Abstract
Ethanol abuse can lead to addiction, brain damage and premature death. The cycle of alcohol addiction has been described as a composite consisting of three stages: intoxication, withdrawal and craving/abstinence. There is evidence for contributions of both genotype and sex to alcoholism, but an understanding of the biological underpinnings is limited. Utilizing both sexes of genetic animal models with highly divergent alcohol withdrawal severity, Withdrawal Seizure-Resistant (WSR) and Withdrawal Seizure-Prone (WSP) mice, the distinct contributions of genotype/phenotype and of sex during addiction stages on neuroadaptation were characterized. Transcriptional profiling was performed to identify expression changes as a consequence of chronic intoxication in the medial prefrontal cortex. Significant expression differences were identified on a single platform and tracked over a behaviorally-relevant time course that covered each stage of alcohol addiction; i.e., after chronic intoxication, during peak withdrawal, and after a defined period of abstinence. Females were more sensitive to ethanol with higher fold expression differences. Bioinformatics showed a strong effect of sex on the data structure of expression profiles during chronic intoxication and at peak withdrawal irrespective of genetic background. However, during abstinence, differences were observed instead between the lines/phenotypes irrespective of sex. Confirmation of identified pathways showed distinct inflammatory signaling following intoxication at peak withdrawal, with a pro-inflammatory phenotype in females but overall suppression of immune signaling in males. Combined, these results suggest that each stage of the addiction cycle is influenced differentially by sex vs. genetic background and support the development of stage- and sex-specific therapies for alcohol withdrawal and the maintenance of sobriety.
Keywords: prefrontal cortex, ethanol, sexual dimorphism, inflammation, astrocytes, low level response to alcohol
1. Introduction
Alcohol use disorder, a chronic relapsing disease, is a well-recognized public health problem and is one of the leading preventable causes of death worldwide (Rehm et al., 2009). Unfortunately, ethanol is the most common substance of abuse in the US, and abuse can lead to physical dependence and addiction. Alcoholism is a complex disease with multiple risk factors, and there is evidence for both sexual-dimorphism and a genetic contribution in both the risk to develop the disorder and in the detrimental responses that result from alcohol abuse. In general, the alcohol addiction cycle has been described by Koob et. al. as consisting of three stages: intoxication, withdrawal and craving/abstinence (Koob and Volkow, 2010). However, understanding of the biological underpinnings of alcohol addiction is limited. In particular, the impact of sex/gender vs. genetic background/phenotype on each stage of the addiction cycle has not been characterized.
Previous reports have demonstrated that sex/gender influences the response to alcohol (Ceylan-Isik et al., 2010). Specific sex differences have been reported in terms of alcohol handling (Addolorato et al., 1999), with body composition differences influencing ethanol partitioning between lipid and water compartments. In addition, females have reduced levels of alcohol dehydrogenase, a key liver enzyme involved in alcohol metabolism and removal (Baraona et al., 2001). Most notably, sex differences have been characterized in numerous alcohol-related behaviors in which females show: reduced risk for the development of ethanol dependence and addiction; increased ethanol sensitivity; increased ethanol consumption; reduced withdrawal severity; and increased risk of recidivism and relapse (Brady and Randall, 1999, Devaud and Chadda, 2001, Sershen et al., 2002, Wang et al., 2003, Carroll et al., 2004, Prescott et al., 2005, Becker and Hu, 2008, Potts et al., 2013). Alcohol may also be more rewarding in females (Blanchard et al., 1993, Torres et al., 2014). Combined, such data stress the importance of sex-specific analysis and place a particular emphasis on improved understanding of mechanisms underlying such responses in female alcoholics (see Wiren, 2013).
In addition to sex differences, the genetic contribution to the overall development of alcoholism is well described (Magnusson et al., 2010). However, understanding of the risks associated with specific polymorphisms/mutations or biological pathways remains quite limited (for review see Han et al., 2013). Given that alcoholism is a disorder of complex genetics with multiple genetic loci that are influenced by interactions with the environment, dissecting genetic contributions to the disease in diverse human populations has proven difficult. Thus, various animal models have been developed that show increased or decreased sensitivity to ethanol as phenotypes. The use of selectively bred rodent lines provides a genetically rich model where the various alleles present in the initial heterogeneous population related to the selection phenotype become differentially segregated with selection pressure in the respective lines. Thus, differences observed in selected lines provide evidence of the genetic underpinnings that influence the trait of interest. Our studies have employed lines of mice with highly divergent withdrawal severity after chronic intoxication derived by selective breeding from heterogeneous stock; the low response to alcohol withdrawal Withdrawal-Seizure Resistant (WSR) and high response Withdrawal-Seizure Prone (WSP) mouse lines (Kosobud and Crabbe, 1986). Evidence of physical dependence is considered a hallmark of alcoholism; one measure of physical dependence is increased neuronal excitability including seizures. Such hyperexcitability during withdrawal is thought to reflect neuroadapations that occur with chronic ethanol intoxication, including changes in gene expression and brain structure which enable an organism to function in the presence of this central nervous system (CNS) depressant. Thus, the WSR and WSP lines are of interest because the large differences in response to chronic alcohol, as evidenced by divergent withdrawal severity, are believed to reflect distinct neuroadaptive responses between the phenotypes that occur with chronic intoxication. Furthermore, WSR and WSP lines also demonstrate sex differences similar to humans since females consume more alcohol yet exhibit reduced withdrawal severity in these lines (Kosobud and Crabbe, 1986).
Current treatments for alcohol dependence are at best only modestly effective (Olive, 2010, Zindel and Kranzler, 2014). Since individuals with alcohol dependence represent a clinically heterogeneous and genetically diverse population, it has been proposed that effective treatments should target specific phenotypes at distinct stages of addiction rather than employ a generic approach to all patients (Kuehn, 2009). Although sex and genotype both contribute to risks of dependence and addiction, the distinct contribution that each may subserve during the stages of the addiction cycle remains uncharacterized as no systematic analyses of the impact of sex vs. genotype on neuroadaptive responses during the addiction cycle following chronic intoxication have been reported. In this work, the WSR and WSP selected lines of mice were employed as preclinical models of genomically-rich widely divergent “response to alcohol” phenotypes and both sexes were examined. Analysis was done using tissue from the medial prefrontal cortex (mPFC). In addition to involvement in the addition cycle (Koob and Volkow, 2010), the mPFC participates in hyperexcitability circuitry during withdrawal from chronic exposure (Chen et al., 2009), is important in executive function and inhibitory control (Fuster, 2002, Kroener et al., 2012), and is associated with cognitive dysfunction and damage in alcoholics (Zahr et al., 2011). Analysis was performed at biologically relevant endpoints during the addiction cycle, i.e., after chronic exposure, during peak withdrawal and after a defined period of abstinence following chronic ethanol intoxication, to identify important contributors in the neuroadaptive response. For this comprehensive addiction stage analysis, we employed the same animal models, the same chronic ethanol exposure paradigm and the same array platform, to compare and contrast expression differences over an addiction time course in a defined system. Results demonstrate for the first time that sex and genotype/phenotype have distinct and varying influences on neuroadapation and result in divergent biological response pathways during each stage of the addiction cycle.
2. Materials and methods
2.1. Animal subjects
Two independently derived replicate WSP and WSR lines (Kosobud and Crabbe, 1986) were generated by selective breeding for divergent withdrawal severity from genetically heterogeneous HS/Ibg mice. Female and male mice from both replicates (i.e., WSR-1, -2 and WSP-1, -2) were tested for expression differences. To identify phenotype-specific differences, expression analysis was collapsed on replicate for each selected line. As these lines are employed to identify genetic underpinnings of the selected phenotype, comparisons between the WSR and WSP mice are referred to as either phenotype, genotype or line differences. Mice were maintained under a light/dark cycle of 0600–1800 light with water and Purina Lab Diet chow available ad libitum. Room temperatures were maintained at 22 ± 1° C. Ethanol (20% v/v) was mixed with 0.9% saline and injected intraperitoneally (i.p.) or introduced without mixing as a vapor into the chambers. All animal procedures were carried out in accordance with the US National Institutes of Health guide for the care and use of laboratory animals (NIH Publications No. 8023, revised 1978). All procedures were approved by the Portland Oregon VA Medical Center Institutional Animal Care and Use Committee, which mandates that all efforts are made to minimize animal suffering, to reduce the number of animals used, and to utilize alternatives to in vivo techniques, if available.
2.2. Chronic ethanol intoxication and brain harvest
Mice were made dependent upon ethanol using a method of vapor inhalation in chambers manufactured in-house, with modifications previously published (Beadles-Bohling and Wiren, 2006, Hashimoto and Wiren, 2008, Hashimoto et al., 2011). This paradigm highlights vulnerability to the effects alcohol, consisting of a single chronic exposure followed by a single synchronized withdrawal. Drug-naïve adult mice from selected generation 26 (filial generations G87 - G116) were used. Mice were injected i.p. with ethanol at 1.5 g/kg for WSP-1, WSR and WSR-2 and 1.75 g/kg for WSP-2 animals, necessary to maintain similar blood ethanol concentrations (BECs) between the selected lines (Terdal and Crabbe, 1994), and 68.1 mg/kg pyrazole HCl (Pyr; an alcohol dehydrogenase inhibitor used to maintain constant blood ethanol levels). Briefly, control animals were placed into air chambers and received Pyr only; a saline-only air control was not included because previous data has shown there is no difference between saline and Pyr treated animals with respect to broad profiles of gene expression analyzed by mRNA differential display (Wiren unpublished observations and Schafer et al., 1998). Ethanol exposed mice had 20 μl of blood taken from the tail daily and following 72 h of constant ethanol vapor exposure for BEC determination by gas chromatography as previously described (Beadles-Bohling and Wiren, 2006). Administration of ethanol via inhalation allows for synchronized withdrawal after high levels of chronic intoxication, which are difficult to achieve in human populations or using voluntary drinking approaches. All animals used in these experiments were purposely not handled to limit the effects of handling-induced withdrawal seizures per se on measurements of gene expression. During withdrawal these mice typically show decreased activity, dysphoria, and mild tremor, but without handling do not show convulsions. Brain tissue was harvested for RNA analysis from animals after chronic intoxication (0 h), at peak withdrawal (8 h) and after a defined period of abstinence (21 d). The mPFC was harvested after careful dissection as previously described (Hashimoto and Wiren, 2008, Hashimoto et al., 2011). The isolated mPFC weighed on average 20 mg. Tissues were snap frozen in liquid nitrogen and stored at −80°C until processing.
2.3. RNA Isolation and GeneFilter microarray processing
Microarray analysis was performed as described in detail previously (Hashimoto and Wiren, 2008, Hashimoto et al., 2011). Briefly, total RNA was DNase treated and probe labeling was performed by linear synthesis with 33P-dATP incorporation using the Array Advantage kit (Ambion, Austin, TX). Microarray hybridization with complex labeled RNA probe was performed overnight with the final post-hybridization wash at 50°C in 0.5X SSC, 0.5% SDS, similar to standard Northern blot procedures. Research Genetics GF400 mouse microarrays (Invitrogen, Carlsbad, CA) employed for these studies contained 4696 clones with unique mouse accession numbers (both genes and ESTs) that average 1500 bp in length, generally with one spot per cDNA. The GF400 mouse microarray represents an enriched population of sequences with known biological associations, thus providing a more useful analysis than sequences with unknown function. Bioinformatic analysis of the clones represented on the array showed enriched proportions of “Integrin Signaling”, “Actin Nucleation by ARP-WASP Complex”, “B Cell Receptor Signaling”, “Glioblastoma Multiforme Signaling” and “Chronic Myeloid Leukemia Signaling” genes compared to the mouse genome as identified by Ingenuity Pathway Analysis (IPA) (Qiagen, Valencia, CA). Notably, for all enrichment analyses, the GF400 genes were used as the reference gene set to reduce bias associated with genes present on the array. Spots with hybridization values below background hybridization values were removed from analyses. Raw expression values were normalized and analyzed using Vector Xpression 3.1 (Invitrogen, Carlsbad, CA). Expression differences were analyzed using a Student’s t-test (n = 3–4 for each group, two-tailed, α = 0.05) to identify a large group of differentially regulated transcripts.
2.4. Bioinformatic data analyses
2.4.1. Unsupervised hierarchical clustering analysis
Unsupervised hierarchical clustering was performed using the group of significantly regulated genes at each stage of addiction. Clusters were generated (average linkage, Euclidian distance) with 100 resampling iterations to determine Bootstrap values for each node using the TIGR Multiple Experiment Viewer (TMEV) software from The Institute For Genomic Research (Saeed et al., 2003). Limited characterization of some of these data sets have been published previously (Hashimoto and Wiren, 2008, Hashimoto et al., 2011). The analyses performed in this report provide for the first time analysis for chronic intoxication and for a combined comparison over the addiction time course. All datasets have been deposited at GEO with the reference numbers GSE23165 for abstinence, GSE56249 for peak withdrawal and GSE56247 for intoxication data.
