SUMMARY
Many cancer cells consume large quantities of glutamine to maintain TCA cycle anaplerosis and support cell survival. It was therefore surprising when RNAi screening revealed that suppression of citrate synthase (CS), the first TCA cycle enzyme, prevented glutamine-withdrawal-induced apoptosis. CS suppression reduced TCA cycle activity and diverted oxaloacetate, the substrate of CS, into production of the nonessential amino acids aspartate and asparagine. We found that asparagine was necessary and sufficient to suppress glutamine-withdrawal-induced apoptosis without restoring the levels of other nonessential amino acids or TCA cycle intermediates. In complete medium, tumor cells exhibiting high rates of glutamine consumption underwent rapid apoptosis when glutamine-dependent asparagine synthesis was suppressed and expression of asparagine synthetase was statistically correlated with poor prognosis in human tumors. Coupled with the success of L-asparaginase as a therapy for childhood leukemia, the data suggest that intracellular asparagine is a critical suppressor of apoptosis in many human tumors.
INTRODUCTION
One of the hallmarks of proliferating cells is their dependency on aerobic glycolysis, which is known as the Warburg effect (Vander Heiden et al., 2009). The enhanced uptake of glucose sustains energy production and the synthesis of macromolecular precursors. In addition to glucose-dependency, mammalian cells also rely on extracellular glutamine to support cell survival and growth (DeBerardinis et al., 2007). Analysis of primary tumor cells has suggested that the in vivo accumulation of tumor cells is limited by the depletion of glutamine in the tumor environment (Roberts et al., 1956). The dependency on glutamine is enhanced by expression of the proto-oncogene MYC that drives a transcriptional program to coordinate the expression of genes involved in glutamine metabolism (Wise et al., 2008; Yuneva et al., 2007). Therefore, targeting glutamine metabolism is being actively pursued as an approach to inhibit tumor cell growth and transformation (Le et al., 2012; Timmerman et al., 2013; Wang et al., 2010; Willems et al., 2013).
Once imported into the cells, glutamine is the major source of cellular nitrogen for de novo biosynthesis of nucleotides and other nonessential amino acids (Wise and Thompson, 2010). In all 5 reactions of purine and pyrimidine biosynthesis, glutamine exclusively donates its amino group at the gamma position and is converted to glutamate. In turn, glutamate is the primary source of nitrogen for other nonessential amino acids by providing the amino group that originates from the alpha position of glutamine. Those reactions are catalyzed by different transaminases that use different α-ketoacids as recipients for the nitrogens and convert glutamate to α-ketoglutarate (α-KG), a key intermediate of the TCA cycle.
The glutamine-derived α-KG has been proposed to be an essential component of glutamine-dependent cell survival. A cell permeable form of α-KG rescues MYC-transformed cells from cell death upon glutamine withdrawal (Wise et al., 2008). This effect correlates with the ability of α-KG to replenish the TCA cycle by providing oxaloacetate (OAA) that condenses with acetyl-CoA to produce citrate to maintain the TCA cycle and support de novo fatty acid biosynthesis. It was also demonstrated that increased expression of pyruvate carboxylase (PC) that can generate OAA from pyruvate can also render MYC-transformed cells resistant to glutamine depletion-induced cell death (Cheng et al., 2011). These data have been interpreted to mean that glutamine maintains cell survival primarily by supporting the anaplerosis of the TCA cycle.
In addition to providing carbons and nitrogens for biosynthesis of macromolecules, glutamine is also involved in other cellular processes, including anti-oxidative stress, mTOR signaling and autophagy (Duran et al., 2012; Nicklin et al., 2009; Son et al., 2013; van der Vos et al., 2012). Therefore, the biological consequences following the inhibition of glutamine metabolism are complex. To further dissect the specific pathway by which glutamine catabolism suppresses apoptosis, we performed an RNAi-based high throughput screen to seek factors whose loss of function protect MYC-transformed cells from apoptosis following glutamine withdrawal. In addition to RNAi against MYC and the proapoptotic protein BAX, we identified citrate synthase (CS) siRNA as a suppressor of cell death upon glutamine withdrawal. This suggested that glutamine’s role in supporting TCA anaplerosis is not the mechanism by which cells utilize glutamine metabolism to suppress apoptosis. We found that knockdown of CS resulted in redirection of OAA into aspartate and asparagine biosynthesis. Subsequent studies demonstrated that asparagine can maintain the viability of glutamine-deprived cells without restoration of anaplerosis or the levels of other nonessential amino acids. Knockdown of asparagine synthetase (ASNS), the enzyme that synthesizes asparagine de novo from aspartate and glutamine, leads to cell death even in the presence of glutamine, which can be reversed by addition of exogenous asparagine. Clinically, the expression of ASNS correlates with the progression of disease and poor prognosis of glioma and neuroblastoma patients. Finally, asparagine suppresses the induction of the apoptotic regulators of the unfolded protein response (UPR) without affecting the transcriptional induction of genes implicated in adaptation/survival. Together, these results demonstrate an unexpected role for asparagine in regulating cell survival.
RESULTS
Glutamine Depletion Induces Apoptosis through BAX/BAK
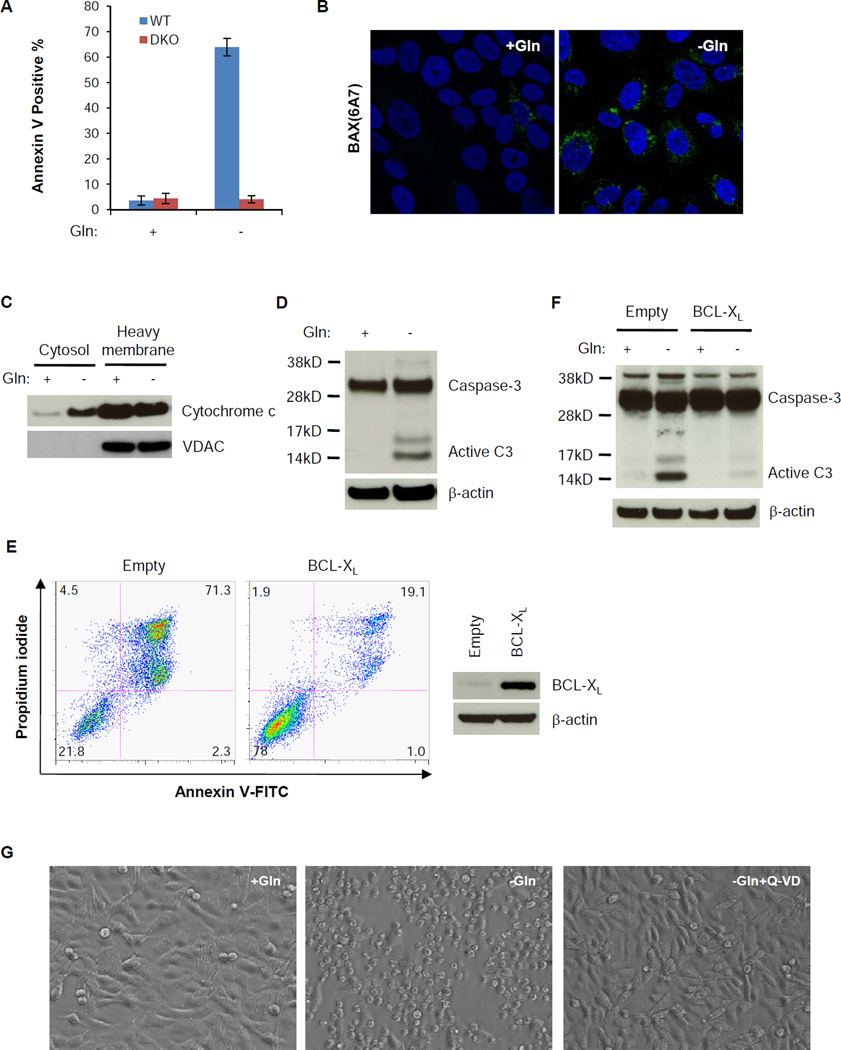
Glutamine depletion induces apoptosis through the upregulation of pro-apoptotic BCL2 family proteins in N-Myc amplified neuroblastoma cell lines (Qing et al., 2012). To genetically confirm the involvement of BCL2 family proteins in regulating glutamine depletion-induced cell death, we withdrew glutamine from immortalized wild type or Bax−/−;Bak−/− mouse embryonic fibroblast cells (MEFs). After 4 days of glutamine depletion, the majority of wild type MEFs died, while Bax−/−;Bak−/− MEFs were still alive (Figure 1A), suggesting the engagement of the intrinsic apoptotic machinery. Consistent with this, when SF188 human glioblastoma cells with MYC amplification were tested, glutamine withdrawal induced BAX activation, cytochrome c release into the cytosol and caspase-3 activation (Figure 1B, 1C and 1D). The caspase-3 activation and cell death following glutamine depletion were completely blocked by forced expression of the anti-apoptotic BCL2 protein, BCL-XL (Figure 1E and 1F). To test whether the death signal downstream of BCL2 protein was dependent on caspase activation, we treated the cells with a pan-caspase inhibitor Q-VD, which also prevented glutamine depletion-induced cell death (Figure 1G).
