Abstract
Purpose:
Accurate prostate segmentation is necessary for maximizing the effectiveness of radiation therapy of prostate cancer. However, manual segmentation from 3D CT images is very time-consuming and often causes large intra- and interobserver variations across clinicians. Many segmentation methods have been proposed to automate this labor-intensive process, but tedious manual editing is still required due to the limited performance. In this paper, the authors propose a new interactive segmentation method that can (1) flexibly generate the editing result with a few scribbles or dots provided by a clinician, (2) fast deliver intermediate results to the clinician, and (3) sequentially correct the segmentations from any type of automatic or interactive segmentation methods.
Methods:
The authors formulate the editing problem as a semisupervised learning problem which can utilize a priori knowledge of training data and also the valuable information from user interactions. Specifically, from a region of interest near the given user interactions, the appropriate training labels, which are well matched with the user interactions, can be locally searched from a training set. With voting from the selected training labels, both confident prostate and background voxels, as well as unconfident voxels can be estimated. To reflect informative relationship between voxels, location-adaptive features are selected from the confident voxels by using regression forest and Fisher separation criterion. Then, the manifold configuration computed in the derived feature space is enforced into the semisupervised learning algorithm. The labels of unconfident voxels are then predicted by regularizing semisupervised learning algorithm.
Results:
The proposed interactive segmentation method was applied to correct automatic segmentation results of 30 challenging CT images. The correction was conducted three times with different user interactions performed at different time periods, in order to evaluate both the efficiency and the robustness. The automatic segmentation results with the original average Dice similarity coefficient of 0.78 were improved to 0.865–0.872 after conducting 55–59 interactions by using the proposed method, where each editing procedure took less than 3 s. In addition, the proposed method obtained the most consistent editing results with respect to different user interactions, compared to other methods.
Conclusions:
The proposed method obtains robust editing results with few interactions for various wrong segmentation cases, by selecting the location-adaptive features and further imposing the manifold regularization. The authors expect the proposed method to largely reduce the laborious burdens of manual editing, as well as both the intra- and interobserver variability across clinicians.
Keywords: interactive segmentation, prostate, feature selection, semisupervised learning, manifold regularization
1. INTRODUCTION
Prostate cancer is one of the top leading causes of cancer-related death for males in the USA. Radiation therapy with high-energy beams or particles is commonly used to cure the early stage cancer. During the radiation therapy, the high-dose radiation should be accurately delivered to the prostate, since the false delivery could lead to undertreatment for the prostate cancer and also the severe side effects for the patient. Therefore, accurate segmentation of the prostate from the surrounding healthy tissues is critical for providing better guidance to deliver radiation beams to the prostate. However, manual segmentation of the prostate in a 3D image is very time-consuming and often causes large intra- and interobserver variability across clinicians.1
Accordingly, many automatic prostate segmentation methods have been proposed to obtain reliable prostate segmentation results. One representative stream is the atlas-based approach.1–5 In this kind of method, the relevant training images and labels are first aligned to the target image and then used to determine the labels with voting schemes or estimation algorithms.1,2,5 These methods are intuitive and easy to be applied to various image modalities, but they often cannot deal well with large shape or appearance changes of the prostate across different subjects. Model-based approach is another stream. Unlike those atlas-based methods that directly use the aligned labels, the statistical shape or appearance model is constructed from the training data in this type of approach.6–13 Recently, the performance of model-based methods was further improved by imposing the object and structure constraints6,9,11 or utilizing the level set framework.10 Although these model-based methods can effectively constrain the possible shape and appearance variations, a large number of training set are needed in order to learn the entire variations of samples. In addition, for most cases, the tedious manual placement of landmarks is often required in the training step. Feature-based classification methods14–16 have also been proposed to reflect the midlevel cues such as appearance pattern or context information. For example, Li et al.15 proposed a location-adaptive classifier to integrate both image appearance and context features. Chowdhury et al.14 extracted features from both MR and CT images and used classification results to fit the statistical shape model. Shi et al.16 divided the image into grids and selected the informative features on each grid to adaptively emphasize the useful local features. Although the classification methods can reflect the midlevel cues beyond the voxel-wise intensity or gradient information, the extraction of good features for capturing all statistical information or anatomical properties is a nontrivial task.
To sum up, it is difficult to develop a fully automatic prostate segmentation method that can address various issues, such as weak boundary between prostate and its surrounding tissues, large shape variations across subjects, and variations of appearance pattern around the prostate due to the uncertainty of bowel gas filling in different treatment days. Although automatic segmentation methods can largely alleviate the burden of clinicians, their results are often not sufficiently good to be directly used for clinical practice. Thus, the heuristic postprocessing or manual editing is often needed for editing the wrong parts after automatic segmentation process.
So far, many interactive segmentation methods17–19 have been proposed in the computer vision field. These methods can generate fast and practical editing results by optimizing the energy model18 based on the observation from user interaction and also the requirement of smoothness between nearby voxels. However, these methods cannot obtain the flexible editing results when a few user interactions are provided, since the constructed models often depend only on the intensity or gradient information of the annotated voxels. Several other methods20–23 have been developed to alleviate the dependency of user interaction. For example, Rother et al. proposed a GrabCut method21 which requires the user to provide only the bounding box of the target object. Kim et al. proposed a multilayer graph based method,20 which requires small quantities of respective foreground and background scribbles or dots. Wang et al.23 and Top et al.22 proposed some active learning based methods to effectively receive user delineations on the ambiguous regions. However, all these methods still rely on the image appearance features, without utilizing prior knowledge from the training data.
