SUMMARY
Proinflammatory stimuli elicit rapid transcriptional responses via transduced signals to master regulatory transcription factors. To explore the role of chromatin-dependent signal transduction in the atherogenic inflammatory response, we characterized the dynamics, structure and function of regulatory elements in the activated endothelial cell epigenome. Stimulation with tumor necrosis factor alpha prompted a dramatic and rapid global redistribution of chromatin activators to massive de novo clustered enhancer domains. Inflammatory super enhancers formed by NF-κB accumulate at the expense of immediately decommissioned, basal endothelial super enhancers, despite persistent histone hyperacetylation. Mass action of enhancer factor redistribution causes momentous swings in transcriptional initiation and elongation. A chemical genetic approach reveals a requirement for BET bromodomains in communicating enhancer remodeling to RNA polymerase and orchestrating the transition to the inflammatory cell state, demonstrated in activated endothelium and macrophages. BET bromodomain inhibition abrogates super enhancer mediated inflammatory transcription, atherogenic endothelial responses and atherosclerosis in vivo.
INTRODUCTION
Precise control of inflammation is essential for host defense. Defense against pyogenic infection requires rapid activation of tissue and circulating leukocytes and their recruitment by activated endothelium. However, inflammation is also integral to the pathophysiology of many common and life-threatening illnesses. Acute, high-grade inflammation accompanying sepsis features systemic inflammatory cell activation and contributes to multi-system organ failure and death (Medzhitov et al., 2012). Chronic, low-grade inflammation is a pathogenic feature of autoimmune disorders as well as highly prevalent and morbid conditions such as diabetes mellitus and atherosclerosis (Libby et al., 2011). As such, there is a pressing need to dissect inflammatory signaling for the elucidation of pathologic mechanisms of disease and the identification of targeted therapeutic interventions.
In inflammation a primary mode of bidirectional cellular communication involves one set of cells releasing cytokines to activate surface receptors on effector cells. Transduced signals converge on activation and translocation of inflammatory transcription factors (Barnes and Karin, 1997). A central pathway common to the interaction between activated leukocytes and endothelium is tumor necrosis factor alpha (TNFα)-mediated signal transduction to nuclear factor-kappa B (NF-κB) – a family of master regulatory, proinflammatory transcription factors canonically defined by the p50/p65 heterodimer (Baltimore, 2011). Following entry into the nucleus, NF-κB binds to DNA cis-regulatory elements at enhancers and promoters, prompting proinflammatory transcription (Pierce et al., 1988).
Genome-bound nuclear NF-κB interacts with transcriptional co-activators to stimulate transcription at multiple steps including the remodeling of chromatin as well as the initiation and elongation of RNA Polymerase II (RNA Pol II) (Barboric et al., 2001; Kaikkonen et al., 2013; Natoli, 2009). NF-κB recruits and interacts with defined chromatin regulators including histone acetyltransferases (P300), histone deacetylases (HDAC1), and epigenetic reader proteins, such as BRD4 (Ashburner et al., 2001; Huang et al., 2009; Zhong et al., 2002). Through these interactions NF-κB is able to engage in crosstalk with chromatin remodeling machinery.
BRD4 is a member of the bromodomain and extra-terminal domain (BET) family of transcriptional co-activators and elongation factors (BRD2, BRD3, BRD4 and BRDT) (Dey et al., 2000; LeRoy et al., 2008). At active genes, BET bromodomains recruit the positive transcription elongation factor complex (P-TEFb) (Jang et al., 2005; Yang et al., 2005), and chromatin remodeling factors including the SWI/SNF complex (Shi et al., 2013) via molecular recognition of poly-acetylated histone tails (Mujtaba et al., 2007). Mechanistically, TNFα or lipopolysaccharide (LPS) stimulation promotes direct acetylation of the p65 subunit (Lys310) of NF-κB by P300 (Chen et al., 2001), promoting a direct interaction with BRD4 through twin acetyl lysine recognizing bromodomains (Huang et al., 2009). This interaction is needed for productive NF-κB transactivation (Huang et al., 2009), suggesting a central role for BRD4 in inflammatory transcriptional signaling.
Prior research from our group and others has identified that BET bromodomains localize genome-wide to promoter and enhancer regions (Anand et al., 2013; Chapuy et al., 2013; Loven et al., 2013; Nicodeme et al., 2010). The majority of enhancer-bound BRD4 is found within a small number of massive enhancer regions termed super enhancers (SE). Like locus control regions or stretch enhancers, SEs concentrate chromatin-bound co-activators to genes essential for specialized cellular function (i.e. immunoglobulin production in plasma cells), and lineage specification (i.e. germinal center differentiation) (Chapuy et al., 2013; Loven et al., 2013; Parker et al., 2013; Whyte et al., 2013). Disruption of SE function by a first acetylated lysine-competitive small molecule BET bromodomain inhibitor from our group known as JQ1 (Filippakopoulos et al., 2010) suggests the mutability of these large chromatin structural elements (Chapuy et al., 2013). However, the role of SEs in the control of dynamic cell state transitions remains unknown. Recently, BET bromodomain inhibition has been shown to abrogate global, maladaptive transcriptional programs during sepsis and heart failure, implicating BRD4 in stress-induced cell state transitions (Anand et al., 2013; Nicodeme et al., 2010). These data provided a rationale for investigating the collaborative roles of NF-κB and BRD4 in regulating SEs during proinflammatory activation.
The endothelium is critical to the initiation and propagation of inflammation. Endothelial cells (ECs) prompt leukocyte recruitment, adhesion, and trafficking into tissues, thus mediating responses essential for many inflammatory disorders, including atherogenesis, in which activation of ECs is pathogenic (Gimbrone et al., 1990; Ley et al., 2007). Despite these important roles in disease, global studies of chromatin structure and function in vascular endothelium have to date not been undertaken. In this study, we investigate the role of BRD4 in determining the inflammatory activation of ECs through NF-κB and SE formation. Here, we provide evidence for the first time that EC activation by the archetypal proinflammatory stimulus TNFα rapidly deploys NF-κB to enhancers and promoters genome-wide, where it recruits BRD4. Through the recruitment of BRD4, NF-κB establishes new SEs coincident with the surprising, rapid redistribution of BRD4 away from endothelial resting state SEs. Newly established NF-κB SEs are proximal to and drive canonical genes of the inflammatory response in ECs, including key effectors of chemotaxis, adhesion, migration and thrombosis. BRD4 depletion from chromatin through small molecule BET bromodomain inhibition impedes NF-κB directed SE reorganization. The failure to form proinflammatory SEs preferentially suppresses SE dependent proinflammatory gene transcription translating into functional suppression of key TNFα-induced endothelial responses of leukocyte rolling, adhesion, and transmigration. In vivo, we find that BET bromodomain inhibition suppresses atherogenesis – a pathogenic process predicated on inflammatory endothelial activation. Together these data establish BET bromodomain-containing proteins as key effectors of the integrated mammalian inflammatory response through their rapid, dynamic, global reorganization of SEs during NF-κB activation and suggest SE targeting during inflammatory cell state transitions as a novel therapeutic approach.