Figure 1. Glutamine depletion induces apoptotic cell death through the BCL2 proteins and caspase.
(A) Wild type (WT) and Bax−/−;Bak−/− (DKO) MEFs were cultured in DMEM without glutamine for 4 days. Percentage of dead cells was defined by Annexin V positive staining. Data are represented as mean +/− S.D., n=3.
(B) SF188 cells were cultured with or without glutamine for 48 hours in the presence of 20 µM Q-VD. Cells were stained with an antibody recognizing N-terminally exposed, activated BAX (Green) and Hoechst nuclear stain (Blue).
(C, D) Cytochrome C release and caspase-3 activation in SF188 cells 24 hours after glutamine depletion. VDAC was used as a marker of mitochondrial fraction.
(E, F) SF188 cells with constitutively overexpressed BCL-XL were starved of glutamine for 48 hours. Cells were collected for Annexin V and PI staining or for western blotting of caspase-3 activation.
(G) SF188 cells were deprived of glutamine with or without pan-caspase inhibitor Q-VD (20 µM) for 48 hours. Images were captured with a Leica DM IRBE fluorescence microscope with a bright field at 40X.
RNAi-based Screen to Identify Mediators, Whose Loss of Function Confers Resistance to Glutamine Depletion
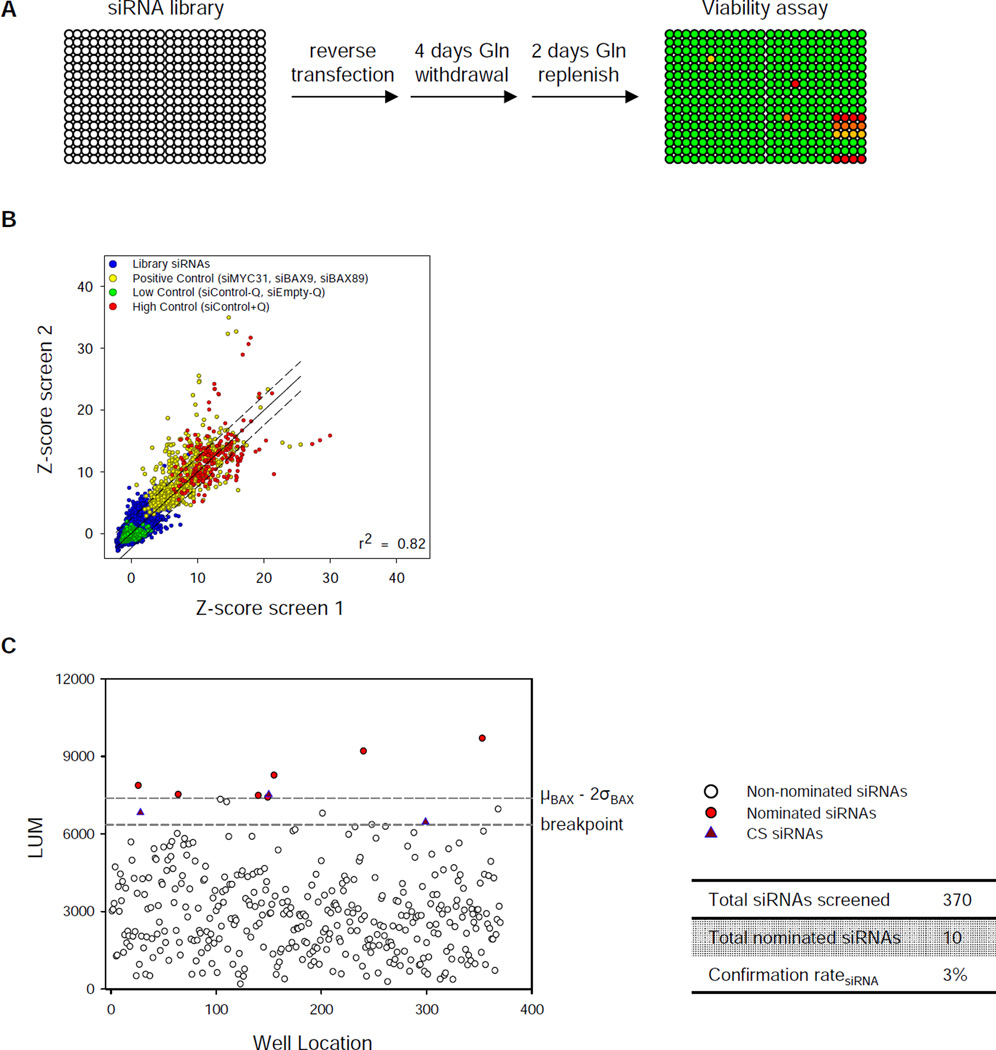
To identify the positive regulators that transduce a signal from glutamine depletion to BAX/BAK- and caspase-dependent apoptosis, we performed siRNA-based high throughput screen in SF188 cells across 9102 genes in the human druggable genomic library (Figure 2A). Each gene has 4 siRNA duplexes with 2 siRNAs pooled per well. To establish positive controls for the screen, we depleted MYC, the driving mutation that causes glutamine addiction in SF188 cells (Wise et al., 2008), or BAX with siRNAs (Figure S1A). Despite a similar degree of protection from death by MYC- or BAX-depletion upon glutamine withdrawal (Figure S1B), siRNA depletion of MYC significantly inhibited cell proliferation in complete medium (Figure S1C). The screen was highly reproducible between duplicates (Figure 2B). All positive control siRNA showed significant protection from cell death, with average value of the entire library equal to the negative controls (Figure S1D). Based on average z-score and reproducibility, we prioritized 119 candidate hits that were filtered through a secondary screen by using ≥ 3 new siRNAs against each gene. Using a stringent threshold of greater than µBax-2σBax, citrate synthase (CS) was the only hit that was confirmed by all 3 siRNAs (Figure 2C and S1E).
Figure 2. High throughput RNAi screen identifies citrate synthase (CS) siRNA as a suppressor of glutamine withdrawal-induced apoptosis.
(A) Schematic representation of the screening strategy in MYC-amplified SF188 human glioblastoma cell line. The right lower corner of each plate is where the positive and negative controls are spotted.
(B) Scatter plot of robust z-scores of the primary screen performed in duplicate. Blue: total library siRNAs without glutamine; Green: negative control siRNAs or no siRNA without glutamine; Yellow: positive control siRNAs against MYC or BAX without glutamine; Red: positive control of scrambled siRNA with glutamine. “−” or “+” Q denotes glutamine status. Dashed line: ± 2 times of the standard deviation (σ) of difference of each data between the real value and the regression value.
(C) Secondary screen of 119 candidates. See extended experimental procedures for the definition of breakpoint cutoff.
See also Figure S1.