Recently, interactive methods using the label information of training data have been proposed. For example, Barnes et al.24,25 proposed an image completion and reshuffling method based on PatchMatch, which can seek for the corresponding image patches efficiently by first randomly searching for a similar patch and then expanding correspondences to the adjacent regions. This method has been also applied to superresolution of cardiac image26 and also hippocampus segmentation.27 Park et al.28 further enforced the spatial relationship of adjacent patches to constrain the specific shapes of organs. These methods improved the editing performances for many applications by using the label information of similar patches to constrain possible variations. However, for segmenting the prostate from the challenging CT images, simple label and intensity information cannot deal with large shape variations as well as weak boundaries around the prostate.
In this paper, we propose a new interactive method, which can (1) flexibly generate the editing results with a few scribbles or dots provided by a clinician, (2) fast deliver intermediate results to the clinician, and (3) sequentially correct the segmentations from any type of automatic or interactive segmentation methods. Unlike those existing interactive methods that directly use the label or intensity information as priors, we formulate the labeling problem as the semisupervised learning problem which can utilize a priori knowledge of training data with user interactions and also the manifold configuration between voxels in the target image. Specifically, in our proposed method, informative features are adaptively extracted from each local editing region according to the training labels, which are matched well with user interactions. Then, the manifold configuration, which reflects the anatomical relationships between target voxels, is computed in the feature space and further enforced in the semisupervised learning algorithm. Since the possible shape variations have been constrained by the selected training labels, the editing result is robust to the amount as well as the locations of user interactions. Furthermore, the location-adaptive features can effectively represent the appearance patterns of local region beyond the simple voxel-wise intensity or gradient values in identifying the prostate. Finally, the semisupervised manifold regularization method can guide the labeling of unconfident voxels by well-reflecting the anatomical relationship between confident and unconfident voxels.
The remainder of this paper is organized as follows: First, the materials and proposed interactive segmentation framework are described in Sec. 2. Then, our experimental setting and results are presented in Sec. 3, to demonstrate both efficiency and robustness of the proposed method. We discuss the properties of the proposed method and several clinical issues by analyzing the experimental results in Sec. 4. Finally, in Sec. 5, we conclude our paper with some possible future research directions.
2. MATERIALS AND METHOD
2.A. Materials
The proposed method was evaluated on a challenging prostate CT image dataset acquired from University of North Carolina Cancer Hospital. The dataset included 73 planning images, scanned from different patients by using a Siemens Somatom CT scanner. The typical dose and field-of-view were set as 200–300 cGy and 50 cm, respectively. The image size was 512 × 512 × (61–81) with 0.94 × 0.94 × 3.00 mm3 voxel spacing. The dataset included various subjects with large variations of shape and appearance pattern around the prostate as shown in Fig. 1. Prostate boundary in each image was manually delineated by a radiation oncologist and used as the ground truth for measuring the segmentation performance.
FIG. 1.
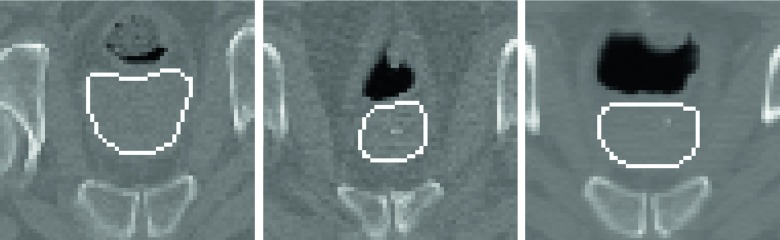
Example of 2D axial images showing a similar prostate region of three different subjects in our dataset. Contours denote prostate boundaries manually delineated by a radiation oncologist. These images show large variations of prostate shape and appearance pattern.
2.B. Method
The proposed method is conducted whenever a clinician’s interaction is inserted into a wrong part of previous segmentation Lt−1, where t is the interaction time. If there are many wrong parts on previous segmentation, multiple corrections are repeatedly conducted until obtaining satisfactory result. Each correction consists of the following procedure. First, the appropriate training labels, which are well matched with user interaction and the previous segmentation, are selected near the interaction from the training set. Based on the selected training labels, both the confident regions (e.g., prostate and background voxels) and the unconfident regions are estimated. Then, the informative features, which are important for separating the prostate and the background on the confident regions, are selected. Next, we formulate the labeling problem as a semisupervised learning problem with manifold regularization, by using the confident and unconfident voxels as the labeled and unlabeled voxels, respectively. Through the manifold regularization method, the labels of unconfident voxels are predicted by considering likelihood of the confident voxels and the voxel-wise similarities defined by the selected informative features. The overall algorithm is described in Fig. 2, with details presented in Subsection 2.B.1–2.B.4.
FIG. 2.
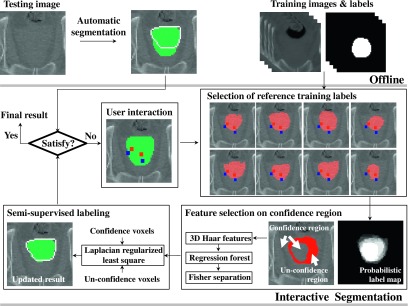
Framework of our proposed interactive method. Initial segmentation label, the selected training labels, and the corrected segmentation label are shown on the top-middle, middle-right, and bottom-left sides, respectively. The ground-truth prostate contour is shown with white line (note that the ground-truth label is actually not shown in the test stage). Whenever foreground and background user interactions (small squares) are inserted on a wrong region, the procedure of interactive segmentation is conducted.
2.B.1. Initial segmentation and preprocessing
The initial segmentation L0 can be obtained by any kind of automatic or interactive methods. In this paper, we use the regression-based automatic segmentation method29 to obtain L0.
All training images and their labels are aligned onto the target image, to allow the rapid search of the appropriate training data during the online editing procedure. We use the MRF based nonrigid registration method (Drop)30 for the intensity-based alignment. It takes roughly 1 min for aligning each training image, but this time-consuming procedure can be done without clinician’s effort before interactive editing. The interactive approach, a main part of our proposed algorithm, is detailed in Subsection 2.B.2.
