Abstract
Background
The aetiology of systemic sclerosis (SSc) is not clear, but there is an emerging evidence of gene-specific epigenetic dysregulation in the pathogenesis of SSc.
Methods
We performed a genome-wide DNA methylation study in dermal fibroblasts in six diffuse cutaneous SSc (dSSc) patients, six limited cutaneous SSc (lSSc) patients compared with 12 age-matched, sex-matched and ethnicity-matched healthy controls. Cytosine methylation was quantified in more than 485 000 methylation sites across the genome. Differentially methylated CpG sites between patients and controls with a fold difference ≥1.2 were identified. Quantitative real-time RT-PCR was performed to assess correlation between DNA methylation changes and gene expression levels.
Results
We identified 2710 and 1021 differentially methylated CpG sites in dSSc and lSSc, respectively. Of the differentially methylated sites, 61% in dSSc and 90% in lSSc were hypomethylated. There were only 203 CpG sites differentially methylated in both dSSc and lSSc, representing 118 hypomethylated and 6 hypermethylated genes. Common hypomethylated genes include ITGA9, encoding an α integrin. Other relevant genes such as ADAM12, COL23A1, COL4A2 and MYO1E, and transcription factors genes RUNX1, RUNX2 and RUNX3 were also hypomethylated in both dSSc and lSSc. Pathway analysis of differentially methylated genes in both dSSc and lSSc revealed enrichment of genes involved in extracellular matrix–receptor interaction and focal adhesion. We demonstrate significant correlation between DNA methylation status and gene expression in the majority of genes evaluated.
Conclusions
Our data highlight common and subset-specific aberrancies in dSSc and lSSc fibroblasts at the epigenomic levels and identify novel candidate genes in SSc.
INTRODUCTION
Scleroderma (systemic sclerosis (SSc)) is a complex multisystem autoimmune disease characterised by vascular damage, autoimmunity and activation of fibroblasts leading to excessive deposition of collagen in the skin and internal organs.1 Depending on the extent of skin involvement, SSc is categorised as limited cutaneous (lSSc) if skin thickening is confined to the extremities distal to the elbows and knees with or without facial involvement, whereas SSc is categorised as diffuse cutaneous (dSSc) if skin thickening involves areas proximal to the elbows and knees, including the trunk.2 The aetiology of SSc remains unclear despite vigorous research efforts. However, there is growing evidence of gene-specific epigenetic alterations in SSc.3–5 Supported by the observation of striking geographic clustering of SSc,6,7 these epigenetic alterations might indicate a possible role for environmental–epigenetic factors in the pathogenesis of SSc.
Tissue fibroblasts are considered the core synthetic cells of the extracellular matrix (ECM) components, including collagen.8 Fibroblasts also play a role in activation of the immune system via production of numerous cytokines and chemokines and upregulation of adhesion and costimulatory molecules. It is well established that SSc fibroblasts differ from normal fibroblasts in several ways that include, but not limited to, persistently activated phenotype characterised by excessive production of collagen,9 increased proliferation and decreased apoptosis in vitro.10 Therefore, studying SSc fibroblasts is of paramount importance in identifying ‘fibroblasts-mediated’ biological pathways, which might serve the relentless search for disease-modifying therapy for this often-fatal disease.
DNA methylation is considered the core epigenetic mechanism that regulates gene expression by altering transcriptional accessibility of regulatory regions within gene sequences.11 In this study, we performed a genome-wide DNA methylation study in dermal fibroblasts from patients with dSSc and lSSc compared with fibroblasts from control subjects. Our aim was to gain a fundamental understanding of the pathogenesis of SSc through identifying the differentially methylated genes in SSc and characterising biological pathways enriched by these genes. We identified aberrant methylation in key common genes and pathways between dSSc and lSSc, as well as subset-specific genes and pathways that are pertinent to the pathogenesis of SSc. Moreover, we demonstrate good correlation between gene-specific DNA methylation status and gene expression in the majority of genes that we evaluated in this study.
METHODS
Systemic sclerosis patients and controls
We studied 6 patients with dSSc, 6 patients with lSSc and 12 healthy controls. Patients and controls were matched for age (±5 years), sex and ethnicity. Demographic features of the study participants are shown in table 1. All patients had skin thickening of the fingers extending proximal to the metacarpophalangeal joints and fulfilled classification criteria of SSc.12–14 The clinical features of patients with SSc included in this study are shown in table 2. The institutional review board at the University of Michigan approved this study. All study participants signed a written informed consent prior to participation.
Table 1.
Demographic characteristics of the study participants
| SSc patients
|
Controls
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | Type | Age (years) | Sex | Ethnicity | Immunosuppressive medications | ID | Age (years) | Sex | Ethnicity |
| P1 | dSSc | 60 | Male | EA | None | C1 | 65 | Male | EA |
| P2 | dSSc | 25 | Female | EA | None | C2 | 23 | Female | EA |
| P3 | dSSc | 71 | Male | EA | None | C3 | 67 | Male | EA |
| P4 | dSSc | 30 | Female | AA | None | C4 | 29 | Female | AA |
| P5 | dSSc | 45 | Female | EA | None | C5 | 44 | Female | EA |
| P6 | dSSc | 65 | Female | EA | None | C6 | 67 | Female | EA |
| P7 | lSSc | 63 | Female | EA | None | C7 | 64 | Female | EA |
| P8 | lSSc | 38 | Female | EA | None | C8 | 38 | Female | EA |
| P9 | lSSc | 61 | Male | EA | None | C9 | 61 | Male | EA |
| P10 | lSSc | 62 | Female | EA | Mycophenolate mofetil | C10 | 62 | Female | EA |
| P11 | lSSc | 61 | Female | EA | Prednisone (5 mg daily) | C11 | 58 | Female | EA |
| P12 | lSSc | 74 | Female | EA | None | C12 | 70 | Female | EA |
Mean age is 49.3 years for patients with dSSc, 49.1 years for their controls, and 59.8 years for lSSc, and 58.8 years for their controls. p Value=0.99 and 0.88, respectively.
