Abstract
This study reports a comparative and mechanistic genotoxicity assessment of four engineered nanomaterials (ENMs) across three species, including E. coli, yeast, and human cells, with the aim to reveal the distinct potential genotoxicity mechanisms among the different nanomaterials and their association with physiochemical features. Both the conventional phenotypic alkaline comet test and the newly developed quantitative toxicogenomics assay, that detects and quantifies molecular level changes in the regulation of six DNA damage repair pathways, were employed. The proposed molecular endpoints derived from the toxicogenomics assays, namely TELI (Transcriptional Effect Level Index) and PELI (Protein Effect Level Index), correlated well with the phenotypic DNA damage endpoints from comet tests, suggesting that the molecular genotoxicity assay is suitable for genotoxicity detection. Temporal altered gene or protein expression profiles revealed various potential DNA damage types and relevant genotoxic mechanisms induced by the tested ENMs. nTiO2_a induced a wide spectrum of DNA damage consistently across three species. Three carbon-based ENMs, namely carbon black, single wall carbon nanotube (SWCNT) and fullerene, exhibited distinct, species and ENM property-dependent DNA damage mechanisms. All carbon based ENMs induced relatively weak DNA damage repair response in E. coli, but more severe DNA double strand break in eukaryotes. The differences in cellular structure and defense systems among prokaryotic and eukaryotic species lead to distinct susceptibility and mechanisms for ENM uptake and, thus, varying DNA damages and repair responses. The observation suggested that eukaryotes, especially mammalian cells, are likely more susceptible to genotoxicity than prokaryotes in the ecosystem when exposed to these ENMs.
Introduction
The production and application of engineered nanomaterials (ENMs) is exponentially increasing in various fields, and it has led to the concomitant rise in public concern of the potential toxicological impact and risks associated with these nanoparticles.1 Nanomaterial-induced toxicity has been observed in human and other lives via mechanisms related to oxidative stress,2 inflammatory response,3−5 protein binding,6 ion influx disturbance,7 and dissolution and release of toxic ions. It is currently accepted that oxidative stress and inflammation are the major mechanisms of nanotoxicity.8,9 While most nanotoxicity assessment has been focusing on phenotypic endpoints-based cytotoxicity at relatively higher doses, chronic and lower concentration effects, such as their genotoxic potential on long-term environmental risk and human health have not been extensively investigated.10 Genotoxicity is caused by agents interacting with DNA and other cellular targets that control the integrity of the genetic materials, including induction of DNA adducts, strand breaks, point mutations, and structural and numerical chromosomal changes.11 Genotoxicity can not only initiate cancer development, but also has an impact on fertility and the health of subsequent generations, and contributes to the potential heritable changes in eco-systems after long-term accumulation.10,12
Current genotoxicity assays are generally based on detection of induced damages, including Ames test, chromosome aberration test, and comet assay in vitro, and chromosome aberration, mutation of endogenous genes, and rodent carcinogenicity bioassay in vivo.12 On the basis of these conventional tests, many ENMs have been reported to be genotoxic, including metal nanoparticles (e.g., nanosilver and gold nanoparticles), metal-oxide nanoparticles (e.g., nanotitanium dioxide), and carbon based ENMs (e.g., carbon black (CB), carbon nanotube (CNT), and fullerene).2,3,5,10,13 However, many inconsistencies have been reported among different nanogenotoxicity studies, which are likely related to differences in ENMs preparation, quantification, and characterization, and inherent limitations in the genotoxicity assays, such as assay-specific target and species-dependent results.14−16 Most in vitro assays only detect one or limited types of damage, which may lead to false negative results, or can be overly sensitive.12,17 One uniquely challenging aspect of nanotoxicology is that it is highly property- and structure-dependent.5,18 The challenges in evaluating the large number of ENMs and their complex variations using resource-intensive and time-consuming in vivo assays motivate research in developing faster, more economical, and reliable genotoxicity assays.
Genetic biomarkers-based molecular assays have been demonstrated to be promising for rapid and sensitive high throughput screening (HTS) evaluation of genotoxicants.19 Particularly, toxicogenomics that monitor many biomarkers simultaneously have been proven to be rapid and sensitive for genotoxicity testing with mechanism identification and adverse health effects prediction.12,19−21 Major challenges in application of these molecular assays are the needs to employ battery and/or tiered in vitro and in vivo genotoxicity assays in order to reliably detect all genotoxic effects and, to derive quantitative molecular endpoints that can be linked to meaningful in vivo phenotypic adverse effects.22 Despite the degree of biological organizational level, cells respond to potential DNA damage through a common architecture with a number of rather conserved specific response and repair pathways in an effort to repair the DNA damage to restore homeostasis.23−25 On the basis of this understanding, we recently proposed and developed a quantitative toxicogenomics-based comprehensive genotoxicity assessment approach with DNA damage and repair pathway ensemble-based cell arrays for multiple species including E. coli, yeast and human cells.26−30 Furthermore, we proposed a quantitative molecular endpoints-PELI that correlated well with conventional phenotypic genotoxicity endpoints, demonstrating that the proposed quantitative toxicogenomics-based genotoxicity assay can be a promising and alternative tool for mechanistic genotoxicity screening and evaluation in vitro (data not shown).
In this study, we performed a comprehensive and comparative genotoxicity evaluation of a number of ENMs using the newly developed quantitative toxicogenomics-based genotoxicity testing approach as well as conventional genotoxic assay (alkaline comet assay), and across three different cell lines including E. coli, yeast and human cells. The tested ENMs included nTiO2_a,14 carbon black (CB),5 single wall carbon nanotube (SWCNT),16 and fullerene (C60).10 The study aimed to detect the potential genotoxic effect of these common-used ENMs for both eco-system and human health and, to reveal differences in their genotoxic mechanisms among organisms of different taxonomic order, as well as potential association with their physicochemical properties.
Materials and Methods
Nanomaterials Used in This Study
Four engineered nanomaterials (ENMs): nTiO2_a (Sigma-Aldrich), Carbon Black N110 (CB; Cabot Corp.), Short Single Wall CNTs (SWCNT; Cheap Tubes Inc.) and Purified fullerene (C60; M.E.R. Co.) were tested in this study (detailed information in SI Table S1). Detailed physical and chemical characterization for these ENMs was previously described by Bello et al.31 ENMs were prepared as 20× stock in PBS with 1% BSA (Acros, NJ, U.S.A.) as a dispersant, which is the currently common approach for bioassays.32,33 The stock solutions were cup-sonicated at ∼90 W for 15 min to maintain a better dispersion before tests, and were immediately diluted in the corresponding media (M9, SD or F12) for the following test. Cytotoxicity of ENMs, mitomycin C (MMC, a model genotoxicant as a positive control) and bisphenol A (BPA, reported to be genotoxicity negative) were evaluated by growth inhibition in E. coli, yeast and by cell counting in A549 cells (human lung cancer cells) respectively, with details in SI Figure S1 and LC5 (concentration that has 95% cell survived) in SI Table S1. Since the molecular assay intends to detect subtle and cellular level response to toxicants, three subcytotoxic doses (≤LC5) for ENMs, and six for MMC and BPA were applied for E. coli and yeast, one concentration was applied for human cells for PCR and comet test (SI Table S1 for details in dose concentrations). PBS with 1%BSA was used as the vehicle control. For library tests, blank wells with medium (with or without chemicals) were used as medium controls for data processing.32,34,35
Characterization Considerations
Extensive physicochemical and morphological characterization of ENMs was performed as previously published.31 In addition, aggregation sizes of ENMs in different media for various exposure times for this study were analyzed by dynamic light scattering (DLS) analyzer (Malvern Zetasizer Nano ZS90) (SI Table S1).