2.B.2. Region of interest (ROI) determination from user interaction
The proposed method receives foreground and background scribbles or dots as the user interactions for the wrong part of previous segmentation Lt−1. The subsequent editing is conducted on a local region near the user interaction, with assumption that the region near the interaction has segmentation errors, while the other regions of Lt−1 are assumed to be correct temporarily. Note that, if there are errors on the other regions, the errors can be sequentially corrected in the next interactions. The local ROI φt is determined as a bounding box that includes the user interaction, but with a small margin, so that the possible local variations can be covered. We set the margin as 9 × 9 × 2 voxels, considering the large slice thickness of our CT images (e.g., 3 mm). The following procedure was conducted on this ROI by using the label image Ut of the tth round interaction, the previous segmentation Lt−1, and the training labels. Here, if a voxel v is interactively labeled as foreground, while if v is interactively labeled as background. Otherwise, .
2.B.3. Selection of reference training data
To use the prior knowledge encoded in the training data, we select appropriate training labels which are well matched with the user interactions on the local ROI region. Although the entire images and labels are aligned to the target image by the Drop registration30 before the online editing step, the aligned labels may not match well with the user interactions on the local ROI region. To alleviate this issue related to the registration error, we first refined the aligned training labels near the user interactions. Specifically, for each training label, we locally searched the best matching of training label by shifting the ROI along different directions within a range. We computed similarity scores on all translated ROIs within the range of 7 × 7 × 3 voxels and then selected the best shifting that led to the highest similarity score.
Here, the similarity can be computed by the appearance-based similarity measurements, such as sum of absolute distances, or normalized cross correlation (NCC), but they cannot reflect the actual similarity between a training label M and the user interaction label Ut. Therefore, we define a label-based similarity to take the user interaction into account. Here, we assume that the appropriate training data should have the training label M which is well matched with (1) Ut on the annotated voxels and also (2) Lt−1 on the other voxels. The label-based similarity S(M, Ut, Lt−1) is thus defined as
| (1) |
where δ is the Kronecker delta. The first term represents the label similarity on user interaction region, while the second term represents the label similarity on other noninteraction region. The more M is consistent with Ut and Lt−1, the higher similarity measure will be obtained. wU is a parameter used to balance between the two terms. Since the number of annotated voxels (Ut(v) ≠ 0) is relatively smaller than that of unlabeled voxels (Ut(v) = 0), wU is set as a small value (i.e., wU = 0.01) in the experiments below.
After computing the similarity scores for all training data, totally nr labels with the highest scores will be selected as the reference training labels. The examples of reference training labels are shown in Fig. 2.
2.B.4. Prostate segmentation by semisupervised labeling
Although the segmentation of target image can be obtained by using the majority voting or weighted voting of the selected reference labels, the simple label fusion technique could not deal with intra- and intersubject appearance variability, because the weight (an important factor for final segmentation) is often computed by the global appearance similarity between training image and target image. To reflect the local appearance relationship between voxels within the target image, we first estimate the confident prostate and background regions in φt by using the reference labels, and then carefully determine the labels of unconfident voxels by using the knowledge of the confident voxels. To simultaneously consider the likelihood of the confident voxels and also the manifold configurations between the confident and unconfident voxels, we formulate the labeling problem as a semisupervised learning problem as below.
Specifically, a probabilistic label map P can be first computed by averaging the reference training labels to estimate the confident regions. For example, if P(v) is higher than τh, v is estimated as a confident prostate voxel (; if P(v) is lower than τl, v is estimated as a confident background voxel (. These confident voxels are regarded as labeled voxels (i.e., with nl as their total number) and all others are regarded as unlabeled voxels ( (i.e., with nu as their total number).
The labeling problem can be then formulated by a Laplacian regularized least square (LapRLS) equation31 as follows:
| (2) |
where w = {w1, …, wnl+nu} is a parameter set to be optimized. The first term penalizes the deviation of prostate likelihood prediction on the labeled voxels, while the second term imposes the smoothness regularization of w. The last term enforces the similarity between the labeled and unlabeled voxels during the prostate likelihood estimation. J = diag(1, …, 1, 0, …, 0) is a diagonal matrix with the first nl diagonal entries as 1 and the rest as 0. y = {y1, …, ynl, ynl+1, …, ynl+nu} is a label set with the labels of the unlabeled voxels initially set to zero. K is the (nl + nu) × (nl + nu) gram matrix with the elements K(vi, vj) defined by the Gaussian RBF kernel as , where f(v) is the feature vector of voxel v. L is the (nl + nu) × (nl + nu) Laplacian matrix, defined across all labeled and unlabeled voxels. The Gaussian RBF kernel is similarly used to define the voxel relationship. γ1 and γ2 are the weighting parameters for the second and third terms, respectively. If γ2 = 0, Eq. (2) becomes the supervised least square problem which only considers the likelihood of confident voxel without taking into account the similarities between confident and unconfident voxels during the prostate likelihood estimation.
The optimal parameter of Eq. (2) can be computed as
| (3) |
where I is the identity matrix. After estimating , the likelihood of all voxels can be computed as . If is larger than 0, the voxel v is classified into the prostate (; otherwise, it is classified into the background (.
