Abstract
Additive interactions can have public health and etiological implications but are infrequently reported. We assessed departures from additivity on the absolute risk scale between 9 established breast cancer risk factors and 23 susceptibility single-nucleotide polymorphisms (SNPs) identified from genome-wide association studies among 10,146 non-Hispanic white breast cancer cases and 12,760 controls within the National Cancer Institute's Breast and Prostate Cancer Cohort Consortium. We estimated the relative excess risk due to interaction and its 95% confidence interval for each pairwise combination of SNPs and nongenetic risk factors using age- and cohort-adjusted logistic regression models. After correction for multiple comparisons, we identified a statistically significant relative excess risk due to interaction (uncorrected P = 4.51 × 10−5) between a SNP in the DNA repair protein RAD51 homolog 2 gene (RAD51L1; rs10483813) and body mass index (weight (kg)/height (m)2). We also compared additive and multiplicative polygenic risk prediction models using per-allele odds ratio estimates from previous studies for breast-cancer susceptibility SNPs and observed that the multiplicative model had a substantially better goodness of fit than the additive model.
Keywords: additive interactions, breast cancer, genome-wide association studies, single-nucleotide polymorphisms
Genome-wide association studies (GWAS) have identified at least 74 independent susceptibility loci for breast cancer (1–12) or for specific subtypes, such as estrogen receptor (ER)-negative breast cancer (13–16). Characterization of the joint effects of multiple susceptibility loci and established risk factors such as number of livebirths and body mass index (BMI; weight (kg)/height (m)2) may shed light on biological mechanisms of breast cancer etiology and help improve risk prediction, potentially influencing public health guidelines. To date, 4 large-scale studies have explored gene-environment interactions between GWAS-identified single-nucleotide polymorphisms (SNPs) and breast cancer risk (17–20). While 3 of these studies found no evidence for departure from multiplicativity after correction for multiple comparisons, the fourth found evidence of effect-measure modification between the lymphocyte-specific protein 1 gene (LSP1) variant rs3817198 and breast cancer risk by parity (20).
Epidemiologists most frequently report departure from multiplicativity while characterizing interactions (21), and they assume multiplicative joint effects on the absolute risk scale while predicting risk (22–24). It is important to consider other measures of interaction between risk factors as well, such as additive joint effects on the absolute risk scale, in order to benefit from the additional information obtained from them (25, 26). To our knowledge, tests for additive interaction between confirmed susceptibility SNPs and established breast cancer risk factors have not been reported.
Additive interactions—departures from additivity on the absolute risk scale—have public health as well as etiological implications (25, 27, 28). Additive interactions between an environmental risk factor (exposure) and a genetic risk factor (SNP) test whether the excess in absolute risk of the disease due to presence of the exposure is the same in persons who carry the SNP and those who do not. They help identify subgroups of individuals for whom a targeted intervention designed to reduce a modifiable exposure could have the highest impact. Assessment of changes in risk differences between exposed and unexposed subjects is straightforward when genotype and exposure data are available on a full cohort of subjects with prospective disease follow-up. However, departures from additivity on the absolute risk scale can also be estimated from case-control data. Several measures of additive interaction that can be calculated from case-control data have been introduced (29–31). The relative excess risk due to interaction (RERI) may be the most useful in terms of assessing synergism between 2 binary risk factors (32, 33).
GWAS SNPs have also been used to improve existing breast cancer risk prediction models (22, 24, 34–37). In these studies, polygenic models have generally assumed multiplicative joint effects of SNPs on the relative risk scale. Under this multiplicative model, the absolute risk in the tails of the distribution can be strikingly elevated relative to the average risk. As more markers are identified and added to the risk model, the increase in risk in the tails may be clinically useful—for example, by identifying women who would benefit from earlier screening or chemoprevention. However, for most women (those in the middle of the risk distribution), their estimated risks under additive and multiplicative risk models are quite similar, while the estimated absolute risks in the tails under an additive risk model are much less extreme (women in the top 1% of the distribution have a 1.3-fold higher risk of breast cancer than average) (38). Thus, how well the multiplicative model fits the observed risk in the tails is an important practical question that has not yet been extensively studied.
Our objective in this study was to test for departures from additivity on the absolute risk scale between 9 established risk factors for breast cancer and 23 GWAS-identified breast cancer susceptibility SNPs within the National Cancer Institute's Breast and Prostate Cancer Cohort Consortium (BPC3). We also assessed whether an additive joint-effects model or a multiplicative joint-effects model based on external individual-marker odds ratio estimates was a better fit for the combined effects of 19 independent GWAS-identified breast cancer susceptibility SNPs (excluding 4 ER-negative SNPs).
METHODS
Study population
Cases and matched controls were drawn from 8 prospective cohort studies (39–46) conducted in the United States, Europe, and Australia, as described in the Web Appendix (available at http://aje.oxfordjournals.org/). We restricted analysis to 10,146 cases of invasive breast cancer and 12,760 controls of European ancestry, matched on age and additional criteria specific to the cohorts.
SNP selection and genotyping
We selected for analysis 23 independent SNPs from 23 distinct risk loci that had been identified in GWAS published before November 2012. Details on the selection process and genotyping are given in the Web Appendix.
Assessment of established nongenetic breast cancer risk factors
Risk factor data were obtained prior to disease diagnosis (at the time of inclusion in the cohort or at blood draw) using structured questionnaires. These data were sent to the German Cancer Research Center (Deutsches Krebsforschungszentrum, Heidelberg, Germany) for systematic, centralized data checks, removal of inconsistencies, and harmonization. We had information on established breast cancer risk factors, including age, height, weight, BMI, age at menarche, number of full-term pregnancies, age at first full-term pregnancy, age at menopause, history of oral contraceptive use, smoking status (never, former, or current smoker), alcohol intake (g/day), and family history of breast cancer in first-degree relatives (Web Table 1).
Statistical analysis
For analyses of gene-gene and gene-environment interaction, we converted all genetic and exposure variables into binary forms to allow for easy interpretability and to draw possible mechanistic conclusions within the sufficient-cause framework (47). Furthermore, since these SNPs and exposures were established risk factors for breast cancer, it was possible (at least for very strong risk factors such as family history) to code our variables so that their expected marginal odds ratios, based on previous literature, would be greater than 1.0. This is consistent with the monotonicity assumption for synergism, but we note that this assumption cannot be verified empirically, as it is based on individuals’ disease outcomes under counterfactual exposure patterns (48). Therefore, all SNPs were assumed to have a dominant effect of the risk allele and were represented in models by a coding of 0 or 1, based on whether the study participant had at least 1 risk allele for the SNP. The 9 exposure variables were transformed to their binary forms based on previously used cutpoints (19)—height (≤1.63 m vs. >1.63 m), BMI (<30 vs. ≥30), age at menarche (>11 years vs. ≤11 years), ever having a full-term pregnancy (yes vs. no), age at first full-term pregnancy (<30 years vs. ≥30 years), age at menopause (<50 years vs. ≥50 years), smoking (never vs. ever), alcohol intake (<14 g/day vs. ≥14 g/day), and family history of breast cancer in a first-degree relative (no vs. yes).
