Abstract
Background
Bacteria of the genus Leptospira, the causative agents of leptospirosis, are categorized into pathogenic and non-pathogenic species. However, the benefit of using a clinical diagnostic that is specific for pathogenic species remains unclear. In this study, we present the development of a real-time PCR (rtPCR) for the detection of pathogenic Leptospira (the pathogenic rtPCR), and we perform a comparison of the pathogenic rtPCR with a published assay that detects all Leptospira species [the undifferentiated febrile illness (UFI) assay] and a reference 16S Leptospira rtPCR, which was originally designed to detect pathogenic species.
Methodology/Principal Findings
For the pathogenic rtPCR, a new hydrolysis probe was designed for use with primers from the UFI assay, which targets the 16S gene. The pathogenic rtPCR detected Leptospira DNA in 37/37 cultured isolates from 5 pathogenic and one intermediate species. Two strains of the non-pathogenic L. biflexa produced no signal. Clinical samples from 65 patients with suspected leptospirosis were then tested using the pathogenic rtPCR and a reference Leptospira 16S rtPCR. All 65 samples had tested positive for Leptospira using the UFI assay; 62 (95.4%) samples tested positive using the pathogenic rtPCR (p = 0.24). Only 24 (36.9%) samples tested positive in the reference 16S rtPCR (p<0.0001 for comparison with the pathogenic rtPCR and UFI assays). Amplicon sequencing confirmed the detection of pathogenic Leptospira species in 49/50 cases, including 3 cases that were only detected using the UFI assay.
Conclusions/Significance
The pathogenic rtPCR displayed similar sensitivity to the UFI assay when testing clinical specimens with no difference in specificity. Both assays proved significantly more sensitive than a real-time molecular test used for comparison. Future studies are needed to investigate the clinical and epidemiologic significance of more sensitive Leptospira detection using these tests.
Introduction
Leptospirosis is a potentially fatal systemic illness resulting from infection with spirochetes of the genus Leptospira [1], [2]. Humans are accidental hosts and typically acquire the infection from direct contact with water contaminated with the urine of small mammals [1]–[3]. A wide range of clinical manifestations can occur following human infection with Leptospira, spanning asymptomatic infection to severe disease, multi-system organ failure and death [1], [2], [4]–[6]. It is estimated that 873,000 severe infections occur annually, with 49,000 deaths [3]. However, the non-specific disease presentation and limitations in available diagnostics for Leptospira likely render these disease estimates inaccurate [1], [2], [7], [8]. While debate exists regarding the efficacy of antibiotics in severe leptospirosis, accurate and early diagnosis may improve patient outcomes by allowing for the timely administration of antibiotic therapy [9], [10].
Currently, the reference standards for the diagnosis of leptospirosis remain bacterial culture and serological microscopic agglutination testing (MAT) [3], [11]. These tests are both resource intensive and cannot provide results in a clinically meaningful timeframe. The culture of Leptospira requires up to four weeks for results and is insensitive compared to other techniques [1], [2], [11], [12]. MAT requires the maintenance of a local reference panel of live bacterial cultures for assay performance and the use of acute and convalescent samples to provide a confirmed diagnosis [2], [3], [11], [13]. Point-of-care serological diagnostics have been developed for Leptospira and allow earlier detection of IgM than MAT [3], [14]. Such assays are less clinically sensitive than MAT, however, and only provide a presumptive diagnosis [14].
Many different molecular diagnostics for leptospirosis have been developed [11], [15]–[25]. These tests can offer sensitive Leptospira detection while also providing a definitive diagnosis in acute disease [3], [13], [20], [26], [27]. Many of these tests have been designed to specifically detect pathogenic Leptospira species [8], [21], [22], [28]. However, the use of such assays for testing clinical specimens has not consistently resulted in improved diagnostic accuracy [2], [8], [19], [20]. This may have resulted from the decreased clinical sensitivity of specific assays [8] combined with the absence of detection of non-pathogenic species in sterile specimens [20]. Recently, our group reported the development of a real-time PCR (rtPCR) for the detection of all Leptospira species (pathogenic and non-pathogenic), which is included in a multiplex assay for the diagnosis of dengue, leptospirosis, and malaria [termed the undifferentiated febrile illness (UFI) assay]. The UFI assay proved significantly more sensitive than conventional PCRs for the detection of flaB and lipL41 [29]. However, the use of conventional PCRs for comparison may have accounted for the increased sensitivity of the UFI assay, which, to date, has not been evaluated against another real-time molecular diagnostic. Furthermore, it was unclear if similar results could be obtained using an assay specific for pathogenic Leptospira species.
In the current study, we report the development and analytical characterization of an rtPCR for the detection of pathogenic Leptospira species (referred to as the pathogenic rtPCR). The pathogenic rtPCR combines the Leptospira primers from the UFI assay with a new hydrolysis probe that targets sequence found in pathogenic Leptospira species. This assay was performed as a monoplex reaction, which differs from the multiplex, internally-controlled design of the UFI assay. Using 65 clinical samples from suspected leptospirosis cases in Brazil, we then performed a comparison of the UFI assay and pathogenic rtPCR with a reference16S rtPCR [22].