RESULTS
p65 and BRD4 Establish Super Enhancers During Proinflammatory Stimulation
To explore the role of NF-κB, BRD4, and SEs in the acute inflammatory activation of ECs, we activated NF-κB in primary human umbilical vein ECs with TNFα, a canonical proinflammatory stimulus, for one hour. As expected, TNFα resulted in NF-κB nuclear translocation (Figure 1A, B), a rapid change in EC state characterized by increased monocyte adhesion (Figure 1C) and induction of adhesion molecule gene expression including E-selectin (SELE) and vascular cell adhesion molecule (VCAM1) (Figure 1D) (Ley et al., 2007). The recognized capacity of BRD4 to bind acetylated NF-κB (Huang et al., 2009), suggests a co-activator role for the BET family in the robust p65-mediated EC inflammatory response observed. We therefore used chromatin immunoprecipitation coupled with high-throughput genome sequencing (ChIP-Seq) to define p65 and BRD4 genomic occupancy in ECs before and following acute proinflammatory activation.
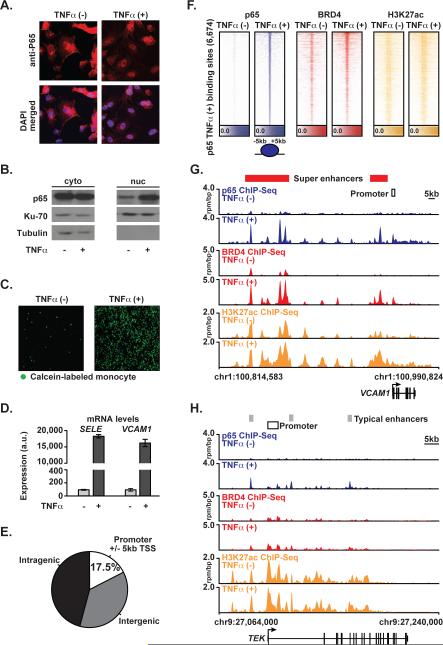
Figure 1. p65 and BRD4 Genome Binding During Proinflammatory Activation in ECs.
(A) Images of ECs ± TNFα stained for p65 (red) or DAPI (blue) (25 ng/mL, 1 hr) cells. (B) Western blot for p65, Ku-70, and Tubulin in cytosolic (left) and nuclear (right) protein fraction lysates in ECs±TNFα. (C) Images showing adhesion of calcein-labeled THP-1 monocytes to ECs ± TNFα (25 ng/mL, 3 hrs). (D) Bar plots showing cell count normalized expression levels of SELE and VCAM1 in ECs±TNFα (25 ng/mL, 3 hrs). Error bars are standard error of the mean (SEM). (E) Pie chart of p65 binding site distribution in EC genome in TNFα(+). (F) Heatmap of p65 (blue), BRD4 (red) and H3K27ac (yellow) levels in resting ECs and after TNFα (25 ng/mL, 1 hr). Each row shows ± 5kb centered on p65 peak. Rows are ordered by max p65 in each region. ChIP-Seq signal (rpm/bp) is depicted by color scaled intensities. (G,H) Gene tracks of ChIP-Seq signal for p65, BRD4, and H3K27ac at the VCAM1 and TEK gene loci in untreated (top) or TNFα(+) (bottom) ECs. Y-axis shows ChIP-Seq signal (rpm/bp). The x-axis depicts genomic position with TNFα gained typical enhancers (TE, gray) and SEs (SE, red) and promoter regions (white) marked. See also Figure S1.
In TNFα-stimulated ECs, p65 enrichment was evident at promoters (17.5%), intragenic (45.8%) and intergenic regulatory sequences (36.7%) (Figure 1E,F). Striking co-localization of BRD4 and p65 was observed by global enrichment alignment and binding site motif analysis (Figure 1F, S1A). TNFα treatment prompts dynamic co-localization of p65 and BRD4 to enhancer and promoter regions marked by H3K27ac, which are significantly enriched for p65 consensus sequences (Figure S1A) (Matys et al., 2006). At the exemplary VCAM1 locus, TNFα stimulation of resting ECs for one hour increased p65 occupancy at both promoters and upstream enhancer elements marked by acetylated chromatin (H3K27ac) (Figure 1G). Coincident with these events, we identified recruitment of exceptionally high levels of BRD4 at discrete hyperacetylated enhancer elements (Figure 1F, G, Figure S1B), consistent with the formation of de novo SEs (SEs). Focal BRD4 co-localization with p65 was observed at each discrete peak, with complete concordance. Comparable evidence is provided at the SELE locus, where TNFα stimulation recruits p65 and high levels of BRD4 to a gene regulatory region completely devoid of p65 and BRD4, augmenting regional hyperacetylation (Figure S1C). The dramatic remodeling of these loci in one hour in TNFα-stimulated ECs corroborates the robust transcriptional activation of these canonical EC inflammatory gene products (Figure 1D). Notably, typical enhancers are found at most other EC genes as exemplified by endothelial tyrosine kinase (TEK) and serpin peptidase inhibitor clade H1 (SEPINH1), where the levels of p65 and BRD4 are an order of magnitude lower compared to the VCAM1 or SELE SE; and TNFα-does not induce mRNA expression (Figure 1H, Figure S1D, E).
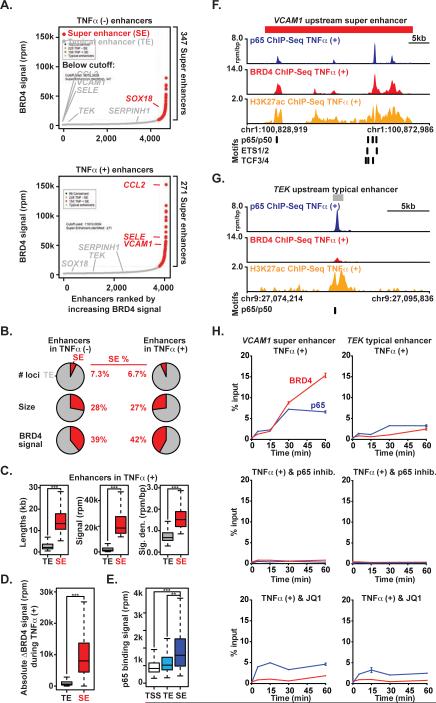
To assess the genome-wide distribution of SEs during the EC inflammatory cell state transition, we characterized and compared the enhancer landscape in resting and TNFα-activated ECs using BRD4 ChIP-Seq datasets. When ranked by increasing BRD4 enrichment, 347 and 271 SEs were identified in resting and TNFα-activated ECs, respectively. These SEs comprised ~ 7% of the total number of discrete EC enhancer loci (Figure 2A, Figure S2A), but represented more than a quarter of the total amount of enhancer size and more than a third of enhancer-bound BRD4 (Figure 2A, B, Figure S2A). Compared to typical enhancers, SE loci are significantly larger in DNA length, total BRD4 signal and signal density and share less overlap between resting and TNFα-activated ECs (Figure 2C, Figure S2A,B). Following TNFα stimulation, the absolute change in BRD4 total signal and density at SEs was greater compared to typical enhancers (Figure 2D, Figure S2C). We observed higher p65 total binding signal and density at SE loci when compared to either typical enhancer regions or active gene transcriptional start sites (TSS) (Figure 2E, Figure S2D). As exemplified by the VCAM1 SE locus and also observed globally, ECs feature dense clustering of multiple regulatory transcription factor binding sites known to be involved in EC proinflammatory responses including p65, p50, ETS1/2, and transcription factor 3/4 (TCF3/4) (Figure 2F, Figure S2E) (De Val et al., 2008; Masckauchan et al., 2005). In contrast, typical enhancer sites typified by the TEK locus possess a much lower density of these motifs (Figure 2G, Figure S2E).
