SUMMARY
Inhibition of muscleblind-like (MBNL) activity due to sequestration by microsatellite expansion RNAs is a major pathogenic event in the RNA-mediated disease myotonic dystrophy (DM). Although MBNL1 and MBNL2 bind to nascent transcripts to regulate alternative splicing during muscle and brain development, another major binding site for the MBNL protein family is the 3′ untranslated region of target RNAs. Here, we report that depletion of Mbnl proteins in mouse embryo fibroblasts leads to mis-regulation of thousands of alternative polyadenylation events. HITS-CLIP and minigene reporter analyses indicate that these polyadenylation switches are a direct consequence of MBNL binding to target RNAs. Mis-regulated alternative polyadenylation also occurs in skeletal muscle in a mouse polyCUG model and human DM resulting in the persistence of neonatal polyadenylation patterns. These findings reveal a novel developmental function for MBNL proteins and demonstrate that DM is characterized by mis-regulation of pre-mRNA processing at multiple levels.
INTRODUCTION
The dynamic processing of primary RNA transcripts, including alternative splicing (AS) and alternative polyadenylation (APA), generates the functional mammalian RNA repertoire essential for normal tissue development and maintenance (Di Giammartino et al., 2011; Kornblihtt et al., 2013). For protein-coding genes, AS produces a vast diversity of isoforms that may vary in coding and non-coding regions. APA may also alter coding regions but primarily creates alternative 3′ untranslated regions (3′ UTRs) that display variable interactions with trans-acting factors, including RNA-binding proteins (RBPs) and microRNAs, leading to modulation of RNA localization, translation and turnover (Tian and Manley, 2013).
Regulation of AS and APA requires the recruitment of numerous RBPs and RBP-RNA complexes to nascent transcripts and alterations in the activities of these transacting factors are associated with a number of human diseases (Danckwardt et al., 2008; Singh and Cooper, 2012). For example, global defects in AS due to SMN loss-of-function have been implicated in spinal muscular atrophy and abnormal splicing due to TARDBP mutations occurs in amyotrophic lateral sclerosis (Arnold et al., 2013; Zhang et al., 2013). Similarly, the expansion of a GCG microsatellite in PABPN1, which encodes the major nuclear poly(A) tail binding protein, results in oculopharyngeal muscular dystrophy (OPMD) and enhanced selection of proximal polyA sites (pAs) during 3′-end processing (de Klerk et al., 2012; Jenal et al., 2012). Interestingly, widespread 3′ UTR shortening due to APA is also observed in cancer cell lines and tissues while progressive 3′ UTR lengthening occurs during mouse embryonic development (Ji et al., 2009; Mayr and Bartel, 2009).
The muscleblind-like (MBNL) protein family (MBNL1, MBNL2, MBNL3) plays a prominent role in the regulation of AS during development (Poulos et al., 2011). MBNL1 and MBNL2 repress embryonic stem cell (ESC), and promote differentiated cell, splicing patterns and knockdown of these proteins increases the expression of key pluripotency genes required for induced pluripotent stem cell generation (Han et al., 2013). Loss of MBNL function is also a central pathological event in the neuromuscular disease myotonic dystrophy (DM). DM is caused by either CTG microsatellite expansions (CTGexp) in the DMPK 3′ UTR (DM1) or CCTG expansions (CCTGexp) in CNBP intron 1 (DM2). Transcription of these expansions yields C(C)UGexp RNAs that sequester the MBNL proteins, which function as alternative splicing factors responsible for switching the splicing of target gene transcripts during postnatal development. While Mbnl1 knockout (KO) mice show salient features of DM muscle pathology, Mbnl3 KOs develop a progressive decline in skeletal muscle regenerative capacity (Kanadia et al., 2003; Poulos et al., 2013). In contrast, Mbnl2 KOs display characteristic central nervous system defects associated with DM (Charizanis et al., 2012). Although Mbnl1; Mbnl2 double knockout mice are embryonic lethal, Mbnl1−/−; Mbnl2+/− mice are viable and develop severe muscle wasting and heart conduction defects (Lee et al, 2013). Thus, we have proposed that DM results from a compound loss of Mbnl activity.
RBPs often perform multiple regulatory functions. Indeed, MBNL proteins function in AS, mRNA localization and pre-miR-1 processing (Charizanis et al., 2012; Ho et al., 2004; Kanadia et al., 2003; Rau et al., 2011; Wang et al., 2012). Here, we test the possibility that MBNL loss-of-function also affects the developmental regulation of APA. PolyA-seq analysis of mouse embryonic fibroblasts (MEFs) showed widespread APA dysregulation following loss of Mbnl proteins. High throughput sequencing-crosslinking immunopurification (HITS-CLIP) and minigene reporter analyses demonstrated that Mbnl binding directs alternative pA selection. Mbnl-mediated APA regulation during postnatal development was confirmed in a transgenic mouse DM1 model and in human DM1 and DM2. These results indicate that Mbnl proteins are required for normal developmental regulation of alternative polyadenylation and multiple pre-mRNA processing steps are adversely affected in DM.
RESULTS
Widespread Shifts in Polyadenylation Site Selection Following Mbnl Depletion
Since previous studies indicate that major Mbnl binding sites exist in 3′ UTRs (Charizanis et al., 2012; Poulos et al., 2013; Wang et al., 2012), we investigated the possibility that Mbnl proteins function in APA regulation. As an initial step, primary MEFs were isolated from WT and Mbnl1ΔE3/ΔE3; Mbnl2ΔE2/ΔE2 (DKO) E13.5 littermates (Lee et al., 2013). Mbnl2 RNA levels were 1.6-fold higher, and Mbnl3 5-fold lower, than Mbnl1 in WT primary MEFs (Figure S1A). Immunoblot analysis of WT primary MEF subcellular fractions confirmed that Mbnl1, Mbnl2 and Mbnl3 were predominantly localized in the nucleus similar to Elavl1, another nuclear RNA-binding protein (Figure 1A). WT MEFs expressed all three Mbnl proteins while Mbnl1 was undetectable in Mbnl1ΔE3/ΔE3 (Mbnl1 KO), Mbnl2 was undetectable in Mbnl2ΔE2/ΔE2 (Mbnl2 KO) and neither Mbnl1 nor Mbnl2 were detectable in DKO MEFs (Figure 1B). In contrast, Mbnl3 protein was constitutively expressed in DKOs. To obtain MEFs deficient in all Mbnl activity, Mbnl3 protein expression was reduced by >80% in DKO MEFs, compared to WT, following siRNA-mediated knockdown (siMbnl3) (Figure 1B, DKO/3KD). DKO/3KD MEFs were viable although their doubling time was reduced ~18% compared to WT cells (Figure S1B).
Figure 1. MEF PolyA Site Shifts Following Mbnl Depletion.
(A) Mbnl proteins are localized primarily in the nucleus in MEFs. Immunoblot analysis of nuclear (Nuc) and cytoplasmic (Cyt) subcellular fractions demonstrated that Mbnl proteins were predominantly nuclear. Elavl1/HuR was included as a nuclear RBP control and LDHA as a cytoplasmic (Cyt) marker.
(B) Immunoblot validation of MEF Mbnl knockouts and knockdowns. Mbnl1-3 protein levels were analyzed by immunoblotting of whole cell lysates isolated from wild type (WT), Mbnl1ΔE3/ΔE3; Mbnl2ΔE2/ΔE2 double KO (DKO), Mbnl1ΔE3/ΔE3 (Mbnl1 KO), Mbnl2ΔE2/ΔE2 (Mbnl2 KO) (left panel). Right panel shows a comparison of Mbnl3 levels in WT (longer exposure compared to left panel) and DKO MEFs following treatment with the siRNAs targeting Mbnl3 (siMbnl3). Gapdh was included as a loading control.
(C) Scatter plots illustrating the shift of 3′ UTR poly(A) sites to shorter (proximal, red) versus longer (distal, blue) sites relative to the coding region in WT versus DKO (left) and WT versus DKO/3 KD MEFs (right) based on FDR <0.001 and −0.15>dI>0.15. The number of shifts (n) were: 1) DKO, proximal (n = 3466), distal (1131), total (4597), no shift (53568); 2) DKO/3KD, proximal (3626), distal (1481), total (5107), no shift (53060).