Here, the feature vector f(v), used in the RBF kernel, can be defined by any type of appearance or context features, but the discriminative power of f(v) is highly related to the segmentation performance. Since simple appearance or context features have low discriminative power, we adopt the regression forest method,32 which has recently been used for anatomy detection, to select the informative features. Since the most unconfident voxels are positioned near the boundary, we sample the voxels near the prostate boundary (in a Gaussian distribution way) from the training data. For each sampled voxel, we extract its various randomized 3D Haar-like features from the 30 × 30 × 10 intensity patch centered at this voxel to capture the midlevel appearance characteristics. The informative features are determined as those selected by the regression forest model32 in estimating the distance of each image voxel to its nearest target boundary. Among all the extracted features obtained from the training data, we use confident voxels to adaptively select the features again, which have the high discriminative power within the local ROI of the target image. (Note that the selected discriminative features could vary from different local ROI regions.16) The discriminative power of each feature is measured by the Fisher separation criterion (FSC) score SFSC33 as
| (4) |
where μp and μb denote the mean feature values of prostate and background voxels, respectively. Similarly, σp and σb denote the variances of prostate and background voxels, respectively. Among all features, nf features with the largest FSC scores, i.e., the highest discriminative power on the local ROI, are finally selected as f(v).
The overall algorithm is summarized in Algorithm I.
ALGORITHM I.
Algorithm of the proposed framework.
| Input: A target image and a number of training data (along with labels). |
| (Offline) Automatic Segmentation and Training Label Alignment |
| 0a: Generation of initial segmentation L0 by using an existing automatic method (Ref. 29). |
| 0b: Alignment of all training data to the target image by using an existing registration method (Drop) (Ref. 30). |
| (Online) Interactive Segmentation |
| Iterate, |
| 1: On a wrong part of previous segmentation, input user scribbles or dots denoted by Ut. |
| 2: Determination of ROI φt. |
| 3: Selection of the nr most similar training labels to ROI φt, according to the similarity score defined in Eq. (1). |
| 4: Fusion of the aligned training labels, and estimation of both confident and unconfident voxels. |
| 5: Selection of the nf discriminative features based on those confident voxels by using Eq. (4). |
| 6: Label prediction by optimizing the semisupervised learning problem as defined in Eq. (2). |
| Loop until the editing result is satisfied. |
3. EXPERIMENTAL RESULTS
We randomly divided the dataset into four sets and conducted fourfold cross validation. First, we applied the regression-based automatic segmentation method29 to 73 images and measured its segmentation performance. The performance was measured by the Dice similarity coefficient (DSC) as (2 × |Lf∩Mg|)/(|Lf| + |Mg|), where Lf and Mg are the final segmentation label and the manual ground-truth label, respectively. We obtained the satisfactory results for 43 images which all have more than 0.85 DSC scores, but obtained the average 0.78 DSC score for the remaining 30 images. The proposed interactive method was then applied to correct the automatic segmentation results of those 30 images. The parameters of the proposed method were empirically determined. For τh and τl, false positive and false negative labels increased on the confident region if the thresholds were too loose, while the performance of semisupervised learning method decreased if the thresholds were too strict. We measured the ratio of true positive and true negative labels on the confident regions with respect to the thresholds from 0.5 to 0.9 for τh and from 0 to 0.3 for τl with an interval of 0.1 for the training data. For the experiments in this paper, τh and τl were set as 0.7 and 0.1, respectively, which make the ratio of true labels over 0.98. Other parameters were similarly set as follows: nf = 1000, nr = 7, γ1 = 10−2, γ2 = 10−1.
The proposed LapRLS method with discriminative feature selection (LapRLS + FS) was compared with (1) manual editing method, (2) weighted voting method based on the appearance similarity (WVA), (3) weighted voting method based on the label-based similarity (WVL), (4) LapRLS method without feature selection (LapRLS + noFS), and (5) LapRLS method with random feature selection (LapRLS + RFS).
-
•
Note that, only the labels of voxels annotated by the user interaction are changed in the manual editing method; thus, the result of the manual editing method shows only the amount of the interactions.
-
•
The weighted voting methods provide results reflecting the information from the training labels. To show the effect of label-based similarity as defined in Eq. (1), we provide both the weighted voting result obtained based on the appearance similarity (WVA) and the result obtained based on the label-based similarity (WVL). Specifically, the training labels that have the highest intensity-based normalized cross correlation (NCC) are selected for the WVA. The selected training labels are then averaged with the weights that are computed by the NCC. The voxels with the averaged label value more than 0.5 are determined as the prostate. Similarly, the training labels that have the highest label-based similarity are selected for the WVL, and the voxels with the averaged label value more than 0.5 are determined as the prostate.
-
•
We also provide the results of LapRLS + noFS and LapRLS + RFS to demonstrate the effectiveness of feature selection. All intensity values in 30 × 30 × 10 patch were used as features for the LapRLS + noFS method, while 1000 randomly selected Haar features were used as features for the LapRLS + RFS method.
To evaluate the efficiency, we corrected the wrong parts of automatic segmentation results in 3D space with several user interactions. The proposed software provided 2D views on coronal, axial, sagittal directions to a user, in order to check the segmentation. The user could choose both the view direction and the brush size of interaction, and then insert the scribbles or dots on the erroneous regions in the 2D view. For the most cases, we set the view as axial direction and the brush size as 3 × 3 × 1, and used several dots as user interactions. The dots were inserted to the wrong parts, according to the manual ground truth provided by a radiation oncologist. To evaluate the robustness, we conducted the above experiment three times, with different user interactions conducted at different time periods, and then computed the DSC scores between the corrected results. Here, the type of different user interactions was similar, but the amount and locations of interactions were different as shown in Fig. 3.
FIG. 3.
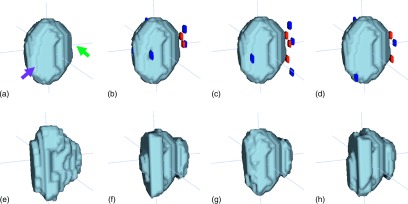
Interactive segmentation results with different user interactions conducted at different time periods. (a) and (e) show automatic segmentation result and ground truth, respectively. Here, the front left part of the automatic segmentation result is over-segmented (left arrow), while the right part is under-segmented (right arrow). (b), (c), and (d) show three different user interactions, respectively. (f), (g), and (h) show the corresponding interactive segmentation results of (b), (c), and (d).
