Summary
Most bacterial species make transitions between habitats, such as switching from free-living to symbiotic niches. We provide evidence that a galaxin protein, EsGal1, of the squid Euprymna scolopes participates in both: (i) selection of the specific partner Vibrio fischeri from the bacterioplankton during symbiosis onset and, (ii) modulation of V. fischeri growth in symbiotic maintenance. We identified two galaxins in transcriptomic databases and showed by qRT-PCR that one (esgal1) was dominant in the light organ. Further, esgal1 expression was upregulated by symbiosis, a response that was partially achieved with exposure to symbiont cell-envelope molecules. Confocal immunocytochemistry of juvenile animals localized EsGal1 to the apical surfaces of light-organ epithelia and surrounding mucus, the environment in which V. fischeri cells aggregate before migration into the organ. Growth assays revealed that one repeat of EsGal1 arrested growth of Gram-positive bacterial cells, which represent the cell type first ‘winnowed’ during initial selection of the symbiont. The EsGal1-derived peptide also significantly decreased the growth rate of V. fischeri in culture. Further, when animals were exposed to an anti-EsGal1 antibody, symbiont population growth was significantly increased. These data provide a window into how hosts select symbionts from a rich environment and govern their growth in symbiosis.
Introduction
A current interest in ecology focuses on how microbial communities are structured in natural habitats, and how microbes within those communities make transitions between different niches (Fuhrman, 2009; Needham et al., 2013). One common niche transition occurs when marine microbes move from planktonic habitats to persistent associations with animal partners (Gallo and Hooper, 2012; Bulgarelli et al., 2013). The light-organ symbiosis between Euprymna scolopes, the Hawaiian bobtail squid, and Vibrio fischeri, a luminescent gram-negative marine bacterium, is a horizontally transmitted association, i.e., acquired each generation (for review, see Stabb and Visick, 2013). The symbionts colonize deep light-organ tissues, or crypts, within hours of the host’s hatching, taking up residence along the apical surfaces of the crypt epithelia. The host responds to a variety of cues presented by V. fischeri in the crypts, including luminescence and microbe-associated molecular patterns (MAMPs), notably cell envelope molecules (for review see McFall-Ngai et al., 2010, and McFall-Ngai et al., 2012). Transitions of V. fischeri between membership in the bacterioplankton to the symbiotic niche occur both at the onset of the symbiosis, when V. fischeri is recruited into animal tissues, and each day of the association with the venting of ~90% of the symbiont population out of the light organ at dawn, when V. fischeri cells cycle into the planktonic niche.
How the bacterioplankton is winnowed in the selection of the specific symbiotic partner at onset of the association has been a continuing focus of studies of the squid-vibrio system. Soon after hatching, juvenile E. scolopes begin to ventilate, bringing environmental microbial communities, as well as the products they release, over superficial ciliated epithelial fields that are specific to the juvenile light organ. The epithelia begin to shed mucus within minutes in response to environmental peptidoglycan (PGN) (Nyholm et al., 2002). V. fischeri is then enriched along these tissues by first attaching to the cilia and then aggregating (Altura et al., 2013). Whereas Gram-positive environmental bacteria appear not to associate with this ciliated field, all tested Gram-negative bacterial cells have the capacity to adhere (Nyholm and McFall-Ngai, 2003; Altura et al., 2013). Through the recruitment process, however, all non-specific Gram-negative bacteria are gradually lost from the aggregating cells such that, by about 3 h, only a small population of V. fischeri cells remains (Altura et al., 2013).
While this stepwise dominance of V. fischeri on the ciliated surface is not well understood, recent research has provided some insight. The shed mucus is acidic (Kremer et al., 2013) and contains abundant biomolecules typically associated with ‘antimicrobial’ activity, including nitric oxide (Davidson et al., 2004) and a peptidoglycan-recognition protein, which has an amidase activity that breaks down PGN (Troll et al., 2010). In addition, a recent comparison of the host transcriptome in 3-h animals exposed or not exposed to V. fischeri revealed robust changes in gene expression in response to the presence of the symbiont (Kremer et al., 2013). The data suggest the possibility that some of the upregulated genes encode proteins with activities that shape the chemical environment of the mucus to favor V. fischeri. A role for host biomolecules as critical determinants of host-symbiont interactions is not without precedent; they have been widely studied as features that can structure and modulate behaviors of host-associated microbial communities (de Oliveira et al., 2012, Shnit-Orland et al., 2012). For example, host factors function in symbiont attraction in the legume-rhizobia symbioses (Redmond et al., 1986), in the structuring of symbiont communities in the mammalian gut (Cash et al., 2006), and in maintaining specificity in nematode-microbe associations (Bulgheresi et al., 2011).
In this study, we have explored the possible roles of another host biomolecule, a protein known as galaxin, in shaping the transition to a symbiotic habitat by V. fischeri. Galaxins were first reported in other symbiotic associations. For example, several studies on corals have correlated galaxin activity with the process of calcification of the exoskeleton (Fukuda et al., 2003, Watanabe et al., 2003), and galaxin transcripts have also been identified in the body wall of Riftia pachyptila (Sanchez et al., 2007), although their function remains unexplored. In the squid-vibrio system, previous studies of symbiont-induced changes in host gene expression showed that transcripts encoding putative galaxin proteins are upregulated at first colonization of the light organ by the symbionts (Chun et al., 2008), and are regulated over the day/night cycle in the adult light organ (Wier et al., 2010). Here we characterize in depth one of the host-squid galaxins, EsGalaxin1 (EsGal1), the prominent isoform in the host light organ, during the selection of the symbiont in onset of the partnership and during the maintenance of the mature association as a balanced system.
Results
Features of the esgal genes and of EsGalaxin1 (EsGal1), the principal galaxin of the light organ
We identified two esgal sequences, esgal1 and esgal2, in the E. scolopes transcriptomic databases. Using qRT-PCR to examine expression in different tissues of the animal, we confirmed that the esgal1 gene is the most highly expressed galaxin gene in both the juvenile and adult light organs. Thus, for this study, we explored aspects of esgal1 gene expression and the EsGal1 protein patterns in the squid-vibrio symbiosis. An unexpected finding was that esgal1 expression is ~250X higher in the light organ than in the accessory nidamental gland (ANG), another symbiotic organ, but one that is less well studied than the light organ. The ANG contains a consortium of bacteria thought to play a role in protection of the eggs during host development (Collins et al., 2012). The expression of esgal2 showed the opposite pattern in that it was ~250X higher in the ANG than in the light organ (Fig. S1).
To examine the biochemical characteristics of the EsGal1 protein more fully, we verified the full-length coding sequence for EsGal1 by 5’RACE. esgal1 encodes a 13.1 kD protein with a pI of ~10. RADAR analysis of the sequence predicted that the protein has three tandem repeats (Fig. 1A) anchored by dicysteine pairs. SignalP analysis predicted a signal peptide, suggesting that the protein is secreted. The high predicted pI, repeat structure, and abundant cysteines (12.3%) provide evidence that the protein is antimicrobial (Fedders and Leippe, 2008).
Fig. 1.

Biochemical properties of the EsGal1 protein. (A) The structure of the protein. Left, major regions, with amino acid position indicated by the numbers below. Mr, molecular mass; pI, predicted isoelectric point; R1-3, predicted repeats and their pIs; SP, predicted signal peptide. Right, the derived amino acid sequence of EsGal1 R1-R3, with the RADAR-predicted repeat structure; predicted signal peptide, underlined; predicted N-glycosylation site, boxed; cysteines, gray. (B) MUSCLE alignment of portions of galaxin sequences from Acropora palmata, Balanoglossus clavigerus, E. scolopes, and Galaxea fasicularis, with the consensus sequence and graphical representation of conservation at each amino acid residue shown below the alignment. Gray shading denotes the classes of the labeled amino acids.
