Abstract
The spatial resolution of chemical images acquired with cluster secondary ion mass spectrometry (SIMS) is limited not only by the size of the probe utilized to create the images, but also by detection sensitivity. As the probe size is reduced to below 1 µm, for example, a low signal in each pixel limits lateral resolution due to counting statistics considerations. Although it can be useful to implement numerical methods to mitigate this problem, here we investigate the use of image fusion to combine information from scanning electron microscope (SEM) data with chemically resolved SIMS images. The advantage of this approach is that the higher intensity and, hence, spatial resolution of the electron images can help to improve the quality of the SIMS images without sacrificing chemical specificity. Using a pan-sharpening algorithm, the method is illustrated using synthetic data, experimental data acquired from a metallic grid sample, and experimental data acquired from a lawn of algae cells. The results show that up to an order of magnitude increase in spatial resolution is possible to achieve. A cross-correlation metric is utilized for evaluating the reliability of the procedure.
Introduction
Molecule specific imaging is arguably the quintessential operating modality of time-of-flight secondary ion mass spectrometry (SIMS) experiments. The combination of high chemical selectivity and sub-micron spatial resolution are unique in materials characterization methods, particularly among those that target biomolecules. Yet, many challenges remain to be overcome for this approach to reach its full potential. Sensitivity is particularly problematic since the number of molecules available for mass spectrometric detection is fundamentally limited. There are 106 molecules per layer per 1 micron pixel, for example, and the ionization efficiency is typically between 10−3 and 10−8. Even though the recent implementation of molecular depth profiling via cluster ion beams allows multiple layers to be summed for increased sensitivity [1], more often than not, chemical image quality suffers from low count rates. Reduced signal-to-noise ratios not only introduce uncertainties in image interpretation, but also severely limit potential lateral resolution due to counting statistics [2].
As a consequence of these issues, numerical chemometric methods have been extensively explored with varying degrees of success to optimize the information content of chemical images. Multivariate image analysis (MVA), such as, principal component analysis (PCA) and maximum autocorrelation factors (MAF) are perhaps the most widely-used approaches [3, 4]. Many other filtering and de-noising schemes including wavelet transform [5] and boxcar averaging [6] have been proposed. However, a chemical image characterized by low intensity is fundamentally limited by a lack of information and image enhancement can only be expected to provide incremental improvements.
Another approach, which involves the combination or fusion of images with different information, may overcome this fundamental limitation in some cases. Disciplines that have benefited from image fusion techniques include remote sensing [7], computer vision [8], satellite imagery [7], night vision [8, 9], weather forecasting [8], forensic science [10], and medical imaging [11, 12]. Multiple sensors are used to acquire two or more images of the same scene, where different, more specific information is contained in each of the images. [13, 14] In surface analysis, correlation of data sets from multiple techniques, such as X-ray photoelectron spectroscopy (XPS), atomic force microscopy (AFM) and secondary electron microscopy (SEM), has been explored using MVA methods, [15, 16] including ToF-SIMS [17]. While previous examples utilize MVA, another successful subset of image fusion is pan-sharpening, a technique that merges one or more colored images that lack spatial resolution with a higher resolution panchromatic image. [18] Pan-sharpening has previously been used to fuse XPS and AFM data, by intensity substitution, resulting in a merging of elemental and topographical information. [19]
Here we explore the use of pan-sharpening to combine a lower resolution SIMS image, where chemical information is mapped by color, with a higher resolution, panchromatic SEM image, to yield a hybrid image that combines several desired properties. Most SIMS instruments are equipped with both an electron detector for acquiring SEM images at high spatial resolution and ion detectors which provide the chemical specificity of mass spectrometry, generally at lower spatial resolution. These properties, including topography, high spatial resolution, pixel intensity and chemical information, produce an image that has greater image quality and provides more information to the viewer than any of the input images would alone.
To demonstrate the efficacy and capabilities of image fusion with pan sharpening in this work, we first create synthetic images to illustrate the basic idea of image fusion when applied to SEM and SIMS input. Next, images of a gold-coated copper mesh grid are shown to reveal the effectiveness of the pan-sharpening technique using experimental data. Most importantly, the approach is applied to the SIMS imaging of B. braunii algal cells to answer a novel question of biological significance: the elucidation of its chemicals’ relation to the delicate and diverse morphological features of colony organization. The algal cells grow in large colonies surrounded by a chemically and morphologically complex extracellular matrix where the purpose of morphological attributes and how they relate to the distribution of its molecular components’ distributions are not well-understood. [20, 21] This strain of algae is a leading candidate for biofuel use because of their ability to synthesize and accumulate excessive long-chain hydrocarbons, therefore, great interest has arisen in trying to elucidate its colonial organization to promote advance in chemical and genetic engineering. [22] Finally, cross correlation, a statistical measure of the change in spatial resolution of each color channel, [23] is applied to the grid and algae data sets to quantify the improved visual quality of a fused image.
Experimental
Preparation of simulated images
A simulated SEM image was created with a vertical solid red line 32 pixels wide down the center of a larger image of 512 × 512 pixels. All pixels inside the line have a relative intensity of 1. Simulated SIMS images were constructed with a vertical line of variable thickness down the center of a larger image consisting of 256 × 256 pixels.
Gold/Copper mesh grid preparation
A half-coated, 600 mesh copper grid (Electron Microscopy Sciences, Ft. Washington, PA) was the basis for the first image fusion experiment. Initially, half of the grid was protected with foil and, subsequently, approximately 75 nm of gold was vapor deposited onto the grid. After removal of the foil, the grid consisted of a gold region and a copper region, delineated by a sharp interface.