Inhibition of Glycolytic Flux into the TCA Cycle Protects from Glutamine Depletion-induced Apoptosis
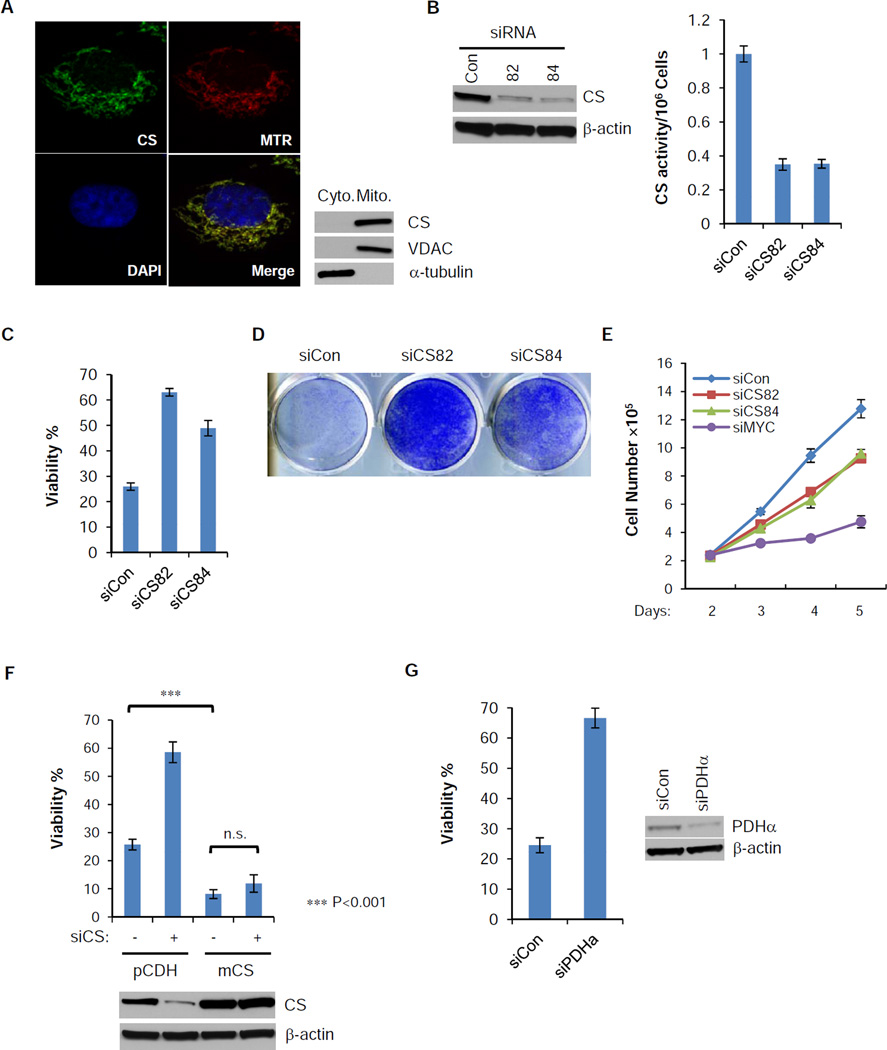
Citrate synthase is a TCA cycle enzyme that catalyzes the formation of citrate from oxaloacetate (OAA) and acetyl-CoA. Immunofluorescence staining and subcellular fractionation confirmed its mitochondrial localization (Figure 3A). siRNA depletion of CS significantly reduces the protein level and enzymatic activity (Figure 3B). In the absence of glutamine, CS knockdown significantly increased the percentage and the total number of viable cells (Figure 3C and 3D). Unlike MYC knockdown, siRNA knockdown of CS only modestly inhibited cellular proliferation in complete DMEM (Figure 3E). In the absence of glutamine, siRNA reduction of CS did not restore cellular proliferation, which is similar to siRNA suppression of BAX or overexpression of BCL-XL (Figure S2A and S2B). To rule out the possibility of off-target effect of siRNA, we transduced a mouse CS gene or a vector control (pCDH) into SF188 cells. Mouse CS overexpression did not affect cellular growth and survival in complete DMEM (data not shown), but sensitized SF188 cells to glutamine depletion and completely reversed the protection by siRNA targeting human CS (Figure 3F). The effect of CS knockdown was not unique to SF188 cells, as RNAi of CS in Kelly and NBLS, two neuroblastoma cell lines, and SV40-transformed MEFs, also protected cell from glutamine depletion-induced apoptosis (Figure S2C and S2D). To test whether consumption of oxaloacetate is critical for glutamine depletion-induced apoptosis, we inhibited pyruvate dehydrogenase (PDH) by siRNA, thereby depleting mitochondrial acetyl-CoA which is required for oxaloacetate consumption by CS. siRNA of PDH phenocopied CS knockdown by protecting from apoptosis following glutamine withdrawal (Figure 3G).
Figure 3. Loss of function of CS protects cells from glutamine withdrawal-induced apoptosis.
(A) SF188 cells were stained with mitoTracker Red (Red), and then fixed and stained with antibody against CS (Green). DAPI (Blue) was applied for nuclear staining. Cytosolic and heavy membrane fractions of SF188 cells were isolated. Distribution of CS was confirmed by western blotting, with VDAC and α-tubulin as mitochondrial and cytosolic markers.
(B) siRNA suppression of CS for 48 hours in SF188 cells reduces both the protein level and the enzymatic activity.
(C,D) SF188 cells were transfected with control or CS siRNA for 2 days and glutamine was deprived for another 48 hours. Viability was measured by Annexin V & PI staining. Crystal violet staining was used to measure total viable cells after 4 days of glutamine depletion followed by re-addition of glutamine for an additional 2 days.
(E) SF188 cells were transfected with control, CS or MYC siRNA in DMEM medium with glutamine, cell numbers were recorded from day 2 to 5 post-transfection.
(F) SF188 cells with constitutively expressed mouse CS (mCS) or empty vector (pCDH) were transfected with control or human CS siRNA for 2 days, and then deprived of glutamine for 48 hours. Viability was measured by Annexin V and PI staining, and expression of total CS was measured by western blotting. p-value was determined by using Student’s 2-tailed t-test.
(G) SF188 cells were transfected with control or pyruvate dehydrogenase subunit α (PDHα) siRNA for 2 days, and then deprived of glutamine for 48 hours. Viability was measured by Annexin V and PI staining.
The data in Figure 3 (B, C, E, F and G) are shown as mean +/− S.D., n=3. See also Figure S2.
Inhibition of CS Redirects Oxaloacetate to Aspartate and Asparagine Biosynthesis
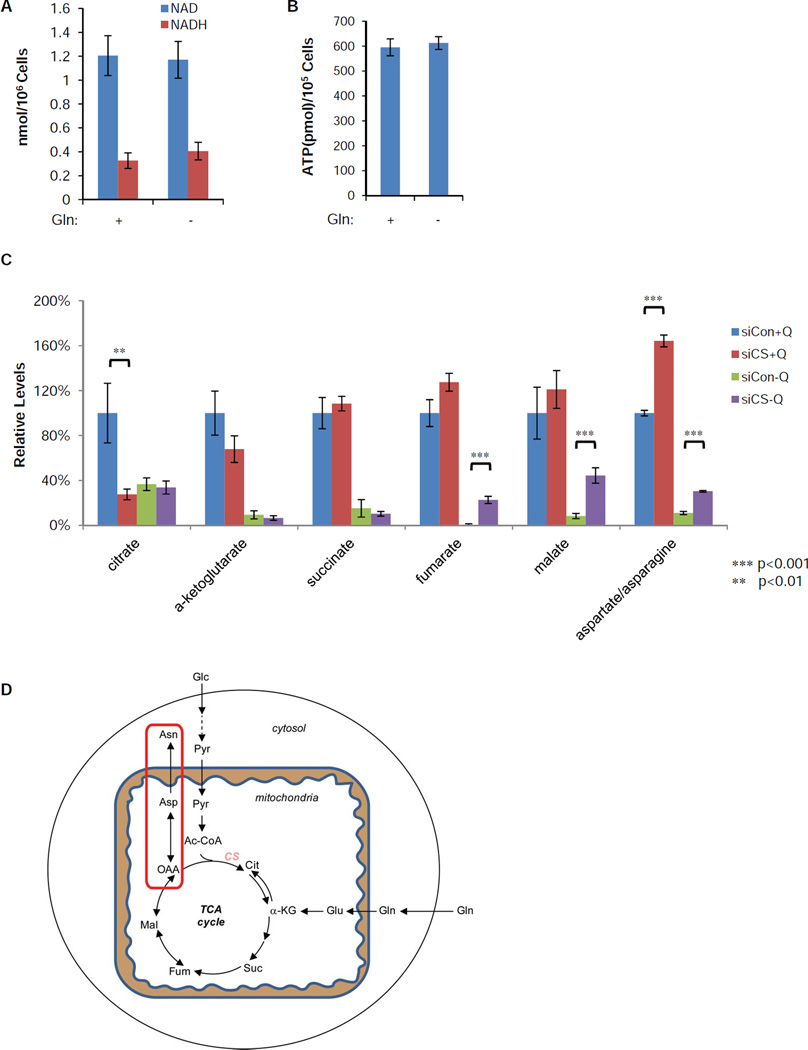
Previously, we and others have suggested that glutamine promotes cell survival by maintaining the mitochondrial supply of α-ketoglutarate (α-KG) (Wise et al., 2008; Yuneva et al., 2007). The ability of CS knockdown to prevent cell death upon glutamine withdrawal suggests that maintaining TCA cycle flux is not the primary mechanism by which glutamine metabolism suppresses apoptosis. Consistent with this, glutamine withdrawal altered neither NAD/NADH ratio nor cellular ATP pool at 16 hours following withdrawal (Figure 4A and 4B). An ATP level decline was observed only if we deprived SF188 cells with constitutively overexpressed BCL-XL of glutamine for longer period (Figure S3A). As a control, inhibition of the respiration chain by antimycin A or disruption of the proton gradient with the uncoupling reagent FCCP caused a rapid reduction of intracellular ATP when glucose was absent (Figure S3A). Glutamine is also reported to be a positive regulator of mTOR activation and a suppressor of autophagy (Nicklin et al., 2009). To examine whether CS knockdown affects mTOR signaling and/or autophagy, we evaluated mTOR downstream signaling, p70S6K, S6 and 4EBP1 phosphorylation and LC3 conjugation by western blotting. No difference was observed between control and CS knockdown following glutamine withdrawal (Figure S3B). There was also no observable increase in autophagy in cells treated with CS siRNA.