BLAST analysis (NCBI) of the derived amino-acid sequence of the protein revealed 25 proteins with some similarity to EsGal1 (Table S2), with the closest match being to a galaxin occurring in the coral Acropora millepora (E-value = 4e-13). Although A. millepora galaxin was the closest BLAST hit to EsGal1, we used the A. palmata galaxin sequence for alignment and biochemical analysis; A. palmata galaxin was chosen because its repeats were predicted to have a higher antimicrobial activity. Examination of full-length sequences revealed a protein family with an array of sizes, from 116 to 828 amino acids (Table S2). Our alignment with portions of other galaxin proteins (Figs. 1B and S2) revealed that EsGal1 shares with the other family members the conserved repeat structure and a number of residues, which suggests that these regions of the protein are critical to structure and function of galaxins.
Patterns of esgal1 expression
To characterize the regulation of expression of esgal1 over early light-organ development, we performed qRT-PCR on both aposymbiotic and symbiotic light organs collected at six time points over the first 2 d of colonization of the juvenile squid (Fig. 2A). These points were chosen to determine when the regulation of esgal1 begins after colonization and then how long after colonization the regulation persists. The esgal1 mRNA levels were higher in the symbiotic than the aposymbiotic light organs; significant upregulation began at 10 h post-hatching and persisted over the first two days of the symbiosis. In addition, symbiotic light-organ expression of esgal1 increased two fold between 10 and 14 h post-hatching, whereas we observed no difference in expression in aposymbiotic light organs, suggesting that symbiosis is increasing expression rather than expression being downregulated in aposymbiotic animals. To determine if galaxin expression is more prevalent in symbiotic tissues than other portions of the squid, we analyzed esgal1 expression in six different tissues of the adult squid and found that relative expression of esgal1 is 100-1000 fold higher in V. fischeri-containing tissue than in any other tissue sampled (Fig. 2B).
Fig. 2.
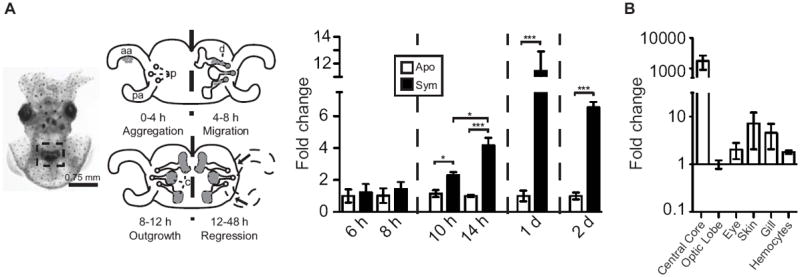
Patterns of esgal1 expression. (A) Top, a ventral view of the juvenile animal, with relevant features. Left, the light organ (box), seen through translucent ventral mantle tissue. Right, stages of early symbiosis development. The acquisition of symbionts from the seawater is facilitated by anterior (aa) and posterior (pa) epithelial ‘appendages’ of a superficial ciliated epithelium. Symbionts (gray) accumulate along these epithelia in the first hours after exposure (Aggregation). They then migrate into pores (p), through the ducts (d) into the crypt spaces (c) (Migration). In the crypts, the symbionts proliferate to fill the spaces (Outgrowth). At 12 h, the symbionts deliver an irreversible morphogenic signal that leads to the eventual loss of the ciliated epithelia (Regression). Bottom, the expression of esgal1 in the light organ at time points over the first 2 d in apo- and symbiotic animals. Significant differences (*, <0.05; ***, <0.001) between conditions by an ANOVA followed by a Tukey’s pairwise comparison. (B) Expression of esgal1 in six tissues of adult animals. Relative qRT-PCR values (+/- SEM). All data are normalized to the condition of lowest level of expression; n=3 biological replicates and 2 technical replicates per condition.
In the squid-vibrio symbiosis, certain symbiont-induced host phenotypes are reversible upon depletion of symbionts from a colonized organ after 12 h, i.e, they require persistent interaction with the symbiont (Lamarcq and McFall-Ngai, 1998; Nyholm et al., 2002), while some are irreversibly triggered by a 12-h colonization (Doino and McFall-Ngai 1995; Davidson et al., 2004). To address whether persistent colonization of the light organ by symbionts was necessary for esgal1 regulation, we depleted colonized animals of their symbionts with antibiotics at 24 h post-hatching and then compared esgal1 expression levels in these depleted light organs 24 h later, i.e, at 48 h post hatching, to those of symbiotic and aposymbiotic animals. Depletion of symbionts did not significantly alter the expression of the esgal1 transcript from the symbiotic level of expression under the conditions used (Fig. 3A), suggesting that the persistence of viable symbionts in the light organ is not necessary for the regulation of esgal1, or that any change in expression requires significantly longer than other reversible phenotypes.
Fig. 3.
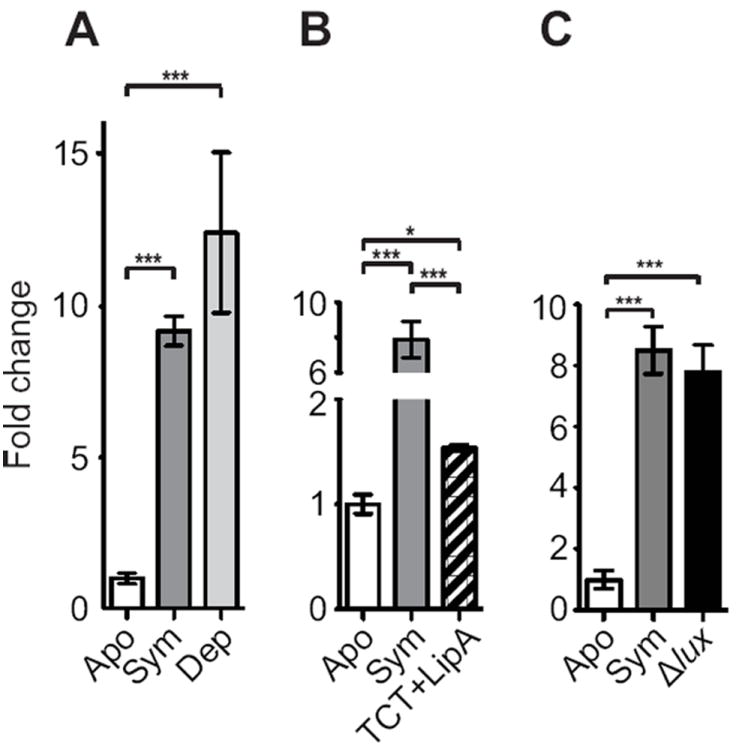
The influence of experimental manipulation on esgal1 expression. Expression, as measured by qRT-PCR, at 48 h in the light organ of animals remaining uncolonized (Apo) or colonized with V. fischeri strain ES114 (Sym) compared with: (A) animals colonized with V. fischeri for 24 h and then antibiotically depleted of their symbionts (Dep); (B) treated with the bacterial products TCT and lipid A (TCT+LipA) (see Materials and methods for details); (C) animals colonized with mutants defective in light production (Δlux). Data are normalized to the time point of lowest expression. Values +/- SEM; Significant differences (* = p< 0.05, ** = p<0.01, *** = p<0.001) between conditions by an ANOVA followed by a Tukey’s pairwise comparison; n = 3 to 4 biological replicates and 2 technical replicates per condition.