2.4.2. Weighed Gene Coexpression Network Analysis (WGCNA)
Networks were constructed for correlation of normalized gene expression over samples specified by variables of sex, selected line/genotype, ethanol treatment and time/addiction stage using the Weighed Gene Coexpression Network Analysis (WGCNA) method (Zhang and Horvath, 2005). Data for the two replicate lines of mice (i.e., WSR-1, -2 and WSP-1, -2) were collapsed to focus on specific phenotype for WSR (low response) or WSP (high response). Four replicates from each sex (M or F), genotype (WSR or WSP), stage (0h, 8h, or 21d) and treatment (control (C) or ethanol (E)) were used for a total of 96 arrays. The resulting data set had 95 arrays, with one array excluded from analysis based on correlation coefficients values below 90% for ‘replicate’ arrays and additionally because of poor clustering in WGCNA analysis. For WGCNA analysis, all genes were hierarchically clustered based on a dissimilarity measure of topological overlap which measures inter-connectedness for a pair of genes (Zhang and Horvath, 2005). The resulting gene dendrogram was used for module detection with the dynamic tree cut method (minimum module size, 27; cutting height, 0.99; and deepSplit = True). Gene modules generated from this detection were individually labeled in a unique color, with unassigned groups labeled in gray.
2.4.3. Overrepresentation analysis and functional annotation of genes
Several complementary approaches were employed to characterize the biological consequences of identified expression differences and for the significant gene modules observed in network analysis. To identify cellular pathways and biological themes that were affected, we used DAVID (the Database for Annotation, Visualization and Integrated Discovery v6.7 (http://david.niaid.nih.gov/david/ease.htm) (Huang et al., 2009a, b) to assign genes to the categories of the Gene Ontology (GO) Consortium (www.geneontology.org) and to test statistically for overrepresentation. GeneCards (http://www.genecards.org) was employed to identify diseases and/or super-pathway associations. Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) was used to identify sets of genes overrepresented in other complex diseases or treatments. GSEA compares user input datasets to curated sets of genes previously identified as related, either by function, location, process, experimental manipulation, disease or other variables. Significant overlap in the user input data and curated gene sets suggests an underlying relationship between the two experiments. Input was the set of ethanol regulated genes for each comparison at each stage, resulting in 12 independent analyses. Each analysis was compared against the GSEA curated gene set (c2, v4.0). Finally, functional association networks were constructed to identify interactions or relationships between regulated genes that may not have formal GO annotations using IPA. IPA was also used to identify significant overlap between our data and genes known to be expressed in specific CNS cell types. These approaches were employed to better characterize transcriptional differences and to generate mechanistic hypotheses underlying neuroadaptive responses during the stages of addiction and the effects of sex vs. genotype.
2.5. Characterization of inflammatory signaling
Characterization of the inflammatory response that follows chronic intoxication and withdrawal was performed using focused qPCR arrays for mouse Th1 & Th2 Responses (SABiosciences, Frederick, MD). These focused arrays survey for transcriptional differences in T cell response. RNA was isolated from the mPFC of control or alcohol-exposed male and female WSR-1 mice at peak withdrawal, and focused qPCR analysis was carried out using a Bio-Rad iCycler system (Bio-Rad Laboratories, Inc. Hercules, CA). Gene expression levels were calculated by the comparative ΔΔCt method and normalized as previously described (Hashimoto and Wiren, 2008, Hashimoto et al., 2011). Expression differences were determined using fold change and significance tested using an uncorrected Student’s t-test (n = 3 for each group, except one control male sample was lost due to a technical issue).
2.6. Statistical analyses
Data were analyzed using Prism v6.04 software (GraphPad Software, Inc., San Diego, CA), SYSTAT 13 (SYSTAT Software Inc., Point Richmond, CA), Microsoft Excel 2010 (Microsoft Corp., Redmond, WA) and Vector Xpression 3.1 (Invitrogen, Carlsbad, CA). Differences of p < 0.05 were considered statistically significant. Results are presented as the mean ± SEM. Comparison of overall BECs between males and females and between WSR and WSP lines were analyzed by two-tailed Student’s t-test. Individual differences in gene expression were also analyzed by two-tailed t-test between ethanol treated and control samples from each sex and line.
3. Results
3.1. Modeling the addiction cycle time course: chronic ethanol exposure, synchronized withdrawal and defined period of abstinence
Neuroadaptive gene expression changes following chronic ethanol exposure were identified in mPFC. Analysis included both sexes and both genotypes/lines at three addiction cycle stages: during high intoxication after chronic ethanol exposure at 0h (with the development of physical dependence); followed by synchronized peak withdrawal hyperexcitability at 8h; and after a defined period of abstinence from alcohol at 21d.
At the onset of the experiments, animals were 70.0 ± 0.63 d old, and body weights were 24.4 ± 0.22 g. During exposure, BECs were monitored daily and ethanol vapor levels were modified to achieve a level with high intoxication targeted at BECs of 2.1 – 2.4 mg/ml (46 – 52 mM). This paradigm provided an average BEC of 2.27 ± 0.04 mg/ml (49.27 mM) over the course of these experiments, with BECs (mean over the 72 h exposure period) in animals tested at 0h of 2.02 ± 0.35 mg/ml, at 8 h of 2.41 ± 0.05 mg/ml and 21d of 2.30 ± 0.08 mg/ml. Comparisons between the sexes by t-test showed no difference in overall BECs between the sexes (mean female BEC: 2.25 ± 0.041 vs. male BEC: 2.28 ± 0.08; p < 0.74); however as a group WSR mice had slightly lower BECs than WSP mice (WSR BEC: 2.16 ± 0.055 vs. WSP BEC: 2.36 ± 0.057; p < 0.05). Such modest differences in BEC do not reliably impact behavior, as previous studies have shown similar withdrawal profiles between WSP mice (and also between WSR mice) with a wide range of BECs after chronic intoxication (0.38 mg/ml – 2.88 mg/ml), as assessed with handling-induced convulsions (see Terdal and Crabbe, 1994, Finn and Crabbe, 1999). Importantly, these blood levels are within the range seen (2.0–3.0 mg/ml) in most alcoholic patients (Adachi et al., 1991) and are above the legal limit for driving (17 mM, 0.8 mg/ml or 0.08% in most US states).
3.2. Expression differences identified over the course of the addiction cycle
The array approach employed here has been shown repeatedly to reliably detect expression differences following chronic alcohol treatment (Hashimoto and Wiren, 2008, Hashimoto et al., 2011). Neuroadaptation, i.e. those specific changes in brain structure and gene expression patterns that result from high intoxication, remains poorly characterized. This may be a reflection of the very modest effects of ethanol on gene expression levels (Treadwell and Singh, 2004, Hashimoto and Wiren, 2008, Hashimoto et al., 2011). In addition, little biological confirmation of the important pathways altered as a consequence of alcohol exposure as identified in array analysis has been reported to date. Instead of analysis of neuroadapation, much transcriptional profiling has focused on inherent genetic differences observed between alcohol-related phenotypes (see Iancu et al., 2013, McBride et al., 2013). Compared with RNA-seq, hybridization arrays have reduced dynamic range but improved sensitivity (see Malone and Oliver, 2011). Improved sensitivity helps reduce stochastic noise and is advantageous particularly for subtle phenotypes and the relatively modest fold expression differences observed with alcohol exposure. The array technology employed here, with longer hybridization targets, linear synthesis of probe and sensitive hybridization and wash conditions, has improved signal-to-noise for reliable detection of such modest expression differences. Most importantly, the biological pathways altered as a result of chronic alcohol exposure and withdrawal identified using this platform have been repeatedly confirmed (Hashimoto and Wiren, 2008, Hashimoto et al., 2011).
Significantly regulated transcripts were identified at each stage of addiction in both male and female WSR and WSP mice. The transcripts that were the most robustly regulated by ethanol in females and males are listed in Tables 1 and 2, respectively. We used an uncorrected p-value to decrease the chance of excluding regulated transcripts (i.e., false negatives) as we (Hashimoto and Wiren, 2008, Hashimoto et al., 2011), and others (for example, see Rodd et al., 2007) have employed. Prior confirmation of expression differences identified by hybridization using qPCR was ~88% (Hashimoto et al., 2011), similar to our previous finding of an ~80% confirmation rate (Hashimoto and Wiren, 2008). This rate of confirmation is high relative to other array platforms and analytical approaches (see Treadwell and Singh, 2004, Tang et al., 2006). Overall, nearly 20% of genes examined demonstrated significant ethanol regulation during any time point in sex- and line-specific comparisons (819 of 4696 genes queried). Expression differences were modest, with the absolute value of most ratios less than 1.3.
Table 1.
Largest expression differences in females as a consequence of chronic intoxication
| Symbol | Clone Accession | Gene ID | Ethanol Regulation | Significant Comparison |
|---|---|---|---|---|
| Arl5a | AI465424 | 75423 | −2.251 | 0hr FR |
| Pnpla6 | AI415470 | 50767 | −1.930 | 8hr FP |
| Qdpr | AI325507 | 110391 | −1.628 | 0hr FR |
| Kif3a | AI429298 | 16568 | −1.493 | 8hr FP |
| Nck2 | AI324113 | 17974 | −1.363 | 8hr FP |
| Sh3yl1 | AI447735 | 24057 | −1.327 | 21d FP |
| Wdr81 | AI413469 | 192652 | −1.224 | 8hr FP |
| Prpf4 | AI413933 | 70052 | −1.187 | 0hr FP |
| Lgals1 | AI465143 | 16852 | −1.186 | 21d FP |
| Grpel2 | AI449184 | 17714 | −1.180 | 0hr FR |
| Tnfsf10 | AI427583 | 22035 | −1.179 | 8hr FP |
| Ncald | AI415396 | 52589 | −1.153 | 0hr FR |
| Spo11 | AI449549 | 26972 | −1.139 | 21d FP |
| Hfe2 | AI414844 | 69585 | −1.135 | 8hr FP |
| Ube2j2 | AI449455 | 140499 | −1.129 | 21d FP |
| Tfdp2 | AI451300 | 211586 | −1.114 | 0hr FR |
| Hbxip | AI448871 | 68576 | −1.109 | 21d FP |
| 1600014C10Rik | AI428873 | 72244 | −1.108 | 0hr FR |
| Rere | AI450210 | 68703 | −1.088 | 21d FP |
| Pi16 | AI448241 | 74116 | −1.081 | 21d FP |
|
| ||||
| Hexim1 | AI426204 | 192231 | 1.212 | 21d FP |
| Fam135a | AI465387 | 68187 | 1.259 | 0hr FP |
| Ccdc88a | AI450288 | 108686 | 1.382 | 0hr FR |
| Lepr | AI323343 | 16847 | 1.393 | 0hr FP |
| Naa50 | AI427986 | 72117 | 1.401 | 0hr FR |
| Alyref | AI465506 | 21681 | 1.405 | 0hr FR |
| Klhdc5 | AI451576 | 232539 | 1.413 | 0hr FR |
| Lfng | AI451068 | 16848 | 1.416 | 0hr FR |
| 9430038I01Rik | AI413095 | 77252 | 1.424 | 0hr FP |
| Ttc27 | AI450085 | 74196 | 1.448 | 0hr FR |
| 4930422G04Rik | AI450087 | 71643 | 1.451 | 0hr FR |
| Fam193a | AI414696 | 231128 | 1.452 | 0hr FR |
| Rhoc | AI324259 | 11853 | 1.453 | 0hr FR |
| Tgfbi | AI661287 | 21810 | 1.463 | 0hr FR |
| Taz | AI661004 | 66826 | 1.525 | 0hr FR |
| Pdzrn3 | AI429718 | 55983 | 1.528 | 0hr FR |
| Hnrnpa1 | AI413150 | 15382 | 1.540 | 0hr FR |
| Mphosph8 | AI660999 | 75339 | 1.641 | 0hr FR |
| Tnks1bp1 | AI429725 | 228140 | 1.710 | 0hr FR |
Significantly up- and down-regulated genes were identified for female WSR and WSP mice at each addiction stage. This table shows the most highly regulated genes ranked by ethanol regulation with the most down-regulated genes listed at the top of the table. Ethanol regulation values denote log2 fold regulation (EtOH/control).
Table 2.
