Abstract
Cytoplasmic dynein is a molecular motor that transports a large variety of cargoes (e.g., organelles, messenger RNAs, and viruses) along microtubules over long intracellular distances. The dynactin protein complex is important for dynein activity in vivo, but its precise role has been unclear. Here, we found that purified mammalian dynein did not move processively on microtubules in vitro. However, when dynein formed a complex with dynactin and one of four different cargo-specific adapter proteins, the motor became ultraprocessive, moving for distances similar to those of native cargoes in living cells. Thus, we propose that dynein is largely inactive in the cytoplasm and that a variety of adapter proteins activate processive motility by linking dynactin to dynein only when the motor is bound to its proper cargo.
Cytoplasmic dynein 1 (dynein), a member of the AAA adenosine triphosphatase (ATPase) family, is the major minus-end–directed microtubule (MT)motor in most eukaryotic cells (1). Several adapter proteins control the recruitment of a soluble pool of cytoplasmic dynein to transport cargos at the right place and time in the cell (2–4). Dynein motor activity also appears to be governed by two general regulatory factors—the Lis1-NudEL complex and the dynactin complex (5). Lis1 may act as a “clutch” that suppresses dynein motility and causes it to form a tight binding complex with the MT (6, 7). Whether dynein motility requires just the detachment of Lis1 or needs an additional activation process has been unclear.
Yeast cytoplasmic dynein, the best characterized dynein in terms of its motility, moves processively on its own (run length of 1 to 2 µm) (8), and dynactin only increases its run length by ~twofold (9). Mammalian cytoplasmic dynein is also generally thought to be constitutively active for motility, as it produces movement when attached to solid surfaces such as glass slides (10), plastic beads (11), or quantum dots (12). However, surface binding of kinesin activates this normally autoinhibited motor (13). Without direct visualization of the motor itself, it also can be difficult to determine whether one or multiple motors are contributing to movement. Prior studies of fluorescently labeled mammalian dynactin, but with unlabeled dynein, have reported processive run lengths of <2 µm in both directions on the MT (14).
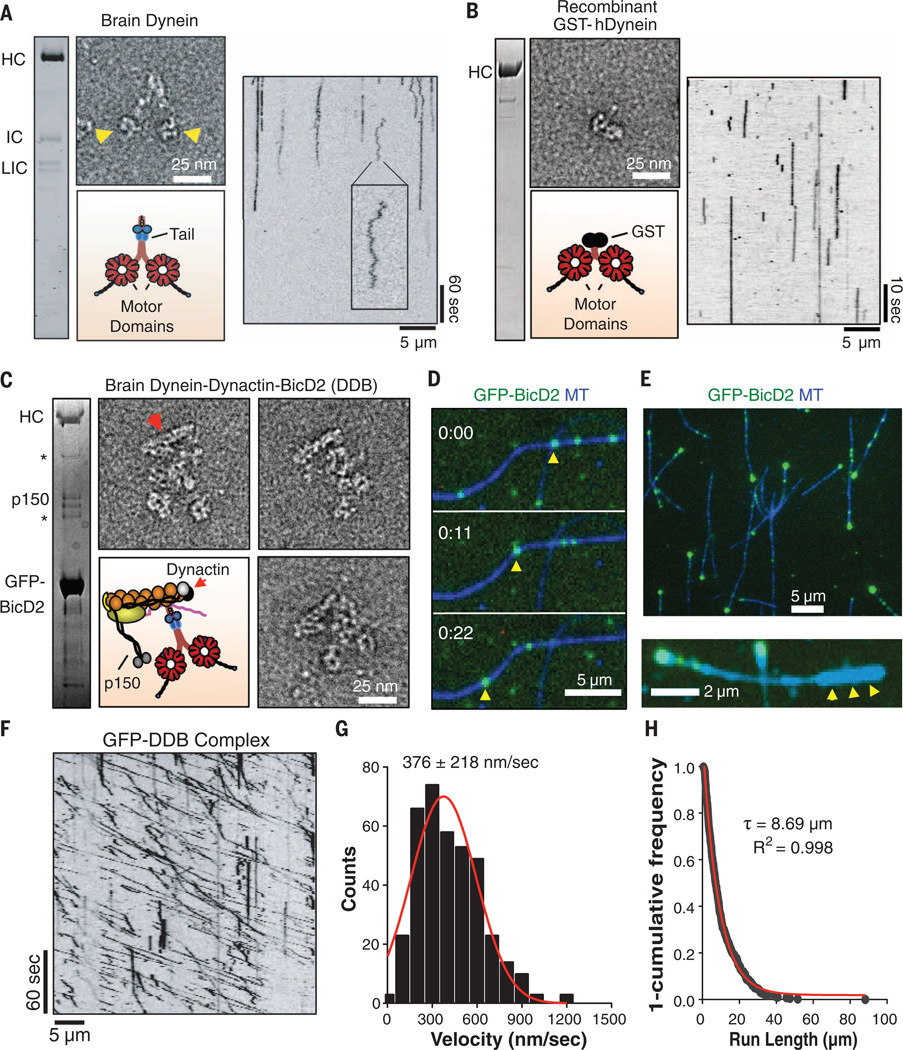