In addition to the 9 exposure variables, we created a risk score based on the Gail model (49), henceforth called the “modified Gail score,” because we did not have information on number of biopsies for all study participants. The modified Gail score was set to missing for 7,085 study participants who had missing values on any of the remaining variables used to calculate the Gail score (age, age at menarche, age at first livebirth, and number of first-degree relatives with breast cancer). The modified Gail score was converted to its binary form by using the 90th percentile among controls as the cutoff. We created a similar polygenic risk score (PRS) for SNPs, which was the count of the number of risk alleles in 19 SNPs (excluding 4 SNPs identified in ER-negative breast cancer GWAS scans), weighted by their log odds ratios as reported in external GWAS (Web Table 2). Missing values for the 19 SNPs were imputed on the basis of allele frequencies in cases and controls. The observed count of risk alleles ranged from 9 to 30, with a mean of 18.3 among controls and 19.1 among cases. The PRS (weighted count of risk alleles) was used to dichotomize participants into 2 groups using the 75th percentile among controls, thus creating a binary variable named the “polygenic score.”
Unconditional logistic regression models were used to test for associations between each of the 23 SNPs (Table 1) and breast cancer, as well as ER-negative breast cancer. All models adjusted for age at baseline (5-year interval groups) and for cohort using indicator variables.
Table 1.
Associations of Selected Single-Nucleotide Polymorphisms With the Risks of Breast Cancer and Estrogen Receptor-Negative Breast Cancer in the Breast and Prostate Cancer Cohort Consortium
| SNPa | Locus and/or Gene | Chr | Position | First Author, Year (Reference No.) | All Breast Cancer |
ER-Negative Breast Cancer |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| ORb | 95% CI | P Value | ORb | 95% CI | P Value | |||||
| rs11249433 | NOTCH2/1p11.2 | 1 | 120,982,136 | Thomas, 2009 (4) | 1.13 | 1.06, 1.20 | 0.0002 | 1.04 | 0.92, 1.17 | 0.534 |
| rs1045485 | CASP8 | 2 | 201,857,834 | Cox, 2007 (8) | 1.29 | 1.02, 1.64 | 0.035 | 1.69 | 0.98, 2.92 | 0.059 |
| rs13387042 | 2q35 | 2 | 217,614,077 | Stacey, 2007 (2) | 1.18 | 1.10, 1.26 | 7.31 × 10−6 | 1.10 | 0.96, 1.26 | 0.178 |
| rs4973768 | 3p24.1/SLC4A7 | 3 | 27,391,017 | Ahmed, 2009 (11) | 1.13 | 1.05, 1.20 | 0.0006 | 1.01 | 0.89, 1.15 | 0.870 |
| rs10069690c | TERT | 5 | 1,332,790 | Haiman, 2011 (14) | 1.05 | 0.99, 1.12 | 0.089 | 1.13 | 1.01, 1.27 | 0.032 |
| rs10941679 | 5p12 | 5 | 44,742,255 | Stacey, 2008 (3) | 1.16 | 1.09, 1.23 | 7.20 × 10−7 | 1.02 | 0.90, 1.14 | 0.798 |
| rs889312 | MAP3K1 | 5 | 56,067,641 | Easton, 2007 (1) | 1.13 | 1.07, 1.20 | 4.78 × 10−5 | 1.04 | 0.93, 1.16 | 0.531 |
| rs17530068c | 6q14.1 | 6 | 82,249,828 | Siddiq, 2012 (15) | 1.09 | 1.02, 1.15 | 0.006 | 1.09 | 0.96, 1.24 | 0.165 |
| rs2046210 | 6q25.1/ESR2 | 6 | 151,990,059 | Zheng, 2009 (10) | 1.11 | 1.05, 1.18 | 0.0005 | 1.14 | 1.01, 1.28 | 0.030 |
| rs1562430 | 8q24.21 | 8 | 128,457,034 | Turnbull, 2010 (5) | 1.16 | 1.07, 1.25 | 0.0004 | 1.03 | 0.87, 1.23 | 0.699 |
| rs1011970 | CDKN2BAS | 9 | 22,052,134 | Turnbull, 2010 (5) | 1.08 | 1.02, 1.15 | 0.012 | 1.14 | 1.00, 1.30 | 0.055 |
| rs865686 | 9q31.2 | 9 | 109,928,299 | Fletcher, 2011 (6) | 1.19 | 1.09, 1.30 | 5.87 × 10−5 | 1.32 | 1.09, 1.61 | 0.005 |
| rs2380205 | 10p15.1 | 10 | 5,926,740 | Turnbull, 2010 (5) | 1.05 | 0.97, 1.13 | 0.209 | 1.04 | 0.89, 1.22 | 0.592 |
| rs10995190 | 10q21.2/ZNF365 | 10 | 63,948,688 | Turnbull, 2010 (5) | 1.30 | 1.04, 1.62 | 0.021 | 1.28 | 0.79, 2.09 | 0.312 |
| rs1250003d | ZMIZ1 | 10 | 80,516,820 | Turnbull, 2010 (5) | 1.08 | 1.02, 1.15 | 0.014 | 0.98 | 0.86, 1.11 | 0.744 |
| rs2981582 | FGFR2 | 10 | 123,342,307 | Easton, 2007 (1); Hunter, 2007 (7) | 1.25 | 1.18, 1.33 | 1.87 × 10−12 | 1.04 | 0.92, 1.17 | 0.533 |
| rs909116 | LSP1 | 11 | 1,898,522 | Turnbull, 2010 (5) | 1.06 | 0.98, 1.14 | 0.133 | 1.04 | 0.89, 1.21 | 0.625 |
| rs614367 | 11q13 | 11 | 69,037,945 | Turnbull, 2010 (5) | 1.16 | 1.09, 1.24 | 7.91 × 10−6 | 1.07 | 0.93, 1.23 | 0.339 |
| rs10483813d | RAD51L1 | 14 | 68,101,037 | Thomas, 2009 (4) | 1.28 | 1.11, 1.47 | 0.0005 | 0.99 | 0.77, 1.28 | 0.947 |
| rs3803662 | TNRC9/TOX3 | 16 | 51,143,842 | Easton, 2007 (1); Stacey, 2007 (2) | 1.23 | 1.16, 1.31 | 6.12 × 10−12 | 1.11 | 0.99, 1.24 | 0.079 |
| rs6504950 | COX11 | 17 | 50,411,470 | Ahmed, 2009 (11) | 1.10 | 0.98, 1.24 | 0.091 | 1.11 | 0.89, 1.40 | 0.344 |
| rs8170c | C19ORF62 | 19 | 17,250,704 | Antoniou, 2010 (13) | 1.01 | 0.95, 1.07 | 0.780 | 1.25 | 1.11, 1.41 | 0.0002 |
| rs2284378c,d | 20q11.22/RALY | 20 | 32,051,756 | Siddiq, 2012 (15) | 1.00 | 0.94, 1.05 | 0.878 | 1.11 | 0.98, 1.25 | 0.087 |
Abbreviations: CASP8, caspase 8, apoptosis-related cysteine peptidase gene; CDKN2BAS, CDKN2B antisense RNA (non-protein coding) gene; Chr, chromosome; CI, confidence interval; C19ORF62, chromosome 19 open reading frame 62 gene; COX11, cytochrome C oxidase assembly homolog 11 gene; ER, estrogen receptor; ESR2, estrogen receptor 2 gene; FGFR2, fibroblast growth factor receptor 2 gene; LSP1, lymphocyte-specific protein 1 gene; MAP3K1, mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein gene; NOTCH2, Notch (Drosophila) homolog 2 gene; OR, odds ratio; RAD51L1, DNA repair protein RAD51 homolog 2 gene; RALY, RALY heterogeneous nuclear ribonucleoprotein gene; SLC4A7, solute carrier family 4, sodium bicarbonate cotransporter, member 7 gene; SNP, single-nucleotide polymorphism; TERT, telomerase reverse transcriptase gene; TNRC9, trinucleotide repeat-containing 9 gene; TOX3, TOX high mobility group box family member 3 gene; ZMIZ1, zinc finger, MIZ-type containing 1 gene; ZNF365, zinc finger protein 365 gene.