Largest expression differences in males following chronic intoxication
| Symbol | Clone Accession | Gene ID | Ethanol Regulation | Significant Comparison |
|---|---|---|---|---|
| Dcaf11 | AI414484 | 28199 | −0.960 | 21d MP |
| Mterfd3 | AI447436 | 74238 | −0.941 | 8hr MP |
| Hmha1 | AI430932 | 70719 | −0.902 | 21d MP |
| Atl3 | AI428126 | 109168 | −0.894 | 21d MP |
| Rpl38 | AI327009 | 67671 | −0.886 | 21d MP |
| Cbr1 | AI323923 | 12408 | −0.864 | 21d MP |
| Zbed3 | AI428386 | 72114 | −0.844 | 8hr MP |
| Tbcd | AI427676 | 108903 | −0.836 | 21d MP |
| Gpr56 | AI604830 | 14766 | −0.821 | 8hr MP |
| Sema6a | AI414953 | 20358 | −0.819 | 21d MP |
| Gnmt | AI326151 | 14711 | −0.817 | 21d MP |
| Yeats2 | AI447638 | 208146 | −0.813 | 8hr MR |
| Rbm25 | AI451252 | 67039 | −0.796 | 8hr MR |
| Pacs2 | AI429435 | 217893 | −0.795 | 21d MP |
| Slc10a7 | AI662162 | 76775 | −0.794 | 8hr MR |
| Fez1 | AI415253 | 235180 | −0.794 | 8hr MR |
| Nudt12 | AI323961 | 67993 | −0.784 | 21d MP |
| Thsd1 | AI448749 | 56229 | −0.783 | 8hr MP |
| Glb1 | AI528555 | 12091 | −0.782 | 21d MP |
| Rras | AI573426 | 20130 | −0.767 | 8hr MP |
|
| ||||
| Sclt1 | AI449339 | 67161 | 0.742 | 8hr MR |
| Zswim7 | AI426230 | 69747 | 0.756 | 21d MR |
| Dkkl1 | AI414495 | 50722 | 0.761 | 21d MR |
| Psma1 | AI325451 | 26440 | 0.762 | 8hr MR |
| Exoc3l2 | AI414503 | 74463 | 0.767 | 21d MR |
| Svop | AI415691 | 68666 | 0.781 | 21d MR |
| Nkiras2 | AI596353 | 71966 | 0.789 | 21d MR |
| Ppp6r3 | AI449653 | 52036 | 0.790 | 0hr MR |
| Cbr1 | AI323923 | 12408 | 0.791 | 0hr MP |
| Rras | AI573426 | 20130 | 0.796 | 21d MR |
| Atp6v0c | AI385732 | 11984 | 0.802 | 21d MR |
| Cx3cl1 | AI413869 | 20312 | 0.806 | 0hr MR |
| 4931428F04Rik | AI426165 | 74356 | 0.817 | 21d MR |
| Bin3 | AI464318 | 57784 | 0.842 | 21d MP |
| BC006965 | AI413436 | 217294 | 0.849 | 21d MR |
| Acvrl1 | AI427544 | 11482 | 0.854 | 8hr MP |
| Toe1 | AI464326 | 68276 | 0.867 | 21d MP |
| Zswim7 | AI426230 | 69747 | 0.936 | 8hr MR |
| Alg6 | AI451111 | 320438 | 0.999 | 8hr MR |
Up- and down-regulated genes were identified at each stage for male WSR and WSP mice. This table shows the most highly regulated genes ranked by ethanol regulation with the most down-regulated genes at the top of the table. Ethanol regulation values denote log2 fold regulation (EtOH/control).
Notably, the most highly regulated transcripts were observed almost exclusively in females in either line or time comparisons, suggesting that females are more sensitive to ethanol. In the list of most responsive genes, there were no genes that were regulated in both males and females in any comparison. Zswim7, involved in repair of damaged DNA (Liu et al., 2011), was significantly up-regulated by ethanol in male WSR mice at two time points; during peak withdrawal and also during abstinence.
Functional enrichment analysis was performed using the online software DAVID to derive an overview of pathways that may be especially sensitive ethanol targets. Unsurprisingly given the lack of overlap in the gene lists, sexually dimorphic responses were observed. The top annotation cluster for the most highly regulated genes in females (enrichment score = 1.73) involved female sexual differentiation/gonadal development while in males, the top annotation cluster included genes involved in transmembrane action/signal peptide (enrichment score = 0.58). Characterization of related super-pathways and diseases associated with the most robustly regulated genes using GeneCards also identified sexually dimorphic patterns. Females showed regulation of genes associated with gonadal/breast/prostate diseases (Arl5a, Spo11, Hfe2, Pi16, Rhoc, Pdzrn3; Tnks1bp1); cell death/neurodegeneration (Wdr81, Prpf4, Lgals1, Ncald, Rere, Tfdp2, Ccdc88a, Klhdc5, Pnpla6); and in inflammation/immune function (Kif3a, Sh3yl1, Hbxip, Whsc2, Hexim1). In contrast in males, ethanol influenced genes involved in nervous system disorders/development (Atl3, Cbr1, Gpr56, Sema6a, Rbm25, Fez1, Dkkl1, Psma1); inflammation/immune function (Hmha1, Nkiras2, Cx3cl1); mitochondria or oxidoreductase/hydroxylase activities (Mterfd3, Pacs2, Cbr1); lysosome/peroxisome (Nudt12, Glb1, Atp6v0c). Although inflammation/immune function was a prominent response in both males and females in this analysis, distinct targets were regulated between the sexes.
3.3. Characterization of alcohol-regulated expression patterns and pathways in WSR and WSP male and female mice
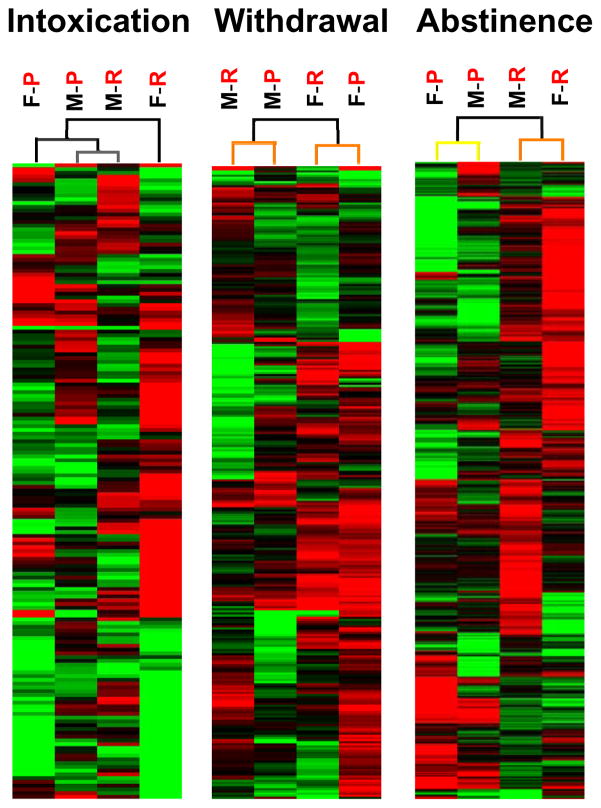
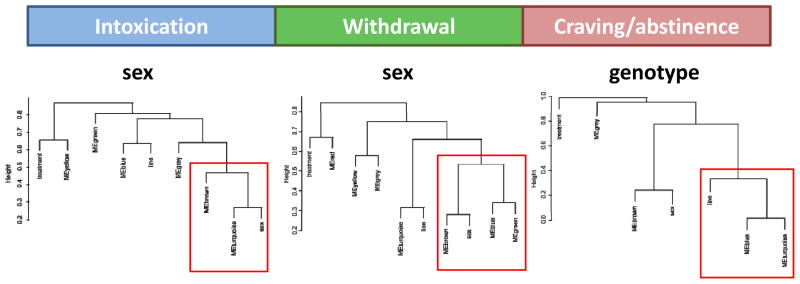
A variety of bioinformatic approaches were employed to characterize the structure of expression differences identified in array analysis. First, the overall impact of sex, line and time was identified using unsupervised hierarchical cluster analysis of the set of significantly regulated genes. As shown in Figure 1, genotype vs. sex had disparate impacts on the neuroadaptive ethanol response at each stage of the addiction cycle. In this analysis, sex determines the clustering of differential gene expression at 8h (peak withdrawal) and has an important influence during chronic intoxication (0h), while genotype most strongly influenced gene expression during abstinence (21d). Bootstrap analysis of clustering using 100 iterations is indicated by the color of the lines in the dendrogram shown above the heat map, with separation of nodes colored black indicating100% recovery, grey indicating 90–100% recovery, yellow indicating 60–70% recovery, and orange indicating 50–60% recovery. Thus, nodes representing clustering during intoxication indicate 90–100% recovery on repeated testing, those during withdrawal at 50–100% recovery and during abstinence between 50–100% recovery. Of note, during withdrawal the separation based on sex and during abstinence the separation based on line are strongly supported with 100% recovery.
Figure 1.
Hierarchical clustering (HCL) at each stage of addiction shows distinct influence of sex vs. line at different stages. Characterization of structural hierarchy for genes that were significantly regulated after chronic ethanol treatment. During the early stages of intoxication and withdrawal, sex is the most important influence on expression differences while during abstinence, selected phenotype/genotype was the strongest influence on expression differences. Boot-strap analysis (100 iterations) of clustering is indicated in the color of the lines in the dendrogram shown above the heat map; with black indicating 100% recovery, grey indicating 90–100% recovery, yellow indicating 60–70% recovery, and orange indicating 50–60% recovery. Each column represents combined analysis from 8 arrays with two mice per array. Gene expression is indicated by color (green (up-regulated), or red (down-regulated), with the intensity of the color proportional to the intensity of regulation. F, female. M, male. P, WSP. R, WSR.
To identify important signaling cascades altered by chronic ethanol administration, IPA was employed at each stage of addiction using genes that were significantly regulated following ethanol treatment. To leverage the structure identified in unsupervised hierarchical clustering, analysis was focused on sex-specific changes at intoxication and during withdrawal, while selected line/genotype differences during abstinence were characterized. With chronic intoxication, analysis in females identified Sphingosine-1-phosphate Signaling as the top canonical pathway (p < 0.01) while in males ethanol-regulated genes were associated with Telomere Extension by Telomerase (p < 0.01). During peak withdrawal, the top canonical pathway was p38 MAPK Signaling in females (p < 0.05) while Retinoate Biosynthesis (p < 0.05) was the top pathway in males. During abstinence, the top pathway in the low response WSR model was Atherosclerosis Signaling (p < 0.05) while in the WSP model the top pathway was Phosphatidylethanolamine Biosynthesis II (p < 0.05). Thus at each time point, disparate canonical pathways were targeted by chronic ethanol exposure and were differentially influenced by sex or genotype.
GSEA was next employed to identify biological pathways associated with neuroadaptation after chronic intoxication. GSEA analysis can identify differential enrichment of sets of genes rather than individual genes, to discover common sets of dysregulated genes overrepresented in complex diseases. Using associative statistics through a comparison to other published datasets, gene sets are ranked according to expression level. GSEA analysis was performed at each stage of addiction (i.e., 0h, 8h and 21d) to focus on the biology underlying the addiction process. Notably, during intoxication, an association with gene expression levels and the adaptive immune system reactome was observed (Table 3). The Reactome (http://www.reactome.org) is a systems-based curated open source bioinformatics database of human pathways. Identification of ethanol-induced perturbations in gene expression that overlap with the adaptive immune system reactome pathway strongly suggests that intoxication disrupts immune signaling early during exposure. In contrast, the developmental biology reactome was identified as associated with neuroadaptive responses observed during abstinence.
Table 3.
