Abstract
Purpose.
Retinitis pigmentosa (RP) is a genetically heterogeneous disease with over 60 causative genes known to date. Nevertheless, approximately 40% of RP cases remain genetically unsolved, suggesting that many novel disease-causing genes are yet to be identified. In this study, we aimed to identify the causative mutation for a large autosomal dominant RP (adRP) family with negative results from known retinal disease gene screening.
Methods.
Linkage analysis followed by whole-exome sequencing was performed. Stringent variant filtering and prioritization was carried out to identify the causative mutation.
Results.
Linkage analysis identified a minimal disease region of 8 Mb on chromosome 10 with a peak parametric logarithm (base 10) of odds (LOD) score of 3.500. Further whole-exome sequencing identified a heterozygous missense mutation (NM_000188.2:c.2539G>A, p.E847K) in hexokinase 1 (HK1) that segregated with the disease phenotype in the family. Biochemical assays showed that the E847K mutation does not affect hexokinase enzymatic activity or the protein stability, suggesting that the mutation may impact other uncharacterized function or result in a gain of function of HK1.
Conclusions.
Here, we identified HK1 as a novel causative gene for adRP. This is the first report that associates the glucose metabolic pathway with human retinal degenerative disease, suggesting a potential new disease mechanism.
Keywords: HK1, hexokinase, autosomal dominant retinitis pigmentosa (adRP), novel disease-causing gene, next-generation sequencing (NGS)
In this study, we identified for the first time HK1 as a novel disease-causing gene for autosomal dominant retinitis pigmentosa, implying the importance of glucose metabolic pathway in human retinal degenerative diseases.
Introduction
Retinitis pigmentosa (RP) is an inherited retinal degenerative disease affecting 1 in 4000 people around the world.1 The typical early symptoms of RP include night blindness and tunnel vision due to the loss of peripheral rod photoreceptor cells. As the disease progresses, cone photoreceptor cells start to degenerate, which may eventually lead to complete blindness.
The genetic basis of RP is highly heterogeneous. To date, mutations in more than 60 genes are known to cause RP.2 These genes function in strikingly diverse biological pathways, including phototransduction, transcription regulation, pre-mRNA splicing, and photoreceptor structure maintenance. Various inheritance patterns have also been observed, including autosomal dominant (ad), autosomal recessive (ar), x-linked (xl), digenic, and even mitochondrial modes.3–6 Despite the implementation of current next-generation sequencing technologies, approximately 40% of all adRP cases still cannot be explained by mutations in known genes, suggesting that novel disease-causing genes likely exist and remain to be identified.7–11 In this study, we aimed to identify novel RP disease-causing genes by studying a large adRP family with negative results from known retinal disease gene screening using linkage mapping followed by whole-exome sequencing.
Materials and Methods
Clinical Diagnosis of RP
Approval from the Institutional Review Board of the University of California-San Diego was obtained for this study, and informed consent was obtained from all patients. Twenty-five individuals from one big family participated in the study, and blood samples were obtained. A visual history was obtained from all patients, and best-corrected visual acuities were assessed. Ophthalmoscopic examination was performed on all patients, and fluorescein angiography was done on three affected patients. This study adhered to the Declaration of Helsinki.
Linkage Analysis
Genome-wide linkage analysis was performed using 5913 single nucleotide polymorphism (SNP) sites (Illumina Linkage-24 panel; Illumina, San Diego, CA, USA). PLINK was used to filter SNP sites of either low genotyping rate or Mendelian errors.12 Linkage analysis was then performed by Merlin using a rare dominant model with 90% penetrance.13
Library Preparation and Whole-Exome Sequencing
The Illumina pair-end DNA library preparation followed the standard manufacturer's instructions. Briefly, DNA was quantified using NanoDrop (Thermo Scientific, Wilmington, DE, USA). Total DNA (1 μg) was sheared into fragments of 300 to 500 bp, end-repaired, and had a 3′ adenosine base added. Then Illumina Y-shaped adapters were added to the DNA fragments, and 10 cycles of PCR reactions were used to amplify the libraries. The library DNA was further quantified using a PicoGreen assay (Life Technologies, Carlsbad, CA, USA). Six library DNA samples were pooled together before the capture step. In total, 3 μg pooled DNA was enriched by the NimbleGen SeqCap EZ Hybridization and Wash kit (NimbleGen SeqCap EZ Human Exome Library v.2.0; NimbleGen, Madison, WI, USA) following the manufacturer's protocols. After that, the postcapture libraries were quantified using PicoGreen assay and then sequenced on an Illumina Hiseq2000 machine.