Toxicogenomics-Based Genotoxicity Assay Library Construction
Homologous genes specific in the DNA repair pathways that are conserved across species were selected for E. coli, yeast, and human cells based on the current knowledge of DNA damage and repair literatures as shown in Table 1.
Table 1. DNA Damages and Indicative Genes/Proteins Involved in Different DNA Damage and Repair Pathways in Both Prokaryotes and Eukaryotes.
| genes
indicative of specific DNA repair pathway in different species |
|||||
|---|---|---|---|---|---|
| DNA damage type | DNA repair pathway | E. coli | yeast36 | human | |
| general damage | SOS response/DNA damage signaling (DDS) | recA, lexA(34)(SOS response) | RAD9,CHK1 (DDS) | not tested | |
| base alkylation | direct reversal repair (DRR) | ada(34) | PHR1 | not tested | |
| base oxidation | base excision repair (BER) | not available in GFP library | OGG1 | OGG137 | |
| base alkylation and deamination | mutT,(34) mutY, mutM, mug, ung, xthA38 | NTG1, NTG2, UNG1, MAG1, RAD27, APN1, APN2 | MPG37 | ||
| single strand break | |||||
| cross-links | nucleotide excision repair (NER) | uvrA,(34) uvrC, mfd39 | RAD1, RAD2, RAD4, RAD14,RAD16,RAD23, RAD34 | XPC39 | |
| pyrimidine dimers | |||||
| bulky adduct | |||||
| mismatches | mismatch repair (MMR) | mutS, mutH, dam(40) | MSH1, MSH2, MSH3, MSH6, PMS1, MLH1, MLH2 | MSH240 | |
| double strand break (DSB) | DSB repair | general DSB response | Not tested | XRS2, MRE11 | Not tested |
| homologous recombination (HR) | recA,(34) recE,38 recX,34 recN,34 ruvA,38 umuD34 | RFA1, RFA2, RFA3, HTA1, HTA2, RAD51, RAD52, RAD54 | Rad5141 | ||
| nonhomologous end joining (NHEJ) | not available in E. coli | LIF1, YKU70 | Ku7041 | ||
Measurement of DNA Damage and Repair Related Gene Expression in E. coli Library
A library of 20 transcriptional fusions of GFP (Open Biosystem, Huntsville, AL) that includes promoters controlling the expression of genes involved in DNA damage and repair in E. coli K12, MG1655 (Table 1) was employed in this study. A more detailed protocol description for gene expression analysis in the E. coli library is available in our previous study.32,34 Briefly, E. coli strains selected were grown with M9 medium in clear bottom black 384-well plates (Costar) for 4–6 h at 37 °C to reach early exponential growth (OD600 about 0.15 to 0.3). Freshly prepared ENM stock solutions or controls dissolved in PBS-1%BSA were added 10 μL per well to obtain the final concentrations (SI Table S1). Three dose concentrations were tested for each ENM (SI Table S1). Plates were then read in a Micro plate Reader (Synergy HT Multi-Mode, Biotech, Winooski, VT) continuously for absorbance (OD600 for cell growth) and GFP signal (filters with 485 nm excitation and 535 nm emission for gene expression) every 5 min for 2 h. All tests were performed in dark in triplicate.
Measurement of DNA Damage and Repair Related Protein Expression in Yeast Library
A library of 37 in-frame GFP fusion proteins involved in DNA damage and repair of S.cerevisiae (Invitrogen, no. 95702, ATCC 201388) (Table 1), constructed by oligonucleotide-directed homologous recombination to tag each open reading frame (ORF) with Aequorea victoria GFP (S65T) in its chromosomal location at the 3′ end, was employed as described in our previous publications.36,42 Yeast strains selected were grown in clear bottom black 384-well plates (Costar) with SD medium for 4–6 h at 30 °C to reach early exponential growth (OD600 about 0.2 to 0.4). ENM stock solutions dissolved in PBS-1%BSA or control were added 10 μL per well to obtain the final concentrations. Three dose concentrations were tested for each ENM (SI Table S1). The plates were then placed in a Micro plate Reader (Synergy H1Multi-Mode, Biotech, Winooski, VT) for absorbance (OD600 for cell growth) and GFP signal (filters with 485 nm excitation and 535 nm emission for protein expression) measurements every 5 min for 2 h after fast shake for 1 min. All tests were performed in the dark in triplicate.
Gene and Protein Expression Profiling Data Processing and Quantitative Molecular Endpoints Derivation
Gene/protein expression profiling data in E. coli and yeast libraries were processed as described previously.32,34−36 For the E. coli library that monitors promoter activities, all data were corrected for various controls, including blank with medium control (with and without NM/chemical) and promoterless bacterial controls (with and without NM/chemical). The alteration in gene expression for a given gene at each time point due to NM/chemical exposure relative to the vehicle control condition without any NM/chemical exposure, also referred as induction factor I, was represented by I = Pe/Pc, where, Pe = (GFP/OD) experiment as the normalized gene expression GFP level in the experiments condition with NM/chemical exposure, and Pc = (GFP/OD) vehicle in the vehicle control condition without any NM/chemical exposure. For the yeast library, OD and GFP raw data were corrected by background OD and GFP signal of medium control (with or without NM/chemical). The protein expression P for each measurement was then normalized by cell number (ODcorrected) as P = (GFPcorrected/ODcorrected). The P level was corrected with vehicle internal control (housekeeping gene PGK143) for plate normalization.
To quantify the chemical-induced gene/protein expression level changes of a treatment, Transcriptional Effect Level Index (TELI)35 for E. coli and Protein Effect Level Index (PELI)36 for yeast were proposed and derived as quantitative molecular endpoints (details described in the SI, part 3). TELI and PELI quantify the accumulative altered gene or protein expression change over the exposure period for a given gene (TELIgene, PELIgene), pathway (TELIpathway, PELIpathway), or for the overall DNA damage and repair pathway ensemble library (TELIgeno, PELIgeno). For a given chemical, TELIgeno or PELIgeno based dose–response patterns (SI Figure S2) were modeled using Four Parameter Logistic (4PL) nonlinear regression model. Genotoxicity positive or negative was determined based on the comparison of the maximal TELIgeno/PELI geno value derived from the TELI or PELI-dose response curve with a threshold value, which was predetermined based on results with postive and negative genotoxic chemicals, as well as the standard deviation range of our testing systems (data not shown).