Methods
Ethics
The Stanford University IRB waived review of this study. All samples were pre-collected as part of routine clinical care and de-identified.
Assay Design
The pathogenic rtPCR utilizes primers that were developed for the UFI assay. These primers target a region of the Leptospira 16S rRNA (rrs) gene (Figure 1), and their design has been described previously [29]. To design the pathogenic probe, sequences that matched conserved regions of available L. biflexa sequences and differed from L. interrogans sequences were removed from an alignment of 704 sequences of the Leptospira 16S rRNA gene. These were re-aligned as non-pathogenic species using MegAlign software (DNASTAR, Madison, WI). This alignment included all available sequences for L. biflexa (n = 12) and the following 22 sequences: L. meyeri (n = 15); L. wolbachii (n = 2); L. vanthielii (n = 2); L. idonii, L. terpstrae, and L. yanagawae (1 each; accession numbers available on request). The remaining 670 sequences from pathogenic species were re-aligned. These pathogenic and non-pathogenic consensus sequences were then aligned using BLAST to identify targets for a pathogen specific probe (Figure 1). The pathogenic probe (5′ – Cal Fluor 560 – GCRATGTGATGATGGTACCTGCCT – BHQ-1 – 3′) was designed using Primer-BLAST [30].
Figure 1. BLAST alignment for the targeted 16S rRNA gene consensus sequences from pathogenic (top) and non-pathogenic (bottom) strains of Leptospira.
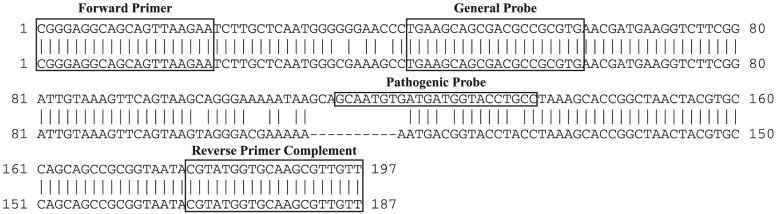
The forward primer sequence and the complement of the reverse primer sequence used in the pathogenic rtPCR and UFI assay are labeled, as are the general (UFI assay) and pathogenic probe sequences.
Assay Performance and Reference PCRs
Pathogenic rtPCR reactions were performed on the Rotor-Gene Q instrument (Qiagen, Germantown, MD) using 25 µL reaction volumes and the SuperScript III Platinum One-Step qRT-PCR kit (Invitrogen, Carlsbad, CA). Each reaction contained 5 µL of nucleic acid template; forward and reverse primers as well as the probe were used at final concentrations of 400 nM. Cycling conditions were identical to conditions described for the UFI assay [29]. During analysis, the first five cycles were cropped to improve baseline normalization. Results were evaluated on the linear scale with slope correction and a threshold of 0.05. A positive result was considered any exponential curve with a cycle threshold (CT) prior to cycle 45.
The 16S rtPCR developed by Smythe, et al., was used as reference for the testing of cultured Leptospira strains and clinical samples [22]. This assay was performed on the Rotor-Gene Q instrument using 25 µL reaction volumes of the TaqMan Universal PCR Master Mix (Life Technologies, Grand Island, NY). Each reaction contained 5 µL of nucleic acid template. Primer and probe concentrations were used according to Thaipadunpanit, et al [8]. Cycling conditions were the following: 95°C for 10 min and 45 cycles of 95°C for 15 sec and 60°C for 60 sec. Detection was performed in the green channel at 60°C; the gain was set at 10 following optimization. During analysis, the first five cycles were cropped; results were evaluated on the linear scale with slope correction and a threshold of 0.05. A positive result was considered any exponential curve with a CT prior to cycle 45. A no-template control was included on each run of the UFI assay, pathogenic, and reference 16S rtPCRs. No signal was observed from the no-template control on any run.
Clinical samples were originally sent to the Laboratório de Zoonoses Bacterianas, Instituto Oswaldo Cruz, Fundação Oswaldo Cruz (Fiocruz) and tested with PCRs for flaB and lipL41 as described [16], [25], [29].
Control Nucleic Acids and Reference Material
The analytical validation of the pathogenic rtPCR was performed using quantitated plasmid DNA. Leptospira interrogans serovar Copenhageni, strain Fiocruz L1-130 (ATCC Number BAA-1198D-5; ATCC, Manassas, VA) genomic DNA was used during plasmid production, as previously described [29]. This is referred to as the Leptospira reference strain. Extracted genomic DNA from 39 cultured Leptospira isolates was tested using the pathogenic and 16S reference rtPCRs. These included strains from 7 species and 23 different serovars of Leptospira (Table 1).