Figure 4. CS knockdown redirects oxaloacetate (OAA) to aspartate and asparagine synthesis.
(A, B) SF188 cells were deprived of glutamine for 16 hours. Total NAD, NADH and ATP levels in viable cells were measured and normalized to cell number. Data are shown as mean +/− S.D., n=3.
(C) SF188 cells were transfected with control or CS siRNA for 2 days, and then cultured in DMEM medium with (+Q) or without (−Q) glutamine for 16 hours. Intracellular citrate, α-ketoglutarate, succinate, fumarate, malate and aspartate/asparagine were quantified by LC-MS and normalized to cellular protein content. Data are represented as mean +/−S.D., n=3, and the p-values are determined by using Student’s 2-tailed t-test.
(D) Schematic diagram shows that CS knockdown redirects OAA to aspartate and asparagine biosynthesis. Abbreviations: Glc, glucose; Pyr, pyruvate; Ac-CoA, acetyl-CoA; Gln, glutamine; Glu, glutamate; Cit, citrate; α-KG, α-ketoglutarate; Suc, succinate; Fum, fumarate; Mal, malate; OAA, oxaloacetate; Asp, aspartate; Asn, asparagine.
See also Figure S3.
To confirm that glutamine removal and CS knockdown had their expected effects on levels of TCA cycle intermediates, we performed liquid chromatography mass spectrum (LC-MS) analysis of intracellular compounds of TCA cycle intermediates and aspartate/asparagine, two nonessential amino acids that can be derived from TCA cycle intermediates, with or without CS knockdown in the presence or absence of glutamine. Glutamine withdrawal dramatically depleted all measured TCA cycle intermediates, as well as aspartate/asparagine levels. In the presence of glutamine, CS knockdown significantly suppressed the cellular citrate level, confirming that CS is the major enzyme that contributes to the citrate pool (Figure 4C). In contrast, combined levels of the two amino acids produced de novo from oxaloacetate (Figure 4D), aspartate and asparagine increased significantly when CS was targeted by siRNA (Figure 4C). Other TCA cycle intermediates did not change significantly when CS was inhibited in the presence of glutamine. However, the levels of fumarate, malate and aspartate/asparagine were partially restored when glutamine was absent (Figure 4C). In part, this reflects the decreased consumption of TCA cycle intermediates by citrate synthase. However, tracing experiments using 13C6-glucose also revealed a significant increase (from 7.6 ± 13 relative units/standard to 480 ± 81 relative units/standard, p<0.01) in the M+3 labeled aspartate suggesting that pyruvate carboxylase activity also plays a role in the increase of these TCA cycle intermediates. Earlier TCA cycle intermediates were not restored presumably because unlike the later reactions in the TCA cycle the reaction catalyzed by succinate dehydrogenase is not reversible. Consistent with acetyl-CoA being consumed with oxaloacetate by the catalytic reaction of CS, siRNA depletion of CS also increased the cellular pool of acetyl-CoA (Figure S3C).
The above data suggested that CS knockdown might lead to enhanced OAA diversion from the TCA cycle into aspartate and asparagine biosynthesis. To demonstrate diversion of OAA from TCA cycle into aspartate biosynthesis, we performed 13C5-glutamine tracing experiment with control or CS siRNA to measure labeling of aspartate. The labeling of the M+4 species was significantly increased when CS was depleted (Figure S3D), demonstrating diversion of OAA to aspartate biosynthesis.
Asparagine Restores Viability, but not Anaplerosis, Other Nonessential Amino Acids, or Proliferation
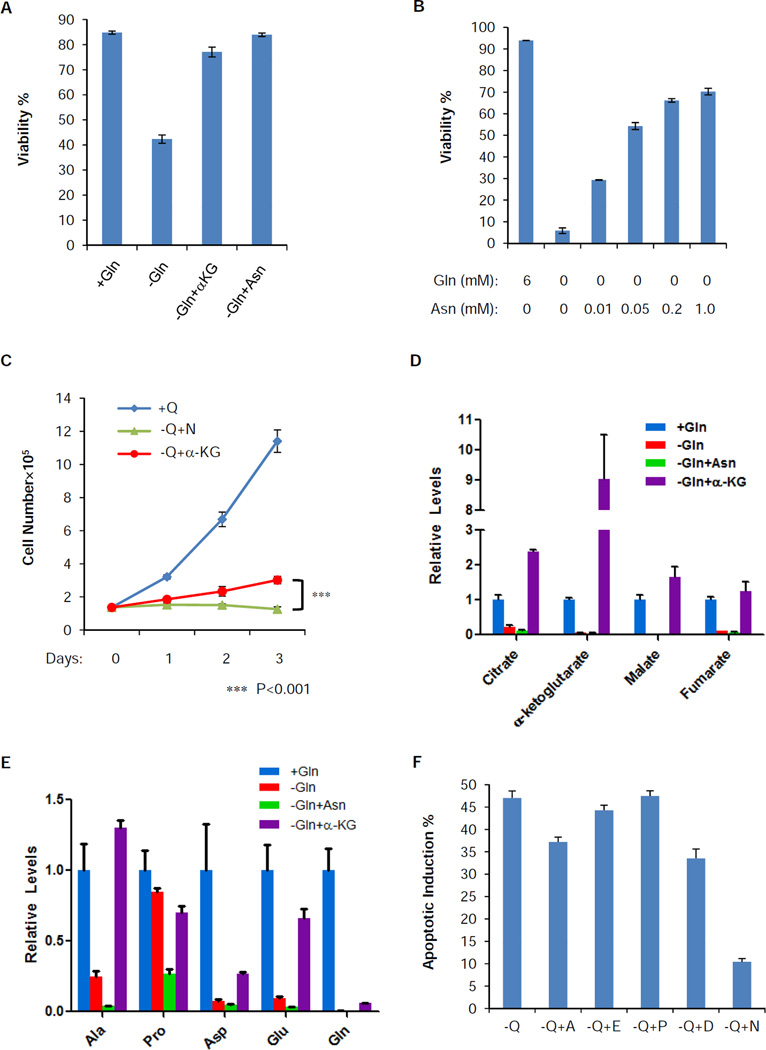
To evaluate whether the increased aspartate and asparagine biosynthesis contributes to suppression of apoptosis following glutamine depletion, we tested the effect of their addition to the medium and found that asparagine blocked glutamine withdrawal-induced apoptosis (Figure 5A). A titration experiment showed that asparagine blocked apoptosis in a dose-dependent manner and 0.2 mM asparagine was sufficient to support survival during long term glutamine starvation (Figure 5B). Asparagine addition did not rescue the ATP level decline during long term glutamine starvation (Figure S4A), suggesting that the ATP level decline is not the cause of cell death. Furthermore, the rescue of survival by asparagine was specific to depletion of the nonessential amino acid, glutamine. Exogenous asparagine had no effect on cell death induced either by depletion of essential amino acids, including leucine, threonine or phenylalanine (Figure S4B), or by glucose depletion or DNA topoisomerase inhibition (Figure S4C). Addition of asparagine completely suppressed cell death in glutamine-deprived cells, but it did not restore proliferation (Figure 5C). Similarly to CS knockdown, asparagine could rescue survival in Kelly, NBLS and SV40 transformed MEFs in response to glutamine depletion (Figure S4D). To distinguish whether the cell survival rescue is dependent on asparagine or the metabolism of asparagine, we tested the extent to which exogenous asparagine is deaminated to replenish the TCA cycle by gas chromatography mass spectrum (GC-MS) analysis. In contrast to α-KG, which was as effective as asparagine at promoting cell survival (Figure 5A), asparagine substitution failed to restore any TCA cycle intermediate (Figure 5D). To test whether asparagine was utilized to synthesize other nonessential amino acids, we quantified the levels of other nonessential amino acids under those conditions by LC-MS. Here we only analyzed the level of alanine (Ala), proline (Pro), aspartate (Asp), and glutamate (Glu), since they are not present in the DMEM and therefore have to be synthesized de novo from glutamine (Gln). Similar to the TCA cycle intermediates, we observed neither any restoration of Ala, Pro, Asp and Glu nor any restoration of Gln itself by asparagine addition (Figure 5E and Figure S4E). Consistently, substitution with Ala, Pro, Asp or Glu failed to suppress apoptosis as well as asparagine (Figure 5F). As a positive control we showed that α-KG can also rescue cells from glutamine deprivation (Figure 5A). In contrast to asparagine, α-KG also enhanced the level of TCA cycle intermediates and nonessential amino acids and restored cell proliferation albeit with a prolonged cell doubling time (Figure 5C, 5D and 5E).