Measurement of DNA Repair Related Gene Expression in Human Cells by RT-qPCR and Data Processing
2 ×105/well of human lung epithelial A549 cells were seeded in 6-well plates (Costar) for 24h in F12 medium with 10% FBS. ENMs, MMC or BPA prepared in F12 medium with 1% BSA were added in PBS-washed cells (2 mL in each well). Dose concentrations were listed in SI Table S1. Cell samples were harvested at four time points after ENMS exposure (concentrations in SI Table S1) in the dark (0.5 h, 2 h, 4 h, or 24 h). RNA was extracted by RNeasy Mini Kit (QIAGEN, Valencia, CA, U.S.A.) and reverse transcribed to cDNA by iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, U.S.A.). Q-PCR was performed in duplicate using SYBR Green Supermix on iQ5 Multicolor Real-Time PCR detection system (Bio-Rad). PCR Primers targeting the six selected genes in Table 1 were obtained from the database at NCBI with the housekeeping gene GAPDH as internal control (Primers listed in SI Table S2). Fold change that reflects relative gene expression change due to treatment compared to vehicle control, also referred to as induction factor I, was determined by comparative CT method (2–ΔΔCT).44 Similar to the E. coli library, the transcriptional effect in human cells was integrated into the Transcriptional Effect Level Index of genotoxicity (TELIgeno).
DNA Damage Alkaline Comet Assay in Human A549 Cells for Phenotypic Confirmation
The alkaline comet assay in human A549 cells upon exposure to ENMs or controls for 24 h was carried out according to the protocol of the ITRC.45 Dose concentrations were similar to those applied for molecular toxicity assay (SI Table S1). All the procedures were performed in the dark with triplicates. The slides were stained with ethidium bromide (2 μg/mL) and checked by florescence microscope (Olympus IX51, camera DP70). Fifty cells of each treatment were measured by software CASP randomly (University of Wroclaw, Institute of Theoretical Physics), and the damage was valued as % Tail DNA. Genotoxicity positive was defined as the significant increase of tail DNA % compared to vehicle control with p < 0.05.
Results
Characterization of Engineered Nanomaterials in Different Media for Genotoxicity Assays
ENMs tested in this study were generally stable along the exposure period (2 h/24 h), but had different sizes in different media (see SI Table S1). For nTiO2_a and CB, larger aggregation sizes were observed in SD media for the yeast assay, while smaller sizes were observed in M9 and F12 media for E. coli and human cells, respectively. C60 shared similar sizes in three different media. Particle size is not available for linear SWCNT.
DNA Damage and Repair Profiles Induced by Nanomaterials are Temporally Dynamic, Concentration- and Species-Dependent
The temporal altered gene/protein expression profiles provided a holistic view of the DNA damage and repair pathways and, reflected dynamic responses of E. coli, yeast, and human cells exposed to ENMs over the exposure time at different dose concentrations. Altered molecular (gene or protein) expression changes were quantified by induction factors as shown in Figure 1 for one representative dose condition and in SI Figure S3 for all other doses. The DNA-damaging chemical MMC (positive control, SI Figure S3) induced significant up-regulation of biomarkers in most of DNA repair pathways in E. coli, yeast, and human cells, with time- and dose-dependent patterns. As expected, no significant up-regulation was observed in the three libraries for the negative control bisphenol A (SI Figure S3).
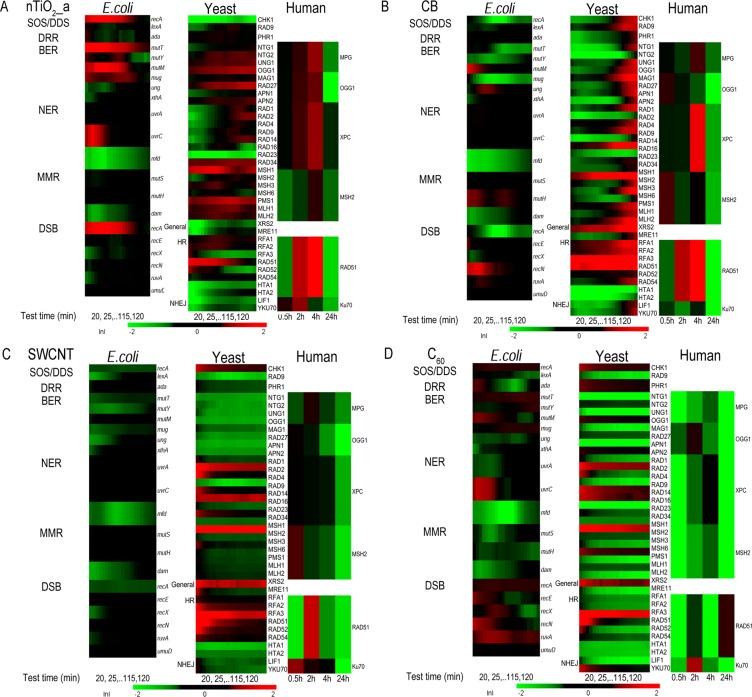
Figure 1.
Temporal gene/protein expression profiles of biomarkers indicative of different DNA damage repair pathways upon exposure to four nanomaterials (A: nTiO2_a, 50 μg/mL, B: carbon black (CB), 5 μg/mL, 50 μg/mL for human cells), C: single wall carbon nanotube (SWCNT, 8 μg/mL for E. coli, 10 μg/mL/L for yeast and human cells) and D: purified fullerene (C60, 50 μg/mL)) across three species. Results for other dose concentrations are provided in the SI (Figure S3). The mean natural log of induction factor (ln I) indicates the magnitude of altered gene/protein expression (represented by a green−black−red color scale at bottom. Red spectrum colors indicate up regulation, green spectrum colors indicate down regulation. Values beyond ±2 are shown as ±2). X-axis bottom: for E. coli and yeast: testing time in minutes, the first data point shown is at 20 min after exposure due to data smoothing with moving average of every five data points; for human cells: testing time in hours. Y-axis left: clusters of genes/proteins by DNA damage repair pathways. Y-axis right for each species: list of genes or proteins (ORFs) tested, with details in Table 1. For yeast and E. coli, n = 3, for human cells, n = 2. For CB exposure, corrections are made to the gene expression analysis for the physical interference of CB on the fluorescence signal reading (details in SI Figure S4).
The DNA damage potential and mechanisms for different ENMs seemed to vary and were concentration- and species-dependent, as indicated by the distinct altered molecular expression profiles (Figure 1 and SI Figure S3). nTiO2_a (Figure 1A) induced expression changes in biomarkers for all the three species. The three carbon based ENMs induced distinct DNA damage repair pathways activation profiles, mainly in eukaryotes, suggesting structure-dependent genotoxicity. CB (Figure 1B) seemed to exert more severe DNA damaging potential than the other two carbon nanomaterials, namely SWCNT (Figure 1C) and C60 (Figure 1D). See more discussion in later genotoxicity mechanisms section.
Comparison of Molecular Genotoxicity Assay Results with Other in Vitro and in Vivo Genotoxicity Assays
Quantitative toxicogenomics assay endpoints, namely TELIgeno, PELIgeno, for different dose concentrations were derived for each ENM and they exhibited dose response curves for the four ENMs examined (SI Figure S2). The maximum values of TELIgeno for E. coli assay and PELIgeno for yeast assay, derived from dose response model fitting, were then compared to predetermined endpoint threshold for molecular genotoxicity assay as positive or negative (see description in methods and SI). The molecualr genotoxicity assay endpoints from our assay were compared to those from conventional phenotypic genotoxicity assays, including alkaline comet assay in A549 cells (SI Figure S6) and other in vitro and in vivo assays from literature (Table 2). Endpoints comparison in Table 2 suggested that our molecular assays were sensitive for detecting DNA damage and for capturing the potential genotoxicity of the ENMs tested. The DNA-damaging model chemical MMC was positive, while BPA was negative for all assays, as expected. nTiO2 _a and C60 were genotoxicity positive in all eukaryotic cell-based assays, as well as in vivo carcinogenesis test, and were negative in prokaryotic assays, suggesting likely species-specific DNA damage mechanisms. Varying results were reported for CB and SWCNT among different in vitro genotoxicity assays. This is likely due to the varying and limited type of DNA damages that can be detected by conventional methods. For example, Ames test targets on frame shift or point mutations, comet assay detects strand breaks and, micronucleus assay detects chromosome damages. DNA repair pathway biomarkers ensemble based assay proposed in this study seemed to be more accurate compared to these commonly used in vitro assays (assuming in vivo assay produces the most reliable endpoint).