Table 1. Reference Leptospira isolates tested with the pathogenic and reference 16S rtPCRs.
| Leptospires Obtained from the Leptospira Collection (CLEP) – Fiocruz (Rio de Janeiro, Brazil) | |||
| Species | Serovar | Strain | CLEP Code |
| L. biflexa | Semaranga | Patoc 1 | 00015 |
| L. biflexa | Andamana | CH11 | 00021 |
| L. borgpetersenii | Tarassovi | Perepelitsin | 00016 |
| L. borgpetersenii | Javanica | Veldrat Batavia 46 | 00010 |
| L. borgpetersenii | Castellonis | Castellon 3 | 00008 |
| L. fainei | Hurstbridge | But 6 | 00026 |
| L. interrogans | Icterohaemorrhagiae | RGA | 00001 |
| L. interrogans | Copenhageni | M20 | 00002 |
| L. interrogans | Canicola | Hond Utrecht IV | 00003 |
| L. interrogans | Pomona | Pomona | 00005 |
| L. interrogans | Australis | Ballico | 00006 |
| L. interrogans | Autumnalis | Akiyami A | 00017 |
| L. interrogans | Pyrogenes | Salinem | 00012 |
| L. interrogans | Lai | Lai | 00028 |
| L. kirshneri | Grippotyphosa | Moskva V | 00004 |
| L. kirshneri | Mozdok | 5621 | 00091 |
| L. noguchii | Panama | CZ214K | 00011 |
| L. weilii | Vughia | LT 89–68 | 00040 |
Analytical Characterization
The pathogenic rtPCR was analytically characterized according to published recommendations [31]. To establish the dynamic range of the pathogenic rtPCR, four replicates of serial 10-fold dilutions of plasmid DNA from 7.0 log10 copies/µL to 1 copy/µL were tested in a single run. The dynamic range of the reference 16S rtPCR was evaluated by testing four replicates of serial 10-fold dilutions of genomic DNA from the Leptospira reference strain from 5.0 to 1.0 log10 copies/µL. The concentration, in copies/µL, of the Leptospira reference strain was calculated from the standard curve generated during the pathogenic rtPCR dynamic range evaluation. The dynamic range was established for each assay by fitting a best-fit line to the data by regression analysis and included the range where the R2 value for this line was ≥0.99.
To establish the lower limit of 95% detection (95% LLOD) for the pathogenic rtPCR, ten replicates of four, two-fold dilutions were tested on a single run. The 95% LLOD was then calculated using probit analysis. The dilutions began at a concentration 2-fold higher than the lowest concentration at which all replicates were detectable during the dynamic range study.
The specificity of the pathogenic rtPCR was evaluated by testing serum samples from 99 Nicaraguan dengue cases. These samples have been described previously [32]. In addition, specificity was evaluated by testing extracted DNA from clinical strains of Staphylococcus aureus and coagulase-negative Staphylococcus (three strains each); and cultured strains of Salmonella enterica subsp. arizonae, S. enterica serovar Typhi, Treponema denticola (ATCC strain 35405), and Borrelia burgdorferi strain B31 (ATCC number 35210).
Clinical Samples
Archived samples (63 serum, 2 plasma) collected from 65 patients in Brazil with suspected leptospirosis were included in this study. These samples have been described in detail previously [29]. Samples were collected between April 2009 and November 2013. Fifty-five acute samples were evaluated using MAT. Convalescent samples were not available for testing. The extraction of DNA was performed on-site in Brazil prior to the shipment of samples for testing. DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD) according to the manufacturer's recommendations. All extracted nucleic acids were stored at −80°C until use. Samples were tested using the UFI assay, pathogenic rtPCR, and the reference 16S rtPCR during a single freeze-thaw cycle. Fifty samples that tested positive in the UFI assay had amplicons were sequenced as described [29]; this included 47 samples that tested positive in the pathogenic rtPCR,
Statistics
Assay comparisons were performed using two-tailed Fisher's exact tests. Comparisons of mean CT values were performed using Welch t-tests. Fisher's exact tests and Welch t-tests were performed with GraphPad software (GraphPad; La Jolla, CA). Probit analysis was performed using SPSS (IBM; Armonk, NY).
Results
rtPCR Analytical Evaluation
The dynamic range for the pathogenic rtPCR extended from 7.0 to 2.0 log10 copies/µL, and the 95% LLOD was 29 copies/µL. Extracted DNA from 39 cultured Leptospira isolates was tested (Table 1). Two non-pathogenic L. biflexa isolates produced no signal in the pathogenic assay (Figure 2A); these samples had CT values of 6.26 and 10.80 in the UFI assay (Figure 2B). The remaining 37 isolates were detected with the pathogenic assay with CT values from 2.81 to 11.88. No amplification was detected when extracted nucleic acids from 99 serum samples from Nicaraguan patients with dengue were tested. Extracted nucleic acids from Staphylococcal isolates and cultured strains of S. enterica subsp. arizonae, S. enterica serovar Typhi, T. denticola, and B. burgdorferi produced no amplification when tested in the pathogenic rtPCR.