Bioinformatics Analysis
Data were processed using an in-house bioinformatics pipeline as previously described.10,11,14 In particular, raw variants were called using Atlas2 Suite.15 Considering a dominant model, variants with an occurrence of one or more in any of the variant databases queried, including 1000 Genome,16 dbSNP135,17 NHLBI Exome Sequencing database,18 NIEHS Exome Sequencing database,19 and an internal control database of 11,000 exomes, were filtered out. dbNSFP v2.3 was used to functionally predict the effects of missense variants.20,21
Sanger Validation
For each identified mutation, a 500-bp flanking sequence at both sides was obtained from the UCSC genome browser (hg19 assembly; https://genome.ucsc.edu/ [in the public domain].). RepeatMasker was used to mask the repetitive sequence in human genome.22 Primer 3 was used to design a pair of primers for generating a 400- to 600-bp PCR product to sequence the mutation site and at least 50 bp surrounding it.23 After PCR amplification, the amplicons were sequenced on an ABI 3730xl or 3500xl Genetic Analyzer (Life Technologies).
Biochemical Assays
Hexokinase 1 cDNA was cloned, and the site mutation was generated by Quikchange II XL Site Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). The expression and purification followed the protocol described in the manual. Hexokinase activities were measured using HK colorimetric assay kit (BioVision, Milpitas, CA, USA) with 0.1 μg purified HK1 proteins. Diagram of the crystal structure of recombinant human hexokinase type I with 1,5-anhydroglucitol 6-phosphate was obtained from the National Center for Biotechnology Information (NCBI) protein database (URL [in the public domain]) with PDB ID 4FPB and analyzed by Cn3D4.3.1, also downloaded from NCBI (http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=111440). HEK-293T cell line was purchased from American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. DNA transfection was performed using lipofectamine 2000 from Invitrogen (Invitrogen, Life Technologies, Carlsbad, CA, USA) and following the manual instructions. Total RNA was isolated by RNeasy Mini Kit (Qiagen, Venlo, Limburg, Netherlands), and mRNA was reverse transcribed to cDNA (SuperScript II kit; Invitrogen). This cDNA was then used for real-time PCR with specific primers complementary to HK1 (forward primer: 5′-CACCTGTGAGGTTGGACTCA-3′; reverse primer: 5′-CCACCATCTCCACGTTCTTC-3′). Human beta-actin (primers 5′-GTACTTGCGCTCAGGAGGAG-3′ and 5′-ACTCTTCCAGCCTTCCTTCC-3′) was simultaneously processed in the same sample as an internal control. A StepOne quantitative PCR (qPCR) machine (Life Technologies) and SYBR Green qPCR kit (Invitrogen) was used for real-time PCR reaction according to the manufacturer's procedure. Cell lysates with 20 μg total proteins were resolved by SDS-PAGE and then blotted with specific antibody for human HK1 (Cell Signaling Technology, Danvers, MA, USA) at a 1:1000 dilution. Student's t-test was used in all statistical analyses, and statistical significance was accepted for P values less than 0.05.
