Abstract
With the goal to contribute for the understanding of satellite DNA evolution and its genomic involvement, in this work it was isolated and characterized the first satellite DNA (PSUcentSat) from Phodopus sungorus (Cricetidae). Physical mapping of this sequence in P. sungorus showed large PSUcentSat arrays located at the heterochromatic (peri)centromeric region of five autosomal pairs and Y-chromosome. The presence of orthologous PSUcentSat sequences in the genomes of other Cricetidae and Muridae rodents was also verified, presenting however, an interspersed chromosomal distribution. This distribution pattern suggests a PSUcentSat-scattered location in an ancestor of Muridae/Cricetidae families, that assumed afterwards, in the descendant genome of P. sungorus a restricted localization to few chromosomes in the (peri)centromeric region. We believe that after the divergence of the studied species, PSUcentSat was most probably highly amplified in the (peri)centromeric region of some chromosome pairs of this hamster by recombinational mechanisms. The bouquet chromosome configuration (prophase I) possibly displays an important role in this selective amplification, providing physical proximity of centromeric regions between chromosomes with similar size and/or morphology. This seems particularly evident for the acrocentric chromosomes of P. sungorus (including the Y-chromosome), all presenting large PSUcentSat arrays at the (peri)centromeric region. The conservation of this sequence in the studied genomes and its (peri)centromeric amplification in P. sungorus strongly suggests functional significance, possibly displaying this satellite family different functions in the different genomes. The verification of PSUcentSat transcriptional activity in normal proliferative cells suggests that its transcription is not stage-limited, as described for some other satellites.
Keywords: Rodentia, satellite DNA, Phodopus sungorus, chromosomal distribution, copy number, transcriptional activity
Introduction
The genomes of higher eukaryotes harbor large amounts of repeated sequences. According to their organization, two major classes can be distinguished, interspersed and tandem repeats. Satellite DNAs (satDNAs) are classified as highly tandem repeated sequences, located not only in heterochromatic regions preferentially around centromeres but also at chromosome interstitial and terminal positions (reviewed by Adega et al. 2009). Structurally these sequences are commonly formed by long arrays of up to 100 Mb, composed of monomers (or repeat units) in a sequential arrangement one after the other (e.g., Plohl et al. 2008).
SatDNAs follow principles of concerted evolution, a nonindependent mode of monomer sequence evolution within a genome and in a population (e.g., Palomeque and Lorite 2008; Plohl et al. 2008). According to this evolutionary model, mutated monomers could be spread or eliminated in the satellite arrays, leading to homogenization of repeats. This is achieved by mechanisms of nonreciprocal transfer within and between chromosomes, as gene conversion, unequal crossing-over, rolling circle replication/reinsertion, and transposon-mediated exchange (Walsh 1987; Elder and Turner 1995; Dover 2002). The chromosome configuration during the early prophase I (bouquet configuration) may facilitate the homogenization process on nonhomologous chromosomes, by the physical proximity between centromeres of chromosomes with a similar size and morphology (Brannan et al. 2001; Mravinać and Plohl 2010; Cazaux et al. 2011). As consequence of the independent action of homogenization mechanisms in different genomes, orthologous satDNAs can present high differences in its monomer size, nucleotide sequence, copy number, or chromosome organization and location (reviewed by Plohl et al. 2008).
To date, the knowledge about the genomic importance of satDNAs is limited, but several functions have been proposed to this eukaryotic genome fraction. It has been suggested the involvement of satDNAs in functions as diverse as centromeric activity (e.g., Marshall and Clarke 1995), tridimensional organization of the interphase nucleus (Manuelidis 1982), and a driver of genome reorganization during evolution (e.g., Wichman et al. 1991; Garagna et al. 1997). This last role of satDNAs is mainly justified by the high molecular dynamics of these repeats, consequence of its evolution mode. Recent works however, also show that the overexpression of satDNAs is directly associated with the occurrence of chromosomal rearrangements. Centromeric and pericentromeric regions have long been regarded as transcriptionally inert portions of chromosomes. Nevertheless, an increasing number of studies in the past 10 years refute this idea and provide credible evidences that these regions are transcriptionally active in several biological contexts (e.g., Vourc’h and Biamonti 2011; Enukashvily and Ponomartsev 2013). In fact, the transcription of satDNAs seems to be a general phenomenon (reviewed by Ugarković 2005). In accordance to what has been described, satDNA transcripts could act as long noncoding RNAs or as precursors of small interfering RNAs, which have an important role in epigenetic processes of chromatin remodeling/heterochromatin formation and in control of gene expression (reviewed by Vourc’h and Biamonti 2011; Bierhoff et al. 2013). The organismal developmental stage and the tissue-specific expression observed in some satDNAs unequivocally point to a regulatory role for these transcripts (Vourc’h and Biamonti 2011).
The species studied in this work belong to Cricetidae (Cricetus cricetus [CCR], Peromyscus eremicus [PER], Phodopus roborovskii [PRO], and Phodopus sungorus [PSU]) and Muridae (Rattus norvegicus [RNO]) families (Tree of Life web project; http://www.tolweb.org/tree/, last accessed October 27, 2014), the most specious rodent families (Musser and Carleton 2005). Based on molecular data, the divergence time between Muridae/Cricetidae can be estimated at 17 Myr (Robinson et al. 1997). In this work we report the isolation and molecular characterization of a PSU satDNA, PSUcentSat. In situ and Southern blot hybridizations suggest the presence of PSUcentSat orthologous sequences in the other studied rodent species. The transcriptional activity of this sequence was verified in normal proliferative fibroblast cells. Our data strongly suggest a functional significance of PSUcentSat in the studied genomes.
Materials and Methods
Chromosome Preparations and Genomic DNA Extraction
Fixed chromosome preparations from CCR, PER, PRO, PSU, and RNO were obtained from fibroblast cell cultures, using standard procedures described elsewhere (Chaves et al. 2004). Genomic DNA of different species was obtained from these fibroblast cell cultures using the JETQUICK DNA kit (Genomed).
Isolation, Cloning, and Sequencing of PSUcentSat Sequence
PSU genomic DNA was digested with the restriction endonuclease (RE) MboI, according to the manufacturers’ instructions (Invitrogen Life Technologies), resulting in a smear with DNA fragments ranging between 3 kb and 100 bp. The restriction products were later cloned using routine procedures (FERMENTAS Life Science, Invitrogen Life Technologies). A part of the obtained colonies were transferred onto a nylon membrane Hybond-N+ (Amersham, GE Healthcare) and the DNA on the membrane probed to MboI restriction products labeled with digoxigenin-11-dUTP, using DIG DNA labeling Kit (Roche Diagnostics). Hybridization was performed at 68 °C. The positive signals were visualized using the chemiluminiscent CDP-Star system (Roche Diagnostics). Plasmidic DNA of the positive clones was isolated using the High Pure Plasmid Isolation kit (Roche Diagnostics) and sequenced in both directions using M13 primers.
