Abstract
The precise and unambiguous elucidation and characterization of interactions between a high affinity recognition entity and its cognate protein provides important insights for the design and development of drugs with optimized properties and efficacy. In oncology, one important target protein has been shown to be the epidermal growth factor receptor (EGFR) through the development of therapeutic anticancer antibodies that are selective inhibitors of EGFR activity. More recently, smaller protein derived from the tenth type III domain of human fibronectin termed an adnectin has also been shown to inhibit EGFR in clinical studies. The mechanism of EGFR inhibition by either an adnectin or an antibody results from specific binding of the high affinity protein to the extracellular portion of EGFR (exEGFR) in a manner that prevents phosphorylation of the intracellular kinase domain of the receptor and thereby blocks intracellular signaling. Here the structural changes induced upon binding were studied by probing the solution conformations of full length exEGFR alone and bound to a cognate adnectin through hydrogen/deuterium exchange mass spectrometry (HDX MS). The effects of binding in solution were identified and compared with the structure of a bound complex determined by X-ray crystallography.
Keywords: protein-protein interactions, Hydrogen/Deuterium exchange, mass spectrometry, protein binding, biopharmaceutical, electron transfer dissociation
Introduction
The epidermal growth factor receptor (EGFR) is a key molecular target in oncology. EGFR is overexpressed or mutated in many cancers and its activation is important in tumor growth and progression [1]. EGFR is composed of a large extracellular ligand-binding region, a single transmembrane domain, an intracellular juxtamembrane region, a cytoplasmic tyrosine kinase domain and a C-terminal regulatory domain [2]. The extracellular region of EGFR (exEGFR) contains two homologous ligand binding domains (domain I and III) and two cysteine rich domains (domains II and IV) [3] (see also Figure 1A). Upon binding to epidermal growth factor (EGF), exEGFR forms a homodimer through its dimerization arm which projects from the cysteine-rich domain II [4]. Dimerization positions the intracellular kinase domains in proximity so that transphosphorylation can occur [5,6]. When the kinase domain of EGFR becomes phosphorylated, it can lead to activation of pathways that are involved in regulating cellular processes [7]. Activation of EGFR may contribute to tumor growth including promotion of proliferation, invasion, and metastasis [8,9]. Therefore, from a medical point of view, blocking signaling can modulate cancer progression.
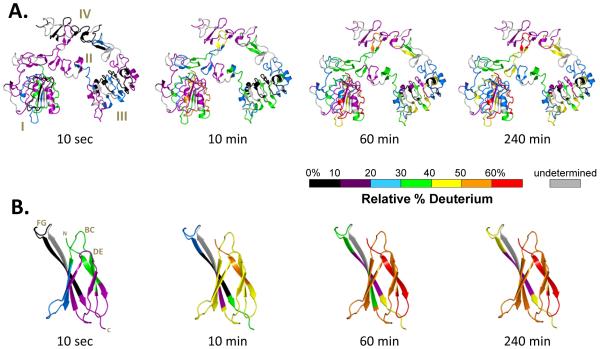
Figure 1.
Summary of all HDX MS data for (A) extracellular EGFR (exEGFR) free in solution and (B) Adnectin 1 free in solution. The HDX MS data are not corrected for back-exchange (see Methods) and are therefore reported as relative. PDB entry 3QWQ [19] was colored using the color code indicated for the deuteration times shown at the bottom of each image.
To inhibit EGFR activation, molecules have been developed that block binding of ligands to exEGFR. For example, monoclonal antibodies (mAbs) directed against exEGFR physically block EGFR binding and thereby inhibit EGFR signaling pathways [1]. Amongst the mAbs directed against EGFR, Cetuximab (Erbitux), for example, is successfully used for the treatment of tumors, such as breast, cervix, colon, head and neck [10]. While mAbs are effective, designing and manufacturing full length mAbs is challenging and the cost of treatment can be prohibitive. Less complex molecules that elicit the same extracellular blocking effects are therefore desirable, including, for example, antibody mimetics [11]. Adnectins are a type of antibody mimetic that have shown tight and specific target binding with low toxicity, high thermal stability, good solubility, and relative ease of manufacturing [12]. Adnectins are derived from the 10th fibronectin type III domain (10Fn3) [13,14] containing complementarity-determining regions (CDRs) (BC, DE and FG loops) that are structurally analogous to the antibody heavy chain CDRs H1, H2 and H3 [11,14-16]. During drug discovery, adnectins can be designed to bind with high affinity (low nM range) and specificity to relevant targets [11,17,18], such as exEGFR.
To better understand binding interactions with exEGFR, biophysical characterizations have been performed with an anti-EGFR adnectin (Adnectin 1). Chief among studies of the bound state is a crystal structure of the exEGFR:adnectin complex wherein the binding interface was described [19]. While X-ray crystallography and NMR structural analyses of complexes are desirable, especially for providing information about binding interactions with atomic level resolution, it is not always possible to obtain such data. As we have and others have pointed out before [20-33] there are many properties of protein:protein interactions in solution that can be elucidated by hydrogen/deuterium exchange mass spectrometry (HDX MS). In the present study, we used HDX MS to measure the flexibility and solution dynamics of both unbound exEGFR and unbound Adnectin 1. Then, the interactions between exEGFR and Adnectin 1 were investigated by HDX MS at the peptide level and, for selected regions, at the amino acid level using directed HDX MS measurements involving electron transfer dissociation (ETD). The results were interpreted in light of the crystallographic structure of the complex, allowing us to draw further conclusions, including a better understanding of what information can and cannot be revealed by HDX MS analysis of protein:protein complexes. In a related paper [34], the same complex was probed with oxidative labeling, a technique that can provide complementary information. In total, the results provide a clearer picture of the solution conformation and the conformational properties of this biologically important complex.
Methods
Proteins
Human exEGFR (residues 1-642) was expressed in Sf9 cells with a C-terminal His tag and purified as described previously [19]. The final purified protein was in phosphate buffer saline (PBS) consisting of 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.2. The expression and purification of Adnectin 1 was previously described [19]. Purified Adnectin 1 was also in PBS buffer (pH 7.2).