a Associations in the Breast and Prostate Cancer Cohort Consortium were determined for each SNP, assuming a dominant effect of risk alleles reported in previous studies.
b ORs and 95% CIs were calculated using unconditional logistic regression models adjusting for age at baseline (5-year intervals) and cohort.
c SNP identified from an ER-negative genome-wide association study scan.
d Some cohort studies genotyped proxy SNPs for the reported SNP. For example, rs704010 was a proxy for rs1250003 (r2 = 1 in the HapMap CEU population (Utah residents with Northern and Western European ancestry) (65)); rs999737 was a proxy for rs10483813 (r2 = 1 in the HapMap CEU population); and rs6059651 was a proxy for rs2284378 (r2 = 1 in the HapMap CEU population).
We tested for departure from additivity on the absolute risk scale for the joint association of each SNP-exposure combination, by estimating the RERI and computing 95% confidence intervals around the estimated RERI, using the delta method described by Hosmer and Lemeshow (29). RERIs and 95% confidence intervals were calculated for all pairwise combinations of 24 genetic variables (23 SNPs and polygenic score) with 10 exposure variables (including modified Gail score), using unconditional logistic regression models adjusting for age at baseline (5-year interval groups) and for cohort. We accounted for multiple comparisons through family-wise error rate correction of P values (Bonferroni correction); that is, adjusted P values were calculated by multiplying unadjusted P values by 240 (the number of tests).
When the adjusted P value for the RERI was less than 0.05, we also calculated point estimates and confidence intervals for other measures of additive interaction—the attributable proportion and the synergy index. For these SNP-exposure combinations, we also further characterized interactions after stratifying participants by menopausal status and by restricting analysis to ER-positive or ER-negative cases.
We next tested whether an additive or multiplicative model was a better fit for the simultaneous combination of 19 SNPs known to be associated with overall breast cancer. To this end, we calculated expected cell counts in the cross-tabulation of case-control status by risk allele count, conditional on the marginal distribution of genotypes and disease indicators and under the assumption of multiplicative combined effects of SNPs (PRSM) and of additive combined effects (PRSA). For calculation of PRSM and PRSA, we weighted each risk allele by the strength of the association (relative risk estimate from log additive models) obtained from external, independent GWAS or GWAS validation studies (Web Table 2). The expected number of cases from multiplicative and additive models was compared with the observed number of cases within each observed count of risk alleles. Participants with fewer than 10 risk alleles (n = 9) were assigned a value of 10, and participants with more than 28 risk alleles (n = 4) were assigned a value of 28, to ensure adequate numbers in each cell. We assessed the goodness of fit of the multiplicative model by comparing a model containing counts of risk alleles as indicator variables with a model in which the β estimate for log(PRSM) was constrained to 1 using the likelihood ratio test. The goodness of fit for the additive model was calculated similarly.
We estimated allele-count-specific 1-year risks via Bayes’ theorem (50, 51) and 95% confidence intervals for 50-year-old non-Hispanic white females, using relative risks estimated from the BPC3 case-control data and assuming the average annual breast cancer incidence rate to be 200 per 100,000 women (approximated from Surveillance, Epidemiology, and End Results data (52)). These values were then compared with predicted absolute risks for each study participant, calculated under the assumptions of multiplicative joint effects (using PRSM) and additive joint effects (using PRSA).
All statistical inferences were made using 2-sided tests. Statistical analyses were conducted using R, version 2.13.1 (R Foundation for Statistical Computing, Vienna, Austria), and STATA/SE, version 11.2 (StataCorp LP, College Station, Texas).
RESULTS
Web Table 1 shows average age, stage at diagnosis, and tumor characteristics (ER status and progesterone receptor status) for the 10,146 case participants, by cohort. The average age at diagnosis was lower in the Nurses’ Health Study II cohort (47.6 years, as compared with 61.3 years for all cohorts combined), because it is predominantly a cohort of premenopausal females (67% premenopausal, compared with 23% in all other cohorts combined). Our study population included a total of 2,540 cases of advanced breast cancer and 1,563 cases of ER-negative breast cancer.
Table 1 shows the associations between the 23 selected SNPs and breast cancer and ER-negative breast cancer. Seventeen SNPs (including 1 of the 4 ER-negative SNPs) were statistically significantly associated with breast cancer. All statistically significant associations were comparable in magnitude and direction with those reported in previous literature. We did not see a statistically significant association for 3 of the 19 breast cancer SNPs (rs2380205/10p15.1 (intergenic); rs909116/LSP1; rs6504950/cytochrome C oxidase assembly homolog 11 (COX11)). Five SNPs, including 2 of the 4 ER-negative SNPs, showed an association with ER-negative breast cancer (Table 1). The associations of the selected nongenetic risk factors with breast cancer and with ER-negative breast cancer are reported in Web Table 3.