We explored signals from V. fischeri that might regulate esgal1 expression. Because the bacterial products lipid A and TCT induce many host phenotypes in the squid-vibrio system (reviewed in McFall-Ngai et al., 2010), we determined whether these products are also responsible for the regulation of esgal1. While exposure to TCT and lipid A increased expression of esgal1 in the light organ on average ~1.75-fold (Fig. 3b), the response was only ~20% of the increase observed in symbiotic relative to aposymbiotic organs. One other inducer of host phenotypes is symbiont light production (McFall-Ngai et al., 2012). However, V. fischeri mutants defective in light production showed no defect in esgal1 upregulation (Fig. 3B).
Localization and abundance of EsGal1 in the juvenile animal
The polyclonal antibody to two EsGal1 peptides recognized apical cytoplasmic and perinuclear sites in the epithelia of the light organ, including the appendages and ducts, but did not label internal musculature (Fig. 4A, A’). The apical localization, in conjunction with the presence of a signal peptide in the esgal1-derived amino acid sequence, suggested that the protein is secreted, which was confirmed by localization of EsGal1 to the mucus outside of the light organ (Fig. 4B). In quantification of labeling of EsGal1 antibody in apo- and symbiotic animals, although the two conditions had the same signal intensity in the duct cells, the epithelial appendages of symbiotic light organs had about twice the signal intensity (Fig. 4A’, inset). We also observed antibody cross reactivity in the symbiont-containing crypts (Fig. 4C). It localized to perinuclear sites inside of the epithelial cells and to the extracellular crypt spaces where the symbionts reside, suggesting secretion of the protein into symbiont-containing regions. We observed no staining in samples exposed to rabbit IgG as a negative control (Fig. S3A-C). When we performed a western blot analysis on squid proteins using the EsGal1 antibody, we observed several specific bands, one of which corresponded to the predicted molecular weight (Fig. S3D). The existence of multiple bands may be due to the putative N-glycosylation site on the EsGal1 protein (Fig. 1A), the modification of which could alter the migration of the protein on an SDS gel, or due to the abundant dicysteine bonds that may not have been fully reduced before protein preparation.
Fig. 4.
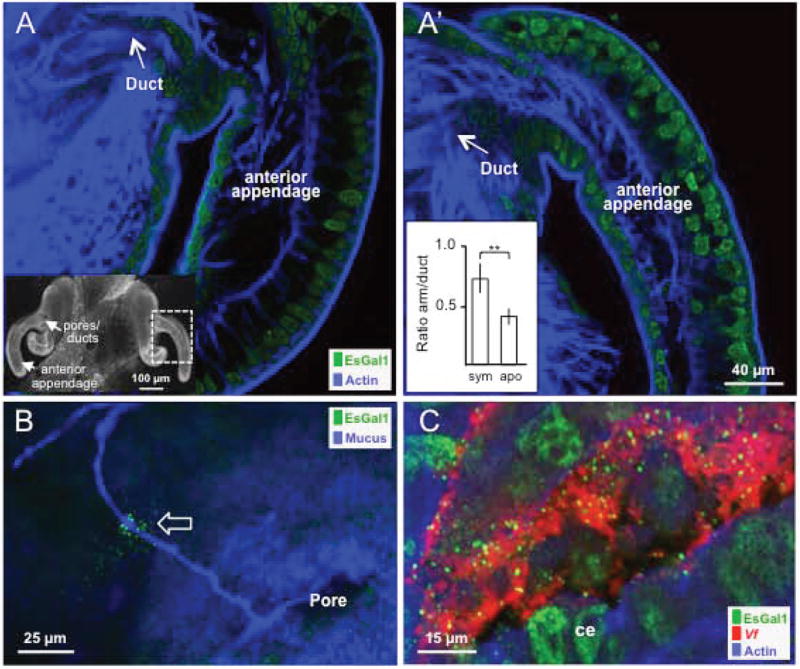
Localization and abundance of EsGal1 in the juvenile light organ. Light organs from 24 h uncolonized (A) and colonized (A’) juveniles probed with the EsGal1 antibody, showing the relative abundance of EsGal1 in each condition. Labeling of tissue actin (phalloidin) was used as a counterstain. Inset in A, confocal micrograph of the entire organ showing regions explored by ICC. Inset graph in A’, relative fluorescent intensity; ** = p<0.01 by an unpaired t-test after log transformation to ensure normality. (B) EsGal1 localization to the mucus outside of the juvenile light organ. Open arrow, EsGal1 in contact with a mucus strand outside of the light-organ pore. (C) EsGal1 secretion into crypt spaces of the colonized organ. ce, crypt epithelium; Vf, Vibrio fischeri.
To determine whether epithelial localization of EsGal1 protein was unique to the light organ, we performed immunocytochemistry with the antibody on other host tissues (Fig. S4). The antibody recognized cytoplasmic and perinuclear sites in all epithelial tissues exposed to seawater, including the tentacles and gills, but did not label internal tissues, such as the white body (the haematopoietic organ of the animal) or musculature of the tentacles. Symbiotic state of the light organ did not detectably alter the localization of EsGal1 in any non-symbiotic tissue.
Evidence for antimicrobial activity of EsGal1
Due to the small size, basic pI, presence of a secretion signal, and numerous cysteines in EsGal1, we hypothesized that the protein acts as an anti-microbial effector, a class of molecules that tend to share all of these properties (Fedders and Leippe, 2008). To determine whether the protein might have such activity, we synthesized one of the three repeats present in the EsGal1 protein (EsGal1R3, Table 1) that, by in silico analysis, was predicted to be the most likely region of the protein to be antimicrobial, and characterized its ability to inhibit bacterial growth. The third repeat of EsGal1 was chosen because of its high net charge and hydrophobic residue ratio, as well as its significant sequence similarity to a known oyster defensin, all determined with the APD2 antimicrobial peptide calculator and predictor (see Experimental Procedures). EsGal1R3 affected the growth of Gram-positive bacteria at concentrations as low as 27 nM (Table 1), but was about 10 to 100 times more effective against non-marine species under the assay conditions used. However, EsGal1R3 did not inhibit the growth of Gram-negative marine species, including V. fischeri, in MIC assays. To determine whether antimicrobial activity was a shared characteristic of galaxin proteins from other species, we also synthesized a peptide corresponding to the predicted repeat 2 from Acropora palmata galaxin (ApGalR2), which we chose for its basic pI and predicted antimicrobial capabilities, and determined its ability to inhibit bacterial growth under the same conditions used for assays with EsGal1R3. ApGalR2 did not exhibit any capacity to inhibit the growth of marine bacteria and was, at most effective, 10-fold less active than EsGal1R3 against non-marine strains.
TABLE 1.