Sequence Analysis of PSUcentSat Sequence
PSUcentSat was analyzed with different sequence database tools and bioinformatic softwares: National Center for Biotechnology Information (NCBI) BLAST (http://www.ncbi.nlm.gov/Blast/, last accessed October 27, 2014), RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker, last accessed October 27, 2014), EMBOSS CpG plot (http://www.ebi.ac.uk/Tools/emboss/cpgplot/, last accessed October 27, 2014), EMBOSS einverted (http://emboss.bioinformatics.nl/cgi-bin/emboss/einverted, last accessed October 27, 2014), Tandem repeats Finder (Benson 1999, version 4.00, free download in http://tandem.bu.edu/trf/trf.html, last accessed October 27, 2014), and vector NTI advance 11 (Invitrogen Life Technologies). A BLAST search of PSUcentSat sequence against nucleotide sequences of GenBank and RepBase was accomplished using NCBI BLAST and RepeatMasker tools. Sequence alignments were performed with the software vector NTI advance 11 that applies the ClustalW algorithm (Thompson et al. 1994) to determine sequence similarities. The search for direct or inverted repeats within PSUcentSat sequence was done using Tandem Repeats Finder and the EMBOSS einverted tool, respectively. EMBOSS einverted tool was used with a minimum score threshold of 20%. The EMBOSS CpG plot allowed the identification of CpG islands. Sequence data from the PSUcentSat clone were deposited in the NCBI Nucleotide database with the following accession number: KJ649148.
Physical Mapping of PSUcentSat Sequence
Physical mapping of PSUcentSat in the chromosomes of the studied species (CCR, PER, PRO, PSU, and RNO) was carried out following fluorescent in situ hybridization (FISH) procedures described by Schwarzacher and Heslop-Harrison (2000). PSUcentSat sequence was labeled with digoxigenin-11-dUTP (Roche Molecular Biochemicals) by polymerase chain reaction (PCR). The most stringent posthybridization wash was 50% formamide/2 × SSC at 42 °C. Digoxigenin-labeled probes were detected with antidigoxigenin-5′-TAMRA (Roche Molecular Biochemicals).
CBP-Banding Sequential to Physical Mapping of PSUcentSat Sequence
After distaining the slides, CBP-banding (C-bands by barium hydroxide with propidium iodide) was performed according to the standard procedure of Sumner (1972) with slight modifications, as in Paço et al. (2013).
Capture and Preparation of Images
Chromosomes were observed in a Zeiss Axioplan Z1 microscope, and images were captured using an Axiocam MRm digital camera with LSM 510 software (version 4.0 SP2). Digitized photos were prepared in Adobe Photoshop (version 7.0); contrast and color optimization were the functions used and affected the whole image equally. The chromosomes of PSU were identified according to Romanenko et al. (2007) and RNO chromosomes according to Levan (1974).
Southern Hybridization Analysis
Genomic DNA from PSU was digested with the endonucleases AluI, HhaI, and MboI. Genomic DNA of the other studied species (CCR, PER, PRO, and RNO) was digested with AluI and MboI. The resulting fragments were separated in an agarose gel and blotted onto a Nylon membrane Hybond-N+ (Amersham, GE Healthcare). The membranes were then probed with the cloned PSUcentSat sequence, previously labeled by PCR with digoxigenin-11-dUTP (Roche Diagnostics). Hybridization was performed at 42 °C in hybridization solution (Roche Diagnostics). The positive signals were visualized using chemiluminiscent CDP-Star system (Roche Diagnostics). Selection of REs was performed using the CLC Sequence Viewer software (version 6.2, http://www.clcbio.com/index.php?id=28, last accessed October 27, 2014).
satDNA Copy Number Quantification (Absolute and Relative) by TaqMan Assay
For PSUcentSat quantification, a quantitative real-time PCR approach was performed (as in Louzada S, Vieira-da-Silva A, Mendes-da-Silva A, Kubickova S, Rubes J, Adega F, Chaves R, submitted for publication). TaqMan specific assay mix (primers/probe) was designed using Primer Express Software v3.0 (Life Technologies Applied Biosystems) based in PSUcentSat sequence. PCR primers PSUcentSat F (5′145-GCTACACTGCGCAAGAGAGATAAG-3′) and PSUcentSat R (5′209-GAGACGCTTT TCGCGAATGCTGTC3′) locate between the positions 146 and 210 bp of PSUcentSat sequence, allowing the amplification of a 64-bp product. The probe (5′170[6-carboxy-fluorescein, FAM]-CACTGTGAGAGTAAAGAG-3′[nonfluorescent quencher, NFQ]) had the fluorescent reporter dye, FAM, located at the 5′-end and the NFQ located at the 3′-end.
For PSUcentSat absolute quantification in PSU genome, the standard curve method was performed. A 10-fold serial dilution series of the plasmid DNA standard, ranging from 1 × 109 to 1 × 105 copies, was used to construct the standard curve (5 points series dilutions). The concentration of the plasmid was measured using the NanoDrop ND-1000 (NanoDrop Technologies) equipment and the corresponding plasmid copy number was calculated using the following equation:
In the respective formula: Avogadros number = 6.023 × 1023 molecules (copy number/1 mol); Average molecular weight of a double-stranded DNA molecule = 600 g/mol/bp and the plasmid DNA length is 3,014 bp (pUC 19 vector plus the insert).
CT values in each dilution were measured using quantitative real-time PCR (qPCR) with the TaqMan-specific assay described above to generate the standard curve for PSUcentSat. Briefly, the standard curve includes a plot of the CT values versus the log concentration of the plasmid DNA standard. For PSU genomic DNA, the unknown total DNA sample was obtained by interpolating its CT value against the standard curve. We used 1 and 5 ng of PSU genomic DNA in the PCR reactions. These reactions were performed for a total of 20 μl with 1.25 µl of the primer/probe assay mixture and 12.5 μl of TaqMan Genotyping Master Mix. This experiment was carried out in StepOne real-time PCR system (Life Technologies Applied Biosystems), where the samples were subjected to an initial denaturation at 95 °C (10 min), and then to 40 cycles at 95 °C for 15 s followed by 60 °C for 1 min. All reactions were performed in triplicate, and negative controls (without DNA) were also run. The StepOne software (version 2.2.2, Life Technologies Applied Biosystems) was used to generate the standard curve and to analyze the data. Only standard curves with the following parameters were considered to be typically acceptable: R2 > 0.99 and slopes between −3.1 and −3.6 giving reaction efficiencies between 90% and 110%. The absolute quantification of PSUcentSat allowed determining the copy number of this sequence in PSU genome to 1 and 5 ng, which comprises 333 and 1,667 haploid genomes, respectively.