RERI point estimates and 95% confidence intervals for the 240 pairwise SNP-exposure combinations are shown in Web Table 4, and Q-Q plots are shown in Web Figure 1. Five SNP-exposure additive interactions with unadjusted P values less than 0.005 were observed, of which 3 involved a SNP in the DNA repair protein RAD51 homolog 2 gene (RAD51L1), rs10483813. After family-wise error rate correction, departure from additivity on the absolute risk scale was observed for this RAD51L1 SNP and BMI (BMI <30 vs. BMI ≥30; RERI = 0.49 (95% confidence interval: 0.26, 0.73); unadjusted P = 4.51 × 10−5, family-wise error rate-corrected P = 0.011). To preserve consistency of interpretation across several measures of additive interaction (53), we recoded the reference category based on the lowest risk stratum in the joint-effects model (Web Table 5). We observed main-effects odds ratios greater than 1 for the TA/TT genotype as well as BMI <30; however, the estimated increase in absolute risk of breast cancer among persons with BMI <30 and the TA/TT genotype was less than expected by addition of the excess risk due to BMI reduction alone or by TA/TT genotype alone: RERI = −0.95 (95% confidence interval: −1.77, −0.12). We observed an inverse association between BMI and breast cancer risk among women with the rs10483813 AA genotype but not among women with the TA or TT genotype (Tables 2 and 3). This interaction pattern did not change substantially within strata of menopausal status or family history (Web Table 6). We note that the risk allele for rs10483813 is quite common (minor allele frequency = 0.23), so the reference category (women homozygous for the reference allele and with BMI ≥30) was rather small (n = 148; 0.9% of total). This suggests that this interaction may have been a false-positive finding due to low power or to statistical properties of the RERI in small samples.
Table 2.
Relative Excess Risk of Breast Cancer Due to Interactiona Between the RAD51L1 Single-Nucleotide Polymorphism rs10483813 and Body Mass Index in the Breast and Prostate Cancer Cohort Consortium
| rs10483813b Genotype | Genotype Frequency | Body Mass Indexc |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <25 |
25–<30 |
≥30 |
|||||||||||
| No. of Cases | No. of Controls | OR | 95% CI | No. of Cases | No. of Controls | OR | 95% CI | No. of Cases | No. of Controls | OR | 95% CI | ||
| AA | 0.056 | 180 | 287 | 1.00 | Referent | 110 | 186 | 0.92 | 0.68, 1.24 | 34 | 114 | 0.50 | 0.33, 0.78 |
| TA | 0.347 | 1,153 | 1,725 | 1.05 | 0.86, 1.29 | 774 | 1,086 | 1.12d | 0.90, 1.38 | 378 | 560 | 1.07e | 0.85, 1.35 |
| TT | 0.598 | 2,122 | 2,933 | 1.15 | 0.95, 1.40 | 1,286 | 1,755 | 1.15f | 0.94,1.40 | 724 | 971 | 1.18g | 0.95, 1.46 |
Abbreviations: CI, confidence interval; OR, odds ratio; RAD51L1, DNA repair protein RAD51 homolog 2 gene; RERI, relative excess risk due to interaction.
a The RERI is a measure of departure from additivity on the absolute risk scale. RERIs and 95% CIs were calculated using unconditional logistic regression models adjusting for age at baseline (5-year intervals) and cohort.
b Some cohort studies genotyped the proxy single-nucleotide polymorphism rs999737 for rs10483813 (r2 = 1 in the HapMap CEU population (65)). The T allele is the risk allele for rs10483813, and the A allele is the protective allele (4). The P value for multiplicative interaction between rs10483813 and body mass index was 0.002. Synergy indices for interaction between rs10483813 and body mass index were undefined using this reference category.
c Weight (kg)/height (m)2; categorized into 3 levels based on the World Health Organization classification (66).
d RERI = 0.15 (95% CI: −0.15, 0.45); P-RERI = 0.335.
e RERI = 0.52 (95% CI: 0.25, 0.79); P-RERI = 0.000168.
f RERI = 0.08 (95% CI: −0.21, 0.38); P-RERI = 0.603.
g RERI = 0.52 (95% CI: 0.22, 0.82); P-RERI = 0.000621.
Table 3.
Relative Excess Risk of Breast Cancer Due to Interactiona Between the RAD51L1 Single-Nucleotide Polymorphism rs10483813 and Body Mass Index After Designation of the Lowest Risk Category (Body Mass Index ≥30 and AA Genotype) as the Reference Category in the Breast and Prostate Cancer Cohort Consortium
| rs10483813b Genotype | Body Mass Indexc |
|||||
|---|---|---|---|---|---|---|
| ≥30 |
≥25–<30 |
<25 |
||||
| OR | 95% CI | OR | 95% CI | OR | 95% CI | |
| AA | 1.00 | Referent | 1.81 | 1.15, 2.85 | 1.98 | 1.29, 3.04 |
| TA | 2.13 | 1.42, 3.20 | 2.21d | 1.48, 3.29 | 2.08e | 1.41, 3.09 |
| TT | 2.33 | 1.57, 3.47 | 2.27f | 1.54, 3.37 | 2.29g | 1.55, 3.38 |
Abbreviations: CI, confidence interval; OR, odds ratio; RAD51L1, DNA repair protein RAD51 homolog 2 gene; RERI, relative excess risk due to interaction.
a The RERI is a measure of departure from additivity on the absolute risk scale. RERIs and 95% CIs were calculated using unconditional logistic regression models adjusting for age at baseline (5-year intervals) and cohort.
b Some cohort studies genotyped the proxy single-nucleotide polymorphism rs999737 for rs10483813 (r2 = 1 in the HapMap CEU population (65)). The T allele is the risk allele for rs10483813, and the A allele is the protective allele (4). The P value for multiplicative interaction between rs10483813 and body mass index was 0.002.
c Weight (kg)/height (m)2; categorized into 3 levels based on the World Health Organization classification (66).
d RERI = −0.73 (95% CI:−1.6, 0.14); P-RERI = 0.097; synergy index = 0.62 (95% CI: 0.45, 0.86).
e RERI = −1.02 (95% CI: −1.95, −0.101); P-RERI = 0.030; synergy index = 0.51 (95% CI: 0.40, 0.67).
f RERI = −0.87 (95% CI: −1.76, 0.015); P-RERI = 0.054; synergy index = 0.59 (95% CI: 0.45, 0.78).
g RERI = −1.03 (95% CI: −1.91, −0.14); P-RERI = 0.023; synergy index = 0.56 (95% CI: 0.43, 0.73).