Antimicrobial activity of EsGal1R3 and ApGal R2
| Strain Characteristics and Strain | EsGal1 R3 MIC (μM) -RCCRYNVYNNSQYALCCAGRVTMKPTKRS | ApGal R2 MIC (μM) - VQRRSGLSPACCGTRPYDAKFRMCCGGT | Source |
|---|---|---|---|
| Gram-positive | |||
| Non-marine | |||
| B. megaterium | 0.027 | 0.287 | This Study |
| B. subtilis | 0.5375 | 4.76 | This Study |
| Marine | |||
| B. algicola CNJ 803 | 4.3 | >152.29 | (Gontang et al., 2007) |
| B. megaterium-like CNJ 778 | 6.88 | >152.29 | (Gontang et al., 2007) |
| E. aestuorii CNJ 771 | 3.44 | >152.59 | (Gontanget al., 2007) |
| Gram-negative | |||
| Non-marine | |||
| E. coli | 1.72 | 76.145 | (Blattner et al., 1997) |
| Marine | |||
| P. leiognathi | >137.5 | >152.29 | (Dunlap, 1985) |
| V. fischeri ES114 | 34.4 | >152.29 | (Boettcher and Ruby, 1990) |
| V. parahaemolyticus | 137.5 | >152.29 | (Nyholm et al., 2000) |
Because of the influence of day/night cycles on the symbiont growth in the association (Nyholm and McFall-Ngai, 2004; Wier et al., 2010) and the potential that EsGal1 has to modulate bacterial growth, we also measured esgal1 gene expression at three time points over the day/night cycle in both juvenile light organs and adult central cores, i.e., the epithelial tissue of the adult light organ that supports the symbiont. In both sample groups, we found an upregulation of the esgal1 gene 2 h after dawn (Fig. 5), at a time point coincident with the rapid symbiont growth in the organ that immediately follows the venting of 90% of the symbionts (Graf and Ruby, 1998; Nyholm and McFall-Ngai, 1998). These data suggest that in both juvenile and adult animals esgal1 is upregulated when the bacterial symbionts are rapidly dividing in the light organ.
Fig. 5.
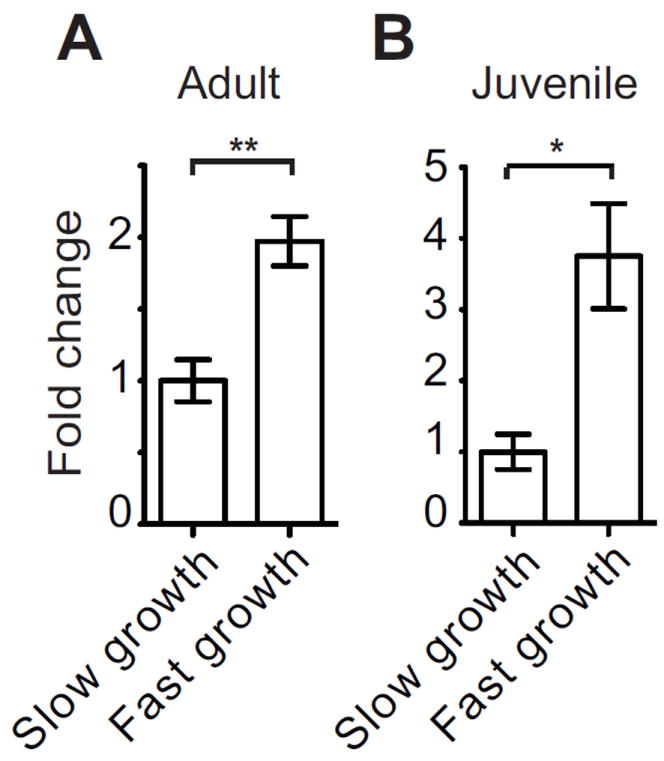
Daily changes in esgal1 expression with variation in light-organ symbiont growth rate. esgal1 expression in wild-caught adult (A) and juvenile (B) light organs during periods of slow and rapid bacterial growth, approximately 12-14 h before dawn and 2 h after dawn, respectively. All data are normalized to the condition of lowest expression. Error bars denote the standard error of the mean. * = p<0.05 and ** = p<0.01 by an unpaired t-test after log transformation to ensure normality.
Although the MIC assay showed that EsGal1R3 does not inhibit growth of V. fischeri except at high concentrations, we hypothesized that in lower concentrations, the peptide could modulate the rate of bacterial growth in the light organ. We first tested the growth response of V. fischeri to sub-inhibitory concentrations of the peptide under in vitro culture conditions. We observed that exposure to EsGal1R3 over 18 h both delayed the start of exponential growth of V. fischeri by approximately 2 h and decreased its growth rate 1.5-fold once in exponential phase growth (Fig. 6A, 6B). While the most robust result was obtained using 17.4 μM EsGal1R3 (half of the concentration determined to be inhibitory), the peptide showed a dose-dependent ability to modulate the growth of V. fischeri at a concentration as low as 8.2 μM (Fig. S5).
Fig. 6.
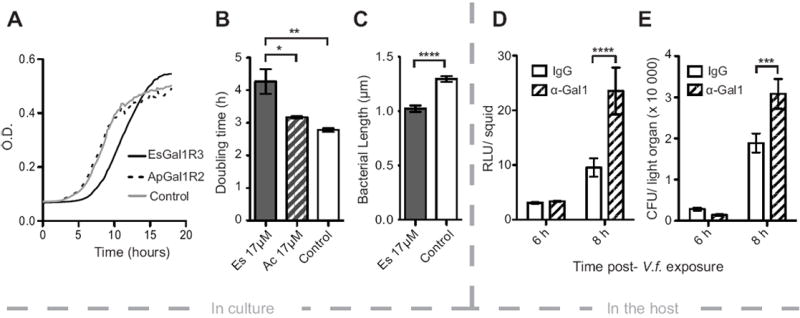
The effect of EsGal1R3 on V. fischeri growth in vitro and in vivo. (A) Growth curve of V. fischeri cells exposed to 17.4 μM EsGal1R3 (solid line), 17.4 μM ApGal1R2 (dashed line), or to no peptide (gray line). (B) Quantification of V. fischeri doubling times in experiment shown in (A). Experiment in (A) and (B) was performed with three biological replicates and two technical replicates. (C) Length of V. fischeri cells exposed to 17.4 μM EsGal1R3 (gray bar) or no peptide (open bar). N= 117 and 124, respectively. (D) Effect of α-EsGal1 antibody or rabbit IgG exposure on animal luminescence at 6 or 8 h after exposure to bacteria. (E) Effect of α-EsGal1 antibody or rabbit IgG exposure on the number of colony-forming units (CFU) in the juvenile light organ at 6 and 8 h after exposure to V. fischeri. N= 23 to 27 for each condition. Error bars denote the standard error of the mean. * = p< 0.05, ** = p<0.01, *** = p<0.001, **** = p<0.0001 by an ANOVA with a post-hoc Tukey’s pairwise comparison.
The growth curve (Fig. 6a) had a higher terminal optical density (OD) for the EsGal1R3 exposed cells, which suggested that either smaller cells or more numerous cells had resulted from the conditions. Measurements of the cells showed an average decrease in their length of about 20% (Fig. 6C), similar to the decrease in size noted in V. fischeri cells adapted to the crypts of the light organ (Ruby and Asato, 1993).
To determine whether EsGal1 might affect V. fischeri growth during onset of the association, we co-incubated the animals during colonization with the EsGal1 antibody to adsorb the protein from the environment, a proven approach to the study of interactions of the symbiont with host biomolecules (Aeckersberg et al., 2001, Kremer et al., 2013). In the treated hatchlings, we observed a 2.5-fold increase in symbiont luminescence (Fig. 6D) and a 1.6-fold increase in bacterial density (Fig. 6E) at 8 h post-bacterial exposure as compared to the negative control. In experiments with addition of the peptide, we found no evidence for a depression of V. fischeri growth in the crypts, suggesting that EsGal1 activity reaches an asymptote during symbiosis initiation. Due to the lack of an accessory nidamental gland in juvenile animals, we were unable to assess the affect of the EsGal1R3 peptide on the ANG-associated bacterial community.
Discussion
In this study, we investigated the role of a host protein, EsGal1, in the habitat transition of Vibrio fischeri from the seawater to the light organ of the squid Euprymna scolopes. We provide evidence for the activity of this protein in the selection of V. fischeri from the complex bacterioplankton, as well as in the long-term maintenance of this symbiont in a cooperative, binary association.