SIMS Gold/Copper mesh grid analyses
Chemical imaging was performed using the J105 – 3D Chemical Imager (Ionoptika Ltd., UK), described previously. [24] In both SEM and SIMS image acquisitions, a 40 keV C60+ primary ion beam was used. Three acquisitions were performed:
High resolution SIMS image: 150 µm × 150 µm field of view, 512 × 512 pixels, 300 nm primary ion beam focus and beam dose of 1.46 × 1013 ions/cm2
Low resolution SIMS image: 150 µm × 150 µm field of view, 128 × 128 pixels, approximately 1 µm primary ion beam focus and beam dose of 9.10 × 1012 ions/cm2
SEM image: 150 µm × 150 µm field of view, 512 × 512 pixels, approximately 300 nm primary ion beam focus
Because both the electron and ion detectors are located in the same analysis chamber, the images are all acquired without moving the sample. Thus, SEM and SIMS images are of the same location and registration is not necessary.
Algal Culture and preparation
B. braunii race A (UTEX 2441) was purchased from The Culture Collection of Algae, The University of Texas at Austin. The alga was grown in sterile Bold 3N Medium (The Culture Collection of Algae). The culture vessels (25-mL cell culture flasks with filter cap) were agitated by a shaker operating at 33 rpm inside an incubator kept at 20 °C and exposed to 115 µEm−2s−1 light intensity with a 16-hour light: 8-hour dark cycle.
A 3-mL aliquot of an algal suspension was centrifuged at 3,000 rpm for 3 minutes to remove growth media. The algal cell pellet was washed three times with 0.15 M ammonium formate (Alfa Aesar, 97%) solution to remove excess media materials. A Si-shard cleaned by sonication 3× with methanol and water solutions, respectively. 10 µL of the algal cell pellet was deposited on the shard and dried in a desiccator before introduction into the mass spectrometer.
SEM and SIMS Algal analysis
Room temperature analysis of algal cells was also performed with the J105 – 3D Chemical Imager. For SEM and SIMS, a primary ion beam of 40 keV C60+ at a beam current of 0.5pA was focused to approximately a 300 nm beam diameter, as measured by line scan.
First, the extracellular area of the algal cell colonies was analyzed. A pair of SEM (512 × 512 pixels) and SIMS (256 × 256 pixels) images was acquired to fuse over the same 150 µm × 150 µm field of view, resulting in a pixel size of 290 and 590 nm, respectively. No field-induced shift in the primary ion beam was observed, so the images are of the same spatial location and, thus, image registration was not necessary. The SIMS image was acquired with an ion fluence of 4.55 × 1012 ions/cm2.
To analyze interior features of the algal cell colonies, the extracellular matrix and cell membranes first must be removed. The C60 primary ion beam was used to etch a 200 µm × 200 µm crater partially through the algal cell colonies (2.33 × 1014 ions/cm2 primary ion dose, approximately 1 µm deep) before acquiring an SEM and SIMS image pair. The SEM (512 × 512 pixels) and SIMS (256 × 256 pixels) images were acquired over a 100 µm × 100 µm field of view for a pixel size of 195 nm and 390 nm, respectively. A field-induced shift of the primary ion beam was observed and thus the SEM and SIMS images were slightly offset from each other. Manual overlap of features present in both images determined the beam shift to be approximately 4% and the overlapping areas of the two images were cropped, resulting in an SEM image that was 468 × 468 pixels and a SIMS image that was 234 × 234 pixels. The field of view for the cropped images was 91 µm × 91 µm. The ion dose for the SIMS image was 7.17 × 1013 ions/cm2.
Pan-sharpening algorithm
Pan-sharpening is a pixel-level image fusion technique [25] aimed toward preserving the integrity of color in the SIMS images. Here, we employ a method that converts pixel color from RGB (red, green, blue) to HSL (hue, saturation, lightness) color space in order to separate the brightness (lightness) of the pixel from the color. The original lightness is then combined with intensity information from the corresponding pixel in a higher resolution panchromatic, or grayscale, image to create an adjusted lightness. Finally, the adjusted color is converted back to RGB color space to create a final, fused bitmap image with the same pixel resolution as the input panchromatic image [7].
Before performing pan-sharpening could be applied, both images must be of the same location so that signal from both input images exactly correlates. If necessary, image registration must be performed to align features in the pair of images. If the images are shifted, manual overlapping of distinguishable features is feasible to crop the area present in both input images. If more complex transformations are necessary, such as rotation and scaling, several automatic techniques exist for which detect features, regions and edges in images and selects the overlapping areas. [26] The pan-sharpening algorithm applied herein then up-converts the SIMS image to match the pixel resolution of the corresponding SEM image by expanding pixels using bilinear interpolation. Next, the panchromatic SEM image is converted to a single grayscale value. Each pixel of the colored SIMS image is broken into its RGB components and converted to HSL color space. Once the conversion is finished, the means and standard deviations for the lightness at each pixel for both the SEM and the SIMS image are calculated.
Following the conversions, an adjusted lightness is calculated for each pixel using the following equation, adapted from a viable and successful pan-sharpening formula based on both visual perception and quantitative metrics [23]:
| (1) |
µSIMS and µSEM are the means of the lightness and panchromatic intensity values, respectively; σSIMS and σSEM are the standard deviations of the lightness and panchromatic intensity values, respectively; and LSEM is the panchromatic intensity at the specific pixel. Finally, the adjusted lightness value is used when converting the SIMS image from HSL back to RGB color space. Since only the lightness value is modified, the algorithm preserves information from the original SIMS pixel, thus, the final color is considered to be representative of both input images. Additionally, the fused image has the same pixel count (512 × 512 pixels) as the input SEM image.