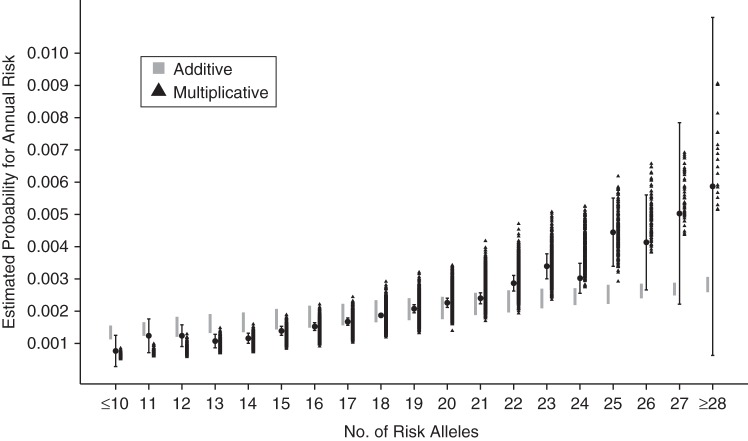
When comparing risks predicted on the basis of a PRS using external weights with the observed data in the BPC3, there was evidence of departure of the observed data from additivity (P < 10−15) as well as from multiplicativity (P = 0.0027) (Table 4). This departure was in the direction of submultiplicative but supra-additive effects (Figure 1), and it did not change substantially within the 3 strata of modified Gail score (Web Figures 2–4). Therefore, based on the comparison of the assessed goodness of fit with respect to a full model containing counts of risk alleles as indicator variables, the multiplicative model was a substantially better fit for the empirical data than the additive model.
Table 4.
Expected and Observed Numbers of Breast Cancer Cases Under Additive and Multiplicative Joint-Effects Model Assumptions in the Breast and Prostate Cancer Cohort Consortium
| No. of Risk Alleles | No. of Cases Expected (Additive Model) | No. of Cases Observed | No. of Cases Expected (Multiplicative Model) |
|---|---|---|---|
| ≤10 | 15.5 | 11 | 9.3 |
| 11 | 30.4 | 28 | 19.8 |
| 12 | 73.5 | 67 | 50.9 |
| 13 | 152.4 | 124 | 111.5 |
| 14 | 329.7 | 268 | 255.9 |
| 15 | 559.0 | 495 | 460.1 |
| 16 | 873.3 | 796 | 757.9 |
| 17 | 1,158.8 | 1,085 | 1,061.0 |
| 18 | 1,382.9 | 1,354 | 1,328.8 |
| 19 | 1,398.8 | 1,414 | 1,412.0 |
| 20 | 1,298.5 | 1,340 | 1,369.5 |
| 21 | 1,087.7 | 1,132 | 1,196.1 |
| 22 | 808.5 | 896 | 924.0 |
| 23 | 480.3 | 561 | 569.2 |
| 24 | 275.9 | 299 | 337.1 |
| 25 | 126.5 | 161 | 159.9 |
| 26 | 58.8 | 69 | 75.4 |
| 27 | 24.7 | 32 | 33.0 |
| ≥28 | 10.8 | 14 | 14.7 |
Figure 1.
Estimated absolute risk of breast cancer according to number of risk alleles among 50-year-old non-Hispanic white females, derived using case-control data from the Breast and Prostate Cancer Cohort Consortium (19, 39) and population incidence rates from the Surveillance, Epidemiology, and End Results Program (52), and comparison with expected risk assuming multiplicative and additive joint-effects models. Bars, 95% confidence intervals.
DISCUSSION
In this large breast cancer consortium study with prospectively obtained risk factor data, we investigated departure from additive combined effects on an absolute risk scale of 23 susceptibility SNPs and 9 established risk factors in the risk of breast cancer. We observed evidence of additive interaction between BMI and a SNP in RAD51L1, rs10483813. BMI was inversely associated with breast cancer risk among women with the AA genotype but not among women who carried the T allele at rs10483813.
The biological mechanisms behind such an interaction, particularly the inverse association between BMI and breast cancer among persons with the AA genotype, are unclear. There is limited information about the functional significance of rs10483813 alleles. This SNP has been not been shown to have significantly different associations with different breast cancer subtypes (54) or to interact with ionizing radiation exposure (55). Vachon et al. (56) recently reported an inverse association between the rs10483813 A allele and mammographic density, after adjusting for age and BMI. Mammographic density—a measure of the proportion of nonfat breast tissue—is one of the strongest known risk factors for breast cancer and is also known to be inversely associated with BMI. Because this is the first report of an additive interaction between rs10483813 and BMI, to our knowledge, and because of the small numbers of cases and controls in some strata, these intriguing results could possibly be false-positive and should be replicated.
Our results regarding the RAD51L1 locus cannot be directly compared with other studies that have previously examined risk ratio heterogeneity between confirmed GWAS-identified SNPs and breast cancer risk, including 1 from the BPC3 (17–20). Campa et al. (19) and Nickels et al. (20) reported no departure from multiplicativity between BMI and the RAD51L1 locus. The other studies did not examine SNPs at this locus.
Although most large studies testing for interaction between GWAS-identified breast cancer risk alleles have not found any statistically significant multiplicative interactions, Nickels et al. (20) recently showed multiplicative interactions between SNPs at the LSP1 locus and number of births. Specifically, they found that the per-risk-allele odds ratio for rs3817198 increased with increasing number of births, from 1.03 among women with 1 birth to 1.26 among women with 4 or more births (Ptrend = 2.4 × 10−6). We did not genotype rs3817198, but we did genotype another SNP at LSP1, rs909116 (r2 = 0.33). In our sample, the trend was reversed and nonsignificant: The per-risk-allele odds ratios for women with 4 or more births and women with 1 birth were 1.03 and 1.14, respectively (Ptrend = 0.43). The multiplicative trend interaction odds ratios (1.06 for Nickels et al. (20) and 0.98 for this study) were statistically significantly different (P < 0.001). These differences may be due to the relatively modest linkage disequilibrium between these 2 SNPs, or the interaction observed by Nickels et al. may not generalize to our sample.
Overall, the paucity of heterogeneity in odds ratios or risk differences for the combined effects of genes and exposures in breast cancer risk is consistent with supra-additive and submultiplicative effects of SNP-exposure combinations, which would reduce the power to detect differences from either side. Moreover, we tested only for 2-level interactions—pairwise for each genetic variant and each exposure—and specific higher-order interactions may have been missed because they were not investigated (although power to detect such interactions will typically be lower than for pairwise interactions). Due to the lack of strongly statistically significant interactions and the violation of monotonicity assumptions, strong public health recommendations as well as sufficient component-cause inferences could not be made from this study.