For the experiments with three different user interactions, it took averagely 59, 55, and 57 interactions for correcting all 30 images, with 1.9 interactions per image. Since the improvement of the editing results could be relatively small if considering the entire prostate although the improvement on the local ROI could be very large, the DSC scores were measured both on the local ROI and on the entire prostate image for each user interaction. The final DSC scores of the accumulated interactive editing results were also measured on the entire prostate image. The average and standard deviation of DSC scores of all comparison methods are presented in Table I. Furthermore, we provide the DSC distributions of all training based methods for the three different user interactions in Figs. 4–6, respectively. Figure 7 shows the qualitative editing results for several wrong cases.
TABLE I.
The average (standard deviation) of DSC on local ROI (Local) and on the entire prostate image (Global) for all separate user interactions, and DSC of the accumulated interactive segmentation results on the entire prostate image for 30 images (Final). Ex. 1, Ex. 2, and Ex. 3 represent three experiments with three different user interactions. The numbers in round brackets represent the numbers of interactions. The best performance among the comparison methods is highlighted as boldface.
| Initial | Manual | WVA | WVL | LapRLS + noFS | LapRLS + RFS | LapRLS + FS | ||
|---|---|---|---|---|---|---|---|---|
| Ex. 1 | Local | 0.564 | 0.593 | 0.794 | 0.821 | 0.788 | 0.779 | 0.837 |
| (59) | (0.2026) | (0.1871) | (0.1459) | (0.1019) | (0.1344) | (0.1291) | (0.0988) | |
| Global | 0.775 | 0.781 | 0.805 | 0.813 | 0.809 | 0.808 | 0.815 | |
| (0.0472) | (0.0429) | (0.0465) | (0.0373) | (0.0376) | (0.037) | (0.0374) | ||
| Final | 0.78 | 0.788 | 0.842 | 0.861 | 0.855 | 0.851 | 0.865 | |
| (0.0508) | (0.0472) | (0.0488) | (0.0133) | (0.0133) | (0.015) | (0.012) | ||
| Ex. 2 | Local | 0.555 | 0.587 | 0.783 | 0.824 | 0.815 | 0.809 | 0.846 |
| (55) | (0.2041) | (0.1848) | (0.1514) | (0.1005) | (0.0984) | (0.0945) | (0.0811) | |
| Global | 0.773 | 0.778 | 0.805 | 0.812 | 0.81 | 0.809 | 0.814 | |
| (0.0463) | (0.0435) | (0.0441) | (0.0388) | (0.0394) | (0.0391) | (0.0386) | ||
| Final | 0.78 | 0.789 | 0.845 | 0.859 | 0.86 | 0.858 | 0.865 | |
| (0.0508) | (0.0471) | (0.0283) | (0.0154) | (0.0121) | (0.0133) | (0.0119) | ||
| Ex. 3 | Local | 0.606 | 0.636 | 0.827 | 0.848 | 0.832 | 0.824 | 0.867 |
| (57) | (0.1902) | (0.1706) | (0.1106) | (0.0838) | (0.0915) | (0.0859) | (0.0650) | |
| Global | 0.773 | 0.779 | 0.808 | 0.814 | 0.811 | 0.81 | 0.817 | |
| (0.0488) | (0.0462) | (0.0467) | (0.0436) | (0.0431) | (0.0424) | (0.0424) | ||
| Final | 0.78 | 0.79 | 0.853 | 0.863 | 0.862 | 0.858 | 0.872 | |
| (0.0508) | (0.047) | (0.0349) | (0.0254) | (0.0159) | (0.0181) | (0.0138) |
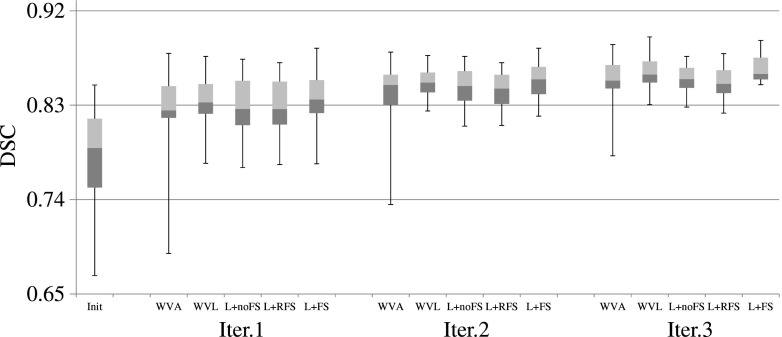
FIG. 4.
DSC scores of the training based methods for the 30 testing images, using the first user interactions. The three horizontal lines of the box represent the upper quartile (75%), median (50%), and lower quartile (25%), while the whiskers indicate the min and max DSC scores. Note that, in the figure, “L” denotes “LapRLS.”
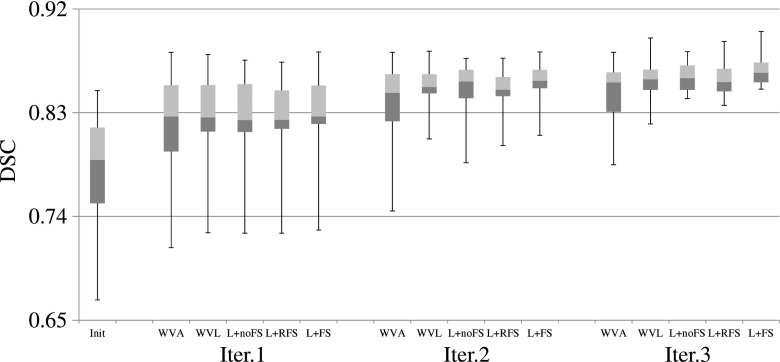
FIG. 5.