Table 2. Comparison of Molecular Genotoxicity Assay Resultsa with Those from Conventional Genotoxicity and Carcinogenesis Assays Across Speciesb.
| molecular
assay |
in vitro
assay |
in vivo assayc | |||||
|---|---|---|---|---|---|---|---|
| E. coli | yeast | human cells | Ames (bacteria) | Comet (A549) | micronucleus assay | carcinogenesis | |
| MMC | + | + | + | + | + | + | + |
| nTiO2_a | + | + | + | – | + | + | + |
| CB | + | + | + | – | + | + | + |
| SWCNT | – | + | ± | – | + | – | – |
| C60 | + | + | + | – | + | + | not available |
| BPA | – | – | – | – | – | – | – |
Genotoxicity positive (+) was defined as maximum TELIgeno or PELIgeno value greater than 1.5 for E. coli and yeast assay, 4.113 for human cell assay based on positive and negative controls, as described in methods section. Maximum TELIgeno and PELIgeno values were determined based on model fitting to dose response curves (details in SI, Table S1 and Figure S2).
Information of Ames and micronucleus assay in vitro and two-year carcinogenesis in vivo was collected from literature for MMC,46−49 BPA47,50 and the same nanomaterials.5,10,51,52
Carcinogenesis data are based on two-year rodent study, except for SWCNT, which is reported to be negative in short to medium-term (6 months or less) carcinogenesis study, because 2-year study is not available.52
Toxicogenomics-Based Molecular Endpoints Quantitatively Correlated with Conventional Phenotypic Endpoint
For the first time, we explored and demonstrated a quantitative correlation of the molecular endpoints derived from the molecular assay with ENM-induced DNA damage (measured by alkaline comet assay in human cell as % Tail DNA), as shown in Figure 2. The phenotypic genotoxicity endpoint based on 24 h comet assay in human cells correlated well with the molecular endpoint TELIgeno for human cell (rp = 0.8147, P = 0.0483), as well as for the endpoint PELIgeno in yeast cell (rp = 0.9073, P = 0.0125). The higher correlation coefficient values of eukaryotes than those of prokaryotic E. coli cells (rp = 0.6102, P = 0.1983) suggest that eukaryotic cells shared more similar responses than prokaryotic cells. Note that yeast cell assay incorporated much more key biomarkers of DNA damage repair pathways than the human cell assay and therefore captures more comprehensive DNA damage information, which may explain the better correlation of comet assay results with yeast cells than with human cells. These results suggest that the applied toxicogenomics genotoxicity assay and proposed quantitative molecular endpoints are adequate for genotoxicity assessment across multiple species and, they can be potentially linked to phenotypic effects quantitatively.
Figure 2.
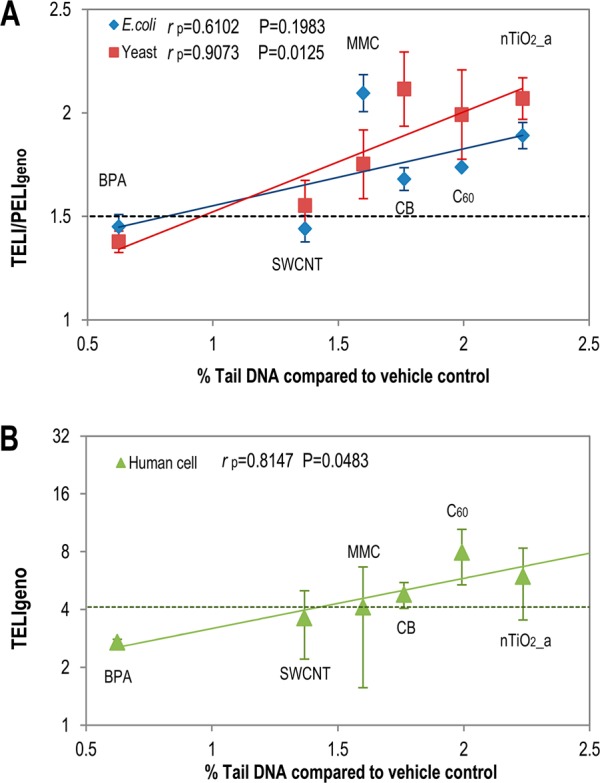
Correlation of TELIgeno and PELIgeno in E. coli, yeast (A) and human cells (B) with DNA damage induced (measured by % Tail DNA compared to vehicle control tested by alkaline comet test) in a human A549 cell line for selected concentrations shown in SI Table S1. The black dashed line indicates the cutoff line (1.5) for E. coli and yeast cells, and the green dashed line indicates the cutoff line for human cells (4.113 based on TELIgeno of MMC for human cell). X-axis bottom: 24-h DNA damage measured by % Tail DNA compared to vehicle control in human A549 cells (details in SI Figure S5); Y-axis: TELI/PELIgeno in E. coli, human cells, or yeast array. rP indicated Pearson correlation coefficient of endpoints from three cell arrays to DNA damage comet assay phenotypic endpoints (% Tail DNA). Mean ± SD. For E. coli, yeast, and comet assay, n = 3, for human cells, n = 2.
Genotoxic Mechanisms of ENMs Revealed by DNA Damage Repair Pathways Profiling and Their Association with ENM Properties
To gain further insights into the detailed DNA damage repair pathway activities induced by the tested ENMs, the magnitude of expression changes in each pathway was quantified by TELI pathway or PELI pathway (see SI, part 3) and compared among ENMs at multiple concentrations (Figure 3). The model genotoxicant MMC induced severe DNA damage, as indicated by the SOS response activation in E. coli and strong pathway activation for double strand break and base damage repair in all three species. In addition, it seemed to cause a potential DNA cross-link, as suggested by the nucleotide excision repair biomarkers in yeast and human cells. These DNA damage responses induced by MMC were consistent with its known high alkylation reactivity to produce base alkylation and cross-link to DNA.53 Meanwhile, negative control BPA only induced a weak activation of direct reversal repair in E. coli and, no pathway activation in yeast but oxidative damage repair (indicated by OGG1 up-regulation) in human cells, which was consistent with previous report that BPA induced oxidative stress in mammals.54
Figure 3.