Here, we examined the motility of purified rat brain cytoplasmic dynein (termed “brain dynein”) by single-molecule fluorescence without attachment to surfaces. Brain dynein, which exhibited a characteristic two-headed shape by electron microscopy (EM) (Fig. 1A), produces fast motility (~0.6 µm/s) of MTs in a multiple motor gliding assay (15). However, individual Cy3-labeled native dynein molecules, examined by total internal reflection (TIRF) microscopy in the presence of 1 mM adenosine 5´-triphosphate (ATP), mostly either bound statically to MTs or exhibited short back-and-forth movements (Fig. 1A, fig. S1A, and movie S1), which are likely due to thermal-driven diffusion, as they persist after addition of the ATPase inhibitor vanadate (fig. S1B) (16). Directional movements were only occasionally observed (<1% of MT-bound dynein); those movements were very slow (~90 nm/s, fig. S1A) and inhibited by vanadate (fig. S1B). A recombinant, glutathione S-transferase (GST)–dimerized human motor domain construct (fig. S1C) also did not show fast, processive motion (Fig. 1B and movie S2), consistent with recent studies of a recombinant human cytoplasmic dynein holoenzyme (17). Thus, in contrast to yeast dynein, purified mammalian dynein rarely displays processive motility.
Fig. 1. Formation of a dynein-dynactin-BicD2 complex induces processive dynein motility.
(A) SDS–polyacrylamide gel electrophoresis (SDS-PAGE) of purified rat brain dynein and negative-stain EM. Arrowheads denote motor domains. Kymograph analysis of Cy3-labeled rat brain dynein on MTs (100 µM ATP). Inset: back-and-forth motion, which is likely diffusion-driven. HC: heavy chain, LIC: light intermediate chain. (B) SDS-PAGE of recombinant human GST-dynein and negative-stain EM. Kymograph analysis of TMR-labeled GST-dynein on MTs revealing no movement (100 µM ATP). (C) SDS-PAGE and negative-stain EM of purified pig brain DDB complex. Asterisks mark nonspecific bands. Red arrowhead denotes dynactin’s Arp1 filament. (D) Movie frames showing a single GFP-labeled DDB complex (yellow arrow-head) moving processively along a MT. (E) DDB complexes accumulate at one end of MTs. Below: polarity-marked MTs (arrowheads mark brightly labeled plus-end) reveal that accumulation occurs at the minus-end. (F) Kymograph analysis of pig brain DDB reveals diagonal lines reflecting long, unidirectional movements (100 µM ATP). (G) Velocity histogram(1 mM ATP) with a Gaussian fit (mean ± SD: 376 ± 218 nm/s, n = 379 molecules). (H) A “1-cumulative frequency distribution plot” run-lengths with fit to a one-phase exponential decay (red). Decay constant (τ, run length) and R2 value of the fit are shown (n = 379molecules, two independent preparations).