For PSUcentSat quantification within the other species genomes, a relative quantification real-time PCR approach was used, being PSU genome the control sample. The same PSUcentSat TaqMan assay described for the absolute quantification and the 18 S gene (HS99999901_s1; Life Technologies Applied Biosystems) was used as the reference assay. For this comparative analysis, PCR reactions were performed with 5 ng of genomic DNA. Mixture reactions and real-time PCR conditions were the same already described. All reactions were performed in triplicate, and negative controls (without template) were run for each master mix. StepOne software version 2.2.2 (Life Technologies Applied Biosystems) was applied for comparative analysis, and the quantification was normalized with 18 S gene. The 2−ΔΔCT method (Livak and Schmittgen 2001) was used to calculate fold changes in the amount of PSUcentSat in the different species. Results are shown as the log10 of 2−ΔΔCT PSUcentSat copy number in CCR, PER, PRO, and RNO relatively to PSU (control sample). Student’s t-test was used to compare the data obtained. Values were expressed as the mean ± SD, and differences were considered statistically significant at P < 0.05, representing the 95% confidence interval.
As it is not yet available information about the genome size (bp) and mass (pg) of PSU genome, we considered that the haploid PSU genome presents approximately 3 × 109 bp and weights 3 pg, according to the size and mass of other Cricetidae genomes in the Animal Genome Size database (http://www.genomesize.com/, last accessed October 27, 2014). The same was considered for PRO haploid genome. The genome mass of CCR, PER, and RNO is approximately 3.44, 3.3, and 3.2, respectively.
RNA Isolation and Reverse Transcription Quantitative Real-Time PCR
Total and small RNA from PER, PSU, and RNO fibroblast cell lines was isolated using mirVana Isolation Kit (Ambion, Invitrogen Life technologies), following manufacturer’s recommendations. Expression experiments were performed using the TaqMan RNA-to-CT 1-Step Kit (Life Technologies Applied Biosystems). The same PSUcentSat TaqMan assay described previously was used as target and as reference assay the glyceraldehyde-3-phosphate dehydrogenate (GAPDH, Rn01749022_g1; Life Technologies Applied Biosystems). The 20 µl reactions included 250 ng of total or small RNA, 1 µl of the primer/probe assay mixture, 10 µl of PCR Master Mix, 0.5 µl of RT enzyme mix (Life Technologies Applied Biosystems), and 3.5 µl of DEPC-treated water. This experiment was carried out in StepOne real-time PCR system (Life Technologies Applied Biosystems), where the samples were subjected to 48 °C for 15 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. All reactions were performed in triplicate, and negative controls (without template) were run for each master mix. StepOne software version 2.2.2 (Life Technologies Applied Biosystems) was applied for comparative analysis, and the relative expression level was normalized with GAPDH gene expression. The 2−ΔΔCT method (Livak and Schmittgen 2001) was used to calculate fold changes in the expression levels of PSUcentSat sequence in different genomes, using the expression in PSU as control. Besides, the fold changes in the expression levels of total and small RNA in each species were calculated using total RNA as control.
Results
Molecular Analysis of PSUcentSat
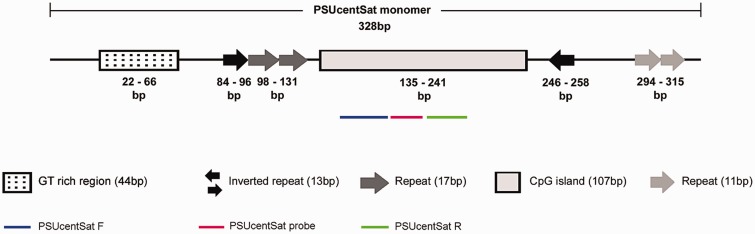
In this work it was isolated, sequenced, and molecularly characterized a novel satDNA from the genome of PSU (PSUcentSat). BLAST search revealed no significant similarity between this sequence and any other described and deposited in GenBank or in RepBase databases. As can be observed in figure 1, direct and inverted short internal repeats were detected within PSUcentSat sequence (PSUcentSat monomer whose length was determined by Southern blot analysis as described below). Namely, two different short direct repeats with 11 and 17 bp, a GT rich region presenting 19 tandem GT repeats and an inverted short repeat with 13 bp. A CpG island with 107 bp, between the positions 135 and 241 bp (fig. 1), was also indentified in this sequence.
Fig. 1.—
Organization of PSUcentSat. Schematic representation of PSUcentSat molecular features and monomer length. Colored lines indicate the region for which TaqMan-specific assay mix (primers/probe) was designed, and was used for copy number quantification and transcription analysis. Blue line corresponds to the PSUcentSat forward primer and the green line corresponds to the PSUcentSat reverse primer.
Physical Distribution of the PSUcentSat in Chromosomes of Five Rodent Species
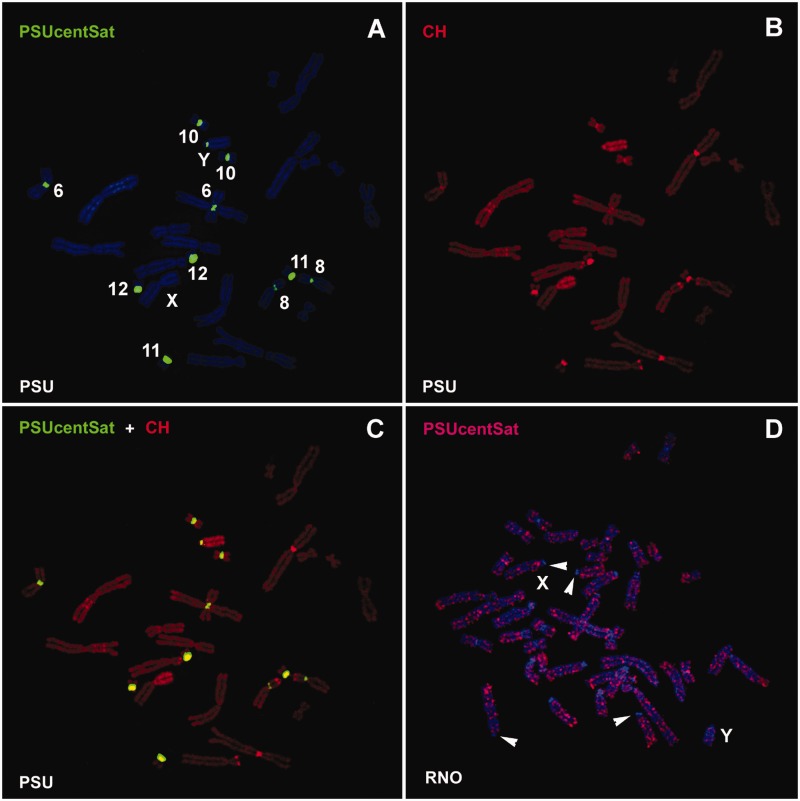
Physical mapping of PSUcentSat was performed by FISH in the studied species CCR, PER, PRO, PSU, and RNO genomes. RNO was used as outgroup for this analysis as it is the only species outside Cricetidae. In PSU genome, PSUcentSat presents a chromosome distribution characteristic of a tandem repeat sequence, organized as large blocks at the (peri)centromeric region of five autosomal pairs and in the Y-chromosome, PSU6, PSU8, PSU10, PSU11, PSU12, and PSUY (fig. 2A). C-banding sequential to FISH (fig. 2B) evidenced a colocalization of this sequence with constitutive heterochromatin, as can be seen in figure 2C. In the other four species, PSUcentSat presents a scattered distribution along all the chromosomes of the complement (as can be observed in fig. 2D for RNO chromosomes). Besides, the majority of the (peri)centromeric regions in these four species presents a depletion of the sequence (some of these regions are evidenced by arrowheads in fig. 2D).