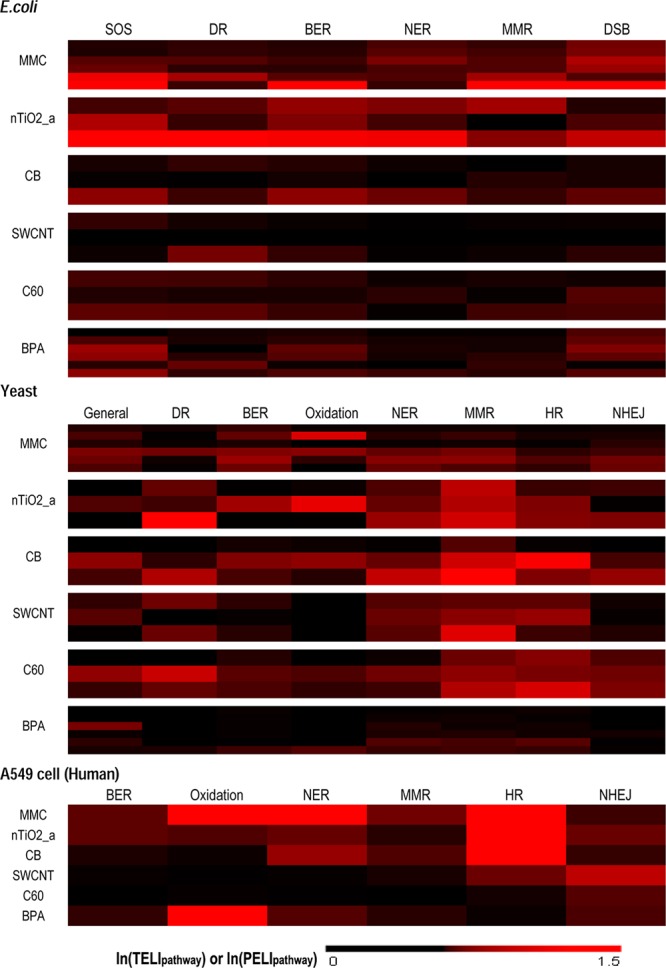
DNA damage repair pathway responses profiles reveal distinct potential DNA damage mechanisms among different ENMs. The mean natural log value of TELIpathway or PELIpathway indicates the magnitude of pathway responses (represented by a black–red color scale at bottom. Values over 1.5 are shown as 1.5). MMC: mitomycin C; CB: carbon black; SWCNT: single wall carbon nanotube; C60: purified fullerene; BPA: bisphenol A. X-axis top: pathways of DNA damage repair (see Table 1 for details). Y-axis left: chemical/ENM and concentration(s) from lowest to highest from top to bottom (see concentrations in SI Table S1).
nTiO2_a induced wide range of pathway activation suggesting strong DNA damage potential consistently among all three species. In E. coli, nTiO2_a led to activation of SOS system indicating severe DNA damage. In yeast and human cells, a wide range of repair pathway activation, including severe double strand break repair, was observed and the up-regulation of OGG1 specific for oxidative damage repair (Imax = 3.4673 for yeast, Imax = 1.9740 for human cells). OGG1 up-regulation in both yeast and human cells suggested that oxidative damage might play an important role in DNA damage of nTiO2_a, which was agreeable with the current understanding that oxidative damage through reactive oxygen species (ROS) production is its main toxicity mechanism.32,55 Note that in this study, the ROS production from photocatalytic reaction was not expected to be significant under our test conditions because all bioassays were performed in the dark and the wavelength of excitation used for OD and fluorescence reading (600 and 485 nm) was out of the active photoactive range for nTiO2_a (385 nm). Several reports were supportive of nonphoto-induced ROS generation by unknown mechanism(s).56 In addition, the dissolution of titanium ions at neutral pH within 2 h was considered to be negligible for nano TiO2,56 and the dissolved ion was reported to exhibit no damage to the cellular DNA.57
Three carbon-based ENMs showed structure-dependent and discernible DNA damage response profiles. Carbon black (CB) seemed to lead to little response in E. coli, likely due to the uptake limit in prokaryotic cells.58,59 In contrast, a wide range of DNA repair pathway activities, including nucleotide excision repair and double strand break repair, were observed in yeast and human cells, suggesting potential damage induced by CB exposure. The up-regulation of OGG1 in yeast (Imax = 3.3050) suggested that oxidative damage may contribute to the genotoxicity of CB.60 However, no OGG1 up-regulation was observed in human cells, suggesting that some mechanisms other than oxidative stress may contribute to the severe DNA damage of CB in human cells, which needs further investigation.
SWCNT exposure did not lead to any DNA repair pathway activation in E. coli, but seemed to induce strong double strand DNA damage in eukaryotes. In contrast to CB, oxidative damage may not be the main mechanism since OGG1 up-regulation was not observed in either yeast or human cells. DNA double strand break in eukaryotes may be a result of membrane damage induced apoptosis/necrosis with DNA cleavage,59,61,62 which has been reported to be a main target for CNTs.63,64 Although membrane damage of SWCNTs has also been observed in E. coli,63 it may not lead to DNA damage since E. coli lacks membrane related cell death process.65
For C60 exposure, specific double strand break response was observed in human cells, but multiple DNA damage pathway activities were detected in yeast. Oxidative damage may not be the main mechanism either, as OGG1 up-regulation was not observed in either yeast or human cells. Another specific repair pathway activation, NHEJ (nonhomologous end-joining) was observed in both yeast and human cells, which repairs DNA double strand break by direct ligation, suggesting some distinguishable mechanism involved in C60 induced DNA damage, such as direct cleavage on both strands. C60 has been reported as an electron-poor photosensitizer, which can induce DNA photocleavage selectively for G base by visible light.66,67 The direct cleavage may break strands with little DNA loss, which can be repaired by direct ligation of NHEJ pathway. As previously mentioned, although the bioassays were conducted in the dark, intermittent excitation during fluorescence reading at 485 nm can potentially irritate the photocleavage of C60, which has a moderate energy gap of about 1.8 eV (∼690 nm).67−69
Discussion
This study performed a comprehensive and comparative genotoxicity assessment for four commonly-used ENMs across multiple species using both conventional phenotypic and newly proposed quantitative toxicogenomics approaches. The results demonstrated that the toxicogenomics assay based on conserved DNA damage repair pathways across three species could detect various DNA damages induced by ENMs. And, for the first time, the molecular endpoints were quantitatively correlated to phenotypic endpoints and generally consistent with those based on conventional in vitro and in vivo genotoxicity assays, suggesting that the toxicogenomics-based assays can be promising for potential genotoxicity assessment of ENMs. Compared to current genotoxicity assay which takes several days (e.g., over two days for Ames test) or even years (two years for in vivo carcinogenesis test), our assay is faster (2 h. for E. coli and yeast, 24 h. for human cells), more sensitive (at subcytotoxic doses), more comprehensive (capture all DNA damage types instead of single or a few biomarkers or specific targets as in other assays), and more cost-effective (the gfp-fused whole cell libraries can be easily grown with less complex protocol). In addition, the molecular profiles can provide mechanistic information regarding specific DNA damage type and repair pathways.
The comparative study of the selected ENMs provided more insights for a better understanding of their genotoxic mechanisms, which was poorly understood previously. Oxidative damage has been considered the main effect of ENMs leading to the genotoxicity for nTiO2_a,32,55 CB,70 fullerene,71,72 and SWCNT.60,73 In this study, oxidative damage was confirmed to play an important role in DNA damage for nTiO2_a and CB, which is consistent with previous studies. However, our results indicated that the other two carbon ENMs (SWCNT and C60) might induce DNA double strand break in eukaryotes through some other mechanisms, implying more structure-and/or property-dependent genotoxicity. Although they both lead to DNA double strand break in eukaryotes, the differences in the activation of pathway-specific genes, for example, Rad51 (for homologous recombination) and Ku70 (for nonhomologous end-joining by direct ligation) in human cells revealed different mechanisms of DSB repair. The results demonstrated the specificity and sensitivity of the gene/protein expression methods for detecting and differentiating DNA damage and related mechanisms, such as among those carbon-based yet structurally distinct ENMs.