The poor single-molecule motility by mammalian dynein might be due to the absence of an activator in our purified preparations. BicD2 is a conserved, dimeric adapter protein that links dynein to Rab6 GTPase on membrane organelles (2, 18); the BicD N-terminal coiled coil (19) facilitates an interaction between dynein and dynactin, forming a stable dynein-dynactin-BicD2 ternary complex (DDB) that can be purified (20). Using this well-characterized green fluorescence protein (GFP)–tagged, N-terminal construct of BicD2 (20), we isolated the DDB complex from pig brain (Fig. 1C and fig. S2A); this preparation did not contain detectable kinesin-1, Lis1, or Nudel (fig. S2B). The DDB complex eluted as a single peak by size-exclusion chromatography (fig. S2C), and EM revealed a single dynein dimer with its tail bound to a single dynactin, identifiable by its 37 nm Arp1 filament (Fig. 1C and fig. S3, A to C). The DDB complex appeared highly flexible, as evidenced by the variable orientations of dynactin’s Arp1 filament (fig. S3B) and variable separation of the two motor domains (fig. S3D).
Next, we examined the motility of single DDB complexes by TIRF microscopy, using the GFP-tag on BicD2. In contrast to brain dynein, the DDB complexes moved robustly and processively along MTs (Fig. 1D and movie S3), although a small fraction (~15%) exhibited back-and-forth diffusive motion (fig. S2D). The DDB complexes also accumulated at MT minus-ends (Fig. 1E), indicating tenacious binding upon reaching the end of the MT track. Similar processive motility and minus-end accumulation was observed recently for dynein-driven transport of purified mRNA particles in vitro (21). By kymograph analysis, DDB processive movement appeared as extended diagonal lines (Fig. 1F). At 2 mM ATP,DDB moved at a mean velocity of 376 nm/s (Fig. 1G) and run length of 8.7 µm(Fig. 1H), considerably longer than yeast dynein (8).When measured at physiological temperature (37°C), the velocity was 892 nm/s (fig. S2E), very close to speeds of retrograde transport in vivo (12). Processive motility was completely abolished by the ATPase inhibitor vanadate (fig. S2F). The addition of BicD2 to purified brain dynein did not stimulate processive motility, indicating a requirement for dynactin (fig. S2G). To further confirm that motile DDB complexes contained single BicD2 dimers and not oligomers or aggregates, we added two fluorescently labeled BicD2 preparations (TMR- and 505-Star–labeled) to pig brain lysates followed by DDB complex purification. Two-color kymographs revealed that moving DDB complexes contained either TMR- or 505-Star-BicD2 but not a mixture (fig. S2H and movie S4), confirming that each DDB complex contains only a single BicD2 molecule. Thus, BicD2 links dynein and dynactin to form a stable complex that moves toward the MT minus-end with ultraprocessive run-lengths.
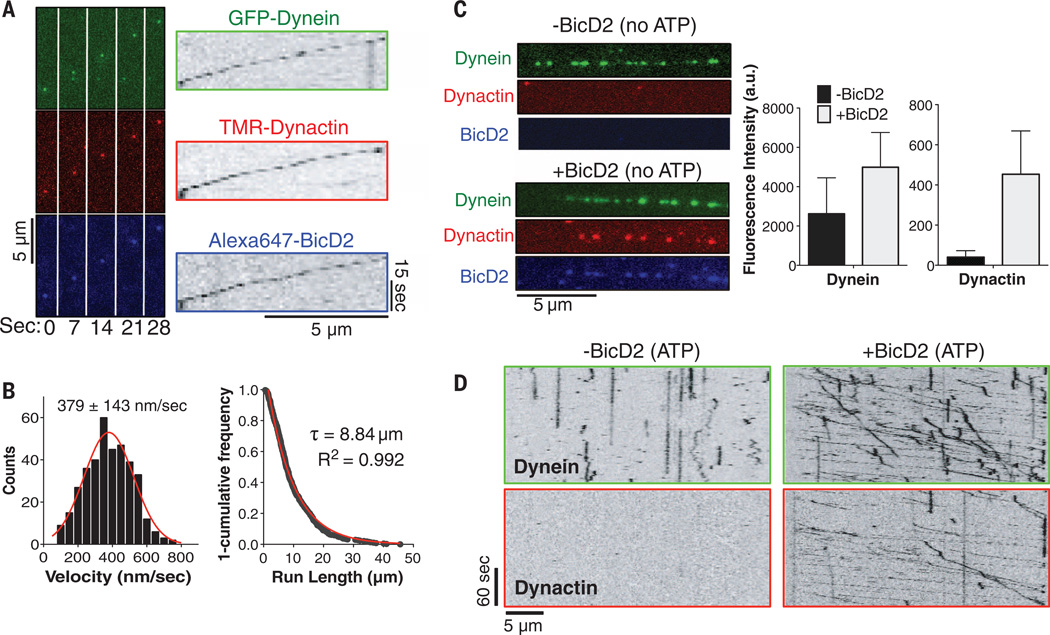