Fig. 2.—
Physical mapping of PSUcentSat on chromosomes of PSU and RNO. (A) Representative in situ hybridization presenting the chromosomal localization of PSUcentSat on chromosomes of PSU. The sequence was labeled with digoxigenin-11-dUTP and detected with 5′-TAMRA (red), but here it is presented in the pseudocolor green. Chromosomes were counterstained with DAPI (blue). (B) Same metaphase after sequential CBP-banding. Chromosomes were counterstained with propidium iodide (red). (C) Overlapping of PSUcentSat hybridization signals with C-bands. (D) Representative in situ hybridization presenting the chromosomal localization of PSUcentSat on chromosomes of RNO. Arrowheads evidence a depletion of PSUcentSat at the (peri)centromeric regions of some RNO chromosomes.
Genomic Organization of PSUcentSat
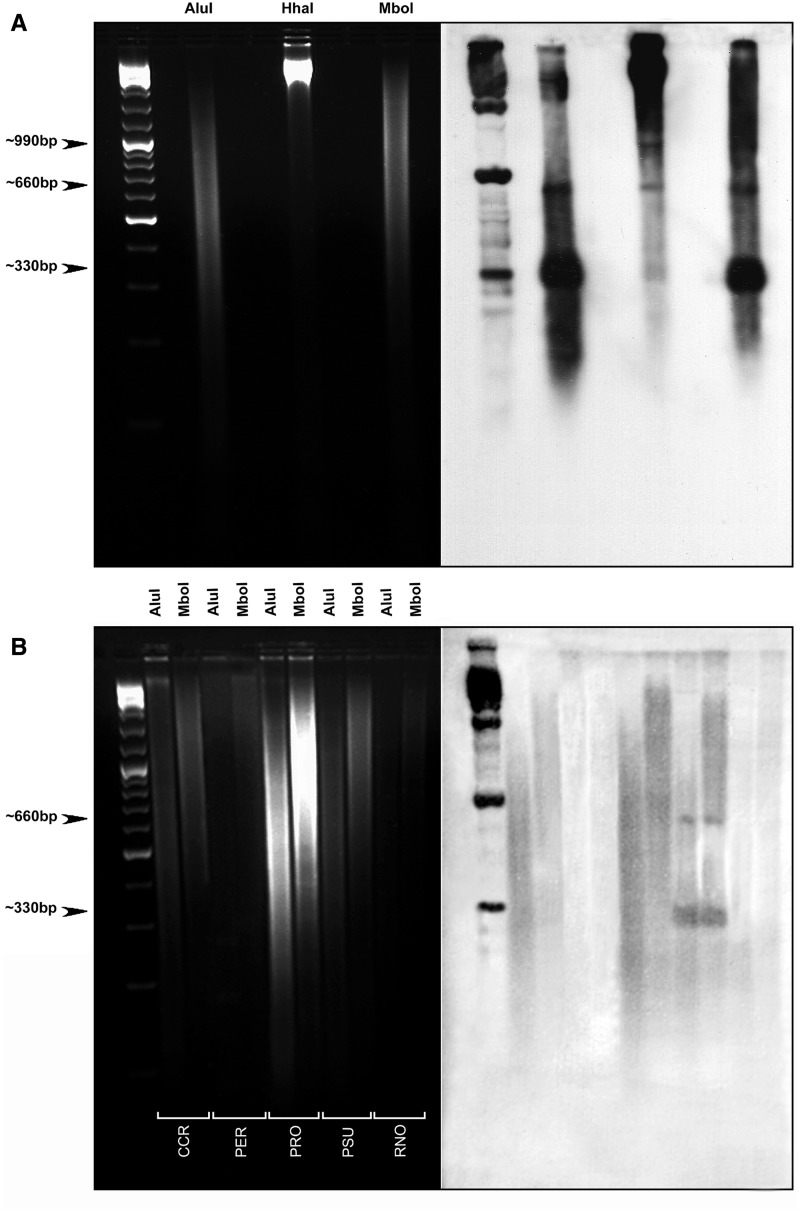
In order to investigate the genomic organization of PSUcentSat in the five studied rodent species, Southern blot analyses were carried out. The ladder hybridization pattern obtained for PSUcentSat in PSU using AluI, HhaI, and MboI enzymes (fig. 3A) indicates a tandem organization characteristic of a satellite sequence. A common band with approximately 330 bp was obtained with all the enzymes used (monomer), and other bands were also observed showing a 330-bp periodicity: 660 bp (dimmer) and 990 bp (trimmer). The enzymes used in these analyses cut only once PSUcentSat monomer, allowing the determination of its length (bp). According to similarities in size (bp), we assumed that PSUcentSat monomers present a very similar sequence with the PSUcentSat clones isolated here, presenting a length of 328 bp. As can be seen in the figure 3B, the Southern hybridization pattern obtained for CCR, PER, PRO, and RNO species is not indicative of a tandem organization for the PSUcentSat in these genomes. Contrary to what occurs in PSU, a scattered pattern of hybridization was observed for these four species (fig. 3B).
Fig. 3.—
Southern blot analysis. (A) Electrophoresis separation of PSU genomic DNA after digestion with AluI, HhaI, and MboI (shown on the left). The corresponding Southern blot obtained after hybridization with PSUcentsat is shown on the right. (B) Electrophoresis separation of CCR, PER, PRO, and RNO genomic DNA after digestion with AluI and MboI (shown on the left). The corresponding Southern blot obtained after hybridization with PSUcentsat is shown on the right.