DSC scores of the training based methods for the 30 testing images, using the second user interactions. The three horizontal lines of the box represent the upper quartile (75%), median (50%), and lower quartile (25%), while the whiskers indicate the min and max DSC scores. Note that, in the figure, L denotes LapRLS.
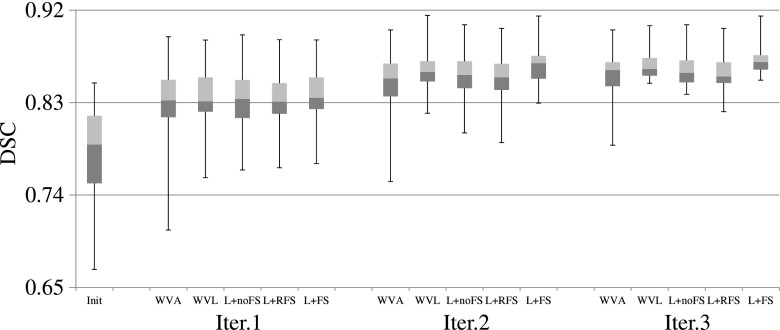
FIG. 6.
DSC scores of the training based methods for the 30 testing images, using the third user interactions. The three horizontal lines of the box represent the upper quartile (75%), median (50%), and lower quartile (25%), while the whiskers indicate the min and max DSC scores. Note that, in the figure, L denotes LapRLS.
FIG. 7.
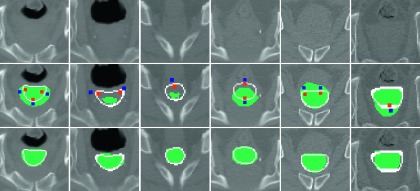
Qualitative interactive editing results for several wrong cases. Testing images are shown in the top row. Initial results, ground-truth boundary (line), and user interactions (small squares) are shown in the middle row. The corrected segmentation results by the proposed method are shown in the bottom row.
To show the robustness of our proposed method, the DSC scores between any two segmentation results, obtained by three different user interactions, were also measured. Specifically, DSC scores between the results obtained by the first and second interactions, between the results by the second and third interactions, and between the results by the first and third interactions were computed. The average and standard deviation of DSC scores of the training based methods are described in Table II. Figure 8 shows the robustness of the proposed method with respect to various interactions.
TABLE II.
The average (standard deviation) of DSC between the results obtained with three different user interactions for the 30 testing images. The best performance among the comparison methods is highlighted as boldface.
| WVA | WVL | LapRLS + noFS | LapRLS + RFS | LapRLS + FS | |
|---|---|---|---|---|---|
| Ex.1 with Ex.2 | 0.924 | 0.937 | 0.938 | 0.928 | 0.942 |
| (0.0487) | (0.0286) | (0.0312) | (0.0316) | (0.0259) | |
| Ex.2 with Ex.3 | 0.92 | 0.926 | 0.928 | 0.918 | 0.935 |
| (0.0545) | (0.0465) | (0.0423) | (0.043) | (0.0397) | |
| Ex.1 with Ex.3 | 0.915 | 0.926 | 0.929 | 0.914 | 0.932 |
| (0.0445) | (0.0362) | (0.0361) | (0.0391) | (0.0321) |
FIG. 8.
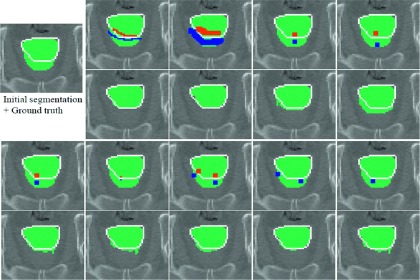
Interactive editing results with respect to various user interactions. Initial segmentation and its manual ground truth (line) are shown in the left-top. Different user interactions (first and third rows) and their corresponding correction results (second and fourth rows) are shown, to demonstrate the robustness of our method.
The experiments were performed on a PC with a 3.5 GHz Intel quad-core i7 CPU, and 16GB of RAM. For our current experimental setting, with a single core implementation, the computational time was less than 14 s for automatic segmentation and 3 s for each interactive editing. For the interactive editing procedure, it took less than 0.1 s for selecting the training labels, 0.2 s for extracting the location-adaptive features, and 0.5–2.5 s for the matrix operation of LapRLS regularization.
4. DISCUSSION
As we can observe, the proposed method outperformed the comparison methods for the most cases in terms of both accuracy and robustness. Since small numbers of dots were used for the interactions, the DSC gain of manual editing method was less than 0.01 from the original automatic segmentation result. On the other hand, the DSC gains of all training based methods were over 0.06 because the selected training labels can constrain the irregular shape variations. For the training based methods, the performance of WVA was lower than that of WVL, since the NCC score could not reflect the similarity between training label and user interactions. The performances of LapRLS + noFS and LapRLS + RFS were comparable with WVL. Even though the manifold regularization algorithm was used in LapRLS + noFS and LapRLS + RFS methods, the uninformative appearance features used in these methods still could not effectively reflect the manifold configurations between voxels. On the other hand, since the location-adaptive features (learned for separation of prostate and background in the local ROI) can enlarge the interclass distance in the feature space, the proposed method (LapRLS + FS) consistently obtained higher DSC accuracies with low standard deviations than all other training based methods. As shown in Fig. 7, the proposed method produced reasonable segmentation results even if few dots were provided as the user interaction.