Figure 2. p65 and BRD4 Establish Super Enhancers During Proinflammatory Stimulation.
(A) Ranked plots of enhancers defined in resting (top) or TNFα(+) (bottom) ECs ranked by increasing BRD4 signal (units rpm). Enhancers are defined as regions of BRD4 ChIP-Seq binding not contained in promoters. The cutoff discriminating TEs from SEs is shown as a dashed line. Genes associated with enhancers that are considered typical or super are colored gray and red respectively. (B) Pie charts displaying characteristics of TE and SE regions including number of loci, size and BRD4 signal. (C) Boxplots of median enhancer length (kb), signal (rpm) and density (rpm/bp) in TNFα-gained enhancers. Significance of the difference between distributions determined using a two-tailed t test. ** p < 1e-5, *** p < 1e-10. (D) Boxplot of absolute change in BRD4 signal in response to TNFα measured at all enhancers in TNFα(-) and TNFα(+). Significance of the difference between distributions determined using a two-tailed t test. ** p < 1e-5, *** p < 1e-10. (E) Boxplot of p65 binding signal (rpm) at all active gene promoters (TSS), TEs and SEs in TNFα treated ECs. Significance of the difference between distributions determined using a two-tailed t test. ** p < 1e-5, *** p < 1e-10. (F,G) Schematic of transcription factor motif binding sites at the VCAM1 SE (red box) (F) or TEK TE (grey box) (G) loci in ECs treated with TNFα. (H) Line plots of kinetic ChIP-PCR showing enrichment (% input normalized to time 0) of p65 and BRD4 at an NF-κB binding site in the VCAM1 (left) SE and TEK TE (right) in ECs treated with TNFα (25 ng/mL; 0, 5, 15, 30, 60 min). The effect of co-treatment with vehicle (top), BAY (NF-kB inhibitor, middle) and JQ1 (bottom) are shown. See also Figure S2.
In order to dissect the temporal relationship between p65 and BRD4 localization to enhancers, we next performed time-ranging chromatin binding studies. ChIP for p65 and BRD4 followed by real time-PCR centered on the most prominent NF-κB binding site in the 5’ VCAM1 and CCL2 SEs revealed enrichment of p65 five minutes after TNFα stimulation, with peak occupancy detected by 30 minutes (Figure 2H, Figure S2F). BRD4 recruitment followed the identical temporal pattern of recruitment at these sites. Inhibition of NF-κB phosphorylation and function by IκB kinase inhibition (BAY 11-7082, “BAY”) (Pierce et al., 1997) completely abrogated TNFα-induced p65 and BRD4 accumulation at both NF-κB sites at all time points, while also suppressing VCAM1 gene induction (Figure S2G). As expected, co-treatment of ECs with JQ1 reduced TNFα-induced enrichment of BRD4 at these NF-κB binding sites. But, in contrast to BAY, JQ1 had no effect on immediate early p65 recruitment (Figure 2H, Figure S2F). Compared to SEs, typical enhancers featured less TNFα-induced recruitment of both NF-κB and BRD4 (Figure 2H, Figure S2F). Collectively, these results establish that p65 is required for TNFα-induced recruitment of BRD4 to SE regions in ECs.
NF-κB Provokes Rapid, Global Redistribution of BRD4 at Super Enhancers
We next explored the dynamics and function of SEs in inflammation in both ECs and macrophages. One hour following TNFα stimulation, we observed the formation of pronounced, prototypical SEs at canonical inflammatory genes, such as the CCL2 chemokine (Figure 3A). There, a density of upstream and intragenic enhancer elements was identified, characterized by regional hyperacetylation (H3K27ac) and peaks of BRD4 enrichment coinciding with focal p65 binding sites (Figure 3A). Functionally, rapid SE formation was associated with recruitment of RNA Pol II (Figure 3A) and marked transcriptional activation (Figure S3A). These data provide a first demonstration that the canonical proinflammatory stimulus TNFα rapidly induces de novo formation of SEs established by p65 at proinflammatory target genes, also including VCAM1 and SELE (Figure 1G, S1C).
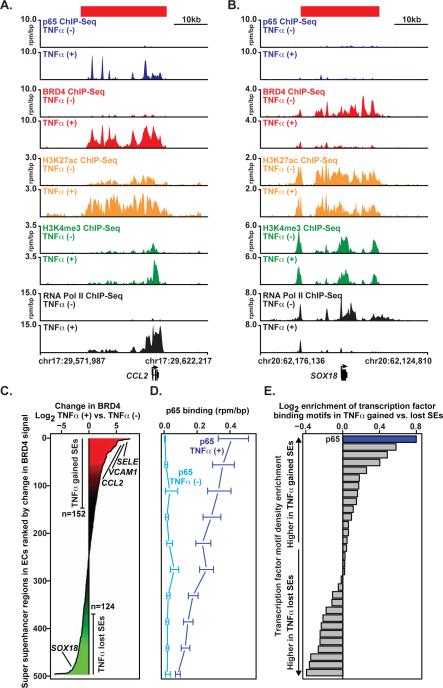
Figure 3. NF-κB Provokes Rapid, Global Redistribution of BRD4.
(A,B) Gene tracks of ChIP-Seq signal (rpm/bp) for p65, BRD4, H3K27ac, H3K4me3, and RNA Pol II at the CCL2 gene (A) or SOX18 (B) locus in TNFα(-) (top) and TNFα(+) (bottom) ECs. (C) All genomic regions containing a SE in TNFα(-) and TNFα(+) ECs are shown ranked by log2 change in BRD4 signal (treated vs. untreated). X-axis shows the log2 fold change in BRD4 signal. Change in BRD4 levels at SEs are colored by intensity of change (green to red). (D) Line plot showing the median levels of p65 binding (rpm/bp) at SEs in either TNFα (-) light blue or TNFα (+) dark blue conditions. SEs were ranked by change in BRD4 and binned (50/bin). The median p65 level was calculated in each bin. Error bars represent 95% confidence intervals of the median determined by empirical resampling. (E) Horizontal bar plot showing the ratio of transcription factor motif density between TNFα-gained and TNFα-lost SEs. Twenty-one transcription factors are displayed whose motifs occur more frequently than expected based on dinucleotide background model. The transcription factor motifs are ranked by log2 fold change in density between TNFα-gained vs. TNFα-lost SEs. See also Figure S3.
Unexpectedly, the enrichment of BRD4 at inflammatory EC SEs was associated with the rapid, reciprocal depletion of BRD4 at resting EC SEs. ECs grown in culture feature 347 canonical SEs, including SEs associated with genes critical for non-inflammatory EC function, such as the SOX18 transcription factor implicated in vasculogenesis (Matsui et al., 2006). In such resting ECs, regional enrichment for H3K27ac is observed at the SOX18 locus, accompanied by characteristic BRD4 occupancy and evident enrichment for RNA Pol II throughout the gene body. In the inflammatory EC state, TNFα fails to drive NF-κB to the SOX18 locus, RNA Pol II enrichment is markedly diminished and transcription is muted (Figure 3B, Figure S3A). Only one hour following TNFα stimulation, BRD4 is effectively depleted, despite persistent hyperacetylation (H3K27ac) of the SOX18 SE (Figure 2B).