AA, African-American; dSSc, diffuse cutaneous systemic sclerosis; EA, European-American; lSSc, limited cutaneous systemic sclerosis.
Table 2.
Clinical and laboratory characteristics of SSc patients and healthy controls included in this study
| Clinical characteristic | dSSc (n=6) | lSSc (n=6) | Controls (n=12) |
|---|---|---|---|
| Age (mean±SD) | 49.3±19.0 | 59.8±11.7 | 54.0±16.2 |
| Sex (M/F) | 2/4 | 1/5 | 3/9 |
| Ethnicity (AA/EA) | 1/5 | 0/6 | 1/11 |
| Disease duration (years) | 3.9±6.0 | 7.6±8.7 | NA |
| MRSS (mean±SD) | 20.6±10.7 | 7.0±4.8 | NA |
| Organ involvement | |||
| Interstitial lung disease | 4 | 2 | NA |
| Raynaud’s phenomena | 6 | 6 | NA |
| Gastrointestinal involvement | 2 | 3 | NA |
| Scleroderma renal crisis | 1 | 0 | NA |
| Pulmonary hypertension | 2 | 2 | NA |
| Serological characteristics | |||
| Anti-topoisomerase I (Scl-70) | 4 | 0 | NA |
| Anti-centromere | 0 | 1 | NA |
| Anti-RNA polymerase III | 1 | 0 | NA |
| Anti-RNP | 0 | 0 | NA |
AA, African-American; EA, European-American; MRSS, modified Rodnan skin score; NA, not applicable.
Fibroblasts isolation
Two 4 mm Punch skin biopsies were obtained under strict aseptic technique from the dorsum of the forearm, distal to the elbow, from all subjects involved in this study. Skin specimens were incubated briefly in RPMI media with an Antibiotic-Antimycotic (Life technologies, Carlsbad, California, USA), then treated with a mix of digestion enzymes that include 2.4 units/mL dispase (Sigma-Aldrich, Carlsbad, California, USA), 650 units/mL type II collagenase (Life Technologies) and 10 000 Dornase units/mL DNase (Millipore, Temecula, California, USA), and incubated for 2 h at 37°C. After digestion, cells were resuspended with complete growth media (EGM-2MV, New Bullet, Lonza) and plated on a gelatin-coated plate. Cells were harvested after 7–10 days. All fibroblasts used in the DNA methylation study were isolated from primary cell culture (without subculturing or cell passage, passage 0) by negative selection after depleting the endothelial cells from cultured cells using CD31 Microbead kit (Miltenyi Biotec, Cambridge, Massachusetts, USA), as described by the manufacturer’s protocol. Isolation of genomic DNA was performed using the DNeasy Kit (Qiagen, Valencia, California, USA) as described in the manufacturer’s protocol.
Gene expression analysis
Total RNA was extracted from fibroblasts for gene expression analysis by using RNeasy kits (Qiagen) as previously described.15 cDNA was prepared using Verso cDNA synthesis kits (Thermo Scientific). Quantitative PCR was performed using SYBR Green PCR master mix (Applied Biosystems). All samples were run in duplicate using Applied Biosystems Real-Time PCR System and analysed using 7500 Applied Biosystems software. Gene quantification cycle values were normalised to β-actin expression using a standard curve methodology to obtain relative cell equivalents. We evaluated expression levels of eight genes that were hypomethylated in both diffuse and limited SSc (ADAM12, COL23A1, COL4A2, ITGA9, MYO1E, PAX9, RUNX2 and RUNX3) in 24 samples that were mostly independent samples (n=8 dSSc-, 8 lSSc-, and 8 control fibroblasts). Primers for RT-PCR are available upon request.
DNA methylation studies
Genome-wide DNA methylation in fibroblasts from SSc patients and controls included in this study was assessed using the Illumina Infinium HumanMethylation450 BeadChip array, which allows for the interrogation of over 485 000 methylation sites within the entire genome. This array covers 99% of RefSeq genes, with an average of 17 CpG sites per gene across the promoter region, 5′ untranslated region (5′-UTR), first exon, gene body and 3′-UTR. It also covers 96% of CpG islands. Non-CpG-methylated sites recently identified in human stem cells are also covered as well as microRNA promoter regions.
DNA methylation analysis was performed using the GenomeStudio methylation analysis package (Illumina) as previously described.16,17 The average level of DNA methylation (β) on each CpG site was compared between patients and controls. To identify differentially methylated CpG sites between cases and controls, we used three data filtering criteria: (i) CpG site with an average difference in DNA methylation level of at least 1.2-fold; (ii) differential methylation score of ≥22 (p≤0.01) after adjusting for multiple testing using a false discovery rate (FDR) of 5%; and (iii) exclusion of CpG sites assessed by probes with a genetic variant located within 10 bp of the 3′ end of the probe. The p value for differential methylation between patients and controls was calculated by the following equation:
where ‘s’ represents the SD estimate and ‘z’ represents the two-sided tail probability of the standard normal distribution. The SD estimate ‘s’ is a function of β and was calculated by the following equation:
where A, B and C values were derived by Illumina from repeatedly measuring loci with known methylation fractions ranging from 0 to 1 and fitting a parabola to the SD as a function of β. A differential methylation scores for each probe was calculated as
To systematically highlight the most over-represented biological terms, we performed gene ontology (GO), network and pathway analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) V.6.7.18 In pathway and GO analysis, we used Expression Analysis Systematic Explorer (EASE) Score threshold of <0.1 for the detection of gene enrichment analysis (EASE score represents a modified Fisher’s exact p value, which is considered a measure to examine the significance of gene-term enrichment), in addition to fold enrichment of 1.5 and FDR for correction of multiple testing <10%.19
RESULTS
We evaluated DNA methylation changes in dermal fibroblasts from two independent sets consisting of dSSc and lSSc patients and age-matched, sex-matched and ethnicity-matched controls. Of the 485 000 CpG sites that were evaluated at a genome-wide level, we identified a total of 3528 differentially methylated CpG sites in fibroblasts from patients with SSc compared with healthy controls. A total of 2710 CpG sites were identified in dSSc fibroblasts, the majorities were hypomethylated (1653 CpG sites) compared with healthy matched controls, representing 818 hypomethylated and 513 hypermethylated gene regions. On the other hand, we identified 1021 differentially methylated CpG sites in lSSc fibroblasts; the majorities are hypomethylated (916 CpG sites), representing 496 hypomethylated and 56 hypermethylated gene regions. Surprisingly, there were only 203 (~6%) common differentially methylated CpG sites between diffuse and limited SSc, representing 118 hypomethylated and 6 hypermethylated genes (table 3, see online supplementary table S1 and figure S1). Further, there were 57 hypomethylated and 7 hypermethylated additional common genes, with unique differentially methylated CpG sites, in both dSSc and lSSc (see online supplementary table S2). This finding highlights an interesting diversity of DNA methylation profile in dermal fibroblasts, at the genome-wide level, between diffuse and limited SSc subsets.
