Abstract
Purpose.
To test the association between the CTG18.1 trinucleotide repeat expansion of TCF4 gene and Fuchs' endothelial corneal dystrophy (FECD) in a Chinese population.
Methods.
The trinucleotide repeat polymorphism CTG18.1 was genotyped using short tandem repeat and triplet repeat primed polymerase chain reaction assays in 57 Chinese subjects with FECD and 121 controls. Statistical association of the expanded CTG18.1 allele and 18 single nucleotide polymorphisms (SNPs) across TCF4 with FECD was evaluated. To investigate the linkage disequilibrium structure of the TCF4 region, haplotype analysis was performed on our study subjects and compared with genotyping data of 97 Han Chinese and 85 Caucasians in the 1000 Genomes Project.
Results.
The expanded CTG18.1 allele was associated with FECD (P = 4.7 × 10−14), with the odds ratio of each copy of the expanded allele estimated to be 66.5 (95% confidence interval: 12.6–350.1). Five TCF4 SNPs showed association with FECD at a nominal level (P < 5.0 × 10−2); however, conditional on the expanded CTG18.1 polymorphism, none of the SNPs showed association with FECD. The only haplotype associated with the disease was the one with the expansion at the CTG18.1 locus.
Conclusions.
Transethnic replication of the association between the CTG18.1 repeat expansion in the TCF4 gene and FECD suggests it is a common, causal variant shared in Eurasian populations conferring significant risk for the development of FECD. Our data suggest that the expanded CTG18.1 allele is the main, if not sole, causal variant at this gene locus in the Chinese population.
Keywords: Fuchs' corneal dystrophy, genetic diseases, TCF4, CTG18.1 trinucleotide repeat
Transethnic replication of the association between the CTG18.1 repeat expansion in the TCF4 gene and Fuchs' endothelial corneal dystrophy suggests it is a common, causal variant shared in Eurasian populations conferring significant risk for the development of the disease.
Introduction
Fuchs' endothelial corneal disorder (FECD) is the most common inherited corneal endothelial disorder in the United States, impacting 1 in 20 Caucasians aged older than 40 years.1 Of the 72,736 corneal transplants performed in the United States in 2013, Fuchs' dystrophy was the leading indication, accounting for 14,153 cases with an additional 12,356 procedures done for related cases of corneal endothelial cell failure.2 Although large-scale prevalence studies are limited, existing cross-sectional data suggest that the prevalence of FECD is higher in European countries compared with other parts of the world. Population-based studies in Iceland documented corneal guttae in 11% of females and 7% of males.3 The disorder, however, is not uncommon in East Asian populations. A comparative study of the prevalence of guttae found a higher rate in Singapore (8.5%) versus Japan (5.5%).4
In this disorder, the endothelium undergoes accelerated senescence and apoptosis.5–9 Descemet's membrane, the underlying basement membrane of the endothelium, becomes diffusely thickened and also develops focal excrescences apparent on slit-lamp biomicroscopy as guttae.5 With progressive decline in the endothelial cell density, the cornea becomes edematous resulting in loss of vision. With advanced disease, bullous keratopathy can cause significant pain and corneal scarring that markedly impacts visual acuity.