We also demonstrated the robustness of the proposed method with Table II and Fig. 8. The DSC between results obtained by the proposed method was 0.005–0.02 higher than by other training based methods. We observed that some errors occurred when the interaction was not tightly provided on the false boundary, as shown in the upper-right of Fig. 8. However, the overall corrected results were still very similar to each other, and also robust to both the locations and the amount of user interactions. We note that this property helps reduce inter-rate variability from different clinicians and different edits of the same clinician.
For the computational time, the proposed method assumed that the automatic segmentation and initial registration processes could be done offline. Note that the computation of initial registration could be reduced by several ways to meet clinical setting. First of all, the registration can be conducted on a local region near the prostate, instead of the entire image as done in the current method. Another way is to use an affine registration method, instead of the nonrigid registration method. Since the appropriate training labels will be searched in the local region again in the online procedure, the affine registration should be enough for our proposed method. For the online editing process, the computational time of selection of reference training label highly depended on the size of training set. However, it took less than 0.1 s for the 54 training images, because only the local translations were considered for adjusting the registration in Subsection 2.B.3. The most computational time was consumed for the matrix operation of LapRLS regularization. When the number of unlabeled voxels nu was over 5000 for each editing, it took around 2.5 s. However, nu was less than 2000 for most cases because the unconfident region was constrained by the local ROI and the probabilistic label map in the proposed method. We expect that the computational time of matrix operation will be reduced to less than a second, if code is optimized or the algorithm is parallelized.
5. CONCLUSION
We have proposed a novel interactive method for editing the prostate segmentation in CT images. The proposed method obtains the robust editing results with a few user interactions for various wrong parts, by selecting the location-adaptive features and imposing the manifold configuration. We expect that the proposed method could largely reduce the tedious manual editing, as well as both the intra- and interobserver variability across clinicians. In the future, we will incorporate the context features to represent the relationship between the user interactions and the correction results. It is expected that the context feature based on the user interactions will complement the appearance features extracted from the confident voxels, when the reference training labels are not found well. In this way, the performance of segmentation could be further improved.
ACKNOWLEDGMENT
This work was supported in part by NIH Grant No. CA140413. Yinghuan Shi was supported by the NSFC (61305068) and Jiangsu NSF (BK20130581) grants.
REFERENCES
- 1.Davis B. C., Foskey M., Rosenman J., Goyal L., Chang S., and Joshi S., “Automatic segmentation of intra-treatment CT images for adaptive radiation therapy of the prostate,” Med. Image Comput. Comput. Assist. Interv. 8(Pt. 1), 442–450 (2005). 10.1007/11566465_55 [DOI] [PubMed] [Google Scholar]
- 2.Klein S., van der Heide U. A., Lips I. M., van Vulpen M., Staring M., and Pluim J. P., “Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information,” Med. Phys. 35(4), 1407–1417 (2008). 10.1118/1.2842076 [DOI] [PubMed] [Google Scholar]
- 3.Liao S., Gao Y., Lian J., and Shen D., “Sparse patch-based label propagation for accurate prostate localization in CT images,” IEEE Trans. Med. Imaging 32(2), 419–434 (2013). 10.1109/TMI.2012.2230018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao S. and Shen D., “A feature-based learning framework for accurate prostate localization in CT images,” IEEE Trans. Image Process. 21(8), 3546–3559 (2012). 10.1109/TIP.2012.2194296 [DOI] [PubMed] [Google Scholar]
- 5.Dowling J. A., Fripp J., Chandra S., Pluim J. P. W., Lambert J., Parker J., Denham J., Greer P. B., and Salvado O., “Fast automatic multi-atlas segmentation of the prostate from 3D MR images,” in Prostate Cancer Imaging: Image Analysis and Image-Guided Interventions (Springer, Germany, 2011), Vol. 6963, pp. 10–21. [Google Scholar]
- 6.Chen S., Lovelock D. M., and Radke R. J., “Segmenting the prostate and rectum in CT imagery using anatomical constraints,” Med. Image Anal. 15(1), 1–11 (2011). 10.1016/j.media.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 7.Costa M. J., Delingette H., Novellas S., and Ayache N., “Automatic segmentation of bladder and prostate using coupled 3D deformable models,” Med. Image Comput. Comput. Assist. Interv. 10(Pt. 1), 252–260 (2007). 10.1007/978-3-540-75757-3_31 [DOI] [PubMed] [Google Scholar]
- 8.Feng Q., Foskey M., Chen W., and Shen D., “Segmenting CT prostate images using population and patient-specific statistics for radiotherapy,” Med. Phys. 37(8), 4121–4132 (2010). 10.1118/1.3464799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin S., Troccaz J., and Daanenc V., “Automated segmentation of the prostate in 3D MR images using a probabilistic atlas and a spatially constrained deformable model,” Med. Phys. 37(4), 1579–1590 (2010). 10.1118/1.3315367 [DOI] [PubMed] [Google Scholar]
- 10.Toth R. and Madabhushi A., “Multifeature landmark-free active appearance models: Application to prostate MRI segmentation,” IEEE Trans. Med. Imaging 31(8), 1638–1650 (2012). 10.1109/TMI.2012.2201498 [DOI] [PubMed] [Google Scholar]