To explore the relevance of changes in SEs provoked by proinflammatory activation across the genome, a systematic analysis of dynamic alterations in SE formation was undertaken. BRD4 enrichment was selected here as a preferred marker for SE identity owing to a concern that H3K27ac, or other more biochemically stable enhancer modifications (such as H3K4me1/2), may lag behind BRD4 redistribution globally in a dynamic cell state change, as above at the SOX18 locus. We identified a dramatic redistribution of genomic BRD4 occupancy following TNFα stimulation (Figure 3C). Differential enhancer analysis revealed an evident global balance in chromatin-associated BRD4, but importantly TNFα stimulation resulted in the reclassification of multiple enhancers from typical enhancer to SE (n=152) and from SE to typical enhancer (n=124) (Figure 3C). Gains in BRD4 occupancy were strongly and directly associated with site-specific increases in p65 binding occupancy genome-wide (Figure 3D). As expected, TNFα-gained SEs are characterized by coordinate increases in BRD4 and H3K27ac enrichment at regions of increased p65 occupancy, which correlates directly with SE formation (Figure 3C and Figure S3B). Consistent with the SOX18 regulatory region, lost SEs are defined by a significant reduction of BRD4 occupancy following TNFα-stimulation that is disproportionate to minimal changes in p65 and modest reductions in H3K27ac (Figure 3D and Figure S3C). Finally, the p65 motif was found at much higher density in TNFα-gained versus TNFα-lost SEs regions in ECs, further suggesting p65 direct binding to clustered sites in TNFα-gained SEs as a causal event in BRD4 SE redistribution during inflammatory activation (Figure 3E), akin to comparable “stretch enhancers” as described by Francis Collins and colleagues (Parker et al., 2013).
NF-κB Formed Super Enhancers Drive Proinflammatory Transcription
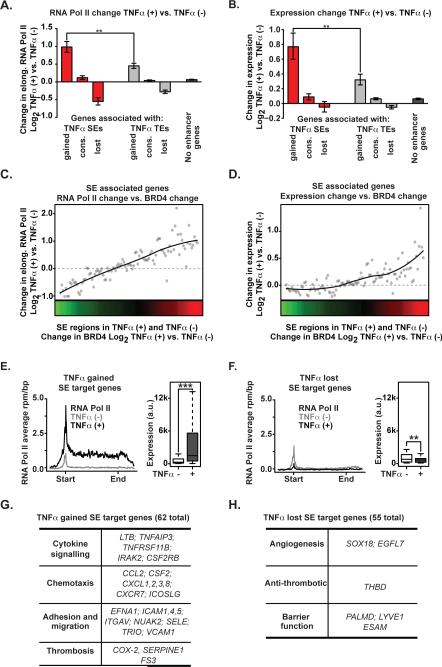
To assess the consequences of SE redistribution on transcriptional output, we integrated genome-wide ChIP-Seq data with cell count-normalized, array-based absolute gene expression profiling measurements obtained before and one hour after TNFα stimulation. Compared to typical and conserved enhancer regions, the gain or loss of SEs provoked by TNFα resulted in the largest changes to RNA Pol II occupancy and expression of adjacent genes (Figure 4A,B). This global relationship was also evident when specifically comparing the change in the levels of SE constituent BRD4 versus change in elongating RNA Pol II and gene expression (Figure 4C,D). Those genes positioned near TNFα-gained SEs, which also feature BRD4 enrichment at their promoters suggestive of promoter-enhancer communication, demonstrated marked induction of transcriptional initiation, elongation, and gene expression (Figure 4E, Figure S4A). Functional classification of TNFα-gained SE marked genes reveals known drivers of key functional facets of EC inflammatory responses: cytokine signaling, chemotaxis, adhesion and migration, and thrombosis (Figure 4G, Figure S4F). In contrast, the reciprocal loss of BRD4 at resting EC SEs resulted in a proportionate decrease in transcription and expression of nearby genes (Figure 4A, B, F), such as anti-thrombin and genes involved in angiogenesis and endothelial barrier function (Figure 4H, Figure S4G). Notably, the cohort of lost SE genes featured marked decreases in transcriptional initiation and elongation by RNA Pol II enrichment metagene analysis in advance of changes in promoter modification (H3K4me3 enrichment). (Figure S4B). Genes with conserved enhancers show minimal change in expression and serve pathways that govern homeostatic function in ECs (Figure 4A, B, Figure S4H). Globally, this chromatin restructuring results in a strong induction of proinflammatory SE driven transcription compared to typical enhancer associated genes. The 62 TNFα-gained, SE associated genes comprise only ~8% of all genes with > 2 fold increase in mRNA expression, but account for ~60% of the total increase in gene expression and ~20% of the increase in cellular mRNA within 3 hours of TNFα stimulation (Figure S4C-E). These data provide discrete examples and global evidence of dynamic, functional remodeling of enhancer factors even preceding the structural decommissioning of abandoned enhancers.
Figure 4. NF-κB Formed Super Enhancers Drive Proinflammatory Transcription.
(A,B) Bar plot of change in elongating RNA Pol II (A) or mRNA expression (B) at genes associated with TNFα-gained, TNFα-lost or TNFα-conserved SEs (red), TEs (gray) and no enhancers (black). Significance of the difference between distributions determined using a two-tailed t test. ** p < 1e-5, *** p < 1e-10. (C, D) Change in elongating RNA Pol II in the gene body region of genes (4C, y-axis) or change in mRNA levels (4D, y-axis) are plotted ranked by change in BRD4 at proximal SEs (x-axis). Dots represent median change sampled across 50 evenly distributed bins with a loess fitted line overlaid. Change in BRD4 levels at proximal SEs are colored by intensity of change (green to red). (E,F) Metagene representations of average RNA Pol II ChIP-Seq signal (grey untreated and black TNFα treated) in units of rpm/bp at a meta composite of target genes of SEs that are gained (E) or lost (F) in response to TNFα treatment. Boxplots of cell count normalized expression levels are shown to the right of each metagene in arbitrary units for genes with associated SEs (grey untreated and black TNFα treated) that are gained (E) or lost (F) in response to TNFα treatment. Significance of the difference between distributions determined using a two-tailed t test. ** p < 1e-5, *** p < 1e-10. (G, H) Table showing the functional categories of selected genes that are targets of SEs gained (G) or lost (H) in response to TNFα treatment. See also Figure S4.
NF-κB Formed Super Enhancers Drive Proinflammatory Gene Expression in a BET Bromodomain-Dependent Manner
Disruption of SEs by disrupting enhancer factors, such as BRD4, has been observed to selectively influence the expression of genes associated with SEs (Loven et al., 2013). In cancer, we have observed that competitive displacement of BET bromodomains from nuclear chromatin provokes coordinated inhibition of the MYC transcriptional program, often associated with downregulation of MYC itself (Chapuy et al., 2013; Delmore et al., 2011; Zuber et al., 2011). Using a chemical genetic approach, we assessed the role of BET bromodomains in the rapid transcriptional response of SE associated, proinflammatory genes in TNFα-stimulated ECs. Small molecule probes, such as the BET bromodomain inhibitor JQ1, are particularly appealing in the study of dynamic processes, as they offer precise temporal perturbation of the biological system (Frye, 2010; Strausberg and Schreiber, 2003). We therefore performed a dynamic, genome-wide analysis of BET bromodomain inhibition on inflammatory SE integrity, global chromatin structure, gene expression, and EC post-inflammatory function.