Results and Discussion
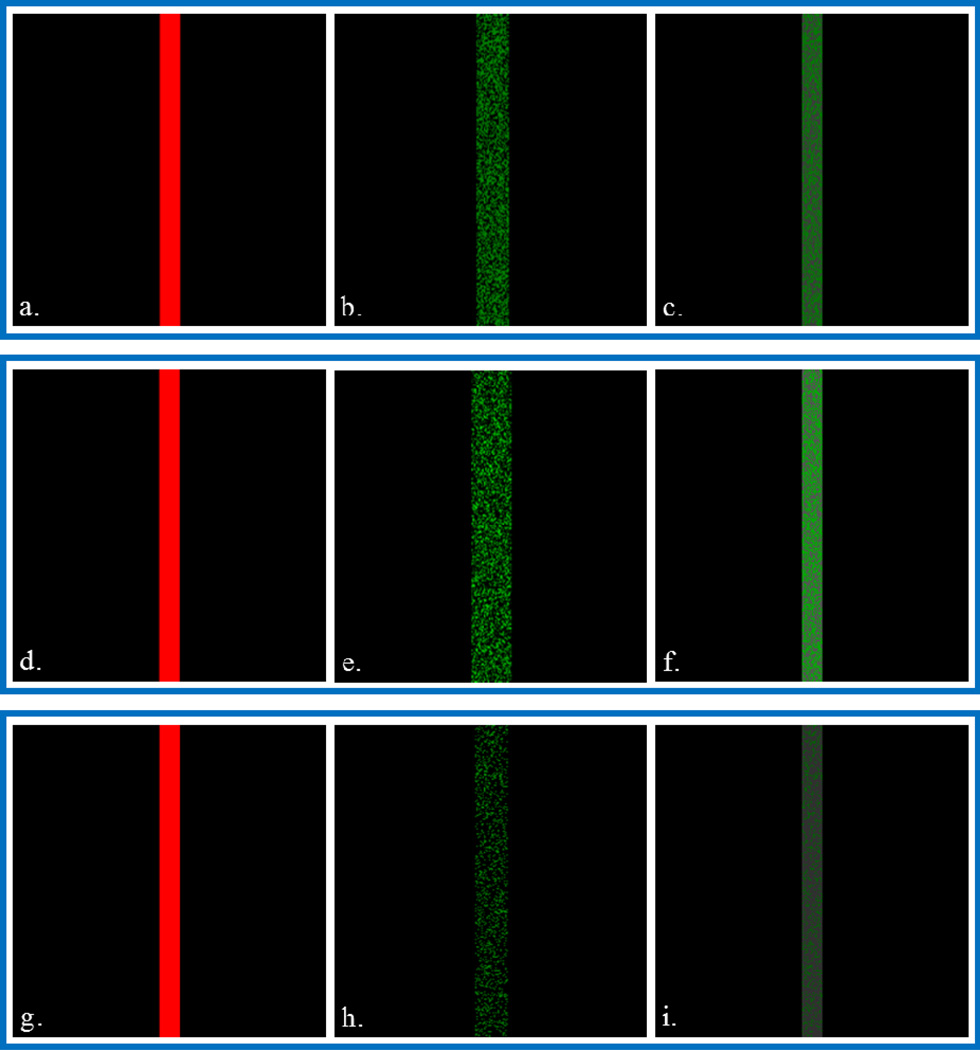
To demonstrate the capabilities of image fusion, simulated data sets each containing a SEM and a SIMS image were created using a custom algorithm. Model images, as well as the corresponding fused image, are shown in Figure 1. Since the intensity of a pixel inside the line and outside of the line is 1 and 0, respectively, the contrast at the pixel edge of the line is considered infinite.
Figure 1.
Left column is simulated SEM images (512 × 512 pixels, solid line is 32 pixels wide), middle column is simulated SIMS images (256 × 256 pixels), and right column is the corresponding fused image (512 × 512 pixels). For the SIMS images: (b) line is 50% fill, 26 pixels wide; (e) line is 50% fill, intensity of filled pixels is 1–100, 32 pixels wide; (h) line is 20% fill, 26 pixels wide. All pixel values are given with respect to the image’s resolution (512 or 256 pixels).
The line in Figure 1b is 26 pixels wide with a 50% fill, where the pixels were randomly filled with a one-in-two probability of having an intensity of 1 while the rest have an intensity of 0. Figure 1c is the resulting fused image, in which the line is 32 pixels wide, the same as in the SEM image in Figure 1a. No bleeding of the green color is observed in the range of pixels that are filled in the SIMS image but empty in the SEM image.
When interpreting the fused image in Figure 1c, there are four possible scenarios based on the combination of input data from the SEM image and the SIMS image. The first scenario is the simplest: both of the input pixels have 0 intensity, resulting in a black output pixel. The next scenario applies to pixels which are outside of the SEM line but do contain signal from the blurred SIMS line. In this case, the lightness of the SIMS image color is adjusted against an SEM intensity of 0. The mean intensities of the first SEM and SIMS image series, 0.0645 and 0.0524, respectively, are similar. However, the variance for the SEM is more than twice the SIMS image: 30.7 and 12.7, respectively. This variance results in a very small, nonzero adjusted lightness calculated from the pan-sharpening equation. Converting back to RGB color space results in black, regardless of whether the pixel in the SIMS image contained signal or not because the lightness is negligible.