EsGal1 in V. fischeri habitat transition
In marine habitats, a given microbial species such as V. fischeri can be found in a wide variety of environments; for example, they can occur as members of the bacterioplankton, in marine snow, sediments, and symbiotic associations (Fuhrman 2009; Stabb and Visick, 2013). As such, habitat transitions are often a critical feature of their ecology. Because the association between V. fischeri and E. scolopes is a horizontally transmitted symbiosis (Bright and Bulgheresi, 2010), the microbial partner must be capable of a robust response both to environmental stressors in the planktonic environment and to the selective pressures imposed by the host niche (Wang et al., 2010). The host, in turn, must provide conditions that: 1) promote selection of the coevolved symbiont from thousands of microbial species in the bacterioplankton during symbiosis onset; and, 2) once the symbiosis is established, mediate the maintenance of the association so that it persists throughout the life of the animal (Visick and Ruby 1996; Nyholm et al., 2002).
Over the 3-4 h following initial exposure to seawater with natural levels of environmental bacteria, including the symbiont, V. fischeri cells become the competitive dominant among bacterial cells associating with the epithelial cell surface of the host’s light organ. The first step in this process of ‘winnowing’ is the elimination of Gram-positive bacteria (for review, see Nyholm and McFall-Ngai, 2004). We found that the host protein EsGal1 is produced on the surface of the epithelial field and secreted into the mucus (Fig. 4C), the environment where V. fischeri attaches and then aggregates (Altura et al., 2013). The lack of symbiont-dependent regulation of esgal1 transcription during time points coincident with aggregation demonstrates that provision of EsGal1 stores is “hard-wired” during embryogenesis to be in place, ready for secretion when the animal hatches from the egg (Fig. 2A). This provision does not appear to be relegated to the light organ in juvenile animals, as all epithelia tested that had contact with the seawater also produced EsGal1 (Fig. S4), suggesting that the protein may be used as a growth modulator in other sites on the squid’s body. Further, a peptide predicted to occur on the protein surface, generated to a single repeat of EsGal1, was active against Gram-positive bacteria in in vitro assays. While the peptide shows robust activity, we recognize that the whole protein may behave differently. The protein itself has three repeats predicted to have antimicrobial activity, so it is possible that the whole protein is more antimicrobial than the derived peptide. However, we feel it is necessary to exercise caution in interpreting how the intact galaxin protein behaves in the symbiosis. Earlier studies provided evidence for the involvement of other proteins that are active against Gram-positives, specifically the findings that a peptidoglycan-recognition protein (EsPGRP2,Troll et al., 2010) is also secreted into the mucus and the gene encoding a lysozyme isoform is upregulated during initial interactions with V. fischeri (Kremer et al., 2013). In addition, other antimicrobials, such as nitric oxide (Davidson et al., 2004), occur in abundance in the mucus. From these studies, a scenario is emerging in which the host provides a selective environment that V. fischeri can withstand and to which it can adapt, and that the symbiont cells participate in the creation of this environment through signal exchange with the host.
Growth modulation by host molecules
Our data also provide evidence that EsGal1 modulates the growth of symbiont populations that have already colonized the host. An earlier study demonstrated that colonization of the crypts causes a significant increase in expression of the gene encoding EsGal1 (Chun et al., 2008) at about 18 h following colonization. In the present study, we showed that this upregulation is detectable as early as 10 h, during the initial symbiont outgrowth to fill the crypt spaces. We confirmed that EsGal1 is secreted into the crypts, where it can act directly on symbiont cells, and co-incubation of juvenile squid during initial colonization with the EsGal1 antibody increased the growth rate of V. fischeri, such that the crypts are colonized more quickly. Once the symbiosis is established, we show that EsGal1 gene expression changes over the day/night cycle, being highest during the rapid growth of V. fischeri following the daily venting of cells into the environment in response to the dawn light cue (Graf and Ruby, 1998). The upregulation of esgal1 during bacterial growth in the light organ is reminiscent of the provision of the antimicrobial coleoptericin by weevils into regions where their intracellular symbionts occur. This protein controls the growth and limits the location of the symbionts to specific host cells, or bacteriocytes (Login et al., 2011). Taken together, these data suggest that the symbiont participates in creating an environment that imposes a governor on symbiont population growth.
The precise mechanisms by which the symbiont signals the host are not fully understood. Although our data show that bacterial cell-envelope molecules may play a role (Fig. 3B), they could not induce the same responses as the presence of the symbiont, suggesting that they may work in synergy with other factors. For some squid-host responses, light works in synergy with these molecules (Heath-Heckman et al., 2013). However, in this study, Δlux mutants had no defect in inducing normal expression of esgal1, so the evidence suggests another factor. Alternatively, under normal conditions these molecules may induce these host responses, but the cell envelope molecules, when added as pharmacological agents, are not presented to host tissue in the same manner as the intact bacterial cell (e.g., concentration, configuration); or, direct interactions with the bacterial cell enable host responses, as has been noted before for other host responses (Chun et al., 2008; Heath-Heckman et al., 2013)
The selection of symbionts by host biomolecules in the squid-vibrio system has parallels in other mutualisms. For example, in Hydra spp., overexpression of periculin, a secreted antimicrobial protein, changes the microbial community density along the host surface and the composition of the community in mature polyps (Fraune et al., 2010). Additionally, Hydra spp. lacking the capability to express antimicrobial peptides, such as arminin, lose the capacity to select for a species-specific microbiota (Franzenburg et al., 2013). In the mammalian gut, exposure of the host to gram-negative bacteria during initial gut colonization induces the secretion of RegIIIγ, a lectin that is anti-gram-positive (Cash et al., 2006). This protein alters microbial community structure in the gut by creating a zone of exclusion between host cells and the microbiota through its bacteriocidal activity. In so doing, it reduces potential symbiont-mediated inflammatory processes in the intestine (Cash et al., 2006; Vaishnava et al., 2011). The mammalian gut also expresses α-defensin, an antimicrobial peptide that has been shown to structure the microbial community to promote gut homeostasis (Salzman et al., 2010). Collectively, these studies suggest that the bacteriostatic property of EsGal1 against V. fischeri may benefit the host by restricting symbionts to the light organ, and maintaining immune homeostasis. Further, our finding that esgal1 and esgal2 have opposite patterns of expression in the adult light organ and accessory nidamental gland suggests that galaxins are a family of molecules that can be used to manage different microbial communities in the same animal. Further studies will be required to explore this possibility. Taken together, these examples demonstrate that antimicrobial peptide expression by animal hosts perform a conserved function of population or community structuring across multiple stages of symbioses.
Galaxin structure and function
Our in silico analysis of galaxin sequences in available databases identifies proteins with sequence similarity to EsGal1 in all major groups of the animal kingdom, except the arthropods and vertebrates (Table S2). Selection and alignment of a subset of widely divergent representatives showed that certain conserved residues are maintained through evolution (Fig. 1B). The conserved residues may serve as a structural scaffold for the protein with the remaining amino acids diverging to confer alternate functions within an animal or in different species. The wide range of sizes and isoelectric points of galaxin proteins supports the hypothesis of other functions for the various members of the protein family.
The mode of action of EsGal1R3 was not determined in the present study, but the results provide some clues about its molecular behavior. Antimicrobial peptides often compromise the integrity of bacterial membranes. The finding that EsGal1R3 was more inhibitory to Gram-positive species suggests that it targets the inner bacterial membrane or that it has a mode of action other than attack of the bacterial membrane (Epand and Epand, 2011). EsGal1 treatment reduced the size of V. fischeri cells, but it did not induce filamentation, suggesting that the peptide does not directly affect septation, as antimicrobial peptides do in some bacteria (Login et al., 2011).