Results
A Large adRP Family With Negative Results From Known Retinal Disease Gene Screening Was Recruited
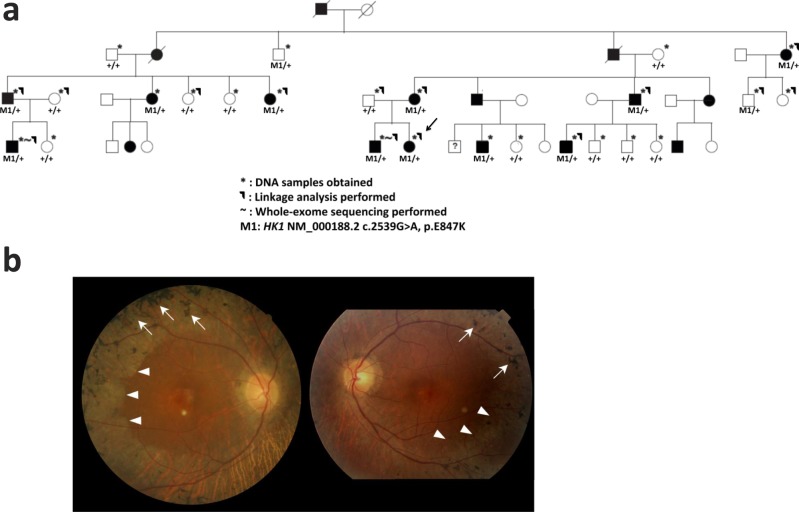
The Caucasian family is from the Midwestern United States with northern European ancestry. Out of the 44 members, 18 are affected. DNA samples were obtained from 25 members, including 11 affected individuals. Among the patients, the onset of reduction in night and peripheral vision ranged from age 5 to 23 (Supplementary Table S1). The disease severity ranged from mild pigmentary changes to severe retinal pigment epithelium and retinal degeneration. In addition, there was concomitant cone photoreceptor degeneration in older patients manifested by photophobia, color vision changes, and decreased central vision. Fundus images of the proband showed an abrupt border of chorioretinal degeneration outside the posterior pole, sclerosis of choroidal vessels, and pigmentary changes in the macula (Fig. 1b).
Figure 1.
(a) Pedigree of the large adRP family. DNA samples were obtained from 25 family members (*). Linkage analysis was performed on 14 family members (–⌉). Whole-exome sequencing was performed on two affected members (~). The HK1 E847K mutation segregated with the disease phenotype across the entire family with 85% penetrance. (b) Fundus photos of adRP proband (age 59, decimal visual acuity 0.1 in both eyes, marked by arrow in [a]) demonstrate typical bone spicule pigmentation (arrows) and extensive retina and RPE atrophy (arrowheads). An abrupt border of chorioretinal degeneration outside the posterior pole, sclerosis of choroidal vessels, and pigmentary changes in the macula region were observed. The pale circles near the center of the fundus are camera artifacts.
Using the retinal disease gene sequencing panel developed in our group,24 mutation screening in all known retinal disease-associated genes, including 23 adRP genes known to date, was performed for one proband. Despite good coverage (~30×), no pathogenic mutations in known retinal disease genes were identified for the patient, suggesting that the potential cause may reside in a novel gene (data not shown).