Top GSEA results at each stage of addiction
| Name | Size | Abs NES | Comparison |
|---|---|---|---|
| Exposure | |||
| BERENJENO_TRANSFORMED_BY_RHOA_UP | 19 | 1.7849 | MRA0 |
| CHARAFE_BREAST_CANCER_LUMINAL_VS_BASAL_UP | 15 | 1.7717 | MRE0 |
| MARTORIATI_MDM4_TARGETS_FETAL_LIVER_DN | 38 | 1.5766 | FRE0 |
| MIKKELSEN_ES_ICP_WITH_H3K4ME3 | 19 | 1.5370 | MPA0 |
| IVANOVA_HEMATOPOIESIS_EARLY_PROGENITOR | 17 | 1.5305 | FPA0 |
| BYSTRYKH_HEMATOPOIESIS_STEM_CELL_QTL_TRANS | 34 | 1.5283 | FPA0 |
| KRIGE_RESPONSE_TO_TOSEDOSTAT_24HR_UP | 26 | 1.5254 | MPA0 |
| REACTOME_ADAPTIVE_IMMUNE_SYSTEM | 19 | 1.5120 | MPE0 |
| FLECHNER_BIOPSY_KIDNEY_TRANSPLANT_REJECTED_VS_OK_DN | 19 | 1.5098 | FRA0 |
| MULLIGHAN_NPM1_SIGNATURE_3_UP | 15 | 1.5025 | FPA0 |
| Withdrawal | |||
| BROWNE_HCMV_INFECTION_48HR_DN | 18 | 1.6251 | FPE8 |
| LEE_BMP2_TARGETS_UP | 21 | 1.5897 | FRA8 |
| LEE_BMP2_TARGETS_UP | 21 | 1.5700 | MPA8 |
| CREIGHTON_ENDOCRINE_THERAPY_RESISTANCE_1 | 17 | 1.5666 | MRA8 |
| GENTILE_UV_HIGH_DOSE_DN | 17 | 1.5531 | FPE8 |
| KIM_ALL_DISORDERS_CALB1_CORR_UP | 21 | 1.5507 | MPA8 |
| MASSARWEH_TAMOXIFEN_RESISTANCE_UP | 20 | 1.5442 | FRE8 |
| NIKOLSKY_BREAST_CANCER_17Q21_Q25_AMPLICON | 15 | 1.5416 | FPE8 |
| ACEVEDO_METHYLATED_IN_LIVER_CANCER_DN | 20 | 1.5379 | FRE8 |
| GEORGES_TARGETS_OF_MIR192_AND_MIR215 | 31 | 1.5357 | MPA8 |
| ROME_INSULIN_TARGETS_IN_MUSCLE_UP | 18 | 1.5235 | MPA8 |
| SENESE_HDAC3_TARGETS_DN | 20 | 1.5230 | MPA8 |
| REACTOME_DEVELOPMENTAL_BIOLOGY | 20 | 1.5119 | FRA8 |
| WAKABAYASHI_ADIPOGENESIS_PPARG_RXRA_BOUND_8D | 31 | 1.5118 | MPA8 |
| WONG_ADULT_TISSUE_STEM_MODULE | 33 | 1.5110 | MPA8 |
| SANSOM_APC_TARGETS_DN | 19 | 1.5100 | MPA8 |
| SCHLOSSER_SERUM_RESPONSE_DN | 25 | 1.5066 | MRE8 |
| MARTENS_TRETINOIN_RESPONSE_DN | 18 | 1.5031 | MPA8 |
| Abstinence | |||
| PUJANA_ATM_PCC_NETWORK | 52 | 1.7171 | MRE21 |
| PUJANA_BRCA1_PCC_NETWORK | 61 | 1.7011 | MRE21 |
| RICKMAN_METASTASIS_UP | 15 | 1.6639 | MPA21 |
| MILI_PSEUDOPODIA_HAPTOTAXIS_DN | 21 | 1.6363 | MRE21 |
| SWEET_LUNG_CANCER_KRAS_DN | 16 | 1.6335 | FPE21 |
| MULLIGHAN_MLL_SIGNATURE_2_DN | 16 | 1.6196 | MRE21 |
| BLALOCK_ALZHEIMERS_DISEASE_UP | 70 | 1.6024 | MRE21 |
| MATSUDA_NATURAL_KILLER_DIFFERENTIATION | 22 | 1.6012 | MRE21 |
| CHEN_METABOLIC_SYNDROM_NETWORK | 37 | 1.5881 | MPA21 |
| SENESE_HDAC3_TARGETS_DN | 20 | 1.5805 | FRE21 |
| PILON_KLF1_TARGETS_UP | 20 | 1.5713 | MRE21 |
| REACTOME_DEVELOPMENTAL_BIOLOGY | 20 | 1.5662 | MRE21 |
| PUJANA_CHEK2_PCC_NETWORK | 28 | 1.5608 | MRE21 |
| GRAESSMANN_APOPTOSIS_BY_DOXORUBICIN_UP | 48 | 1.5584 | MRE21 |
| STARK_PREFRONTAL_CORTEX_22Q11_DELETION_DN | 17 | 1.5529 | MRE21 |
| ONKEN_UVEAL_MELANOMA_UP | 28 | 1.5457 | MRE21 |
| BAKKER_FOXO3_TARGETS_DN | 25 | 1.5455 | MRE21 |
| WONG_ADULT_TISSUE_STEM_MODULE | 33 | 1.5314 | MRE21 |
| PHONG_TNF_RESPONSE_NOT_VIA_P38 | 16 | 1.5189 | MPA21 |
| LIM_MAMMARY_STEM_CELL_UP | 24 | 1.5144 | FRE21 |
| BLALOCK_ALZHEIMERS_DISEASE_DN | 43 | 1.5142 | MRE21 |
| DODD_NASOPHARYNGEAL_CARCINOMA_UP | 34 | 1.5125 | MRE21 |
| BONOME_OVARIAN_CANCER_SURVIVAL_SUBOPTIMAL_DEBULKING | 20 | 1.5116 | FPE21 |
| KIM_ALL_DISORDERS_OLIGODENDROCYTE_NUMBER_CORR_UP | 33 | 1.5063 | MRE21 |
| SPIELMAN_LYMPHOBLAST_EUROPEAN_VS_ASIAN_UP | 22 | 1.5058 | MRE21 |
| DANG_BOUND_BY_MYC | 42 | 1.5048 | MRE21 |
| KIM_BIPOLAR_DISORDER_OLIGODENDROCYTE_DENSITY_CORR_UP | 27 | 1.5014 | MRE21 |
GSEA categories with absolute NES (Abs NES) scores greater than 1.5.
3.4. Gene coexpression networks identify correlated expression sets
To better understand the biological impact of alcohol abuse in the CNS and further characterize important gene interactions, networks were then constructed using weighted gene coexpression network analysis (WGCNA). WGCNA is a systems biology method for modeling the correlation patterns among expression levels of genes across many samples, to identify repeated directional changes consistent across the samples. To construct networks and characterize coexpression patterns, all ethanol regulated genes over the addiction time course from both males and females and both WSR and WSP preclinical models were evaluated. In an overall addiction consensus network using all arrays for WGCNA analysis, there was no major effect of ethanol treatment in the mPFC in any module (data not shown). These results suggest that neuroadapation as a result of chronic ethanol exposure does not reflect a consistent effect of ethanol over all time points and conditions. Instead, the influence of sex, line and addiction stage each play important roles in the CNS response to abuse. This lack of overarching pathways influenced strictly by ethanol exposure highlights the need to interpret the effects of ethanol in the context of the sex, genotype/phenotype and/or addiction stage being studied.
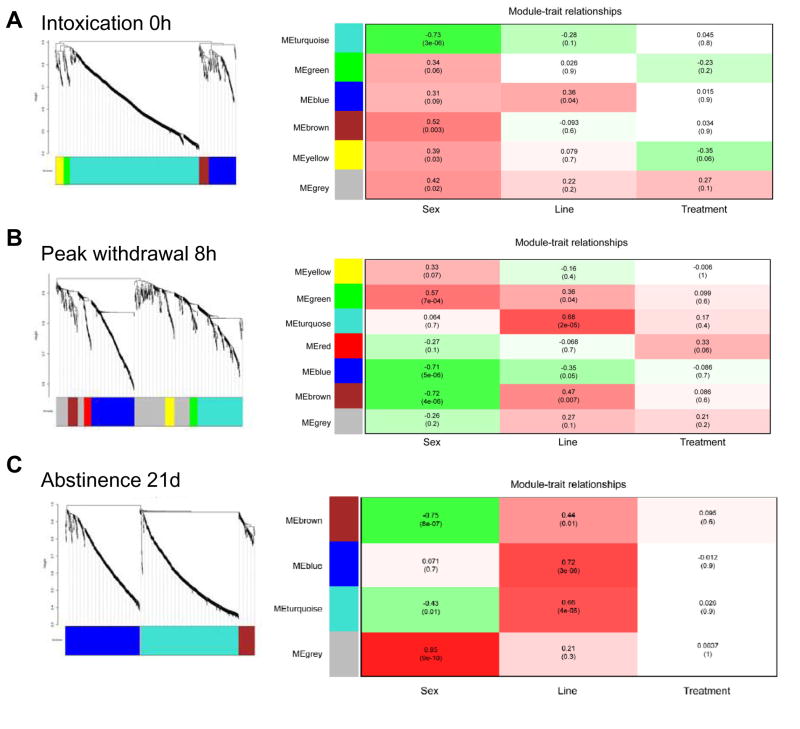
Given the lack of significant treatment effects over the addiction time course, WGCNA was then performed at each stage of addiction to identify significant modules of correlated expression. Overall eigengene dendrograms identified in WGCNA analysis (shown in Figure 2) demonstrate distinct influences of genotype vs. sex at each addiction stage, with the lowest point in the dendrogram identifying the experimental variable associated with the mostly highly correlated modules. This result is consistent with the data structure identified in unsupervised hierarchal clustering illustrated in Figure 1.
Figure 2.
Network analysis of gene coexpression after chronic intoxication identifies clustering of distinct module eigengenes and traits at each stage of addiction. From the WGCNA analysis, module eigengenes and traits were clustered using an unsigned correlation matrix so that modules with high absolute value correlations grouped together. Consistent with hierarchical clustering analysis, coexpression modules demonstrate the influence of sex during the early stages of intoxication and withdrawal, while selected phenotype/genotype is most influential during abstinence.
The WGCNA at each time point generated three data sets with modules that have significant associations with sex, line or treatment (shown in Figure 3). Notably, effects of ethanol treatment alone were again subtle, suggesting that other experimental variables (sex, genotype and time) interact uniquely with ethanol intoxication, highlighting the complexity of the response to alcohol. To characterize the most important modules at each addiction stage, bioinformatics using IPA analysis was employed. During chronic intoxication (Figure 3A), analysis of the genes in the most robustly regulated module in females (brown module) identified the Role of Lipids/Lipid Rafts in the Pathogenesis of Influenza (p < 0.01), while the Mevalonate Pathway I (p < 0.05) was identified as the top canonical pathway in males (turquoise module). Previous analysis of the transcriptional response associated with ethanol exposure has identified modules with strong associations with specific CNS cell types (Ponomarev et al., 2012). Cell type analysis of our data has extended these findings by identifying genes enriched for expression in astrocytes (yellow module; p < 0.01) as an ethanol target in females associated with chronic intoxication (Table 4). During peak withdrawal (Figure 3B), the top canonical pathway in females (green module) was Gαs Signaling (p = 0.122). In males, the Role of Lipids/Lipid Rafts in the Pathogenesis of Influenza (p < 0.01) was identified (brown module). Finally during abstinence (Figure 3C), the top pathway was Death Receptor Signaling (p < 0.05), with increased expression in the low response WSR model (blue module). Treatment effect was the least robust difference with regard to statistical significance. Analysis at each stage thus identified novel modules/pathways influenced by sex and time, likely reflecting a distinct biology that underlies the development of addiction.
Figure 3.
WGCNA analysis at each stage of addition identified distinct modules of coexpressed genes for sex and for line differences. A. Intoxication (0 h). B. Peak withdrawal hyperexcitability (8 h). C. Abstinence (21 d). Significant association between modules and the traits of sex and line were observed, but treatment effects (the consequences of chronic ethanol exposure) were modest in the combined datasets. In the Module-Trait relationships table, red color indicates higher expression in females, higher expression in WSR, and higher expression with ethanol while green indicates higher expression in males, higher expression in WSP, and lower expression with ethanol. For each block in the module-trait relationship table, values represent Pearson’s correlation coefficients, while numbers in parentheses below indicate the p-value for the significance of the relationship.
Table 4.
Modules with significant overlap with CNS cell types
| Group/Module | Name | p-value | Ratio |
|---|---|---|---|
| 0hr Yellow | Astrocyte Genes - All | 0.005 | 12/2551 |
| Astrocytes - Cahoy 2008 | 0.031 | 10/2413 | |
|
| |||
| 8hr Red | Neuron - Cahoy 2008 | 0.025 | 7/1953 |
| Neuron Expressed Genes | 0.049 | 10/3575 | |
|
| |||
| 21d Blue | Astrocytes - Daginakatte 2008 | 0.017 | 3/39 |
|
| |||
| 21d Brown | Neurons - Oldham 2008 | 0.020 | 12/1985 |
IPA was used to identify significant overlap between our data and genes which are known to be expressed in specific CNS cell types (Cahoy et al., 2008, Daginakatte et al., 2008, Oldham et al., 2008, Obayashi et al., 2009).
3.5. Functional changes associated with chronic intoxication identified by a systems approach
By employing analysis at behaviorally-relevant time points and using both sexes of selected line models of response to ethanol, we created an improved functional framework for interpretation of neuroadaptive differential expression between sex vs. genotype over the stages of addiction. Combined results demonstrated that sex had the strongest influence on neuroadapation with intoxication and during early withdrawal stages. Although pathways characterized in expression difference analysis can identify candidate signaling mechanisms important in the phenotype under study, additional analyses of these candidate pathways is required to provide independent confirmation of the underlying biological differences. Because of the importance of sex in the transcriptional response, we next focused our efforts on confirmation of sexual dimorphism during withdrawal, the addiction stage with the strongest dimorphic response.
