn TS, Noor MA. Evaluation of the genomic extent of effects of fixed inversion differences on intraspecific variation and interspecific gene flow in Drosophila pseudoobscura and D. persimilis. Genetics. 2007;175:1289–1306. doi: 10.1534/genetics.106.064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, et al. The Drosophila melanogaster genetic reference panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet MA, Bouchard JM, Kimber CM, Chippindale AK. Experimental mutation-accumulation on the X chromosome of Drosophila melanogaster reveals stronger selection on males than females. BMC Evol Biol. 2011;11:156. doi: 10.1186/1471-2148-11-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Axelsson E, Ellegren H. Fast-X on the Z: rapid evolution of sex-linked genes in birds. Genome Res. 2007;17:618–624. doi: 10.1101/gr.6031907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Nam K, Ellegren H. Faster-Z evolution is predominantly due to genetic drift. Mol Biol Evol. 2010;27:661–670. doi: 10.1093/molbev/msp282. [DOI] [PubMed] [Google Scholar]
- Mank JE, Vicoso B, Berlin S, Charlesworth B. Effective population size and the faster-X effect: empirical results and their interpretation. Evolution. 2010;64:663–674. doi: 10.1111/j.1558-5646.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. Evolution in sexual and asexual populations. Am Nat. 1968;102:469–473. [Google Scholar]
- McGaugh SE, et al. Recombination modulates how selection affects linked sites in Drosophila. PLoS Biol. 2012;10:e1001422. doi: 10.1371/journal.pbio.1001422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh SE, Noor MAF. Genomic impacts of chromosomal inversions in parapatric Drosophila species. Philos Trans R Soc Lond B Biol Sci. 2012;367:422–429. doi: 10.1098/rstb.2011.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Presgraves DC. Little evidence for demasculinization of the Drosophila X chromosome among genes expressed in the male germline. Genome Biol Evol. 2012;4:1007–1016. doi: 10.1093/gbe/evs077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP. Towards a more nuanced understanding of the relationship between sex-biased gene expression and rates of protein-coding sequence evolution. Mol Biol Evol. 2011;28:1893–1900. doi: 10.1093/molbev/msr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Connallon T. The faster-X effect: integrating theory and data. Trends Genet. 2013;29:537–544. doi: 10.1016/j.tig.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Malone JH, Clark AG. Faster-X evolution of gene expression in Drosophila. PLoS Genet. 2012a;8:e1003013. doi: 10.1371/journal.pgen.1003013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Malone JH, Clark AG. Disentangling the relationship between sex-biased gene expression and X-linkage. Genome Res. 2012b;22:1255–1265. doi: 10.1101/gr.132100.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. Bearing of the Drosophila work on systematics. In: Julian Huxley., editor. The new systematics. Oxford (United Kingdom): Oxford University Press; 1940. pp. 185–268. [Google Scholar]
- Noor MA, Garfield DA, Schaeffer SW, Machado CA. Divergence between the Drosophila pseudoobscura and D. persimilis genome sequences in relation to chromosomal inversions. Genetics. 2007;177:1417–1428. doi: 10.1534/genetics.107.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Johnson NA, Hey J. Gene flow between Drosophila pseudoobscura and D. persimilis. Evolution. 2000;54:2174–2175. doi: 10.1111/j.0014-3820.2000.tb01262.x. discussion 2176-2177. [DOI] [PubMed] [Google Scholar]
- Orr HA. The rate of adaptation in asexuals. Genetics. 2000;155:961–968. doi: 10.1093/genetics/155.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Betancourt AJ. Haldane’s sieve and adaptation from the standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri N, Kosiol C, Schlötterer C. The life cycle of Drosophila orphan genes. eLife. 2014;3:e01311. doi: 10.7554/eLife.01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JC, Harrison PW, Mank JE. The ontogeny and evolution of sex-biased gene expression in Drosophila melanogaster. Mol Biol Evol. 2014;31:1206–1219. doi: 10.1093/molbev/msu072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, et al. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 2012;8:e1003080. doi: 10.1371/journal.pgen.1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW, et al. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics. 2008;179:1601–1655. doi: 10.1534/genetics.107.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ND, Davis JC, Petrov DA. X-linked genes evolve higher codon bias in Drosophila and Caenorhabditis. Genetics. 2005;171:145–155. doi: 10.1534/genetics.105.043497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ND, Larracuente AM, Clark AG. Contrasting the efficacy of selection on the X and autosomes in Drosophila. Mol Biol Evol. 2008;25:454–467. doi: 10.1093/molbev/msm275. [DOI] [PubMed] [Google Scholar]
- Singh ND, Macpherson JM, Jensen JD, Petrov DA. Similar levels of X-linked and autosomal nucleotide variation in African and non-African populations of Drosophila melanogaster. BMC Evol Biol. 2007;7:202. doi: 10.1186/1471-2148-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996. [Internet]. [cited 2014 Oct 22]. Available from: http://www.repeatmasker.org. [Google Scholar]
- Smith NG, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH. The genetics of Drosophila simulans. Carnegie Inst Washington Publ. 1929;399:1–62. [Google Scholar]
- Thornton K, Bachtrog D, Andolfatto P. X chromosomes and autosomes evolve at similar rates in Drosophila: no evidence for faster-X protein evolution. Genome Res. 2006;16:498–504. doi: 10.1101/gr.4447906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson DG, Singh RS. Enhanced adaptive evolution of sperm-expressed genes on the mammalian X chromosome. Heredity (Edinb) 2006;96:39–44. doi: 10.1038/sj.hdy.6800749. [DOI] [PubMed] [Google Scholar]
- True JR, Mercer JM, Laurie CC. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics. 1996;142:507–523. doi: 10.1093/genetics/142.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Effective population size and the faster-X effect: an extended model. Evolution. 2009;63:2413–2426. doi: 10.1111/j.1558-5646.2009.00719.x. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Haddrill PR, Charlesworth B. A multispecies approach for comparing sequence evolution of X-linked and autosomal sites in Drosophila. Genet Res. 2008;90:421–431. doi: 10.1017/S0016672308009804. [DOI] [PubMed] [Google Scholar]
- Welch JJ. Estimating the genomewide rate of adaptive protein evolution in Drosophila. Genetics. 2006;173:821–837. doi: 10.1534/genetics.106.056911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Bachtrog D. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science. 2012;337:341–345. doi: 10.1126/science.1225385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.