We also investigated whether an additive joint-effects model or a multiplicative joint-effects model is a better fit for empirical data on the combined effects of 19 confirmed breast cancer susceptibility loci. We found a statistically significant deviation from both the additive model and the multiplicative models, in the direction of submultiplicativity and supra-additivity. The heterogeneity of the observed data from the additive joint-effects model was very strong in magnitude and highly statistically significant.
Polygenic models used in disease risk prediction have generally assumed multiplicative joint effects of SNPs on the absolute risk scale. We find that a multiplicative model for the joint effects of multiple risk SNPs is substantially better than an additive model. This provides some support for the use of a multiplicative model based on individual-SNP marginal odds ratios, as has been suggested in the context of risk screening and implemented by several consumer genetics companies (24, 57–60).
However, the statistically significant submultiplicative deviation of empirical data from multiplicative effects leaves some scope for improvement. This deviation may reflect the fact that the external odds ratios used to define the multiplicative risk score were overestimated due to the “winner's curse” (61, 62). In sensitivity analyses where we adjusted all per-allele β coefficients by a constant deflation factor (adjusted β = 0.9 × unadjusted β) or by a factor proportional to the per-allele odds ratio (ranging from 0.92 to 0.997 (63)), the multiplicative model was a much better fit (goodness of fit: P = 0.99 for constant adjustment and P = 0.45 for proportional adjustment). This suggests that multiplicative risk models constructed using odds ratios from initial discovery publications are likely to be overestimates (or underestimates) in the upper (respectively lower) tails. However, when accurate odds ratio estimates from large studies independent of the original discovery samples are used, a multiplicative polygenic risk model may be a good fit, and this model could be utilized to identify a small number of women at markedly increased risk who would benefit from risk-reducing interventions or more intensive screening. We stress that risk estimates for women in the tails of the risk distribution are projections beyond the support of the bulk of the training data and that we had limited statistical power to detect departures from multiplicativity in the tails; note the wide confidence interval for the count ≥28 group in Figure 1.
Tests for pairwise interactions between the 23 SNPs failed to show statistically significant departures from additivity between these low-penetrance variants. This may reflect the low power to detect small departures from a pairwise additive risk model, even with our large sample size. The departure from an additive joint-effects model from the observed fitted risks for the combination of 19 SNPs suggests the presence of additive interactions between the SNPs and breast cancer risk.
This study had several other weaknesses. First, although data on exposure variables were prospectively collected in all of the cohorts, the instruments used to collect the information were not uniform across cohorts, potentially inducing exposure misclassification during harmonization. It has been demonstrated that nondifferential misclassification of 2 independent exposures preserves the validity of tests for additive interaction (64). Furthermore, for ease of interpretation of additive effects, these exposure variables were represented using dichotomized categories, which may not be the optimal modeling strategy for reflecting their biological associations with breast cancer. However, in most cases the cutoffs for exposure variables were not arbitrary, and a priori information was used to choose them. Similarly, genetic variants were also dichotomized assuming a dominant effect for the risk allele, without a priori evidence for the same. Our results for marginal effects of most of the SNPs were not substantially different from results of previous studies that assumed log-additive SNP effects. Moreover, adjustment for age and cohort may not have sufficiently controlled for the variability among the included studies. The calculation of exposure risk score (Gail score) could not be done in a manner that would allow for comparison with previous studies, because we did not have information on the number of biopsies for all study participants.
The results from our study are generalizable only to females of European descent, since participants who self-reported ethnicities other than non-Hispanic white were excluded from the analyses due to the limited sample size.
To our knowledge, this is one of the largest studies (10,146 cases and 12,760 controls) to have investigated interactions between known breast cancer risk factors and GWAS-identified susceptibility loci, and the first study to find additive interactions. In this study we also replicated previous associations of 23 known SNPs with breast cancer risk using the risk-dominant model. We did not observe strong evidence of 2-level additive interactions between SNPs and the established breast cancer risk factors, except for a statistically significantly protective association with rs10483813 in RAD51L1 among obese females. This study also showed that the multiplicative joint-effects model is a substantially better fit than the additive joint-effects model in the prediction of absolute risk of breast cancer.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Program in Genetic Epidemiology and Statistical Genetics, Harvard School of Public Health, Boston, Massachusetts (Amit D. Joshi, Sara Lindström, David J. Hunter, Peter Kraft); Division of Cancer Epidemiology, German Cancer Research Center (Deutsches Krebsforschungszentrum), Heidelberg, Germany (Anika Hüsing, Myrto Barrdahl, Rudolf Kaaks); Department of Epidemiology, Harvard School of Public Health, Boston, Massachusetts (Peter Kraft, Tyler J. VanderWeele, Dimitrios Trichopoulos); Department of Biology, University of Pisa, Pisa, Italy (Daniele Campa); Department of Biostatistics, Harvard School of Public Health, Boston, Massachusetts (Tyler J. VanderWeele); Genomic Epidemiology Group, German Cancer Research Center (Deutsches Krebsforschungszentrum), Heidelberg, Germany (Daniele Campa, Federico Canzian); Genetic Epidemiology Unit, American Cancer Society, Atlanta, Georgia (Mia M. Gaudet); Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, Maryland (Jonine D. Figueroa, Stephen J. Chanock, Robert N. Hoover, Regina G. Ziegler); Cancer Epidemiology Centre, Cancer Council Victoria, Carlton South, Victoria, Australia (Laura Baglietto, Graham G. Giles, Gianluca Severi); Centre for Molecular, Environmental, Genetic and Analytic Epidemiology, School of Population Health, University of Melbourne, Melbourne, Victoria, Australia (Laura Baglietto, Graham G. Giles, Gianluca Severi); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Christine D. Berg); Division of Preventive Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, Massachusetts (Julie E. Buring, I-Min Lee, Shumin Zhang); Department of Epidemiology, Murcia Regional Health Council, Murcia, Spain (María-Dolores Chirlaque); Consorcio de Investigación Biomédica de Epidemiología y Salud Pública, Madrid, Spain (María-Dolores Chirlaque); Epidemiology Research Program, American Cancer Society, Atlanta, Georgia (W. Ryan Diver, Michael J. Thun); Institut National de la Santé et de la Recherche Médicale, Centre for Research in Epidemiology and Population Health, U1018, Institut Gustave Roussy, Villejuif, France (Laure Dossus); Paris South University, Unité Mixte de Recherche en Santé 1018, Villejuif, France (Laure Dossus); Faculty of Medicine, Monash University, Melbourne, Victoria, Australia (Graham G. Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California (Christopher A. Haiman, Fredrick Schumacher, Daniel O. Stram, Brian E. Henderson); Division of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst, Massachusetts (Susan E. Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Claudine Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu, Hawaii (Laurence N. Kolonel); Epidemiology and Prevention Unit, Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy (Vittorio Krogh); University of Hawaii Cancer Center, Honolulu, Hawaii (Loic Le Marchand); Institute of Community Medicine, University of Tromsø, Tromsø, Norway (Eiliv Lund); Essentia Institute of Rural Health, Duluth, Minnesota (Catherine A. McCarty); Section for Epidemiology, Department of Public Health, Aarhus University, Aarhus, Denmark (Kim Overvad); Department of Epidemiology, Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht, the Netherlands (Petra H. Peeters); Department of Epidemiology and Biostatistics, School of Public Health, Faculty of Medicine, Imperial College London, London, United Kingdom (Petra H. Peeters, Elio Riboli); Department of Surgery, Umeå University Hospital, Umeå, Sweden (Malin Sund); Cancer Epidemiology Unit, Nuffield Department of Clinical Medicine, University of Oxford, Oxford, United Kingdom (Ruth C. Travis); Hellenic Health Foundation, Athens, Greece (Dimitrios Trichopoulos); Bureau of Epidemiologic Research, Academy of Athens, Athens, Greece (Dimitrios Trichopoulos); and Department of Nutrition, Harvard School of Public Health, Boston, Massachusetts (Walter C. Willett).