- 11.Zhan Y. and Shen D., in Medical Image Computing and Computer-Assisted Intervention - MICCAI 2003, edited by Ellis R. and Peters T. (Springer, Berlin Heidelberg, 2003), Vol. 2878, pp. 688–696. [Google Scholar]
- 12.Zhan Y., Shen D., Zeng J., Sun L., Fichtinger G., Moul J., and Davatzikos C., “Targeted prostate biopsy using statistical image analysis,” IEEE Trans. Med. Imaging 26(6), 779–788 (2007). 10.1109/TMI.2006.891497 [DOI] [PubMed] [Google Scholar]
- 13.Shen D., Lao Z., Zeng J., Zhang W., Sesterhenn I. A., Sun L., Moul J. W., Herskovits E. H., Fichtinger G., and Davatzikos C., “Optimized prostate biopsy via a statistical atlas of cancer spatial distribution,” Med. Image Anal. 8(2), 139–150 (2004). 10.1016/j.media.2003.11.002 [DOI] [PubMed] [Google Scholar]
- 14.Chowdhury N., Toth R., Chappelow J., Kim S., Motwani S., Punekar S., Lin H., Both S., Vapiwala N., Hahn S., and Madabhushi A., “Concurrent segmentation of the prostate on MRI and CT via linked statistical shape models for radiotherapy planning,” Med. Phys. 39(4), 2214–2228 (2012). 10.1118/1.3696376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W., Liao S., Feng Q., Chen W., and Shen D., “Learning image context for segmentation of prostate in CT-guided radiotherapy,” Med. Image Comput. Comput. Assist. Interv. 14(Pt. 3), 570–578 (2011). 10.1007/978-3-642-23626-6_70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y., Liao S., Gao Y., Zhang D., and Shen D., “Prostate segmentation in CT images via spatial-constrained transductive lasso,” in IEEE Conference on Computer Vision and Pattern Recognition (IEEE, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai X. and Sapiro G., “A geodesic framework for fast interactive image and video segmentation and matting,” in IEEE International Conference on Computer Vision (IEEE, 2007), pp. 809–816. [Google Scholar]
- 18.Boykov Y., Veksler O., and Zabih R., “Fast approximate energy minimization via graph cuts,” IEEE Trans. Pattern Anal. 23(11), 1222–1239 (2001). 10.1109/34.969114 [DOI] [Google Scholar]
- 19.Grady L., “Random walks for image segmentation,” IEEE Trans. Pattern Anal. 28(11), 1768–1783 (2006). 10.1109/TPAMI.2006.233 [DOI] [PubMed] [Google Scholar]
- 20.Kim T. H., Lee K. M., and Lee S. U., “Nonparametric higher-order learning for interactive segmentation,” in IEEE Conference on Computer Vision and Pattern Recognition (IEEE, 2010), pp. 3201–3208. [Google Scholar]
- 21.Rother C., Kolmogorov V., and Blake A., “‘GrabCut’—Interactive foreground extraction using iterated graph cuts,” Acm. Trans. Graphic. 23(3), 309–314 (2004). 10.1145/1015706.1015720 [DOI] [Google Scholar]
- 22.Top A., Hamarneh G., and Abugharbieh R., “Active learning for interactive 3D image segmentation,” Med. Image Comput. Comput. Assist. Interv. 6893, 603–610 (2011). 10.1007/978-3-642-23626-6_74 [DOI] [PubMed] [Google Scholar]
- 23.Wang D., Yan C., Shan S., and Chen X., “Active learning for interactive segmentation with expected confidence change,” in ACCV (Springer, Germany, 2013), Vol. 7724, pp. 790–802. [Google Scholar]
- 24.Barnes C., Shechtman E., Finkelstein A., and Goldman D. B., “PatchMatch: A randomized correspondence algorithm for structural image editing,” Acm. Trans. Graphic. 28(3), 24:1–24:11 (2009). 10.1145/1531326.1531330 [DOI] [Google Scholar]
- 25.Barnes C., Shechtman E., Goldman D. B., and Finkelstein A., “The generalized PatchMatch correspondence algorithm,” in ECCV (Springer, Germany, 2010), Vol. 6313, pp. 29–43. [Google Scholar]
- 26.Shi W. Z., Caballero J., Ledig C., Zhuang X. H., Bai W. J., Bhatia K., de Marvao A. M. S. M., Dawes T., O’Regan D., and Rueckert D., “Cardiac image super-resolution with global correspondence using multi-atlas PatchMatch,” Med. Image Comput. Comput. Assist. Interv. 8151, 9–16 (2013). 10.1007/978-3-642-40760-4_2 [DOI] [PubMed] [Google Scholar]
- 27.Ta V.-T., Giraud R., Collins D. L., and Coupe P., “Optimized PatchMatch for near real time and accurate label fusion,” Med. Image Comput. Comput. Assist. Interv. 8675, 105–112 (2014). 10.1007/978-3-319-10443-0_14 [DOI] [PubMed] [Google Scholar]
- 28.Park S. H., Yun I. D., and Lee S. U., “Data-driven interactive 3D medical image segmentation based on structured patch model,” Inf. Process Med. Imaging 23, 196–207 (2013). 10.1007/978-3-642-38868-2_17 [DOI] [PubMed] [Google Scholar]
- 29.Gao Y., Wang L., Shao Y., and Shen D., “Learning distance transform for boundary detection and deformable segmentation in ct prostate images,” in Machine Learning in Medical Imaging, MICCAI (Springer, Germany, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glocker B., Komodakis N., Tziritas G., Navab N., and Paragios N., “Dense image registration through MRFs and efficient linear programming,” Med. Image Anal. 12(6), 731–741 (2008). 10.1016/j.media.2008.03.006 [DOI] [PubMed] [Google Scholar]
- 31.Belkin M., Niyogi P., and Sindhwani V., “Manifold regularization: A geometric framework for learning from labeled and unlabeled examples,” J. Mach. Learn. Res. 7, 2399–2434 (2006). [Google Scholar]
- 32.Criminisi A., Robertson D., Konukoglu E., Shotton J., Pathak S., White S., and Siddiqui K., “Regression forests for efficient anatomy detection and localization in computed tomography scans,” Med. Image Anal. 17(8), 1293–1303 (2013). 10.1016/j.media.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 33.Gao Y., Liao S., and Shen D., “Prostate segmentation by sparse representation based classification,” Med. Phys. 39(10), 6372–6387 (2012). 10.1118/1.4754304 [DOI] [PMC free article] [PubMed] [Google Scholar]