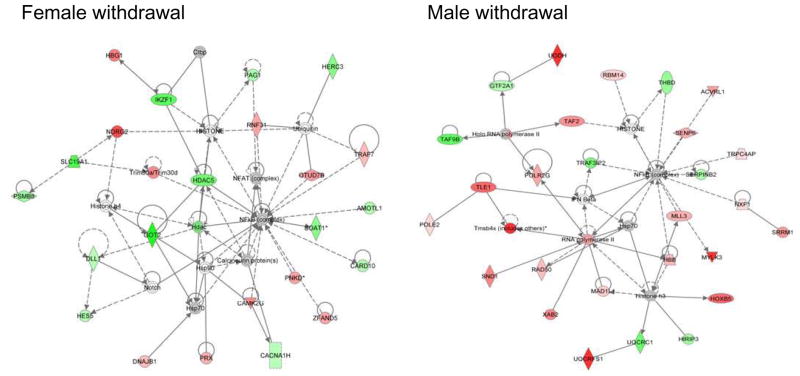
Using data collapsed by line for each sex (i.e. all WSR and WSP male controls vs. all WSR and WSP male ethanol) to identify ethanol regulated genes, the most significant networks of interacting genes at peak withdrawal in males vs. females were identified by IPA analysis. As shown in Figure 4, a central node in both male and female networks was the transcription factor NF-κB; however, there is a striking distinction in the interacting molecules between the sexes. In fact, none of the NF-κB interacting genes is regulated in common between the males and females, indicating a truly dimorphic signaling pattern. NF-κB itself is involved in numerous physiological processes. However, examination of the interacting molecules and their direction of regulation can provide a clearer picture of the processes being affected. In particular several of the regulated genes in females are indicative of a pro-inflammatory response while instead examination of the network of regulated interacting genes in males suggests overall immunosuppression.
Figure 4.
Sexually-dimorphic NF-κB nodes with differentially-expressed interacting gene sets identified in males and females. IPA analysis at 8h in mPFC identified an NF-κB node in both males and females with sex collapsed on line (genotype), with completely different interacting proteins between sexes. Red indicates genes with increased expression; green indicates genes with decreased expression; gray indicates that the gene was present in dataset but was not significantly regulated.
3.6. Molecular evidence for sexually-dimorphic inflammatory response
The combined bioinformatics approaches described here are useful to both analyze and characterize data structure and to generate hypotheses for validation of the results observed. Given the sexually-dimorphic responses observed with respect to inflammatory signaling during the early stages of addiction consistently identified in the various bioinformatic analyses performed (GSEA, GeneCards, WGCNA and IPA analysis), we tested the hypothesis that withdrawal from chronic intoxication differentially activates immune signaling in males vs. females at peak withdrawal. For this analysis, focused qPCR arrays that broadly characterize immune function were interrogated with RNA harvested from male vs. female WSR mPFC at peak withdrawal. These focused arrays were chosen because they allow for the simultaneous characterization of both pro-inflammatory and anti-inflammatory signaling, and because T cell signaling is a major component of the adaptive immune system that was identified as differentially regulated by GSEA analysis.
Consistent with a sexually-dimorphic inflammatory response in the mPFC, ethanol-mediated expression differences were observed between males and females. To visualize the differences between the sexes, a Volcano plot was constructed (Figure 5) where differentially expressed genes were arranged along dimensions of biological impact (fold change) versus statistical significance (for reliability of change). As can be observed, expression differences identified using focused qPCR arrays were strongly dimorphic. There is a notable skew in the male plot toward down-regulation of several co-stimulatory molecules, chemokines and cytokines. By contrast, the female response to ethanol was more balanced, with interleukin 6 (IL-6), tumor necrosis factor (TNF-α), chemokine (C-C motif) ligand 11 (Ccl11) and chemokine (C-C motif) ligand 5 (Ccl5; RANTES) exhibiting regulation consistent with a pro-inflammatory response, though not all differences reached statistical significance (see Table 5, Figure 5). Highlighting the sexually-dimorphic nature of the response, Ccl5 was one of the most strongly down-regulated genes among the male responses, exhibiting a 2.3 fold reduction in expression compared with a 1.6 fold up-regulation in females (see Table 5 for list). Several of the regulated transcripts are also markers of activated and/or polarized glia (Durafourt et al., 2012, Crain et al., 2013). These data provide biological confirmation of the gene network results during synchronized withdrawal from chronic intoxication, indicating an immunosuppressed response in males and instead a pro-inflammatory response in females in the mPFC. Thus, during each stage of addiction, distinct pathways and signaling mechanisms are altered by chronic alcohol exposure that are differentially influenced by sex or genotype.
Figure 5.
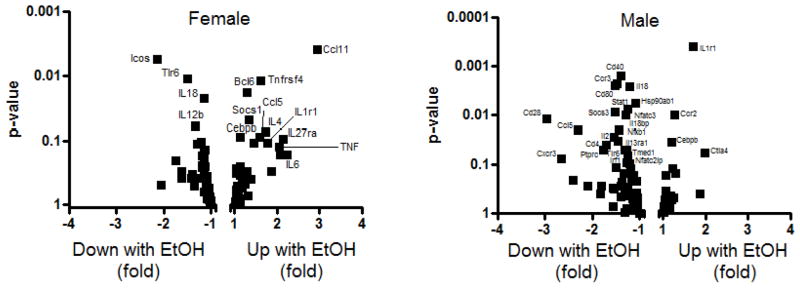
Confirmation of sexually dimorphic immune signaling using focused qPCR immune function arrays. Volcano plots of expression differences in WSR male (on the right) and female (on the left) mPFC. The plots identify sequences that are differentially expressed during withdrawal as a consequence of chronic ethanol intoxication. Females exhibit up-regulation of several key inflammatory mediators such as TNF-α, IL-6, CCL11 and CCL5 while males exhibit a profound down-regulation of several chemokines, cytokines and co-stimulatory molecules indicative of a relative immunosuppression.
Table 5.
Inflammatory focused qPCR array results
| Gene Symbol | Male EtOH vs Male Con | Male t-test (p-value) | Female EtOH vs Female Con | Female t-test (p-value) |
|---|---|---|---|---|
| Bcl6 | 1.01 | 0.8587 | 1.28 | 0.0183 |
| Ccl11 | −1.28 | 0.7918 | 2.95 | 0.0039 |
| Ccl5 | −2.30 | 0.0199 | 1.58 | 0.0891 |
| Ccr2 | 1.27 | 0.0099 | −1.24 | 0.3874 |
| Ccr3 | −1.47 | 0.0023 | 1.28 | 0.3406 |
| Cd28 | −2.97 | 0.0118 | −1.36 | 0.5152 |
| Cd80 | −1.52 | 0.0025 | 1.07 | 0.6829 |
| Cebpb | 1.19 | 0.0353 | 1.12 | 0.0895 |
| Icos | −1.83 | 0.4010 | −2.15 | 0.0056 |
| Il18 | −1.22 | 0.0027 | −1.15 | 0.0225 |
| Il18bp | −1.43 | 0.0198 | 1.17 | 0.5378 |
| Il1r1 | 1.70 | 0.0004 | 1.77 | 0.1088 |
| Il2 | −1.55 | 0.0272 | 1.44 | 0.1079 |
| Nfatc3 | −1.29 | 0.0101 | −1.03 | 0.9494 |
| Nfkb1 | −1.45 | 0.0338 | 1.04 | 0.6115 |
| Ptprc | −1.71 | 0.0395 | −1.10 | 0.4722 |
| Socs1 | −1.14 | 0.5014 | 1.34 | 0.0492 |
| Socs3 | −1.52 | 0.0089 | −1.03 | 0.7428 |
| Stat1 | −1.26 | 0.0077 | −1.15 | 0.2998 |
| Tlr6 | −1.32 | 0.0507 | −1.49 | 0.0110 |
| Tnfrsf4 | −1.04 | 0.5751 | 1.62 | 0.0119 |
| Cd40 | −1.40 | 0.0016 | −1.08 | 0.4140 |
Genes with significant ethanol regulation identified by t-test
4. Discussion
The development of alcohol dependence, continued alcohol abuse and relapse after abstinence remain serious public health concerns that are ineffectively treated with currently available therapies. The analysis presented here for the first time provides insight into differential contributions of genotype/phenotype vs. sex-specific influences at every stage of alcohol addiction cycle. This characterization was performed by identifying the transcriptional response in mPFC, using both males and females and both the WSR and WSP selected lines as preclinical models of alcohol response genotypes. Comprehensive time course analysis of addiction stages over behaviorally-relevant time points indicated that sex has the strongest influence on the early response to chronic intoxication and during the immediate withdrawal period, while genotype/phenotype was more important in abstinence. Affected biological pathways were identified using a variety of complementary bioinformatics approaches, with confirmation of identified pathways that demonstrated distinct inflammatory signaling in mPFC between males and females at peak withdrawal following chronic intoxication. These results are consistent with a pro-inflammatory inflammotoxic phenotype (see Béraud et al., 2013, Chhor et al., 2013) in females, which may be associated with neurodegeneration (Hashimoto and Wiren, 2008). In contrast, immune suppression and a relative neuroprotection was observed in males, again as previously seen in this model (Hashimoto and Wiren, 2008). Such sexually-dimorphic signaling supports the hypothesis that neuroadapation after chronic alcohol exposure is mechanistically distinct between the sexes and importantly that females, uniquely vulnerable to ethanol-induced tissue damage (Loft et al., 1987, Ammendola et al., 2000, Hommer et al., 2001, Fernandez-Sola and Nicolas-Arfelis, 2002, Mann et al., 2005), may be especially at risk during chronic intoxication and early withdrawal.
4.1. Distinct biological response at each stage of addiction
The specific impact of sex vs. alcohol phenotype/genotype over the time course of addiction has not been previously examined. These studies were designed to characterize important biological pathways sensitive to dysregulation by alcohol at each stage, and a variety of bioinformatic approaches were employed to provide a biological context to the changes observed in gene expression. In the characterization of differential gene expression patterns at each stage of addiction in sex vs. genotype comparisons, the most significant canonical pathway identified during intoxication in females was associated with sphingosine-1-phosphate (S1P) signaling while in males the top pathway was telomere extension by telomerase. Notably, a relationship between S1P signaling and immune signaling/inflammation has been observed (Maceyka and Spiegel, 2014). In addition, previous studies have shown increased S1P levels in the developing mouse brain after ethanol treatment, which is associated with neuroapoptosis (Chakraborty et al., 2012). Depending on receptor subtype, activation of S1P receptors can also influence blood brain barrier integrity (Marsolais and Rosen, 2009). Disruption of S1P signaling would thus be expected to have profound effects in females. In males in contrast, changes in telomerase activity identified here are indicative of protection of telomere ends from degradation, and thus may play a crucial role in cellular aging. Consistent with these findings, ethanol treatment has been shown to elongate telomeres (Romano et al., 2013). Overall, sex-specific activation of these disparate pathways is consistent with vulnerability to neurodegeneration in females but not males following chronic alcohol exposure, as described below. In our observations of changes identified during the intoxication stage, gene expression was altered strictly by chronic alcohol administration and highlight an advantage of the vapor inhalation paradigm in mechanistic terms, as the synchronized withdrawal procedure had not been initiated at the time of tissue harvest. During peak withdrawal, the top canonical pathway was p38 MAPK signaling in females. Although p38 MAPK signaling is complex and involved in many cascades, increased signaling is observed with TLR4/Type I IL-1 receptor activation after ethanol treatment and is known to be involved in the induction of inflammatory mediators associated with cell death (Blanco et al., 2005). Instead in males, retinoate biosynthesis was targeted, likely reflecting the increased levels of retinoic acid observed after chronic ethanol treatment (Kane et al., 2010), and the involvement of alcohol and aldehyde dehydrogenase in retinol catalysis (see Pino-Lagos et al., 2008). Interestingly, retinoic acid reduces production of inflammatory mediators from peritoneal macrophages in response to lipopolysaccharide (Mehta and Guidot, 2012). During the abstinence phase, analysis identified atherosclerosis signaling as the top pathway in the low response WSR model. In contrast, in the WSP model phosphatidylethanolamine biosynthesis was significant during abstinence, which may reflect the complex effects of alcohol toxicity on myelin synthesis (de la Monte and Kril, 2014). Combined, these results suggest that optimal targets for the treatment of alcohol dependence are complex and likely to be distinct in males and females and also at each stage of addiction.