Few functional studies of galaxins are available. Although BLAST analysis revealed other family members, our study is only the second outside of the Cnidaria to report a sequence as belonging to the galaxin family of proteins, the first being the identification of a partial sequence in the vent tubeworm Rifita pachyptila (Sanchez et al., 2007). Three in-depth studies of galaxins in corals (Phylum Cnidaria) have localized the proteins to the calcium carbonate exoskeleton, and larvae of the coral Acropora millepora express galaxins and galaxin-like transcripts in portions of the larva that secrete the exoskeleton during settlement (Reyes-Bermudez et al., 2009). The close association of the galaxins to the coral exoskeleton, their high abundance, as well as the timing of their production, has led researchers to link the proteins with biomineralization of the exoskeleton. However, galaxins were not found to bind calcium, and direct involvement of galaxins with production of the coral exoskeleton has not been demonstrated. Interestingly, galaxin proteins were not found in the exoskeleton of sun corals (Tubastrea spp.), which coincidentally do not host the photosynthetic partner of most corals, the zooxanthellae (Watanabe et al., 2003). These data do not preclude the involvement of galaxins in exoskeletal formation in some corals, but do suggest alternative functions. The pattern of expression in coral larvae mirrors the symbiont-dependent developmental upregulation and pan-epithelial localization we observed in the juvenile squid, and suggests that coral galaxins may be involved in the first contact between host and microbe, as corals associate with both algal and bacterial symbionts (Krediet et al., 2013). Although the peptide derived from Acropora palmata was not antimicrobial under the conditions of our experiments, the protein in its native conformation, or with the addition or subtraction of additional amino acids on either end, may be active or may serve another function in this species. Alternatively, these data suggest that the ability to modulate bacterial growth through potent antimicrobial activity is not conserved among all galaxin proteins; rather it is a character restricted to particular animals or galaxin types. The various roles of galaxins in other animal species present a fruitful area of future research.
Finally, galaxins may also function as part of a general response to either biotic (e.g., symbiosis) or abiotic (e.g., temperature, heavy metals) stress. In a wide array of symbioses, including the squid-vibrio system, plant-legume associations, and coral-zooxanthellae partnership, various stress responses (e.g., oxidative, nitrosylative) are a part of the normal activity of the symbiotic state. In support of this possibility, transcription is downregulated by copper exposure in coral Montastrea franksi, and galaxin SNP variation is correlated with water temperature in A. millepora (Schwarz et al., 2012; Lundgren et al., 2013). Since corals also undergo profound biochemical and physiological changes due to their symbionts (Davy et al., 2012), it is possible that galaxin transcription may instead be due to a stressor’s effect on algal endosymbionts.
In conclusion, our study proposes a new function for galaxin proteins as antimicrobial agents used by the squid host to select and maintain a specific symbiont, and identifies a new taxonomic group in which galaxin proteins occur in abundance. Future studies will determine whether the biochemical role of galaxins in the squid-vibrio system is a widespread attribute of these proteins across animal taxa. In the greater context of host-microbe interactions, our data support the growing paradigm that antimicrobial proteins do not serve to simply exclude bacteria from sites on an animal’s body. Rather, they are selective forces imposed by hosts on microbial communities to favor the acquisition and maintenance of coevolved symbiotic partnerships.
Experimental procedures
General methods
Adult Euprymna scolopes were collected and maintained as previously described (Wollenberg and Ruby, 2012). Juveniles from the breeding colony were collected within 15 min of hatching. Aposymbiotic (Apo) animals were maintained in V. fischeri-free, unfiltered seawater, while other juveniles were exposed to ~5,000 colony-forming units (CFU) /mL of the V. fischeri strain ES114 (Boettcher and Ruby, 1990) overnight to produce the symbiotic (Sym) condition. Colonization of the symbiotic juvenile squid was determined by measuring luminescence output of the symbionts with a TD 20/20 luminometer (Turner Designs, Sunnyvale, CA); aposymbiotic squid were also analyzed to ensure that their light organs had not been colonized. In experiments with V. fischeri surface molecules, the lipid A and the peptidoglycan monomer (also called tracheal cytotoxin, or TCT) were prepared as previously described (Foster et al., 2000; Koropatnick et al., 2004) and exposed to animals at 10 ng/mL and 10 μM, respectively for 48 h with the water + MAMPs replaced every 24h. Symbiont depletion from the light organ was performed with antibiotics as previously described (Doino and McFall-Ngai, 1995). In experiments examining the role of symbiont light production in esgal1 upregulation, animals were colonized with the light-deficient V. fischeri strain EVS102 (Δlux) (Bose et al., 2008) as described above and a subset of the exposed squid were plated to ensure colonization. All reagents were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. All animal experiments conform to the relevant regulatory standards established by the University of Wisconsin – Madison.
Identification of galaxin gene sequences from the EST database and subsequent analyses
Two galaxin gene sequences were identified by a tblastn search against the expressed sequence tag (EST) database of the juvenile-host light organ (Chun et al., 2006) using Galaxea fasicularis galaxin as the query sequence (Fukuda et al., 2003). The identified sequences in the EST database sequence were used for primer design for subsequent rapid amplification of cDNA ends (RACE) to obtain the full sequence of the open-reading frame (for details, see Supplementary Methods).
Sequences obtained by RACE were assembled into contigs using the CAP3 sequence assembly program (http://pbil.univ-lyon1.fr/cap3.php). The resulting full-length sequence was analyzed by BLAST searches of GenBank using the default parameters. The cDNA sequence was translated using the ExPASy Translate tool (http://web.expasy.org/translate/), and the resulting derived amino acid sequence was examined for a putative repeat structure using RADAR (Rapid Automatic Detection and Alignment of Repeats), http://www.ebi.ac.uk/Tools/pfa/radar/). Signal peptide prediction was performed using the SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/), and a potential N-glycosylation site was identified using the NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/). Potential for antimicrobial activity was assayed in silico with the APD2 antimicrobial peptide predictor (http://aps.unmc.edu/AP/prediction/prediction_main.php). The Galaxea fasicularis, Acropora palmata, and Balanoglossus clavigerus sequences used in this study were derived from the NCBI protein database. Amino acids from the first three putative repeats of the proteins (shown in Fig. S1) were aligned using MUSCLE (Edgar, 2004) and the subsequent alignment was visualized with the CLC Sequence viewer (CLC Bio, Cambridge, MA, http://clcbio.com/index.php?id=28).
RNA and cDNA preparation and quantitative-reverse-transcriptase PCR (qRT-PCR)
Whole juvenile animals were stored in RNALater RNA stabilization reagent (Qiagen, Valencia, CA) for 24h at 4°C and then transferred to -80°C for extended storage until RNA extraction. Extracted RNA was prepared by standard methods (for details, see Supplementary Methods).
All qRT-PCR assays were performed in compliance with the MIQE guidelines (Bustin et al., 2009). Gene-specific primers were designed for esgal1 and 2, and for the Euprymna scolopes 40S ribosomal RNA sequence, which was used as a control for equal well loading (Table S1) (for details, see Supplementary Methods).
Western blotting and immunocytochemistry (ICC)
A polyclonal antibody to EsGal1 was produced in rabbit (Genscript, Piscataway, NJ) to two unique peptides within the EsGal1 sequence (GNRTYDPQFQIC and CKYRAYDTDNFR), chosen for their predicted antigenicity, surface exposure, and lack of similarity to other known, predicted E. scolopes or V. fischeri proteins. Protein samples for western blotting were prepared as described previously (Troll et al., 2010) (for details, see Supplementary methods).