Figure 2. The pathogenic rtPCR does not amplify DNA from cultured L. biflexa isolates.
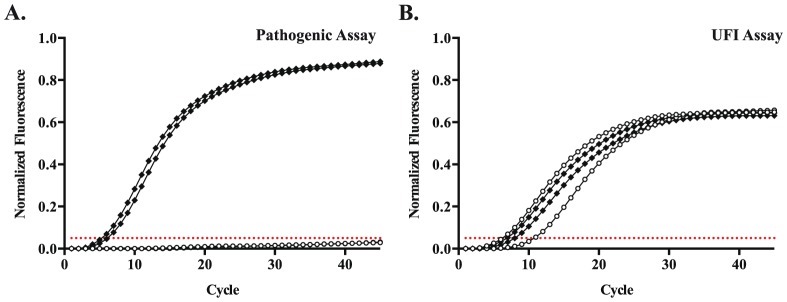
Amplification curves for cultured isolates of L. interrogans (solid diamonds) and L. biflexa (open circles) in the (A) pathogenic rtPCR and (B) UFI assay. Results are displayed for two isolates of each species. The threshold for positivity is set at 0.05 normalized fluorescence units for both assays (dotted red line).
The dynamic range of the reference 16S rtPCR extended from 5.0 to 1.0 log10 copies/µL. Extracted DNA from the 39 cultured Leptospira isolates were also evaluated using this assay. Thirty-five isolates were positive in the reference 16S rtPCR and demonstrated early CT values (range 12.26–21.74). One strain each of L. biflexa, L. weilii, and L. fainei, tested positive but had late CT values of 34.19, 34.48, and 36.03. The remaining L. biflexa isolate demonstrated no amplification.
Clinical Samples
Available clinical data for the 65 cases of suspected leptospirosis is shown in Table 2. The day of illness (DOI) of sample collection was recorded for 45 patients; the median DOI was 8 (interquartile range 5–12). Of the 28 patients for whom the presenting signs and symptoms were available, the most common findings were fever (n = 26; 93%), myalgia (n = 19; 68%), and jaundice (n = 13; 46%).
Table 2. Reported clinical data for 65 samples from patients with suspected leptospirosis.
| Patients, n | 65 |
| Day of Illness | |
| Patients, n (%) | 45 (69.2) |
| Median (IQR) | 8.0 (5–12) |
| Clinical Presentation | |
| Patients, n (%) | 28 (43.1) |
| Fever | 26 |
| Myalgia | 19 |
| Jaundice | 13 |
| Gastrointestinal Complaints1 | 13 |
| Headache | 12 |
| Respiratory Complaints2 | 8 |
| Hemorrhage | 8 |
| Renal Failure | 6 |
| Conjunctival congestion | 2 |
Includes nausea, vomiting, abdominal pain, diarrhea, anorexia, and hemorrhage.
Includes cough, shortness of breath, and hemoptysis.
All 65 samples were positive for Leptospira using the UFI assay, and no co-infections with dengue or malaria were detected (Table 3). Sixty-two (95.4%) samples were positive using the pathogenic rtPCR, and 24 (38.1%) samples were positive using the reference 16S rtPCR. The rates of Leptospira DNA detection using the UFI assay and pathogenic rtPCR did not differ significantly (p = 0.24). However, both assays detected Leptospira DNA in significantly more samples than the reference 16S rtPCR (p<0.0001 for both comparisons). The differences in rates of Leptospira detection using the reference 16S rtPCR (38.1%) and prior results using PCRs for flaB and lipL41 (16/65 samples detected, 25.4%) did not reach statistical significance (p = 0.13).
Table 3. Pairwise comparisons of Leptospira PCR diagnostics.
| Pathogenic rtPCR | ||||
| Positive | Negative | Total | ||
| UFI Assay | Positive | 62 | 3 | 65 |
| Negative | 0 | 0 | 0 | |
| Reference 16S rtPCR | Positive | 24 | 0 | 24 |
| Negative | 39 | 2 | 41 | |
| flaB/lipL41 | Positive | 15 | 1 | 16 |
| Negative | 47 | 2 | 49 | |
| Composite Reference | Positive | 28 | 1 | 29 |
| Negative | 34 | 2 | 36 | |
Comparison of the pathogenic rtPCR with the UFI assay, reference 16S rtPCR, conventional PCRs for flaB/lipL41, and a composite reference that takes into account flaB/lipL41 and the reference 16S rtPCR. Samples that tested positive by at least one of these assays were considered positive, while those that tested negative by both assays were considered negative for this composite reference. Samples that tested negative in the pathogenic rtPCR had CT values of 34.77, 35.26, and 36.11 in the UFI assay.