PSUcentSat DNA Copy Number Analysis
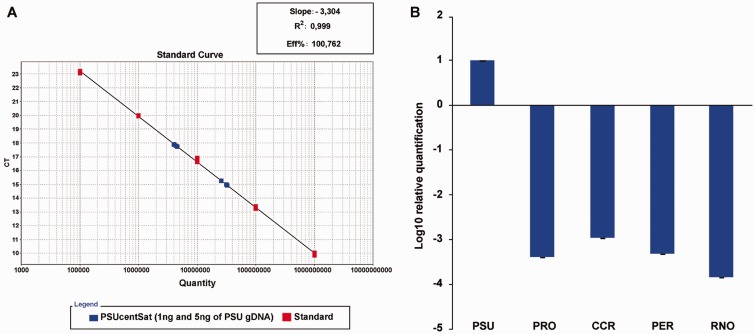
The satellite copy number quantification, performed by a new methodology based in real-time qPCR allied to TaqMan chemistry (as described in Louzada S, Vieira-da-Silva A, Mendes-da-Silva A, Kubickova S, Rubes J, Adega F, Chaves R, submitted for publication), shows significant differences in the copy number of PSUcentSat in the five studied genomes. Absolute quantification using a standard curve (fig. 4A) revealed that at least 0.2% of PSU haploid genome is comprised by PSUcentSat, corresponding to at least 17,895 copies per haploid genome. Considering that PSUcentSat can present several monomer variants and we have only analyzed one, the copy number estimated by this approach for PSU is the minimal number of copies that this satDNA present in this genome. Relative quantification showed that the amount of PSUcentSat in the other species is lower than in PSU (940–7,000 times lower) (fig. 4B), presenting all the results statistically significant values (P < 0.05). From the other analyzed species, it is the RNO genome that presents the lower number of copies of PSUcentSat ( ∼ 7,000 times lower) in comparison with the genome of PSU.
Fig. 4.—
PSUcentsat copy number quantification. (A) Standard calibration curve used in the absolute quantification of PSUcentSat copy number in the genome of PSU. For this analysis were used 1 and 5 ng of genomic DNA (gDNA PSU). The two groups of blue cubes indicate the copy number estimated for 1 and 5 ng of gDNA. (B) Relative quantification (represented as log10) of PSUcentSat in CCR, PRO, PER, and RNO using PSU as control. Error bars represent ± SD.
Transcription Analysis of PSUcentSat Satellite Sequence
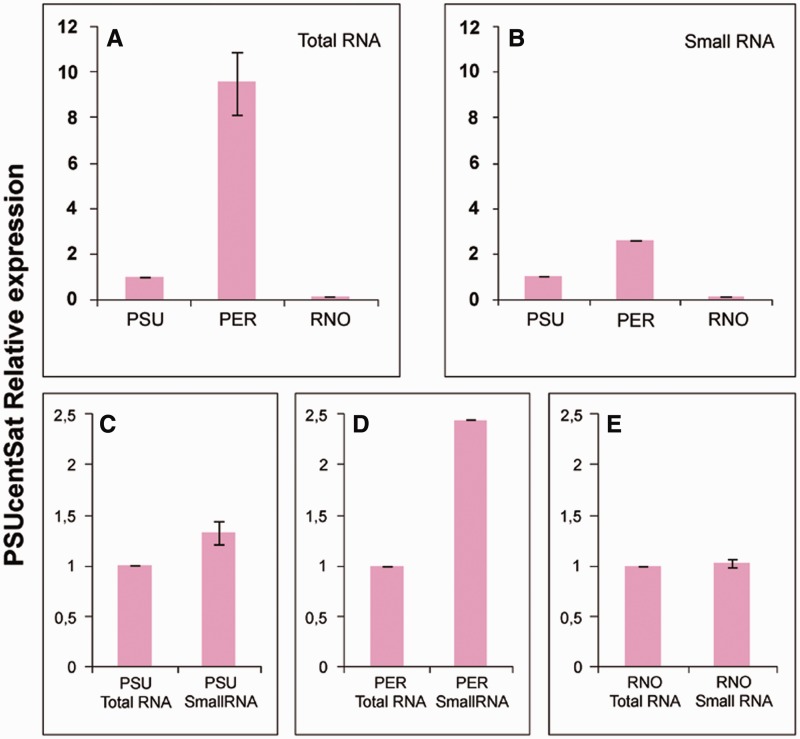
We have also verified the transcription of PSUcentSat in total and small RNA isolated from normal proliferative fibroblast cells of PER, PSU, and RNO. Figure 5 resumes the results of the relative reverse transcription quantitative real-time PCR (RT-qPCR) quantification, in terms of the fold change in PSUcentSat RNA expression, normalized using GAPDH gene expression, and calculated relatively to PSU PSUcentSat expression (expression in different genomes) or relatively to total RNA PSUcentSat expression (expression in each genome). The levels of PSUcentSat transcription in both total and small RNA is higher in PER and lower in RNO, relatively to what happens in PSU (fig. 5A and B). In PER, PSUcentSat transcription in small RNA is higher relatively to the transcription in total RNA (fig. 5D). The expression values presented in figure 5A, 5B, and 5D were considered statistically significant following analyses using Student’s t-test with a P value < 0.05. In PSU and RNO, the differences in the transcription level for both RNA fractions were considered statistically nonsignificant (fig. 5C and 5E).
Fig. 5.—
Relative expression analysis of PSUcentSat in fibroblast cells of PSU, PER, and RNO. (A) Relative expression analysis of PSUcentSat in total RNA from fibroblast cells of PSU, PER, and RNO. (B) Relative expression analysis of PSUcentSat in small RNA from fibroblast cells of PSU, PER, and RNO. Expression results were obtained by RT-qPCR, normalized with the expression of the reference gene GAPDH and the PSUcentSat expression in PER and RNO genomes compared with the expression in PSU genome (control). (C) Relative expression analysis of PSUcentSat in total and small RNA from a fibroblast cells of PSU, (D) PER, (E) and RNO. Expression results were obtained by RT-qPCR, normalized with the expression of GAPDH gene and PSUcentSat expression in small RNA compared with the expression in the total RNA (control). Data are presented as mean corresponding to fold change relative to the control sample (P < 0.05). Error bars represent ± SD.
Discussion
As far as we know, this report corresponds to the first study describing a satDNA sequence (PSUcentSat) from the genome of the rodent PSU. BLAST search revealed no significant similarity between PSUcentSat and any other described DNA sequence, both in GenBank or in RepBase databases, indicating that this sequence corresponds to a novel described satellite.