This study was funded by the National Cancer Institute, US National Institutes of Health (grant CA148065 and cooperative agreements U01-CA98233 and U19-CA148065 to D.J.H., U01-CA98710 to M.J.T., U01-CA98216 to E.R. and R.K., and U01-CA98758 to B.E.H.) and the Intramural Research Program of the National Institutes of Health/National Cancer Institute, Division of Cancer Epidemiology and Genetics.
Conflict of interest: none declared.
REFERENCES
- 1.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 3.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40(6):703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 4.Thomas G, Jacobs KB, Kraft P, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41(5):579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbull C, Ahmed S, Morrison J, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fletcher O, Johnson N, Orr N, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst. 2011;103(5):425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 7.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox A, Dunning AM, Garcia-Closas M, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39(3):352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 9.Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105(11):4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng W, Long J, Gao YT, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41(3):324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmed S, Thomas G, Ghoussaini M, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41(5):585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michailidou K, Hall P, Gonzalez-Neira A, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–361. doi: 10.1038/ng.2563. 361e351–361e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antoniou AC, Wang X, Fredericksen ZS, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42(10):885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haiman CA, Chen GK, Vachon CM, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet. 2011;43(12):1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siddiq A, Couch FJ, Chen GK, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21(24):5373–5384. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Closas M, Couch FJ, Lindstrom S, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45(4):392–398. doi: 10.1038/ng.2561. 398e391–398e392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Travis RC, Reeves GK, Green J, et al. Gene-environment interactions in 7610 women with breast cancer: prospective evidence from the Million Women Study. Lancet. 2010;375(9732):2143–2151. doi: 10.1016/S0140-6736(10)60636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milne RL, Gaudet MM, Spurdle AB, et al. Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the Breast Cancer Association Consortium: a combined case-control study. Breast Cancer Res. 2010;12(6):R110. doi: 10.1186/bcr2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campa D, Kaaks R, Le Marchand L, et al. Interactions between genetic variants and breast cancer risk factors in the Breast and Prostate Cancer Cohort Consortium. J Natl Cancer Inst. 2011;103(16):1252–1263. doi: 10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nickels S, Truong T, Hein R, et al. Evidence of gene-environment interactions between common breast cancer susceptibility loci and established environmental risk factors. PLoS Genet. 2013;9(3):e1003284. doi: 10.1371/journal.pgen.1003284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knol MJ, Egger M, Scott P, et al. When one depends on the other: reporting of interaction in case-control and cohort studies. Epidemiology. 2009;20(2):161–166. doi: 10.1097/EDE.0b013e31818f6651. [DOI] [PubMed] [Google Scholar]
- 22.Pharoah PD, Antoniou AC, Easton DF, et al. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358(26):2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 23.Gail MH. Value of adding single-nucleotide polymorphism genotypes to a breast cancer risk model. J Natl Cancer Inst. 2009;101(13):959–963. doi: 10.1093/jnci/djp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mealiffe ME, Stokowski RP, Rhees BK, et al. Assessment of clinical validity of a breast cancer risk model combining genetic and clinical information. J Natl Cancer Inst. 2010;102(21):1618–1627. doi: 10.1093/jnci/djq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothman KJ. Synergy and antagonism in cause-effect relationships. Am J Epidemiol. 1974;99(6):385–388. doi: 10.1093/oxfordjournals.aje.a121626. [DOI] [PubMed] [Google Scholar]
- 26.Siemiatycki J, Thomas DC. Biological models and statistical interactions: an example from multistage carcinogenesis. Int J Epidemiol. 1981;10(4):383–387. doi: 10.1093/ije/10.4.383. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Closas M, Rothman N, Figueroa JD, et al. Common genetic polymorphisms modify the effect of smoking on absolute risk of bladder cancer. Cancer Res. 2013;73(7):2211–2220. doi: 10.1158/0008-5472.CAN-12-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han SS, Rosenberg PS, Garcia-Closas M, et al. Likelihood ratio test for detecting gene (G)-environment (E) interactions under an additive risk model exploiting G-E independence for case-control data. Am J Epidemiol. 2012;176(11):1060–1067. doi: 10.1093/aje/kws166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosmer DW, Lemeshow S. Confidence interval estimation of interaction. Epidemiology. 1992;3(5):452–456. doi: 10.1097/00001648-199209000-00012. [DOI] [PubMed] [Google Scholar]
- 30.Nie L, Chu H, Li F, et al. Relative excess risk due to interaction: resampling-based confidence intervals. Epidemiology. 2010;21(4):552–556. doi: 10.1097/EDE.0b013e3181e09b0b. [DOI] [PubMed] [Google Scholar]
- 31.Assmann SF, Hosmer DW, Lemeshow S, et al. Confidence intervals for measures of interaction. Epidemiology. 1996;7(3):286–290. doi: 10.1097/00001648-199605000-00012. [DOI] [PubMed] [Google Scholar]
- 32.VanderWeele TJ. A word and that to which it once referred: assessing “biologic” interaction. Epidemiology. 2011;22(4):612–613. doi: 10.1097/EDE.0b013e31821db393. [DOI] [PubMed] [Google Scholar]
- 33.VanderWeele TJ, Robins JM. The identification of synergism in the sufficient-component-cause framework. Epidemiology. 2007;18(3):329–339. doi: 10.1097/01.ede.0000260218.66432.88. [DOI] [PubMed] [Google Scholar]