Fuchs' endothelial corneal dystrophy is a common, complex trait with genetic heterogeneity. Rare heterozygous mutations in COL8A2, SLC4A11, TCF8, LOXHD1, CLU, and AGBL1 have been implicated in FECD, but account for a small fraction of the genetic burden.10–19 In 2010, a genome-wide association study (GWAS) identified association between alleles in the transcription factor 4 gene (TCF4), encoding a member of the E-protein family (E2-2), with typical FECD, whereby the association increased the odds of having FECD by a factor of 30 in homozygous persons, defying the norm for GWAS with common variants.20 In this study, the most significant signal was detected with an intronic single nucleotide polymorphism (SNP), rs613872.20
While the association between rs613872 and FECD was replicated in other Caucasian populations,21,22 in a Chinese FECD cohort from Singapore, instead of rs613872—which was monomorphic—two other TCF4 intronic SNPs (rs17089887 and rs17089925) were significant experiment-wide (P = 7.34 × 10−5 and 4.5 × 10−4, respectively).23 Interestingly, the vicinal region of TCF4 on chromosome 18 was previously linked to FECD in three large Caucasian pedigrees.24 However, the risk allele rs613872 was found not to cosegregate with the trait in these three families.21 Sequencing of TCF4 in 96 late-onset FECD cases also failed to identify any probable pathogenic variants in coding regions of TCF4.21
In 2012, a strong association between a CTG trinucleotide repeat locus (CTG18.1) in intron 3 of the TCF4 and FECD was reported, which displayed greater specificity than rs613872.25 The locus CTG18.1 was initially discovered by the repeat expansion detection assay and was found to be expanded in 3% of subjects in Caucasian pedigrees without known associated phenotype.26 Expanded alleles with greater than 37 CTG repeats at this locus were found to be unstable.26 Our group replicated the association with 120 FECD cases and 100 controls, finding that one copy of the expanded CTG18.1 allele increased risk of disease by 32.3-fold (95% confidence interval [CI], 13.4–77.6).27 Additionally, we showed the expanded allele cosegregated with the trait with complete penetrance in 11 out of 29 Caucasian kindreds examined and cosegregated with incomplete penetrance in an additional 9 of these 29 families.27
Transethnic mapping has been suggested as a powerful tool to detect novel disease susceptibility loci and to locate the disease causal variants.28–30 Empirical observations for many traits show high transethnic replicability of GWAS results,31 which support a model that the underlying causal variants at many loci are shared across ancestry groups. Therefore, transethnic analysis can increase the detection power through meta-analysis with large sample sizes.32 By exploiting the differences in the linkage disequilibrium (LD) structure between diverse populations, transethnic comparison can also amplify the association signal at the causal variant and thus locate either a small region harboring the causal mutation or the functional mutation itself.31,33 Considering rs613872, the most significant SNP in Caucasians, was absent in the Chinese FECD cohort,23 we hypothesize this variant is not causal. Instead, here we show transethnic replication of the association of the expanded CTG18.1 trinucleotide repeat polymorphism, which is in the same LD block as rs613872, with FECD in the ethnic Chinese population.
Methods
Study Participants
The study protocol had the approval of the institutional review boards of University of Texas Southwestern Medical Center and the Singapore National Eye Centre and was in compliance with the tenets of the Declaration of Helsinki. The subjects of this study (57 cases and 121 controls) were the same as in the previous report of an association of TCF4 gene polymorphisms with Fuchs' dystrophy in ethnic Chinese from Singapore.23 All subjects had undergone a complete ophthalmic examination including slit lamp examination, confocal specular microscopy, and fundoscopy. Inclusion criteria for FECD cases were the presence of greater than 5 mm of confluent central corneal endothelial guttae in each eye (Krachmer grade 4 or higher) on slit lamp examination or histopathologic confirmation of the diagnosis after corneal transplantation.34 A detailed history was recorded for all subjects, including any family history and duration of onset of symptoms. Informed consent was obtained from each study participant. Chinese subjects without central corneal guttae (Krachmer grade 0) on slit lamp examination and confocal specular microscopy were recruited as control subjects. Individuals in whom the corneal endothelium could not be evaluated were excluded.
Genotyping
Genomic DNA was extracted from leukocytes of peripheral blood samples with a blood extraction kit (Nucleon; Amersham Biosciences, Buckinghamshire, UK). Previously published genotyping results of 18 SNPs across TCF4 from this cohort were reanalyzed in this study in conjunction with the CTG18.1 allele.23
The trinucleotide repeat polymorphism CTG18.1 was genotyped using short tandem repeat (STR) and triplet repeat primed polymerase chain reaction (TP-PCR) assays as previously described.25,27,35 The STR assay was performed on genomic DNA samples from all study subjects. On samples where the STR assay detected only one allele or no alleles, TP-PCR assay was performed to confirm the presence of an expanded CTG18.1 allele.