Genome-Wide Linkage Analysis Revealed a Minimal Disease Haplotype Region of 8 Mb on Chromosome 10 With Peak LOD Score of 3.500
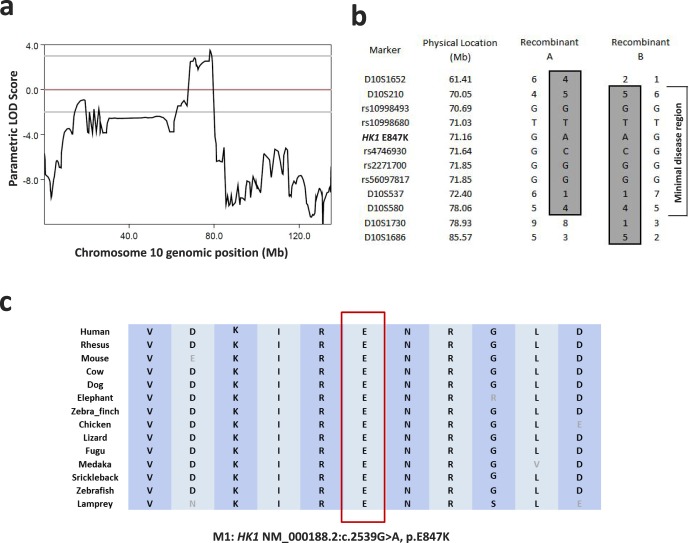
In order to narrow down the causative loci, we performed genome-wide linkage analysis using 5913 SNP sites on 14 family members, nine of whom were affected (Fig. 1a). Using PLINK,12 125 SNP sites were removed from further analysis due to either low genotyping rate or Mendelian errors. The remaining 5788 sites were then analyzed by Merlin using a rare dominant model with 90% penetrance.13 A peak parametric logarithm (base 10) of odds (LOD) score of 2.974 was observed on chromosome 10, with a minimal linkage region (LOD score > 0) of 10.4 Mb ranging from marker rs1925622 (chr10: 68769638) to marker rs158421 (chr10: 79182034). To further confirm the linkage mapping and reconstruct the disease haplotype, short tandem repeat (STR) typing was performed across all 25 samples. The combined genotyping boosted the peak parametric LOD score to 3.500 with a minimal disease haplotype region of 8 Mb (Figs. 2a, 2b; Supplementary Fig. S1). It is worth noting that the minimal disease region does not include any known adRP genes. This further excluded the possibility that the causative mutations reside in the known genes.
Figure 2.
(a) A peak parametric LOD score of 3.500 was observed on chromosome 10. (b) A minimal disease haplotype region of 8 Mb was obtained. (c) The HK1 E847 is highly conserved across vertebrates.
Whole-Exome Sequencing Identified Missense Allele E847K in HK1 as the Top Candidate Causative Mutation
To identify the causative mutation, we performed whole-exome capture sequencing of two affected family members (Fig. 1a). High-quality sequencing results were obtained for both samples with a mean coverage of 90× and 83×, respectively. An in-house bioinformatics pipeline was used to process the raw reads and perform variant calling, filtering, and annotation as described in Material and Methods. A total of only four rare coding-change variants were shared by the two affected members. Surprisingly, three of them were within the linkage region. Among the three candidates within the linkage region, one was a heterozygous missense variant (NM_000188.2: c.2539G>A, p.E847K) in HK1. The two other variants (NM_030625: c.598A>G, p.K200E and c.5996A>C, p.H1999P) were in TET1. The only shared rare coding-change variant outside the linkage region was a missense change (NM_032292: c.1565T>C, p.M522T) in GON4L. All four variants were completely absent in control databases totaling approximately 18,500 individuals (1000 Genomes, ESP6500, and our internal exome sequencing database of 11,000 individuals).
To further prioritize the candidate list, information such as gene expression and variant pathogenicity prediction was utilized. As showed in Supplementary Table S2, HK1 had significantly higher mRNA expression in the human retina than TET1 and GON4L.25 Meanwhile, in a proteomic study of bovine retina, HK1 protein was detected in the outer segment of photoreceptor cells, as well as multiple subcellular compartments of the retina, while TET1 and GON4L protein was not detected at all.26 In addition, the in silico prediction tool, dbNSFP version 2.3,20,21 predicted that the HK1 variant was likely deleterious to the protein's function, while TET1 and GON4L variants were predicted to be benign (Supplementary Table S3).