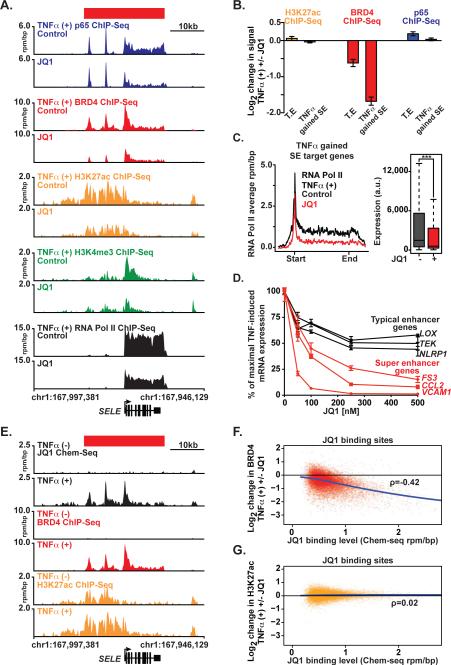
First we characterized the effect of JQ1 on EC chromatin structure and function immediately following TNFα exposure. ECs were treated with JQ1 (500 nM) to displace BET bromodomains, then stimulated with TNFα for one hour. Chromatin from treated and untreated ECs was subjected to ChIP-Seq for promoters (H3K4me3), enhancers (H3K27ac), RNA Pol II, the p65 transcription factor and the BRD4 co-activator. At the SELE locus, BET inhibition had no effect on TNFα-mediated recruitment of p65 (Figure 5A, B). However, JQ1 depleted BRD4 resulting in abrogated SELE expression assessed by decreased RNA Pol II enrichment (Figure 5A and cell surface protein levels assessed by flow cytometry (Figure S5E). Comparable observations are evident at the CCL2 and IRAK2 loci, where BET inhibition selectively displaces BRD4 leading to impaired transcription induction and elongation by RNA Pol II (Figure S5A, B).
Figure 5. NF-κB Formed Super Enhancers Drive Proinflammatory Gene Expression in a BET Bromodomain-Dependent Manner.
(A) Gene tracks of ChIP-Seq signal (rpm/bp) for p65, BRD4, H3K27ac, H3K4me3, and RNA Pol II at the SELE locus in TNFα treated ECs co-treated with vehicle (top) or JQ1 (bottom). (B) The mean log2 fold change in H3K27ac (yellow), BRD4 (red) and p65 (blue) ChIP-Seq signal in TNFα treated cells ±JQ1 at either TEs or SEs gained in response to TNFα treatment. Error bars represent 95% confidence intervals of the mean determined by empirical resampling. (C) Metagene representations of average RNA Pol II ChIP-Seq signal (black TNFα treated and red JQ1 treated) in units of rpm/bp at a meta composite of target genes of SEs gained in response to TNFα treatment. Boxplots (right) show cell count normalized expression levels in TNFα (25 ng/mL, 3 hours) treated ECs ± JQ1. Significance of the difference between distributions determined using a two-tailed t test. ** p < 1e-5, *** p < 1e-10. (D) Line plots of mRNA levels (qRT-PCR) of 3 representative genes associated with TEs (LOX, TEK, NLRP1 in black) and SEs (FS3, CCL2, VCAM1 in red) in response TNFα and JQ1 (50, 100, 250, 500 nM). The mRNA levels from TNFα + VEH (10 ng/mL, 3 hrs) treated ECs were set to 100%. Results displayed as the %reduction from maximum. Error bars represent SEM. Representative results from 2 independent experiments are shown. (E) Gene tracks from Chem-Seq (JQ1) and ChIP-seq (BRD4, H3K27ac) datasets of the SELE SE locus (rpm/bp) for JQ1, BRD4 and H3K27ac from TNFα (-) or TNFα (+) stimulated ECs. (F,G) Scatter plot of JQ1 genome-wide binding levels on x-axis compared to the log2 change in BRD4 (F) or H3K27ac (G) ChIP-Seq signal on y-axis. The change in BRD4 and H3K27ac signal was determined comparing TNFα + JQ1 vs. TNFα. See also Figure S5 and S6.
Global analysis of TNFα-stimulated ECs revealed preferential loss of BRD4 at TNFα-gained SEs compared to typical enhancers, with minimal effect on TNFα-induced p65 binding or H3K27ac levels (Figure 5B). TNFα-gained SEs possessed a diminished capacity to drive proinflammatory transcription initiation and elongation in the presence of the BRD4 inhibitor, JQ1, (Figure 5C), and these dynamic effects on RNA Pol II enrichment were independent of alterations in promoter modification by differential metagene analysis of H3K4me3 enrichment at the TSS (Figure S5C). Functionally, preferential JQ1-induced loss of transcription at TNFα-gained SE associated genes culminated in more potent suppression of their proinflammatory gene expression program relative to genes driven by typical enhancers (Figure 5C, Figure S5D). Treatment of ECs with TNFα and JQ1 suppressed the maximal mRNA induction of SE associated genes (FS3, CCL2, VCAM1) at a lower concentration of JQ1 and to a greater degree compared to TE-associated genes (LOX, TEK, NLRP1) (Figure 5D). Together, these data support a model where BET bromodomains mediate dynamic and immediate inflammatory EC response gene transcription, by facilitating chromatin-dependent signal transduction from NF-κB to RNA Pol II.
To determine whether JQ1 transcriptional effects resulted from specific engagement of BETs at nuclear chromatin, we spatially localized the JQ1 molecule genome-wide using a new biotechnology called Chem-Seq(Anders et al., 2014). This technique maps the interactions of small molecules with chromatin in the human genome, in this instance by using a retrievable synthetic derivative of JQ1 (biotin-JQ1). Notably, despite high levels of acetylated HK27ac at enhancers, we detected no biotin-JQ1 occupancy by Chem-Seq at SELE and other SEs (Figure 5E, Figure S5F-G) in resting ECs. Rather, at these loci, biotin-JQ1 binding to chromatin perfectly colocalized with BRD4 spatially and temporally following TNFα-stimulation. Genome-wide analysis demonstrated a dose-dependent relationship between biotin-JQ1 localization and the decrease in genomic BRD4 occupancy, not H3K27ac, in ECs treated with TNFα+□□ compared to TNFα alone (Figure 5F,G). Taken together, these data reveal that JQ1 directly targets BRD4 during proinflammatory activation in ECs. Preferential loss of BRD4 at inflammatory SEs by BET bromodomain inhibition serves mechanistically to underscore the observed selective effects on transcription.
To assess the generalizability of these observations, we tested whether the BRD4-dependent formation of proinflammatory SEs is relevant for other immune effector cells. We integrated robust, publically available acetyl-histone data (H4K12ac) with RNA Pol II ChIP-Seq data in LPS stimulated macrophages (Nicodeme et al., 2010), to map SEs and explore the effect of BET inhibition on enhancer-mediated transcriptional signaling. H4K12ac is a histone mark defining active enhancers that can be used to identify SEs in the absence of specific enhancer coactivator data (Hnisz et al., 2013; Whyte et al., 2013).