The study of PSUcentSat genomic organization in PSU shows that this sequence presents a monomer length of approximately 330 bp. As revealed by sequence analysis, different short direct and inverted repeat submotifs were identified within PSUcentSat monomer (fig. 1). satDNAs from different organisms, as primates, cattle, rodents, nematodes, and insects, also present internal short repeats (e.g., Miklos and Gill 1982; Singer 1982; Modi 1992, 1993; Castagnone-Sereno et al. 2000; Modi et al. 2003; Lorite et al. 2004; Mravinac et al. 2004). The functional significance of these internal repeats is unclear but it has been assumed that it is associated with the conformation of chromatin (Modi 1993; Plohl 2010), as DNA secondary and tertiary structures can be induced by particular distributions of nucleotides (reviewed by Plohl 2010). According to this, the identified repeats’ submotifs found in PSUcentSat might be contributing to the conformation of chromatin in the satellite sequence, most probably affecting its accessibility for transcription. These internal repeats could also influence the homogenization of the satellite by favoring recombinational mechanisms.
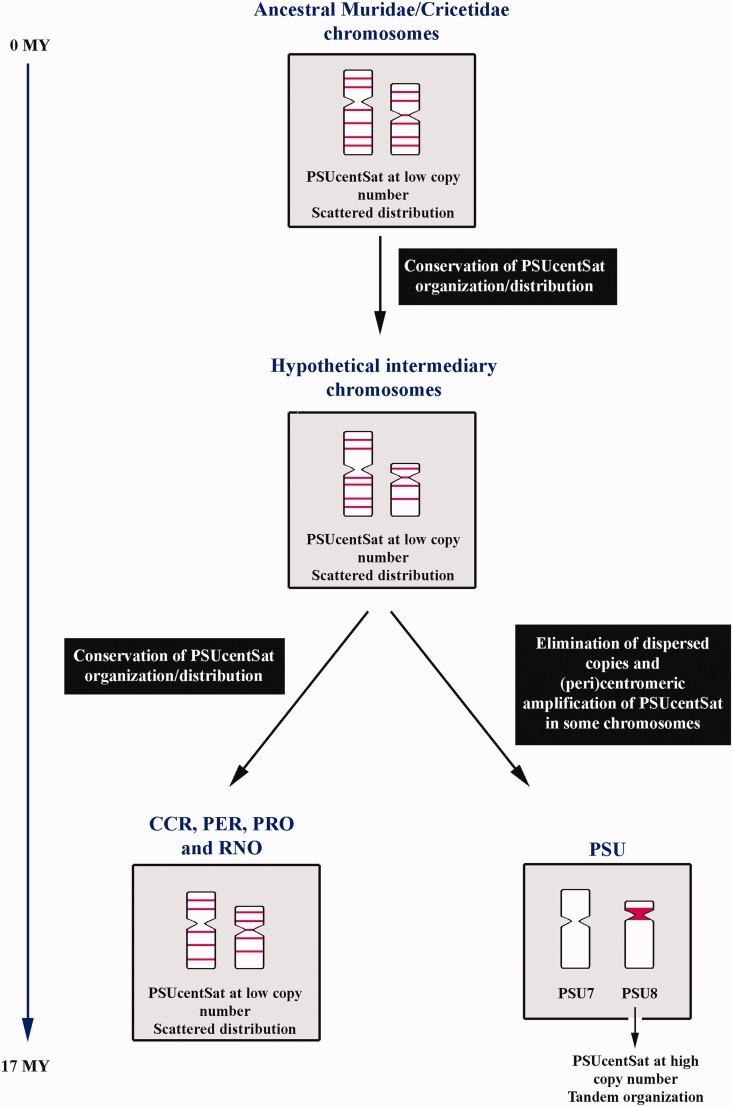
Physical mapping of PSUcentSat in PSU chromosomes showed large arrays of this sequence at the heterochromatic (peri)centromeric region of five autosomal pairs and in the Y-chromosome (fig. 2A). Southern blot and FISH indicated the presence, in an interspersed fashion, of orthologous PSUcentSat sequences in the genomes of four other rodent species belonging to Cricetidae: CCR, PER, and PRO; and Muridae: RNO (fig. 2D). A new methodology based in real-time quantification allowed estimating the copy number of PSUcentSat, revealing a 940 - to 7,000-fold lower number of copies in the analyzed genomes in comparison to PSU. Regarding PSUcentSat chromosomal distribution in the considered outgroup species (RNO) and contrarily to what would be expectable by parsimony, we can conclude that in an ancestor Muridae/Cricetidae (divergence at ∼ 17 Myr according to Robinson et al. 1997) this satDNA sequence presented, most probably a low copy number repeat with scattered distribution. For some reason, during evolution, this sequence changed its genomic organization, from initially interspersed to tandemly repeated (satDNA). Specifically, we can now observe the presence of PSUcentSat large repeat arrays at the (peri)centromeric regions of a few chromosomes in PSU. Figure 6 presents a schematization for the hypothetical evolution mode of PSUcentSat in the studied genomes.
Fig. 6.—
Hypothetical model explaining the evolution of PSUcentSat. In this figure is schematized the most parsimonious evolutionary pathway for PSUcentSat during the evolution of the studied genomes, CCR, PER, PRO, PSU, and RNO in two chromosomes, as example. Red blocks correspond to PSUcentSat location. Time estimates are according to Robinson et al. (1997).
The high level of PSUcentSat amplification in PSU chromosomes may have been mediated through different recombinational mechanisms, as unequal crossing-over and rolling circle amplification/reinsertion. Most probably, in a more ancestral version of PSU karyotype, not all chromosomes displayed this sequence at the (peri)centromeric region, as it is currently observed in the other studied species (fig. 2D), what may restricted its amplification to only a few chromosomes. The bouquet chromosome configuration (during early prophase I) possibly has also played an important role in PSUcentSat selective amplification, as it provides physical proximity of the centromeric regions of chromosomes with similar size and/or morphology. In this stage, all chromosomes migrate to one area of the nucleus and adopt an orientation in which all telomeres attach to the nuclear membrane (Scherthan et al. 1996). All the acrocentric chromosomes, independently of their size, exhibit greater proximity between the (peri)centromeric regions during the bouquet stage, favoring the occurrence of recombinational events in these regions (if homology exists). In PSU genome, all acrocentric chromosomes (PSU11, PSU12, and PSUY) present large arrays of PSUcentSat repeats at the (peri)centromeric region, explaining why PSUcentSat was amplified in the Y but not in the X-chromosome. Moreover, the analysis of synaptonemal complexes between the sex chromosomes of PSU (Spyropoulos et al. 1982) shows that the (peri)centromeric regions of the sex chromosomes do not pair, supporting our theory and justifying the apparent absence of this satellite in the X-chromosome. Simultaneously to PSUcentSat amplification in some (peri)centromeric regions, the dispersed PSUcentSat sequences initially present in PSU chromosomes were probably reduced in its copy number or eliminated (as these were not detected by the FISH analysis [fig. 2A]). This may happen due to selective pressure to keep the genome size, as proposed by Nijman and Lenstra (2001), when explaining the life history of satellite sequences.