4.2. Female vulnerability to the damaging consequences of alcohol abuse
During the early stages of addiction, sex has a powerful influence on neuroadaptive responses in the mPFC. This result is consistent with previous analysis that has revealed a strong gender bias for the development of many different brain disorders (Holden, 2005), including drug abuse and alcoholism (Becker and Hu, 2008). Sex differences are noted in many alcohol-related problems, but increased vulnerability to the toxic effects of alcohol may be one of the most devastating. Thus, the degree of cardiomyopathy (Fernandez-Sola and Nicolas-Arfelis, 2002), peripheral neuropathy (Ammendola et al., 2000) and cirrhosis (Loft et al., 1987) is worse in female alcoholics than in males. Females also demonstrate the phenomena of “telescoping” with increased damage after a shorter period of ethanol abuse compared to males (Brady and Randall, 1999, Mann et al., 2005). Thus, accumulating data indicate that brain damage may be enhanced in alcohol dependent females in both animal models (Hashimoto and Wiren, 2008, Alfonso-Loeches et al., 2013) and in humans (Hommer et al., 2001, Mann et al., 2005). In this report, genes associated with cell death/neurodegeneration were significantly regulated in females. Ethanol targeting of genes involved in neurodegeneration and cell death is particularly problematic given the finding that females are more sensitive to the effects of ethanol intoxication (compare Table 1 to Table 2), consistent with previous reports (Sershen et al., 2002). In contrast to female sensitivity, males appear to be protected from early damage in our model of alcohol vulnerability. Consistent with this, males are relatively resistant to injury immediately following withdrawal evaluated in slice cultures (Walls et al., 2013) and glial activation observed after alcohol administration in males appears protective (Marshall et al., 2013). Alcohol-induced brain damage is a vitally important issue, as a majority of alcoholics have impaired cognitive function (Vetreno et al., 2011). Damage to mPFC, observed in chronic alcoholics (Zahr et al., 2011), is of particular concern given the important role this brain region subserves in executive function and inhibitory control (Fuster, 2002, Kroener et al., 2012). The most devastating consequence of alcohol abuse is excess mortality (Rehm et al., 2009), and a high death rate (premature death) in individuals with alcohol use disorder has been well documented (Timko et al., 2006, Campos et al., 2011). Consistent with enhanced female vulnerability to the toxic effects of alcohol (Ceylan-Isik et al., 2010), annualized death rates for female alcoholics are substantially elevated relative to male alcoholics (Batty et al., 2009, John et al., 2013). Combined, these results demonstrate that response pathways and underlying molecular mediators may exacerbate ethanol-induced neurodegeneration in the mPFC in females during the early stages of addiction. Given repeated and consistent studies demonstrating higher mortality, worse outcomes, a distinct psychopathology (Helzer and Pryzbeck, 1988, Brady and Randall, 1999, Alonso et al., 2004, Dawson et al., 2010), and greater sensitivity to the detrimental effects of alcohol among women, future research is warranted toward identifying sexually-dimorphic effects of alcohol and characterizing mechanisms that underlie the increased sensitivity to alcohol-induced tissue damage among women (see Wiren, 2013).
4.3. The importance of genotype and alcohol response phenotype during abstinence
One characteristic of alcoholism is uncontrolled excessive consumption of alcohol, characterized by an inability to remain abstinent. Approximately 90% of alcoholics experience at least one relapse over a 4-year period following treatment (Polich, 1981). Abstinence from alcohol drinking is an important goal for the treatment of alcoholism, but there is limited understanding of factors that influence risk of relapse and as a result few effective treatments (Olive, 2010, Zindel and Kranzler, 2014). Unfortunately, there are no controlled studies that have definitively shown either a single or combined intervention reliably prevents relapse to drinking. The scientific literature addressing the heritable component of alcoholism has demonstrated a significant genetic contribution to the disease (Magnusson et al., 2010), consistent with genetic risk factors. Although controversial (Morean and Corbin, 2010), one group of individuals with a greater risk for the development of alcohol use disorders typically have a lower subjective response to the effects of ethanol (Schuckit et al., 2009). Schuckit et al. have proposed that a low level of response (low LR) reflects differences in sensitivity to the pharmacological effects of alcohol (Mayfield et al., 2008, Schuckit et al., 2008, Schuckit et al., 2009). The low LR to alcohol phenotype, with increased voluntary ethanol consumption and a low level of ataxic response to alcohol, can be a strong predictor of developing alcohol dependency in both males and females (Schuckit et al., 2000). Interestingly, similarities exist between the WSR line and characteristics of this low LR to alcohol intermediate phenotype that include: decreased response during withdrawal from chronic ethanol exposure (Kosobud and Crabbe, 1986), increased voluntary ethanol consumption (Kosobud et al., 1988) particularly in replicate-2 mice, decreased sensitivity to ethanol-induced conditioned place preference (Crabbe, 1992) and most importantly increased voluntary relapse drinking, as we have demonstrated (see Hashimoto et al., 2011). Due to the heterogeneity and genetic diversity of individuals with alcohol dependence, it has been proposed that effective treatments should target specific phenotypes at distinct stages of addiction rather than employ a generic approach to all patients (Kuehn, 2009). Although no animal model fully duplicates alcoholism, analyses employing models like the WSR line may provide insight into changes observed in the specific subset of LR to alcohol patients. In this context it is notable that during abstinence in the low response WSR model, Death Receptor Signaling was identified as increased in WGCNA analysis, while IPA analysis identified Atherosclerosis Signaling as significantly regulated and GSEA analysis identified the developmental biology reactome. The importance of these pathways during abstinence is supported by published reports. For example, we have reported increased cell death in mPFC during abstinence in WSR mice (Hashimoto et al., 2011) that is distinct from the neurotoxicity observed during the early stages of intoxication and withdrawal (Hashimoto and Wiren, 2008, Hashimoto et al., 2011). Furthermore, supporting the importance of the developmental biology reactome in this model, bioinformatics analysis in human populations representing the low LR to alcohol phenotype identified Nervous System Development/Function as a top enriched function (Joslyn et al., 2010). Consistent with changes in atherosclerosis signaling during abstinence, alcohol abuse is associated with a 2- to 3-fold increase in mortality risk for cardiovascular disease at least in some genotypes (Roerecke and Rehm, 2014) and previous analysis has identified angiogenesis and the platelet-derived growth factor signaling pathways as molecular targets associated with overall ethanol toxicity (Wang et al., 2007, Wang et al., 2010). It is possible that expression differences determined by heritable factors as reflected in the selected phenotype, combined with plastic neuroadapative changes that persist after chronic intoxication and withdrawal, increase the risk of relapse (see Petronis, 2010). We previously reported that an intervention based on pathways identified in WSR mice during abstinence can reduce voluntary relapse drinking (Hashimoto et al., 2011). Thus, analysis using preclinical animal models may provide targets for therapeutic intervention particularly for the low LR patient population.
4.4. Sexually-dimorphic inflammatory signaling in the mPFC after alcohol treatment
One of the most consistent and significant differences between males and females overall was seen in immune signaling/inflammation following chronic intoxication. At each level of analysis, from single gene through superpathway analyses, alterations in immune signaling were noted. Immune alterations were most strongly associated with the early stages of addiction, i.e., after chronic intoxication and during peak withdrawal hyperexcitability. Pathway confirmation studies were undertaken using qPCR that was focused on immune function.to characterize changes in gene expression in mPFC during withdrawal. Again consistent with bioinformatics, changes in inflammatory signaling were seen, with a pro-inflammatory response in female mice while instead changes consistent with immunosuppression were seen in males. Similar to other studies in females (Alfonso-Loeches et al., 2010, Lippai et al., 2013) increased expression of TNF-α, interleukin 6 (IL-6) and CCL5/RANTES were observed following ethanol exposure. Increased TNF-α expression may reflect activation of the bacterial endotoxin sensor toll-like receptor 4 (TLR4) (for review, see Hanisch, 2002) which is associated with classical microglial activation. Similar to microglia, astrocytes are also capable of polarization and release both pro- and anti-inflammatory mediators such as TNF-α and IL-10 in response to danger signals (Jang et al., 2013). Notably, bioinformatic analysis to identify cell-type-specific gene expression indicates involvement of astrocytes in females during intoxication. Evidence of glial activation and some aspects of neuroinflammation have been observed in postmortem brain tissue from alcoholics (Zou and Crews, 2012, Crews et al., 2013, McClintick et al., 2013). Chronic alcohol exposure can result in immune dysfunction and immunosuppression, at least in some tissues (Kaphalia and Calhoun, 2013, Khocht et al., 2013, Mehta et al., 2013, Parlet et al., 2014), and alcoholics frequently develop severe respiratory infections which result in increased hospital stays and a higher likelihood of intensive care unit admission (Saitz et al., 1997). Typical of most studies examining alcohol abuse, the study by Saitz et al. (1997) was underpowered to examine the influence of gender (~20% of alcoholic patients were female), as female alcoholics make up a smaller (though quickly increasing) segment of alcohol abusers, leaving many important sexually-dimorphic responses poorly characterized (for discussion, see Schuckit et al., 2012). Thus, the importance of sex/gender has not been carefully examined with respect to inflammation and is particularly poorly characterized in females. Furthermore, while the prefrontal cortex is a primary target for ethanol-related brain damage (e.g. Kril et al., 1997), very little is known about specific inflammatory changes in this brain region in human alcoholics. The anti-inflammatory signaling observed in males in our studies compared to others may also reflect differences between alcohol exposure paradigms; our approach focuses on vulnerability to alcohol-induced changes and not long-term toxicity. Females may exhibit a hyper-inflammatory response to a danger molecule such as the TLR4 agonist lipopolysaccharide (LPS), perhaps due to priming by alcohol or via changes in the blood brain barrier, and thus increased vulnerability to neurodegeneration. Consistent with this model, we have previously shown brain damage and neurotoxicity in females, but not in males, 10 days after chronic ethanol exposure (Hashimoto and Wiren, 2008).
In summary, significant gene expression differences that result from chronic intoxication were identified that were differentially influenced by sex or genotype during a distinct time course of addiction. Data structure indicated a strong effect of sex on neuroadaptive responses, with high similarity between expression profiles of males and females irrespective of genetic background during chronic intoxication and at peak withdrawal, associated with sex-specific engagement of divergent biological pathways and processes due to alcohol exposure. However, during abstinence, analysis showed striking differences instead between the lines/phenotypes irrespective of sex. The time points in our analysis were chosen based on identified stages of addiction: intoxication, peak withdrawal and abstinence. Intoxication and early withdrawal were characterized by pathways that were sexually-dimorphic in the mPFC, particularly with respect to inflammation/immune response where females showed a pro-inflammatory inflammotoxic cascade while male expression patterns indicated a strong overall immunosuppressive response. During abstinence, pathways that were altered were more strongly influenced by genotype and include developmental pathways and alterations in atherosclerosis signaling. Surprisingly, both sex and genetic background critically influence the neuroadaptive response to alcohol, far outweighing any overarching impact of alcohol per se, suggesting that similar patterns in humans are likely to confound experimental findings unless sex and genotype are taken into account. Improved understanding of sexually-dimorphic responses and disparate molecular underpinnings involved in adult neurodamage can lead to novel targeted treatment options for the addicted, as current therapy has little effect on survival (John et al., 2013). Furthermore, the impact of genotype/family history is known to have an important influence on the development of alcoholism but is also likely to play a significant role in mechanisms that underlie the risk of relapse. Ultimately, characterization of these distinct influences will be important in determining effective therapeutic approaches during the stages of alcohol addiction, since successful treatment may depend on an individualized approach. Combined, these results have implications for targeted therapeutic approaches in the treatment of alcohol addiction and the maintenance of sobriety that reflect the distinct involvement of both sex and genotype.
Sex and genetics distinctly influence gene expression at each addiction stage
NF-κB signaling is sexually dimorphic at peak ethanol withdrawal
Females exhibit an inflammotoxic phenotype in mPFC at peak withdrawal
Males exhibit an immune suppressed phenotype at peak withdrawal in mPFC
Targeted therapies may be needed depending on addiction stage, sex and genotype
Acknowledgments
This research was supported by grants from the Department of Veterans Affairs (BX001172 (KMW) and BX001294 (CJW)) and from the NIH/NIAAA (R01AA021468 (KMW)). Additionally, this material is the result of work supported with resources and the use of facilities at the Portland VA Medical Center (KMW). We also acknowledge support from NIAAA for the Portland Alcohol Research Center (P60AA010760) and for the maintenance of colonies of WSR and WSP mice (R24AA020245) used in the present studies and thank Melissa Andrew for assistance with the vapor exposure procedure and Casia Wardzala for comments provided after careful reading of the manuscript.