Table 3.
Summary of the differentially methylated CpG sites and genes in diffuse and limited systemic sclerosis fibroblasts compared with healthy control fibroblasts
| Increased methylation | Decreased methylation | Total | |
|---|---|---|---|
| dSSc | |||
| CpG sites | 1057 | 1653 | 2710 |
| Fold change | 1.20–21.40 | 1.20–11.58 | |
| (Average) | 1.59 | 1.52 | |
| Differential score | 22.0 to 338.5 | −22.0 to −336.2 | |
| (Average) | 41.2 | −46.8 | |
| Genes | 513 | 818 | 1269* |
| lSSc | |||
| CpG sites | 105 | 916 | 1021 |
| Fold change | 1.20–21.20 | 1.22–6.53 | |
| (Average) | 2.35 | 1.52 | |
| Differential score | 22.0 to 337.0 | −22.0 to −183.9 | |
| (Average) | 81.7 | −36.5 | |
| Genes | 56 | 496 | 548* |
| Common between dSSc and lSSc | |||
| CpG sites | 15 | 188 | 203 |
| Fold change | 1.20–13.11 | 1.21–4.56 | |
| (Average) | 2.37 | 1.62 | |
| Differential score | 22.4 to 338.4 | −22.1 to −268.6 | |
| (Average) | 74.2 | −44.2 | |
| Genes | 6 | 118 | 121* |
This total represents total differentially methylated genes, some with both hypomethylated and hypermethylated CpG sites.
dSSc, diffuse cutaneous systemic sclerosis; lSSc, limited cutaneous systemic sclerosis.
None of the patients included in this study were taking any medications that are known to affect DNA methylation. One patient in the lSSc group was taking mycophenolate mofetil and another patient was on a small dose of prednisone (table 1). The mean DNA methylation levels in the differentially methylated CpG sites were not different between these two patients and the rest of the patients included in the lSSc group who are not on any medications. This was further confirmed using hierarchal clustering analysis (not shown). Therefore, it is unlikely that medication exposure is affecting the DNA methylation differences observed between patients and controls in this study.
Common differentially methylated genes in both dSSc and lSSc
As demonstrated above, around 6% of all differentially methylated CpG sites in fibroblasts were shared between dSSc and lSSc subsets; the vast majority of these were hypomethylated in reference to control fibroblasts. Hypomethylated common genes between diffuse and limited SSc include ITGA9, encoding for integrin α 9, which is an integral membrane glycoprotein that mediates cell–cell and cell–matrix adhesion. We identified one hypomethylated CpG site in the body of ITGA9. Other relevant differentially methylated genes in both diffuse and limited SSc include ADAM12, which encodes for ADAM metallopeptidase domain 12, and has extracellular metalloprotease and cell-binding functions. We identified three hypomethylated CpG sites in the body of ADAM12 in dSSc, two of which were also hypomethylated in lSSc.
Other genes hypomethylated in both dSSc and lSSc include COL4A2 and COL23A1, which encode for collagen type IV α2 and collagen type XXIII α1, respectively. Of note, we identified other genes that encode for collagens, which were hypomethylated in SSc subsets; specifically, COL8A1, COL16A1 and COL29A1 were hypomethylated in dSSc fibroblasts, whereas COL1A1, COL6A3 and COL12A1 were hypomethylated in lSSc fibroblasts. Moreover, PAX9 was hypomethylated in both dSSc and lSSc (four and two CpG sites, respectively). This gene encodes the pro-α2 chain of type I collagen, which is found in most connective tissues. Among genes encoding for ECM glycoproteins, we identified hypomethylation of TNXB in diffuse and limited SSc.
Among the transcription factors, RUNX1, RUNX2 and RUNX3 were all hypomethylated in fibroblasts from both dSSc and lSSc patients. Table 4 lists some of the top-rank relevant differentially methylated common genes between dSSc and lSSc.
Table 4.