For samples that only tested positive using the UFI assay and pathogenic rtPCR, the mean CT value was significantly later (32.62; standard deviation, 2.21) than the mean CT value for samples that also tested positive in the reference 16S rtPCR, 28.33 (standard deviation, 4.05; p<0.0001) (Table 4). When results in the pathogenic rtPCR were plotted against DOI of sample collection, no change in the CT value was observed (Figure 3). MAT results were positive for 6 of 55 acute samples tested by this method. Leptospira DNA was only detectable in these six samples using the UFI assay and pathogenic rtPCR (Table 5).
Table 4. Amplicon sequencing results and CT values for select clinical samples.
| n | Pathogenic rtPCR Positive, n (%) | UFI Assay Mean CT (Standard Deviation) | Amplicons Sequenced | Sequence Matches1 (n) | |
| Reference 16S rt PCR Positive 1 | 24 | 24 (100) | 28.33 (4.05) | 13 | L. interrogans, L. kirschneri, or L. noguchii (9) |
| L. borgpetersenii, L. santarosai, or L. weilii (3) | |||||
| L. meyeri, L. biflexa, or L. wobachii (1) | |||||
| flaB/lipL41 PCR and Reference 16S rt PCR Negative | 36 | 34 (94.4) | 32.62 (2.21) | 34 | L. interrogans, L. kirschneri, or L. noguchii (34) |
Given the highly conserved nature of this region, final species determinations cannot be made from amplicon sequences.
Figure 3. Leptospira DNA levels do not correlate with the day of illness of sample collection.
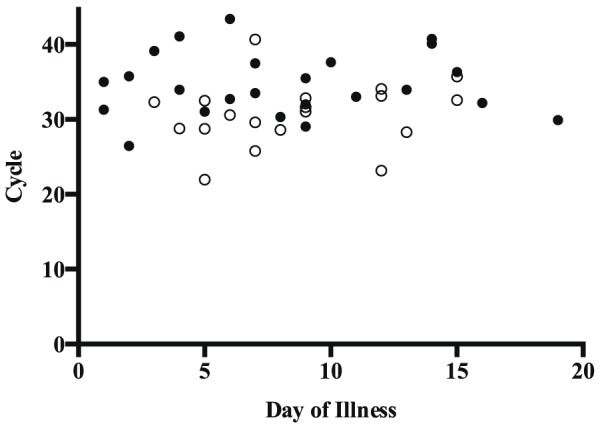
The cycle threshold in the pathogenic rtPCR was compared to the day of illness of sample collection. Results are shown for the 43 samples detected in the pathogenic rtPCR with available day of illness information. Samples positive in the reference 16S rtPCR or conventional PCRs (open circles) and samples positive only in the UFI assay and pathogenic rtPCR (closed circles) are displayed.
Table 5. Amplicon sequencing and CT values for select clinical samples evaluated by MAT.
| n | Pathogenic rtPCR Positive, n (%) | UFI Assay Mean CT (Standard Deviation) | Amplicons Sequenced | Sequence Matches1 (n) | |
| MAT Positive | 6 | 6 (100) | 32.51 (2.27) | 6 | L. interrogans, L. kirschneri, or L. noguchii (6) |
| MAT Negative | 49 | 47 (95.9) | 30.49 (3.90) | 39 | L. interrogans, L. kirschneri, or L. noguchii (35) |
| L. borgpetersenii, L. santarosai, or L. weilii (3) | |||||
| L. meyeri, L. biflexa, or L. wobachii (1) |
Given the highly conserved nature of this region, final species determinations cannot be made from amplicon sequences.
For 50 clinical samples, bidirectional sequence was obtained using the forward and reverse Leptospira primers. This included 34/36 samples that were only detected using the pathogenic rtPCR or UFI assay. Forty-nine sequences matched publicly available sequences from pathogenic Leptospira species (Table 4). A single sequence matched the 16S gene from non-pathogenic species: L. meyeri, L. biflexa, or L. wobachii. This sample was detected in all molecular tests. However in the pathogenic rtPCR, the amplification curve was flattened compared to curves from pathogenic species (Figure 4A) and compared to the shape of the curve when the same strain was amplified in the UFI assay (Figure 4B).
Figure 4. The pathogenic rtPCR shows blunted amplification of a clinical sample with 16S sequence consistent with a non-pathogenic Leptospira strain.
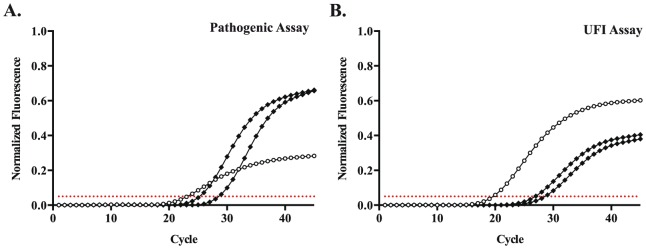
Amplification curves for a clinical sample with sequence matching the non-pathogenic Leptospira, L. meyeri, L. biflexa, and L. wolbachii (open circles) in the (A) pathogenic rtPCR and (B) UFI assay. Two clinical samples with 16S sequence matching pathogenic strains (solid diamonds) are shown for comparison. The threshold for positivity is set at 0.05 normalized fluorescence units for both assays (dotted red line).