For the STR assay, a 5′ FAM-labeled primer was utilized for the PCR. After polymerase chain reaction, 5 μL DNA was mixed with 10 μL internal lane standard 600 (ILS600; Promega Corp., Madison, WI, USA). Sequencing was carried out using a DNA analyzer (ABI 3730XL; Applied Biosystems, Foster City, CA, USA) and the data were analyzed using genotyping software (ABI GeneMapper 4.0; Applied Biosystems).
We performed the TP-PCR assay to detect the expanded CTG18.1 allele(s) as previously described.27,35 Locus-specific fluorescent primer P1 is designed to an upstream region flanking the CTG18.1 polymorphism. The companion repeat-specific reverse primer P4 on the complementary strand includes five units of the CTG repeat and a 5′ tail to serve as an anchor for a second reverse primer P3, which prevents progressive shortening of the amplicons with subsequent cycles. The 5′ tail of primer P4 and the “common” flag primer P3 share no homology with the human genome. Polymerase chain reaction was performed with the following parameters: 200 ng of genomic DNA, 1 μmol/L of primer P1, and 0.03 μmol/L of primer P4 and 1 μmol/L of primer P3, 200 μmol/L dNTPs, 1.5 mmol/L MgCl2, and 1 U of Taq DNA polymerase (5 Prime, Gaithersburg, MD, USA). The cycling conditions were an initial denaturation of 9 minutes at 95°C, followed by 10 cycles of 95°C for 30 seconds, 62°C for 30 seconds, and 72°C for 4 minutes; and then 30 cycles of 95°C for 45 seconds, 62°C for 45 seconds, and 72°C for 4 minutes with a 15-second extension at each cycle. The final extension step was 72°C for 10 minutes. We analyzed the TP-PCR amplicons on the DNA analyzer (Applied Biosystems).
Characteristic tracing patterns of the TP-PCR electropherograms were used to distinguish samples that were homozygous for a CTG18.1 allele from those that had an expanded CTG18.1 allele not detected by STR assay. The tracings of TP-PCR were also used to detect the presence of two expanded CTG18.1 alleles in samples where the STR analysis did not detect any allele.
Statistical Analysis
Comparisons of demographic features between cases and controls were performed by a two-sample t-test for age and by Fisher's exact test for sex. For the trinucleotide repeat CTG18.1 polymorphism, we dichotomized alleles such that CTG ≥40 was considered an expanded allele, denoted as X, and CTG <40 was considered a normal allele, denoted as S. An expanded CTG18.1 allele was defined as ≥40 CTG repeats as a conservative cutoff value, as we did in a previous report,27 which is slightly higher than the 37 CTG repeats reported as the threshold of instability at this locus.26 Hardy-Weinberg equilibrium (HWE) was examined in cases and controls, separately, by an exact test. Logistic regression models were fit to test association between genotype and FECD affection status by the likelihood ratio test.36 First, each variant was tested adjusting for risk factors age and sex; second, the analysis was repeated by further conditioning on the CTG18.1 genotype. The genotypic value was coded in an additive manner—that is, 0, 1, and 2 denoted TT, TG, and GG genotypes, respectively, for SNPs, and SS, SX, XX genotypes, respectively, for the CTG18.1 polymorphism.
To investigate the LD structure of the TCF4 region, we extracted the genotype of 97 Han Chinese (CHB) subjects and 85 Caucasian (CEU) subjects from the 1000 Genomes Project.37 Specifically, the genotype of 14 polymorphic SNPs in the original report and SNP rs613872 were extracted.23 Haplotype analyses were performed using haplotype analysis software Haploview38 (Broad Institute, Cambridge, MA, USA) and haplo.stats.39
Results
The demographic information of the subjects was previously described.23 Specifically there were more females in cases than in controls (79% vs. 58%, P = 7.0 × 10−3), but there was no age difference between the two groups (Table 1). The dichotomized CTG18.1 trinucleotide repeat polymorphism was in HWE in both cases and controls (P > 5.0 × 10−2). It was associated with FECD (P = 4.7 × 10−14) with the odds ratio (OR) of each copy of the expanded allele estimated to be 66.5 (95% CI: 12.6–350.1).