Hydrogen-deuterium exchange
For hydrogen exchange labeling, 50 pmol of EGFR protein were incubated with Adnectin 1 for a final EGFR:Adnectin 1 concentration ratio of 1:2 during deuterium labeling. Under these conditions, >99.94% of the EGFR protein molecules were bound based on 2nM Kd (as reported elsewhere [11,15,19]). Mixtures were incubated for 30 min at room temperature (21 °C) before deuterium labeling. As a control, EGFR and Adnectin 1 were incubated alone in PBS buffer (pH 7.2) and treated exactly as the EGFR:Adnectin 1 complex. Deuterium exchange was initiated by dilution of each protein with 15-fold PBS buffer made with 99% D2O, (pD 7.2) at room temperature. At each deuterium exchange time point (from 10 s to 4 hours) an aliquot from the exchange reaction was removed and labeling was quenched by adjusting the pH to 2.5 with an equal volume of quench buffer [4M GnHCl, 0.5M Tris (2-carboxyethyl) phosphine hydrochloride (TCEP-HCl), 200mM sodium phosphate, H2O]. Quenched samples were immediately frozen on dry ice and stored at −80 °C until analysis.
Chromatography and Mass Spectrometry
Each frozen sample was thawed rapidly and injected into a custom Waters nanoACQUITY UPLC HDX ManagerTM [35] and analyzed on a Xevo-G2 mass spectrometer (Waters Corp., Milford, MA, USA) as previously described [36]. The average amount of back-exchange using this experimental setup was 18% to 25%, based on analysis of highly deuterated peptide standards. All comparison experiments were done under identical experimental conditions such that deuterium levels were not corrected for back-exchange and are therefore reported as relative [31]. All experiments were performed in duplicate. The error of measuring the mass of each peptide was ± 0.20 Da in this experimental setup, consistent with previously obtained values [37,38], see also [39] for a review. Deuteration uptake was calculated by subtracting the centroid of the isotopic distribution for peptide ions from undeuterated protein from the centroid of the isotopic distribution for peptide ions from the deuterium labeled sample. The resulting relative deuterium levels were plotted versus the exchange time using Waters DynamX 2.0TM software. Identification of the peptic peptides was accomplished through a combination of exact mass analysis and MSE [40] using Identity Software (Waters Corp., Milford, MA, USA), as previously described [41,42]. All assignments, deuterated spectra, and data processing were manually checked and verified.
Targeted ETD fragmentation
For ETD fragmentation, free exEGFR and Adnectin 1-bound exEGFR were prepared, labeled (a single labeling time of 30 min was chosen), and quenched as described above. All analyses (undeuterated controls and deuterated samples for both free and bound EGFR) were run in triplicate. A Waters Synapt G2-S HDMS system in resolution mode was used for ETD. The conditions were derived from similar experiments in this instrumental format, as described previously [43]: source capillary 3.0 kV, cone 20 V, offset 10; triwave trap wave height 0.3 V; RF setting trap ETD 450 V; gas control for trap 22.0 mL/min and gas control for transfer 0.7 mL/min. The reagent used for ETD fragmentation was 1,3-Dicyanobenzene. Targeted ETD on exEGFR peptic peptide 1-19 (+4 charge state) was performed with a selection mass of 520 m/z for unlabeled and 522 m/z for the labeled peptide. Control experiments for ETD fragmentation and validation of low-scrambling conditions were performed with the P1 peptide [43,44], +3 charge state, with a set mass of 517 m/z for unlabeled and 518 m/z for labeled P1 peptide (see Supplemental Figure S5). The c and z ions were assigned using BioLynx software and deuterated ETD spectra were manually processed using HX-Express [45,46].
Data Visualization
Peptic maps were obtained with the help of DynamX 2.0™. Pymol [47] was used to map the conformational changes on the crystal structure of the exEGFR (PDB id: 3QWQ).
Results and Discussion
Characterization of exEGFR by HDX MS
To form the basis for comparison of the exchange into the Adnectin 1 bound form, we first measured the incorporation of deuterium into exEGFR (residues 1-642, 69 kDa) free in solution and not bound to Adnectin 1. Undeuterated exEGFR was digested with pepsin using the same experimental conditions that were used later for deuterium labeling. Peptides constituting 84% linear coverage were identified (Supplemental Figure S1) and deuteration of exEGFR was then followed in 76 peptic peptides covering 80.3% of the exEGFR sequence (Supplemental Figure S2). The hydrogen exchange results for exEGFR are summarized in graphical form in Figure 1A (deuterium incorporation graphs for exEGFR alone are presented in Supplemental Figure S3).
Deuterium labeling of exEGFR alone showed that the majority of the molecule was resistant to rapid labeling, with most regions becoming only 40% deuterated after 4 hours of labeling and only a few mobile or dynamic regions becoming 60% deuterated. These HDX MS data are consistent with the structure observed by crystallography (i.e., regions where exchange is predicted to be slow based on the secondary and tertiary stricter exchanged slowly, etc.) and provide insight into the solution dynamics of full length exEGFR.
Interestingly, domain I and III share similar β-helix solenoid structures and have only 37% sequence homology [3], however their exchange patterns are somehow different with most of the domain III peptides being protected from exchange even after 4 h of labeling. On the contrary, some of the domain I peptides exchanged more than 50% of their available amide hydrogens after 4 h, indicating different backbone amide hydrogen environments despite the structural similarity. Both of these domains are known for their ability to recognize and bind ligands [2].
In vivo, upon epidermal growth factor (EGF) binding, EGFR undergoes dimerization accompanied by a conformational change: EGFR transitions from a tethered into an extended conformation [2,3]. All contacts across the dimer interface are mediated through domain II. This domain contains a β-hairpin (the “dimerization arm”) located between amino acids 240-260. HDX MS data showed that in the monomeric form, the dimerization arm is solvent exposed and after 4 h of labeling more than 60% of this region becomes deuterated. However, some studies [48-50] have indicated that in the absence of the ligand (epidermal growth factor, EGF), EGFR could also exist as preformed homodimers, and that would be consistent with the observed lack of deuteration in other regions encompassing domain II, but excluding the dimerization arm.