Discussion
In this study, we describe the development of an rtPCR for the detection of pathogenic Leptospira species and present the results from a comparison of molecular diagnostics for Leptospira using samples from 65 suspected leptospirosis cases. The current study expands on earlier findings from our group by demonstrating the increased clinical sensitivity of the UFI assay as well as the pathogenic rtPCR when compared to another real-time nucleic acid amplification test, the reference 16S rtPCR.
The reference 16S rtPCR selected as a comparator in this study was originally reported by Smythe, et al., in 2002 [22], and primer and probe concentrations were later modified by Thaipadunpanit, et al. [8]. While these modifications may have affected the analytical performance of the assay compared to the original, this is unlikely to fully explain the difference in Leptospira detection rates observed here. The 16S rtPCR, as modified by Thaipadunpanit, et al., proved equally sensitive to MAT using acute and convalescent samples [8], [13]. In the current study, the reference 16S rtPCR demonstrated a similar dynamic range compared to the pathogenic rtPCR and results from 39 cultured isolates were consistent with published reports [8], [22], including the late amplification of some non-pathogenic strains.
Many nucleic acid amplification tests for leptospirosis have been developed that preferentially detect Leptospira species categorized as pathogenic [15], [18], [22], [28], [33]–[35]. While such assays may prove useful in other contexts, such as testing environmental samples [28], [36], [37], the results of this evaluation highlight concerns regarding such a testing approach for human specimens. Of the clinical samples that were successfully sequenced, 49/50 yielded sequence consistent with pathogenic Leptospira species. This includes 34/34 samples that were not detected using the reference 16S rtPCR or conventional PCRs for flaB and lipL41, and 3/3 samples that were negative in the pathogenic rtPCR. One sample most closely matched the 16S sequence of several Leptospira species considered non-pathogenic: L. meyeri, L. biflexa, and L. wobachii. However, it should be noted that some Leptospira strains originally identified as L. meyeri have been reclassified as pathogenic species [38]. This specimen was obtained from a patient who presented with fever, conjunctival congestion, vomiting and jaundice, and the sample tested positive in all assays evaluated. This case suggests that distinguishing pathogenic from non-pathogenic strains may not be clinically relevant in symptomatic patients with suspected leptospirosis. It also underscores the difficulty of designing an assay for pathogenic Leptospira species based on an evolving classification system. Interestingly, this sequence contained a single base difference compared to the consensus sequence of non-pathogenic strains, which resulted in an extra three bases that matched the 5′ end of the pathogenic probe. Such a difference may have allowed sufficient binding to generate a clear but blunted signal in the pathogenic rtPCR (Figure 4A), while even very high concentrations of reference isolates of L. biflexa produced no detectable signal (Figure 2A).
Leptospiremia occurs during the acute phase of clinical illness, though the duration remains poorly defined. It has been reported that nucleic acid amplification methods for the diagnosis of leptospirosis may only be useful during the first week of illness, though in untreated cases, Leptospira DNA has been detected past day 15 [2], [3], [8], [12], [26]. In a study by Agampodi, et al., the sensitivity of a 16S rtPCR was not affected by the length of time between the onset of symptoms and sample collection [26]. Consistent with that finding, we detected Leptospira DNA in samples collected up to 19 days after the reported onset of symptoms. Also, there was no apparent relationship between the DOI of sample collection and CT values in the pathogenic rtPCR (Figure 3). This finding warrants further study, including an evaluation of serial samples from individual patients, as the duration of leptospiremia or the rate of change in bacterial load may be predictive of patient outcomes.
While the sample size was sufficient to demonstrate improved Leptospira detection using the UFI assay and pathogenic rtPCR, the number of clinical samples and available information were insufficient to evaluate correlations between disease severity and the level of leptospiremia. The current study also involved clinical samples from a single geographical location, and our findings will need to be confirmed in other regions. A limitation to the pathogenic rtPCR, which is common to all nucleic acid amplification tests, involves a concern regarding the emergence of divergent bacterial strains with mutations in the sequences targeted by the primers and probes. We have attempted to address this limitation by targeting highly conserved regions of available Leptospira sequences, but this concern cannot be eliminated.
In conclusion, we present the development of a pathogenic rtPCR for Leptospira. Using a set of 65 clinical samples, the pathogenic rtPCR as well as the UFI assay demonstrated significantly improved clinical sensitivity compared to the reference 16S rtPCR. Future studies are needed to investigate the clinical and epidemiologic significance of more sensitive Leptospira detection using these assays.