Figure 5. Extracellular asparagine completely restores the viability, but not the TCA cycle intermediates, other nonessential amino acids or proliferation, in the absence of glutamine.
(A) SF188 cells were cultured in DMEM with the following modification: +Gln, −Gln, −Gln+αKG (5 mM) and -Gln+Asn (4 mM) for 48 hours. Viability was measured by Annexin V and PI staining. Data are shown as mean +/− S.D., n=3.
(B) SF188 cells were cultured in DMEM without glutamine in the presence of various concentrations of asparagine for 4 days. Viability was measured by Annexin V and PI staining. Data are shown as mean +/− S.D., n=3.
(C) SF188 cells were grown in DMEM with the following modification: +Q, −Q+N (4 mM) and −Q+αKG (5 mM) for 3 days and cell number was recorded. Data are shown as mean +/− S.D., n=3. The p-value was determined by using Student’s 2 tailed t-test.
(D) SF188 cells were cultured in the same conditions as panel (A) for 16 hours. Intracellular citrate, α-ketoglutarate, malate and fumarate were quantified by GC-MS and normalized to cellular protein content. Data are shown as mean + S.D., n=3.
(E) SF188 cells were cultured in the same conditions as panel (A) for 16 hours. Intracellular levels of alanine (Ala), proline (Pro), aspartate (Asp), glutamate (Glu) and glutamine (Gln) were quantified by LC-MS and normalized to total cell volume. Data are shown as mean + S.D., n=3.
(F) SF188 cells were cultured in DMEM in the absence of glutamine (−Q), individually supplemented with 0.2 mM alanine (−Q+A), glutamate (−Q+E), proline (−Q+P), aspartate (−Q+D), or asparagine (−Q+N). Apoptotic induction percentage was defined by Annexin V positive staining 2 days post medium change and then subtracted by the basal level (cells grown in complete DMEM with glutamine) of apoptotic rates. Data are shown as mean + S.D., n=3.
See also Figure S4.
Asparagine Synthetase is Required for Glutamine-dependent Survival
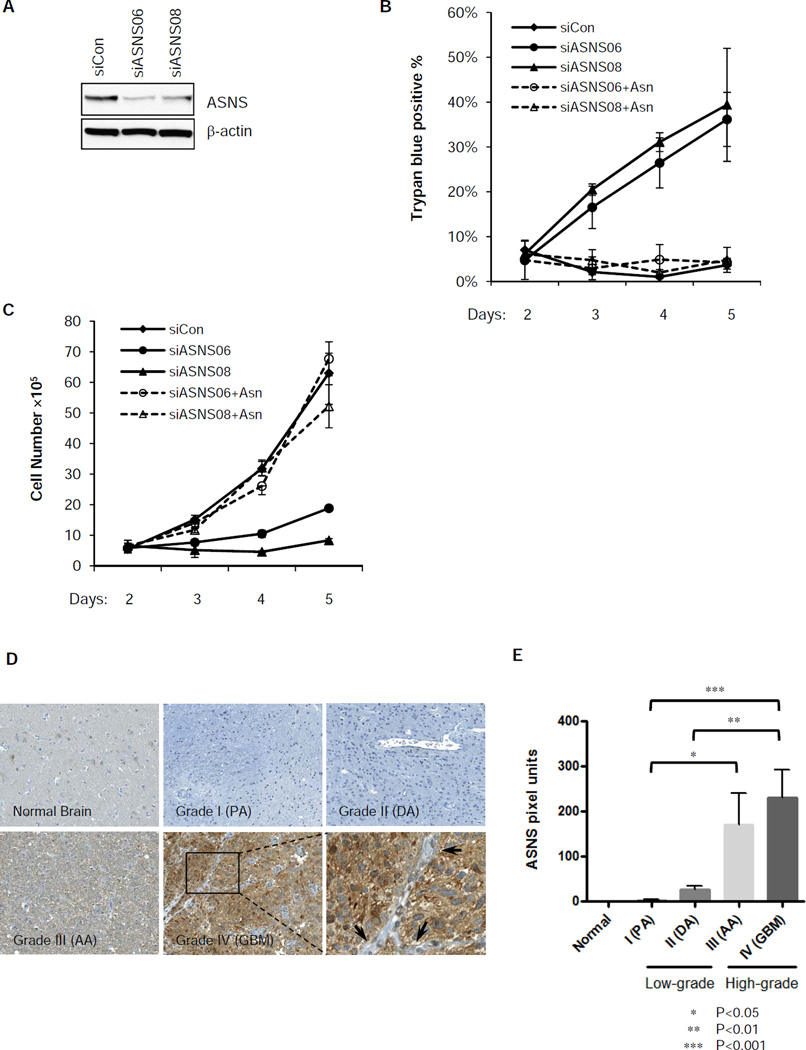
The evidence above suggests that asparagine plays a critical role in suppressing apoptosis. The importance of asparagine to the survival of some tumor cell types has already been established through the use of L-asparaginase as a therapy of childhood/adolescent hematopoietic tumors. Hematopoietic cells lack the ability to synthesize asparagine from glutamine and thus are dependent on exogenous asparagine to maintain their viability (Haskell and Canellos, 1969). In contrast, solid tumors express asparagine synthetase (ASNS) and synthesize asparagine from glutamine. To address the question whether the anti-apoptotic function of glutamine depends on the ability of asparagine synthetase to maintain glutamine-dependent biosynthesis of asparagine, we used siRNA to deplete asparagine synthetase, the enzyme that transfers the gamma amino group of glutamine to aspartate and therefore generates asparagine. ASNS knockdown led to profound apoptosis even in the presence of glutamine (Figure 6A and 6B). Addition of extracellular asparagine completely restored cell survival and proliferation under these conditions (Figure 6B and 6C).
Figure 6. Expression of asparagine synthetase (ASNS) is required for glutamine-dependent survival and is correlated with poor prognosis in human glioma.
(A) SF188 cells were transfected with siRNA targeting ASNS or control for 2 days. Knockdown of ASNS was confirmed by western blotting.