To examine each component of the DDB complex during motility, we purified a triple-colored DDB complex consisting of Alexa647-labeled, SNAPf-tagged BicD2 (expressed in Escherichia coli); GFP-tagged dynein intermediate chain (GFP-IC); and a tetramethylrhodamine (TMR)–labeled, Halo-tagged p62 dynactin subunit [tagged IC and p62 were coexpressed in human RPE-1 cells (22); fig. S4, A and B)]. This triple-color–labeled DDB moved processively, and many moving GFP-dyneins colocalized with a TMR-dynactin and an Alexa647-BicD2 molecule (Fig. 2A and movie S5); the mean velocity and run length of human DDB was 379 nm/s (Fig. 2B, left) and 8.84 µm (Fig. 2B, right), respectively. The fluorescence intensities of the motile GFP-labeled dynein molecules were similar to those of the well-characterized dimeric protein, GFP-kinesin K560 (fig. S4C). Additionally, the fluorescence intensities of isolated TMR-dynactin and Alexa647-BicD2 were similar to the intensities of these components within the motile DDB complex (fig. S4, D and E). Thus, consistent with the EM data (Fig. 1B), a ternary complex, containing single copies of dynein, dynactin, and BicD2, is the active entity that moves processively along MTs.
Fig. 2. Three-color single-molecule analysis of the DDB complex and requirement of BicD2 for motility.
(A) Successive frames from movie S5 showing a processive DDB complex purified from human RPE-1 cells with all three components fluorescently labeled (see text and fig S4A). Right: corresponding kymographs. (B) Histogram of human DDB velocities with a Gaussian fit (mean ± SD; n = 374 molecules). Right: a “1-cumulative frequency distribution plot” of human DDB run-lengths fit to a one-phase exponential decay (red). Decay constant (τ) and R2 value of the fit are shown (n =374 molecules). (C) Binding of fluorescently labeled human dynein and dynactin without ATP to MTs (not visible) in the absence and presence of BicD2. Note that not all DDBs are triple-labeled. Right: quantification of fluorescence along MTs (mean ± SD, n = 16 and 11 MTs before and after BicD2 addition, respectively). (D) Kymograph analysis in the presence of 100 µM ATP shows that BicD2 addition is required for human dynein motility and dynactin binding to the MTs. Note that not all dynactins in the DDB particles (+BicD2) are labeled, owing to incomplete Halo tag labeling and/or photobleaching.
Because dynactin has been suggested to activate dynein motility in the absence of other factors (9, 23), we next examined the single-molecule behavior of dynein and dynactin in the absence of BicD2. GFP-dynein and TMR-dynactin could be separated from BicD2 by salt elution from a BicD2 affinity column (fig. S4F). Without ATP, human GFP-dynein molecules decorated MTs, but MT binding by human TMR-dynactin was not observed (Fig. 2C), as was also reported for yeast dynactin (9). The lack of MT binding by dynactin was surprising, because the recombinant N-terminal half of p150Glued (a subunit in the dynactin complex, Fig. 1B; herein termed “p150”) avidly binds to MTs (24, 25). However, upon addition of Alexa647-BicD2 in the absence of ATP, the intensity of GFP-dynein and TMR-dynactin fluorescence on MTs increased by ~2-and 50-fold, respectively (Fig. 2C). With ATP, GFP-tagged dynein bound statically or diffused back-and-forth on MTs (Fig. 2D), and TMR-dynactin did not bind to the MTs (Fig. 2D). Re-addition of Alexa647-BicD2 strongly stimulated processive motility of TMR-dynactin and GFP-labeled dynein (Fig. 2D and movie S6) without evidence of aggregation (fig. S4G). Thus, dynein and dynactin have very low affinity for one another in the absence of BicD2, in agreement with prior biochemical studies (20), and BicD2 is required for processive motion. Furthermore, MT binding by the p150 subunit appears to be masked in the isolated dynactin complex, perhaps reflecting an autoinhibited state.