The other two possible scenarios represented in Figure 1c involve a SIMS pixel that corresponds to an SEM pixel inside of the line. If a SIMS pixel contains signal, the original color is either increased or decreased, depending upon the intensities of both images. In the other case, the SIMS pixel does not contain signal. Even though there is no SIMS intensity, the information is still merged with the corresponding SEM intensity. Increasing the lightness of a black pixel will result in a gray color; this is observed in half of the pixels shown in Figure 1c. These four scenarios from Figure 1c demonstrate the underlying concepts of image fusion with respect to the information that is observed.
The effect of SIMS intensity on a resulting fused image is shown in Figures 1d–f. As in Figure 1b, each SIMS pixel (Figure 1e) inside of the 32 pixel-wide line has a 50% fill probability. However, here, the intensity of the pixel is assigned a random value from 1 – 100, such as in a typical SIMS experiment. The resulting fused image, Figure 1f, is similar to the fused image in Figure 1c where no bleeding is observed outside of the 32-pixel-wide line. The visible brightness of the line, when compared to Figure 1c, is due to the greater overall intensity of the SIMS image.
In addition, data in Figures 1(d–f) reveal a clear distinction between inside and outside of the line in the fused image. After up-converting the SIMS image to match the pixel resolution of the SEM image, the SIMS line becomes 64 pixels wide, which is double the width of the SEM line. Comparison of several varying SIMS line widths (not pictured) showed that the lack of bleeding exists up to a SIMS line twice the width as the SEM line, as is the case in Figures 1(d–f). As the SIMS line is widened further, low intensity bleeding increases. Recalling the pan-sharpening equation, low intensity green pixels are the consequence of an increased SIMS intensity average term, which is added to the SEM intensity. In the case of pixels outside of the SEM line but within the SIMS line, any green color intensity is decreased since the SEM intensity is still 0. However, the color does not become black due to the higher average SIMS intensity.
Finally, the SIMS image in Figure 1h is again 26 pixels wide. However the fill probability is 20%, which may be more representative of a typical SIMS experiment, and the intensity at a filled pixel is 1. From the resulting fused image in Figure 1i, in which, again, no bleeding is observed, it is clear that the fill percentage of the SIMS line has an important effect. As expected, the number of gray pixels increases relative those shown in Figure 1c since there are more pixels within the SEM line that have a corresponding SIMS pixel with no intensity. Note that the brightness of green pixels in the fused image is relatively dark, due to the lower mean SIMS intensity. Even with these results, however, clear boundaries between pixels inside the line and pixels outside the line are still observed in Figure 1i.
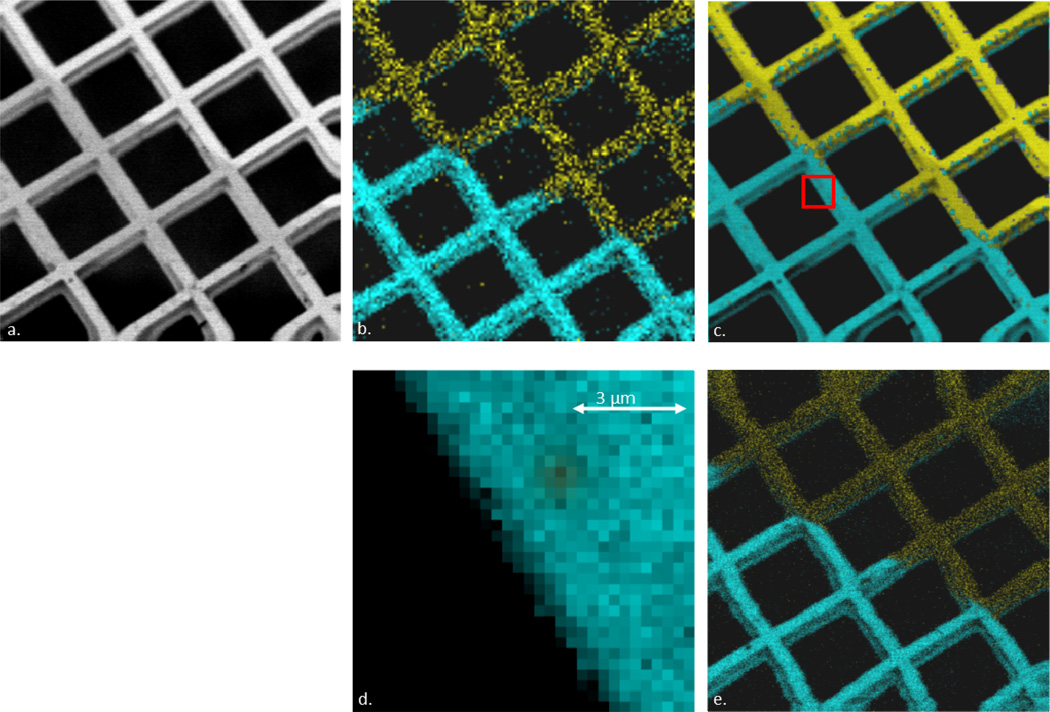
Establishing the applicability of image fusion to a SIMS experiment was accomplished by obtaining SEM and SIMS data at the coated-uncoated interface of a gold-coated copper grid, seen in Figure 2. The SIMS image shown in Figure 2b is obtained by chemically mapping the deposited gold signal and the copper signal from the uncoated portion of the grid, using a C60+ primary ion beam focused to approximately 1 µm diameter. The gold signal is the sum of three signals: Au+ (m/z 196.877) + Au2+ (m/z 393.766) + Au3+ (m/z 590.616). The copper signal is the sum of four signals: 63Cu+ (m/z 62.906), 65Cu+ (m/z 64.902), 63Cu2+ (m/z 125.798) and 65Cu2+ (m/z 129.804). The resulting fused image is shown in Figure 2c, and at higher magnification in Figure 2d. For comparison, Figure 2e is a SIMS image of the same region obtained with a 300 nm C60 probe is shown in Figure 2e. This image was obtained at 512 × 512 pixels, matching the resolution and beam size of the SEM image shown in Figure 2a.