(B) SF188 cells were transfected with control or ASNS siRNA for 48 hours, then cultured in DMEM with glutamine with or without extracellular asparagine (4 mM). Cell death was measured by Trypan blue staining from day 2 to 5 post-transfection. Data are shown as mean +/− S.D., n=3.
(C) SF188 cells were transfected with control or ASNS siRNA and cultured in the same conditions as panel (B). Viable cell number was recorded from day 2 to 5 post-transfection by Trypan blue exclusion. Data are shown as mean +/− S.D., n=3.
(D) Tissues from normal brain or tumors of glioma patients were stained for ASNS by immunohistochemistry. Representative images of each category, including normal brain, grade I (pilocytic astrocytomas, PA), grade II (diffuse astrocytomas, DA), grade III (anaplastic astrocytomas, AA) and grade IV (glioblastomas, GBM), were shown. A magnified region of ASNS staining of a GBM sample is provided, which shows cytoplasmic distribution of the staining. The arrow heads point to absence of staining of the vasculature within the tumor.
(E) The intensity of ASNS staining based on pixel units of each image were quantified and summarized for each category. Data are shown as mean + S.E.M., normal brain (n=3), PA (n=3), DA (n=5), AA (n=10), GBM (n=31). See extended experimental procedures for definition of each category. All the p-values were determined by using Student’s 2 tailed t-test.
See also Figure S5.
Expression of Asparagine Synthetase in Glioma and Neuroblastoma Correlates with Poor Prognosis
The data suggests that asparagine synthetase (ASNS) might play an important role during tumor cell accumulation and progression by maintaining cell viability. To gain insights into whether the expression of ASNS correlates with the prognosis of patients with solid tumors, we performed immunohistochemical staining for ASNS across a panel of primary human glioma and neuroblastoma tissues. ASNS expression was not detected in normal brain tissue, and the expression was marginal in low-grade pilocytic astrocytoma (Grade I) and diffuse astrocytoma (Grade II); however, in high-grade anaplastic astrocytoma (Grade III) and glioblastoma (Grade IV), ASNS expression was significantly increased (p<0.001 between grade IV and I, p<0.01 between grade IV and II, p<0.05 between grade III and I) (Figure 6D and 6E). Similarly, ASNS expression was low in normal adrenal cortex and neuroblastoma cases with favourable prognosis (Figure S5A and S5B). In neuroblastoma with unfavourable prognosis, ASNS expression was significantly higher (p<0.05) (Figure S5A and S5B). Further, ASNS expression was inversely correlated with the degree of differentiation (p<0.05) and positively correlated with MYC amplification (p<0.05) in neuroblastomas. (Figure S5C and S5D).
Asparagine Depletion Converts the ATF4 Stress Response from Adaptation to Apoptosis
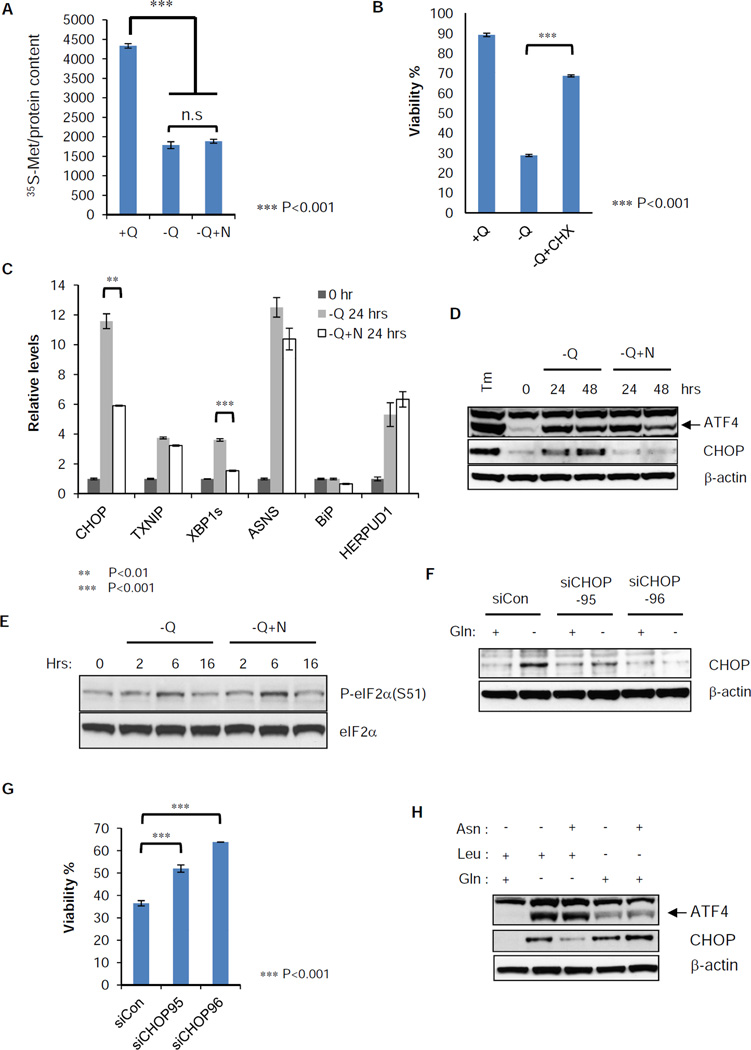
The only known use of asparagine in mammalian cells is in protein synthesis (Ubuka and Meister, 1971). Therefore, the simplest explanation of asparagine rescue of cell death is the ability to rescue protein synthesis. Glutamine withdrawal resulted in a 60% reduction of de novo protein synthesis. Asparagine failed to significantly increase total protein synthesis under these conditions (Figure 7A). Furthermore, complete suppression of protein synthesis by cycloheximide rescued cells from death upon glutamine withdrawal (Figure 7B). Consistent with this, the unbiased pathway analysis from the secondary screen revealed that the RNAi suppression of proteins essential for Cap-dependent translation also suppressed apoptosis following glutamine withdrawal (Table S1). This suggested that asparagine is the regulator of a pathway that induces translation-dependent apoptosis.
Figure 7. Induction of endoplasmic reticulum (ER) stress marker genes by glutamine withdrawal is suppressed by asparagine addition.
(A) SF188 cells were grown in complete medium (+Q) or glutamine-deficient medium with (−Q+N) or without (−Q) asparagine (4 mM) for 16 hours, then switched to the same fresh medium without methionine. 0.1 mCi 35S-methionine was pulsed for 30 minutes and whole cell extract was prepared. Radioactivity of labeled protein was measured in a scintillation counter and normalized to total protein content.
(B) SF188 cells were cultured 48 hours in DMEM with the following modifications: with glutamine (+Q), without glutamine (−Q) or without glutamine in the presence of cycloheximide (1 µg/mL) (−Q+CHX). Viability was measured by Annexin V and PI staining.
(C) SF188 cells were cultured in DMEM without glutamine (−Q) or without glutamine but with asparagine (4 mM) (−Q+N) for 24 hours. Q-VD was added at 20 µM to prevent cell death. mRNA was extracted and Q-PCR was performed to detect relative abundance of ER stress marker genes, CHOP, TNXIP, XBP1s, ASNS, BiP and HERPUD1 normalized to 18s ribosomal RNA.
(D) SF188 cells were cultured as in panel (C) for 48 hours. Protein extracts were prepared at 0, 24 and 48 hours for western blotting of ATF4 and CHOP. Tunicamycin (Tm) was added at 10 µg/ml for 4 hours as a positive control.
(E) SF188 cells were switched to minus glutamine DMEM with (−Q+N) or without (−Q) asparagine (4 mM) for 16 hours. Protein extracts were prepared at 0, 2, 6 and 16 hours after medium change. Western blotting was performed for phospho-eIF2α (S51) and total eIF2α.
(F) SF188 cells were transfected with control or CHOP siRNA for 2 days. Then glutamine was withdrawn for 24 hours in the presence of Q-VD (20 µM). Whole cell extracts were prepared for western blotting of CHOP.
(G) SF188 cells were transfected with control or CHOP siRNA for 2 days. Then glutamine was withdrawn for 48 hours, and viability was measured by Annexin V staining.
(H) SF188 cells were cultured in DMEM without glutamine (Gln) or leucine (Leu) individually. 4 mM asparagine (Asn) was added or not added to each amino acid deficient medium for 24 hours. Q-VD was added at 20 µM to prevent cell death and protein extract was prepared for western blotting of ATF4 and CHOP.