As a result, the HK1 variant was considered the top candidate causative mutation. Further segregation tests by Sanger sequencing indicated that the HK1 mutation segregated with the disease phenotype across the entire family with 85% penetrance (Fig. 1a). Two asymptomatic family members, who had the disease haplotype and the HK1 E847K mutation but did not exhibit the phenotype due to incomplete penetrance, were examined at ages 78 and 37. Their vision was unaffected, and fundus examination did not reveal bone spicule pigmentations. The HK1 mutation affects an amino acid site that is highly conserved across vertebrates (Fig. 2c). Moreover, the mutation resides in a common exon that is shared by all known HK1 isoforms including the retinal-specific one; and indeed, RNA-seq analysis indicated that the mutation site was highly expressed in the human retina.25
Biochemical Assays Showed That HK1 E847K Mutation Did Not Affect Hexokinase Enzymatic Activity or the Protein Stability
To investigate the functional consequences of the HK1 E847K mutation identified in the adRP family, a series of biochemical assays were performed to test if the mutation would affect hexokinase enzymatic activity or the protein stability. Wild-type (WT) and mutant HK1 flag-tag fusion proteins were overexpressed in 293T cells and purified using anti-flag-M2 magnetic beads (Sigma-Aldrich, St. Louis, MO, USA) (Supplementary Fig. S2). The kinase activities of recombinant HK1 proteins were then tested using hexokinase colorimetric assay kit (BioVision). The HK1 mutant protein showed similar hexokinase activity compared to WT protein (Supplementary Fig. S2b). This is consistent with the three-dimensional crystal structure (PDB ID 4FPB) analysis indicating that the mutation site was not located in any kinase activity-associated domain (Supplementary Fig. S2c). In addition to enzymatic activity, mRNA and protein levels of the HK1 mutant were also measured in 293T cells 2 days after transfection, using RT-qPCR and Western blot analysis. Similar mRNA and protein levels were observed for both mutant and WT HK1 (Supplementary Figs. S2d, S2e).
Discussion
In this study, we identified for the first time HK1 as a novel disease-causing gene for adRP, implying the importance of glucose metabolic pathway in human retinal degenerative diseases. One large multigeneration family was found to carry mutation in HK1 after linkage analysis followed by whole-exome sequencing. The identified E847K mutation is extremely rare, affects highly conserved amino acid, and was predicted to be detrimental.
Hexokinase 1 encodes one of the four human hexokinases that play essential roles in glucose metabolism. Among them, HK1 is the most ubiquitously expressed hexokinase gene and is one of the only two that were found to be highly expressed in the human retina. Within the retina, the HK1 protein is highly enriched in the photoreceptor inner segment, outer plexiform layer, inner nuclear layer, inner plexiform layer, and ganglion cell layer.27 The high abundance and broad distribution of HK1 suggest its crucial function in satisfying the energy requirement resulting from the high rate of metabolic activity in the retina.
Because of HK1's essential function in glycolysis, complete loss of function (LOF) of HK1 is hypothesized to be embryonic lethal. Supporting this hypothesis is the fact that only partial LOF mutations in HK1 have been observed in rare autosomal recessive nonspherocytic hemolytic anemia cases. The pathogenesis is primarily due to the hexokinase enzymatic deficiency in red blood cells.28–30 On the other hand, HK1 has also been implicated in the autosomal recessive Russe type of hereditary motor and sensory neuropathy (HMSNR).31 However, in HMSNR patients, the change of hexokinase enzymatic activity was not evident, leaving the possibility that the causality involves a novel function of HK1.31
In our study, biochemical assays showed no obvious difference between the E847K mutant and WT allele in terms of both enzymatic activity and mRNA and protein expression level, suggesting that hexokinase deficiency was not likely to be the underlying mechanism. Indeed, our patients with the E847K mutation did not show any sign of anemia. In addition, no signs of exercise intolerance or brain cognition deficits were observed. Taken together, given the dominant inheritance in nature, we propose that the mutation identified in our study is likely to lead to defect in HK1's retina-specific function or result in a gain of function.