Table 1.
Demographic Information and TCF4 CTG18.1 Genotype Distribution* in Chinese FECD Cases and Controls
|
Characteristic |
Cases, n
= 57 |
Controls, n
= 121 |
P
Value |
| Men/women | 12/45 | 51/70 | 7.0 × 10−3 |
| Age ± SD, y | 67.4 ± 9.9 | 65.1 ± 7.2 | 1.4 × 10−1 |
| CTG18.1* | |||
| XX | 3 | 0 | 4.7 × 10−14† |
| SX | 22 | 2 | |
| SS | 32 | 119 |
Alleles were dichotomized with X denoting CTG≥40 and S denoting CTG<40.
Under an additive genetic model adjusted for age and sex.
Of the 18 previously genotyped TCF4 SNPs, four were monomorphic, including rs613872. Of the remaining 14 SNPs and the CTG18.1 polymorphism, five showed nominal association with FECD (P < 5.0 × 10−2), and the CTG18.1 polymorphism had the most significant signal (P = 4.7 × 10−14). However, conditional on the CTG18.1 polymorphism, none of the SNPs showed association with FECD (Table 2).
Table 2.
Association of TCF4 Polymorphisms With FECD in a Chinese Population
|
Polymorphism |
Position* |
P
Value† |
P
Value‡ |
| rs1348047 | 53050058 | 3.6 × 10−3 | 2.3 × 10−1 |
| rs2919450 | 53084545 | 4.5 × 10−1 | 6.6 × 10−1 |
| rs17089826 | 53126993 | 2.8 × 10−1 | 7.9 × 10−1 |
| rs7233312 | 53143573 | 1.3 × 10−1 | 8.8 × 10−1 |
| rs2123389 | 53201972 | 3.5 × 10−1 | 8.5 × 10−1 |
| rs1452787 | 53207207 | 3.8 × 10−3 | 3.2 × 10−1 |
| rs1319637 | 53207325 | 1.4 × 10−1 | 5.2 × 10−1 |
| rs17089887 | 53208256 | 1.4 × 10−4 | 1.9 × 10−1 |
| rs2123392 | 53214865 | 3.0 × 10−3 | 3.0 × 10−1 |
| CTG18.1 | 53253385 | 4.7 × 10−14 | – |
| rs17089925 | 53400834 | 2.2 × 10−4 | 2.1 × 10−1 |
| rs1477440 | 53696010 | 2.1 × 10−1 | 3.1 × 10−1 |
| rs2286812 | 53717464 | 6.1 × 10−1 | 9.9 × 10−1 |
| rs1945737 | 53744545 | 9.9 × 10−1 | 5.0 × 10−1 |
| rs7235583 | 53747212 | 6.8 × 10−1 | 4.1 × 10−1 |
Physical coordinates on chromosome 18 by human genome build 37.1.
Adjusting for age and sex.
Adjusting for age and sex, and CTG18.1 genotype.