(D) Wiggle plots of PolyA-seq data of two Mbnl target genes (Fosl2, Papola). The figure includes terminal exons with 3′ UTRs (thick black line) and coding regions (black box) showing pA shifts (left) and 3′ RACE gel validation (right) of WT (turquoise) and DKO (orange) MEFs.
(E) Wiggle plots of Calm3 PolyA-seq (left), 3′ RACE (middle) and qRT-PCR bar graphs (right, n = 3 per sample, data represented as mean +/- SEM, **p < 0.01) for WT (turquoise), DKO (orange) and DKO/3 KD (red) MEFs.
A PolyA-seq strategy, which involved Illumina sequencing followed by computational removal of oligod(T) internal priming events at templated A-rich sequences, was pursued to determine if Mbnl proteins were involved in APA regulation (Derti et al., 2012). PolyA-seq libraries were generated from RNA isolated from WT, DKO and DKO/3KD MEFs and the proportional change of polyA site usage and false discovery rate, estimated by the Benjamini method (see Supplemental Experimental Procedures), were used to identify significant pA changes. Interestingly, thousands (4,597) of pA shifts were identified in DKO, while 5,107 shifts occurred in DKO/3KD, MEFs when compared to WT (FDR ≤ 0.001, dI ≥ I0.15I; Table S1). Shifts to more proximal (relative to the coding region termination codon) and distal pAs were routinely observed in WT versus DKO and DKO/3KD MEFs (Figure 1C). Selected APA targets were validated by 3′ RACE using oligo(dT) reverse, and gene-specific forward, primers (Figure 1D). As examples, compound depletion of Mbnl1 and Mbnl2 resulted in profound dysregulation of Fosl2 (distal to proximal shift) and Papola (proximal to distal) APA site selection in DKO compared to WT MEFs. Further depletion of Mbnl3 resulted in more pronounced pA shifts in DKO/3KD MEFs and 3′ RACE and qRT-PCR confirmed APA shifts in Calm3 (Figure 1E). These results indicated that Mbnl proteins are essential for the normal MEF APA signature. To assess if these pA shifts were a direct effect of Mbnl protein binding, HITS-CLIP analysis was performed.
Mbnl Proteins Bind Directly to Target RNAs
HITS-CLIP was done using either WT MEFs or both WT and DKO for Mbnl3 to determine the effects on Mbnl3 binding following compound loss of Mbnl1 and Mbnl2 (Table S2, Figure S2). Libraries, prepared using RNAs purified from low RNase lanes, were sequenced and significant clusters identified after mapping reads to the mouse genome. As shown previously for C2C12 myoblasts, the majority of Mbnl1 MEF CLIP tags were intronic (40%) followed by the 3′ UTR (24%) (Figure 2A) as opposed to mouse tissues where the majority of reads are located in the 3′ UTR (Wang et al., 2012). Similar to mouse brain (Charizanis et al., 2012), most Mbnl2 CLIP tags mapped to the 3′ UTR (35%). In contrast to C2C12 cells and embryonic muscle (Poulos et al., 2013), the majority (71% for WT, 63% for DKO) of MEF Mbnl3 binding sites were intronic (Figure 2A). Considerable, but not complete, overlap was observed between gene transcripts that bound to Mbnl1, Mbnl2 and Mbnl3 (Figure 2B) suggesting that Mbnl proteins, expressed concurrently in the same cellular compartment, recognize both common and unique binding sites possibly due to differences in the various isoforms expressed by each Mbnl gene (Pascual et al., 2006).
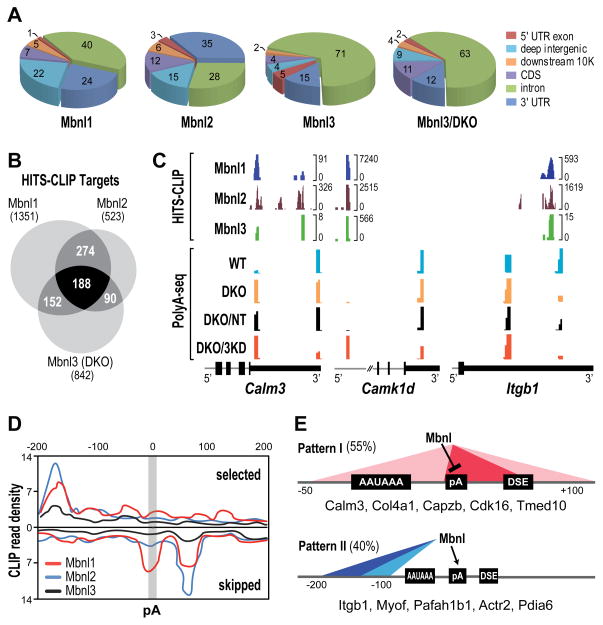
Figure 2. Mbnl Binding Sites and Alternative Polyadenylation.
(A) Pie charts showing genomic distributions of binding sites for Mbnl1, Mbnl2 and Mbnl3 in WT MEFs and Mbnl3 in Mbnl1/2 KO (Mbnl3/DKO) MEFs.
(B) Venn diagram of genes that contain Mbnl binding sites (HITS-CLIP targets) showing overlap of genes that are regulated by Mbnl1-3 in MEFs.
(C) Wiggle plots of HITS-CLIP (top) and PolyA-seq (bottom) highlight Mbnl binding sites that overlap and flank affected pAs in MEFs for 3′ UTRs (Calm3, Itgb1) and introns (Camk1d). For Mbnl3 HITS-CLIP, DKO MEFs were used since loss of Mbnl1 and Mbnl2 led to an increase in Mbnl3 binding to Mbnl targets. HITS-CLIP brackets indicate the number of unique tags at each site. PolyA-seq analysis was performed on WT (turquoise), DKO (orange), DKO treated with non-targeting siRNAs (DKO/NT, black) and DKO treated with Mbnl3 siRNAs (DKO/3KD, red) MEFs.
(D) Mbnl polyadenylation regulatory map showing Mbnl CLIP tag density ± 200bp from the cleavage site and CLIP read density (x103).
(E) Two major functional patterns for Mbnl proteins in alternative polyadenylation.
RNA regulatory maps demonstrate that Mbnl splicing activity is position-dependent comparable to other alternative splicing factors (Charizanis et al., 2012; Du et al., 2010; Licatalosi et al., 2008; Wang et al., 2012). Interestingly, HITS-CLIP and PolyA-seq analysis indicated that Mbnl1, Mbnl2 and Mbnl3 binding sites were often enriched near pAs (Figure 2C). A common pattern emerged as exemplified by Calm3, where the major binding sites for all three Mbnl proteins either overlap a proximal pA or are located upstream of a major WT distal site. Similar to Mbnl intronic binding sites, Mbnl1, Mbnl2 and Mbnl3 recognized YGCY clusters as the preferred binding motif although some sequence variance flanking this core motif was evident (R/YGCY, Figure S2A). Loss of Mbnl1 and Mbnl2 in DKO MEFs led to a large increase in proximal pA use while further depletion of Mbnl3 in the DKO/3KD decreased distal site use further for both 3′ UTR (Calm3) and intronic (Camk1d) alternative pAs (Figure 2C). While this observation suggested that Mbnl binding to sites in the immediate vicinity of a pA tended to silence site utilization, binding further upstream enhanced use. This latter possibility was exemplified by Itgb1 where Mbnl binding sites were restricted to a region ~100 nt upstream of the distal pA and loss of Mbnl1-3 resulted in silencing of the distal site while the proximal site was enhanced. Using the HITS-CLIP and RNA-seq datasets, the MEF Mbnl RNA target PolyA map was generated based on the top 200 pA changes (Figure 2D), which highlighted two major APA patterns (Figure 2E). Mbnl protein binding proximal to a pA showed enhanced utilization in Mbnl1/2 DKO MEFs so Mbnl proteins normally block these sites (Figure 2E, Pattern I) as suggested previously for Nova proteins (Licatalosi et al., 2008). Alternatively, Mbnl binding to distal sites, primarily upstream of an alternative cleavage site (−100 to −200), may facilitate recruitment of the 3′-end processing machinery possibly by stabilizing RNA secondary structures (Pattern II). To determine if Mbnl protein binding proximal to a pA regulated APA directly, a minigene mutational analysis was pursued.