Figure 2.
Fused image series of a Cu mesh grid. (a) SEM image (512 × 512 pixels); (b) SIMS image (~1 µm pixel resolution) of copper [teal; sum of 63Cu+ (m/z 62.906) + 65Cu+ (m/z 64.902) + 63Cu2+ (m/z 125.798) + 65Cu2+ (m/z 129.804)] and gold [yellow; sum of Au+ (m/z 196.877) + Au2+ (m/z, 393.766) + Au3+ (m/z 590.616)] signal (128 × 128 pixels); (c) Fusion image from figures 2a and 2b (512 × 512 pixels); (d) Zoomed area of figure 2c (32 × 32 pixels; approximately 9 µm × 9 µm); (e) High resolution SIMS image (~300 nm pixel resolution) of copper [teal; sum of 63Cu+ (m/z 62.906) + 65Cu+ (m/z 64.902) + 63Cu2+ (m/z 125.798) + 65Cu2+ (m/z 129.804)] and gold [yellow; sum of Au+ (m/z 196.877) + Au2+ (m/z, 393.766) + Au3+ (m/z 590.616)] signal (512 × 512 pixels); Field of view for all images (unless specified) is 150 µm × 150 µm.
The benefit of image fusion is clearly seen in Figure 2. The original SIMS image (Figure 2b) is somewhat pixelated, and the relatively lower pixel intensities, coupled with the lower resolution (128 × 128 pixels versus 512 × 512 pixels in the SEM image, Figure 2a), makes the gridlines appear dark and blurred. In addition, some noise signal is present in the area between gridlines, where there should be no signal present since the grid was placed over a hole in the sample block. The fused image in Figure 2c improves both the intensity and contrast issues observed in the lower resolution SIMS image. In addition, some features, such as defects in the grid and/or the coating, are visible in the SEM image but not in the low resolution SIMS image. These features, however, become discernible in the fused image.
The improved contrast is highlighted in the zoomed area of Figure 2c, shown in Figure 2d, where the gridline region contrasts very well with the black background. The zoomed region also demonstrates the limitation of the fusion algorithm in terms of resolution, as a sharp cutoff does not exist. Instead, a gradual fall-off of copper signal from the gridline to the background is observed. However, that gradual fall is now on the scale of ~1 µm instead of greater than 10 µm in the lower resolution SIMS image.
Finally, a high resolution SIMS image is shown in Figure 2e, which matches the pixel resolution and beam size of the SEM image in Figure 2a (512 × 512 pixels, 300 nm diameter) to support the claim that valuable information missing in lower resolution SIMS experiments could be extracted with image fusion. The desired contrast between the gridlines and the background is achieved in the high resolution SIMS image. However, the pixel intensities are still lower compared to the fusion result. Therefore, while obtaining a high resolution SIMS image at the cost of a large acquisition time is an improvement, the image quality is best in a fused image of a high-resolution SEM image and a low-resolution SIMS image.
A cross-correlation metric was implemented on the image set in order to quantitatively determine the effectiveness of the image fusion algorithm. Cross-correlation is used here to determine if information is lost in the fusion process. The equation used to calculate the cross-correlation value between signal A and signal B is:
| (2) |
where µA and µB are the mean values of each signal and i is the current pixel number. [23] For this application, A is the fused image and B is the SIMS image. Possible values fall within a range of −1 to 1, where 1 suggests that the two images are the same while −1 indicates dissimilar images. [23, 27] This procedure is repeated to allow each of the red, green and blue bands to be compared across the two images. Cross-correlation values for the fused series in Figure 2 are shown in Table 1.
Table 1.
Calculated Cross-correlation Values for Red, Green and Blue Channels between SIMS and Fused Images.
Considering that the range for a cross-correlation value is from −1 to 1, the results certainly favor the upper end of the range. For the gold-copper grid experiment, the cross correlation values of 0.9906, 0.9907 and 0.9825 for the red, green and blue channels, respectively, suggest very strong correlation between the input SIMS image and resulting fused image. In terms of spatial resolution, these numbers support the determination that the fused image quality is improved significantly from the original SIMS image without compromising the integrity of the SIMS data. These values are greater than a separate evaluation of this type of pan-sharpening technique where the method was confirmed. [23] Preserving as much of the original SIMS information is critical when processing images in order to maintain the validity of the data. Taking into account the high correlation and the visibly observable improvement in spatial resolution between the images, the copper grid images in Figure 2 demonstrate the applicability of image fusion to SIMS data.