Large gains of histone acetylation were identified in macrophages following LPS stimulation at the Irak2 proinflammatory gene locus (Figure S6A). LPS treatment provoked an immediate increase in promoter and intergenic acetylation, accompanied by an increase in RNA Pol II enrichment throughout the gene body. Globally, SE analysis identified 122 gained SEs following LPS treatment near proinflammatory genes involved in cytokine signaling, cell adhesion and chemotaxis (Figure S6B,C). Unlike inflammatory ECs, the target genes induced and associated with proinflammatory SEs in macrophages are largely distinct from those found in ECs (Figure S6D). Transcription at SE-associated genes was more strongly induced by LPS compared to genes controlled by typical enhancers, as revealed by composite analysis of RNA Pol II enrichment and elongation (Figure S6E,F). As in activated ECs, BET bromodomain inhibition in macrophages preferentially suppressed transcription of genes driven by proinflammatory SEs as compared to genes controlled by typical enhancers (Figure 6SG,H). The Irak2 locus provides an exemplary illustration of the effect of BET bromodomain inhibition using the structurally analogous I-BET inhibitor (Nicodeme et al., 2010), on depletion of RNA Pol II enrichment (Figure S6A). Collectively, these data from ECs and macrophages demonstrate that proinflammatory SEs drive proinflammatory gene activation in a cell context specific manner.
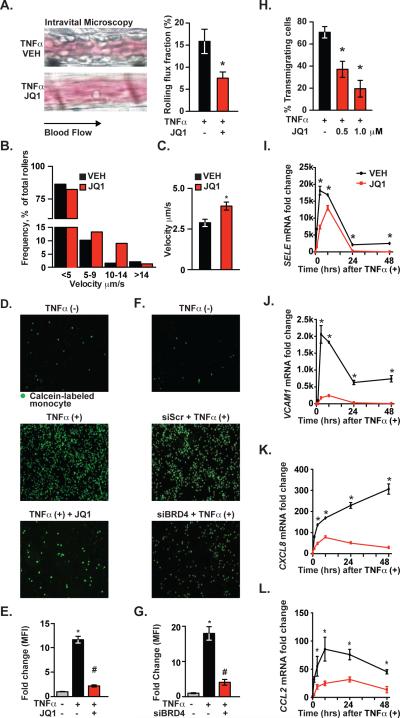
Figure 6. Phenotypic Consequences of BET Bromodomain Inhibition in Endothelium.
(A) Intravital microscopy image (left) and bar plot quantification (right) of leukocyte flux fraction in the cremaster post-capillary venule after TNFα (2 hr, n=7/group) in VEH or JQ1 treated samples. Error bars represent SEM. The statistical significance of the difference between JQ1 treated and VEH treated samples was determined using a two-tailed t test. * p < 0.05. (B) Velocity distribution of leukocytes measured in A. (C) Bar plot showing mean leukocyte velocity in cremaster post-capillary venule in TNFα(+) animals ± BET bromodomain inhibition. Error bars represent SEM using a two-tailed t test. * p < .05. (D,F) Representative fluorescence microscopy images showing adhesion of calcein-labeled THP1 cells to (D) ECs pretreated with JQ1 then activated with TNFα□□□□ as well as (F) TNFα treated ECs after siRNA knockdown of BRD4. (E,G) Bar plots showing quantification of fluorescence from D,F. (H) Bar plots showing quantification of transmigrating neutrophils on TNFα-activated EC monolayers. Results pooled from 3 independent experiments. Data represent mean ± SEM. The statistical significance of the difference between JQ1 and VEH treated samples was determined using a two-tailed t test. * p <.05. (I-L) Line plots of mRNA levels (qRT-PCR) for (I) SELE, (J) VCAM1, (K) CXCL8 and (L) CCL2 measured after stimulation of ECs with TNFα (12.5 ng/mL; 1, 3, 8, 24, 48 hrs) ± JQ1 (500 nM). The statistical significance of the difference in expression between vehicle (VEH) or JQ1 at each time point was determined using a two-tailed t test. *p < 0.05. Data represent mean ± SEM of fold change vs. 0hr. See also Figure S7.
BET Bromodomain Inhibition Suppresses Leukocyte Rolling, Adhesion and Transmigration in Endothelium
In response to proinflammatory stimuli, endothelial-leukocyte interactions follow a sequential cascade involving leukocyte chemoattraction, their slow rolling and subsequent firm adhesion to ECs and culminating in leukocyte endothelial transmigration into tissue (Ley et al., 2007). To explore a phenotypic effect of SE disruption by BET bromodomain inhibition, we tested the functional effects of JQ1 on leukocyte rolling across TNFα-activated endothelium in vivo. C57Bl/6 mice were pretreated with JQ1 (50 mg/kg) twelve hours prior to TNFα injection. As visualized by intravital microscopy of leukocyte rolling in the cremaster post-capillary venule, BET bromodomain inhibition significantly reduced the leukocyte rolling flux (15.8 vs. 7.5, p < 0.01, Figure 6A) and the number of leukocyte rollers/minute (42.8 vs. 25.14, p < 0.05; data not shown), without changing systemic white blood cell count (WBC) or shear stress (Figure S7A). BET bromodomain inhibition also shifted the distribution of leukocyte velocity and increased mean velocity (2.89 μm/second vs. 3.91 μm/second, p < 0.01), consistent with an effect on E-selectin mediated slow rolling (Figure 6B, C).
Next, we examined firm adhesion of the human monocytic cell line (THP1) to activated ECs in vitro. TNFα-stimulated ECs had significantly increased numbers of attached THP1 cells (Figure 6D). JQ1 pretreatment of ECs suppressed THP1 adhesion to TNFα-activated ECs by 70% (Figure 6D, E). Similarly, siRNA inhibition of BRD4 expression in ECs recapitulated JQ1's effects on THP1 adhesion to ECs (Figure 6F, G). Lastly, we tested BET bromodomain inhibitor effects on leukocyte transmigration in a parallel-plate flow chamber. TNFα stimulation of ECs resulted in transmigration of 67% of human neutrophils (Figure 6H). Pretreatment of ECs with JQ1 prior to TNFα stimulation reduced neutrophil transmigration in a concentration-dependent manner (Figure 6H). In kinetic studies of transcription response of TNFα-stimulated ECs, JQ1 demonstrated prolonged inhibitory effects on expression of SE-associated genes (SELE, VCAM1, CXCL8, CCL2), over 48 hours (Figure 6I-L). These data establish BET bromodomain inhibition as a functional suppressor of the phenotypic features of EC proinflammatory activation.