For ICC, light organs were fixed, permeabilized, and blocked as described previously (Troll et al., 2009). A 1:500 dilution of the EsGal1 antibody was used in these experiments (for details, see Supplementary Methods). The light organs were then counterstained with rhodamine phalloidin as previously described (Troll et al., 2009), mounted on glass slides, and examined on a Zeiss LSM 510 confocal microscope. For mucus secretion assays, samples were prepared as previously described (Kremer et al., 2013). Host mucus was stained with Alexa-633 conjugated wheat germ agglutinin (Life Technologies, Carlsbad, CA) (for details, see Supplementary Experimental Procedures). Fluorescence intensities were quantified using the Zeiss LSM 510 software.
Minimum Inhibitory Concentration (MIC) assays
To determine the minimal (growth) inhibitory concentration for the galaxin repeat-derived peptides, we performed micro-dilution susceptibility assays (Fedders and Leippe, 2008). Briefly, a two-fold serial dilution of each peptide was carried out in 96-well plates in 10 mM sodium phosphate buffer, with the addition of 345 mM NaCl for marine strains, pH 8.0. One hundred CFU of a log-phase bacterial culture were added to each peptide dilution and incubated overnight at 28°C for marine bacteria and 37°C for non-marine strains. After incubation, the MIC was defined as the peptide dilution where no visible bacterial growth was detected after incubation. The values are expressed as the median of at least two experiments, each performed in duplicate, with a divergence of not more than one dilution step. The following bacterial strains were used: Bacillus subtilis 1009, Bacillus megaterium 1006, Escherichia coli MG1665 K12 (Blattner et al., 1997), CNJ 771 (Exiguaobacterium aestuarii-like) (Gontang et al., 2007), CNJ 778 (Bacillus megaterium-like) (Gontang et al., 2007), CNJ 803 (Bacillus algicola-like) (Gontang et al., 2007), Vibrio fischeri ES114 (Boettcher and Ruby, 1990), Photobacterium leiognathi Ln1a (Dunlap 1985), and Vibrio parahemolyticus KNH1 (Nyholm et al., 2000). Marine bacteria were grown in SWT medium (Boettcher and Ruby, 1990), whereas B. subtilis, B. megaterium, and E. coli were grown in LB medium with (wt/vol) 1% tryptone, 0.5% yeast extract, and 1% NaCl.
Bacterial growth assays
Overnight cultures of V. fischeri strain ES114 (Boettcher and Ruby, 1990) were diluted 1:500 and then grown to OD600 0.2 at room temperature with shaking in LBS (LB with sodium chloride) medium with (wt/vol) 1% tryptone, 0.5% yeast extract, and 2% NaCl, and 50 mM Tris-HCl, pH 7.5. The cultures were then diluted with LBS 1:9 to reach an OD600 of about 0.02. Ten μL of the bacterial culture were then added to 190 μL of 10% LBS medium in a 96-well microtiter plate containing 17 μM EsGal1R3 or ApGalR2 (Genscript, Piscataway, NJ, sequences can be found in table 1), or no addition as a control. The plate was then placed in a Tecan Genios Pro plate reader (Tecan Group, Männedorf, Switzerland) and incubated at 27°C with shaking for 18 h, with OD600 readings taken every 15 min. Sample wells were also run without added bacteria to ensure that no contamination occurred. Bacterial doubling times were calculated using the equation G= [log(N-n)/log(2)]*(T-t) where N is the OD600 at time T (final) and n is the OD600 at time t (initial).
Measurement of bacterial length
Cultures of RFP-expressing V. fischeri (Strain ES114 containing the plasmid pVSV208 (Dunn et al., 2006)) were grown as for the bacterial growth assays for 18 h, and were then visualized using a Zeiss AxioImager.M2 epifluorescence microscope. Bacterial length was then measured using Zeiss software.
In vivo antibody adsorption and peptide supplementation
Newly hatched juvenile squid were placed in seawater containing 5000 CFU/mL V. fischeri strain ES114 cells/mL filter-sterilized Instant Ocean (FSIO) for 3 h, and were then washed three times in FSIO to remove any non-attached bacteria. The juvenile squid were then either placed into FSIO containing a 1:500 dilution of the α-EsGal1 antibody or FSIO containing an equal concentration of purified rabbit IgG (Genscript, Piscataway, NJ) and then animal luminescence was measured and light organ bacterial density was determined by dilution plating of light organ homogenates on LBS agar at 6 and 8 h post-bacterial exposure. For experiments in which EsGal1R3 was added during colonization, the above conditions were used in the absence of the antibody with the addition of 34.4 μM EsGal1R3. This concentration was chosen based on the activity of the peptide in in vitro analyses.
Statistics
All qRT-PCR data were log transformed to provide a normally distributed data set and then analyzed in R (version 2.12.1; R Foundation for Statistical Computing, Vienna, Austria [http://R=project.org]) by one-way ANOVA followed by a Tukey’s pairwise comparison. Shapiro-Wilk and Levene tests were used to ensure the normal distribution and homoscedasticity of the residuals, respectively.
Supplementary Material
Acknowledgments
The authors would like to thank S. Mazzone, B. Rader, J. Schwartzman, and E. G. Ruby for assistance with data acquisition, figure development, and experimental design. This work was supported by grants from National Institutes of Health (NIH) R01-RR12294 (to EGR) and R01-AI50661 (to MM-N), and National Science Foundation IOS 0817232 (to MM-N & EGR). EACH-H was supported by NRSA T-32 GM07215.
Footnotes
Author contributions: EACH, AAG, and MXG performed the experiments, and EACH performed the data analysis. EACH, AAG, RA, and MJM designed the experiments. WEG contributed reagents. EACH and MJM prepared the manuscript, with help from, and editing by, all co-authors.
Nucleotide accession numbers
EsGal1 has been submitted to Genbank with the submission ID 1681541.