The distinct genotoxic responses between prokaryotic and eukaryotic species revealed in this study were likely related to the different capability and mechanisms of the ENMs’ (particularly carbon based ENMs) uptake among species. For mammalian cells, nanoparticle uptake has been reported to occur through eukaryote-specific processes such as phagocytosis and endocytosis. Endocytosis in yeast also makes it possible for ENM uptake; some unclear mechanism in yeast seems to be employed to internalize nanomaterials too.74 However, the rigid cell wall of bacteria may avoid the direct uptake process.58,59 Moreover, bacteria lack the ability to perform endocytosis, which may make them more resistant to ENMs.10 Adhesion of ENMs to eukaryotic cells may also contribute to a series of membrane damage-related cell death processes, including the DNA damage response.59 As programed cell death is limited in bacteria,65 this adhesion effect may contribute little to the nanogenotoxicity of E. coli.
In conclusion, this study revealed various genotoxicity of the four commonly used ENMs by real-time gene/protein expression profiling, which seemed to be both species and ENM-property-dependent. The differences in cellular structure and defense systems among prokaryotic and eukaryotic species lead to distinct susceptibility and mechanisms for ENMs uptake and, thus varying DNA damages and repair responses. The observation suggested that eukaryotes, especially mammalian cells, are likely more susceptible to genotoxicity than prokaryotes in eco-system when exposed to these ENMs.
Acknowledgments
This study was supported by National Science Foundation (NSF) (EEC-0926284 and CAREER CBET-0953633, CBET-1440764), National Science Foundation Nanoscale Science, Engineering Center (NSEC) for Highrate Nanomanufacturing (0425826), and National Institute of Environmental Health Sciences (NIEHS P42ES017198, Puerto Rico Testsite for Exploring Contamination Threats (PROTECT)). We thank Professor Dhimiter Bello and Anoop K. Pal from University of Massachusetts at Lowell for providing the nanomaterials and physicochemical characterization information.
Supporting Information Available
Cytotoxicity of ENMs/chemicals in E. coli, yeast, and A549 cells; detailed physical and chemical characterization of the nanomaterials tested in this study, as well as the concentrations tested; detailed data processing and quantitative molecular endpoints derivation; the dose responses of model chemicals and ENMs; primers used for in RT q-PCR for human A549 cells, with GenBank accession number or UniSTS number; real-time gene/protein expression profiles for all concentrations tested in this study; the interference of carbon black to florescence signal; results of alkaline comet test in human A549 cells. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Ivask A.; Suarez E.; Patel T.; Boren D.; Ji Z. X.; Holden P.; Telesca D.; Damoiseaux R.; Bradley K. A.; Godwin H. Genome-wide bacterial toxicity screening uncovers the mechanisms of toxicity of a cationic polystyrene nanomaterial. Environ. Sci. Technol. 2012, 4642398–2405. [DOI] [PubMed] [Google Scholar]
- Li Y.; Zhang Y.; Yan B. Nanotoxicity overview: Nano-threat to susceptible populations. Int. J. Mol. Sci. 2014, 1533671–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani M. Y.; Hashim M. A.; Nabi F.; Malik M. A.. Nanotoxicity: Dimensional and morphological concerns. Adv. Phys. Chem. 2011, 2011. [Google Scholar]
- Savolainen K.; Alenius H.; Norppa H.; Pylkkanen L.; Tuomi T.; Kasper G. Risk assessment of engineered nanomaterials and nanotechnologies—A review. Toxicology 2010, 2692–392–104. [DOI] [PubMed] [Google Scholar]
- Landsiedel R.; Kapp M. D.; Schulz M.; Wiench K.; Oesch F. Genotoxicity investigations on nanomaterials: Methods, preparation and characterization of test material, potential artifacts, and limitations—Many questions, some answers. Mutat. Res.—Rev. Mutat. Res. 2009, 6812–3241–258. [DOI] [PubMed] [Google Scholar]
- Zuo G. H.; Kang S. G.; Xiu P.; Zhao Y. L.; Zhou R. H. Interactions Between Proteins and Carbon-Based Nanoparticles: Exploring the Origin of Nanotoxicity at the Molecular Level. Small 2013, 99–101546–1556. [DOI] [PubMed] [Google Scholar]
- Jakubek L. M.; Hurt R. H. Nanomaterials and ion channels: Observed effects and possible mechanisms. Nanomed. Nervous Syst. 2012, 119. [Google Scholar]
- Elsaesser A.; Howard C. V. Toxicology of nanoparticles. Adv. Drug Deliv. Rev. 2012, 642129–137. [DOI] [PubMed] [Google Scholar]
- Li A. N. T. X. L. M. N. Toxic potential of materials at the nanolevel. Science 2006, 311, 6. [DOI] [PubMed] [Google Scholar]
- Singh N.; Manshian B.; Jenkins G. J. S.; Griffiths S. M.; Williams P. M.; Maffeis T. G. G.; Wright C. J.; Doak S. H. NanoGenotoxicology: The DNA damaging potential of engineered nanomaterials. Biomaterials 2009, 3023–243891–3914. [DOI] [PubMed] [Google Scholar]
- Baumstark-Khan C.; Hellweg C. E.; Reitz G. Cytotoxicity and genotoxicity reporter systems based on the use of mammalian cells. Whole Cell Sens. Syst. II 2010, 118, 113–151. [DOI] [PubMed] [Google Scholar]
- Ahn J. M.; Hwang E. T.; Youn C. H.; Banu D. L.; Kim B. C.; Niazi J. H.; Gu M. B. Prediction and classification of the modes of genotoxic actions using bacterial biosensors specific for DNA damages. Biosens. Bioelectron. 2009, 254767–772. [DOI] [PubMed] [Google Scholar]
- Mroz R. M.; Schins R. P. F.; Li H.; Drost E. M.; Macnee W.; Donaldson K. Nanoparticle carbon black driven DNA damage induces growth arrest and AP-1 and NF kappa B DNA binding in lung epithelial A549 cell line. J. Phys. Pharmacol. 2007, 58, 461–470. [PubMed] [Google Scholar]
- Gurr J. R.; Wang A. S. S.; Chen C. H.; Jan K. Y. Ultrafine titanium dioxide particles in the absence of photoactivation can induce oxidative damage to human bronchial epithelial cells. Toxicology 2005, 2131–266–73. [DOI] [PubMed] [Google Scholar]
- Wang J. J.; Wang H.; Sanderson B. J. S. Ultrafine quartz-induced damage in human lymphoblastoid cells in vitro using three genetic damage end-points. Toxicol. Mech. Methods 2007, 174223–232. [DOI] [PubMed] [Google Scholar]
- Kisin E. R.; Murray A. R.; Keane M. J.; Shi X. C.; Schwegler-Berry D.; Gorelik O.; Arepalli S.; Castranova V.; Wallace W. E.; Kagan V. E.; Shvedova A. A. Single-walled carbon nanotubes: Geno- and cytotoxic effects in lung fibroblast V79 cells. J. Toxicol. Environ. Health-Part A-Curr. Issues 2007, 70242071–2079. [DOI] [PubMed] [Google Scholar]