Sample of the top-rank differentially methylated common genes between dSSc and lSSc, including the genes used to evaluate gene expression by RT-PCR*
| dSSc
|
lSSc
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| # | Gene | # CpG | Location | Meth status | Diff score: mean (range) | Fold change: mean (range) | # CpG | Location | Meth status | Diff score: mean (range) | Fold change: mean (range) |
| 1 | NCLN | 1 | Body | HypOMeth | −268.64 | 1.53 | 1 | Body | HypOMeth | −83.55 | 1.32 |
| 2 | SORCS2 | 2 | Body | HypOMeth | −91.76 (−27.17 to −156.34) | 1.96 (1.21–2.72) | 2 | Body | HypOMeth | −23.96 (−23.52 to −24.40) | 1.42 (1.40–1.43) |
| 3 | CAMK1D | 2 | Body | HypOMeth | −101.84 (−76.65 to −127.04) | 2.06 (1.83–2.28) | 2 | Body | HypOMeth | −46.53 (−37.07 to −55.99) | 1.57 (1.42–1.71) |
| 4 | KRT12 | 1 | Body | HypOMeth | −116.64 | 1.93 | 1 | Body | HypOMeth | −28.68 | 1.40 |
| 5 | IMMP2L | 2 | Body | HypOMeth | −81.69 (−53.22 to −110.16) | 2.13 (1.28–2.98) | 2 | Body | HypOMeth | −47.62 (−42.01 to −53.23) | 1.57 (1.32–1.82) |
| 6 | CTDSPL | 1 | Body | HypOMeth | −108.00 | 2.25 | 1 | Body | HypOMeth | −26.56 | 1.50 |
| 7 | NRGN | 1 | Body | HypOMeth | −106.00 | 1.58 | 1 | Body | HypOMeth | −52.06 | 1.28 |
| 8 | RAPGEF4 | 2 | Body | HypOMeth | −83.27 (−64.02 to −102.53) | 2.05 (1.51–2.59) | 1 | Body | HypOMeth | −78.63 | 2.02 |
| 9 | LFNG | 2 | TSS1500; Body | HypOMeth | −77.32 (−56.16 to −98.47) | 1.94 (1.38–2.50) | 2 | TSS1500; Body | HypOMeth | −37.24 (−32.55 to −41.92) | 1.70 (1.58–1.83) |
| 10 | IFT88 | 1 | Body | HypOMeth | −37.37 | 1.41 | 1 | Body | HypOMeth | −93.06 | 1.78 |
| 11 | ODZ4 | 2 | 5′UTR; Body | HypOMeth | −57.31 (−26.94 to −87.68) | 1.58 (1.30–1.86) | 2 | 5′UTR; Body | HypOMeth | −45.07 (−34.86 to −55.28) | 1.49 (1.30–1.67) |
| 12 | WIPF1 | 2 | 5′UTR; TSS1500 | HypOMeth | −53.95 (−27.41 to −80.49) | 1.36 (1.32–1.39) | 1 | 5′UTR; TSS1500 | HypOMeth | −39.91 | 1.23 |
| 13 | RCBTB2 | 1 | Body | HypOMeth | −76.25 | 1.90 | 1 | Body | HypOMeth | −83.20 | 1.89 |
| 14 | COL4A2* | 1 | Body | HypOMeth | −50.34 | 1.35 | 1 | Body | HypOMeth | −28.64 | 1.25 |
| 15 | ESRRG | 1 | 5′UTR | HypOMeth | −47.74 | 2.03 | 1 | 5′UTR | HypOMeth | −87.36 | 2.46 |
| 16 | NRP1 | 3 | Body; 3′UTR | HypOMeth | −62.34 (−43.35 to −81.46) | 1.49 (1.37–1.73) | 1 | Body | HypOMeth | −83.24 | 2.21 |
| 17 | ITGA9* | 1 | Body | HypOMeth | −41.40 | 1.35 | 1 | Body | HypOMeth | −37.29 | 1.80 |
| 18 | COL23A1* | 1 | Body | HypOMeth | −40.35 | 1.25 | 1 | Body | HypOMeth | −136.16 | 1.25 |
| 19 | CACNA1C | 1 | Body | HypOMeth | −32.87 | 1.67 | 1 | Body | HypOMeth | −27.13 | 1.55 |
| 20 | PAX9* | 4 | 5′UTR; TSS1500; body | HypOMeth | −31.95 (−27.35 to −39.98) | 1.48 (1.34–1.77) | 2 | 5′UTR | HypOMeth | −29.97 (−26.44 to −33.50) | 1.35 (1.24–1.49) |
| 21 | NID2 | 2 | TSS1500; Body | HypOMeth | −32.01 (−22.60 to −41.43) | 2.10 (1.36–2.85) | 2 | TSS1500 | HypOMeth | −46.09 (−23.02 to −69.16) | 1.58 (1.46–1.71) |
| 22 | RUNX2* | 2 | 5′UTR; 1st Exon; body | HypOMeth | −36.41 (−29.99 to −42.83) | 1.30 (1.25–1.35) | 1 | 5′UTR; 1st Exon | HypOMeth | −46.20 | 1.27 |
| 23 | ADAM12* | 3 | Body | HypOMeth | −38.31 (−23.04 to −47.87) | 1.38 (1.22–1.75) | 2 | Body | HypOMeth | −42.41 (−32.80 to −52.01) | 1.58 (1.39–1.77) |
| 24 | MYO1E* | 3 | Body | HypOMeth | −59.18 (−53.72 to −63.75) | 1.85 (1.49–2.17) | 2 | Body | HypOMeth | −26.38 (−23.51 to −29.25) | 1.37 (1.21–1.58) |
| 25 | RUNX3* | 6 | 5′UTR; TSS200; TSS1500; 1st Exon | HypOMeth | −62.37 (−31.45 to −95.09) | 1.27 (1.21–1.48) | 1 | TSS1500 | HypOMeth | −55.14 | 1.32 |
| 26 | MIR154 | 1 | TSS1500 | HypOMeth | −63.76 | 1.90 | 1 | TSS1500 | HypOMeth | −24.18 | 1.51 |
| 27 | RUNX1 | 3 | 5′UTR; TSS1500; 1st Exon; Body | HypOMeth | −71.94 (−38.88 to −92.95) | 1.79 (1.46–2.09) | 2 | Body | HypOMeth | −36.88 (−28.59 to −45.16) | 1.58 (1.53–1.64) |
| 28 | MYOM2 | 2 | Body | HypOMeth | −81.74 (−79.46 to −84.01) | 3.41 (2.27–4.56) | 2 | Body | HypOMeth | −36.21 (−29.06 to −43.36) | 1.70 (1.58–1.83) |
| 29 | WDSUB1 | 1 | 5′UTR;1st Exon | HypERMeth | 26.07 | 9.80 | 1 | 5′UTR;1stExon | HypERMeth | 47.97 | 13.11 |
| 30 | OSR2 | 5 | Body | HypERMeth | 58.17 (39.73 to 91.80) | 1.38 (1.27–1.48) | 1 | Body | HypERMeth | 47.88 | 1.27 |
RT-qPCR performed for these genes.
dSSc, diffuse cutaneous systemic sclerosis; HypERMeth, hypermethylated; HypOMeth, hypomethylated; lSSc, limited cutaneous systemic sclerosis; Meth status, methylation status; TSS1500, within 1500 bps from transcription start site; 5′UTR, 5′ untranslated region.
