Abstract
Introduction:
In the phase IV, open-label, single-arm study NCT01203917, first-line gefitinib 250 mg/d was effective and well tolerated in Caucasian patients with epidermal growth factor receptor (EGFR) mutation-positive non–small-cell lung cancer (previously published). Here, we report EGFR mutation analyses of plasma-derived, circulating-free tumor DNA.
Methods:
Mandatory tumor and duplicate plasma (1 and 2) baseline samples were collected (all screened patients; n = 1060). Preplanned, exploratory analyses included EGFR mutation (and subtype) status of tumor versus plasma and between plasma samples. Post hoc, exploratory analyses included efficacy by tumor and plasma EGFR mutation (and subtype) status.
Results:
Available baseline tumor samples were 1033 of 1060 (118 positive of 859 mutation status known; mutation frequency, 13.7%). Available plasma 1 samples were 803 of 1060 (82 positive of 784 mutation status known; mutation frequency, 10.5%). Mutation status concordance between 652 matched tumor and plasma 1 samples was 94.3% (95% confidence interval [CI], 92.3–96.0) (comparable for mutation subtypes); test sensitivity was 65.7% (95% CI, 55.8–74.7); and test specificity was 99.8% (95% CI, 99.0–100.0). Twelve patients of unknown tumor mutation status were subsequently identified as plasma mutation-positive. Available plasma 2 samples were 803 of 1060 (65 positive of 224 mutation status-evaluable and -known). Mutation status concordance between 224 matched duplicate plasma 1 and 2 samples was 96.9% (95% CI, 93.7–98.7). Objective response rates are as follows: mutation-positive tumor, 70% (95% CI, 60.5–77.7); mutation-positive tumor and plasma 1, 76.9% (95% CI, 65.4–85.5); and mutation-positive tumor and mutation-negative plasma 1, 59.5% (95% CI, 43.5–73.7). Median progression-free survival (months) was 9.7 (95% CI, 8.5–11.0; 61 events) for mutation-positive tumor and 10.2 (95% CI, 8.5–12.5; 36 events) for mutation-positive tumor and plasma 1.
Conclusion:
The high concordance, specificity, and sensitivity demonstrate that EGFR mutation status can be accurately assessed using circulating-free tumor DNA. Although encouraging and suggesting that plasma is a suitable substitute for mutation analysis, tumor tissue should remain the preferred sample type when available.
Keywords: EGFR mutation, Gefitinib, non–small-cell lung cancer, Caucasian, Circulating-free tumor DNA
Successful analysis of epidermal growth factor receptor (EGFR) mutations in advanced non–small-cell lung cancer (NSCLC) has provided many patients with EGFR mutation-positive disease with the opportunity to receive optimal, targeted treatments.1,2 Tumor tissue is considered the preferred definitive standard sample type for EGFR mutation analysis3; however, for many patients, this sample type is not available (~10–15% from author’s clinical experience, and ~23% in the United Kingdom in 2011).4 A molecular-based treatment decision in these patients can therefore be problematic, not only at diagnosis but also at progression, to detect resistance mutations (e.g., T790M) in those who experience disease progression after first-line treatment with EGFR tyrosine kinase inhibitors (TKIs).
Consequently, focus has turned to evaluating the use of surrogate sample types for EGFR mutation analysis, the aim of which is to identify the molecular characteristics of tumors from patients who do not have tumor tissue samples.5–7 One alternative sample type is circulating-free tumor DNA (ctDNA), which is obtained through less invasive methods to source tumor DNA, such as plasma or serum samples. The available evidence for the use of ctDNA for the analysis of EGFR mutations is encouraging, a brief review of which is given below.
In 2009, results were published from a large Spanish study which assessed EGFR mutation detection in 164 ctDNA samples from the serum of patients with EGFR mutation-positive NSCLC as determined from tumor tissue testing.8 Peptide nucleic acid clamp analysis identified EGFR mutations in 97 ctDNA samples, a sensitivity of 59.2%. A Chinese study of 35 matched ctDNA and tumor tissue samples also published in 2009 successfully analyzed all ctDNA samples using digital polymerase chain reaction (PCR).9 Subsequent to the publication of these studies, a review published by Aung et al.10 discussed the current status and future potential of EGFR mutation testing from ctDNA and concluded that plasma ctDNA would be a viable alternative/additional source of DNA, particularly as advances in PCR technology had allowed the analysis of point mutations in EGFR, KRAS, BRAF, and PIK3CA genes from ctDNA isolated from patients’ serum or plasma.11–14 The 2011 publication by Liu et al.15 reported the successful EGFR mutation analysis of 86 matched plasma-derived ctDNA and formalin-fixed, paraffin-embedded samples using a Scorpion-amplification refractory mutation system (ARMS). Of the 40 tumor samples identified as EGFR mutation-positive, 27 were correctly identified in ctDNA, a sensitivity of 67.5%. More recently, EGFR mutation testing of 194 serum-derived ctDNA samples was undertaken as part of a preplanned, exploratory analysis of the Japanese subset of the IPASS study.5 EGFR mutations (tested using the ARMS-based DxS EGFR mutation Test Kit [DxS, Manchester, UK]) were successfully detected in all 194 ctDNA samples, with sensitivity of 43.1%. Moreover, in the 22 patients where mutations were identified in ctDNA and tumor samples, the mutation subtypes were identical in 21 cases. Ongoing real-world studies are also evaluating the utility of plasma ctDNA-based EGFR mutation testing compared with tumor DNA. The noninterventional ASSESS study (Europe and Japan)16 and interventional IGNITE study (Asia-Pacific region and Russia)17 are non-comparative studies of EGFR mutation status in patients with advanced NSCLC of adenocarcinoma and non-adenocarcinoma histologies. Concordance between EGFR mutation status obtained through tissue/cytology and blood (plasma)-based testing is a primary and secondary objective of the ASSESS and IGNITE studies, respectively. Results from these large, international studies will help establish whether plasma is a suitable, less invasive sample type for reliably determining the EGFR mutation status of patients with advanced NSCLC, thus informing the use of this sample type in patients without available/evaluable tumor samples.