Maintaining EGFR in a tethered (down-regulated) conformation is important for its autoinhibitory function. It is believed that interactions between domain II and IV are responsible for maintaining the down-regulated conformation, however small angle X-ray scattering (SAXS) results have indicated that actually interactions between domains II and III contribute to the down-regulated conformation [51]. It was proposed that the region linking domain II and III may be extremely rigid and that this region is actually responsible for maintaining EGFR in a tethered configuration. Interestingly, HDX MS data for the peptide 298-311 encompassing that area shows that this loop is protected from exchange and only partially dynamic at earlier time points (10 sec). After 10 min of labeling it reaches its “full” deuteration potential of 40% and then it remains unchanged throughout the 4 h labeling time.
Domains II and IV are cysteine-rich domains, each of them having 10 intra-domain disulfide bonds. These domains also contain sites of N-linked glycosylation [3]. While these domains have a structure comprised mostly of β-hairpins and loops, the deuterium incorporation results (Figure 1A) indicated that they exchange slowly, with the exception of several peptides that incorporated more than 50% deuterium after 4 h of labeling. Amongst these, regions in domain II between amino acids 245-253 (a β-hairpin loop corresponding to the dimerization arm, which is in close proximity to domain IV) and 254-267 incorporated more than 50% deuterium at the longest time point. Peptides between amino acids 518-539 from domain IV also were heavily labeled after 4 h, also exchanging 50% of their available backbone amides.
Analysis of Adnectin 1 by HDX MS
Hydrogen/deuterium exchange MS data for Adnectin 1 alone were also obtained. Upon pepsin digestion, a total of 13 peptides covering 90.3% of the amino acid sequences were identified (Supplemental Figure S4A) and exchange into all of these peptides was followed. The results for HDX MS of Adnectin 1 alone are shown in Figure 1B (see also Supplemental Figure S4B for deuterium incorporation graphs). Adnectin’s higher-order structure is mainly formed of anti-parallel β-sheets, with connecting loops (BC, DE and FG) that are equivalent to the CDRs of an antibody heavy chain [13,14,16]. After 10 sec of deuteration, most parts of Adnectin 1 were protected from exchange with the exception of the BC loop that exchanged up to 40% of its available backbone amide hydrogens (NHs). In time, after 4 h of labeling, most of the molecule was more than 50% labeled, including all three loops FG, BC and DE, indicating a dynamic solution conformation. Only one β-strand (peptide 91-97) remained protected from exchange after 4 hours of labeling, displaying only 20% deuterium incorporation.
HDX MS of exEGFR upon Adnectin 1 binding
To characterize the complex, we next incubated exEGFR with Adnectin 1 and labeled the complex with deuterium. The concentrations were set up such that we monitored only the impact of Adnectin 1 on exEGFR and not the opposite; therefore, no data regarding changes in deuteration of Adnectin 1 when bound to exEGFR were obtained. In the protein complex experiment, 76 exEGFR peptic peptides were followed by HDX MS (Supplemental Figures S2, S3). Overall, deuterium incorporation, and therefore protein dynamics, was the same (up to 4 hours) for most of the peptides when comparing unbound and bound exEGFR. However, several important regions showed differences in deuterium uptake and provided insight into the impact of Adnectin 1 binding on both protection from deuteration in exEGFR and on exEGFR dynamics. A summary of all of differences in all the 76 peptic peptides at all the time points is provided in Figure 2A.
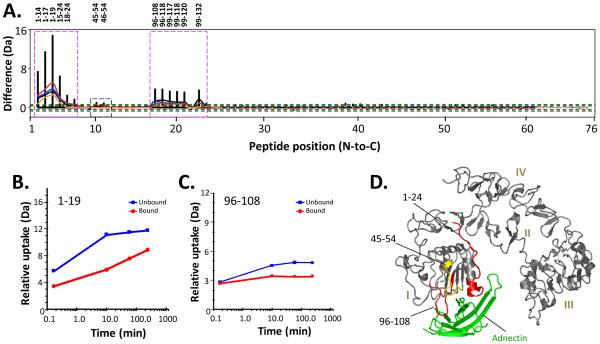
Figure 2.
Differences in HDX in exEGFR upon binding to Adnectin 1. A. Comparability profile [38]. The difference (y-axis) is [D levelexEGFR – D levelexEGFR with Adnectin 1]. Orange, red, cyan, blue, and black lines correspond to data acquired at 10 sec, 10, 60, and 240 min of deuteration, respectively. The black dotted lines at y-axis values ±0.4 Da represent the 98% confidence limits for differences between each single exchange time point while the green dotted lines at y-axis values ±1.0 Da indicate the boundary of significance for the sum of all differences from all time points (see also Ref. [38] for more details). Peptides displaying obvious differences are marked with magenta boxes and the corresponding peptide numbers are given above. A region with subtle differences (45-54) in uptake is marked with a black dotted box. B,C. Comparison of deuterium exchange in two exEGFR peptides where obvious differences were found: (B) residues 1-19; (C) 96-108; exEGFR (blue lines) and exEGFR bound to Adnectin 1 (red lines). Deuterium uptake graphs for all peptides are in Supplemental Figure S4. D. Location of the exEGFR peptides showing less deuterium uptake upon Adnectin 1 binding, mapped onto the structure of exEGFR in the exEGFR:Adnectin 1 co-crystal (PDB entry 3QWQ Ref. [19]). Adnectin 1 has been colored green. Obvious changes in deuterium levels upon binding (colored red) were defined as a difference between deuterium exchange-in curves of 1.0 Da or more. Subtle changes in deuterium levels upon binding (colored yellow) were 0.5-1.0 Da. No changes (colored gray) were differences of 0.0-0.5 Da.