Direct Role for Mbnl Proteins in PolyA Site Selection
Minigene APA reporters were constructed using an IRES-driven firefly luciferase ORF upstream of either the WT (Calm3WT, Itgb1WT) or mutant (Calm3MUT, Itgb1MUT) mouse Calm3 and Itgb1 3′ UTRs, which contain two major pAs (pA1, pA2) (Figure 3A and Table S3). Mbnl binding sites were identified as crosslinking-induced mutation sites (CIMS) by HITS-CLIP (Zhang and Darnell, 2011) and the GC dinucleotide core of the Mbnl-binding motifs was mutated to GG, a strategy that has been shown to block Mbnl AS activity (Du et al., 2010). COSM6 cells were selected for this analysis since they express primarily MBNL1 (Figure 3B) and mouse Calm3 and Itgb1 primers did not amplify COSM6 endogenous RNAs. The level of MBNL1 expression was assessed by transfecting cells with WT or MUT plasmids with, or without, exogenous myc-Mbnl1 expression driven by pcDNA3.1-Mbnl1myc (Figure 3C) and the effects of Calm3 and Itgb1 mutations on distal/proximal (D/P) site ratios was determined by 3′ RACE (Figure 3D). As anticipated (Figure 2E), mutation of the Calm3 (Calm3MUT) proximal, or Itgb1 distal (Itgb1MUT), Mbnl-binding sites led to an increase in pA1 site use with a corresponding 40% (Calm3) to 60% (Itgb1) decrease in D/P ratio (Figure 3D). While increased Mbnl1 expression following pcDNA3.1-Mbnl1myc transfection enhanced Calm3 WT distal pA selection, presumably due to increased silencing of Calm3 pA1, there were no significant effects on Calm3MUT D/P ratios. The Calm3MUT 3′ UTR plasmid showed elevated luciferase activity compared to Calm3WT and Mbnl1myc expression significantly reduced Calm3WT, but not Calm3MUT, luciferase expression (Figure 3E). In contrast, Itgb1 MBNL-binding site mutations upstream of the pA2 site reduced luciferase activity suggesting enhanced use of Itgb1 pA1 decreased mRNA stability. This result supported a recent study that identified two ELAVL1/HuR binding sites between pA1 and pA2 (Figure 3A) which inhibit Itgb1 mRNA turnover (Naipauer et al., 2013) and ELAVL1 was expressed at a relatively high level in COSM6 cells (Figure 3B).
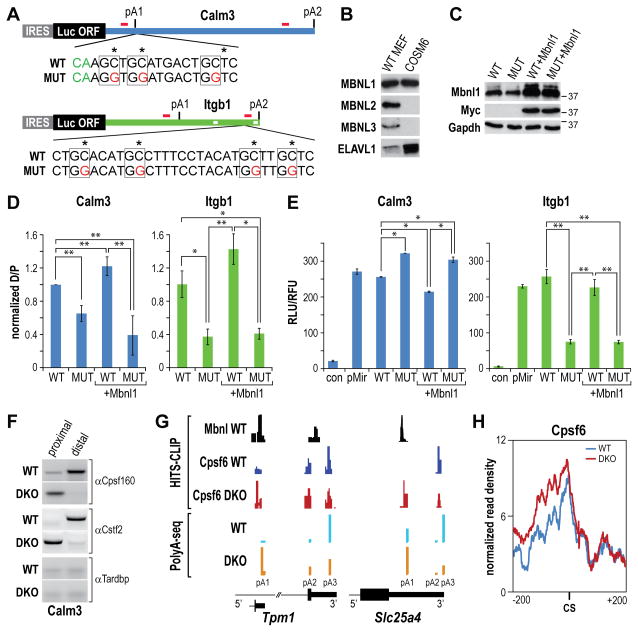
Figure 3. Minigene Polyadenylation Reporter Analysis.
(A) Minigene reporters composed of the Calm3 and Itgb1 3′UTRs (thick green lines) with the proximal (pA1) and distal (pA2) polyadenylation sites cloned downstream of the IRES-driven (grey box) luciferase coding region (turquoise box). The primer binding sites for RT-PCR and 3′ RACE (red bars), the wild type (WT) and mutant (MUT, red letters) sequences downstream (Calm3) or upstream (Itgb1) of the cleavage sites (green CA indicated for Calm3) and the AU-rich binding sites for ELAVL1/HuR (white boxes in Itgb1 3′ UTR) are also indicated.
(B) Relative protein levels for MBNL1, MBNL2, MBNL3 and ELAVL1/HuR in WT MEFs compared to COSM6 cells were determined by immunoblotting.
(C) Immunoblots showing myc-tagged Mbnl1 overexpression following transfection of COSM6 cells with pcDNA3.1-Mbnl1myc. Antibodies to Mbnl1, Myc and Gapdh (loading control) are shown for cells transfected with Calm3WT (WT) and Calm3MUT (MUT) polyadenylation reporters without/with co-transfection with pcDNA3.1-Mbnl1myc (+Mbnl1).
(D) Mbnl proteins repress Calm3 proximal pA (pA1), and activate Itgb1 distal pA (pA2), site use. The ratio of distal to proximal pA selection (D/P) was determined by qRT-PCR for COSM6 cells transfected with the Calm3WT (WT), Calm3MUT (MUT), Itgb13WT (WT), Itgb1MUT (MUT) reporters without/with Mbnl1myc overexpression (+Mbnl1) (data represented as mean +/- SEM, *p < 0.05, **p < 0.01).
(E) Elevated Calm3 pA1 site use leads to enhanced, while loss of Itgb1 distal (pA2) results in decreased, luciferase activity. COSM6 cells were mock transfected (con) or transfected as described in (D) (data represented as mean +/- SEM, *p < 0.05, **p < 0.01).
(F) RNA immunoprecipitation assay with anti-Cpsf160, anti-Cstf2 and anti-Tardbp (control) antibodies.
(G) Examples (Tpm1, Slc25a4) of HITS-CLIP (top) and PolyA-Seq (bottom) showing overlap of Mbnl and Cpsf6 binding sites in wild type (WT), but not Mbnl DKO, MEFs. Enhanced Cpsf6 binding and activation of upstream pA sites were observed following loss of Mbnl activity in DKO MEFs.
(H) RNA map of Cpsf6 normalized CLIP tag density (± 200 bp from the cleavage site, CS) in WT versus DKO MEFs. Polyadenylation sites without additional proximal (±300 bp) sites were selected to avoid interference.
Several approaches were used to determine if Mbnl binding directly influenced endogenous pA site selection by modifying site-specific recruitment of 3′ end processing factors (3′ PF). First, an RNA immunoprecipitation protocol (RIP) was used to monitor binding of Cpsf160 and Cstf2, two core components of the 3′ PF machinery that recognize the polyadenylation signal and U/GU-rich downstream region, respectively (Di Giammartino et al., 2011). As an example, both Cpsf160 and Cstf2 bound to the Calm 3 distal (pA2) site in WT MEFs but switched to proximal (pA1) site binding following loss of Mbnl activity in DKOs (Figure 3F). Since this result suggested that Mbnl proteins block 3′ PF recruitment to normally cryptic upstream sites, HITS-CLIP analysis was performed on WT versus DKO MEFs using an antibody against another 3′ PF, Cpsf6 (CF Im68). This global analysis revealed a common reciprocal pattern of Mbnl and Cpsf6 binding to polyadenylation sites. For example, Mbnl proteins bound proximal to Tpm1 and Slc25a pA1 sites that were poorly utilized in WT MEFs and these binding sites overlapped Cpsf6 binding, which was activated in Mbnl DKOs (Figure 3G). Indeed, the RNA map of Cpsf6 normalized CLIP tag density indicated increased Cpsf6 binding upstream of pA sites that were activated following loss of Mbnl activity.