Several studies of hexokinase already suggest possible hypotheses for HK1's molecular pathogenesis in retinal degeneration. Studies in the mouse retina show that mitochondrial metallochaperone Cox11, chaperone protein Ranbp2, and Hk1 interact with each other in equilibrium to maintain metabolic activity and normal retina function.32 It is possible that mutations in HK1 could abolish such interaction, causing disturbance of the equilibrium and resulting in retinal degeneration. Another hypothesis comes from a recent study showing that light activation of insulin receptors can regulate the hexokinase–mitochondria interaction and inhibit apoptosis of photoreceptors.33 It is possible that mutations in HK1 could disrupt the regulation from insulin receptors, causing dissociation between hexokinase and mitochondria that would eventually trigger photoreceptor degeneration. If that is the case, enhancing the hexokinase–mitochondria association may be a potential therapeutic approach.
In summary, we report, for the first time, HK1 as a novel disease-causing gene for adRP. The identified missense mutation causes a glutamate-to-lysine change at position 847. Surprisingly, an identical mutation (E847K) in HK1 was identified as the top candidate for another large adRP family by an independent group, and there is no sign that the two families are related (Daiger SP, written communication, 2014). This further strengthens our conclusion that the E847K mutation in HK1 is a rare cause for adRP.
Web Resources
PLINK: http://pngu.mgh.harvard.edu/~purcell/plink/.
Merlin: http://www.sph.umich.edu/csg/abecasis/Merlin/index.html.
RetNet: http://www.sph.uth.tmc.edu/Retnet.
1000 Genome: http://www.1000genomes.org/.
ESP6500: http://evs.gs.washington.edu/EVS/.
Acknowledgments
We thank the adRP family for participating in the study. We thank Zachry T. Soens for reviewing and editing the manuscript.
Supported by grants from the Retinal Research Foundation, Foundation Fighting Blindness (BR-GE-0613-0618-BCM), and the National Eye Institute (R01EY022356) (RC). FW is supported by a predoctoral fellowship: The Houston Laboratory and Population Sciences Training Program in Gene-Environment Interaction, The Burroughs Wellcome Fund. Next-generation sequencing was conducted at the Functional Genomic Core (FGC) facility at Baylor College of Medicine supported by National Institutes of Health Shared Instrument Grant 1S10RR026550 (RC).
Disclosure: F. Wang, None; Y. Wang, None; B. Zhang, None; L. Zhao, None; V. Lyubasyuk, None; K. Wang, None; M. Xu, None; Y. Li, None; F. Wu, None; C. Wen, None; P.S. Bernstein, None; D. Lin, None; S. Zhu, None; H. Wang, None; K. Zhang, None; R. Chen, None
References
- 1. Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis. 2006; 1: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. RetNet. http://www.sph.uth.tmc.edu/Retnet. Accessed March 25, 2014. [Google Scholar]
- 3. Fahim AT, Daiger SP, Weleber RG. Retinitis pigmentosa overview. In: Pagon RA, MP Adam, Ardinger HH, et al. , eds. GeneReviews. Seattle, WA: University of Washington; 1993. [Google Scholar]
- 4. Kajiwara K, Berson EL, Dryja TP. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science. 1994; 264: 1604–1608. [DOI] [PubMed] [Google Scholar]
- 5. Dryja TP, Hahn LB, Kajiwara K, Berson EL. Dominant and digenic mutations in the peripherin/RDS and ROM1 genes in retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1997; 38: 1972–1982. [PubMed] [Google Scholar]
- 6. Mansergh FC, Millington-Ward S, Kennan A, et al. Retinitis pigmentosa and progressive sensorineural hearing loss caused by a C12258A mutation in the mitochondrial MTTS2 gene. Am J Hum Genet. 1999; 64: 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neveling K, Collin RW, Gilissen C, et al. Next-generation genetic testing for retinitis pigmentosa. Hum Mutat. 2012; 33: 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O'Sullivan J, Mullaney BG, Bhaskar SS, et al. A paradigm shift in the delivery of services for diagnosis of inherited retinal disease. J Med Genet. 2012; 49: 322–326. [DOI] [PubMed] [Google Scholar]