- Knight A. W.; Little S.; Houck K.; Dix D.; Judson R.; Richard A.; McCarroll N.; Akerman G.; Yang C.; Birrell L. Evaluation of high-throughput genotoxicity assays used in profiling the US EPA ToxCast chemicals. Regul. Toxicol. Pharmacol. 2009, 552188–199. [DOI] [PubMed] [Google Scholar]
- Verma A.; Stellacci F. Effect of surface properties on nanoparticle-cell interactions. Small 2010, 6112–21. [DOI] [PubMed] [Google Scholar]
- Mahadevan B.; Snyder R. D.; Waters M. D.; Benz R. D.; Kemper R. A.; Tice R. R.; Richard A. M. Genetic toxicology in the 21st century: Reflections and future directions. Environ. Mol. Mutagen. 2011, 525339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroz R. M.; Schins R. P. F.; Li H.; Jimenez L. A.; Drost E. M.; Holownia A.; MacNee W.; Donaldson K. Nanoparticle-driven DNA damage mimics irradiation-related carcinogenesis pathways. Eur. Respir. J. 2008, 312241–251. [DOI] [PubMed] [Google Scholar]
- Zhu L.; Chang D. W.; Dai L. M.; Hong Y. L. DNA damage induced by multiwalled carbon nanotubes in mouse embryonic stem cells. Nano Lett. 2007, 7123592–3597. [DOI] [PubMed] [Google Scholar]
- McKim J. M. Building a tiered approach to in vitro predictive toxicity screening: A focus on assays with in vivo relevance. Comb. Chem. High Throughput Screening 2010, 132188–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., W G., Siede W, Wood R. D., Schultz R. A., Ellenberger T. DNA Repair and Mutagenesis; ASM Press: Washington, DC, 2006. [Google Scholar]
- Sancar A.; Lindsey-Boltz L. A.; Unsal-Kacmaz K.; Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004, 73, 39–85. [DOI] [PubMed] [Google Scholar]
- Taylor E. M.; Lehmann A. R. Conservation of eukaryotic DNA repair mechanisms. Int. J. Radiat. Biol. 1998, 743277–286. [DOI] [PubMed] [Google Scholar]
- Prakash S.; Prakash L. Nucleotide excision repair in yeast. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2000, 4511–213–24. [DOI] [PubMed] [Google Scholar]
- Milanowska K.; Krwawicz J.; Papaj G.; Kosinski J.; Poleszak K.; Lesiak J.; Osinska E.; Rother K.; Bujnicki J. M. REPAIRtoire—A database of DNA repair pathways. Nucleic Acids Res. 2011, 39, D788–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard S.; Schurman S. H.; Harboe C.; de Souza-Pinto N. C.; Bohr V. A. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis 2009, 3012–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M. R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A.; Lindsey-Boltz L. A.; Ünsal-Kaçmaz K.; Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Ann. Rev. Biochem. 2004, 73139–85. [DOI] [PubMed] [Google Scholar]
- Bello D.; Hsieh S. F.; Schmidt D.; Rogers E. Nanomaterials properties vs. biological oxidative damage: Implications for toxicity screening and exposure assessment. Nanotoxicology 2009, 33249–U114. [Google Scholar]
- Gou N.; Onnis-Hayden A.; Gu A. Z. Mechanistic toxicity assessment of nanomaterials by whole-cell-array stress genes expression analysis. Environ. Sci. Technol. 2010, 44155964–5970. [DOI] [PubMed] [Google Scholar]
- Schultz-Cherry S.; Turpin E.; Luke M.; Jones J.; Tumpey T.; Konan K. Influenza virus infection increases p53 activity: Role of p53 in cell death and viral replication. J. Virol. 2005, 79148802–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onnis-Hayden A.; Weng H. F.; He M.; Hansen S.; Ilyin V.; Lewis K.; Gu A. Z. Prokaryotic real-time gene expression profiling for toxicity assessment. Environ. Sci. Technol. 2009, 43124574–4581. [DOI] [PubMed] [Google Scholar]
- Gou N.; Gu A. Z. A new transcriptional effect level index (TELI) for toxicogenomics-based toxicity assessment. Environ. Sci. Technol. 2011, 45125410–5417. [DOI] [PubMed] [Google Scholar]
- O’Connor S. T. F.; Lan J.; North M.; Loguinov A.; Zhang L.; Smith M. T.; Gu A. Z.; Vulpe C. Genome-wide functional and stress response profiling reveals toxic mechanism and genes required for tolerance to benzo[a]pyrene in S. cerevisiae. Front. Genet. 2012, 3, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan H. E.; Nilsen H.; Skorpen F.; Otterlei M.; Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000, 4761–273–77. [DOI] [PubMed] [Google Scholar]
- Kanehisa M.; Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28127–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C. X.; Tang T. S.; Friedberg E. C. SnapShot: Nucleotide excision repair. Cell 2010, 1405754–U169. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A.; Erie D. A. DNA mismatch repair. Annu. Rev. Biochem. 2005, 74, 681–710. [DOI] [PubMed] [Google Scholar]
- Karran P. DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev. 2000, 102144–150. [DOI] [PubMed] [Google Scholar]
- Huh W. K.; Falvo J. V.; Gerke L. C.; Carroll A. S.; Howson R. W.; Weissman J. S.; O’Shea E. K. Global analysis of protein localization in budding yeast. Nature 2003, 4256959686–691. [DOI] [PubMed] [Google Scholar]
- Nakatani Y.; Yamada R.; Ogino C.; Kondo A. Synergetic effect of yeast cell-surface expression of cellulase and expansin-like protein on direct ethanol production from cellulose. Microb. Cell Fact. 2013, 12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D.; Livak K. J. Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 2008, 361101–1108. [DOI] [PubMed] [Google Scholar]
- Dhawan A.; Bajpayee M. M.; Pandey A. K.; Parmar D.. Protocol for the single cell gel electrophoresis/comet assay for rapid genotoxicity assessment. In ITRC: THE SCGE/ COMET ASSAY PROTOCOL, Vol. 1077, p 1. [Google Scholar]
- Levin D. E.; Hollstein M.; Christman M. F.; Schwiers E. A.; Ames B. N. A new Salmonella tester strain (TA102) with AXT base pairs at the site of mutation detects oxidative mutagens. Proc. Natl. Acad. Sci. U. S. A. 1982, 79237445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L. S.; Slone T. H.; Bernstein L. Summary of carcinogenic potency and positivity for 492 rodent carcinogens in the carcinogenic potency database. Environ. Health Perspect. 1989, 79, 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.; Niu Y. J.; Do H. R.; Cao X. W.; Shi D.; Hao Q. L.; Zhou Y. L. A stable and sensitive testing system for potential carcinogens based on DNA damage-induced gene expression in human HepG2 cell. Toxicol. In Vitro 2009, 231158–165. [DOI] [PubMed] [Google Scholar]