Mis-Regulation of Developmental Polyadenylation in a DM1 Model
To determine if APA was also affected by expression of CUGexp RNA in vivo, we next analyzed HSALR mice, a model for DM1 muscle disease that express a CUGexp in the 3′ UTR of a human skeletal actin transgene (Mankodi et al., 2000). Immunoblot analysis demonstrated that Mbnl1 is the most highly expressed member of the Mbnl family in adult FVB quadriceps with lower expression of Mbnl2 and no detectable Mbnl3 (Figure 4A). Immunofluorescence of transverse sections of 4 month old FVB quadriceps showed a diffuse nucleoplasmic localization pattern for Mbnl1 in WT FVB while Mbnl1 re-localized into nuclear CUGexp foci in HSALR muscle (Figure 4B) as described previously (Lin et al., 2006). Mbnl2 protein expression in myonuclei was below the immunofluorescence detection threshold in WT mice but Mbnl2 sequestration in nuclear foci was evident in HSALR while Mbnl3 proteins were not observed in either WT or HSALR adult muscle.
Figure 4. APA dysregulation in the HSALR mouse DM1 model.
(A) Mbnl1 and Mbnl2, but not Mbnl3, are expressed in WT and HSALR adult muscle. Immunoblots showing that Mbnl1 protein was expressed at equivalent levels in both WT and HSALR quadriceps while Mbnl2 expression was upregulated in HSALR compared to WT adult and Mbnl3 was not expressed at a detectable level.
(B) Mbnl1 and Mbnl2 proteins colocalize with CUGexp RNAs in HSALR muscle. Immunofluorescence (green), using anti-Mbnl1 (α-Mbnl1) and anti-Mbnl2 (α-Mbnl2) antibodies, and RNA-FISH (red), using Cy3-conjugated (CAG)10 to detect CUGexp RNA, of quadriceps muscle sections (nuclear DNA is stained with DAPI, blue). The Mbnl2 nuclear/cytoplasmic ratio was unchanged in WT versus HSALR so the fluorescence signal in HSALR (α-Mbnl2) versus no signal in WT reflects increased Mbnl2 concentration in HSALR RNA foci.
(C) Venn diagram of overlapping pA changes in WT neonatal (WT P1) versus WT adult (light blue) and WT adult versus HSALR adult muscle (green) and the overlap (dark blue).
(D) Scatter plots illustrating 3′ UTR shortening (proximal, red) and lengthening (distal, blue) in WT neonatal (P1) and HSALR quadriceps muscles versus WT adult based on FDR < 0.05 and −0.15>dI>0.15. The number of shifts (n) were: 1) P1, proximal (2588), distal (1360), total (3948), no shift (42601); 2) HSALR, proximal (n = 641), distal (342), total (983); no shift (45566).
(E) Concordance of APA patterns between WT neonatal and HSALR adult muscle. The distal to proximal pA site (D/P) ratios were determined for Fxr1, Ndrg3, Pdlim5, Sgcg and Tuba4a by PCR for WT adult (blue), WT neonates (P1, green) and HSALR adult (red) muscles within the top 10 genes having pA shifts and Mbnl1 CLIP tags (data represented as mean +/- SEM, **p < 0.01, ***p < 0.001). The majority of Mbnl RNA targets, except Sgcg and Pdlim5, show significant concordant shifts between WT P1 and HSALR.
To investigate APA regulation in HSALR, PolyA-seq was performed for WT and HSALR quadriceps muscle and 532 different genes showing pA changes were uncovered in HSALR compared to WT (FDR < 0.05, dI ≥I0.15I) (Table S1 and Figure 4C). Remarkably, and similar to AS, APA generally reverted to an embryonic pattern in HSALR adult muscle. We found 1,848 genes that showed pA changes between WT P1 neonate and adult quadriceps and 349 (~66% of total HSALR genes) of the genes showing pA shifts in HSALR muscle also showed consistent changes in P1 muscle. Similar to MEFs, pA shifts to both distal and proximal sites were observed in HSALR and WT P1 although proximal shifts predominated (Figure 4D) (FDR < 0.05, dI ≥ I0.15I). The majority of these switch events showed a similar pattern in the distal/proximal ratio between WT P1 neonatal and adult HSALR muscles indicating that loss of Mbnl activity by sequestration of CUGexp RNAs generally resulted in a reversion to the fetal polyadenylation pattern although some genes showed the opposite pattern (e.g., Sgcg) (Figure 4E).
To determine which polyadenylation changes were directly regulated by Mbnl proteins, Mbnl1 HITS-CLIP was performed using adult FVB quadriceps muscle and the Mbnl binding consensus was identified using the top 500 clusters. Nearly half (44%) of the total Mbnl1 binding sites were located in the 3′ UTR and many of these Mbnl peaks were concentrated around annotated pA sites (Figure 5A). HITS-CLIP clusters were identified for more proximal (Calm3) and distal (Tuba4a) pA shifts (Figure 5B). Similar to MEFs, when Mbnl binding overlapped a pA that site was blocked but when the binding site was predominantly upstream then use of the downstream site was enhanced.
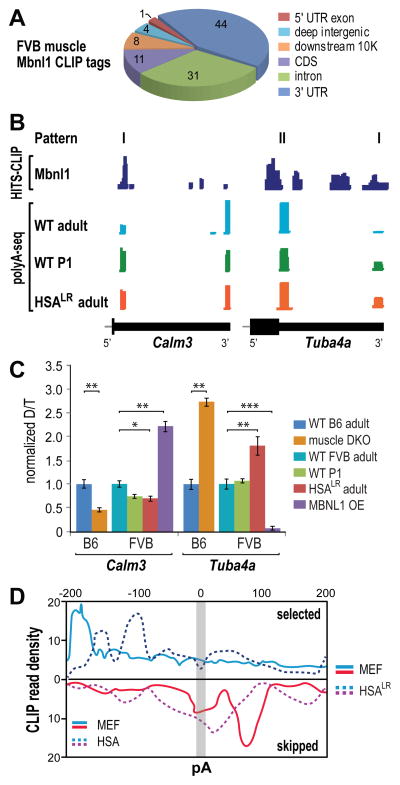
Figure 5. Reversion to Fetal APA Patterns in HSALR Mice.
(A) Pie chart of Mbnl1 CLIP tag distribution in WT FVB quadriceps muscle.
(B) Mbnl-regulated alternative polyadenylation patterns in vivo. Wiggle plots of Mbnl1 HITS-CLIP (dark blue) together with PolyA-seq of WT adult (light blue), WT P1 (green) and HSALR adult (red) muscle.
(C) Normalized Calm3 and Tuba4a distal/total (D/T) pA ratios were determined for WT B6 adult (dark blue), DKO (orange), WT FVB adult (light blue), WT P1 neonate (green), HSALR adult (red) and MBNL1 overexpression (MBNL1 OE, purple) mouse skeletal muscle (data represented as mean +/- SEM, *p < 0.05, **p < 0.01, ***p < 0.001).
(D) Polyadenylation regulatory map comparing MEF Mbnl1-3 and HSALR patterns.
The effects of altering Mbnl levels on APA in vivo were also studied. Calm3 and Tuba4a skeletal muscle RNAs were analyzed by qRT-PCR from either WT, Mbnl1−/−; Mbnl2c/c; Myo-Cre+ (muscle DKO) or MBNL1 overexpression (OE) mice. For DKOs, the genetic background was C57/BL6 (B6) while FVB was the background for WT P1, HSALR and MBNL1 OE mice (Chamberlain and Ranum, 2012; Lee et al., 2013; Mankodi et al., 2000). For Calm3, loss of Mbnl1 and Mbnl2 proteins in DKO muscle resulted in a significant increase in proximal pA selection, and a corresponding ~50% decrease in the distal to total (D/P+D) pA ratio, similar to that observed for WT P1 and HSALR (Figure 5C). In contrast, MBNL1 OE repressed Calm3 pA1 and led to enhanced distal site selection. The opposite pattern was observed for Tuba4a where the DKO showed an increase, and Mbnl1 OE a large decline, in distal pA use. The Mbnl RNA target PolyA map showed similar patterns to that observed in MEFs although Mbnl1 binding sites that either promoted pA selection or skipping were shifted to sites closer to the affected pA (Figure 5D). These results indicate that APA of Mbnl target RNAs is responsive to Mbnl protein levels in vivo and that Mbnl sequestration by a CUGexp often leads to reversion to the fetal APA pattern.