Even though differences in genomic organization/location of this satDNA between genomes, the fact is that it was preserved for at least approximately 17 Myr in different Muridae and Cricetidae species (CCR, PER, PRO, PSU, and RNO) pointing to a probable functional significance. Nevertheless, the reasons for the different PSUcentSat amount, chromosomal location, and genomic organization in the studied genomes are unknown. Most probably, the amplification and maintenance of PSUcentSat as large arrays located at the (peri)centromere of some PSU chromosomes provided an adaptive advantage to this species, possibly in the centromeric function.
Therefore, to get some more insights about the probable functional significance of PSUcentSat, the transcriptional activity of PSUcentSat was also investigated in this work; namely in normal proliferative fibroblast cells of some of the species in analysis, PSU, PER, and RNO. Interestingly, we have demonstrated the presence of PSUcentSat transcripts in the total and small RNA fraction of these species’ cells, being the level of transcription significantly higher in PER in comparison to PSU. These interesting results show that not all PSUcentSat copies are transcriptionally active in PSU, as we found an increase of 2,090 times in copy number of this sequence in this species, in comparison to PER, showing in contrast the later a 9.6 times (total RNA) or 2.6 times (small RNA) higher transcription level of PSUcentSat in comparison to the reference genome PSU. The finding of a CpG island spreading from position 135 to 241 bp in PSUcentSat (fig. 1) indicates a possible DNA methylation transcription mediator, acting as a simple triggering mechanism, identical to that of the majority of gene promoters. Approximately, 60% of all gene promoters in human and mouse colocalize with CpG islands (Antequera 2003). This can explain the differences found between the number of DNA copies versus PSUcentSat transcripts, and why not all copies of this sequence in PSU are being transcribed in the analyzed cells. Furthermore, it has been often described in the literature, satDNAs temporally transcribed at a particular developmental stage or in different cell types, tissues or organs (reviewed by Ugarković 2005), being the methylation status an easy way to modulate its transcription. An example of this kind of satDNAs is the major satellite of mouse, which is differently expressed during development of the central nervous system, as well as in the adult liver and testis (Rudert et al. 1995). PSUcentSat transcription, in contrast, seems to occur commonly, as we detected PSUcentSat transcripts in normal fibroblast-like proliferative cells from PER, PSU, and RNO, which point to basic/constitutive cellular functions displayed by these RNAs. To unveil these, the different chromosomal locations of the sequence must also be taken into account. We believe that in PSU PSUcentSat might have a centromeric role as it was highly amplified and maintained in this region. But in PER and RNO genomes, where this sequence is highly interspersed, PSUcentSat might be involved in the regulation of gene expression, most probably, by the RNA interference mechanism, as the transcription analysis here conducted shows a significantly higher level of transcription in the small RNA relatively to the total RNA fraction in PER (fig. 5D), which could, in turn, result in small interfering RNAs (e.g., Pezer and Ugarković 2012; Enukashvily and Ponomartsev 2013).
Future works focusing in a complete characterization of PSUcentSat RNAs (transcripts length, occurrence of single- or double-stranded transcripts, and subcellular localization) will certainly enlighten the functional significance of this repeated sequence in each of the studied genomes. Nevertheless, in this context it is important to emphasize that the role of PSUcentSat cannot be restricted to its transcripts functionality, as the apparently (or temporarily, as these PSUcentSat copies may be active in specific cell types or during different development stages) inactive copies of the sequence that are maintained in the genomes might have a structural role. Several works point to structural functions for centromeric satellites sequences, namely its involvement in the loading of histone H3-like proteins (centromeric chromatin mark) and in the establishment of a favorable chromatin environment for sister chromatid cohesion (reviewed by Plohl et al. 2012). This might be the case for some PSUcentsat copies in the genome of PSU given its (peri)centromeric location.
Acknowledgments
This work was supported by the project POCI/BIA-BCM/58541/2004 and a PhD grant (SFRH/BD/41574/2007); all from the Science and Technology Foundation (FCT) from Portugal.
Literature Cited
- Adega F, Guedes-Pinto H, Chaves R. Satellite DNA in the karyotype evolution of domestic animals—clinical considerations. Cytogenet Genome Res. 2009;126:12–20. doi: 10.1159/000245903. [DOI] [PubMed] [Google Scholar]
- Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff H, Postepska-Igielska A, Grummt I. Noisy silence: Non-coding RNA and heterochromatin formation at repetitive elements. Epigenetics. 2013;9(1):1–8. doi: 10.4161/epi.26485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan CI, Disteche CM, Park LS, Copeland NG, Jenkins NA. Autosomal telomere exchange results in the rapid amplification and dispersion and Csf2ra genes in wild-derived mice. Mamm Genome. 2001;12:882–886. doi: 10.1007/s00335-001-2084-0. [DOI] [PubMed] [Google Scholar]
- Castagnone-Sereno P, Leroy F, Abad P. Cloning and characterization of an extremely conserved satellite DNA family from the root-knot nematode Meloidogyne arenaria. Genome. 2000;43:346–353. doi: 10.1139/g00-007. [DOI] [PubMed] [Google Scholar]
- Cazaux B, Catalan J, Veyrunes F, Douzery EJP, Britton-Davidian J. Are ribosomal DNA clusters rearrangement hotspots? A case study in the genus Mus (Rodentia, Muridae) BMC Evol Biol. 2011;11:124. doi: 10.1186/1471-2148-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves R, Frönicke L, Guedes-Pinto H, Wienberg J. Multidirectional chromosome painting between the Hirola antelope (Damaliscus hunteri, Alcelaphini, Bovidae), sheep and human. Chromosome Res. 2004;12:495–503. doi: 10.1023/B:CHRO.0000034751.84769.4c. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive. Trends Genet. 2002;18:587–589. doi: 10.1016/s0168-9525(02)02789-0. [DOI] [PubMed] [Google Scholar]