Abbreviations
- BEC
blood ethanol concentration
- Ccl5
chemokine (C-C motif) ligand 5
- Ccl11
chemokine (C-C motif) ligand 11
- CNS
central nervous system
- GO
Gene Ontology
- GSEA
Gene Set Enrichment Analysis
- IPA
Ingenuity Pathway Analysis
- Low LR
low level of response
- mPFC
medial prefrontal cortex
- NLRP3
nod-like receptor family, pyrin domain containing 3
- TNF-α
tumor necrosis factor
- WSR
Withdrawal Seizure-Resistant selected line
- WSP
Withdrawal Seizure-Prone selected line
- WGCNA
Weighted Gene Coexpression Network Analysis
Footnotes
Conflicts of interest
None of the authors have any knowledge of potential conflicts of interest regarding the research presented in this report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi J, Mizoi Y, Fukunaga T, Ogawa Y, Ueno Y, Imamichi H. Degrees of alcohol intoxication in 117 hospitalized cases. J Stud Alcohol. 1991;52:448–453. doi: 10.15288/jsa.1991.52.448. [DOI] [PubMed] [Google Scholar]
- Addolorato G, Capristo E, Caputo F, Greco AV, Ceccanti M, Stefanini GF, Gasbarrini G. Nutritional status and body fluid distribution in chronic alcoholics compared with controls. Alcohol Clin Exp Res. 1999;23:1232–1237. doi: 10.1111/j.1530-0277.1999.tb04283.x. [DOI] [PubMed] [Google Scholar]
- Alfonso-Loeches S, Pascual M, Guerri C. Gender differences in alcohol-induced neurotoxicity and brain damage. Toxicology. 2013;311:27–34. doi: 10.1016/j.tox.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Alonso J, Angermeyer MC, Bernert S, Bruffaerts R, Brugha TS, Bryson H, de Girolamo G, Graaf R, Demyttenaere K, Gasquet I, Haro JM, Katz SJ, Kessler RC, Kovess V, Lepine JP, Ormel J, Polidori G, Russo LJ, Vilagut G, Almansa J, Arbabzadeh-Bouchez S, Autonell J, Bernal M, Buist-Bouwman MA, Codony M, Domingo-Salvany A, Ferrer M, Joo SS, Martinez-Alonso M, Matschinger H, Mazzi F, Morgan Z, Morosini P, Palacin C, Romera B, Taub N, Vollebergh WA EsemeD/Mhedea Investigators ESotEoMDP. 12-Month comorbidity patterns and associated factors in Europe: results from the European Study of the Epidemiology of Mental Disorders (ESEMeD) project. Acta Psychiatr Scand Suppl. 2004:28–37. doi: 10.1111/j.1600-0047.2004.00328.x. [DOI] [PubMed] [Google Scholar]
- Ammendola A, Gemini D, Iannaccone S, Argenzio F, Ciccone G, Ammendola E, Serio L, Ugolini G, Bravaccio F. Gender and peripheral neuropathy in chronic alcoholism: a clinical-electroneurographic study. Alcohol Alcohol. 2000;35:368–371. doi: 10.1093/alcalc/35.4.368. [DOI] [PubMed] [Google Scholar]
- Baraona E, Abittan CS, Dohmen K, Moretti M, Pozzato G, Chayes ZW, Schaefer C, Lieber CS. Gender differences in pharmacokinetics of alcohol. Alcohol Clin Exp Res. 2001;25:502–507. [PubMed] [Google Scholar]
- Batty GD, Hunt K, Emslie C, Lewars H, Gale CR. Alcohol problems and all-cause mortality in men and women: predictive capacity of a clinical screening tool in a 21-year follow-up of a large, UK-wide, general population-based survey. J Psychosom Res. 2009;66:317–321. doi: 10.1016/j.jpsychores.2008.09.021. [DOI] [PubMed] [Google Scholar]
- Beadles-Bohling AS, Wiren KM. Anticonvulsive effects of kappa-opioid receptor modulation in an animal model of ethanol withdrawal. Genes, brain, and behavior. 2006;5:483–496. doi: 10.1111/j.1601-183X.2005.00200.x. [DOI] [PubMed] [Google Scholar]
- Becker JB, Hu M. Sex differences in drug abuse. Front Neuroendocrinol. 2008;29:36–47. doi: 10.1016/j.yfrne.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béraud D, Hathaway H, Trecki J, Chasovskikh S, Johnson D, Johnson J, Federoff H, Shimoji M, Mhyre T, Maguire-Zeiss K. Microglial Activation and Antioxidant Responses Induced by the Parkinson’s Disease Protein α-Synuclein. J Neuroimmune Pharmacol. 2013;8:94–117. doi: 10.1007/s11481-012-9401-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard BA, Steindorf S, Wang S, Glick SD. Sex differences in ethanol-induced dopamine release in nucleus accumbens and in ethanol consumption in rats. Alcohol Clin Exp Res. 1993;17:968–973. doi: 10.1111/j.1530-0277.1993.tb05650.x. [DOI] [PubMed] [Google Scholar]
- Blanco AM, Valles SL, Pascual M, Guerri C. Involvement of TLR4/type I IL-1 receptor signaling in the induction of inflammatory mediators and cell death induced by ethanol in cultured astrocytes. J Immunol. 2005;175:6893–6899. doi: 10.4049/jimmunol.175.10.6893. [DOI] [PubMed] [Google Scholar]
- Brady K, Randall C. Gender differences in substance use disorders. Psychiatr Clin North Am. 1999;22:241–252. doi: 10.1016/s0193-953x(05)70074-5. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J, Roca L, Gude F, Gonzalez-Quintela A. Long-term mortality of patients admitted to the hospital with alcohol withdrawal syndrome. Alcohol Clin Exp Res. 2011;35:1180–1186. doi: 10.1111/j.1530-0277.2011.01451.x. [DOI] [PubMed] [Google Scholar]
- Carroll M, Lynch W, Roth M, Morgan A, Cosgrove K. Sex and estrogen influence drug abuse. Trends Pharmacol Sci. 2004;25:273–279. doi: 10.1016/j.tips.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Ceylan-Isik AF, McBride SM, Ren J. Sex difference in alcoholism: who is at a greater risk for development of alcoholic complication? Life Sci. 2010;87:133–138. doi: 10.1016/j.lfs.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty G, Saito M, Shah R, Mao R-F, Vadasz C, Saito M. Ethanol triggers sphingosine 1-phosphate elevation along with neuroapoptosis in the developing mouse brain. J Neurochem. 2012;121:806–817. doi: 10.1111/j.1471-4159.2012.07723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Reilly MT, Kozell LB, Hitzemann R, Buck KJ. Differential activation of limbic circuitry associated with chronic ethanol withdrawal in DBA/2J and C57BL/6J mice. Alcohol. 2009;43:411–420. doi: 10.1016/j.alcohol.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhor V, Le Charpentier T, Lebon S, Oré M-V, Celador IL, Josserand J, Degos V, Jacotot E, Hagberg H, Sävman K, Mallard C, Gressens P, Fleiss B. Characterization of phenotype markers and neuronotoxic potential of polarised primary microglia in vitro. Brain Behav Immun. 2013;32:70–85. doi: 10.1016/j.bbi.2013.02.005. http://dx.doi.org/10.1016/j.bbi.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe J. Selected line differences. Behav Genet. 1992;22:21–23. doi: 10.1007/BF01066788. discussion 35–42. [DOI] [PubMed] [Google Scholar]
- Crain JM, Nikodemova M, Watters JJ. Microglia express distinct M1 and M2 phenotypic markers in the postnatal and adult central nervous system in male and female mice. J Neurosci Res. 2013;91:1143–1151. doi: 10.1002/jnr.23242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews FT, Qin L, Sheedy D, Vetreno RP, Zou J. High mobility group box 1/Toll-like receptor danger signaling increases brain neuroimmune activation in alcohol dependence. Biological psychiatry. 2013;73:602–612. doi: 10.1016/j.biopsych.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daginakatte GC, Gadzinski A, Emnett RJ, Stark JL, Gonzales ER, Yan P, Lee JM, Cross AH, Gutmann DH. Expression profiling identifies a molecular signature of reactive astrocytes stimulated by cyclic AMP or proinflammatory cytokines. Experimental neurology. 2008;210:261–267. doi: 10.1016/j.expneurol.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Dawson DA, Goldstein RB, Moss HB, Li TK, Grant BF. Gender differences in the relationship of internalizing and externalizing psychopathology to alcohol dependence: likelihood, expression and course. Drug Alcohol Depend. 2010;112:9–17. doi: 10.1016/j.drugalcdep.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Monte SM, Kril JJ. Human alcohol-related neuropathology. Acta Neuropathol. 2014;127:71–90. doi: 10.1007/s00401-013-1233-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaud L, Chadda R. Sex differences in rats in the development of and recovery from ethanol dependence assessed by changes in seizure susceptibility. Alcohol Clin Exp Res. 2001;25:1689–1696. [PubMed] [Google Scholar]
- Durafourt BA, Moore CS, Zammit DA, Johnson TA, Zaguia F, Guiot MC, Bar-Or A, Antel JP. Comparison of polarization properties of human adult microglia and blood-derived macrophages. Glia. 2012;60:717–727. doi: 10.1002/glia.22298. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sola J, Nicolas-Arfelis J. Gender differences in alcoholic cardiomyopathy. J Gend Specif Med. 2002;5:41–47. [PubMed] [Google Scholar]
- Finn D, Crabbe J. Chronic ethanol differentially alters susceptibility to chemically induced convulsions in Withdrawal Seizure-Prone and -Resistant mice. J Pharmacol Exp Ther. 1999;288:782–790. [PubMed] [Google Scholar]
- Fuster JM. Frontal lobe and cognitive development. J Neurocytol. 2002;31:373–385. doi: 10.1023/a:1024190429920. [DOI] [PubMed] [Google Scholar]
- Han S, Yang BZ, Kranzler HR, Liu X, Zhao H, Farrer LA, Boerwinkle E, Potash JB, Gelernter J. Integrating GWASs and human protein interaction networks identifies a gene subnetwork underlying alcohol dependence. Am J Hum Genet. 2013;93:1027–1034. doi: 10.1016/j.ajhg.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch UK. Microglia as a source and target of cytokines. Glia. 2002;40:140–155. doi: 10.1002/glia.10161. [DOI] [PubMed] [Google Scholar]
- Hashimoto JG, Forquer MR, Tanchuck MA, Finn DA, Wiren KM. Importance of genetic background for risk of relapse shown in altered prefrontal cortex gene expression during abstinence following chronic alcohol intoxication. Neuroscience. 2011;173:57–75. doi: 10.1016/j.neuroscience.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto JG, Wiren KM. Neurotoxic consequences of chronic alcohol withdrawal: expression profiling reveals importance of gender over withdrawal severity. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2008;33:1084–1096. doi: 10.1038/sj.npp.1301494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helzer JE, Pryzbeck TR. The co-occurrence of alcoholism with other psychiatric disorders in the general population and its impact on treatment. J Stud Alcohol. 1988;49:219–224. doi: 10.15288/jsa.1988.49.219. [DOI] [PubMed] [Google Scholar]
- Holden C. Sex and the suffering brain. Science. 2005;308:1574. doi: 10.1126/science.308.5728.1574. [DOI] [PubMed] [Google Scholar]
- Hommer D, Momenan R, Kaiser E, Rawlings R. Evidence for a gender-related effect of alcoholism on brain volumes. Am J Psychiatry. 2001;158:198–204. doi: 10.1176/appi.ajp.158.2.198. [DOI] [PubMed] [Google Scholar]
- Huang D, Sherman B, Lempicki R. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Iancu OD, Oberbeck D, Darakjian P, Metten P, McWeeney S, Crabbe JC, Hitzemann R. Selection for drinking in the dark alters brain gene coexpression networks. Alcohol Clin Exp Res. 2013;37:1295–1303. doi: 10.1111/acer.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang E, Kim JH, Lee S, Kim JH, Seo JW, Jin M, Lee MG, Jang IS, Lee WH, Suk K. Phenotypic polarization of activated astrocytes: the critical role of lipocalin-2 in the classical inflammatory activation of astrocytes. J Immunol. 2013;191:5204–5219. doi: 10.4049/jimmunol.1301637. [DOI] [PubMed] [Google Scholar]
- John U, Rumpf HJ, Bischof G, Hapke U, Hanke M, Meyer C. Excess mortality of alcohol-dependent individuals after 14 years and mortality predictors based on treatment participation and severity of alcohol dependence. Alcohol Clin Exp Res. 2013;37:156–163. doi: 10.1111/j.1530-0277.2012.01863.x. [DOI] [PubMed] [Google Scholar]
- Joslyn G, Ravindranathan A, Brush G, Schuckit M, White RL. Human variation in alcohol response is influenced by variation in neuronal signaling genes. Alcohol Clin Exp Res. 2010;34:800–812. doi: 10.1111/j.1530-0277.2010.01152.x. [DOI] [PubMed] [Google Scholar]
- Kane MA, Folias AE, Wang C, Napoli JL. Ethanol elevates physiological all-trans-retinoic acid levels in select loci through altering retinoid metabolism in multiple loci: a potential mechanism of ethanol toxicity. The FASEB Journal. 2010;24:823–832. doi: 10.1096/fj.09-141572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaphalia L, Calhoun WJ. Alcoholic lung injury: metabolic, biochemical and immunological aspects. Toxicol Lett. 2013;222:171–179. doi: 10.1016/j.toxlet.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khocht A, Schleifer S, Janal M, Keller S. Neutrophil function and periodontitis in alcohol-dependent males without medical disorders. Journal of the International Academy of Periodontology. 2013;15:68–74. [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosobud A, Bodor A, Crabbe J. Voluntary consumption of ethanol in WSP, WSC and WSR selectively bred mouse lines. Pharmacol Biochem Behav. 1988;29:601–607. doi: 10.1016/0091-3057(88)90026-3. [DOI] [PubMed] [Google Scholar]