- 9. Shanks ME, Downes SM, Copley RR, et al. Next-generation sequencing (NGS) as a diagnostic tool for retinal degeneration reveals a much higher detection rate in early-onset disease. Eur J Hum Genet. 2012; 21: 274–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fu Q, Wang F, Wang H, et al. Next-generation sequencing-based molecular diagnosis of a Chinese patient cohort with autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2013; 54: 4158–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang F, Wang H, Tuan HF, et al. Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet. 2014; 133: 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002; 30: 97–101. [DOI] [PubMed] [Google Scholar]
- 14. Koenekoop RK, Wang H, Majewski J, et al. Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nat Genet. 2012; 44: 1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Challis D, Yu J, Evani US, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012; 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Genomes Project Consortium, et al. A map of human genome variation from population-scale sequencing. Nature. 2010; 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. National Center for Biotechnology Information, National Library of Medicine. Database of Single Nucleotide Polymorphisms (dbSNP Build 135). [Google Scholar]
- 18. NHLBI GO Exome Sequencing Project (ESP). Seattle, WA: http://evs.gs.washington.edu/EVS/. ESP6500SI. Accessed March 12, 2013. [Google Scholar]
- 19. NIEHS Environmental Genome Project. Seattle, WA: http://evs.gs.washington.edu/niehsExome/. NIEHS95. Accessed March 12, 2013. [Google Scholar]
- 20. Liu X, Jian X. Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011; 32: 894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu X, Jian X. Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013; 34: E2393–E2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 1996-2010. http://www.repeatmasker.org. Accessed October 10, 2013. [Google Scholar]
- 23. Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000; 132: 365–386. [DOI] [PubMed] [Google Scholar]
- 24. Wang X, Wang H, Sun V, et al. Comprehensive molecular diagnosis of 179 Leber congenital amaurosis and juvenile retinitis pigmentosa patients by targeted next generation sequencing. J Med Genet. 2013; 50: 674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Farkas MH, Grant GR, White JA, Sousa ME, Consugar MB, Pierce EA. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics. 2013; 14: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kwok MC, Holopainen JM, Molday LL, Foster LJ, Molday RS. Proteomics of photoreceptor outer segments identifies a subset of SNARE and Rab proteins implicated in membrane vesicle trafficking and fusion. Mol Cell Proteomics. 2008; 7: 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reidel B, Thompson JW, Farsiu S, Moseley MA, Skiba NP, Arshavsky VY. Proteomic profiling of a layered tissue reveals unique glycolytic specializations of photoreceptor cells. Mol Cell Proteomics. 2011; 10:M110.002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rijksen G, Akkerman JW. van den Wall Bake AW, Hofstede DP, Staal GE. Generalized hexokinase deficiency in the blood cells of a patient with nonspherocytic hemolytic anemia. Blood. 1983; 61: 12–18. [PubMed] [Google Scholar]
- 29. Bianchi M, Magnani M. Hexokinase mutations that produce nonspherocytic hemolytic anemia. Blood Cells Mol Dis. 1995; 21: 2–8. [DOI] [PubMed] [Google Scholar]
- 30. van Wijk R, Rijksen G, Huizinga EG, Nieuwenhuis HK, van Solinge WW. HK. Utrecht: missense mutation in the active site of human hexokinase associated with hexokinase deficiency and severe nonspherocytic hemolytic anemia. Blood. 2003; 101: 345–347. [DOI] [PubMed] [Google Scholar]
- 31. Hantke J, Chandler D, King R, et al. A mutation in an alternative untranslated exon of hexokinase 1 associated with hereditary motor and sensory neuropathy – Russe (HMSNR). Eur J Hum Genet. 2009; 17: 1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aslanukov A, Bhowmick R, Guruju M, et al. RanBP2 modulates Cox11 and hexokinase I activities and haploinsufficiency of RanBP2 causes deficits in glucose metabolism. PLoS Genet. 2006; 2: e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rajala A, Gupta VK, Anderson RE, Rajala RV. Light activation of the insulin receptor regulates mitochondrial hexokinase. A possible mechanism of retinal neuroprotection. Mitochondrion. 2013; 13: 566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]