One study which has explored per-protocol the use of plasma ctDNA alongside the more traditional tumor tissue for EGFR mutation analysis is the phase IV, prospective, open-label, multicenter, single-arm, first-line study of the EGFR TKI gefitinib in Caucasian patients with EGFR mutation-positive, advanced NSCLC (NCT01203917). The efficacy and tolerability results of this study have been published previously.18 Briefly, first-line gefitinib was effective in this patient population, as assessed by objective response rate (ORR; 70%, 95% confidence interval [CI], 60.5–77.7%), and supported by disease control rate (91%), median progression-free survival (PFS, 9.7 mo), and median overall survival (19.2 mo).18 First-line gefitinib was well tolerated, with adverse events consistent with the characterized tolerability/safety profile for gefitinib in previous studies.19–22
Patients with NSCLC initially entered the screening phase of the study (n = 1060), and eligible patients were progressed to enrollment on the basis of tumor tissue availability and a positive EGFR mutation status. Here, we report a comparison of baseline tumor EGFR mutation status in all screened patients with evaluable results for baseline plasma. Further analyses compared plasma-derived ctDNA EGFR mutation status in duplicate baseline plasma samples from the same patient to evaluate reliability of methodology in non-tumor samples. Post hoc, exploratory analyses of efficacy (ORR and PFS) according to tumor-derived DNA and plasma-derived ctDNA EGFR mutation status (and mutation subtype) are also reported.
MATERIALS AND METHODS
Study Design and Patients
Full details of this study (NCT01203917) have been published previously.18 Briefly, the gefitinib follow-up measure study was a prospective, open-label, multicenter, single-arm study to characterize the efficacy, safety, and tolerability of gefitinib (250 mg/d) as first-line treatment of Caucasian patients with activating, sensitizing, EGFR mutation-positive, locally advanced, or metastatic NSCLC. Patient eligibility criteria have been published previously18 and included mandatory provision of tumor samples and duplicate plasma samples for EGFR mutation testing at baseline; in addition, optional plasma samples were collected at disease progression (see below for EGFR mutation subtype eligibility criteria).
All patients provided written, informed consent, including provision for collection of tumor and plasma samples for biomarker analyses. Study approval was obtained from independent ethics committees at each institution. The study was conducted in accordance with the Declaration of Helsinki, the International Conference on Harmonisation/Good Clinical Practice, applicable regulatory requirements, and AstraZeneca’s policy on bioethics.
Assessments
Tumor assessment by computed tomography scan was performed every 6 weeks. The primary end point of the study, ORR (complete response plus partial response) by investigator assessment, was determined by the Response Evaluation Criteria In Solid Tumors23 version 1.1. PFS (secondary end point; time from start of study treatment to date of objective tumor progression [excluding clinical deterioration without evidence of objective progression]) was also determined by Response Evaluation Criteria In Solid Tumors 1.1. Preplanned exploratory objectives included a comparison of baseline tumor EGFR mutation status with evaluable results for baseline plasma, a comparison of plasma-derived ctDNA EGFR mutation status in duplicate baseline plasma samples (plasma 1 and 2 samples), and a comparison of baseline and progression plasma samples, from the same patient. Post hoc exploratory analyses included efficacy of gefitinib according to tumor and plasma EGFR mutation (and mutation subtype) status.
DNA Extraction and EGFR Mutation Analysis
One tumor sample (mandatory) and two plasma samples (mandatory; plasma 1 and plasma 2) were collected from each patient at baseline (screening) for DNA extraction and EGFR mutation analysis. An optional plasma sample was collected at disease progression.
Whole blood samples were collected into venous blood collection tubes using ethylenediaminetetraacetic acid as anticoagulant. Samples were mixed thoroughly and plasma isolated within 2 to 4 hours of sample collection by centrifugation at approximately 2000 g for 10 minutes (at 4°C or room temperature for pre-chilled samples). Once isolated, plasma samples were centrifuged again (as above), before transferring into cryovials and freezing at −70°C within 4 hours of collection. Samples were not thawed until the time of processing.
A central laboratory (LabCorp, Durham, NC) performed DNA extraction and mutation analysis of both the tumor and plasma samples. Tumor DNA was extracted using the Qiagen QIAamp DNA Mini Kit (Qiagen, Crawley, UK). whereas ctDNA was extracted from plasma using the Qiagen QIAamp Circulating Nucleic Acid Kit (Qiagen). Sample processing details, including modifications to processes, are shown in Supplementary Appendix Table 1 (Supplementary Digital Content 1, http://links.lww.com/JTO/A638).
EGFR mutation status of all samples was assessed using a Scorpion ARMS-based EGFR mutation detection kit (Therascreen EGFR RGQ PCR kit; Qiagen, Crawley, UK), which detects 29 mutations across the EGFR gene. For tumor samples, all mutations in the kit were analyzed. Methodological and technical details for EGFR mutation testing, including modifications to tumor/ctDNA analysis kit instructions and data quality control details, are shown in Supplementary Appendix Table 1 (Supplementary Digital Content 1, http://links.lww.com/JTO/A638). A comparison of the Qiagen kit used for ctDNA EGFR mutation analysis in the current study and the previous version of the DxS kit used in the IPASS study19,22 are shown in Supplementary Appendix Table 2 (Supplementary Digital Content 1, http://links.lww.com/JTO/A638).
EGFR mutation status was assigned to baseline tumor samples according to agreed eligibility criteria: positive, more than or equal to one activating, sensitizing EGFR mutation with no ineligible mutations; positive ineligible, more than or equal to one ineligible mutation (exon 20 point mutations T790M; S768I; exon 20 insertions, either alone or in combination with another ineligible mutation or a sensitizing mutation [see Douillard et al.17 appendix]); negative, no mutations detected; and unknown, no mutation results available (exhaustion of samples, poor quality, or low DNA yield).