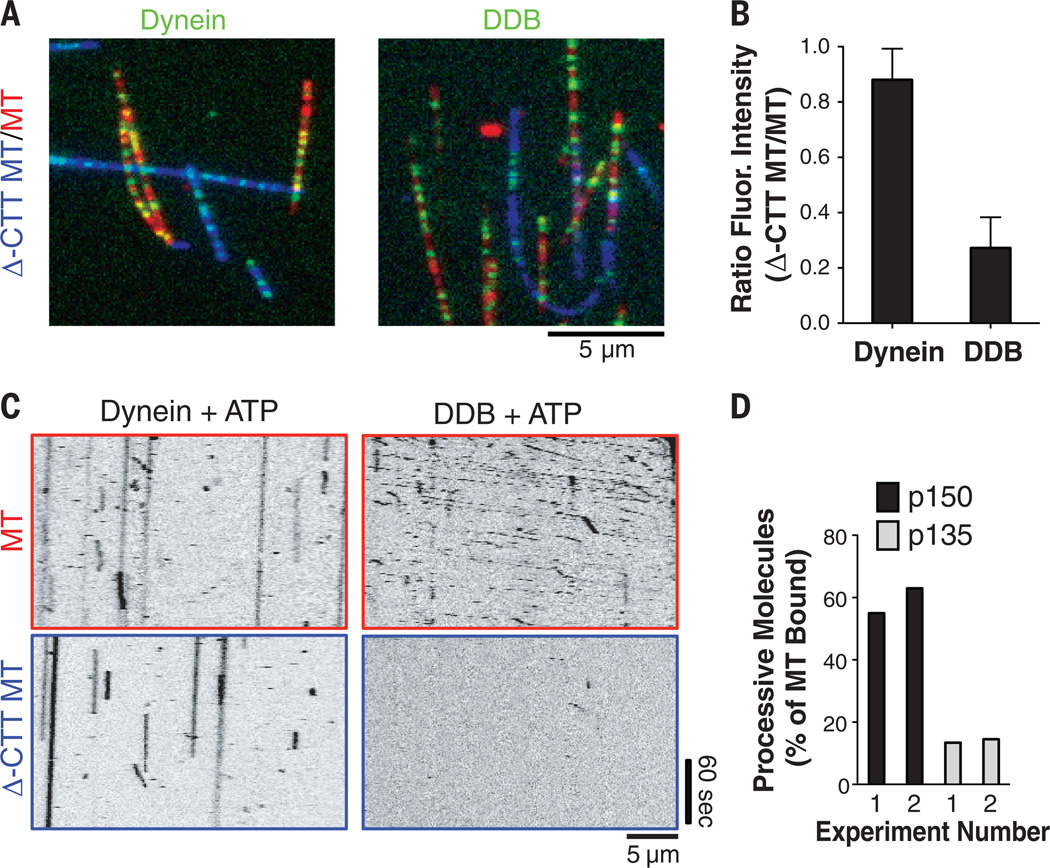
The above experiments raised the question of whether MT binding by p150 plays a role in DDB motility. Previous studies showed that removal of tubulin’s C-terminal tails with the protease subtilisin (Δ-CTT MTs) markedly reduced MT binding of a purified p150 construct (25), which we confirmed (fig. S5, A and B). In the absence of ATP, we found that GFP-human dynein bound almost equally well to both control and Δ-CTT MTs, whereas human DDB complex exhibited greatly reduced binding to Δ-CTT MTs, thus behaving more like p150 than dynein (Fig. 3, A and B, and fig. S5, A and B). The difference was even more pronounced in the presence of ATP. GFP-human dynein bound to both types of MTs (but did not move), whereas DDB moved robustly on untreated MTs, but rarely moved on Δ-CTT MTs that were placed on the same slide (Fig. 3C and movie S7). Similar findings were made for DDB from bovine brain (fig. S5, A to C). In contrast, Saccharomyces cerevisiae dynein, which is not regulated by BicD2, moves almost as well on Δ-CTT as on untreated MTs (26). p150 contains a well-defined MT binding site (a CAP-Gly domain flanked by a basic rich region) at its N terminus (24). To test the role of this domain, we overexpressed Halo-tagged versions of p150 or p135, a naturally occurring splice form lacking the CAP-Gly domain, in RPE cells (27), and then isolated DDB complexes and fluorescently labeled them with Halo-TMR. MT-bound p135-containing DDB complexes displayed one-third as many processive movements versus p150-containing complexes (Fig. 3D). Of the TMR-p135 complexes that moved processively, their velocity (498 ± 226 nm/s) and run-length (8.9 µm) were similar to those of moving TMR-p150 complexes (417 ± 147 nm/s and 12.19 µm; fig. S5, D and E). The significant fraction (~15%) of DDB-p135 complexes that exhibited ultraprocessivity suggests either that the CAP-Gly domain is not absolutely required for motility or that the residual motion observed with TMR-p135 could be due to heterodimerization with endogenous p150. Further work is required to understand the complex interplay between dynactin’s CAP-Gly domain and dynein activation (9, 28, 29).