BET Bromodomain Inhibition Suppresses Atherogenesis in Hypercholesterolemic Mice
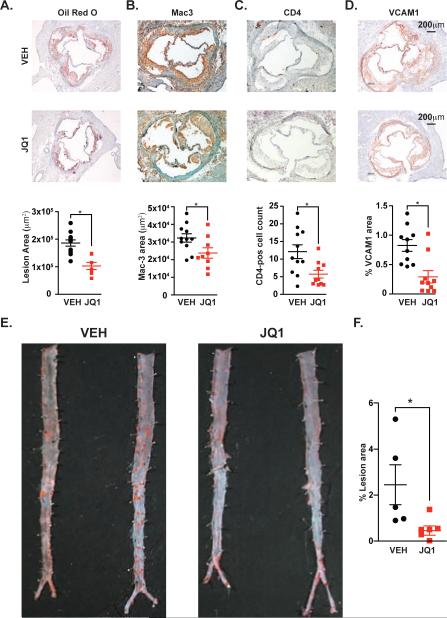
Proinflammatory activation of ECs is a seminal, early event in atherogenesis, a process driven by vascular inflammation that also involves monocytes/macrophages and precedes atherosclerosis (Cybulsky et al., 2001). The transcriptional and functional effects of BET bromodomain inhibition in ECs prompted us to examine their role in atherogenesis using the well-established LDL receptor-deficient (Ldlr−/−) mouse model. Vehicle-treated mice fed a cholesterol-enriched diet (ten weeks) developed atherosclerosis, as measured by Oil Red O staining (Figure 7A). Once-daily JQ1 treatment (50 mg/kg) reduced aortic plaque area by 40% (Figure 7A). Notably, there was no difference in LDL and HDL cholesterol between the treatment groups (168 vs. 164 mg/dL and 56 vs. 59 mg/dL, respectively). Early atherosclerotic lesions are comprised of macrophages (98-99%) with lesser amounts of T lymphocytes (1-2%). Mac-3 staining JQ1 treatment significantly lowered total macrophage staining area, CD4-positive T lymphocytes and levels of VCAM1 protein were all significantly reduced by JQ1 treatment (Figure 7B-D). Oil Red O staining of en face thoracoabdominal aortas revealed decreased atherosclerotic plaque beyond the aortic root (Figure 7E,F). Soluble VCAM1 and ICAM1 levels were also significantly reduced in JQ1-treated animals compared to vehicle (Figure S7B,C), suggesting an effect of BET bromodomain inhibition on systemic proinflammatory activation in vivo. We next tested whether BET bromodomain inhibition mitigated the activation of proadhesion pathways in aortic endothelium, which occurs during the first 10 weeks of exposure to atherogenic diet. In ex vivo aortic adhesion assays, the aortas harvested from animals (6 weeks on diet) treated with JQ1 supported less adhesion of fluorescently-labeled monocytes (Figure S7D,E). These data demonstrate BET inhibition significantly decreased atherogenesis and accumulation of inflammatory cells in a well-characterized murine model of atherosclerosis.
Figure 7. BET Bromodomain Inhibition Suppresses Atherogenesis in Ldlr−/− Mice.
(A-D) Photomicrographs of aortic root sections from Ldlr−/− animals treated with VEH or JQ1 stained for (A) oil red o, (B) Mac-3, (C) CD-4, or (D) VCAM1. Quantification of staining is shown below. Results represent mean ± SEM. The statistical significance of the difference between JQ1 and VEH treated samples was determined using a two-tailed t test. * p = .002 for (A); and * p < .05 for (B-D). (E) Oil red o staining of en face aortas prepared from cohort in A-D. (F) Quantification of lesion area (%) between VEH and JQ1 treated en face aortas. The statistical significance of the difference between JQ1 and VEH treated samples was determined using a two-tailed t test. * p < .05. See also Figure S7.
DISCUSSION
The inflammatory response underlies numerous chronic diseases. NF-κB is a master regulatory transcription factor in several dominant inflammatory signaling cascades, which cooperates with chromatin-associated regulatory complexes to direct inflammatory transcription (Natoli, 2009). Enhancer-bound NF-κB arises following nuclear translocation in a manner influenced by pioneer transcription factors (Kaikkonen et al., 2013; Natoli, 2009; Ostuni et al., 2013) and a pre-established topology that places distal enhancer regions and target genes into close spatial proximity (Jin et al., 2013). Here, we explore the role of chromatin in terminal signal transduction from NF-κB to RNA Polymerase, specifically at massive regulatory regions.
Super or stretch enhancers represent less than 5 % of the enhancers in a cell, yet they utilize almost half of all enhancer coactivator proteins (Loven et al., 2013; Parker et al., 2013; Whyte et al., 2013), and are highly transcribed, producing large amounts of enhancer RNA that may itself facilitate target gene activation (Hnisz et al., 2013; Kaikkonen et al., 2013). The SE landscape is remarkably cell-type specific, driving expression of the genes that define and maintain cell identity in different tissues and cell lineages. The present study demonstrates that NF-κB engages most endothelial enhancers following proinflammatory activation, yet significant BRD4 recruitment to form de novo SEs is restricted to a subset of these enhancer regions. NF-κB directed SE formation causes global reorganization of the BRD4 SE landscape and induces the transcription of many canonical proinflammatory endothelial genes. Whereas previous studies have studied the importance of SEs in the maintenance of cell identity (Whyte et al., 2013), here we describe de novo SE formation as a mechanism by which stimulus-coupled master regulatory transcription factors such as NF-κB can coordinate a rapid transcriptional response that drives a dynamic change in cell state.
This study of kinetic transcriptional response during the inflammatory cell state transition in ECs unexpectedly identified a rapid loss of SEs upon cytokine stimulation. As orchestrated by the master regulatory inflammatory transcription factor NF-κB, SE formation comes at the immediate expense of SEs associated with active transcription of genes in unstimulated ECs, including targets involved in cell specification and non-inflammatory cell states. Many of these lost SE associated genes, such as SOX18, will be downregulated. Curiously, we find persistent marks of open, active euchromatin at the SOX18 locus (H3K27ac) yet BRD4 depletion is associated with loss of RNA Pol II transcription. In studies of dynamic cell state transitions, as here, we find that experimental measurements of canonical enhancer marks (i.e. H3K27ac, H3K4me1/2) are inferior to assessments of enhancer coactivators (i.e. BRD4) for characterizing rapid changes in enhancer structure and function. As such, BRD4 may be viewed and utilized as a rheostat for enhancer output, converting typical enhancers into SEs, thereby driving rapid and robust induction of inflammatory transcription.
In our previous studies, genes with the highest occupancy of BRD4 at their proximal SEs were the most selectively downregulated by BET bromodomain inhibition (Chapuy et al., 2013; Loven et al., 2013). However, these studies characterized effects on cells at ground state, where stable, pre-established SEs predominate. During cell state transitions, such as the present study of EC and macrophage activation, we observe potent and selective effects on upregulated genes associated with de novo SEs. Preferential disruption of dynamic SEs by BET bromodomain inhibition abrogates the induction of inflammatory transcription. The direct relationship between BRD4 enrichment and transcription suggests that modulating BRD4 levels at enhancers may influence the pathogenesis of inflammatory diseases. Changes in eRNA levels by NF-κB-directed BRD4 SE formation may also be an important factor in proinflammatory transcription in ECs, but awaits future study. Further granularity on dynamic enhancer remodeling will accompany progress in genome-wide enhancer detection and assignment.
In many disease settings, the degree of host inflammatory response is a key determinant of severity (Medzhitov et al., 2012). Here we show small molecule BET bromodomain inhibition (JQ1) significantly attenuated endothelial activation during acute inflammation in vitro and ex vivo, as revealed by suppression of TNFα induced leukocyte rolling, adhesion and transmigration. Finally, in a hypercholesterolemic murine model of atherosclerosis, in which EC proinflammatory activation is a seminal early event, ten-week treatment with JQ1 suppressed cardinal histopathologic features of atherogenesis. These data in vascular endothelial activation establish a critical and early role for BET bromodomains in dynamic enhancer remodeling. They describe a mechanism for rapid inflammatory gene activation by NF-κB mediated formation of SEs. Taken in the context of prior research in macrophage activation, spermatogenesis, and myocyte hypertrophy (Anand et al., 2013; Delmore et al., 2011; Matzuk et al., 2012; Nicodeme et al., 2010), these data support a model where localization of BET bromodomains to chromatin facilitates cell state transitions. The existence of BET bromodomain inhibitors provides, then, a broad opportunity for inflammatory gene control through modulation of chromatin structure and function.