- Kosobud A, Crabbe J. Ethanol withdrawal in mice bred to be genetically prone or resistant to ethanol withdrawal seizures. J Pharmacol Exp Ther. 1986;238:170–177. [PubMed] [Google Scholar]
- Kril JJ, Halliday GM, Svoboda MD, Cartwright H. The cerebral cortex is damaged in chronic alcoholics. Neuroscience. 1997;79:983–998. doi: 10.1016/s0306-4522(97)00083-3. [DOI] [PubMed] [Google Scholar]
- Kroener S, Mulholland PJ, New NN, Gass JT, Becker HC, Chandler LJ. Chronic alcohol exposure alters behavioral and synaptic plasticity of the rodent prefrontal cortex. PLoS One. 2012;7:e37541. doi: 10.1371/journal.pone.0037541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn BM. Findings on alcohol dependence point to promising avenues for targeted therapies. JAMA. 2009;301:1643–1645. doi: 10.1001/jama.2009.535. [DOI] [PubMed] [Google Scholar]
- Liu T, Wan L, Wu Y, Chen J, Huang J. hSWS1.SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. J Biol Chem. 2011;286:41758–41766. doi: 10.1074/jbc.M111.271080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loft S, Olesen KL, Dossing M. Increased susceptibility to liver disease in relation to alcohol consumption in women. Scand J Gastroenterol. 1987;22:1251–1256. doi: 10.3109/00365528708996472. [DOI] [PubMed] [Google Scholar]
- Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67. doi: 10.1038/nature13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson A, Goransson M, Heilig M. Early onset alcohol dependence with high density of family history is not “male limited”. Alcohol. 2010;44:131–139. doi: 10.1016/j.alcohol.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011;9:34. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Ackermann K, Croissant B, Mundle G, Nakovics H, Diehl A. Neuroimaging of gender differences in alcohol dependence: are women more vulnerable? Alcohol Clin Exp Res. 2005;29:896–901. doi: 10.1097/01.alc.0000164376.69978.6b. [DOI] [PubMed] [Google Scholar]
- Marshall SA, McClain JA, Kelso ML, Hopkins DM, Pauly JR, Nixon K. Microglial activation is not equivalent to neuroinflammation in alcohol-induced neurodegeneration: The importance of microglia phenotype. Neurobiology of disease. 2013;54:239–251. doi: 10.1016/j.nbd.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsolais D, Rosen H. Chemical modulators of sphingosine-1-phosphate receptors as barrier-oriented therapeutic molecules. Nat Rev Drug Discov. 2009;8:297–307. doi: 10.1038/nrd2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD, Harris RA, Schuckit MA. Genetic factors influencing alcohol dependence. Br J Pharmacol. 2008;154:275–287. doi: 10.1038/bjp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hyytia P, Colombo G, Liang T, Edenberg HJ, Lumeng L, Bell RL. Gene expression within the extended amygdala of 5 pairs of rat lines selectively bred for high or low ethanol consumption. Alcohol. 2013;47:517–529. doi: 10.1016/j.alcohol.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintick JN, Xuei X, Tischfield JA, Goate A, Foroud T, Wetherill L, Ehringer MA, Edenberg HJ. Stress-response pathways are altered in the hippocampus of chronic alcoholics. Alcohol. 2013;47:505–515. doi: 10.1016/j.alcohol.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AJ, Guidot DM. Alcohol abuse, the alveolar macrophage and pneumonia. Am J Med Sci. 2012;343:244–247. doi: 10.1097/MAJ.0b013e31823ede77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AJ, Yeligar SM, Elon L, Brown LA, Guidot DM. Alcoholism causes alveolar macrophage zinc deficiency and immune dysfunction. Am J Respir Crit Care Med. 2013;188:716–723. doi: 10.1164/rccm.201301-0061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morean ME, Corbin WR. Subjective response to alcohol: a critical review of the literature. Alcohol Clin Exp Res. 2010;34:385–395. doi: 10.1111/j.1530-0277.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- Obayashi S, Tabunoki H, Kim SU, Satoh J. Gene expression profiling of human neural progenitor cells following the serum-induced astrocyte differentiation. Cellular and molecular neurobiology. 2009;29:423–438. doi: 10.1007/s10571-008-9338-2. [DOI] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nature neuroscience. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive MF. Pharmacotherapies for alcoholism: the old and the new. CNS Neurol Disord Drug Targets. 2010;9:2–4. doi: 10.2174/187152710790966722. [DOI] [PubMed] [Google Scholar]
- Parlet CP, Waldschmidt TJ, Schlueter AJ. Chronic Ethanol Feeding Induces Subset Loss and Hyporesponsiveness in Skin T Cells. Alcohol Clin Exp Res. 2014 doi: 10.1111/acer.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- Pino-Lagos K, Benson MJ, Noelle RJ. Retinoic acid in the immune system. Ann N Y Acad Sci. 2008;1143:170–187. doi: 10.1196/annals.1443.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polich JM. Epidemiology of alcohol abuse in military and civilian populations. American journal of public health. 1981;71:1125–1132. doi: 10.2105/ajph.71.10.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts JR, Howard MR, Verma S. Recurrent severe alcoholic hepatitis: clinical characteristics and outcomes. Eur J Gastroenterol Hepatol. 2013;25:659–664. doi: 10.1097/MEG.0b013e32835d83d9. [DOI] [PubMed] [Google Scholar]
- Prescott C, Caldwell C, Carey G, Vogler G, Trumbetta S, Gottesman I. The Washington University Twin Study of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2005;134:48–55. doi: 10.1002/ajmg.b.30124. [DOI] [PubMed] [Google Scholar]
- Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J. Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. Lancet. 2009;373:2223–2233. doi: 10.1016/S0140-6736(09)60746-7. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bertsch BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E, Jerome RE, Lumeng L, Nurnberger JI, Jr, Edenberg HJ, McBride WJ, Niculescu AB. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J. 2007;7:222–256. doi: 10.1038/sj.tpj.6500420. [DOI] [PubMed] [Google Scholar]
- Roerecke M, Rehm J. Cause-specific mortality risk in alcohol use disorder treatment patients: a systematic review and meta-analysis. Int J Epidemiol. 2014 doi: 10.1093/ije/dyu018. [DOI] [PubMed] [Google Scholar]
- Romano GH, Harari Y, Yehuda T, Podhorzer A, Rubinstein L, Shamir R, Gottlieb A, Silberberg Y, Pe’er D, Ruppin E, Sharan R, Kupiec M. Environmental stresses disrupt telomere length homeostasis. PLoS genetics. 2013;9:e1003721. doi: 10.1371/journal.pgen.1003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Saitz R, Ghali WA, Moskowitz MA. The impact of alcohol-related diagnoses on pneumonia outcomes. Arch Intern Med. 1997;157:1446–1452. [PubMed] [Google Scholar]
- Schafer GL, Crabbe JC, Wiren KM. Identification of neuroendocrine-specific protein as an ethanol-regulated gene with mRNA differential display. Mammalian genome: official journal of the International Mammalian Genome Society. 1998;9:979–982. doi: 10.1007/s003359900910. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Hesselbrock V, Bucholz KK, Bierut L, Edenberg H, Kramer J, Longacre E, Fukukura T, Kalmijn J, Danko GP, Trim R. Clinical implications of tolerance to alcohol in nondependent young drinkers. Am J Drug Alcohol Abuse. 2008;34:133–149. doi: 10.1080/00952990701877003. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Kalmijn J, Tsuang J, Hesselbrock V, Bucholz K. Response to alcohol in daughters of alcoholics: a pilot study and a comparison with sons of alcoholics. Alcohol Alcohol. 2000;35:242–248. doi: 10.1093/alcalc/35.3.242. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Trim R, Fukukura T, Allen R. The overlap in predicting alcohol outcome for two measures of the level of response to alcohol. Alcohol Clin Exp Res. 2009;33:563–569. doi: 10.1111/j.1530-0277.2008.00870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Trim RS, Kuperman S, Kramer J, Hesselbrock V, Bucholz KK, Nurnberger JI, Jr, Hesselbrock M, Saunders G. Sex differences in how a low sensitivity to alcohol relates to later heavy drinking. Drug and alcohol review. 2012;31:871–880. doi: 10.1111/j.1465-3362.2012.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sershen H, Hashim A, Vadasz C. Strain and sex differences in repeated ethanol treatment-induced motor activity in quasi-congenic mice. Genes, brain, and behavior. 2002;1:156–165. doi: 10.1034/j.1601-183x.2002.10303.x. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Zhu ZG, Qu Y, Li JF, Ji YB, Cai Q, Liu BY, Yan M, Yin HR, Lin YZ. The impact of hyperthermic chemotherapy on human gastric cancer cell lines: preliminary results. Oncol Rep. 2006;16:631–641. [PubMed] [Google Scholar]
- Terdal E, Crabbe J. Indexing withdrawal in mice: Matching genotypes for exposure in studies using ethanol vapor inhalation. Alcohol Clin Exp Res. 1994;18:542–547. doi: 10.1111/j.1530-0277.1994.tb00907.x. [DOI] [PubMed] [Google Scholar]
- Timko C, Debenedetti A, Moos BS, Moos RH. Predictors of 16-year mortality among individuals initiating help-seeking for an alcoholic use disorder. Alcohol Clin Exp Res. 2006;30:1711–1720. doi: 10.1111/j.1530-0277.2006.00206.x. [DOI] [PubMed] [Google Scholar]
- Torres OV, Walker EM, Beas BS, O’Dell LE. Female rats display enhanced rewarding effects of ethanol that are hormone dependent. Alcohol Clin Exp Res. 2014;38:108–115. doi: 10.1111/acer.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treadwell JA, Singh SM. Microarray analysis of mouse brain gene expression following acute ethanol treatment. Neurochem Res. 2004;29:357–369. doi: 10.1023/b:nere.0000013738.06437.a6. [DOI] [PubMed] [Google Scholar]
- Vetreno RP, Hall JM, Savage LM. Alcohol-related amnesia and dementia: animal models have revealed the contributions of different etiological factors on neuropathology, neurochemical dysfunction and cognitive impairment. Neurobiol Learn Mem. 2011;96:596–608. doi: 10.1016/j.nlm.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls SA, Rosenwasser AM, Devaud LL. Sex and regional differences in effects of chronic intermittent ethanol exposure on subsequent excitotoxic challenges in hippocampal slice cultures. Neurosci Lett. 2013 doi: 10.1016/j.neulet.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Wang G, Volkow N, Fowler J, Franceschi D, Wong C, Pappas N, Netusil N, Zhu W, Felder C, Ma Y. Alcohol intoxication induces greater reductions in brain metabolism in male than in female subjects. Alcohol Clin Exp Res. 2003;27:909–917. doi: 10.1097/01.ALC.0000071740.56375.BA. [DOI] [PubMed] [Google Scholar]
- Wang J, Gutala R, Sun D, Ma JZ, Sheela RC, Ticku MK, Li MD. Regulation of platelet-derived growth factor signaling pathway by ethanol, nicotine, or both in mouse cortical neurons. Alcohol Clin Exp Res. 2007;31:357–375. doi: 10.1111/j.1530-0277.2006.00331.x. [DOI] [PubMed] [Google Scholar]
- Wang LL, Yang AK, He SM, Liang J, Zhou ZW, Li Y, Zhou SF. Identification of molecular targets associated with ethanol toxicity and implications in drug development. Curr Pharm Des. 2010;16:1313–1355. doi: 10.2174/138161210791034030. [DOI] [PubMed] [Google Scholar]
- Wiren KM. Males and females are just different: sexually dimorphic responses to chronic ethanol exposure in hippocampal slice cultures. Neuroscience letters. 2013;550:1–5. doi: 10.1016/j.neulet.2013.06.030. [DOI] [PubMed] [Google Scholar]
- Zahr NM, Kaufman KL, Harper CG. Clinical and pathological features of alcohol-related brain damage. Nature reviews Neurology. 2011;7:284–294. doi: 10.1038/nrneurol.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article 17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zindel LR, Kranzler HR. Pharmacotherapy of alcohol use disorders: seventy-five years of progress. Journal of studies on alcohol and drugs Supplement. 2014;75(Suppl 17):79–88. doi: 10.15288/jsads.2014.s17.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Crews FT. Inflammasome-IL-1beta Signaling Mediates Ethanol Inhibition of Hippocampal Neurogenesis. Frontiers in neuroscience. 2012;6:77. doi: 10.3389/fnins.2012.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]