Ontologies and pathways enriched by differentially methylated CpG sites common between dSSc and lSSc
We used DAVID software18 to facilitate the systematic identification and grouping of differentially methylated genetic loci into biological networks. Canonical pathway analysis identified ECM–receptor interaction (p=2.92E-03) and focal cell adhesion (p=8.86E-03) as the most significant pathways unifying the differentially methylated genes in fibroblasts from patients with dSSc and lSSc (table 5). GO analysis of hypomethylated common genes between dSSc and lSSc demonstrated enrichment of genes involved in negative regulation of MAP kinase activity (p=2.14E-04), cell projection morphogenesis (p=9.72E-04), axonogenesis (p=1.11E-03), cell part morphogenesis (p=1.29E-03), cell adhesion (p=1.89E-03), cell motility (p=3.97E-03) and positive regulation of transcription, DNA-dependent (p=6.17E-03), among other ontologies (table 5).
Table 5.
Canonical pathways and gene ontologies enriched by differentially methylated genes with common differentially methylated CpG sites between dSSc and lSSc
| Category | ID | Term | p Value | Fold enrichment | FDR |
|---|---|---|---|---|---|
| (A) Canonical pathways | |||||
| KEGG_PATHWAY | hsa04512 | ECM–receptor interaction | 2.92E-03 | 5.95 | 0.03 |
| KEGG_PATHWAY | hsa04510 | Focal adhesion | 8.86E-03 | 3.32 | 0.09 |
| (B) Gene ontologies of hypomethylated common gens between dSSc and lSSc* | |||||
| GOTERM_BP_FAT | GO:0043407 | Negative regulation of MAP kinase activity | 2.14E-04 | 16.63 | 0.00 |
| GOTERM_BP_FAT | GO:0048858 | Cell projection morphogenesis | 9.72E-04 | 4.40 | 0.02 |
| GOTERM_BP_FAT | GO:0007409 | Axonogenesis | 1.11E-03 | 4.96 | 0.02 |
| GOTERM_BP_FAT | GO:0032990 | Cell part morphogenesis | 1.29E-03 | 4.21 | 0.02 |
| GOTERM_BP_FAT | GO:0007155 | Cell adhesion | 1.89E-03 | 2.57 | 0.03 |
| GOTERM_BP_FAT | GO:0003012 | Muscle system process | 2.71E-03 | 4.99 | 0.04 |
| GOTERM_BP_FAT | GO:0030030 | Cell projection organisation | 3.47E-03 | 3.25 | 0.05 |
| GOTERM_BP_FAT | GO:0030029 | Actin filament-based process | 3.88E-03 | 3.97 | 0.06 |
| GOTERM_BP_FAT | GO:0051674 | Localisation of cell | 3.97E-03 | 3.51 | 0.06 |
| GOTERM_BP_FAT | GO:0048870 | Cell motility | 3.97E-03 | 3.51 | 0.06 |
| GOTERM_BP_FAT | GO:0000904 | Cell morphogenesis involved in differentiation | 4.15E-03 | 3.93 | 0.06 |
| GOTERM_BP_FAT | GO:0010604 | Positive regulation of macromolecule metabolic process | 4.64E-03 | 2.24 | 0.07 |
| GOTERM_BP_FAT | GO:0042325 | Regulation of phosphorylation | 5.26E-03 | 2.83 | 0.08 |
| GOTERM_BP_FAT | GO:0006469 | Negative regulation of protein kinase activity | 5.85E-03 | 6.88 | 0.09 |
| GOTERM_BP_FAT | GO:0045893 | Positive regulation of transcription, DNA-dependent | 6.17E-03 | 2.76 | 0.09 |
| GOTERM_BP_FAT | GO:0043405 | Regulation of MAP kinase activity | 6.31E-03 | 5.09 | 0.09 |
| GOTERM_BP_FAT | GO:0051254 | Positive regulation of RNA metabolic process | 6.54E-03 | 2.74 | 0.10 |
| GOTERM_BP_FAT | GO:0033673 | Negative regulation of kinase activity | 6.59E-03 | 6.65 | 0.10 |
Number of hypermethylated common genes between dSSc and lSSc is inadequate to evaluate hypermethylated ontologies.
ECM, extracellular matrix; FDR, false discovery rate; GOTERM_BP_FAT, Gene Ontology term biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Genes, gene ontologies and pathways enriched by differentially methylated genes in dSSc
We identified differential methylation of several genes that are likely related to the pathogenesis of SSc in fibroblasts isolated from patients with dSSc. We identified two hypomethylated CpG sites in CDH11, which encode for cadherin-11, both CpG sites are located in the 5′UTR, with an average differential score of −51.8, and average fold change of 1.30 compared with healthy controls. Additionally, PDGFC, which encode for platelet-derived growth factor isoform C, was hypomethylated in diffuse SSc fibroblasts. PDGFC is a profibrotic factor that is overexpressed in SSc20 and that contributes to fibroblast activation and transformation to myofibroblast in SSc.
The Wnt/β-catenin signalling pathway has been established as a critical pathway in activation of fibroblasts in wound healing and maintenance of fibrogenesis in SSc.21–23 In this study, we demonstrate hypomethylation of CTNNA2 and CTNNB1 in dSSc.
We demonstrate hypermethylation of SOX2OT, which encode for a long non-protein-coding RNA. SOX2OT was represented on the microarray by 91 CpG sites across the gene. We identified 53 hypermethylated CpG sites in fibroblasts from patients with dSSc compared with control fibroblasts, around 1/3 of these CpG sites were located in proximity to transcription start site. Of particular interest, none of these CpG sites in SOX2OTwere differentially methylated in lSSc fibroblasts.