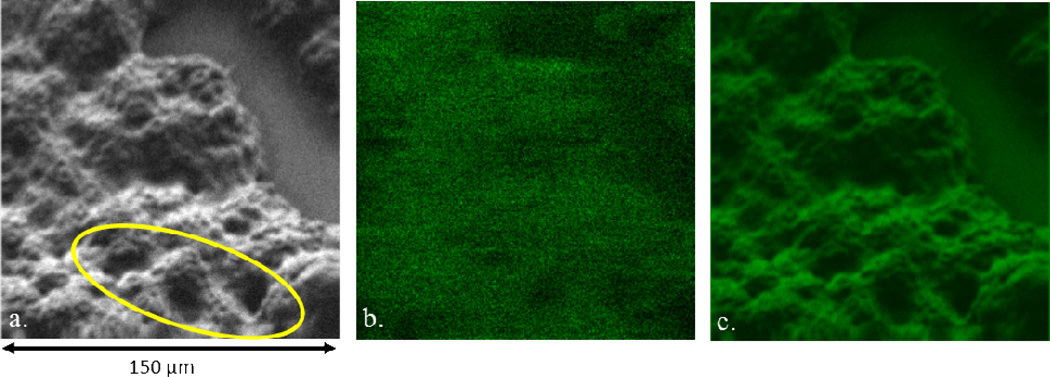
Next, to demonstrate the applicability of image fusion to a bioimaging experiment, the SEM and SIMS images of Botryococcus braunii algal cell colonies were examined, as shown in Figure 3a and b. The fused image is shown in Figure 3c. The UTEX 2441 race A B. braunii cells grow in colonies and the extracellular matrix material allows the algal cells to adhere to each other. [28] Several secondary ions in the range of m/z 400 to 600 localize to the extracellular matrix. The most prominent is m/z 530.5, a wax monoester previously discovered elsewhere. [29] In nature, wax monoesters have the ability to provide a protective function against bacterial infection and desiccation. Here, it is found to be a component of the sturdy and rigid extracellular matrix that is localized to the surface of the algal colony. [22, 30] Unlike the SEM image, the roughness of the extracellular material and the spherical-like protrusions belonging to the apex of the individual cells underneath cannot be distinguished in the SIMS image. Furthermore, the SIMS image is not able to differentiate between a patchy or continuous signal, therefore, it cannot be said whether this signal is located to specific morphological features or is continuously coating the extracellular matrix. This information is crucial to design hydrocarbon extraction techniques able to bypass a hydrophobic distribution of wax monoesters. However, in the fused image in Figure 3c, the m/z 530.5 distribution and roughness is clearly defined from the nearly submicron pitting covering the cell to the edges of the colony. Thus, the chemical information is able to imply a more meaningful significance – the signal is localized exactly to the relatively small and large features of the extracellular matrix.
Figure 3.
Fused image series of algal cells. The image on the left (a) is an SEM (512 × 512 pixels) of the identical field-of-view in the SIMS (256 × 256 pixels) image (b), while the fused image (c) is to the right. The field-of-view for all images is 150 µm × 150 µm.
The continuous coating of wax monoester on the algal colony can be probed further by examining the three prominent craters (approx. 20 µm in diameter) that appear as dark circular features in the highlighted region of Figure 3a. These features are blurred in the SIMS image. In particular, one of the craters is almost completely indistinguishable in the SIMS image over the seemingly un-localized signal inside of the crater. These three craters are prominent and defined in the fused image and reveal that some of the cells appear to protrude more from the colony than others, resulting in these deep craters in the extracellular material between cells. Without the use of the fused image, the SIMS image appears to be depicting a flat surface with large features and areas of low signal and continuity of the coating could not be definitely discerned. Instead, the chemical information is revealed to be indicative of the continuous, highly complex, morphology between cells which coats the protrusions and craters on the surface of colony. This morphological information about the system has been probed to a degree that would not be possible with only the SIMS information.
Worth noting in Figure 3 is that the extracellular area is detected in the SEM image, indicating that the area is not substrate. In addition, the SIMS data suggest that there may be more than just noise in that area and that the m/z used to identify the algal cells may overlap with some hydrocarbon cellular byproduct. Since the SEM image cannot distinguish between signal from the cells and signal from the extracellular matrix, the potential hydrocarbon signal interference in the SIMS image is carried over and appears in the fused image. This observation supplements the evidence discussed earlier to make the claim that image fusion does not simply add color artificially, but rather combines information from both input images.
Cross-correlation values for the algal cell image series were calculated in the same manner as earlier and are presented in Table 1. The results were 1.000, 0.9949 and 1.000 for the red, green and blue channels, respectively. Since the color of the SIMS image is green, and, thus, does not contain any information in the blue and red channels, the cross-correlation values of 1.000 for these two channels is expected, as those channels remain unchanged between the SIMS and fused images. For the green channel cross correlation, the value of 0.9949 supports the claim that the spatial resolution of the fused image has been increased without compromising the original data. As in previous examples, the fused algal image demonstrates the benefits of image fusion with SIMS data while maintaining, statistically, the integrity of the input SIMS data, establishing the applicability of the image fusion to bio-analytical SIMS experiments.