EXPERIMENTAL METHODS
Animal Models
LDL receptor knockout mice (4 week old) on a C57Bl/6 background were purchased from Jackson Laboratories. Mice were fed an atherogenic diet (Clinton/Cybulsky Rodent Diet, D12108 with 1.25% cholesterol, Research Diets) for 10 weeks. While on diet, animals were treated with vehicle (DMSO) or JQ1 (50 mg/kg) by intraperitoneal injection, once daily (n=10/group). Oil Red O staining was used to quantify atherosclerotic plaque lesion area in the aortic root. Macrophage (Mac-3) and T lymphocyte (CD-4) accumulation was assessed in the aortic root and total staining area analyzed using computer-assisted imaging analysis. For CD-4 cells, total cell numbers were counted. Whole aorta from the left subclavian artery to the iliac bifurcation was used for en face preparation and stained with oil red o. All protocols concerning animal use were approved by the Harvard Medical School Institutional Animal Care and Use Committee (IACUC) and conducted in accordance with NIH Guide for the Care and Use of Laboratory Animals. All studies were performed in C57Bl/6J mice (Jackson Laboratories), maintained in a pathogen-free facility with standard light/dark cycling and ad libitum access to food and water.
Reagents and cell culture
ECs from pooled human umbilical cords were cultured in M199 medium supplemented with 20% FBS, 0.1% heparin, 50 μg/mL ECGF (Biomedical Technologies), penicillin/streptomycin on gelatin coated tissue culture plates. U937 cells (ATCC) were maintained in RPMI with 10% FBS and antibiotics. For trans-endothelial migration (see below), HUVEC (subculture 2) were grown on fibronectin-coated glass coverslips (5 mg/mL; BD Biosciences) and treated with JQ1 (500 nM) or vehicle (DMSO) for 1 hour before TNFα stimulation (10 ng/mL, 4 hours). Recombinant human TNFα was obtained from PeproTech (Rocky Hill, NJ). JQ1 was dissolved in DMSO at a concentration of 50 mg/mL. Working stocks of JQ1 were prepared by diluting 1:10 in 10% beta-cyclodextrin solution (Filippakopoulos et al., 2010). Animals were treated at 50 mg/kg once daily by intraperitoneal injection
ChIP-Seq and Data Analysis
ChIP was performed in ECs in the presence or absence of TNFα (1 hour, 25 ng/mL) and JQ1 (3 hour pretreatment, 500 nM). Specific antibodies and detailed methods are described in the extended experimental methods. ChIP was carried out as described in (Loven et al., 2013). All ChIP-Seq datasets were aligned using Bowtie (version 1.0.0) (Langmead et al., 2009) to build version NCBI36/HG18 of the human genome or build version NCBI37/MM9 of the mouse genome. Enhancers and super enhancers were mapped as in (Loven et al., 2013). Additional details are provided in the extended experimental methods.
Supplementary Material
Highlights.
Activated NF-κB prompts rapid formation of super enhancers
Super enhancer bound BRD4 co-activates inflammatory genes
Redistribution of BRD4 results in eviction from basal super enhancers
BET bromodomain inhibition attenuates atherogenesis
ACKNOWLEDGEMENTS
We are grateful to R. Young, P. Rahl, T. Graf and C. Van Oevelen for stimulating discussions; M. Berkeley, Z. Herbert and the late E. Fox (DFCI Microarray Core) for assistance with microarray experiments; and T. Volkert, J. Love, and S. Gupta at the Whitehead Genome Core for assistance with genome sequencing. This research was supported by: NIH-K08 HL105678, The Watkins Discovery Research Award and The Harris Family Award (JDB); U.S. Department of Defense CDMRP CA120184 (CYL); Sarnoff Cardiovascular Research Foundation (GG; SB); NIH PO1 HL36028 (FWL); NIH-K08 CA128972, the Burroughs-Wellcome Fund, the Damon-Runyon Cancer Research Foundation, the Richard and Susan Smith Family Foundation, and the Next Generation Award (JEB); NHLBI P01 HL048743 (JP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
JDB, CYL, QD, JEB and JP designed research. JDB and QD performed in vitro and in vivo endothelial function studies with assistance from GG, GN, AHL, GWL, KC. JDB and CYL performed ChIP studies and analyzed ChIP data. AF developed motif analysis code. AK treated mice with JQ1 for atherogenesis studies. JDB, CYL, JEB, JP wrote manuscript.
ACCESSION NUMBERS
All ChIP-Seq and Chem-Seq data generated in this publication can be found online associated with GEO Publication Reference ID GSE53998 (www.ncbi.nlm.nih.gov/geo/). This Publication Reference ID also includes the microarray data. GEO accession numbers for all analyzed datasets can also be found in File S1. Aligned and raw data can be found online associated with the GEO Accession ID GSE54000.
REFERENCES
- Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, Alaiti MA, Bullard J, Alazem K, Margulies KB, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, Loven J, Sigova AA, Smith WB, Lee TI, et al. Genome-wide localization of small molecules. Nature biotechnology. 2014;32:92–96. doi: 10.1038/nbt.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner BP, Westerheide SD, Baldwin AS., Jr. The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. NF-kappaB is 25. Nat Immunol. 2011;12:683–685. doi: 10.1038/ni.2072. [DOI] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- Barnes PJ, Karin M. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- Cybulsky MI, Iiyama K, Li H, Zhu S, Chen M, Iiyama M, Davis V, Gutierrez-Ramos JC, Connelly PW, Milstone DS. A major role for VCAM-1, but not ICAM-1, in early atherosclerosis. J Clin Invest. 2001;107:1255–1262. doi: 10.1172/JCI11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, et al. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye SV. The art of the chemical probe. Nature chemical biology. 2010;6:159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- Gimbrone MA, Jr., Bevilacqua MP, Cybulsky MI. Endothelial-dependent mechanisms of leukocyte adhesion in inflammation and atherosclerosis. Annals of the New York Academy of Sciences. 1990;598:77–85. doi: 10.1111/j.1749-6632.1990.tb42279.x. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–689. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masckauchan TN, Shawber CJ, Funahashi Y, Li CM, Kitajewski J. Wnt/beta-catenin signaling induces proliferation, survival and interleukin-8 in human endothelial cells. Angiogenesis. 2005;8:43–51. doi: 10.1007/s10456-005-5612-9. [DOI] [PubMed] [Google Scholar]
- Matsui T, Kanai-Azuma M, Hara K, Matoba S, Hiramatsu R, Kawakami H, Kurohmaru M, Koopman P, Kanai Y. Redundant roles of Sox17 and Sox18 in postnatal angiogenesis in mice. Journal of cell science. 2006;119:3513–3526. doi: 10.1242/jcs.03081. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, et al. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- Natoli G. Control of NF-kappaB-dependent transcriptional responses by chromatin organization. Cold Spring Harbor perspectives in biology. 2009;1:a000224. doi: 10.1101/cshperspect.a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Program NCS, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce JW, Lenardo M, Baltimore D. Oligonucleotide that binds nuclear factor NF-kappa B acts as a lymphoid-specific and inducible enhancer element. Proc Natl Acad Sci U S A. 1988;85:1482–1486. doi: 10.1073/pnas.85.5.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, Gerritsen ME. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. The Journal of biological chemistry. 1997;272:21096–21103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, Minder JL, Mercan F, Wang E, Eckersley-Maslin MA, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes & development. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg RL, Schreiber SL. From knowing to controlling: a path from genomics to drugs using small molecule probes. Science. 2003;300:294–295. doi: 10.1126/science.1083395. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.