Three regions of exEGFR were affected by Adnectin 1 binding (Figure 2A). A series of overlapping peptides covering the N-terminal region between amino acids 1-24 of exEGFR domain I had decreased deuteration upon Adnectin 1 binding. Amongst them, overlapping peptides 1-14, 1-17, 1-19 and 15-24 had a difference of more than 3 Da upon binding, indicating that at least three amide hydrogens became protected from exchange in the exEGFR:Adnectin 1 complex. These peptides became protected even at the earliest time points (10 sec labeling) suggesting an immediate occlusion from the solvent [52-54] upon Adnectin 1 binding. The deuterium uptake plot for peptide 1-19 is shown in Figure 2B and a significant decrease in deuterium incorporation in the bound form is clearly seen. Peptides 18-24 and 20-24 (see Figure S3) also had reduced deuterium uptake at longer time points (from 10 min and greater), indicating that the solvent exposure of these peptides was not nearly as affected as the long-term dynamics and breathing of this region. Another region, with overlapping peptic peptide coverage, that exchanged less upon ligand binding was region 96-120 from domain I where at least a 1 Da decrease in uptake at multiple time points was observed upon binding (see Figure S3), mostly from 10 min of labeling onward and concentrated in peptide 96-108 (Figure 2B). exEGFR peptides 45-54 and 46-54 (see Figure 2A and S3) showed very subtle differences in deuteration upon Adnectin 1 binding, indicating that these regions were either not directly involved in binding, that their conformation was only modestly altered in the presence of Adnectin 1, or both. A summary of all regions of exEGFR displaying altered deuterium exchange upon binding is shown in Figure 2C and mapped onto the exEGFR crystal structure [19]. Interestingly, all the peptides that had reduced deuterium uptake upon Adnectin 1 binding were located in domain I of exEGFR and in close proximity to, although not necessarily in direct contact with, Adnectin 1 as observed in the co-crystal.
Targeted ETD fragmentation for binding site refinement
A well-known limitation of peptide-level HDX MS is the inability to locate deuterium to the single residue level [55-57]. This limitation was apparent in the peptide-level results for exEGFR bound to Adnectin 1. The deuterium difference between bound and unbound peptide 1-19 after 30 minutes of labeling was approximately 4-5 deuterium atoms (when considering back exchange, see Figure 2B) but at peptide-level resolution, it was unknown where these 4-5 deuterium atoms were located in these 19 residues. In order to better refine the amino acids that were involved in Adnectin 1 binding, we performed targeted ETD fragmentation [43,44,58] on deuterium labeled exEGFR (at the 30 minute labeling time point) peptide 1-19, from both the free and bound forms of exEGFR. Peptides covering other regions (e.g., 96-108) provided poor ETD fragmentation and no useful data could be obtained. The +4 ionization state of peptide 1-19 provided a good number of ETD fragments (Figure 3A) corresponding to z and c ions [44,59] and there were significant differences in deuterium content between several c and z ions of bound and unbound exEGFR.
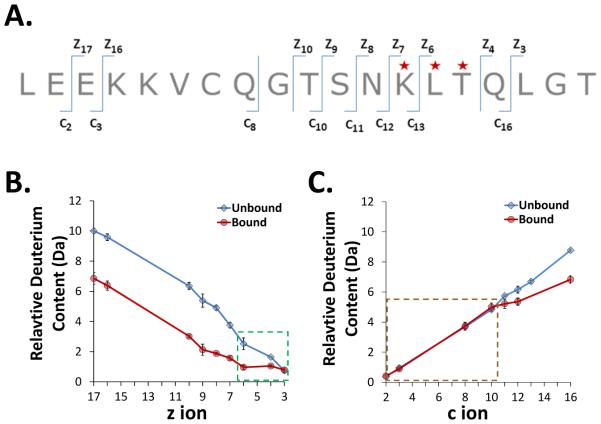
Figure 3.
Targeted ETD analysis of deuterated exEGFR bound to Adnectin 1. A. The sequence of the N-terminal exEGFR peptic peptide (residues 1-19) that was selected for ETD fragmentation, with the c and z ions that could be observed indicated. Residues highlighted with a red star showed significant differences in uptake between bound and unbound forms. Note that peptide backbone cleavage which produces c, z ions occurs on the phi bond, which is to the right (going N- to C-terminally) of the next amino acid amide hydrogen, e.g. c1 contains both the amide hydrogen for amino acid 1 and the amide hydrogen for amino acid 2 [64]. B. Deuterium content of z ions in peptide 1-19 of unbound and bound exEGFR. The green highlighted box indicates ions with differences in deuterium levels between bound and unbound forms. Notice the bound form in this region remained constant whereas uptake in the unbound form increased at higher z ion value. C. Deuterium content of c ions in peptide 1-19 of unbound and bound exEGFR. The brown highlighted box around the ions up to c10 shows the c ions which incorporated the same amount of deuterium between unbound and bound exEGFR.
As shown in Figure 3B, all z ions except z3 showed different deuterium content between bound and unbound exEGFR. Changes in the difference between deuteration levels for bound and free peptide 1-19 occurred for z ions z3 to z6 (highlighted in green box, Figure 3B). While bound exEGFR showed no significant increase in deuteration in this region (z3 to z6) – an indication of protection – unbound exEGFR showed an increasing amount of deuterium when moving from z3 to z6. The change in mass from z3 to z6 for the bound form was only 0.18 Da while the change in mass of the unbound form was 1.78 Da from z3 to z6. From z6 to z8, in bound exEGFR there was a gradual increase in deuterium content while unbound exEGFR showed a greater increase in deuterium, likely resulting from uptake of one more deuterium in the unbound form. Finally, from z9 ion to z17, in both bound and unbound forms, the uptake curve showed a constant increase in deuteration and the difference between deuterium content for the unbound and bound forms (3 deuterium) was constant. The conclusion from analysis of deuterium in z ions was that binding of exEGFR protects primarily the region involving residues 13KLTQ16 of peptide 1-19. Analysis of deuterium levels in the c ions (Figure 3C) confirms the conclusions gleaned from the z ions. The c ions from c2 to c10 showed no difference in deuterium uptake between the bound and unbound exEGFR (highlighted in brown dotted box, Figure 3C) and the uptake difference started to become apparent for ions c11 to c16. Combining c and z ions, the results point to the same region of differences (residues from K13 to Q16). The X-ray crystal structure [19] shows the contact residues of sequence 1-19 to be at L14, T15, Q16, L17 and G18, although the contact residue side-chains are not the same as the backbone amide hydrogens that would be protected by binding. Figure 4 attempts to rationalize the HDX MS ETD data in light of the crystal structure as detailed further below.