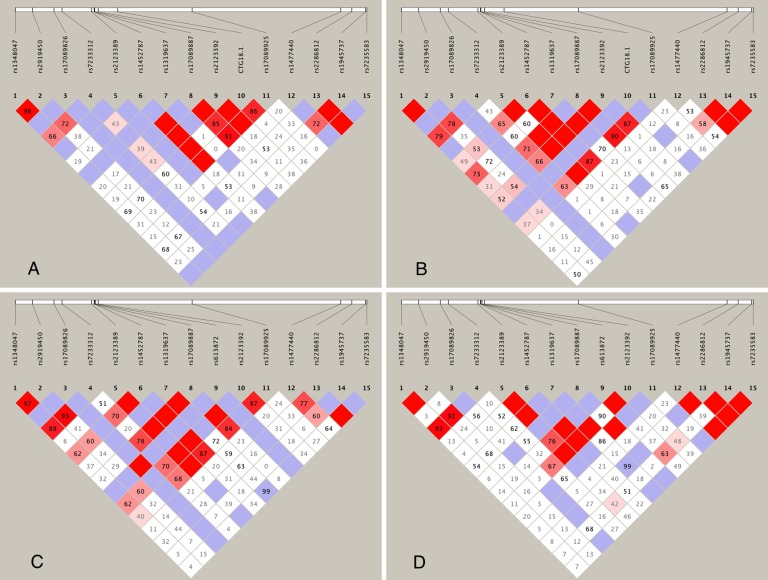
Allele frequencies of 16 TCF4 polymorphisms in the four populations are shown in Supplementary Table S1. According to the LD heat maps (Fig.), there was a haplotype block spanning variants rs1452787, rs1319637, rs17089887, rs2123392, CTG18.1, and rs17089925 in all four populations. Among the six variants, rs1319637 was rare in cases (minor allele frequency [MAF] < 0.035) and it was not associated with FECD by the single-variant test (P = 1.4 × 10−1), whereas the other five were common and significantly associated with FECD. Thus, we further performed haplotype-based association analysis based on these five variants (Table 3). There were five common haplotypes with frequencies greater than 0.01 in either cases or controls. The haplotype A-C-T-S-T was most common in both cases (frequency = 0.394) and controls (frequency = 0.387), and was treated as the baseline. The only haplotype associated with FECD was A-C-T-X-T (P = 2.1 × 10−5) with frequencies of 0.211 and 0.009 in cases and controls, respectively. Note that it was the only common haplotype harboring the expanded CTG18.1 allele, and it was different from the baseline haplotype only at the CTG18.1 locus. A global test resulted in a P value of 1.5 × 10−9.
Table 3.
Haplotype Association* of TCF4 Polymorphisms With FECD in a Chinese Population
|
rs1452787 |
rs17089887 |
rs2123392 |
CTG18.1 |
rs17089925 |
Haplotype Frequencies |
P
Value |
Global
P
Value |
|
|
Cases |
Control |
|||||||
| A | C | T | S | T | 0.394 | 0.387 | – | 1.5 × 10−9 |
| G | T | C | S | C | 0.272 | 0.375 | 2.5 × 10−1 | |
| A | T | T | S | C | 0.050 | 0.148 | 1.7 × 10−1 | |
| A | C | T | X | T | 0.211 | 0.009 | 2.1 × 10−5 | |
| A | C | T | S | C | 0.038 | 0.055 | 5.7 × 10−1 | |
Haplotypes with frequencies greater than 0.01 in either cases or controls were considered. The most common haplotype (A-C-T-S-T) was treated as the reference. A generalized linear model was fit adjusting for age and sex. Both haplotype-specific P values and global P values were reported.
Discussion
Our results show a strong association of the expanded CTG18.1 allele of the TCF4 gene with FECD in ethnic Chinese from Singapore. The genome-wide association study on the FECD trait in Caucasians found strong associations with SNPs (most strongly SNP rs613872) spanning TCF4.20 Association of TCF4 polymorphisms with FECD was replicated in this Singapore Chinese cohort, but the highest scoring SNP in Caucasians rs613872 was absent from this study group.23 Sequencing data from 1000 Genomes Project reveal that this SNP rs613872 is common in Caucasians (CEU MAF = 0.188), but extremely rare in Han Chinese (CHB MAF = 0.005). The disease association originally found in the Chinese with TCF4 SNPs rs1348047, rs2123392, rs17089887, and rs17089925 (generating three independent association signals) was no longer significant when conditioned on the CTG18.1 locus.23 We also found that the only haplotype associated with the disease is the one with the expansion at the CTG18.1 locus. The haplotype association signal (P = 2.1 × 10−5) is weaker than that of single-variant analysis (P = 4.7 × 10−14). On one hand, it can be due to power loss when taking the haplotype ambiguity into account by probabilities; on the other hand, it may indicate that the haplotype analysis did not gain power by tagging nongenotyped causal variants or integrating multiple causal variants, as the method is supposed to do when the causal variant is not genotyped.40 All these data suggest that the expanded CTG18.1 allele is the main, if not sole, functional variant at this gene locus in this population.
Figure.