Plasma samples were analyzed for exon 19 deletions, L858R point mutation, and the T790M point mutation only. The following EGFR mutation status was assigned to individual plasma samples: positive, more than or equal to 1 mutation (L858R, exon 19 deletions [19 different mutations], and T790M); negative, no mutations were detected; and unknown, no mutation results available (no sample or poor quality).
Statistical Analysis
Data were analyzed using a data cutoff (August 15, 2012) at 6 months after the last patient had started study treatment. ORR (primary end point) was calculated from investigator data and summarized in the full analysis set (FAS; all screened patients with an eligible, positive EGFR mutation status who received ≥1 dose of gefitinib), with 95% CIs (Wilson score intervals). PFS was estimated with 95% CIs (FAS) using Kaplan-Meier methods and Greenwood’s formula (Greenwood 1926)24 (PFS rates) and Brookmeyer and Crowley’s method (median PFS).
Baseline tumor and plasma 1 EGFR mutation status (overall and by mutation subtype), in patients in the screened population who were evaluable for both samples, were compared with rates (percentages) and 95% CIs (Clopper-Pearson method) calculated for concordance, sensitivity, specificity, and positive- and negative-predictive value. Duplicate baseline plasma 1 and plasma 2 samples and baseline and progression plasma samples were compared, with rates and 95% CIs calculated.
RESULTS
Patients
Of 1060 patients screened from 13 countries, 1033 (97.5%) provided baseline tumor samples. Of these, EGFR mutation status could be determined for 859 patients (81.0%), and 118 patients (11.1%) were assigned an eligible EGFR mutation-positive status (mutation frequency, 13.7%) and enrolled in the study from September 8, 2010, to February 15, 2012; gefitinib treatment was started in 106 of these patients (FAS population) (Fig. 1). One additional patient of EGFR mutation-positive ineligible status was treated with gefitinib in error. Of the 201 patients (19%) for whom tumor EGFR mutation status could not be determined (unevaluable/unavailable), tissue was not provided from 27 patients (2.6%) who consented, and 174 patients (16.4%) could not be evaluated for EGFR mutation status (100 [9.4%] failed histopathology, 42 [4.0%] yielded insufficient DNA, 2 [0.2%] failed EGFR mutation analysis, and 30 [2.8%] were not shipped from the study site in time). A flow diagram of the accountability of tumor samples is presented in Figure 2.
FIGURE 1.

Patient flow diagram. aAll screened patients. Used to calculate the correlation between clinical characteristics and tumor EGFR mutation status and the comparison of EGFR mutation status between tumor DNA and plasma-derived circulating-free tumor DNA. bOne patient of EGFR mutation-positive ineligible status was treated in error and included in the evaluable-for-safety population. A total of 107 patients therefore started study treatment. cFull analysis set population. Used to summarize efficacy data and for the comparison of EGFR mutation status in plasma and tumor samples. dNumber of patients with EGFR mutation-positive tumors (n = 118) used as the denominator for the percentage calculation. eNumber of patients started on treatment (n = 107) used as the denominator for the percentage calculation. EGFR, epidermal growth factor receptor. Reproduced, in part, from Douillard et al.17 Br J Cancer 2014;110:55–62.
FIGURE 2.
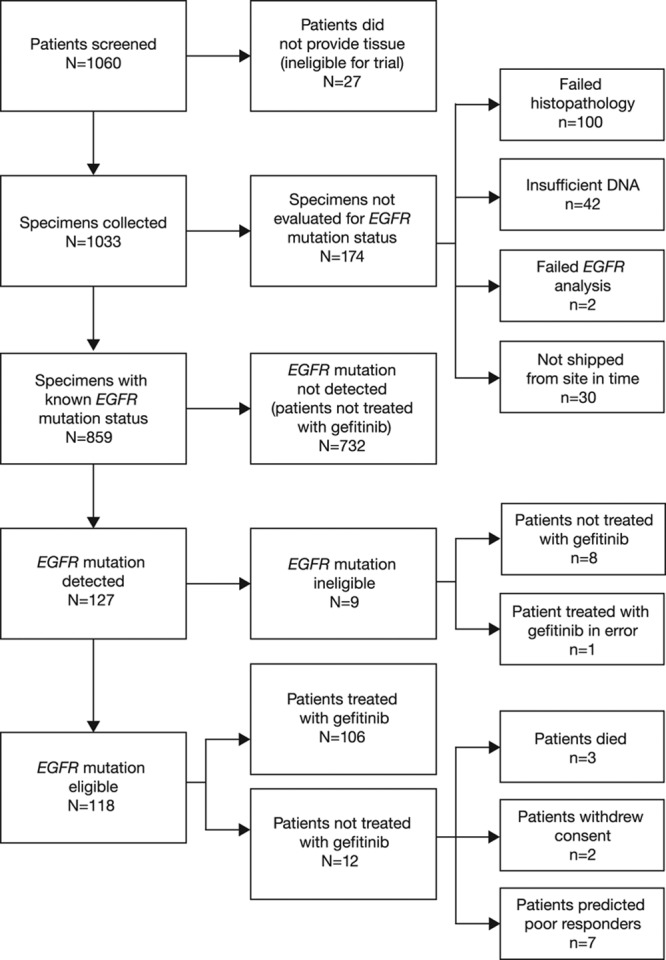
Tumor sample flow diagram. EGFR, epidermal growth factor receptor.
A total of 803 of 1060 screened patients (75.8%) provided duplicate baseline plasma 1 and 2 samples. Of these, EGFR mutation status could be determined using plasma 1 samples for 784 patients (74.0%), and 82 patients (7.7%) were assigned an EGFR mutation-positive status (mutation frequency, 10.5%). To achieve a similar measure of accuracy for analysis of plasma 2 samples and avoid unnecessary sample use, EGFR mutation status was determined for 224 plasma 2 samples whose corresponding plasma 1 sample was EGFR mutation status-known (all EGFR mutation-positive plasma 1 for whom plasma 2 was available, and a matched number of EGFR mutation-negative). Of these 224 plasma 2 samples chosen for analysis, 65 were assigned an EGFR mutation-positive status.