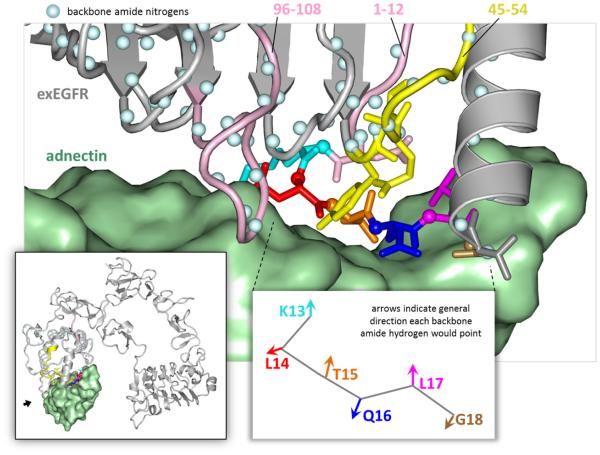
Figure 4.
HDX MS contact areas: the interacting region of exEGFR (grey with color-coded residues) with Adnectin 1 (green) (PBD ID: 3QWQ Ref. [19]). The backbone amide hydrogens are illustrated as blue balls. The peptides that were found to have significant protection from deuteration upon Adnectin 1 binding are shown in pink (1-19, 96-108) and the ones with moderate protection from deuteration are shown in yellow (45-54). Targeted ETD reveals that exEGFR residues cyan (K13), red (L14), orange (T15), blue (Q16), magenta (L17) and bronze (G18) were protected from deuteration when bound to Adnectin 1, overlapping with the same residues previously confirmed by X-ray crystal structure (T15, Q16, L14, and G18) [19]. The view of the main figure is from a spot indicated with a small black arrow in the lower left inset.
Conclusions
The unambiguous characterization of the binding interfaces of a protein to its ligand(s) can play a significant role in the development process of improved biotherapeutic agents. Such information can best be obtained with a crystal or NMR structure wherein the position of the atoms from each member of the complex becomes apparent. However, it is not always possible to obtain a crystal structure of a protein complex and if one can be obtained, defining the interface could be confounded by crystal packing effects. Protein complexes may not be amenable to NMR for various reasons, especially for complexes where the total size exceeds that possible for conventional NMR. As a result, other techniques for determining protein:protein interactions / epitope mapping strategies are also important [60-62]. Some of these techniques, e.g., screening peptide libraries for binding, may not be ideal because they might only detect short, linear stretches of amino acids that are recognized by a certain ligand/ antibody which may or may not adopt the relevant conformation. In the present study, amide hydrogen/deuterium exchange of a protein free and then in complex with a binding partner was monitored with mass spectrometry. HDX MS, like the other techniques, is also not without its limitations. The final output of HDX MS is a description of where in the protein(s) deuterium incorporation became altered by complex formation. The most useful information comes from changes in exchange of highly solvent exposed backbone amide hydrogens, as those that exchange slowly and are solvent inaccessible and protected by hydrogen bonding are generally not affected by complex formation [30,32,53,54]. Because HDX MS samples the local environment of backbone amide hydrogens (NHs), it is not always possible to delineate binding interfaces, particularly those involving primarily side-chain interactions driven by electrostatics or by hydrophobic forces [30]. So what then, if any, is the value of HDX MS measurements of protein:protein interactions, particularly when the structure of a protein complex is already known? The present study of exEGFR bound to Adnectin 1 was undertaken to define some of the insights that can be obtained by HDX MS.
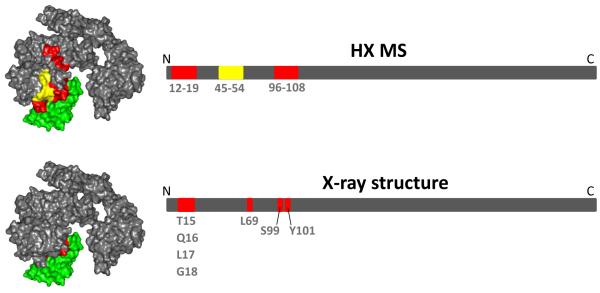
The crystal structure of Adnectin 1 bound to exEGFR has been published and the contact areas are known [19]. All three adnectin loops make contact with exEGFR with interactions occurring through the FG loop (175 Å2 of buried surface), the DE loop (70 Å2) and the BC loop (40 Å2). Three hydrogen bonds are formed between atoms in residues T15, Q16 and L17 of exEGFR and a series of Van der Waals interactions take place between Adnectin 1 and exEGFR primarily involving exEGFR residues L14, L69, S99 and Y101. Based on the structure of the complex, one should be able to predict with some certainty what the HDX MS might be, assuming of course that the crystalline conformation is also found in solution. Not surprisingly, the HDX MS data reveal the regions of exEGFR with reduced deuteration upon binding to be in the same regions as indicated by the crystal structure (Figure 5). Therefore, if a structure were not known, and one were to rely on HDX MS solely to define the interface of exEGFR and Adnectin 1, the conclusions from HDX MS for this particular complex would be mostly correct – not to the same level of detail – but certainly sufficiently defining the interface areas. The HDX MS results from Figure 5 could be used to design other tests to validate the interface, such as informed site-directed mutation or peptide screening.
Figure 5.
Summary of the regions of exEGFR with differences in deuterium uptake as determined by HDX MS (top) and by the x-ray crystal structure [19] (bottom). A surface view of the complex (left) and linear (right) representations of exEGFR regions that are involved in Adnectin 1 binding are both shown. Adnectin 1 is shown in green. The residues participating in complex formation that were identified by each method have been indicated in both views (red); a region (45-54) that was subtly affected, likely as a result of protein stabilization, is shown in yellow.