Fig. 3. MT binding and processivity of DDB requires the C-terminal tails of tubulin.
(A) MT-binding behavior of GFP-tagged dynein (no BicD2), or GFP-DDB from human RPE cells, on normal MTs (red) and Δ-CTT MTs (blue) in the absence of ATP. (B) Quantification of the fluorescence intensity ratios (GFP-Dyn: 24 MTs, 22 Δ-CTT MTs; DDB: 59 MTs, 38 Δ-CTT MTs, mean ± SD). (C) Kymograph analysis of MT binding and motility with ATP (2 mM) on normal MTs (upper row) and on Δ-CTT MTs (lower row). (D)Quantification of processively moving (>2 µm) human DDB complexes with incorporated TMR-labeled p150-Halo or p135-Halo subunits (22) (percent of total MT bound; others were statically bound or diffusing). Results from two independent experiments are shown.
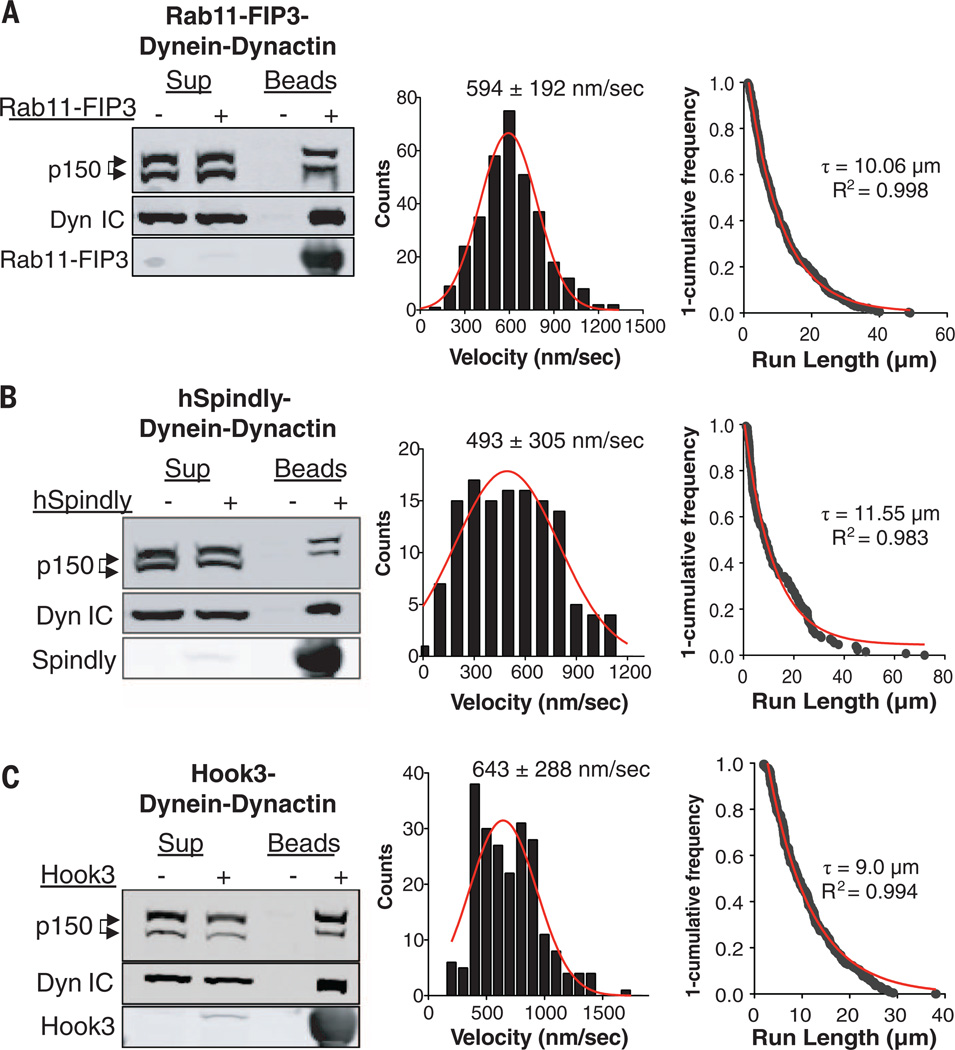
In addition to BicD2, several other coiled-coil proteins have been implicated in linking dynein to cargoes, including Rab11-FIP3 on Rab11-positive recycling endosomes (4), Spindly (hSpindly) on kinetochores (3), and Hook proteins on early endosomes (30, 31). We asked if these cargo adapter proteins might similarly initiate processive motion by increasing dynein’s affinity for dynactin. Recombinant SNAPf-tagged Rab11-FIP3, human Spindly, and Hook3 (fig. S6, A and B) all efficiently coprecipitated dynein and dynactin from pig brain lysates (Fig. 4, A to C). The dynein-dynactin-adapter complexes, labeled with TMR on the SNAPf tag and which did not contain contaminating BicD2 (fig. S6C), all displayed fast (~500 nm/s) and