Works Cited
- Aeckersberg F, Lupp C, Feliciano B, Ruby EG. Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J Bacteriol. 2001;183:6590–6597. doi: 10.1128/JB.183.22.6590-6597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altura MA, Heath-Heckman EA, Gillette A, Kremer N, Krachler AM, Brennan C, et al. The first engagement of partners in the Euprymna scolopes-Vibrio fischeri symbiosis is a two-step process initiated by a few environmental symbiont cells. Environ Microbiol. 2013;15:2937–2950. doi: 10.1111/1462-2920.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Boettcher KJ, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Rosenberg CS, Stabb EV. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol. 2008;190:169–183. doi: 10.1007/s00203-008-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol. 2010;8:218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- Bulgheresi S, Gruber-Vodicka HR, Heindl NR, Dirks U, Kostadinova M, Breiteneder H, et al. Sequence variability of the pattern recognition receptor Mermaid mediates specificity of marine nematode symbioses. ISME J. 2011;5:986–998. doi: 10.1038/ismej.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun CK, Scheetz TE, Bonaldo Mde F, Brown B, Clemens A, Crookes-Goodson WJ, et al. An annotated cDNA library of juvenile Euprymna scolopes with and without colonization by the symbiont Vibrio fischeri. BMC Genomics. 2006;7:154. doi: 10.1186/1471-2164-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun CK, Troll JV, Koroleva I, Brown B, Manzella L, Snir E, et al. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc Natl Acad Sci U S A. 2008;105:11323–11328. doi: 10.1073/pnas.0802369105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AJ, LaBarre BA, Won BS, Shah MV, Heng S, Choudhury MH, et al. Diversity and partitioning of bacterial populations within the accessory nidamental gland of the squid Euprymna scolopes. Appl Environ Microbiol. 2012;78:4200–4208. doi: 10.1128/AEM.07437-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol. 2004;6:1139–1151. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- Davy SK, Allemand D, Weis VM. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev. 2012;76:229–261. doi: 10.1128/MMBR.05014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira LS, Gregoracci GB, Silva GG, Salgado LT, Filho GA, Alves-Ferreira M, et al. Transcriptomic analysis of the red seaweed Laurencia dendroidea (Florideophyceae, Rhodophyta) and its microbiome. BMC Genomics. 2012;13:487. doi: 10.1186/1471-2164-13-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doino JA, McFall-Ngai MJ. A transient exposure to symbiosis-competent bacteria induces light organ morphogenesis in the host squid. Biol Bull. 1995;189:347–355. doi: 10.2307/1542152. [DOI] [PubMed] [Google Scholar]
- Dunlap PV. Osmotic control of luminescence and growth in Photobacterium leiognathi from ponyfish light organs. Arch Microbiol. 1985;141:44–50. doi: 10.1007/BF00446738. [DOI] [PubMed] [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol. 2006;72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand RM, Epand RF. Bacterial membrane lipids in the action of antimicrobial agents. J Pept Sci. 2011;17:298–305. doi: 10.1002/psc.1319. [DOI] [PubMed] [Google Scholar]
- Fedders H, Leippe M. A reverse search for antimicrobial peptides in Ciona intestinalis: identification of a gene family expressed in hemocytes and evaluation of activity. Dev Comp Immunol. 2008;32:286–298. doi: 10.1016/j.dci.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Foster JS, Apicella MA, McFall-Ngai MJ. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev Biol. 2000;226:242–254. doi: 10.1006/dbio.2000.9868. [DOI] [PubMed] [Google Scholar]
- Franzenburg S, Walter J, Kunzel S, Wang J, Baines JF, Bosch TC, et al. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Procs Natl Acad Sci U S A. 2013:E3730–E3738. doi: 10.1073/pnas.1304960110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune S, Augustin R, Anton-Erxleben F, Wittlieb J, Gelhaus C, Klimovich VB, et al. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci U S A. 2010;107:18067–18072. doi: 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA. Microbial community structure and its functional implications. Nature. 2009;459:193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- Fukuda I, Ooki S, Fujita T, Murayama E, Nagasawa H, Isa Y, et al. Molecular cloning of a cDNA encoding a soluble protein in the coral exoskeleton. Biochem Biophys Res Commun. 2003;304:11–17. doi: 10.1016/s0006-291x(03)00527-8. [DOI] [PubMed] [Google Scholar]
- Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontang EA, Fenical W, Jensen PR. Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl Environ Microbiol. 2007;73:3272–3282. doi: 10.1128/AEM.02811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J, Ruby EG. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci U S A. 1998;95:1818–1822. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath-Heckman EAC, Peyer SM, Whistler CA, Apicella MA, Goldman WE, McFall-Ngai MJ. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-Vibrio symbiosis. MBio. 2013;4 doi: 10.1128/mBio.00167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- Krediet CJ, Ritchie KB, Paul VJ, Teplitski M. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc Biol Sci. 2013;280:20122328. doi: 10.1098/rspb.2012.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer N, Philipp EE, Carpentier MC, Brennan CA, Kraemer L, Altura MA, et al. Initial Symbiont Contact Orchestrates Host-Organ-wide Transcriptional Changes that Prime Tissue Colonization. Cell Host Microbe. 2013;14:183–194. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarcq LH, McFall-Ngai MJ. Induction of a gradual, reversible morphogenesis of its host’s epithelial brush border by Vibrio fischeri. Infect Immun. 1998;66:777–785. doi: 10.1128/iai.66.2.777-785.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Login FH, Balmand S, Vallier A, Vincent-Monegat C, Vigneron A, Weiss-Gayet M, et al. Antimicrobial peptides keep insect endosymbionts under control. Science. 2011;334:362–365. doi: 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- Lundgren P, Vera JC, Peplow L, Manel S, van Oppen MJ. Genotype - environment correlations in corals from the Great Barrier Reef. BMC Genet. 2013;14:9. doi: 10.1186/1471-2156-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Nyholm SV, Castillo MG. The role of the immune system in the initiation and persistence of the Euprymna scolopes--Vibrio fischeri symbiosis. Semin Immunol. 2010;22:48–53. doi: 10.1016/j.smim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, Harvie EA. The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol. 2012;24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham DM, Chow CE, Cram JA, Sachdeva R, Parada A, Fuhrman JA. Short-term observations of marine bacterial and viral communities: patterns, connections and resilience. ISME J. 2013;7:1274–1285. doi: 10.1038/ismej.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull. 1998;195:89–97. doi: 10.2307/1542815. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol. 2002;68:5113–5122. doi: 10.1128/AEM.68.10.5113-5122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl Environ Microbiol. 2003;69:3932–3937. doi: 10.1128/AEM.69.7.3932-3937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond JW, Batley M, Djordjevic MA, Innes RW, Kuempel PL, Rolfe BG. Flavones induce expression of nodulation genes in rhizobium. Nature. 1986;323:632–635. [Google Scholar]
- Reyes-Bermudez A, Lin Z, Hayward DC, Miller DJ, Ball EE. Differential expression of three galaxin-related genes during settlement and metamorphosis in the scleractinian coral Acropora millepora. BMC Evol Biol. 2009;9:178. doi: 10.1186/1471-2148-9-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, Asato LM. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S, Hourdez S, Lallier FH. Identification of proteins involved in the functioning of Riftia pachyptila symbiosis by Subtractive Suppression Hybridization. BMC Genomics. 2007;8:337. doi: 10.1186/1471-2164-8-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JA, Mitchelmore CL, Jones R, O’Dea A, Seymour S. Exposure to copper induces oxidative and stress responses and DNA damage in the coral Montastraea franksi. Comp Biochem Physiol C Toxicol Pharmacol. 2012;157:272–279. doi: 10.1016/j.cbpc.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Shnit-Orland M, Sivan A, Kushmaro A. Antibacterial activity of Pseudoalteromonas in the coral holobiont. Microb Ecol. 2012;64:851–859. doi: 10.1007/s00248-012-0086-y. [DOI] [PubMed] [Google Scholar]
- Stabb E, Visick K. Vibrio fisheri: Squid Symbiosis. In: Rosenberg E, DeLong E, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Springer Berlin Heidelberg; 2013. pp. 497–532. [Google Scholar]
- Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, Goldman WE, et al. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell Microbiol. 2009;11:1114–1127. doi: 10.1111/j.1462-5822.2009.01315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troll JV, Bent EH, Pacquette N, Wier AM, Goldman WE, Silverman N, et al. Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol. 2010;12:2190–2203. doi: 10.1111/j.1462-2920.2009.02121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KG, Ruby EG. Construction and symbiotic competence of a luxA-deletion mutant of Vibrio fischeri. Gene. 1996;175:89–94. doi: 10.1016/0378-1119(96)00129-1. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, Ruby EG. Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-Vibrio symbiosis. Mol Microbiol. 2010;78:903–915. doi: 10.1111/j.1365-2958.2010.07376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Fukuda I, China K, Isa Y. Molecular analyses of protein components of the organic matrix in the exoskeleton of two scleractinian coral species. Comp Biochem Physiol B Biochem Mol Biol. 2003;136:767–774. doi: 10.1016/s1096-4959(03)00177-5. [DOI] [PubMed] [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiasse RP, Schaefer AL, Koroleva I, et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci U S A. 2010;107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg MS, Ruby EG. Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME J. 2012;6:352–362. doi: 10.1038/ismej.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