Acknowledgments
We would like to thank Sarah Wynwood at the WHO/FAO/OIE Collaborating Centre for Leptospirosis Reference and Research, Queensland Health, Queensland, Australia for the preparation and shipment of reference Leptospira strains. We also thank Dr. Brian Blackburn for his thoughtful comments on this manuscript, Dr. Rajiv Gaur for providing DNA from reference bacterial isolates, the staff of the Stanford Clinical Virology Laboratory, and the Stanford SPARK program for their support over the course of this project.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This research was supported by the National Institutes of Health (NIH) (grant RC4 TW008781-01) and the Thrasher Research Fund Award (11979). Salary support was provided by the NIH (grant 2T32AI007502-16A1 to JJW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, et al. (2003) Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3: 757–771. [DOI] [PubMed] [Google Scholar]
- 2. Levett PN (2001) Leptospirosis. Clin Microbiol Rev 14: 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Picardeau M, Bertherat E, Jancloes M, Skouloudis AN, Durski K, et al. (2014) Rapid tests for diagnosis of leptospirosis: Current tools and emerging technologies. Diagn Microbiol Infect Dis 78: 1–8. [DOI] [PubMed] [Google Scholar]
- 4. Agampodi SB, Dahanayaka NJ, Bandaranayaka AK, Perera M, Priyankara S, et al. (2014) Regional differences of leptospirosis in sri lanka: observations from a flood-associated outbreak in 2011. PLoS Negl Trop Dis 8: e2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van de Werve C, Perignon A, Jaureguiberry S, Bricaire F, Bourhy P, et al. (2013) Travel-related leptospirosis: a series of 15 imported cases. J Travel Med 20: 228–231. [DOI] [PubMed] [Google Scholar]
- 6. Bruce MG, Sanders EJ, Leake JA, Zaidel O, Bragg SL, et al. (2005) Leptospirosis among patients presenting with dengue-like illness in Puerto Rico. Acta Trop 96: 36–46. [DOI] [PubMed] [Google Scholar]
- 7.Pan American Health O (2011) Case definitions: dengue and leptospirosis. pp.1.
- 8. Thaipadungpanit J, Chierakul W, Wuthiekanun V, Limmathurotsakul D, Amornchai P, et al. (2011) Diagnostic accuracy of real-time PCR assays targeting 16S rRNA and lipL32 genes for human leptospirosis in Thailand: a case-control study. PLoS One 6: e16236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brett-Major DM, Coldren R (2012) Antibiotics for leptospirosis. Cochrane Database Syst Rev 2: CD008264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moon JE, Rivard RG, Griffith ME, Ressner RA, McCall S, et al. (2007) Effect of timing and duration of azithromycin therapy of leptospirosis in a hamster model. J Antimicrob Chemother 59: 148–151. [DOI] [PubMed] [Google Scholar]
- 11. Balassiano IT, Vital-Brazil JM, Pereira MM (2012) Leptospirosis diagnosis by immunocapture polymerase chain reaction: a new tool for early diagnosis and epidemiologic surveillance. Diagn Microbiol Infect Dis 74: 11–15. [DOI] [PubMed] [Google Scholar]
- 12. de Abreu Fonseca C, Teixeira de Freitas VL, Calo Romero E, Spinosa C, Arroyo Sanches MC, et al. (2006) Polymerase chain reaction in comparison with serological tests for early diagnosis of human leptospirosis. Trop Med Int Health 11: 1699–1707. [DOI] [PubMed] [Google Scholar]
- 13. Limmathurotsakul D, Turner EL, Wuthiekanun V, Thaipadungpanit J, Suputtamongkol Y, et al. (2012) Fool's gold: Why imperfect reference tests are undermining the evaluation of novel diagnostics: a reevaluation of 5 diagnostic tests for leptospirosis. Clin Infect Dis 55: 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goris MG, Leeflang MM, Loden M, Wagenaar JF, Klatser PR, et al. (2013) Prospective evaluation of three rapid diagnostic tests for diagnosis of human leptospirosis. PLoS Negl Trop Dis 7: e2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahmed A, Engelberts MF, Boer KR, Ahmed N, Hartskeerl RA (2009) Development and validation of a real-time PCR for detection of pathogenic leptospira species in clinical materials. PLoS One 4: e7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahmed N, Devi SM, Valverde Mde L, Vijayachari P, Machang'u RS, et al. (2006) Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob 5: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koizumi N, Nakajima C, Harunari T, Tanikawa T, Tokiwa T, et al. (2012) A new loop-mediated isothermal amplification method for rapid, simple, and sensitive detection of Leptospira spp. in urine. J Clin Microbiol 50: 2072–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levett PN, Morey RE, Galloway RL, Turner DE, Steigerwalt AG, et al. (2005) Detection of pathogenic leptospires by real-time quantitative PCR. J Med Microbiol 54: 45–49. [DOI] [PubMed] [Google Scholar]