Demographics and baseline characteristics by EGFR mutation status derived from baseline tumor samples (FAS population), plasma 1 samples, and plasma 2 samples are presented in Supplementary Appendix Table 3 (Supplementary Digital Content 1, http://links.lww.com/JTO/A638) and were similar for all populations. Exon 19 deletions and L858R point mutations were the most commonly occurring mutations.
Comparison of EGFR Mutation Status in Matched Baseline Tumor and Plasma 1 Samples (Screened Population)
Fewer patients were identified as having EGFR mutation-positive status using plasma 1-derived ctDNA (detection rate, 10.5%; 82 of 784 patients) than with tumor-derived DNA (detection rate, 13.7%, 118 of 859 patients). Due to technical problems with tumor samples (e.g., low tumor content, poor sample quality, insufficient quantity, poor/inappropriate fixation, no DNA), 201 of 1060 screened patients (19.0%) had an unknown EGFR mutation status; 12 of these patients were subsequently found to have a positive EGFR mutation status in their corresponding plasma 1 samples.
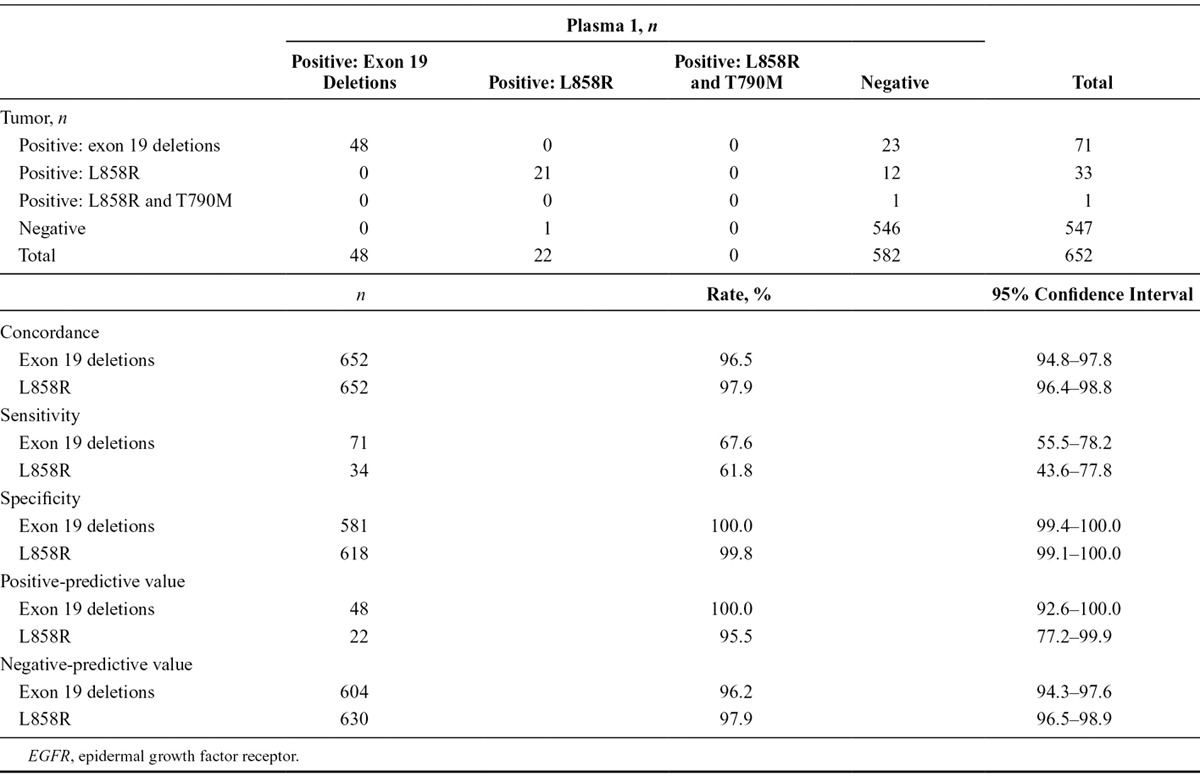
A total of 652 patients provided matched baseline tumor and plasma 1 samples that were evaluable for EGFR mutation status using both sample types. A comparison of EGFR mutation status between tumor and plasma 1 samples is shown in Table 1. Concordance between baseline tumor and plasma 1 EGFR mutation status for patients evaluable for both samples was 94.3% (95% CI, 92.3–96.0), with a sensitivity of 65.7% (95% CI, 55.8–74.7) and specificity of 99.8% (95% CI, 99.0–100.0). Positive-predictive value and negative-predictive value of EGFR mutation status detection between baseline tumor and plasma 1 samples for patients evaluable for both samples are also shown. A comparison of EGFR mutation status by mutation subtype for baseline tumor and plasma 1 samples is presented in Table 2. When analyzed by mutation subtype, concordance between baseline tumor and plasma 1 EGFR mutation status for patients evaluable for both samples was 96.5% (95% CI, 94.8–97.8) for exon 19 deletions and 97.9% (95% CI, 96.4–98.8) for L858R point mutations; sensitivity was 67.6% (95% CI, 55.5–78.2) and 61.8% (95% CI, 43.6–77.8), and specificity was 100.0% (95% CI, 99.4–100.0) and 99.8% (95% CI, 99.1–100.0), respectively.
TABLE 1.