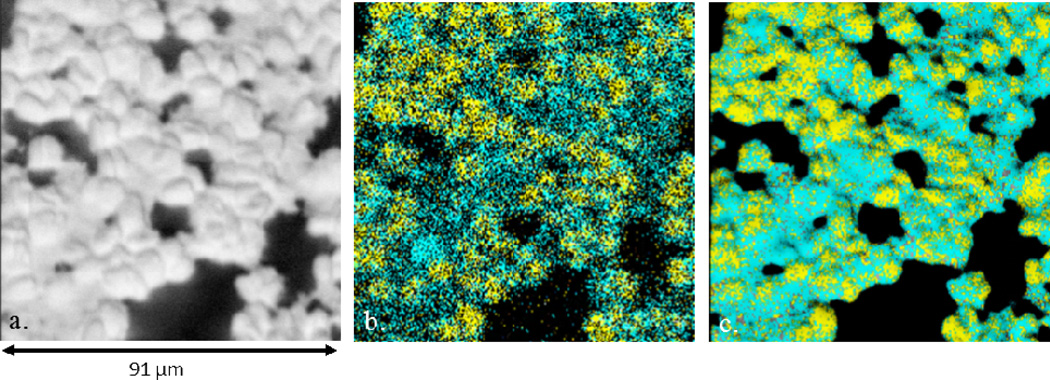
Finally, to explore the effect on image fusion of multiple signals in a biologically complex environment, the surface coating and tops of algal cells were etched away to chemically image the intracellular features of the algal cells. Shown in Figure 4 is an image series of the interior of an algal cell colony after removing approximately 1 micron of the sample. Pictured in the SIMS image in Figure 4b is a salt signal, K3SO4, localized to the cytoplasm in teal and hydrocarbon peaks from inside oil bodies in yellow. The data shows that there are 0 to 3 oil bodies per cell. Image fusion aids to clearly define the cellular boundaries from each other, which is not as noticeable in the chemical image. Furthermore, in the chemical image, how the oil bodies correlate with the morphology is lost, yet, in the fused image, the oil body areas correlate to certain raised features in the SEM. The observed round morphological feature of the oil corresponds to microscopic investigations where circular oil bodies are witnessed.
Figure 4.
Fused image series of the intracellular contents of the algal cells. (a) SEM image (468 × 468 pixels); (b) SIMS image (234 × 234 pixels) of oil bodies containing hydrocarbons [yellow; sum of m/z 410.4 + 424.4 + 425.4 + 439.4] and K3PO4 [teal; m/z 212.8]; (c) Fused image from Figures 4a and 4b (468 × 468 pixels). Field-of-view for all images is 91 µm × 91 µm.
Again, to verify that no information has been lost between the SIMS image and the fused image, cross-correlation values were calculated for each color channel and are shown in Table 1. The values of 0.9493, 0.9912, and 0.9616 for the red, green, and blue channels, respectively, suggest that, as in previous experiments, the two images are highly correlated. Thus, the improvement in resolution and enhanced definition of interior regions and features of the algal cells has not compromised the original SIMS data.
Conclusion
We show the power of an image fusion algorithm and its applicability to SIMS imaging experiments. Through a model set of data and a relatively simple copper grid system, improved observable image quality has been shown. These hybrid images are complemented by a quantitative metric of correlated image information. Cellular images of the chemically and morphologically complex B. braunii algae were also enhanced using the pan-sharpening algorithm, demonstrating the effectiveness of the method to bring together highly complex chemical (SIMS) and morphological (SEM) data in the determination of novel, biologically-significant information. Pan-sharpening, and more generally, image fusion, could prove to be an important tool for the advancement of SIMS as a bioimaging technique for cell characterization.
Acknowledgements
The authors acknowledge financial support from the National Institute of Health under Grant No. VUMC-37846 and the Department of Energy under Grant No. DE-FG-02-06ERER15803. The authors would like to thank Richard Caprioli for suggesting the use of image fusion in SIMS, as well as Jordan Lerach for preparing and Hua Tian for obtaining SEM and SIMS images of gold-coated grid samples.
References
- 1.Fletcher JS, Lockyer NP, Vickerman JC. Molecular SIMS imaging; spatial resolution and molecular sensitivity: Have we reached the end of the road? Is there light at the end of the tunnel? Surface and Interface Analysis. 2011;43:253–256. [Google Scholar]
- 2.Piehowski PD, Davey AM, Kurczy ME, Sheets ED, Winograd N, Ewing AG, Heien ML. Time-of-Flight Secondary Ion Mass Spectrometry Imaging of Subcellular Lipid Heterogeneity: Poisson Counting and Spatial Resolution. Analytical Chemistry. 2009;81:5593–5602. doi: 10.1021/ac901065s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henderson A, Fletcher JS, Vickerman JC. A comparison of PCA and MAF for ToF-SIMS image interpretation. Surface and Interface Analysis. 2009;41:666–674. [Google Scholar]
- 4.Tyler BJ, Rayal G, Castner DG. Multivariate analysis strategies for processing ToF-SIMS images of biomaterials. Biomaterials. 2007;28:2412–2423. doi: 10.1016/j.biomaterials.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyler B. Interpretation of TOF-SIMS images: multivariate and univariate approaches to image de-noising, image segmentation and compound identification. Applied Surface Science. 2003;203:825–831. [Google Scholar]
- 6.Wickes BT, Kim Y, Castner DG. Denoising and multivariate analysis of time-of-flight SIMS images. Surface and Interface Analysis. 2003;35:640–648. [Google Scholar]
- 7.Wang Z, Zhou D, Armenakis C, Li D, Li Q. A Comparative Analysis of Image Fusion Methods. IEEE Transactions on Geoscience and Remote Sensing. 2005;43:1391–1402. [Google Scholar]
- 8.Mumtaz A, Majid A, Mumtaz A. Genetic Algorithms and its Application to Image Fusion. 2008 International Conference on Emerging Technologies, Proceedings; 2008. pp. 6–10. [Google Scholar]