Online Supplementary table S3 provides a list of all differentially methylated CpG sites in diffuse SSc fibroblasts compared with control fibroblasts.
GO analysis of all hypomethylated genes in dSSc demonstrated enrichment of genes involved in cell adhesion (p=1.51E-09), cell morphogenesis involved in transformation (p=3.11E-06), neuron projection development (p=7.71E-06), skeletal system development (p=6.01E-05), blood vessel morphogenesis (p=2.92E-03) and immune system development (p=5.40E-03). Whereas GO ontologies of hypermethylated genes in dSSc include genes involved in embryonic morphogenesis (p=1.93E-04), negative regulation of mitotic cell cycle (p=2.26E-04), actin cytoskeleton organisation (p=9.90E-04), negative regulation of transcription (p=1.26E-03), regulation of cell migration (p=1.76E-03) and actin filament-based process (p=1.87E-03), among others. Pathway analysis of the differentially methylated genes in dSSc demonstrated gene enrichment of pathways involved in focal adhesion (p=2.84E-06), vascular smooth muscle contraction (p=1.34E-04), adherens junction (p=1.60E-03), ECM–receptor interaction (p=3.57E-03) and gap junction (p=5.91E-03) (see online supplementary table S4).
Genes, gene ontologies and pathways enriched by differentially methylated genes in lSSc
The Wnt/β-catenin pathway was represented by hypomethylation of CTNNA3 and CTNND2 in lSSc. In addition, COL1A1, COL6A3 and COL12A1 were hypomethylated in lSSc fibroblasts as mentioned above (see online supplementary table S5 which provides all differentially methylated CpG sites in lSSc fibroblasts).
GO analysis of hypomethylated genes in lSSc demonstrated enrichment of genes involved in embryonic morphogenesis (p=9.20E-05), skeletal system development (p=1.55E-04), cell adhesion (p=1.61E-04), positive regulation of transcription (p=1.66E-04), actin filament-based process (p=1.09E-03) and regulation of cell-substrate adhesion (p=5.60E-03), among other ontologies. Pathway analysis demonstrated enrichment of genes involved in ECM–receptor interaction (p=1.75E-04), and focal adhesion (p=8.85E-04) (see online supplementary table S6).
Correlation between methylation status of representative genes and gene expression
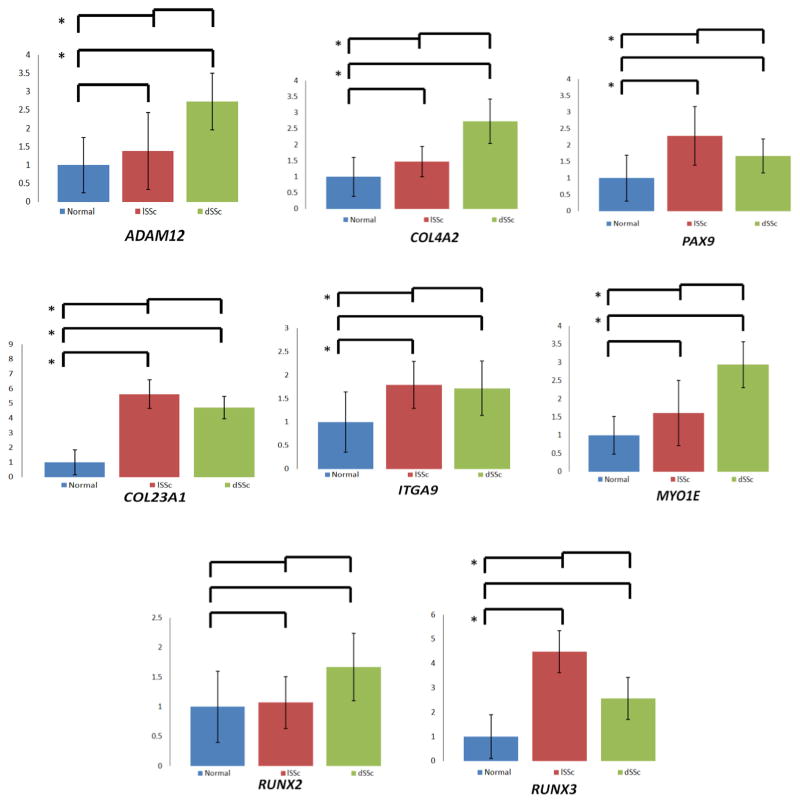
To assess the functional effect of gene methylation status on gene expression, we performed RT-PCR analysis on a sample of genes that were hypomethylated in both dSSc and lSSc (ADAM12, COL23A1, COL4A2, ITGA9, MYO1E, PAX9, RUNX3 and RUNX2). We found significant overexpression of the majority of these genes in SSc fibroblasts compared with control fibroblasts (figure 1).
Figure 1.
Correlation of gene expression and methylation status. To evaluate the functional significance of methylation status on gene expression, we performed RT-qPCR for eight selected genes that were hypomethylated in both diffuse and limited SSc fibroblasts. Overall most of the hypomethylated genes were overexpressed in SSc fibroblasts. The columns in the graph show the average individual gene expression level in limited and diffuse SSc in reference to control fibroblasts, error bars represent the SD. t test was used to compare the mean of individual gene expression levels in each SSc subset, as well as all SSc samples. *p value ≤0.05.
DISCUSSION
In this study, we performed an unbiased genome-wide DNA methylation study in fibroblasts from patients with dSSc and lSSc, and identified several differentially methylated genes and involved pathways compared with healthy matched controls. The most striking finding in this study is the distinct and characteristic methylation pattern displayed in fibroblasts from dSSc and lSSc subsets, as reflected by the number of the differentially methylated CpG sites and corresponding genes in dSSc and lSSc (ratio 2.6:1, respectively). Nevertheless, we also demonstrate an interesting similarity as highlighted by the observation that around 6% (203 out of 3528) of the differentially methylated CpG sites were indeed common CpG sites between diffuse and limited SSc.