Closer inspection of the HDX MS data reveals a few other details. Upon binding, several regions from exEGFR showed reduced deuteration as described in Figure 2. Particularly, the N-terminal loop in exEGFR domain I between amino acids 1-24 underwent reduction in deuterium uptake upon adnectin binding, where more than 3 amide hydrogens were protected in the complex. From the crystal structure, this N-terminal region consists of a long β-hairpin loop (residues 1-19) followed by an α-helix. In unbound exEGFR, the loop is rapidly deuterated while the α-helix shows maximum deuteration after 4 h labeling of only between 20-40%. Upon Adnectin 1 binding, the beginning of the α-helix (residues 20-24) incorporates less deuterium, likely as a result of stabilization of the hydrogens bond network at the beginning of the helix, while residues 1-19 were much more affected.
Fortunately, a multiply charged (+4) peptide ion that covered residues 1-19 provided a strong signal that was compatible with ETD fragmentation. This permitted better delineation of what part of the 1-19 sequence was protected upon Adnectin 1 binding. Bearing in mind that NHs alternate the direction they face along a purely linear sequence of polypeptide chain, binding along one face of such a linear sequence would cause protection of every other NH in the stretch of linear polypeptide chain. Rotation of the phi and psi bonds could change the direction the NHs are oriented and this could then play a role in what is protected and what is not. HDX followed by ETD MS restricted the deuterium protection region to residues K13-Q16 (Figure 3). Close inspection of this area in the crystal structure (Figure 4) showed that the NHs of residues L14, Q16 and G18 would be directly occluded by Adnectin 1 in the complex while the NH of T15 would form a stable hydrogen bond with the backbone carboxyl oxygen of N12 and the NH of L17 would point away from the binding interface. The HDX MS results indicating that exchange in K13-Q16 was reduced are there therefore highly consistent with what is seen in the co-crystal.
Protection from exchange in the peptide covering residues 96-108 (Figure 2) involved stabilization and/or protection of 2-3 NHs. As is clear in the crystal structure (Figure 4), this part of exEGFR forms a loop, originating from one strand of a parallel beta sheet, that comes into close proximity with Adnectin 1. Without further refinement of the position by ETD, which was not possible for this region, one could speculate that the protected NHs reside in the loop that would make most contact with Adnectin 1. Residues 45-54 had somewhat less protection upon complex formation. These residues form another loop that makes contact with Adnectin 1, primarily involving a tyrosine residue (Y45) that points directly towards the Adnectin 1 (Figure 4). Stabilization of this tyrosine via interaction with Adnectin 1 could cause a ripple effect of stabilization that would influence NHs in the other parts of peptide 45-54.
As we have just demonstrated, it is easy to rationalize the HDX MS when the structure is known. What about when the structure is not known? One would not be certain which regions (1-19, 96-108 or 45-54) were the primary determinants of binding/interaction, but it would still be clear that those regions were important. Caution must be taken, however, as not all interactions are as straightforward as A binding to B, and other protein factors such as allostery must be considered [63]. Subsequent experiments such as site-directed mutagenesis would then be required to validate which regions were the most critical for binding, if any, and which were a function of allostery. An initial round of HDX MS followed by mutagenesis and then re-examination of each mutant with HDX MS might be done more quickly than structure determination by other means, especially in cases where the complex is recalcitrant to analysis. The present results on exEGFR and Adnectin 1 emphasize the effectiveness of HDX MS, in addition to other structural methods such as those reported by Yan et al. in an accompanying paper [34], when it comes to characterization of interactions between proteins, and certainly so for those with therapeutic potential. Finally, the data presented in this manuscript are the first that report on the solution conformation of the full-length extracellular domain of EGFR, opening insights into how the dynamics influence the function of this medically important target protein. It is hoped that such knowledge can lead to a better understanding of the structure-function relationship and therefore to new strategies for improved cancer therapeutics.
Supplementary Material
Acknowledgements
The authors would like to thank Prof. Thomas E. Wales for helpful discussions. The authors also thank Dr. Bruce Car, Dr. Morrey Atkinson and Dr. Peter Moesta from Bristol-Myers Squibb Company for their support of this project. This work was supported in part by grants from the National Institute of Health (GM086507 and GM 101135) and a research collaboration with the Waters Corporation.
References
- (1).Harari PM. Epidermal growth factor receptor inhibition strategies in oncology. Endocr Relat Cancer. 2004;11:689–708. doi: 10.1677/erc.1.00600. [DOI] [PubMed] [Google Scholar]
- (2).Lemmon MA. Ligand-induced ErbB receptor dimerization. Exp Cell Res. 2009;315:638–648. doi: 10.1016/j.yexcr.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ferguson KM. Structure-based view of epidermal growth factor receptor regulation. Annu Rev Biophys. 2008;37:353–373. doi: 10.1146/annurev.biophys.37.032807.125829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- (5).Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw DE. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Shan Y, Eastwood MP, Zhang X, Kim ET, Arkhipov A, Dror RO, Jumper J, Kuriyan J, Shaw DE. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149:860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- (7).Sherrill JM, Kyte J. Activation of epidermal growth factor receptor by epidermal growth factor. Biochemistry. 1996;35:5705–5718. doi: 10.1021/bi9602268. [DOI] [PubMed] [Google Scholar]