ultraprocessive (9- to 11-µm run-length) motility (Fig. 4, A to C), although the number of moving Spindly-dynactin-dynein complexes was somewhat lower than for Rab11-FIP3, Hook3, and BicD2 (movie S8). Triple-color single-molecule assays using material from human RPE cells confirmed that Alexa647-labeled Rab11-FIP3, Spindly, and Hook3 colocalized with moving GFP-dynein and TMR-dynactin (fig. S6, D to F; movie S9). Thus, four adapter proteins that link dynein to cargoes can induce the formation of highly processive dynein-dynactin-adapter complexes.
Fig. 4. Rab11-FIP3, hSpindly, and Hook3 activate brain dynein motility by linking together dynein and dynactin.
(A) Recombinant full-length Rab11-FIP3, (B) full-length human Spindly, and (C) human N-terminal Hook3 (amino acids 1 to 552) were prepared with an N-terminal SNAPf tag for TMR labeling (22). Left panels: All three adapters attached to beads specifically bound to dynein and dynactin from pig brain lysate (immunoblots show dynein intermediate chain and dynactin p150). Middle panels: Velocity histograms of the indicated brain dynein-dynactin-adapter complexes [Gaussian fits (red) and mean ± SD velocities are shown]. Right panels: “1-cumulative frequency distribution plot” of run-lengths for the indicated dynein-dynactin-adapter complexes. One-phase exponential decay (red), decay constants (τ) to give the run-length, and R2 values of fit are shown [n values for both middle and right panels: Rab11-FIP3 (333); hSpindly (129); Hook3 (219)].