- 34.Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100(14):1037–1041. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darabi H, Czene K, Zhao W, et al. Breast cancer risk prediction and individualised screening based on common genetic variation and breast density measurement. Breast Cancer Res. 2012;14(1):R25. doi: 10.1186/bcr3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pashayan N, Duffy SW, Chowdhury S, et al. Polygenic susceptibility to prostate and breast cancer: implications for personalised screening. Br J Cancer. 2011;104(10):1656–1663. doi: 10.1038/bjc.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Husing A, Canzian F, Beckmann L, et al. Prediction of breast cancer risk by genetic risk factors, overall and by hormone receptor status. J Med Genet. 2012;49(9):601–608. doi: 10.1136/jmedgenet-2011-100716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moonesinghe R, Khoury MJ, Liu T, et al. Discriminative accuracy of genomic profiling comparing multiplicative and additive risk models. Eur J Hum Genet. 2011;19(2):180–185. doi: 10.1038/ejhg.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunter DJ, Riboli E, Haiman CA, et al. A candidate gene approach to searching for low-penetrance breast and prostate cancer genes. Nat Rev Cancer. 2005;5(12):977–985. doi: 10.1038/nrc1754. [DOI] [PubMed] [Google Scholar]
- 40.Calle EE, Rodriguez C, Jacobs EJ, et al. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer. 2002;94(9):2490–2501. doi: 10.1002/cncr.101970. [DOI] [PubMed] [Google Scholar]
- 41.Riboli E, Hunt KJ, Slimani N, et al. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5(6B):1113–1124. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 42.Kolonel LN, Altshuler D, Henderson BE. The Multiethnic Cohort Study: exploring genes, lifestyle and cancer risk. Nat Rev Cancer. 2004;4(7):519–527. doi: 10.1038/nrc1389. [DOI] [PubMed] [Google Scholar]
- 43.Colditz GA, Hankinson SE. The Nurses’ Health Study: lifestyle and health among women. Nat Rev Cancer. 2005;5(5):388–396. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 44.Hayes RB, Reding D, Kopp W, et al. Etiologic and early marker studies in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials. 2000;21(6 suppl):349S–355S. doi: 10.1016/s0197-2456(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 45.Rexrode KM, Lee IM, Cook NR, et al. Baseline characteristics of participants in the Women's Health Study. J Womens Health Gend Based Med. 2000;9(1):19–27. doi: 10.1089/152460900318911. [DOI] [PubMed] [Google Scholar]
- 46.Giles GG, English DR. The Melbourne Collaborative Cohort Study. IARC Sci Publ. 2002;156:69–70. [PubMed] [Google Scholar]
- 47.VanderWeele TJ, Robins JM. Directed acyclic graphs, sufficient causes, and the properties of conditioning on a common effect. Am J Epidemiol. 2007;166(9):1096–1104. doi: 10.1093/aje/kwm179. [DOI] [PubMed] [Google Scholar]
- 48.Höfler M. Causal inference based on counterfactuals. BMC Med Res Methodol. 2005;5:28. doi: 10.1186/1471-2288-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gail MH, Brinton LA, Byar DP, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81(24):1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 50.Dupont WD. Converting relative risks to absolute risks: a graphical approach. Stat Med. 1989;8(6):641–651. doi: 10.1002/sim.4780080603. [DOI] [PubMed] [Google Scholar]
- 51.Yang Q, Khoury MJ, Botto L, et al. Improving the prediction of complex diseases by testing for multiple disease-susceptibility genes. Am J Hum Genet. 2003;72(3):636–649. doi: 10.1086/367923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Howlader N, Noone AM, Krapcho M, et al. Bethesda, MD: National Cancer Institute; 2014. SEER Cancer Statistics Review (CSR) 1975–2011. http://seer.cancer.gov/csr/1975_2011/ Based on November 2013 SEER data submission Published April 2014 Updated August 27, 2014. Accessed April 14, 2014. [Google Scholar]
- 53.Knol MJ, VanderWeele TJ, Groenwold RH, et al. Estimating measures of interaction on an additive scale for preventive exposures. Eur J Epidemiol. 2011;26(6):433–438. doi: 10.1007/s10654-011-9554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Figueroa JD, Garcia-Closas M, Humphreys M, et al. Associations of common variants at 1p11.2 and 14q24.1 (RAD51L1) with breast cancer risk and heterogeneity by tumor subtype: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20(23):4693–4706. doi: 10.1093/hmg/ddr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhatti P, Doody MM, Rajaraman P, et al. Novel breast cancer risk alleles and interaction with ionizing radiation among U.S. radiologic technologists. Radiat Res. 2010;173(2):214–224. doi: 10.1667/RR1985.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vachon CM, Scott CG, Fasching PA, et al. Common breast cancer susceptibility variants in LSP1 and RAD51L1 are associated with mammographic density measures that predict breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2012;21(7):1156–1166. doi: 10.1158/1055-9965.EPI-12-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Navigenics, Inc. Foster City, CA: Navigenics, Inc; The science behind the Navigenics service. http://www.navigenics.com/static/pdf/Navigenics%20White%20Paper.pdf. Accessed July 1, 2012. [Google Scholar]
- 58.Macpherson M, Naughton B, Hsu A, et al. Mountain View, CA: 23andme; 2007. Estimating genotype-specific incidence for one or several loci. (White Paper 23-01) https://23andme.https.internapcdn.net/res/pdf/HIC-SXIYiYqXreldAxO5yA_23-01_Estimating_Genotype_Specific_Incidence.pdf. Published September 5, 2007. Updated November 18, 2007. Accessed August 19, 2013. [Google Scholar]
- 59.Patel CJ, Sivadas A, Tabassum R, et al. Whole genome sequencing in support of wellness and health maintenance. Genome Med. 2013;5(6):58. doi: 10.1186/gm462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morgan AA, Chen R, Butte AJ. Likelihood ratios for genome medicine. Genome Med. 2010;2(5):30. doi: 10.1186/gm151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008;19(5):640–648. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 62.Kraft P. Curses—winner's and otherwise—in genetic epidemiology. Epidemiology. 2008;19(5):649–651. doi: 10.1097/EDE.0b013e318181b865. [DOI] [PubMed] [Google Scholar]
- 63.Zhong H, Prentice RL. Bias-reduced estimators and confidence intervals for odds ratios in genome-wide association studies. Biostatistics. 2008;9(4):621–634. doi: 10.1093/biostatistics/kxn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanderweele TJ. Inference for additive interaction under exposure misclassification. Biometrika. 2012;99(2):502–508. doi: 10.1093/biomet/ass012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.National Center for Biotechnology Information. Guidelines for referring to the HapMap populations in publications and presentations. http://hapmap.ncbi.nlm.nih.gov/citinghapmap.html. Updated June 20, 2005. Accessed September 5, 2014.
- 66.World Health Organization. BMI classification. http://apps.who.int/bmi/index.jsp?introPage=intro_3.html. Updated May 9, 2014. Accessed September 5, 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.