- (8).Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- (9).Woodburn JR. The epidermal growth factor receptor and its inhibition in cancer therapy. Pharmacol Ther. 1999;82:241–250. doi: 10.1016/s0163-7258(98)00045-x. [DOI] [PubMed] [Google Scholar]
- (10).Ennis BW, Lippman ME, Dickson RB. The EGF receptor system as a target for antitumor therapy. Cancer Invest. 1991;9:553–562. doi: 10.3109/07357909109018953. [DOI] [PubMed] [Google Scholar]
- (11).Xu L, Aha P, Gu K, Kuimelis RG, Kurz M, Lam T, Lim AC, Liu H, Lohse PA, Sun L, Weng S, Wagner RW, Lipovsek D. Directed evolution of high-affinity antibody mimics using mRNA display. Chem Biol. 2002;9:933–942. doi: 10.1016/s1074-5521(02)00187-4. [DOI] [PubMed] [Google Scholar]
- (12).Lipovsek D. Adnectins: engineered target-binding protein therapeutics. Protein Eng Des Sel. 2011;24:3–9. doi: 10.1093/protein/gzq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dickinson CD, Veerapandian B, Dai XP, Hamlin RC, Xuong NH, Ruoslahti E, Ely KR. Crystal structure of the tenth type III cell adhesion module of human fibronectin. J Mol Biol. 1994;236:1079–1092. doi: 10.1016/0022-2836(94)90013-2. [DOI] [PubMed] [Google Scholar]
- (14).Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- (15).Getmanova EV, Chen Y, Bloom L, Gokemeijer J, Shamah S, Warikoo V, Wang J, Ling V, Sun L. Antagonists to human and mouse vascular endothelial growth factor receptor 2 generated by directed protein evolution in vitro. Chem Biol. 2006;13:549–556. doi: 10.1016/j.chembiol.2005.12.009. [DOI] [PubMed] [Google Scholar]
- (16).Koide A, Abbatiello S, Rothgery L, Koide S. Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor. Proc Natl Acad Sci U S A. 2002;99:1253–1258. doi: 10.1073/pnas.032665299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wojcik J, Hantschel O, Grebien F, Kaupe I, Bennett KL, Barkinge J, Jones RB, Koide A, Superti-Furga G, Koide S. A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat Struct Mol Biol. 2010;17:519–527. doi: 10.1038/nsmb.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hackel BJ, Kapila A, Wittrup KD. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J Mol Biol. 2008;381:1238–1252. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ramamurthy V, Krystek SR, Jr., Bush A, Wei A, Emanuel SL, Das Gupta R, Janjua A, Cheng L, Murdock M, Abramczyk B, Cohen D, Lin Z, Morin P, Davis JH, Dabritz M, McLaughlin DC, Russo KA, Chao G, Wright MC, Jenny VA, Engle LJ, Furfine E, Sheriff S. Structures of adnectin/protein complexes reveal an expanded binding footprint. Structure. 2012;20:259–269. doi: 10.1016/j.str.2011.11.016. [DOI] [PubMed] [Google Scholar]
- (20).Tiyanont K, Wales TE, Aste-Amezaga M, Aster JC, Engen JR, Blacklow SC. Evidence for increased exposure of the Notch1 metalloprotease cleavage site upon conversion to an activated conformation. Structure. 2011;19:546–554. doi: 10.1016/j.str.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Malito E, Faleri A, Lo Surdo P, Veggi D, Maruggi G, Grassi E, Cartocci E, Bertoldi I, Genovese A, Santini L, Romagnoli G, Borgogni E, Brier S, Lo Passo C, Domina M, Castellino F, Felici F, van der Veen S, Johnson S, Lea SM, Tang CM, Pizza M, Savino S, Norais N, Rappuoli R, Bottomley MJ, Masignani V. Defining a protective epitope on factor H binding protein, a key meningococcal virulence factor and vaccine antigen. Proc Natl Acad Sci U S A. 2013;110:3304–3309. doi: 10.1073/pnas.1222845110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhang Q, Willison LN, Tripathi P, Sathe SK, Roux KH, Emmett MR, Blakney GT, Zhang HM, Marshall AG. Epitope mapping of a 95 kDa antigen in complex with antibody by solution-phase amide backbone hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2011;83:7129–7136. doi: 10.1021/ac201501z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Pacholarz KJ, Garlish RA, Taylor RJ, Barran PE. Mass spectrometry based tools to investigate protein-ligand interactions for drug discovery. Chem Soc Rev. 2012;41:4335–4355. doi: 10.1039/c2cs35035a. [DOI] [PubMed] [Google Scholar]
- (24).Percy AJ, Rey M, Burns KM, Schriemer DC. Probing protein interactions with hydrogen/deuterium exchange and mass spectrometry-a review. Anal Chim Acta. 2012;721:7–21. doi: 10.1016/j.aca.2012.01.037. [DOI] [PubMed] [Google Scholar]
- (25).Chalmers MJ, Busby SA, Pascal BD, West GM, Griffin PR. Differential hydrogen/deuterium exchange mass spectrometry analysis of protein-ligand interactions. Expert Rev Proteomics. 2011;8:43–59. doi: 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Tsutsui Y, Wintrode PL. Hydrogen/deuterium exchange-mass spectrometry: a powerful tool for probing protein structure, dynamics and interactions. Curr Med Chem. 2007;14:2344–2358. doi: 10.2174/092986707781745596. [DOI] [PubMed] [Google Scholar]
- (27).Kaveti S, Engen JR. Protein interactions probed with mass spectrometry. Methods Mol Biol. 2006;316:179–197. doi: 10.1385/1-59259-964-8:179. [DOI] [PubMed] [Google Scholar]
- (28).Maier CS, Deinzer ML. Protein conformations, interactions, and H/D exchange. Methods Enzymol. 2005;402:312–360. doi: 10.1016/S0076-6879(05)02010-0. [DOI] [PubMed] [Google Scholar]
- (29).Engen JR, Wales TE, Chen S, Marzluff EM, Hassell KM, Weis DD, Smithgall TE. Partial cooperative unfolding in proteins as observed by hydrogen exchange mass spectrometry. Int. Rev. Phys. Chem. 2013;32:96–127. doi: 10.1080/0144235X.2012.751175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Engen JR. Analysis of protein complexes with hydrogen exchange and mass spectrometry. Analyst. 2003;128:623–628. doi: 10.1039/b212800b. [DOI] [PubMed] [Google Scholar]
- (31).Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- (32).Mandell JG, Baerga-Ortiz A, Croy CH, Falick AM, Komives EA. Application of amide proton exchange mass spectrometry for the study of protein-protein interactions. Curr Protoc Protein Sci. 2005 doi: 10.1002/0471140864.ps2009s40. Chapter 20, Unit20.29. [DOI] [PubMed] [Google Scholar]