Heat map of pairwise linkage disequilibrium (D') across 15 TCF4 polymorphisms in multiple populations: (A) Singapore Chinese FECD cases. (B) Singapore Chinese control subjects. (C) Han Chinese (CHB) from the 1000 Genomes Project. (D) Caucasians (CEU) from the 1000 Genomes Project.
It is hypothesized that disease causal variants may have comparable effect size across populations if not masked by other gene-gene or gene-environment interactions.41 In a recent comprehensive survey of GWAS replicability of 28 disorders, the investigators found a “strong and significant correlation of odds ratios across European and East Asian populations, indicating that underlying causal variants are common and shared between the two ancestries.”31 We found the OR of each copy of the expanded allele estimated to be 66.5 (95% CI: 12.6–350.1) in the Chinese population, mirroring its extremely high OR in Caucasians of 32.3 (95% CI: 13.4–77.6).27 The large effect sizes of the expanded CTG18.1 allele across both ethnic groups further implies this polymorphism as a causal variant.41
The strong association of the expanded CTG18.1 allele in the TCF4 gene with FECD in Caucasians,25,27 demonstration of its cosegregation with the trait in Caucasian pedigrees,27 and now transethnic replication of the association with similar effect sizes across populations are all compelling evidence that the repeat expansion is a causal variant. However, functional data are required to prove that the expanded CTG18.1 allele is indeed a causal mutation.
It is known that haploinsufficiency of TCF4 by microdeletions or missense mutations can lead to autosomal dominant Pitt-Hopkins syndrome, a phenotype comprising of microcephaly, encephalopathy, epilepsy, psychomotor delay, and episodic hyperventilation.42,43 This phenotypic spectrum is quite dissimilar to FECD. Therefore, we speculate that haploinsufficiency of the TCF4 gene may be neither sufficient nor necessary to produce the FECD phenotype. The allele CTG18.1 is likely working via complex molecular mechanisms of expanded repeats.
In summary, the transethnic replication of the association between the CTG18.1 repeat expansion in the TCF4 gene and FECD suggests it a common, causal variant shared in Eurasian populations conferring significant risk for the development of FECD.
Acknowledgments
The authors thank the patients for their participation in this study.
Supported by Grants R01EY022161 (VVM) and P30EY020799 from the National Eye Institute, National Institutes of Health, Bethesda, Maryland, United States, and an unrestricted grant from Research to Prevent Blindness, New York, New York, United States. The authors alone are responsible for the content and writing of the paper.
Disclosure: C. Xing, None; X. Gong, None; I. Hussain, None; C.-C. Khor, None; D.T.H. Tan, None; T. Aung, None; J.S. Mehta, None; E.N. Vithana, None; V.V. Mootha, None
References
- 1. Lorenzetti DW, Uotila MH, Parikh N, Kaufman HE. Central cornea guttata. Incidence in the general population. Am J Ophthalmol. 1967; 64: 1155–1158. [PubMed] [Google Scholar]
- 2. Eye Bank Association of America. 2013 Eye Banking Statistical Report. Washington: Eye Bank Association of America; 2013. Available at: http://www.restoresight.org/wp-content/uploads/2014/04/2013_Statistical_Report-FINAL.pdf. Accessed May 15, 2014. [Google Scholar]
- 3. Zoega GM, Fujisawa A, Sasaki H, et al. Prevalence and risk factors for cornea guttata in the Reykjavik Eye Study. Ophthalmology. 2006; 113: 565–569. [DOI] [PubMed] [Google Scholar]
- 4. Kitagawa K, Kojima M, Sasaki H, et al. Prevalence of primary cornea guttata and morphology of corneal endothelium in aging Japanese and Singaporean subjects. Ophthalmic Res. 2002; 34: 135–138. [DOI] [PubMed] [Google Scholar]