- Matsuoka A.; Yamazaki N.; Suzuki T.; Hayashi M.; Sofuni T. Evaluation of the micronucleus test using a chinese-hamster cell-line as an alternative to the conventional invitro chromosomal aberration test. Mutat. Res. 1992, 2723223–236. [DOI] [PubMed] [Google Scholar]
- Fic A.; Sollner Dolenc M.; Filipič M.; Peterlin Mašić L. Mutagenicity and DNA damage of bisphenol A and its structural analogues in HepG2 cells. Arh. Hig. Rada Toksikol. 2013, 642189–199. [DOI] [PubMed] [Google Scholar]
- Heinrich U.; Fuhst R.; Rittinghausen S.; Creutzenberg O.; Bellmann B.; Koch W.; Levsen K. Chronic inhalation exposure of Wistar rats and two different strains of mice to diesel engine exhaust, carbon black, and titanium dioxide. Inhal. Toxicol. 1995, 74533–556. [Google Scholar]
- Tsuda H.; Xu J. G.; Sakai Y.; Futakuchi M.; Fukamachi K. Toxicology of engineered nanomaterials—A review of carcinogenic potential. Asian Pacific J. Cancer Prevent. 2009, 106975–980. [PubMed] [Google Scholar]
- Tomasz M. Mitomycin-C—Small, fast and deadly (but very selective). Chem. Biol. 1995, 29575–579. [DOI] [PubMed] [Google Scholar]
- Chitra K. C.; Latchoumycandane C.; Mathur P. P. Induction of oxidative stress by bisphenol A in the epididymal sperm of rats. Toxicology 2003, 1851–2119–127. [DOI] [PubMed] [Google Scholar]
- Petkovi J.; Žegura B.; Filipi M. In Influence of TiO2 Nanoparticles on Cellular Antioxidant Defense and Its Involvement in Genotoxicity in HepG2 Cells; IOP Publishing: 2011; p 012037. [Google Scholar]
- Dalai S.; Pakrashi S.; Kumar R. S. S.; Chandrasekaran N.; Mukherjee A. A comparative cytotoxicity study of TiO2 nanoparticles under light and dark conditions at low exposure concentrations. Toxicol. Res.-U.K. 2012, 12116–130. [Google Scholar]
- Ortiz A. J.; Fernandez E.; Vicente A.; Calvo J. L.; Ortiz C.. Metallic ions released from stainless steel, nickel-free, and titanium orthodontic alloys: toxicity and DNA damage. In American Journal of Orthodontics and Dentofacial Orthopedics: Official Publication of the American Association of Orthodontists, Its Constituent Societies, And the American Board of Orthodontics; 2011, Vol. 140; 3, e115–22.. [DOI] [PubMed]
- Kasemets K.; Ivask A.; Dubourguier H. C.; Kahru A. Toxicity of nanoparticles of ZnO, CuO and TiO2 to yeast Saccharomyces cerevisiae. Toxicol. In Vitro 2009, 2361116–1122. [DOI] [PubMed] [Google Scholar]
- Nomura T.; Miyazaki J.; Miyamoto A.; Kuriyama Y.; Tokumoto H.; Konishi Y. Exposure of the yeast Saccharomyces cerevisiae to functionalized polystyrene latex nanoparticles: Influence of surface charge on toxicity. Environ. Sci. Technol. 2013, 4773417–3423. [DOI] [PubMed] [Google Scholar]
- Jacobsen N. R.; Pojana G.; White P.; Møller P.; Cohn C. A.; Smith Korsholm K.; Vogel U.; Marcomini A.; Loft S.; Wallin H. Genotoxicity, cytotoxicity, and reactive oxygen species induced by single-walled carbon nanotubes and C60 fullerenes in the FE1-Muta Mouse lung epithelial cells. Environ. Mol. Mutagen. 2008, 496476–487. [DOI] [PubMed] [Google Scholar]
- Dong Z.; Saikumar P.; Weinberg J. M.; Venkatachalam M. A. Internucleosomal DNA cleavage triggered by plasma membrane damage during necrotic cell death—Involvement of serine but not cysteine proteases. Am. J. Pathol. 1997, 15151205–1213. [PMC free article] [PubMed] [Google Scholar]
- Cui D. X.; Tian F. R.; Ozkan C. S.; Wang M.; Gao H. J. Effect of single wall carbon nanotubes on human HEK293 cells. Toxicol. Lett. 2005, 155173–85. [DOI] [PubMed] [Google Scholar]
- Elimelech M.; Kang S.; Herzberg M.; Rodrigues D. F. Antibacterial effects of carbon nanotubes: Size does matter. Langmuir 2008, 24136409–6413. [DOI] [PubMed] [Google Scholar]
- Hu X.; Cook S.; Wang P.; Hwang H.; Liu X.; Williams Q. L. In vitro evaluation of cytotoxicity of engineered carbon nanotubes in selected human cell lines. Sci. Total Environ. 2010, 40881812–1817. [DOI] [PubMed] [Google Scholar]
- Erental A.; Sharon I.; Engelberg-Kulka H. Two programmed cell death systems in Escherichia coli: An apoptotic-like death is inhibited by the mazEF-mediated death pathway. PLoS Biol. 2012, 10, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein R.; Prat F.; Foote C. S. On the mechanism of DNA cleavage by fullerenes investigated in model systems: Electron transfer from guanosine and 8-oxo-guanosine derivatives to C-60. J. Am. Chem. Soc. 1999, 1212464–465. [Google Scholar]
- Yamakoshi Y.; Umezawa N.; Ryu A.; Arakane K.; Miyata N.; Goda Y.; Masumizu T.; Nagano T. Active oxygen species generated from photoexcited fullerene (C60) as potential medicines: O2–* versus 1O2. J. Am. Chem. Soc. 2003, 1254212803–12809. [DOI] [PubMed] [Google Scholar]
- Guldi D. M.; Prato M. Excited-state properties of C-60 fullerene derivatives. Acc. Chem. Res. 2000, 3310695–703. [DOI] [PubMed] [Google Scholar]
- Andersson L. M.; Tanaka H. Empirical evidence for identical band gaps in substituted C-60 and C-70 based fullerenes. Appl. Phys. Lett. 2014, 104, 4. [Google Scholar]
- Hussain S.; Boland S.; Baeza-Squiban A.; Hamel R.; Thomassen L. C.; Martens J. A.; Billon-Galland M. A.; Fleury-Feith J.; Moisan F.; Pairon J. C.; Marano F. Oxidative stress and proinflammatory effects of carbon black and titanium dioxide nanoparticles: Role of particle surface area and internalized amount. Toxicology 2009, 2601–3142–149. [DOI] [PubMed] [Google Scholar]
- Johnston H. J.; Hutchison G. R.; Christensen F. M.; Aschberger K.; Stone V. The biological mechanisms and physicochemical characteristics responsible for driving fullerene toxicity. Toxicol. Sci. 2010, 1142162–182. [DOI] [PubMed] [Google Scholar]
- Orfanopoulos M.; Kambourakis S. Chemical evidence of singlet oxygen production from C60 and C70 in aqueous and other polar media. Tetrahedron Lett. 1995, 363435–438. [Google Scholar]
- Migliore L.; Saracino D.; Bonelli A.; Colognato R.; D’Errico M. R.; Magrini A.; Bergamaschi A.; Bergamaschi E. Carbon nanotubes induce oxidative DNA damage in RAW 264.7 cells. Environ. Mol. Mutagen. 2010, 514294–303. [DOI] [PubMed] [Google Scholar]
- Phillips C. L.; Yah C. S.; Iyuke S. E.; Rumbold K.; Pillay V. The cellular response of Saccharomyces cerevisiae to multi-walled carbon nanotubes (MWCNTs). J. Saudi Chem. Soc. 2012, 10.1016/j.jscs.2012.01.005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.