The data in Figure 7 (A, B, C and G) are shown as mean +/− S.D., n=3, and the p-values are determined by using Student’s 2 tailed t-test. See also Table S1.
Asparagine depletion has previously been reported to lead to an adaptive cellular response in which uncharged tRNA activates the serine threonine kinase GCN2 (Dong et al., 2000; Hao et al., 2005). GCN2 in turn phosphorylates the translation initiation factor eIF2α resulting in increased translation of the transcription factor ATF4 from a downstream open reading frame (Harding et al., 2000). As part of the reported adaptive pathway ATF4 induces asparagine synthetase which results in glutamine-dependent asparagine synthesis from aspartate (Ye et al., 2010). In turn asparagine accumulation then suppresses GCN2 and reduces ATF4. However, in our experiments, cells are deficient in glutamine and this feedback loop is impaired. Asparagine addition to glutamine-deprived cells altered the transcriptional response, suppressing the induction of the reported UPR effectors CHOP and XBP1 while maintaining the transcriptional induction of adaptive components of the UPR-response such as ASNS and HERPUD1 (Figure 7C). At the protein level, exogenous addition of asparagine suppressed CHOP induction without altering ATF4 accumulation or upstream eIF2α phosphorylation (Figure 7D and 7E). Consistent with this effect mediating asparagine-induced suppression of apoptosis, RNAi inhibition of CHOP suppressed apoptosis in response to glutamine withdrawal in the absence of asparagine (Figure 7F and 7G). Asparagine’s ability to promote cell survival was restricted to depletion of glutamine. Asparagine addition had no effect on apoptosis in response to depletion of essential amino acids, including leucine, threonine, and phenylalanine (Figure S4B). The failure of asparagine to rescue death in the absence of leucine correlated with the inability of asparagine to suppress CHOP induction following leucine deprivation (Figure 7H).
DISCUSSION
Proliferating cells utilize glutamine to maintain anaplerosis of the TCA cycle and to support the production of nucleotides and nonessential amino acids (Wise and Thompson, 2010). Many cancer cell lines exhibit dependence on glutamine to maintain cell survival and cell growth (Wise et al., 2008; Yuneva et al., 2007), and glutamine is the most commonly depleted amino acid in primary solid tumors (Roberts et al., 1956). Despite the fact that glutamine is a nonessential amino acid and can be synthesized from glucose-derived α-ketoglutarate, many cancer cells die when depleted of extracellular glutamine. Under these same conditions most non-transformed cells undergo cell cycle arrest and metabolic adaptation. The existing evidence suggests that maintenance of the TCA cycle is critical for glutamine-dependent survival. However, the fact that inhibition of CS, the first enzyme of the TCA cycle, actually prevents glutamine depletion-induced apoptosis challenges this model. Here we show that while glutamine-deprived cancer cells do undergo cell cycle arrest and reduce their rate of protein translation, the translation of stress response RNAs including CHOP result in the activation of apoptosis. Surprisingly, addition of a single amino acid, asparagine, can specifically suppress that apoptosis through the suppression of CHOP induction without altering the upstream signaling cascade, including eIF2α phosphorylation and ATF4 accumulation. The consequence of ATF4 induction in tumor cell survival during metabolic stress is controversial (Qing et al., 2012; Ye et al., 2010). Here we show that the restoration of a single nonessential amino acid, asparagine, is sufficient to suppress the apoptotic function of ATF4 while retaining its ability to control metabolic adaptation.
In support of the model that anaplerosis per se is not required for survival, asparagine addition does not restore the levels of TCA cycle intermediates or other nonessential amino acids. Asparagine-rescued cells undergo cell cycle arrest and exhibit long-term survival. Furthermore, in glutamine-replete cells suppression of glutamine-dependent asparagine synthetase is sufficient to induce apoptosis. Together, the data suggest that asparagine plays a critical role in the regulation of the cellular adaptation to depletion of glutamine. Sustained levels of asparagine suppress apoptosis and promote cellular adaptation to depletion of glutamine and other nonessential amino acids, while intracellular depletion of asparagine induces apoptosis even in the face of an abundant supply of glutamine and other nonessential amino acids.
Extracellular depletion of asparagine using L-asparaginase has been used successfully as a chemotherapeutic treatment of acute lymphoblastic leukemia (ALL) (Avramis, 2012). The leukemic cells lack constitutive expression of asparagine synthetase, and die when deprived of exogenous asparagine (Aslanian et al., 2001; Haskell and Canellos, 1969). In contrast, most solid tumors express glutamine-dependent asparagine synthetase and synthesize asparagine de novo (Balasubramanian et al., 2013), and L-asparaginase has not proven an effective therapy. This has led to the belief that asparagine regulation of leukemic cell survival is a special case. The present data argue that intracellular asparagine plays a more generalized role in regulating cell survival than previously believed.
Together, the past and present data suggest that asparagine has evolved to be a metabolic regulator of cell fate. Unlike the other 19 common amino acids, the only reported use of asparagine in mammalian cells is in protein synthesis. It is not utilized in other synthetic pathways (Ubuka and Meister, 1971), nor is it degraded to aspartate and oxaloacetate at any measurable rate in the cells we have studied. It is the last nonessential amino acid synthesized from glucose metabolism in the glycolytic pathway/ TCA cycle and its amination exclusively depends on glutamine. Asparagine production is therefore an ideal gauge of the availability of TCA cycle intermediates and supply of reduced nitrogen needed to maintain nonessential amino acid synthesis. Consistent with this idea, the intracellular concentration of asparagine is the lowest among the nonessential amino acids synthesized by proliferating cells (Table S2). Asparagine’s ability to directly suppress apoptosis in metabolically compromised cells may also explain its important role as a nutritional/ dietary supplement for patients with inborn errors of metabolism affecting the TCA cycle (Oizumi et al., 1984; Oizumi et al., 1983). Cells from such patients are highly susceptible to apoptosis and the use of asparagine in large quantities may be directly suppressing apoptosis in such patients.
Asparagine synthetase (ASNS) is a transcriptional target of ATF4 in response to amino acid starvation through the GCN2/eIF2α axis. The activation of the GCN2/eIF2α/ATF4 pathway has been reported in primary solid tumors and GCN2- or ATF4-deficient cells failed to give rise to tumors in vivo (Horiguchi et al., 2012; Ye et al., 2010), suggesting that to maintain asparagine production is critical for solid tumor progression in their nutrient-limited environment. The present results also suggest that inhibition of ASNS should be explored as potential therapy in solid tumors. First, the expression of asparagine synthetase correlates with the progression and poor prognosis of tumors from glioma and neuroblastoma patients. Second, inhibition of asparagine synthetase induces cell death in glioblastoma cell line in culture only when exogenous asparagine is absent. Currently such inhibitions have only been tested in L-asparaginase-refractory cases of acute leukemia. Such agents may well have an effective therapeutic index in other cancers when used together with L-asparaginase, as our data suggest that a cell’s sensitivity to apoptosis upon asparagine depletion is dependent on the rate of ongoing protein synthesis. Cell death following glutamine withdrawal can be suppressed by cycloheximide. In conclusion, the present data add asparagine to a growing list of metabolites that cells utilize to coordinate cellular responses with metabolic reserves.
EXPERIMENTAL PROCEDURES
Cell Culture and Media
Human glioblastoma cell line SF188 (UCSF), neuroblastoma cell lines Kelly and NBLS (gift from Dr. Celeste Simon) and mouse embryo fibroblast (MEF) cells are maintained in DMEM with 10% FBS, 25 mM glucose, 6 mM glutamine, 100 units/ml penicillin and 100 ug/ml streptomycin and split every 2–3 days before reaching confluency. For glutamine starvation, cells were briefly rinsed with PBS and cultured in DMEM supplemented with 10% dialyzed FBS (dFBS), 100 units/ml penicillin and 100 ug/ml streptomycin but without glutamine.
siRNA Transfection and Western Blotting
See “Extended Experimental Procedures” for detailed information of siRNAs. SF188 cells were reverse-transfected with siRNA mixed with Lipofectamine RNAiMAX (13778075, Life Technologies) in Opti-MEM® Reduced Serum Medium (31985070, Life Technologies) as described by the manufacturer. Two days post-transfection, protein extracts were prepared and equal amount of total protein was separated on NuPAGE Bis-Tris Gel (Life Technologies), transferred to nitrocellulose membrane and subjected to the incubation with individual primary antibodies (Extended Experimental Procedures).