EGFR Mutation Status Summary, Concordance, Sensitivity, Specificity, and Positive- and Negative-Predictive Value for Tumor vs. Plasma 1 Circulating-Free Tumor DNA Samples by EGFR Mutation Status (Screened Patients Evaluable for Both Samples, n = 652)
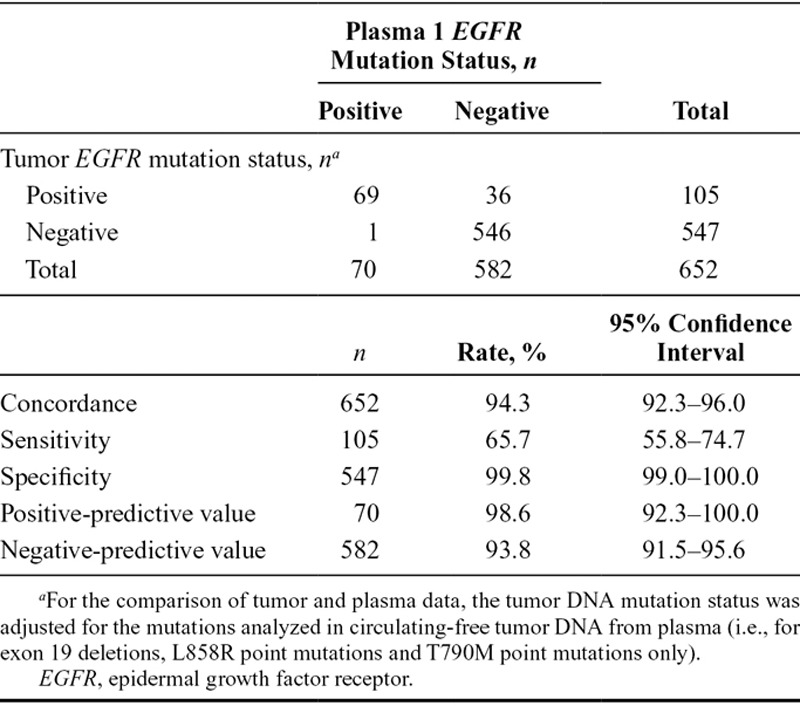
TABLE 2.
EGFR Mutation Status Summary, Concordance, Sensitivity, Specificity, and Positive- and Negative-Predictive Value for Tumor vs. Plasma 1 Circulating-Free Tumor DNA Samples by EGFR Mutation Subtype (Screened Patients Evaluable for Both Samples, n = 652)
Of the 547 patients considered to have an EGFR mutation-negative status by analysis of tumor DNA, one patient was considered to have an EGFR mutation-positive status by analysis of plasma 1 ctDNA, giving a false-positive rate of 0.2% (1 of 547). However, further investigation indicated that this was a false-positive result due to signal drift and was not a genuine positive amplification of the ctDNA (see Supplementary Appendix, Supplementary Digital Content 1, http://links.lww.com/JTO/A638, for further details).
Comparison of EGFR Mutation Status in Duplicate Baseline Plasma Samples (Screened Population)
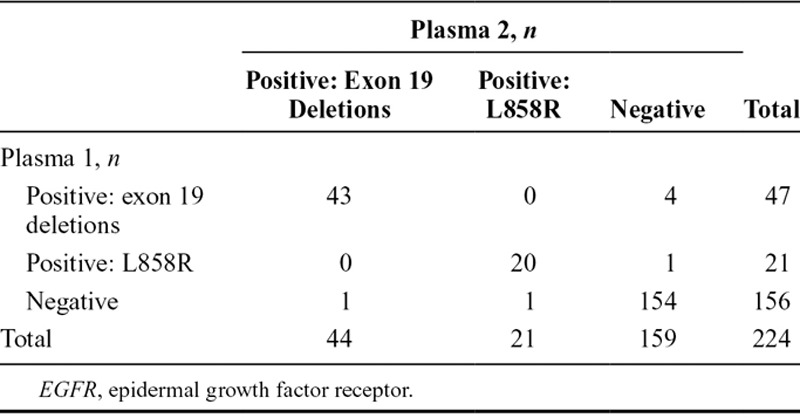
Of the 803 screened patients who provided duplicate baseline plasma samples, 224 were evaluated and known for ctDNA EGFR mutation status. A total of 63 were identified with EGFR mutation-positive status in both samples, seven were EGFR mutation-positive in one plasma sample only, and 154 were negative in both samples (Table 3). Concordance between duplicate baseline plasma sample EGFR mutation status (regardless of mutation status) for patients evaluable for both samples was 96.9% (95% CI, 93.7–98.7) (Table 3). A summary of EGFR mutation status in duplicate baseline plasma samples by mutation subtype for patients evaluable for both samples is presented in Table 4.
TABLE 3.
EGFR Mutation Status Comparisons for Plasma 1 vs Plasma 2 Circulating-Free Tumor DNA Samples by EGFR Mutation Status Summary and Concordance (Screened Patients Evaluable for Both Samples, n = 224)
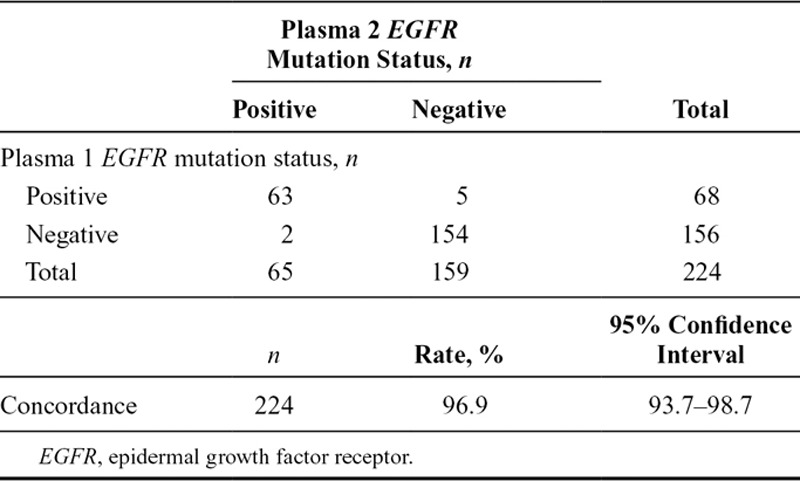
TABLE 4.