Dysregulation of Alternative Polyadenylation in Myotonic Dystrophy
To pursue the possibility that APA dysregulation is also a distinctive feature of myotonic dystrophy, PolyA-seq analysis was performed on human control (non-DM muscle atrophy) and DM1 skeletal muscles and the results compared with the corresponding datasets for HSALR muscle (Figure S3 and Table S4). Similar to MEFs following depletion of Mbnl1-3 proteins as well as WT neonatal (P1) muscle, the majority (59%) of pAs shifted upstream in DM1 (Figure 6A). The highest scoring pA shift mapped to the DHFR gene (Table S1). DHFR contains a major promoter responsible for DHFR pre-mRNA synthesis as well as an upstream minor promoter, which produces a non-coding (nc) RNA that represses the major promoter (Martianov et al., 2007). While multiple pAs in DHFR intron 2 have been documented for the ncRNA, the relevance of these sites to ncRNA repression activity is currently unclear. In control muscle, PolyA-seq indicated that three of these intronic sites were frequently used with the most distal pA the least utilized (Figure 6B). In DM1 muscle, use of the distal pA increased, while the proximal site decreased, and polyadenylation at the major transcript 3′ UTR pA increased. Since the current DHFR regulatory model predicts that lower levels of the ncRNA should relieve transcriptional repression of the major promoter, these results suggested that a control element for ncRNA levels exists between the intron 2 proximal and distal pAs. Indeed, the ncRNA in DM1 decreased, while DHFR mRNA increased, significantly compared to control muscle (Figure 6C).
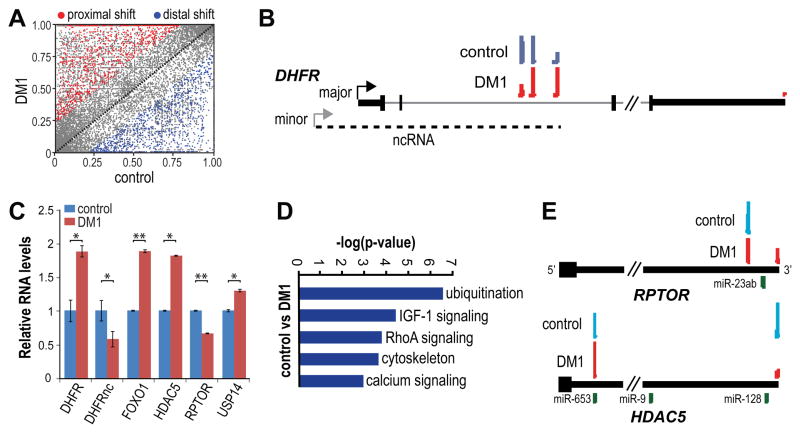
Figure 6. Dysregulation of Alternative Polyadenylation in DM1.
(A) Predominance of proximal pA shifts in DM1 muscle. Scatter plots illustrating 3′ UTR shortening (proximal, red, n=1863) and lengthening (distal, blue, n=1425) in DM1 versus control muscle based on FDR < 0.05 and −0.15>dI>0.15 (no shift, n=20580).
(B) PolyA-seq revealed a proximal to distal pA shift for the ncRNA that modulates transcription of the DHFR coding transcript. A diagram of the DHFR coding (major) and non-coding (ncRNA, minor, dash line) transcriptional units (black box, coding region; thick black line, 3′ UTR) are illustrated together with PolyA-seq wiggle plots of control (blue) and DM1 (red). Arrows indicated the site of transcription initiation and only four exons are included for the major transcript.
(C) Concordance between pA location and RNA level. RNA levels (relative to WT) were determined by qRT-PCR for DHFR (DHFR, DHFRnc) as well as genes involved in the mTOR (FOXO1, HDAC5, RPTOR) and ubiquitination (USP14) pathways (data represented as mean +/- SEM, *p < 0.05, **p < 0.01).
(D) GO analysis based on PolyA-seq analysis of the principle biochemical pathways affected by APA changes in DM1.
(E) PolyA-seq wiggle plots of control (blue) and DM1 (red) of RPTOR and HDAC5 terminal exons (black box, coding region; thick black line, 3′ UTR). Also indicated are the positions of potential miR binding sites (UCSC Genome Browser).
Pathways altered by the DMPK CTGexp mutation were identified by GO analysis using genes with altered 3′-end processing in DM1 versus control muscle. This analysis highlighted genes involved in ubiquitination, IGF-1 signaling and calcium regulation (Figures 6D and S4). Because a debilitating feature of DM1 is muscle weakness/wasting, we focused on IGF-1 and mTOR signaling due to its central role in growth regulation and muscle hypertrophy/atrophy (Laplante and Sabatini, 2012). mTORC1 promotes protein synthesis and hypertrophy and is activated by Akt, which also represses the activator of muscle catabolism FOXO1. In DM1, FOXO1 increased while the mTORC1 component RPTOR decreased which could trigger loss of muscle mass (Figure 6C). PolyA-seq suggested that the decrease in RPTOR RNA levels resulted from an increase in the use of a more distal 3′ UTR pA (Figure 6E). Myogenin, in addition to its role in promoting muscle development, regulates neurogenic atrophy (Moresi et al., 2010). Myogenin expression is controlled by Class II HDACs, which are upregulated following denervation and muscle atrophy (Cohen et al., 2007; Tang et al., 2009). Since Hdac5−/− KO mice are protected against denervation-mediated atrophy due to a failure of myogenin upregulation (Moresi et al., 2010), we also investigated HDAC5 APA and found a large decrease in distal pA selection (Figure 6E) together with an approximately two-fold increase in HDAC5 mRNA (Figure 6C). Therefore, the expression of CUGexp RNA and MBNL protein sequestration led to discrete alterations in alternative polyadenylation that induce key pathways known to inhibit protein synthesis and activate protein catabolism leading to muscle atrophy.
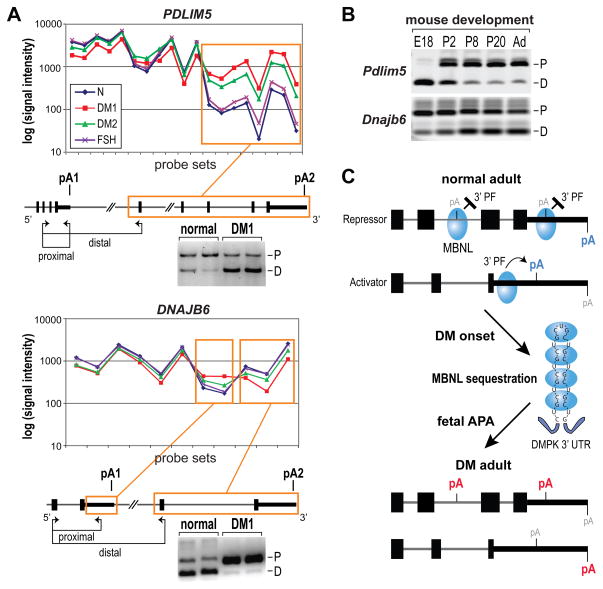
To confirm alternative polyadenylation in DM using a different experimental approach, we analyzed our recent AllExon microarray dataset of human controls (normal and the disease control FSHD), DM1 and DM2 skeletal muscle biopsies (Nakamori et al., 2013). To prioritize candidates for further investigation, probe sets were selected with a >2-fold change in DM1 or DM2 vs. normal controls (p < 0.01 for DM1 vs. normal, p < 0.05 for DM2 vs. normal). Among 858 probe sets selected by these criteria, we eliminated candidates that could result from differences in transcription start site, genes that were represented by ≤5 probe sets, retrogenes and non-protein coding genes leaving 438 probe sets. Among these candidate events, 123 (28%) belonging to 80 genes represented possible alternative 3′ exons or APA sites (Table S5). We examined 24 possible APA examples using RT-PCR with a forward primer in a common exon and two reverse primers positioned in each of the alternative 3′ regions (Table S6). Results of this analysis were consistent with differential 3′-end formation for 17 of the 24 candidates for both DM1 and DM2 muscle compared to normal controls (Figures 7A and S5). Nearly half (8/17) of these events showed similar changes in DKO/3KD MEFs, which indicated that MBNL sequestration strongly impacted these DM-associated RNA processing defects. Interestingly, the majority (10/12) of APA changes observed also occurred during normal muscle development in mice (Figure 7B, Table S7).