The second important finding in this study is the demonstration of several hypomethylated and overexpressed genes that are integral to the pathogenesis of SSc (figure 1). Of particular interest, most of the hypomethylated CpG sites in these genes were detected in gene bodies, but are nonetheless associated with significant expression changes.
We demonstrate hypomethylation and mRNA overexpression of ITGA9 in both dSSc and lSSc fibroblasts. Upregulation of integrins was demonstrated previously in both SSc fibroblasts24–26 as well as in lung fibroblasts in patients with fibrotic lung disease.27 Overexpression of integrins contributes to myofibroblast differentiation28 and activation of TGF-β.29 There is evidence of an extensive bidirectional crosstalk between integrins and TGF-β signalling in fibrosis, with TGF-β inducing integrin expression and integrins modulating TGF-β activation and signalling.30 Our results indicate a possible role of ITGA9 hypomethylation in its overexpression in SSc, which may contribute to TGF-β upregulation.
It has been increasingly apparent that ADAM12 overexpression contributes to the process of fibrosis through enhancing TGF-β signalling pathway.31–34 Knockdown of ADAM12 in fibroblasts has been shown to be sufficient to limit collagen accumulation in vitro.35 We demonstrate hypomethylation of ADAM12 in both dSSc and lSSc fibroblasts compared with control fibroblasts, and we confirmed ADAM12 overexpression, especially in dSSc fibroblasts.
Tissue fibrosis in SSc is orchestrated by fibroblasts via production of excessive amount of ECM components, including collagens. Among the common differentially methylated genes in dSSc and lSSc, we identified hypomethylation of several genes that encode for collagens and demonstrate overexpression of two collagen genes (COL23A1, COL4A2) in dSSc and lSSc fibroblasts compared with control fibroblasts. Moreover, TNXB was hypomethylated in dSSc and lSSc fibroblasts. TNXB encodes a member of the tenascin family of ECM glycoproteins, which are involved in matrix maturation.36
RUNX1 and RUNX2 are transcription factors that induce expression of SOX5 and SOX6, which leads to the induction of type II collagen expression via the direct regulation of promoter activity, and therefore, collagen synthesis.37,38 RUNX3, another member of the Runx family, is also likely to contribute to collagen synthesis in association with RUNX2.39 We identified hypomethylation of RUNX1, RUNX2 and RUNX3 in dSSc and lSSc. Furthermore, we evaluated mRNA expression of RUNX2 and RUNX3 and demonstrate overexpression of RUNX3 in SSc fibroblasts, but no statistically significant difference in the expression of RUNX2 in either dSSc or lSSc compared with controls (figure 1). The observation that the hypomethylated genes that we evaluated are generally overexpressed argues for a possible role of DNA methylation defects in overexpression of these genes and suggests a possible role of DNA methylation in the pathogenesis of SSc. However, although hypomethylated DNA in gene regulatory regions represents an epigenetic mark for accessibility of the transcriptional machinery to the chromatin, DNA hypomethylation should not be viewed as the sine qua none for genomic transcription as other factors, like transcription factors, are needed for active transcription.
There has been tremendous interest to understand the factors that lead to recapitulation of Wnt/β-catenin signalling pathway in SSc,22,40–43 which is generally required for embryonic development, and cell fate determination. We identified aberrant methylation of genes representative of the Wnt/β-catenin pathway in SSc. Specifically, we identified hypomethylation CTNNA2, CTNNB1 in dSSc fibroblasts, and CTNNA3, and CTNND2 in lSSc fibroblasts compared with control fibroblasts. Our observation supports a role of Wnt/β-catenin in SSc pathogenesis and provides evidence for epigenetic alterations in this pathway in SSc fibroblast.
In diffuse SSc fibroblasts, we identified hypomethylation of CDH11, which encode for cadherin-11. Cadherins are a group of transmembrane glycoproteins that mediate cell-to-cell adhesion. It has been established that CDH11 has a role in the process of fibrosis,44 wound healing, lung fibrosis45,46 and SSc.47,48 Indeed, CDH11-deficient mice had decreased fibrotic endpoints in bleomycin-induced fibrosis.45 We postulate that hypomethylation of CDH11 contributes to its overexpression, which facilitates the differentiation of resident tissue fibroblasts into myofibroblasts in SSc.
Multiple studies in a variety of cell types have demonstrated altered DNA methylation patterns in a number of autoimmune and fibrotic diseases, providing important new directions towards understanding the pathogenesis of immune-mediated diseases.17,49–55 Epigenetic studies in fibroblasts in autoimmunity included recent genome-wide DNA methylation profiling of synovial fibroblasts in rheumatoid arthritis.50,55 De la Rica and colleagues55 reported differential methylation of 2571 CpG sites, associated with 1240 different genes in rheumatoid arthritis synovial fibroblasts (RASF). Of interest, 330 of these genes were also differentially methylated in at least one subset of SSc fibroblasts in our study. Out of the 330 common differentially methylated genes, 125 were hypomethylated and 79 were hypermethylated in both RASF and SSc fibroblasts, and the rest showed mixed methylation pattern (see online supplementary table S7). Moreover, we identified 30 hypomethylated and 4 hypermethylated common genes between RASF and both subsets of SSc (lSSc and dSSc) (see online supplementary table S7). Canonical pathway analysis identified ‘tight junction’ (hsa04530, p=2.46E-03, FDR=2.8%) as the most significant pathway representing common differentially methylated genes in RASF and SSc fibroblasts (n=330).