EGFR Mutation Status Comparisons for Plasma 1 vs Plasma 2 Circulating-Free Tumor DNA Samples by EGFR Mutation Status Summary by EGFR Mutation Subtype (Screened Patients Evaluable for Both Samples, n = 224)
Comparison of EGFR Mutation Status in Baseline and Progression Plasma Samples (Screened Population)
A total of 12 patients provided matched baseline (plasma 1) and progression plasma samples. EGFR mutation status concordance rate was 75.0% (95% CI, 42.8–94.5); mutation subtype results agreed in eight patients (caution is advised when interpreting these data due to small n numbers). Four patients had differences in EGFR mutation status: no mutations were detected in progression plasma ctDNA from three patients in which exon 19 deletions were detected in baseline plasma 1 ctDNA; one patient with an exon 19 deletion in baseline plasma 1 ctDNA acquired an additional T790M mutation in progression plasma ctDNA.
Post Hoc Analysis of Efficacy of Gefitinib According to Tumor and Plasma EGFR Mutation Status (FAS Population)
As of data cutoff, an objective response was seen in 74 of 106 patients in the FAS population whose tumors were EGFR mutation-positive, with an ORR of 69.8% (95% CI, 60.5–77.7) based on investigator assessment (n = 2 with complete response; n = 72 with partial response) (Fig. 3).
FIGURE 3.
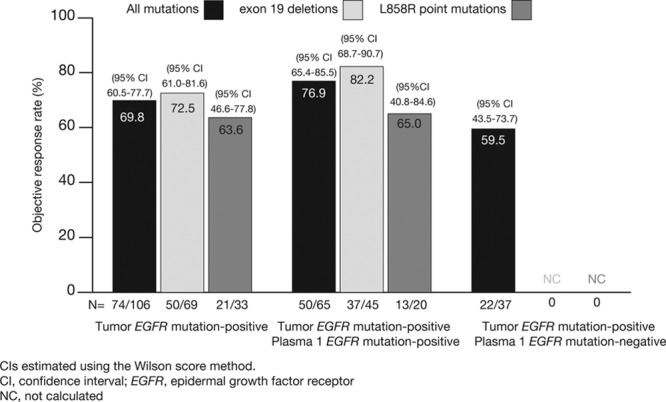
Objective response rate according to EGFR mutation (and subtype) status for patients who were tumor EGFR mutation positive, tumor and plasma 1 EGFR mutation positive, and tumor EGFR mutation positive and plasma EGFR mutation negative. Reproduced, in part, from Douillard et al.17 Br J Cancer 2014;110:55–62.
For patients with matched tumor and plasma 1 samples, an ORR of 76.9% (95% CI, 65.4–85.5) was observed among those with a positive EGFR mutation result in both sample types (n = 50/65) (Fig. 3). An ORR of 59.5% (95% CI, 43.5–73.7) was observed among the 22 of 37 patients found to be mutation-positive by tumor but mutation-negative by plasma. Results were also consistent when analyzed by mutation subtypes: ORR for patients with both EGFR mutation-positive tumor and plasma 1 for exon 19 deletions (n = 45) 82.2% (95% CI, 68.7–90.7) and L858R (n = 20) 65.0% (95% CI, 40.8–84.6).
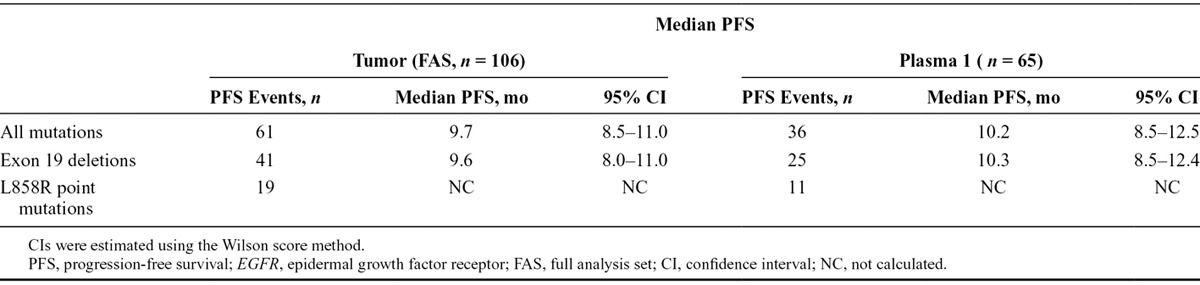
Median PFS for patients with matched tumor and plasma 1 samples that were positive for EGFR mutation in both sample types was 10.2 months (36 events; 95% CI, 8.5–12.5) (Table 5), consistent with those patients who were EGFR mutation-positive by tumor, regardless of plasma ctDNA EGFR mutation status (median PFS, 9.7 mo [95% CI, 8.5–11.0]). When analyzed by mutation subtypes, median PFS for patients with matched tumor and plasma 1 samples that were positive in both sample types for exon 19 deletions (25 events) was 10.3 months (95% CI, 8.5–12.4), consistent with the PFS for patients whose tumors were exon 19 deletion-positive (41 events; median PFS, 9.6 mo; 95% CI, 8.0–11.0). There were an insufficient number of events to assess the PFS of patients with samples that were L858R mutation-positive (tumor or ctDNA).
TABLE 5.
Median PFS for Patients With Tumor and Plasma 1 Circulating-Free Tumor DNA Samples Overall and by EGFR Mutation Subtypes (FAS Population)
DISCUSSION
The gefitinib follow-up measure study18 is, to our knowledge, the first prospective, large-scale study of first-line gefitinib to be conducted in Caucasian patients with EGFR mutation-positive advanced NSCLC. Previously published efficacy and tolerability results from the study show that first-line gefitinib is effective and well tolerated in this patient population.18 The preplanned and exploratory biomarker analyses reported here show a high rate of concordance for EGFR mutation status (including mutation subtypes) between tumor tissue DNA and plasma ctDNA and between duplicate plasma ctDNA samples. Tumor EGFR mutation status could not be determined (unevaluable/unavailable) for 19% of patient samples. Together, these results suggest that a single plasma-derived ctDNA sample may be considered appropriate for assessment of EGFR mutation status when tumor tissue is unavailable or exhausted.