Our results suggest a general model for regulating mammalian cytoplasmic dynein motility (fig. S7). In the cytoplasm, without attachment to a cargo, dynein adopts an inactive conformation that cannot undergo processive motion. Activation of motility requires the simultaneous binding of dynein to an adapter protein that defines a particular cargo for transport (e.g., BicD2, Rab11-FIP3, Spindly, Hook3) and dynactin, a universal activator involved in the transport of many dynein cargos. S. cerevisiae dynein is constitutively active on its own and dynactin only modestly increases its processivity (9), suggesting that the yeast motor may have lost the switch-like regulation of motility displayed by mammalian dynein. Processive motility appears to involve tethering of the DDB complex to the tubulin C-terminal tails, which might be facilitated by the flexibility of dynactin within the DDB complex (Fig. 1C), although allosteric activation mechanisms might also be involved (9, 28, 29). Our results also suggest that dynactin may use a yet undiscovered autoregulatory mechanism, because its MT binding appears to be masked until it becomes incorporated into a dynein-cargo complex. Thus, mammalian dynein and dynactin both become activated for transport in a process that is coupled to cargo selection.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Ori-McKenney for helpful discussions and analysis. This work was supported by NIH F32 postdoctoral grant F32GM096484 (to R.J.M.) and R01GM097312 (to R.D.V.). G.B. is the Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG-2136-12).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/345/6194/337/suppl/DC1
Materials and Methods
Figs. S1 to S7 Movies S1 to S9
References (32–37)
REFERENCES AND NOTES
- 1.Allan VJ. Biochem. Soc. Trans. 2011;39:1169–1178. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- 2.Grigoriev I, et al. Dev. Cell. 2007;13:305–314. doi: 10.1016/j.devcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Griffis ER, Stuurman N, Vale RD. J. Cell Biol. 2007;177:1005–1015. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horgan CP, Hanscom SR, Jolly RS, Futter CE, McCaffrey MW. J. Cell Sci. 2010;123:181–191. doi: 10.1242/jcs.052670. [DOI] [PubMed] [Google Scholar]
- 5.Vallee RB, McKenney RJ, Ori-McKenney KM. Nat. Cell Biol. 2012;14:224–230. doi: 10.1038/ncb2420. [DOI] [PubMed] [Google Scholar]
- 6.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Cell. 2012;150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reck-Peterson SL, et al. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kardon JR, Reck-Peterson SL, Vale RD. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5669–5674. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paschal BM, Vallee RB. Nature. 1987;330:181–183. doi: 10.1038/330181a0. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Khan S, Sheetz MP. Biophys. J. 1995;69:2011–2023. doi: 10.1016/S0006-3495(95)80071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ori-McKenney KM, Xu J, Gross SP, Vallee RB. Nat. Cell Biol. 2010;12:1228–1234. doi: 10.1038/ncb2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coy DL, Hancock WO, Wagenbach M, Howard J. Nat. Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- 14.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Nat. Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 15.Tanenbaum ME, Vale RD, McKenney RJ. eLife. 2013;2:e00943. doi: 10.7554/eLife.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miura M, Matsubara A, Kobayashi T, Edamatsu M, Toyoshima YY. FEBS Lett. 2010;584:2351–2355. doi: 10.1016/j.febslet.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 17.Trokter M, Mücke N, Surrey T. Proc. Natl. Acad. Sci. U.S.A. 2012;109:20895–20900. doi: 10.1073/pnas.1210573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoogenraad CC, et al. EMBO J. 2001;20:4041–4054. doi: 10.1093/emboj/20.15.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuurman N, et al. Eur. J. Cell Biol. 1999;78:278–287. doi: 10.1016/s0171-9335(99)80061-2. [DOI] [PubMed] [Google Scholar]
- 20.Splinter D, et al. Mol. Biol. Cell. 2012;23:4226–4241. doi: 10.1091/mbc.E12-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soundararajan HC, Bullock SL. eLife. 2014;3:e01596. doi: 10.7554/eLife.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Materials and methods are available as supplementary materials on Science Online.
- 23.King SJ, Schroer TA. Nat. Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 24.Culver-Hanlon TL, Lex SA, Stephens AD, Quintyne NJ, King SJ. Nat. Cell Biol. 2006;8:264–270. doi: 10.1038/ncb1370. [DOI] [PubMed] [Google Scholar]
- 25.Lazarus JE, Moughamian AJ, Tokito MK, Holzbaur EL. PLOS Biol. 2013;11:e1001611. doi: 10.1371/journal.pbio.1001611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redwine WB, et al. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokito MK, Howland DS, Lee VM, Holzbaur EL. Mol. Biol. Cell. 1996;7:1167–1180. doi: 10.1091/mbc.7.8.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H, et al. J. Cell Biol. 2007;176:641–651. doi: 10.1083/jcb.200608128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moughamian AJ, Holzbaur EL. Neuron. 2012;74:331–343. doi: 10.1016/j.neuron.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bielska E, et al. J. Cell Biol. 2014;204:989–1007. doi: 10.1083/jcb.201309022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Qiu R, Arst HN, Jr, Peñalva MA, Xiang X. J. Cell Biol. 2014;204:1009–1026. doi: 10.1083/jcb.201308009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.