Figure 7. AllExon Microarray Analysis of APA in DM1 and DM2 muscle.
(A) AllExon array hybridization results for probe sets representing PDLIM5 (top) and DNAJB6 (bottom) for muscle isolated from healthy individuals (blue line, N), DM1 (red), DM2 (green) and FSHD (FSH, violet) patients. Probe sets showing significant differences for both DM1 vs N and DM2 vs N comparisons are indicated by orange frames. Positions of differentially expressed RNA regions are marked on gene maps with experimentally confirmed polyA sites indicated (pA1, pA2; black box, coding region; thick black line, 3′ UTR; thin line, introns). The positions of RT-PCR primers (proximal, distal) are shown by arrows below each gene structure. Also shown are APA-specific RT-PCR gels that confirmed the transition from proximal (P) to distal (D) for PDLIM5, and from distal to proximal for DNAJB6, polyA sites in DM1 versus normal adult muscle.
(B) Both Pdlim5 and Dnajb6 showed pA shifts during mouse muscle development from embryonic day (E)18, postnatal days P2, P8 and P20 to adult (Ad) mice. Note the fetal (E18) and neonatal (P2) patterns are similar to DM1.
(C) Model for MBNL control of alternative polyadenylation. When MBNL protein (blue ovals) binding sites overlap a polyA site, they repress recruitment of 3′ end processing factors (3′ PF) while binding primarily upstream activates 3′ end processing at the downstream site. Depletion of MBNL activity due to sequestration by CUGexp RNAs results in aberrant activation of polyA sites normally expressed during embryogenesis (red pA) and silencing of adult polyA sites (blue pA).
DISCUSSION
The majority of human gene transcripts are regulated by alternative 3′ end formation (Danckwardt et al., 2008; Derti et al., 2012). While the coupled cleavage and polyadenylation reaction is primarily controlled by the 3′ end processing core machinery, additional RNPs and RNP-RNA complexes play critical modulatory roles in pA selection (de Klerk et al., 2012; Di Giammartino et al., 2011; Elkon et al., 2013; Jenal et al., 2012; Tian and Manley, 2013; Yao et al., 2012). Given the importance of alternative 3′-end formation during the transition from embryonic to adult life, it is likely that additional APA modulatory factors are required for normal development.
Mbnl Proteins Modulate Splicing and Polyadenylation during Development
During mouse development, there is a progressive increase in the overall lengths of mRNA 3′ UTRs, which has been attributed to a decline in the activity of the core cleavage and polyadenylation machinery resulting in the skipping of pAs with less conserved core sequence elements (Ji et al., 2009). Analysis of EST, SAGE and microarray data sets shows that 3′ UTR lengthening occurs in most tissues during embryogenesis with <E8 being the most active phase followed by a more gradual increase during E8 to P0 and no overall length changes postnatally although 3′ UTRs become progressively longer and shorter during postnatal life in brain and testis, respectively (Ji et al., 2009).
While MBNL proteins are essential for the alternative splicing of specific RNAs during multiple developmental windows (Han et al., 2013; Poulos et al., 2011), Mbnl proteins are also required for normal mRNA localization. Mbnl1 and Mbnl2 depletion results in impaired trafficking of membrane-associated mRNAs as well as inhibition of translation and protein secretion (Wang et al., 2012). Here we demonstrate that loss of Mbnl function in both cell and animal models leads to widespread APA mis-regulation characterized by a shift to fetal APA patterns in adult tissues. The polyadenylation regulatory map indicates that Mbnl binding within the core 3′ end processing region suppresses, while more distal binding elements positioned predominantly upstream activates, pA selection. Direct binding of Mbnl proteins is required for APA regulation since mutation of Mbnl crosslinking sites suppressed Mbnl-induced regulation. Furthermore, loss of Mbnl activity in vivo by sequestration on CUGexp RNAs, in either a mouse DM1 model or in human DM1 and DM2, led to APA dysregulation and re-expression of fetal polyadenylation patterns in adult muscle. Thus, MBNL proteins regulate both alternative splicing and 3′-end processing of target gene transcripts and loss of MBNL function in DM causes a reversion to fetal pre-mRNA processing events. Importantly, the effects of MBNL loss on downstream events, including altered localization, translation and turnover of MBNL targets, may be impacted by these 3′ UTR structural changes.
Mis-regulation of 3′ End Processing in Disease
Errors in 3′ end cleavage and polyadenylation are associated with a number of human hereditary diseases with both recessive and dominant mutations occurring in the cleavage and polyadenylation core sequence elements (Danckwardt et al, 2008; Tian and Manley, 2013). Although current evidence indicates that variations in the cellular levels of 3′ end core processing factors, such as CstF-64/CSTF2, are a common trigger of pA changes (Di Giammartino et al., 2011), additional factors are also important for APA control. In addition to its role in splice site selection, U1 snRNP suppresses premature cleavage and polyadenylation, possibly by co-transcriptional inhibition of Pol II-associated CPSF (Berg et al., 2012; Kaida et al., 2010). Another factor that influences APA is PABPN1. GCGexp mutations in this gene cause another muscular dystrophy, OPMD, and normal APA patterns are altered in transgenic PABPN1 GCGexp (A17.1) mice with the majority showing a shift to more proximal pA sites (de Klerk et al., 2012; Jenal et al., 2012). Thus, PABPN1 might normally suppress proximal/weak pA selection and loss of this function in OPMD leads to APA global dysregulation (Jenal et al., 2012).
In this study, we demonstrate that MBNL activity is required for a normal developmental APA signature and loss of MBNL function due to sequestration by CUGexp RNAs leads to selective effects on APA regulation. In agreement with a prior study on Nova proteins, Mbnl proteins show position-dependent effects on 3′ end processing although Nova regulates a smaller number of APA events and binds preferentially to unannotated 3′ UTRs within previously classified intergenic regions (Licatalosi et al., 2008). Since Mbnl proteins recognize R/YGCY motifs in both ssRNA and dsRNA contexts, Mbnl-mediated utilization of a polyadenylation site may be attributed to direct recruitment of the core 3′ end processing machinery or Mbnl binding might promote the formation, or stabilization, of RNA secondary structures amenable to pA selection. Based on these findings, we propose that Mbnl proteins regulate developmental APA of its target RNAs and loss of Mbnl function in DM leads to aberrant 3′-end processing of these targets (Figure 7C). Pathological changes in DM muscle may be attributable to specific alterations in 3′ UTR structures and consequent changes in RNA localization and/or protein isoforms and levels. The discovery of APA dysregulation in DM reveals an additional molecular mechanism underlying this disease and provides important new biomarkers to assess the efficacy of ongoing and future therapeutics.
EXPERIMENTAL PROCEDURES
MEF Isolation, Viability, RNA-FISH and RNA Immunoprecipitation
Animal procedures were approved by the University of Florida IACUC. Wild-type (WT), Mbnl1ΔE3/ΔE3 (Mbnl1 KO), Mbnl2ΔE2/ΔE2 (Mbnl2 KO) and Mbnl1ΔE3/ΔE3; Mbnl2ΔE2/ΔE2 DKO MEFs were isolated from E13.5 embryos. For Mbnl3 RNA knockdown, DKO MEFs were either mock-treated or treated with siRNAs directed against Mbnl3 (siMbnl3) (ON-TARGET plus SMARTpool, Thermo Scientific Dharmacon) or a non-targeting siRNA control (siRNAnt) 6 hr post-seeding following the manufacturer’s protocol. Mbnl1-3 protein levels and localization were determined as described previously (Poulos et al., 2013). For analysis of cell viability, WT and DKO MEFs were transfected with siRNAnt and siMbnl3 for 48 hr followed by trypsin treatment and re-seeding in a 96 well plate. Cell viability was analyzed 24 hr post-seeding using PrestoBlue (Life Technologies) following the manufacturer’s protocol. RNA-FISH and RNA immunoprecipitation (RIP) were performed as described (see Supplemental Experimental Procedures for details).