The plasma ctDNA EGFR mutation analysis results reported here demonstrated 94.3% concordance, 99.8% specificity, and 65.7% sensitivity, an improvement on previous analysis of ctDNA, for example, in the IPASS study.5 This improvement may be due to a number of reasons, including different sample type (plasma versus serum), different DNA extraction method (the current study used a bespoke ctDNA extraction method), or a modified ARMS mutation detection kit. With regard to concordance between detection of EGFR mutations in matched tumor and plasma samples, a recent study of 111 Chinese patients with stage I–IV NSCLC reported 71.2% concordance, 35.6% sensitivity, and 95.5% specificity using enriched PCR and sequencing.25 Similarly, Yung et al.9 reported a specificity and sensitivity (using microfluidics digital PCR) of EGFR mutation status with ctDNA and matched tumor samples from 35 Chinese patients with NSCLC of 100% and 92%, respectively.
When considering ctDNA sample EGFR mutation status as patient selection for EGFR TKI therapy, the results we have reported here suggest that use of this alternative sample type may be appropriate in patients without available tumor samples, as patients with EGFR mutation-positive ctDNA, regardless of mutation subtype, had a similar ORR to patients with EGFR mutation-positive tumors (76.9% and 69.8%, respectively). A study of 50 samples (32 pleural fluid; 18 plasma) from Chinese patients with NSCLC previously treated with EGFR TKIs (line of therapy not stated) also found that an EGFR mutation-positive status detected by these surrogate samples was a significant and consistently good indicator of response to EGFR TKIs (pleural fluid-positive ORR 81.3% with direct sequencing, 72.7% with ARMS; plasma-positive ORR 80.0% with ARMS).15
It is widely accepted that there are challenges in EGFR mutation testing practice. In the past, this has been particularly evident in the Asia-Pacific region, where a lack of access and/or adoption of testing was a barrier to large-scale testing.26 A 2011 consensus meeting to discuss EGFR mutation testing in East Asia considered tissue acquisition and pre-test sample evaluation as important steps to increase specificity and sensitivity and to thus help standardize mutation test methodology in the region.26 Data generated from large-scale studies of EGFR mutation frequency, such as the PIONEER study,27 are confirming that large-scale testing across countries is feasible, can be standardized, and can result in a high analysis success rate. It is hoped that data generated from ongoing diagnostic studies such as ASSESS16 and IGNITE17 will add to this growing body of evidence. In addition, patient monitoring through ctDNA testing during treatment and at progression might provide important information on progression mechanisms. Our limited series of 12 patients showed that in one case a resistance mutation (T790M) was identified. Such information might be useful for managing patients with progression. The issue of re-biopsy at progression is presently a major concern and ctDNA testing could potentially be an alternative approach, as recently published28 and more generally discussed.29
A challenge for those undertaking testing is that mutation detection kits are often validated solely on tumor-derived DNA. In the future, kits for use on surrogate sample types such as cytology or plasma may become available as the body of evidence increases. As many patients with NSCLC attending clinic do not have tumor tissue samples available (10–15% from author’s clinical experience), this could limit the number of patients able to benefit from the current mutation testing environment. Progress is being made, however, as highlighted by the Chinese State Food and Drug Administration’s recent approval for the clinical use of three ADx-ARMS mutation tests (including EGFR) produced by Amoy Dx (Xiamen, China) for testing of fresh, frozen, and paraffin-embedded tissue as well as blood and plasma samples, so addressing the current area of unmet need in patients with unevaluable/unavailable tumor samples at diagnosis.
A conundrum challenging the NSCLC community is the apparent lack of detectable mutations in the ctDNA of some patients. As noted above, the EGFR mutation detection rate in the study reported here was 13.7% in tumor DNA and 10.5% in matched ctDNA; it is unclear whether this (commonly reported) lower detection rate in ctDNA is due to patient biology (tumor heterogeneity or biologic evolution of the disease) or technical limitations of current technologies (sampling, extraction, and mutation detection). To help minimize any ctDNA false-negative results due to technical difficulties, a highly sensitive mutation detection kit should be used, for example, ARMS technology,15 or one of the new more sophisticated techniques, for example, Beads, Emulsions, Amplification, and Magentics (BEAMing) or digital PCR. Indeed, the use of ARMS technology in the study reported here helped to achieve a 94.3% concordance and 65.7% sensitivity between matched tumor and plasma samples.
In light of these challenges/gaps in current knowledge, coupled with the unmet medical need in diagnosis, it is hoped that the results of this study, in conjunction with data from ongoing studies, will help to inform the NSCLC community about several aspects of mutation testing, including the use of sample types in diagnostic practice which are surrogates of tumor tissue when tissue is unavailable; EGFR mutation test processes and methodology; the use of multiple sample types in the assessment of EGFR mutation status; and the impact of EGFR mutation status on therapy choice. These data may help drive improvements in the EGFR mutation testing environment, ensuring that patients have access to testing and are treated appropriately on the basis of the molecular features of their disease. To date, there are no published guidelines for the use of ctDNA for EGFR mutation analysis in the absence of NSCLC tumor samples. Such a lack of formal direction has necessitated clinicians to either limit the use of this sample type to the research setting or form their own clinical perspective/opinion on the use of surrogate samples to ascertain mutation status. Therefore, although tumor tissue should be considered the preferred sample type for mutation analysis when available, the encouraging results from ctDNA analysis reported here may in the future help to address this current area of unmet need for those patients who do not have a tumor tissue sample, that is, their sample is unavailable or exhausted during diagnosis.