- 5. Chi HH, Teng CC, Katzin HM. Histopathology of primary endothelial-epithelial dystrophy of the cornea. Am J Ophthalmol. 1958; 45: 518–535. [DOI] [PubMed] [Google Scholar]
- 6. Laing RA, Leibowitz HM, Oak SS, Chang R, Berrospi AR, Theodore J. Endothelial mosaic in Fuchs' dystrophy. A qualitative evaluation with the specular microscope. Arch Ophthalmol. 1981; 99: 80–83. [DOI] [PubMed] [Google Scholar]
- 7. Bigar F. Specular microscopy of the corneal endothelium. Optical solutions and clinical results. Dev Ophthalmol. 1982; 6: 1–94. [PubMed] [Google Scholar]
- 8. Borderie VM, Baudrimont M, Vallee A, Ereau TL, Gray F, Laroche L. Corneal endothelial cell apoptosis in patients with Fuchs' dystrophy. Invest Ophthalmol Vis Sci. 2000; 41: 2501–2505. [PubMed] [Google Scholar]
- 9. Li QJ, Ashraf MF, Shen DF, et al. The role of apoptosis in the pathogenesis of Fuchs endothelial dystrophy of the cornea. Arch Ophthalmol. 2001; 119: 1597–1604. [DOI] [PubMed] [Google Scholar]
- 10. Biswas S, Munier FL, Yardley J, et al. Missense mutations in COL8A2, the gene encoding the alpha2 chain of type VIII collagen, cause two forms of corneal endothelial dystrophy. Hum Mol Genet. 2001; 10: 2415–2423. [DOI] [PubMed] [Google Scholar]
- 11. Zhang C, Bell WR, Sundin OH, et al. Immunohistochemistry and electron microscopy of early-onset Fuchs corneal dystrophy in three cases with the same L450W COL8A2 mutation. Trans Am Ophthalmol Soc. 2006; 104: 85–97. [PMC free article] [PubMed] [Google Scholar]
- 12. Vithana EN, Morgan P, Sundaresan P, et al. Mutations in sodium-borate cotransporter SLC4A11 cause recessive congenital hereditary endothelial dystrophy (CHED2). Nature Genet. 2006; 38: 755–757. [DOI] [PubMed] [Google Scholar]
- 13. Vithana EN, Morgan PE, Ramprasad V, et al. SLC4A11 mutations in Fuchs endothelial corneal dystrophy. Hum Mol Genet. 2008; 17: 656–666. [DOI] [PubMed] [Google Scholar]
- 14. Riazuddin SA, Zaghloul NA, Al-Saif A, et al. Missense mutations in TCF8 cause late-onset Fuchs corneal dystrophy and interact with FCD4 on chromosome 9p. Am J Hum Genet. 2010; 86: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krafchak CM, Pawar H, Moroi SE, et al. Mutations in TCF8 cause posterior polymorphous corneal dystrophy and ectopic expression of COL4A3 by corneal endothelial cells. Am J Hum Genet. 2005; 77: 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mehta JS, Vithana EN, Tan DT, et al. Analysis of the posterior polymorphous corneal dystrophy 3 gene, TCF8, in late-onset Fuchs endothelial corneal dystrophy. Invest Ophthalmol Vis Sci. 2008; 49: 184–188. [DOI] [PubMed] [Google Scholar]
- 17. Riazuddin SA, Parker DS, McGlumphy EJ, et al. Mutations in LOXHD1, a recessive-deafness locus, cause dominant late-onset Fuchs corneal dystrophy. Am J Hum Genet. 2012; 90: 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riazuddin SA, Vasanth S, Katsanis N, Gottsch JD. Mutations in AGBL1 cause dominant late-onset Fuchs corneal dystrophy and alter protein-protein interaction with TCF4. Am J Hum Genet. 2013; 93: 758–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuot A, Hewitt AW, Griggs K, et al. Association of TCF4 and CLU polymorphisms with Fuchs' endothelial dystrophy and implication of CLU and TGFBI proteins in the disease process. Eur J Hum Genet. 2012; 20: 632–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baratz KH, Tosakulwong N, Ryu E, et al. E2-2 protein and Fuchs's corneal dystrophy. The New Eng J Med. 2010; 363: 1016–1024. [DOI] [PubMed] [Google Scholar]
- 21. Riazuddin SA, McGlumphy EJ, Yeo WS, Wang J, Katsanis N, Gottsch JD. Replication of the TCF4 intronic variant in late-onset Fuchs corneal dystrophy and evidence of independence from the FCD2 locus. Invest Ophthalmol Vis Sci. 2011; 52: 2825–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li YJ, Minear MA, Rimmler J, et al. Replication of TCF4 through association and linkage studies in late-onset Fuchs endothelial corneal dystrophy. PLoS One. 2011; 6: e18044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thalamuthu A, Khor CC, Venkataraman D, et al. Association of TCF4 gene polymorphisms with Fuchs' corneal dystrophy in the Chinese. Invest Ophthalmol Vis Sci. 2011; 52: 5573–5578. [DOI] [PubMed] [Google Scholar]