- 19. Merien F, Amouriaux P, Perolat P, Baranton G, Saint Girons I (1992) Polymerase chain reaction for detection of Leptospira spp. in clinical samples. J Clin Microbiol 30: 2219–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Merien F, Portnoi D, Bourhy P, Charavay F, Berlioz-Arthaud A, et al. (2005) A rapid and quantitative method for the detection of Leptospira species in human leptospirosis. FEMS Microbiol Lett 249: 139–147. [DOI] [PubMed] [Google Scholar]
- 21. Slack AT, Symonds ML, Dohnt MF, Smythe LD (2006) Identification of pathogenic Leptospira species by conventional or real-time PCR and sequencing of the DNA gyrase subunit B encoding gene. BMC Microbiol 6: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smythe LD, Smith IL, Smith GA, Dohnt MF, Symonds ML, et al. (2002) A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect Dis 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sonthayanon P, Chierakul W, Wuthiekanun V, Thaipadungpanit J, Kalambaheti T, et al. (2011) Accuracy of loop-mediated isothermal amplification for diagnosis of human leptospirosis in Thailand. Am J Trop Med Hyg 84: 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR (2009) Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis 64: 247–255. [DOI] [PubMed] [Google Scholar]
- 25. Kawabata H, Dancel LA, Villanueva SY, Yanagihara Y, Koizumi N, et al. (2001) flaB-polymerase chain reaction (flaB-PCR) and its restriction fragment length polymorphism (RFLP) analysis are an efficient tool for detection and identification of Leptospira spp. Microbiol Immunol 45: 491–496. [DOI] [PubMed] [Google Scholar]
- 26. Agampodi SB, Matthias MA, Moreno AC, Vinetz JM (2012) Utility of quantitative polymerase chain reaction in leptospirosis diagnosis: association of level of leptospiremia and clinical manifestations in Sri Lanka. Clin Infect Dis 54: 1249–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Merien F, Baranton G, Perolat P (1995) Comparison of polymerase chain reaction with microagglutination test and culture for diagnosis of leptospirosis. J Infect Dis 172: 281–285. [DOI] [PubMed] [Google Scholar]
- 28. Murgia R, Riquelme N, Baranton G, Cinco M (1997) Oligonucleotides specific for pathogenic and saprophytic leptospira occurring in water. FEMS Microbiol Lett 148: 27–34. [DOI] [PubMed] [Google Scholar]
- 29.Waggoner JJ, Abeynayake J, Balassiano I, Lefterova M, Sahoo MK, et al.. (2014) A Multiplex Nucleic Acid Amplification Test for the Diagnosis of Dengue, Malaria, and Leptospirosis. J Clin Microbiol. [DOI] [PMC free article] [PubMed]
- 30. Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, et al. (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burd EM (2010) Validation of laboratory-developed molecular assays for infectious diseases. Clin Microbiol Rev 23: 550–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Waggoner JJ, Abeynayake J, Sahoo MK, Gresh L, Tellez Y, et al. (2013) Development of an internally controlled real-time reverse transcriptase PCR assay for pan-dengue virus detection and comparison of four molecular dengue virus detection assays. J Clin Microbiol 51: 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boonsilp S, Thaipadungpanit J, Amornchai P, Wuthiekanun V, Chierakul W, et al. (2011) Molecular detection and speciation of pathogenic Leptospira spp. in blood from patients with culture-negative leptospirosis. BMC Infect Dis 11: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Branger C, Blanchard B, Fillonneau C, Suard I, Aviat F, et al. (2005) Polymerase chain reaction assay specific for pathogenic Leptospira based on the gene hap1 encoding the hemolysis-associated protein-1. FEMS Microbiol Lett 243: 437–445. [DOI] [PubMed] [Google Scholar]
- 35. Woo TH, Patel BK, Smythe LD, Norris MA, Symonds ML, et al. (1998) Identification of pathogenic Leptospira by TaqMan probe in a LightCycler. Anal Biochem 256: 132–134. [DOI] [PubMed] [Google Scholar]
- 36. Viau EJ, Boehm AB (2011) Quantitative PCR-based detection of pathogenic Leptospira in Hawai'ian coastal streams. J Water Health 9: 637–646. [DOI] [PubMed] [Google Scholar]
- 37. Vital-Brazil JM, Balassiano IT, Oliveira FS, Costa AD, Hillen L, et al. (2010) Multiplex PCR-based detection of Leptospira in environmental water samples obtained from a slum settlement. Mem Inst Oswaldo Cruz 105: 353–355. [DOI] [PubMed] [Google Scholar]
- 38. Slack AT, Galloway RL, Symonds ML, Dohnt MF, Smythe LD (2009) Reclassification of Leptospira meyeri serovar Perameles to Leptospira interrogans serovar Perameles through serological and molecular analysis: evidence of a need for changes to current procedures in Leptospira taxonomy. Int J Syst Evol Microbiol 59: 1199–1203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.