In summary, the high concordance, specificity, and sensitivity reported here between DNA and ctDNA EGFR mutation status demonstrates that EGFR mutation status can be accurately assessed using ctDNA. Although these results are encouraging and suggest that plasma is a suitable substitute for mutation analysis regardless of mutation subtype, tumor tissue should be considered the preferred sample type when available.
Acknowledgments
This study was funded by AstraZeneca. We thank the patients and investigators for their participation in this study. We thank Gael McWalter (AstraZeneca) for the implementation of the biomarker analyses at LabCorp, and Sarah Lewis, from Complete Medical Communications, who provided medical writing support, funded by AstraZeneca. IRESSA is a trademark of the AstraZeneca group of companies.
Footnotes
Disclosure: Dr. Douillard has received advisory board and symposia fees from AstraZeneca, Roche, Merck Serono, Amgen, Boehringer Ingelheim, Pfizer, Sanofi-aventis, GlaxoSmithKline, and Bayer Healthcare Pharmaceuticals and has received a research grant from Merck Serono. Dr. Cole, Ms. McWalter, Dr. Walker, Mr. Dearden, Mr. Webster, Dr. Milenkova, and Dr. McCormack are employees of AstraZeneca and hold shares in AstraZeneca. All other authors declare no conflict of interest.
REFERENCES
- 1.Vallières E, Peters S, Van Houtte P, Dalal P, Lim E. Therapeutic advances in non-small cell lung cancer. Thorax. 2012;67:1097–1101. doi: 10.1136/thoraxjnl-2011-201043. [DOI] [PubMed] [Google Scholar]
- 2.Li T, Kung HJ, Mack PC, Gandara DR. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol. 2013;31:1039–1049. doi: 10.1200/JCO.2012.45.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol. 2013;8:823–859. doi: 10.1097/JTO.0b013e318290868f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Health and Social Care Information Centre NLCA. National Lung Cancer Audit Report 2012. Available at: http://www.hqip.org.uk/assets/NCAPOP-Library/NCAPOP-2012–13/Lung-Cancer-National-Audit-Report-pub-2012.pdf. Accessed September 27, 2013. [Google Scholar]
- 5.Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol. 2012;7:115–121. doi: 10.1097/JTO.0b013e3182307f98. [DOI] [PubMed] [Google Scholar]
- 6.Goto K, Satouchi M, Ishii G, et al. An evaluation study of EGFR mutation tests utilized for non-small-cell lung cancer in the diagnostic setting. Ann Oncol. 2012;23:2914–2919. doi: 10.1093/annonc/mds121. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Lu Y, Zhu G, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol. 2013;66:1065–1069. doi: 10.1136/jclinpath-2013-201728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosell R, Moran T, Queralt C, et al. Spanish Lung Cancer Group. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 9.Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15:2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- 10.Aung KL, Board RE, Ellison G, et al. Current status and future potential of somatic mutation testing from circulating free DNA in patients with solid tumours. Hugo J. 2010;4:11–21. doi: 10.1007/s11568-011-9149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Board RE, Ellison G, Orr MC, et al. Detection of BRAF mutations in the tumour and serum of patients enrolled in the AZD6244 (ARRY-142886) advanced melanoma phase II study. Br J Cancer. 2009;101:1724–1730. doi: 10.1038/sj.bjc.6605371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Board RE, Wardley AM, Dixon JM, et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat. 2010;120:461–467. doi: 10.1007/s10549-010-0747-9. [DOI] [PubMed] [Google Scholar]
- 13.Hodgson DR, Wellings R, Orr MC, et al. Circulating tumour-derived predictive biomarkers in oncology. Drug Discov Today. 2010;15:98–101. doi: 10.1016/j.drudis.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer. 2007;97:778–784. doi: 10.1038/sj.bjc.6603949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Liu B, Li XY, et al. A comparison of ARMS and direct sequencing for EGFR mutation analysis and tyrosine kinase inhibitors treatment prediction in body fluid samples of non-small-cell lung cancer patients. J Exp Clin Cancer Res. 2011;30:111. doi: 10.1186/1756-9966-30-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reck M, Hagiwara K, Tjulandin S, et al. Investigating the utility of plasma-derived circulating-free DNA for the detection of EGFR mutations in European and Japanese patients with advanced NSCLC: ASSESS study design. J Thorac Oncol. 2013:S483. doi: 10.1016/j.jtho.2016.05.036. 8(Supplement 2) [DOI] [PubMed] [Google Scholar]
- 17.Han B, Tjulandin S, Normanno N, et al. A diagnostic study to determine the prevalence of EGFR mutations in Asian and Russian patients with NSCLC of adenocarcinoma and non-adenocarcinoma histology: IGNITE study design. J Thorac Oncol. 2013;8(Supplement 2):S1098 [Google Scholar]
- 18.Douillard JY, Ostoros G, Cobo M, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Cancer. 2014;110:55–62. doi: 10.1038/bjc.2013.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol. 2011;29:2866–2874. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 20.Kim ES, Hirsh V, Mok T, et al. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet. 2008;372:1809–1818. doi: 10.1016/S0140-6736(08)61758-4. [DOI] [PubMed] [Google Scholar]
- 21.Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 22.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 23.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 24.Greenwood M. 1926: The natural duration of cancer. Reports on public health and medical subjects. Her Majesty's Stationery Office. Vol 33:1–26. [Google Scholar]
- 25.Zhao X, Han RB, Zhao J, et al. Comparison of epidermal growth factor receptor mutation statuses in tissue and plasma in stage I-IV non-small cell lung cancer patients. Respiration. 2013;85:119–125. doi: 10.1159/000338790. [DOI] [PubMed] [Google Scholar]
- 26.Salto-Tellez M, Tsao MS, Shih JY, et al. Clinical and testing protocols for the analysis of epidermal growth factor receptor mutations in East Asian patients with non-small cell lung cancer: a combined clinical-molecular pathological approach. J Thorac Oncol. 2011;6:1663–1669. doi: 10.1097/JTO.0b013e318227816a. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9:154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20:1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]