- (33).Lerner EC, Trible RP, Schiavone AP, Hochrein JM, Engen JR, Smithgall TE. Activation of the Src family kinase Hck without SH3-linker release. J Biol Chem. 2005;280:40832–40837. doi: 10.1074/jbc.M508782200. [DOI] [PubMed] [Google Scholar]
- (34).Yan Y, Chen G, Wei H, Huang R, Mo J, Rempel DL, Tymiak AA, Gross ML. Fast photochemical oxidation of proteins (FPOP) maps the epitope of EGFR binding to adnectin. J Am Soc Mass Spectrom this issue, insert pages here. 2014 doi: 10.1007/s13361-014-0993-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Iacob RE, Bou-Assaf GM, Makowski L, Engen JR, Berkowitz SA, Houde D. Investigating Monoclonal Antibody Aggregation Using a Combination of H/DX-MS and Other Biophysical Measurements. J Pharm Sci. 2013;102:4315–4329. doi: 10.1002/jps.23754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Burkitt W, O'Connor G. Assessment of the repeatability and reproducibility of hydrogen/deuterium exchange mass spectrometry measurements. Rapid Commun Mass Spectrom. 2008;22:3893–3901. doi: 10.1002/rcm.3794. [DOI] [PubMed] [Google Scholar]
- (38).Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2011;100:2071–2086. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Iacob RE, Engen JR. Hydrogen exchange mass spectrometry: are we out of the quicksand? J Am Soc Mass Spectrom. 2012;23:1003–1010. doi: 10.1007/s13361-012-0377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Plumb RS, Johnson KA, Rainville P, Smith BW, Wilson ID, Castro-Perez JM, Nicholson JK. UPLC/MS(E); a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun Mass Spectrom. 2006;20:1989–1994. doi: 10.1002/rcm.2550. [DOI] [PubMed] [Google Scholar]
- (41).Wales TE, Eggertson MJ, Engen JR. Considerations in the analysis of hydrogen exchange mass spectrometry data. Methods Mol Biol. 2013;1007:263–288. doi: 10.1007/978-1-62703-392-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Ahn J, Cao MJ, Yu YQ, Engen JR. Accessing the reproducibility and specificity of pepsin and other aspartic proteases. Biochim Biophys Acta. 2013;1834:1222–1229. doi: 10.1016/j.bbapap.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Rand KD, Pringle SD, Morris M, Engen JR, Brown JM. ETD in a traveling wave ion guide at tuned Z-spray ion source conditions allows for site-specific hydrogen/deuterium exchange measurements. J Am Soc Mass Spectrom. 2011;22:1784–1793. doi: 10.1007/s13361-011-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zehl M, Rand KD, Jensen ON, Jorgensen TJ. Electron transfer dissociation facilitates the measurement of deuterium incorporation into selectively labeled peptides with single residue resolution. J Am Chem Soc. 2008;130:17453–17459. doi: 10.1021/ja805573h. [DOI] [PubMed] [Google Scholar]
- (45).Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- (46).Guttman M, Weis DD, Engen JR, Lee KK. Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J Am Soc Mass Spectrom. 2013;24:1906–1912. doi: 10.1007/s13361-013-0727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- (49).Clayton AH, Walker F, Orchard SG, Henderson C, Fuchs D, Rothacker J, Nice EC, Burgess AW. Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. J Biol Chem. 2005;280:30392–30399. doi: 10.1074/jbc.M504770200. [DOI] [PubMed] [Google Scholar]
- (50).Low-Nam ST, Lidke KA, Cutler PJ, Roovers RC, van Bergen en Henegouwen PM, Wilson BS, Lidke DS. ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nat Struct Mol Biol. 2011;18:1244–1249. doi: 10.1038/nsmb.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Dawson JP, Bu Z, Lemmon MA. Ligand-induced structural transitions in ErbB receptor extracellular domains. Structure. 2007;15:942–954. doi: 10.1016/j.str.2007.06.013. [DOI] [PubMed] [Google Scholar]
- (52).Huxford T, Mishler D, Phelps CB, Huang DB, Sengchanthalangsy LL, Reeves R, Hughes CA, Komives EA, Ghosh G. Solvent exposed non-contacting amino acids play a critical role in NF-kappaB/IkappaBalpha complex formation. J Mol Biol. 2002;324:587–597. doi: 10.1016/s0022-2836(02)01149-x. [DOI] [PubMed] [Google Scholar]
- (53).Mandell JG, Baerga-Ortiz A, Akashi S, Takio K, Komives EA. Solvent accessibility of the thrombin-thrombomodulin interface. J Mol Biol. 2001;306:575–589. doi: 10.1006/jmbi.2000.4416. [DOI] [PubMed] [Google Scholar]
- (54).Dharmasiri K, Smith DL. Mass spectrometric determination of isotopic exchange rates of amide hydrogens located on the surfaces of proteins. Anal Chem. 1996;68:2340–2344. doi: 10.1021/ac9601526. [DOI] [PubMed] [Google Scholar]
- (55).Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Smith DL. Local structure and dynamics in proteins characterized by hydrogen exchange and mass spectrometry. Biochemistry (Mosc) 1998;63:285–293. [PubMed] [Google Scholar]
- (58).Rand KD, Zehl M, Jensen ON, Jorgensen TJ. Protein hydrogen exchange measured at single-residue resolution by electron transfer dissociation mass spectrometry. Anal Chem. 2009;81:5577–5584. doi: 10.1021/ac9008447. [DOI] [PubMed] [Google Scholar]
- (59).Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Ladner RC. Mapping the epitopes of antibodies. Biotechnol Genet Eng Rev. 2007;24:1–30. doi: 10.1080/02648725.2007.10648092. [DOI] [PubMed] [Google Scholar]
- (61).Hager-Braun C, Tomer KB. Determination of protein-derived epitopes by mass spectrometry. Expert Rev Proteomics. 2005;2:745–756. doi: 10.1586/14789450.2.5.745. [DOI] [PubMed] [Google Scholar]
- (62).Clementi N, Mancini N, Castelli M, Clementi M, Burioni R. Characterization of epitopes recognized by monoclonal antibodies: experimental approaches supported by freely accessible bioinformatic tools. Drug Discov Today. 2013;18:464–474. doi: 10.1016/j.drudis.2012.11.006. [DOI] [PubMed] [Google Scholar]
- (63).Ahn J, Engen JR. The use of Hydrogen/Deuterium exchange mass spectrometry in epitope mapping. Chemistry today. 2013;31:25–28. [Google Scholar]
- (64).Roepstorff P, Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.