- 9.Khan AM, Khan A. Fusion of visible and thermal images using support vector machines; 10th IEEE International Multitopic Conference 2006, Proceedings; 2006. pp. 146–151. [Google Scholar]
- 10.Wen CY, Chen JK. Multi-resolution image fusion technique and its application to forensic science. Forensic Science International. 2004;140:217–232. doi: 10.1016/j.forsciint.2003.11.034. [DOI] [PubMed] [Google Scholar]
- 11.Ashoori A, Moshiri B, Setarehdan SK. Fuzzy image fusion application in detecting coronary layers in IVUS pictures. 2008 3rd International Symposium on Communications, Control and Signal Processing; 2008. pp. 20–24. [Google Scholar]
- 12.Rubio-Guivernau JL, Gurchenkov V, Luengo-Oroz MA, Duloquin L, Bourgine P, Santos A, Peyrieras N, Ledesma-Carbayo MJ. Wavelet-based image fusion in multi-view three-dimensional microscopy. Bioinformatics. 2012;28:238–245. doi: 10.1093/bioinformatics/btr609. [DOI] [PubMed] [Google Scholar]
- 13.Zhong Z, Blum RS. A categorization of multiscale-decomposition-based image fusion schemes with a performance study for a digital camera application. Proceedings of the IEEE. 1999;87:1315–1326. [Google Scholar]
- 14.Shivsubramani Krishnamoorthy KPS. Implemetation and Comparative Study of Image Fusion Algorithms. International Journal of Computer Applications. 2010;9:25–35. [Google Scholar]
- 15.Artyushkova K, Pylypenko S, Dowlapalli M, Atanassov P. Use of digital image processing of microscopic images and multivariate analysis for quantitative correlation of morphology, activity and durability of electrocatalysts. Rsc Advances. 2012;2:4304–4310. [Google Scholar]
- 16.Artyushkova K, Fulghum JE. Multivariate image analysis methods applied to XPS imaging data sets. Surface and Interface Analysis. 2002;33:185–195. [Google Scholar]
- 17.Lloyd KG, Walls DJ, Wyre JP. Correlating data from multiple surface-specific techniques using multivariate methods: examples and considerations. Surface and Interface Analysis. 2009;41:686–693. [Google Scholar]
- 18.Rokni K, Marghany M, Hashim M, Hazini S. Comparative statistical-based and color-related pan sharpening algorithms for ASTER and RADARSAT SAR satellite data. Computer Applications and Industrial Electronics (ICCAIE), 2011 IEEE International Conference on; 2011. [Google Scholar]
- 19.Artyushkova K, Farrar JO, Fulghum JE. Data fusion of XPS and AFM images for chemical phase identification in polymer blends. Surface and Interface Analysis. 2009;41:119–126. [Google Scholar]
- 20.Simpson AJ, Zang X, Kramer R, Hatcher PG. New insights on the structure of algaenan from Botryoccocus braunii race A and its hexane insoluble botryals based on multidimensional NMR spectroscopy and electrospray-mass spectrometry techniques. Phytochemistry. 2003;62:783–796. doi: 10.1016/s0031-9422(02)00628-3. [DOI] [PubMed] [Google Scholar]
- 21.Tanoi T, Kawachi M, Watanabe MM. Effects of carbon source on growth and morphology of Botryococcus braunii. Journal of Applied Phycology. 2011;23:25–33. [Google Scholar]
- 22.Weiss TL, Chun HJ, Okada S, Vitha S, Holzenburg A, Laane J, Devarenne TP. Raman Spectroscopy Analysis of Botryococcene Hydrocarbons from the Green Microalga Botryococcus braunii. Journal of Biological Chemistry. 2010;285:32458–32466. doi: 10.1074/jbc.M110.157230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chris Padwick MD, Pacifici Fabio, Smallwood Scott. WorldView-2 Pan-Sharpening. ASPRS 2010 Annual Conference.2010. [Google Scholar]
- 24.Fletcher JS, Rabbani S, Henderson A, Blenkinsopp P, Thompson SP, Lockyer NP, Vickerman JC. A New Dynamic in Mass Spectral Imaging of Single Biological Cells. Analytical Chemistry. 2008;80:9058–9064. doi: 10.1021/ac8015278. [DOI] [PubMed] [Google Scholar]
- 25.Pavlic G, Singhroy V, Duk-Rodkin A, Alasset PJ. Satellite Data Fusion Techniques for Terrain and Surficial Geological Mapping. Geoscience and Remote Sensing Symposium, 2008. IGARSS 2008. IEEE International; 2008. p. 314. [Google Scholar]
- 26.Zitova B, Flusser J. Image registration methods: a survey. Image and Vision Computing. 2003;21:977–1000. [Google Scholar]
- 27.Manjusha Deshmukh UB. Image Fusion and Image Quality Assessment of Fused Images. International Journal of Image Processing. 2010;4:484–508. [Google Scholar]
- 28.Weiss TL, Roth R, Goodson C, Vitha S, Black I, Azadi P, Rusch J, Holzenburg A, Devarenne TP, Goodenough U. Colony Organization in the Green Alga Botryococcus braunii (Race B) Is Specified by a Complex Extracellular Matrix. Eukaryotic Cell. 2012;11:1424–1440. doi: 10.1128/EC.00184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kucher A, Jackson LM, Lerach JO, Bloom AN, Popczun NJ, Wucher A, Winograd N. Strong field ionization and imaging of C60 sputtered molecules: overcoming matrix effects and improving sensitivity. Analytical Chemistry, Submitted. 2014 doi: 10.1021/ac501586d. [DOI] [PubMed] [Google Scholar]
- 30.T E. Oner Pretreatment Techniques for Biofuels and Biorefineries. Springer: 2013. pp. 35–36. [Google Scholar]