PolyA-seq and HITS-CLIP
Total RNAs were isolated and PolyA-seq libraries were prepared as described (Derti et al., 2012) with several modifications (see Table S3 for primers). For MEFs and mouse quadriceps muscle, total RNA (2 μg) was used for library preparation while 5 μg total RNA was used for human muscle samples (see Human Subjects below and Table S4). DNA libraries of 200–500 bp were obtained and sequenced (Illumina) using an oligo dT-containing sequencing primer for 35 bp reads. Sequence files were mapped to the mouse genome (mm10) using OLego (Wu et al., 2013), followed by identification, quantification and analysis of differential usage of pA sites using custom methods. HITS-CLIP was performed as described (Charizanis et al., 2012; Jensen and Darnell, 2008) with some modifications (see Supplemental Experimental Procedures).
3′ RACE and RT-PCR Validation
For 3′ RACE, RNA was reverse transcribed using the U1-T10VN primer and Superscript III (Invitrogen) followed by second strand synthesis as described for PolyA-seq library generation and nested PCR with gene specific forward primers and an oligo dT containing reverse primer (Berg et al., 2012). For qRT-PCR experiments, the MyiQ real-time PCR detection system (Biorad) was used to amplify cDNAs and relative RNA levels were calculated to determine distal/proximal (D/P) or distal/total (D/T) site ratios, which were normalized to the corresponding WT values.
Minigene Reporter Assays
For polyadenylation analysis using a minigene reporter, wild type or mutant Calm3 (Calm3WT, Calm3MUT) and Itgb1 (Itgb1WT, Itgb1MUT) 3′UTRs were ligated downstream of luciferase in the pMirTarget vector (Origene). Substitution mutants were generated using Quickchange II site-directed mutagenesis (Agilent) according to the manufacturer’s protocol followed by sequencing. COSM6 cells were transfected with 1 μg of plasmid (Calm3WT, Calm3MUT, Itgb1WT, Itgb1MUT, pCDNA3.1-Mbnl1myc) and Fugene6 (Roche) according to manufacturer’s protocol for 48 hr. For luciferase assays, transfected COSM6 cells (as above, 24 hr) were transferred into a 96 well plate at 2 X 103 cells/well for 24 hr. Luciferase expression was measured using britelite plus (PerkinElmer) according to the manufacturer’s protocol (n = 3 per sample) and luciferase activity was normalized to RFP fluorescence values.
Human Subjects and Microarray Analysis
Signed informed consent was obtained for all study participants under protocols approved by the University of Rochester or University of Florida Human Subjects Review Boards. Participants had DM1, DM2 or FSHD by clinical and genetic criteria (Nakamori et al., 2013) and vastus lateralis samples were obtained by needle biopsy or during autopsy, flash frozen and stored at −80°C. Human AllExon array analysis was performed using biopsied muscle as described for DM1 (n=8), DM2 (n=7), FSHD (n=8, disease control) and non-dystrophic (n=8, normal control) individuals (Nakamori et al., 2013) (see Supplemental Experimental Procedures).
Supplementary Material
Figure S1. Mbnl Knockout MEF RNA Levels and Cell Viability
(A) Transcript abundance in WT primary MEFs was assessed using total reads per million (RPM) mapped to Mbnl polyA sites. RPM data were normalized to Mbnl1 levels.
(B) Cell viability of WT, Mbnl1 KO, Mbnl2 KO, DKO and DKO/3KD MEFs. Related to Figure 1.
Figure S2. Selection of RNA-Mbnl Complexes for HITS-CLIP
(A) Gel analysis of MEF Mbnl-protein complexes followed UV-light induced crosslinking and treatment with high (thick black bar) and low (sloping black bar) RNase A concentrations. The RNA-protein complexes isolated for HITS are indicated by a bracket and both major (38 kDa) and minor (27 kDa) Mbnl3 isoforms are indicated. Although the 27 kDa minor isoform of Mbnl3 (Poulos et al., 2013) was also detectably crosslinked to MEF RNA, RNA-protein complexes for the larger isoform were primarily selected to be consistent with the Mbnl1 and Mbnl2 datasets. Preferred Mbnl binding sites motifs (R/YGCY) are illustrated below each CLIP gel and contain either a single (Mbnl3) or multiple (Mbnl1, Mbnl2) R/YGCY motifs. Related to Figure 2.
(B) As (A) but Mbnl1 binding to wild type (FVB) mouse quadriceps muscle RNA.
Related to Figure 2.
Figure S3. Altered APA in Human DM1 Muscle
(A) Hierarchical clustering of log2(n+1) transformed data of polyA sites (top 600) in control and DM1 human autopsy muscle where n is the read count of each polyA site.
(B) Principal component analysis (PCA) of log2(n+1) transformed data of polyA sites (top 2500) for control and DM1 human autopsy muscle.
(C) Venn diagram of PolyA-seq showing overlap between the mouse HSALR and human DM1 data sets.
(D) Wiggle plots of PolyA-seq results for SGCA and RBFOX1. Related to Figure 6.
Figure S4. Misregulated APA gene networks in DM1 muscle
STRING v9.1 (Franceschini et al., 2013) was used to evaluate gene networks affected at the APA level in DM1 skeletal muscle. Cellular pathways for the regulation of RNA processing, ubiquitination, muscle contraction and cytoskeleton, transcription, lipid metabolism and muscle maintenance are affected. Related to Figure 6.
Figure S5. DM-specific APAs based on AllExon array analysis
(A) Three examples (TNNI1, TPM2, TPM3) of alternative 3′ end formation where 3′ UTR sequence and length are mainly affected.
(B) Example (DTNA) of APA shift that results in a much longer protein coding region.
(C) APA examples (CALM3, LMAN2) selected based on polyA-seq with corresponding changes in AllExon analysis that are statistically significant but failed the restrictive initial selection criteria so are not listed in Table S5.
Related to Figure 7.
Table S1. PolyA-seq Analysis (Related to Figures 2, 4, 5, 6)
(A) MEF WT vs Mbnl1−/−; Mbnl2−/− (DKO)
(B) MEF WT vs DKO/Mbnl3 knockdown (DKO/3KD)
(C) WT adult vs WT postnatal day 1 (P1) quadriceps muscle
(D) WT adult vs HSALR adult muscle
(E) Control vs DM1 muscle
(F) pA-supported Mbnl HITS-CLIP tags (>20 tags/cluster)
(G) pA shifts only in DKO/3KD MEFs
(H) pA shifts in which DKO/3KD > DKO MEFs
Table S2. HITS-CLIP Analysis (Related to Figures 2, 5)
(A) Mbnl1 WT MEF
(B) Mbnl2 WT MEF
(C) Mbnl3 DKO MEF
(D) Mbnl3 WT MEF
(E) Mbnl1 FVB WT muscle
(F) Cpsf6 WT MEF
(G) Cpsf6 DKO MEF
(H) HITS-CLIP summary
(I) Noncoding RNAs: Mir (note Mir1a-1 in Mbnl1 HITS-CLIP of quadriceps)
(J) Noncoding RNAs: snoRNA
Table S4. Human Skeletal Muscle Autopsy Samples (Related to Figure 6)
Table S5. Candidate APA Events Identified by AllExon Microarrays (Related to Figure 7)
Table S6. Human and Mouse Primers for AllExon Array Studies (Related to Figure 7).
Table S7. RT-PCR Verified APAs Selected from AllExon Microarray Dataset
Acknowledgments
We thank H. Baker and C. Lopez for sequencing assistance, C. Chamberlain and L. Ranum for MBNL1 OE mice, C. MacDonald for anti-Cstf2 and UF Research Computing for computational resources. This work was supported by grants from the NIH (AR046799 and NS058901 to M.S.S.; R00GM95713 to C.Z.; NS048843 to C.A.T.) and the Polish National Science Centre (2011/01/B/NZ1/01603 to K.S.).
Footnotes
ACCESSION NUMBERS
PolyA-seq and HITS-CLIP data have been deposited under GEO accession number GSE60487.
Supplemental Information includes five figures, seven tables and Supplemental Experimental Procedures and can be found with this article online at __.