Apoptosis and Cell Proliferation Assay
All glutamine starvation or media change experiments were done 2 days post-transfection or plating. For apoptosis analysis, 2×105 cells (alive plus dead) were collected, stained with Annexin V-FITC and propidium iodide (51-65874X, 51-66211E, BD Biosciences) and analyzed by FACS Calibur using the FL1 and FL3 channels. For proliferation assays, attached cells were collected and counted by using a Multisizer 4 particle analyzer (Beckman Coulter).
GC-Mass Spectrum and LC-Mass Spectrum Analysis of Metabolites
SF188 cells were transfected with siRNA for either control or CS. Two days post-transfection, the culture medium was replaced with fresh complete DMEM with or without glutamine for another 16 hours. Cells were briefly rinsed with PBS and incubated with 80% methanol pre-chilled to −80°C for 30 minutes at −80°C. Supernatant was collected by centrifugation. For GC-MS, the supernatant was dried by spin vacuum, dissolved in 40mg/mL methoxyamine in pyridine and derivatized with MSTFA+1% TMCS (TS-48915, Thermo Scientific). One microliter of trimethylsilyl-derivatized organic acids was analyzed through an Agilent 7890A GC equipped with an HP-5MS capillary column and connected to an Agilent 5975 C mass selective detector. The LC-MS method involved reversed-phase ion-pairing chromatography coupled by negative mode electrospray ionization to a stand-alone orbitrap mass spectrometer (Thermo Scientific) scanning from m/z 85–1000 at 1 Hz at 100,000 resolution (Lemons et al., 2010; Lu et al., 2010; Munger et al., 2008) with LC separation on a Synergy Hydro-RP column (100 mm × 2 mm, 2.5 µm particle size, Phenomenex, Torrance, CA) using a gradient of solvent A (97:3 H2O/MeOH with 10 mM tributylamine and 15 mM acetic acid), and solvent B (100% MeOH). The gradient was 0 min, 0% B; 2.5 min, 0% B; 5 min, 20% B; 7.5 min, 20% B; 13 min, 55% B; 15.5 min, 95% B; 18.5 min, 95% B; 19 min, 0% B; 25 min, 0% B. Injection volume was 10 µL, flow rate 200 µl/min, and column temperature 25 °C. Data were analyzed using the MAVEN software suite (Melamud et al., 2010). All the values of the compounds from mass spectrum were corrected to the internal spiked-in standard of D5-2-hydroxyglutarate (D5-2HG) before further normalization.
Supplementary Material
HIGHLIGHTS.
Glutamine depletion-induced apoptosis results from asparagine depletion
Reducing citrate synthase levels promotes aspartate/asparagine biosynthesis
Asparagine is sufficient to suppress apoptosis in response to glutamine-depletion
Asparagine promotes ATF4-dependent adaptive stress responses
ACKNOWLEDGEMENTS
We thank members of the Thompson laboratory, particularly Jiangbin Ye, for critical discussions and suggestions for the manuscript. We also thank the Flow Cytometry and the Molecular Cytology Core Facilities of MSKCC for their help with flow cytometry and imaging data. The HTS Core Facility is supported by Mr. William H. Goodwin and Mrs. Alice Goodwin, the Commonwealth Foundation for Cancer Research, the ETC of MSKCC, the Lillian S. Wells Foundation, and by a NIH/NCI Cancer Center Support Grant P30 CA008748. J.Z was supported by the Leukemia and Lymphoma Society fellowship. J.F was supported by HHMI international student research fellowship. S.V is supported by NIH K08 CA181475. This work was also supported, in part, by grants from NIH R01 CA105463, the Abramson Family Cancer Research Institute, and Memorial Sloan Kettering Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aslanian AM, Fletcher BS, Kilberg MS. Asparagine synthetase expression alone is sufficient to induce l-asparaginase resistance in MOLT-4 human leukaemia cells. Biochem J. 2001;357:321–328. doi: 10.1042/0264-6021:3570321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramis VI. Asparaginases: biochemical pharmacology and modes of drug resistance. Anticancer Res. 2012;32:2423–2437. [PubMed] [Google Scholar]
- Balasubramanian MN, Butterworth EA, Kilberg MS. Asparagine synthetase: regulation by cell stress and involvement in tumor biology. Am J Physiol Endocrinol Metab. 2013;304:E789–E799. doi: 10.1152/ajpendo.00015.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Mates JM, DeBerardinis RJ. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci U S A. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell. 2012;47:349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- Hao S, Sharp JW, Ross-Inta CM, McDaniel BJ, Anthony TG, Wek RC, Cavener DR, McGrath BC, Rudell JB, Koehnle TJ, et al. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science. 2005;307:1776–1778. doi: 10.1126/science.1104882. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Haskell CM, Canellos GP. l-asparaginase resistance in human leukemia--asparagine synthetase. Biochem Pharmacol. 1969;18:2578–2580. doi: 10.1016/0006-2952(69)90375-x. [DOI] [PubMed] [Google Scholar]
- Horiguchi M, Koyanagi S, Okamoto A, Suzuki SO, Matsunaga N, Ohdo S. Stress-regulated transcription factor ATF4 promotes neoplastic transformation by suppressing expression of the INK4a/ARF cell senescence factors. Cancer Res. 2012;72:395–401. doi: 10.1158/0008-5472.CAN-11-1891. [DOI] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemons JM, Feng XJ, Bennett BD, Legesse-Miller A, Johnson EL, Raitman I, Pollina EA, Rabitz HA, Rabinowitz JD, Coller HA. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 2010;8:e1000514. doi: 10.1371/journal.pbio.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oizumi J, Donnell GN, Ng WG, Mulivor RA, Greene AE, Coriell LL. Congenital lactic acidosis associated with pyruvate carboxylase deficiency. Repository identification No. GM6056. Cytogenet Cell Genet. 1984;38:80. doi: 10.1159/000132035. [DOI] [PubMed] [Google Scholar]
- Oizumi J, Shaw KN, Giudici TA, Carter M, Donnell GN, Ng WG. Neonatal pyruvate carboxylase deficiency with renal tubular acidosis and cystinuria. J Inherit Metab Dis. 1983;6:89–94. doi: 10.1007/BF01800731. [DOI] [PubMed] [Google Scholar]
- Qing G, Li B, Vu A, Skuli N, Walton ZE, Liu X, Mayes PA, Wise DR, Thompson CB, Maris JM, et al. ATF4 Regulates MYC-Mediated Neuroblastoma Cell Death upon Glutamine Deprivation. Cancer Cell. 2012;22:631–644. doi: 10.1016/j.ccr.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E, Simonsen DG, Tanaka KK, Tanaka T. Free amino acids in growing and regressing ascites cell tumors: host resistance and chemical agents. Cancer Res. 1956;16:970–978. [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman LA, Holton T, Yuneva M, Louie RJ, Padro M, Daemen A, Hu M, Chan DA, Ethier SP, van 't Veer LJ, et al. Glutamine Sensitivity Analysis Identifies the xCT Antiporter as a Common Triple-Negative Breast Tumor Therapeutic Target. Cancer Cell. 2013;24:450–465. doi: 10.1016/j.ccr.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubuka T, Meister A. Studies on the utilization of asparagine by mouse leukemia cells. J Natl Cancer Inst. 1971;46:291–298. [PubMed] [Google Scholar]
- van der Vos KE, Eliasson P, Proikas-Cezanne T, Vervoort SJ, van Boxtel R, Putker M, van Zutphen IJ, Mauthe M, Zellmer S, Pals C, et al. Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. Nat Cell Biol. 2012;14:829–837. doi: 10.1038/ncb2536. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems L, Jacque N, Jacquel A, Neveux N, Trovati Maciel T, Lambert M, Schmitt A, Poulain L, Green AS, Uzunov M, et al. Inhibiting glutamine uptake represents an attractive new strategy for treating acute myeloid leukemia. Blood. 2013;122:3521–3532. doi: 10.1182/blood-2013-03-493163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. Embo J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.