- 24. Sundin OH, Broman KW, Chang HH, Vito EC, Stark WJ, Gottsch JD. A common locus for late-onset Fuchs corneal dystrophy maps to 18q21.2-q21.32. Invest Ophthalmol Vis Sci. 2006; 47: 3919–3926. [DOI] [PubMed] [Google Scholar]
- 25. Wieben ED, Aleff RA, Tosakulwong N, et al. A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2-2) gene predicts Fuchs corneal dystrophy. PLoS One. 2012; 7: e49083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Breschel TS, McInnis MG, Margolis RL, et al. A novel, heritable, expanding CTG repeat in an intron of the SEF2-1 gene on chromosome 18q21.1. Hum Mol Genet. 1997; 6: 1855–1863. [DOI] [PubMed] [Google Scholar]
- 27. Mootha VV, Gong X, Ku HC, Xing C. Association and familial segregation of CTG18.1 trinucleotide repeat expansion of TCF4 gene in Fuchs' endothelial corneal dystrophy. Invest Ophthalmol Vis Sci. 2014; 55: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cooper RS, Tayo B, Zhu X. Genome-wide association studies: implications for multiethnic samples. Hum Mol Genet. 2008; 17: R151–R155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010; 11: 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zaitlen N, Pasaniuc B, Gur T, Ziv E, Halperin E. Leveraging genetic variability across populations for the identification of causal variants. Am J Hum Genet. 2010; 86: 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marigorta UM, Navarro A. High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genet. 2013; 9: e1003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Evangelou E, Ioannidis JP. Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet. 2013; 14: 379–389. [DOI] [PubMed] [Google Scholar]
- 33. Marigorta UM, Lao O, Casals F, et al. Recent human evolution has shaped geographical differences in susceptibility to disease. BMC Genomics. 2011; 12: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krachmer JH, Purcell JJ Jr, Young CW, Bucher KD. Corneal endothelial dystrophy. A study of 64 families. Arch Ophthalmol. 1978; 96: 2036–2039. [DOI] [PubMed] [Google Scholar]
- 35. Warner JP, Barron LH, Goudie D, et al. A general method for the detection of large CAG repeat expansions by fluorescent PCR. J Med Genet. 1996; 33: 1022–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xing G, Lin CY, Wooding SP, Xing C. Blindly using Wald's test can miss rare disease-causal variants in case-control association studies. Ann Hum Genet. 2012; 76: 168–177. [DOI] [PubMed] [Google Scholar]
- 37. Genomes Project C Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature. 2010; 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005; 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 39. Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002; 70: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schaid DJ. Evaluating associations of haplotypes with traits. Genet Epidemiol. 2004; 27: 348–364. [DOI] [PubMed] [Google Scholar]
- 41. McCarthy MI. Casting a wider net for diabetes susceptibility genes. Nat Genet. 2008; 40: 1039–1040. [DOI] [PubMed] [Google Scholar]
- 42. Brockschmidt A, Todt U, Ryu S, et al. Severe mental retardation with breathing abnormalities (Pitt-Hopkins syndrome) is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Hum Mol Genet. 2007; 16: 1488–1494. [DOI] [PubMed] [Google Scholar]
- 43. Zweier C, Peippo MM, Hoyer J, et al. Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome). Am J Hum Genet. 2007; 80: 994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]