AUTHOR CONTRIBUTIONS
R.B. and M.S.S. designed the study and wrote the manuscript; R.B., K.C., M.M. and M.G. performed HITS-CLIP, PolyA-seq and 3′ RACE; M.L. performed protein analysis; K.C. and A.M. generated MEF cell lines; R.B., D.J.F., C.Z. designed and contributed the bioinformatics analysis; R.B. and K.C. performed minigene reporter studies; K.S. and C.A.T. contributed the microarray results.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnold ES, Ling SC, Huelga SC, Lagier-Tourenne C, Polymenidou M, Ditsworth D, Kordasiewicz HB, McAlonis-Downes M, Platoshyn O, Parone PA, et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci USA. 2013;110:E736–745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain CM, Ranum LP. Mouse model of muscleblind-like 1 overexpression: skeletal muscle effects and therapeutic promise. Hum Mol Genet. 2012;21:4645–4654. doi: 10.1093/hmg/dds306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charizanis K, Lee KY, Batra R, Goodwin M, Zhang C, Yuan Y, Shiue L, Cline M, Scotti MM, Xia G, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TJ, Waddell DS, Barrientos T, Lu Z, Feng G, Cox GA, Bodine SC, Yao TP. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J Biol Chem. 2007;282:33752–33759. doi: 10.1074/jbc.M706268200. [DOI] [PubMed] [Google Scholar]
- de Klerk E, Venema A, Anvar SY, Goeman JJ, Hu O, Trollet C, Dickson G, den Dunnen JT, van der Maarel SM, Raz V, et al. Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucl Acids Res. 2012;40:9089–9101. doi: 10.1093/nar/gks655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14:153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Li M, Manchanda M, Batra R, Charizanis K, Mohan A, Warren SA, Chamberlain CM, Finn D, Hong H, et al. Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol Med. 2013;5:1887–1900. doi: 10.1002/emmm.201303275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1772. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, Shelton JM, Backs J, Klein WH, Richardson JA, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naipauer J, Gattelli A, Degese MS, Slomiansky V, Wertheimer E, Lamarre J, Castilla L, Abba M, Kordon EC, Coso OA. The use of alternative polyadenylation sites renders integrin β1 (Itgb1) mRNA isoforms with differential stability during mammary gland development. Biochem J. 2013;454:345–357. doi: 10.1042/BJ20130062. [DOI] [PubMed] [Google Scholar]
- Nakamori M, Sobczak K, Puwanant A, Welle S, Eichinger K, Pandya S, Dekdebrun J, Heatwole CR, McDermott MP, Chen T, et al. Splicing biomarkers of disease severity in myotonic dystrophy. Ann Neurol. 2013;74:862–872. doi: 10.1002/ana.23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Vicente M, Monferrer L, Artero R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- Poulos MG, Batra R, Charizanis K, Swanson MS. Developments in RNA splicing and disease. Cold Spring Harb Perspect Biol. 2011;3:a000778. doi: 10.1101/cshperspect.a000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos MG, Batra R, Li M, Yuan Y, Zhang C, Darnell RB, Swanson MS. Progressive impairment of muscle regeneration in muscleblind-like 3 isoform knockout mice. Hum Mol Genet. 2013;22:3547–3558. doi: 10.1093/hmg/ddt209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer MC, Jost B, Dembele D, Gourdon G, Nicole A, Duboc D, et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat Struct Mol Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Macpherson P, Marvin M, Meadows E, Klein WH, Yang XJ, Goldman D. A histone deacetylase 4/myogenin positive feedback loop coordinates denervation-dependent gene induction and suppression. Mol Biol Cell. 2009;20:1120–1131. doi: 10.1091/mbc.E08-07-0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Anczukow O, Krainer AR, Zhang MQ, Zhang C. OLego: fast and sensitive mapping of spliced mRNA-Seq reads using small seeds. Nucl Acids Res. 2013;41:5149–5163. doi: 10.1093/nar/gkt216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Biesinger J, Wan J, Weng L, Xing Y, Xie X, Shi Y. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci USA. 2012;109:18773–18778. doi: 10.1073/pnas.1211101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotech. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Pinto AM, Wan L, Wang W, Berg MG, Oliva I, Singh LN, Dengler C, Wei Z, Dreyfuss G. Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc Natl Acad Sci USA. 2013;110:19348–19353. doi: 10.1073/pnas.1319280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Mbnl Knockout MEF RNA Levels and Cell Viability
(A) Transcript abundance in WT primary MEFs was assessed using total reads per million (RPM) mapped to Mbnl polyA sites. RPM data were normalized to Mbnl1 levels.
(B) Cell viability of WT, Mbnl1 KO, Mbnl2 KO, DKO and DKO/3KD MEFs. Related to Figure 1.
Figure S2. Selection of RNA-Mbnl Complexes for HITS-CLIP
(A) Gel analysis of MEF Mbnl-protein complexes followed UV-light induced crosslinking and treatment with high (thick black bar) and low (sloping black bar) RNase A concentrations. The RNA-protein complexes isolated for HITS are indicated by a bracket and both major (38 kDa) and minor (27 kDa) Mbnl3 isoforms are indicated. Although the 27 kDa minor isoform of Mbnl3 (Poulos et al., 2013) was also detectably crosslinked to MEF RNA, RNA-protein complexes for the larger isoform were primarily selected to be consistent with the Mbnl1 and Mbnl2 datasets. Preferred Mbnl binding sites motifs (R/YGCY) are illustrated below each CLIP gel and contain either a single (Mbnl3) or multiple (Mbnl1, Mbnl2) R/YGCY motifs. Related to Figure 2.
(B) As (A) but Mbnl1 binding to wild type (FVB) mouse quadriceps muscle RNA.
Related to Figure 2.
Figure S3. Altered APA in Human DM1 Muscle
(A) Hierarchical clustering of log2(n+1) transformed data of polyA sites (top 600) in control and DM1 human autopsy muscle where n is the read count of each polyA site.
(B) Principal component analysis (PCA) of log2(n+1) transformed data of polyA sites (top 2500) for control and DM1 human autopsy muscle.
(C) Venn diagram of PolyA-seq showing overlap between the mouse HSALR and human DM1 data sets.
(D) Wiggle plots of PolyA-seq results for SGCA and RBFOX1. Related to Figure 6.
Figure S4. Misregulated APA gene networks in DM1 muscle
STRING v9.1 (Franceschini et al., 2013) was used to evaluate gene networks affected at the APA level in DM1 skeletal muscle. Cellular pathways for the regulation of RNA processing, ubiquitination, muscle contraction and cytoskeleton, transcription, lipid metabolism and muscle maintenance are affected. Related to Figure 6.
Figure S5. DM-specific APAs based on AllExon array analysis
(A) Three examples (TNNI1, TPM2, TPM3) of alternative 3′ end formation where 3′ UTR sequence and length are mainly affected.
(B) Example (DTNA) of APA shift that results in a much longer protein coding region.
(C) APA examples (CALM3, LMAN2) selected based on polyA-seq with corresponding changes in AllExon analysis that are statistically significant but failed the restrictive initial selection criteria so are not listed in Table S5.
Related to Figure 7.
Table S1. PolyA-seq Analysis (Related to Figures 2, 4, 5, 6)
(A) MEF WT vs Mbnl1−/−; Mbnl2−/− (DKO)
(B) MEF WT vs DKO/Mbnl3 knockdown (DKO/3KD)
(C) WT adult vs WT postnatal day 1 (P1) quadriceps muscle
(D) WT adult vs HSALR adult muscle
(E) Control vs DM1 muscle
(F) pA-supported Mbnl HITS-CLIP tags (>20 tags/cluster)
(G) pA shifts only in DKO/3KD MEFs
(H) pA shifts in which DKO/3KD > DKO MEFs
Table S2. HITS-CLIP Analysis (Related to Figures 2, 5)
(A) Mbnl1 WT MEF
(B) Mbnl2 WT MEF
(C) Mbnl3 DKO MEF
(D) Mbnl3 WT MEF
(E) Mbnl1 FVB WT muscle
(F) Cpsf6 WT MEF
(G) Cpsf6 DKO MEF
(H) HITS-CLIP summary
(I) Noncoding RNAs: Mir (note Mir1a-1 in Mbnl1 HITS-CLIP of quadriceps)
(J) Noncoding RNAs: snoRNA
Table S4. Human Skeletal Muscle Autopsy Samples (Related to Figure 6)
Table S5. Candidate APA Events Identified by AllExon Microarrays (Related to Figure 7)
Table S6. Human and Mouse Primers for AllExon Array Studies (Related to Figure 7).
Table S7. RT-PCR Verified APAs Selected from AllExon Microarray Dataset