- Elder JF, Turner BJ. Concerted evolution of repetitive DNA sequences in eukaryotes. Q Rev Biol. 1995;70:297–320. doi: 10.1086/419073. [DOI] [PubMed] [Google Scholar]
- Enukashvily NI, Ponomartsev NV. Mammalian satellite DNA: a speaking dumb. Adv Protein Chem Struct Biol. 2013;90:31–65. doi: 10.1016/B978-0-12-410523-2.00002-X. [DOI] [PubMed] [Google Scholar]
- Garagna S, et al. Genome composition in Venezuelan spiny-rats of the genus Proechimys (Rodentia, Echimyidae). I. Genome size, C-heterochromatin and repetitive DNAs in situ hybridization patterns. Cytogenet Cell Genet. 1997;78:36–43. doi: 10.1159/000134622. [DOI] [PubMed] [Google Scholar]
- Levan G. Nomenclature for G-bands in rat chromosomes. Hereditas. 1974;77:37–52. doi: 10.1111/j.1601-5223.1974.tb01352.x. [DOI] [PubMed] [Google Scholar]
- Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lorite P, Carrillo JA, Tinaut A, Palomeque T. Evolutionary dynamics of satellite DNA in species of the genus Formica (Hymenoptera, Formicidae) Gene. 2004;332:159–168. doi: 10.1016/j.gene.2004.02.049. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Repeated DNA sequences and nuclear structure. In: Dover GA, Flavell RB, editors. Genome evolution. London (United Kingdom): Academic Press; 1982. pp. 263–285. [Google Scholar]
- Marshall LG, Clarke L. A novel cis-acting centromeric DNA elements affects S. pombe chromatin structure at a distance. J Cell Biol. 1995;128:445–454. doi: 10.1083/jcb.128.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos G, Gill AC. Nucleotide sequences of highly repeated DNAs, compilation and comments. Genet Res. 1982;39:1–30. doi: 10.1017/s0016672300020711. [DOI] [PubMed] [Google Scholar]
- Modi WS. Nucleotide sequence and genomic organization of a tandem satellite array from the rock vole Mierotus ehrotorrhinus (Rodentia) Mamm Genome. 1992;3:226–232. doi: 10.1007/BF00355723. [DOI] [PubMed] [Google Scholar]
- Modi WS. Rapid, localized amplification of a unique satellite DNA family in the rodent Microtus chrotorrhinus. Chromosoma. 1993;102:484–490. doi: 10.1007/BF00357104. [DOI] [PubMed] [Google Scholar]
- Modi WS, Serdyukova NA, Vorobieva NV, Graphodatsky AS. Chromosomal localization of six repeated DNA sequences among species of Microtus (Rodentia) Chromosome Res. 2003;11:705–713. doi: 10.1023/a:1025922813756. [DOI] [PubMed] [Google Scholar]
- Mravinać B, Plohl M. Parallelism in evolution of highly repetitive DNAs in sibling species. Mol Biol Evol. 2010;27:1857–1867. doi: 10.1093/molbev/msq068. [DOI] [PubMed] [Google Scholar]
- Mravinać B, Plohl M, Ugarković Đ. Conserved patterns in the evolution of Tribolum satellite DNAs. Gene. 2004;332:169–177. doi: 10.1016/j.gene.2004.02.055. [DOI] [PubMed] [Google Scholar]
- Musser G, Carleton M. Superfamily Muroidea. In: Wilson DE, Reeder DM, editors. Mammal species of the world. Washington (DC): Smithsonian Institution Press; 2005. pp. 894–1522. [Google Scholar]
- Nijman IJ, Lenstra JA. Mutation and recombination in cattle satellite DNA: a feedback model for the evolution of satellite DNA repeats. J Mol Evol. 2001;52:361–371. doi: 10.1007/s002390010166. [DOI] [PubMed] [Google Scholar]
- Paço A, Chaves R, Vieira-da-Silva A, Adega F. The involvement of repetitive sequences in the remodelling of karyotypes: the Phodopus genomes (Rodentia, Cricetidae) Micron. 2013;46:27–34. doi: 10.1016/j.micron.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Palomeque T, Lorite P. Satellite DNA in insects: a review. Heredity. 2008;100:564–573. doi: 10.1038/hdy.2008.24. [DOI] [PubMed] [Google Scholar]
- Pezer Ž, Ugarković Đ. Satellite DNA-associated siRNAs as mediators of heat shock response in insects. RNA Biol. 2012;9(5):587–595. doi: 10.4161/rna.20019. [DOI] [PubMed] [Google Scholar]
- Plohl M. Those mysterious sequences of satellite DNAs. Period Biol. 2010;112(4):403–410. [Google Scholar]
- Plohl M, Luchetti A, Meštrović N, Mantovani B. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene. 2008;409:72–82. doi: 10.1016/j.gene.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Plohl M, Meštrović N, Mravinac B. Satellite DNA evolution. In: Garrido-Ramos MA, editor. Repetitive DNA. Basel (Switzerland): Karger; 2012. pp. 126–152. [DOI] [PubMed] [Google Scholar]
- Robinson M, Catzeflis F, Briolay J, Mouchiroud D. Molecular phylogeny of rodents, with special emphasis on murids: evidence from nuclear gene LCAT. Mol Phylogenet Evol. 1997;8:423–434. doi: 10.1006/mpev.1997.0424. [DOI] [PubMed] [Google Scholar]
- Romanenko SA, et al. Karyotype evolution and phylogenetic relationships of hamsters (Cricetidae, Muroideia, Rodentia) inferred from chromosomal painting and banding comparison. Chromosome Res. 2007;15:283–297. doi: 10.1007/s10577-007-1124-3. [DOI] [PubMed] [Google Scholar]
- Rudert F, Bronner S, Garnier JM, Dollé P. Transcripts from opposite strands of γ satellite DNA are differentially expressed during mouse development. Mamm Genome. 1995;6:76–83. doi: 10.1007/BF00303248. [DOI] [PubMed] [Google Scholar]
- Scherthan H, et al. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol. 1996;134:1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzacher T, Heslop-Harrison JS, editors. Practical in situ hybridization. Oxford: Bios Scientific Publishers; 2000. [Google Scholar]
- Singer MF. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Spyropoulos B, Ross PD, Moens PB, Cameron DM. The synaptonemal complex karyotypes of palearctic hamsters, Phodopus roborovskii Satunin and P. sungorus Pallas. Chromosoma. 1982;86:397–408. [Google Scholar]
- Sumner AT. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 1972;75:304–306. doi: 10.1016/0014-4827(72)90558-7. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarković Đ. Functional elements residing within satellite DNAs. EMBO Rep. 2005;6(11):1035–1039. doi: 10.1038/sj.embor.7400558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourc’h C, Biamonti G. Long non-coding RNAs. Progress in molecular and subcellular biology. In: Ugarkovic D, editor. Transcription of satellite DNAs in mammals. Berlin (Germany): Springer-Verlag; 2011. pp. 95–118. [DOI] [PubMed] [Google Scholar]
- Walsh JB. Persistence of tandem arrays: implications for satellite DNA and simple-sequence DNAs. Genetics. 1987;115:553–567. doi: 10.1093/genetics/115.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman HA, et al. Genomic distribution of heterochromatic sequences in equids: implications to rapid chromosomal evolution. J Hered. 1991;82:369–377. doi: 10.1093/oxfordjournals.jhered.a111106. [DOI] [PubMed] [Google Scholar]