In summary, we identified genome-wide DNA methylation changes for the first time in SSc fibroblasts. Our study demonstrates a striking diversity of DNA methylation profile, as demonstrated by more intense aberrant DNA methylation in dSSc compared with lSSc fibroblasts, which provides a new perspective to the heterogeneity of SSc. We evaluated the effect of aberrant DNA methylation on gene expression in a set of genes that were hypomethylated in both dSSc and lSSc, and established good correlation between DNA methylation pattern and gene expression. These data indicate that abnormal DNA methylation exists in genes involved in integral pathways involved in fibrogenesis, such as the TGF-β and Wnt/β-catenin signaling pathways. Our findings emphasise the potential role of DNA methylation changes in the pathogenesis of SSc, and the potential for epigenome-wide studies to identify novel aspects in complex diseases. Replication studies and studies looking at DNA methylation aberrancies associated with specific features of SSc are warranted.
Supplementary Material
Acknowledgments
The authors are very grateful to Dr. Alisa E. Koch for her contributions to this study.
Funding
This work was supported by funds from the University of Michigan to AHS, and new investigator start-up funds from the School of Medicine at University of Toledo to NA. DK is supported by NIH/NIAMS grant number K24 AR063120-02, P-ST is supported by the Arthritis Foundation and AHS is supported by NIH/NIAID grant number R01AI097134.
Footnotes
Additional material is published online only. To view please visit the journal online (http://dx.doi.org/10.1136/annrheumdis-2014-205303).
Contributors
All authors listed contributed and fulfil authorship criteria as follows: substantial contributions to the conception or design of the work; or the acquisition, analysis or interpretation of data for the work; drafting the work or revising it critically for important intellectual content; final approval of the version to be published; and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Competing Interest
None.
Ethics approval
IRB at the University of Michigan.
Provenance and peer review
Not commissioned; externally peer reviewed.
References
- 1.Abraham DJ, Varga J. Scleroderma: from cell and molecular mechanisms to disease models. Trends Immunol. 2005;26:587–95. doi: 10.1016/j.it.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 2.LeRoy EC, Black C, Fleischmajer R, et al. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol. 1988;15:202–5. [PubMed] [Google Scholar]
- 3.Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 2006;54:2271–9. doi: 10.1002/art.21948. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Kahaleh B. Epigenetic repression of bone morphogenetic protein receptor II expression in scleroderma. J Cell Mol Med. 2013;17:1291–9. doi: 10.1111/jcmm.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Wang Q, Zhao M, et al. Demethylation of ITGAL (CD11a) regulatory sequences in CD4+T lymphocytes of Systemic Sclerosis. San Diego: American College of Rheumatolgoy; 2013. Abstract # 2905. [Google Scholar]
- 6.Valesini G, Litta A, Bonavita MS, et al. Geographical clustering of scleroderma in a rural area in the province of Rome. Clin Exp Rheumatol. 1993;11:41–7. [PubMed] [Google Scholar]
- 7.Silman AJ. Epidemiology of scleroderma. Ann Rheum Dis. 1991;50(Suppl 4):846–53. doi: 10.1136/ard.50.suppl_4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharyya S, Wei J, Tourtellotte WG, et al. Fibrosis in systemic sclerosis: common and unique pathobiology. Fibrogenesis Tissue Repair. 2012;5(Suppl 1):S18. doi: 10.1186/1755-1536-5-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leroy EC. Connective tissue synthesis by scleroderma skin fibroblasts in cell culture. J Exp Med. 1972;135:1351–62. doi: 10.1084/jem.135.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu YS, Kong J, Cheema GS, et al. The immunobiology of systemic sclerosis. Semin Arthritis Rheum. 2008;38:132–60. doi: 10.1016/j.semarthrit.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Altorok N, Sawalha AH. Epigenetics in the pathogenesis of systemic lupus erythematosus. Curr Opin Rheumatol. 2013;25:569–76. doi: 10.1097/BOR.0b013e328364206f. [DOI] [PubMed] [Google Scholar]
- 12.van den Hoogen F, Khanna D, Fransen J, et al. 2013 classification criteria for systemic sclerosis: an american college of rheumatology/european league against rheumatism collaborative initiative. Arthritis Rheum. 2013;65:2737–47. doi: 10.1002/art.38098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Hoogen F, Khanna D, Fransen J, et al. 2013 classification criteria for systemic sclerosis: an American college of rheumatology/European league against rheumatism collaborative initiative. Ann Rheum Dis. 2013;72:1747–55. doi: 10.1136/annrheumdis-2013-204424. [DOI] [PubMed] [Google Scholar]
- 14.Preliminary criteria for the classification of systemic sclerosis (scleroderma) Subcommittee for scleroderma criteria of the American Rheumatism Association Diagnostic and Therapeutic Criteria Committee. Arthritis Rheum. 1980;23:581–90. doi: 10.1002/art.1780230510. [DOI] [PubMed] [Google Scholar]
- 15.Tsou PS, Talia NN, Pinney AJ, et al. Effect of oxidative stress on protein tyrosine phosphatase 1B in scleroderma dermal fibroblasts. Arthritis Rheum. 2012;64:1978–89. doi: 10.1002/art.34336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altorok N, Coit P, Hughes T, et al. Genome-wide DNA methylation patterns in naive CD4T cells from patients with primary Sjogren’s syndrome. Arthritis Rheum. 2014;66:731–9. doi: 10.1002/art.38264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coit P, Jeffries M, Altorok N, et al. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+ T cells from lupus patients. J Autoimmun. 2013;43:78–84. doi: 10.1016/j.jaut.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Tan FK, Hildebrand BA, Lester MS, et al. Classification analysis of the transcriptosome of nonlesional cultured dermal fibroblasts from systemic sclerosis patients with early disease. Arthritis Rheum. 2005;52:865–76. doi: 10.1002/art.20871. [DOI] [PubMed] [Google Scholar]
- 21.Fathke C, Wilson L, Shah K, et al. Wnt signaling induces epithelial differentiation during cutaneous wound healing. BMC Cell Biol. 2006;7:4. doi: 10.1186/1471-2121-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei J, Melichian D, Komura K, et al. Canonical Wnt signaling induces skin fibrosis and subcutaneous lipoatrophy: a novel mouse model for scleroderma? Arthritis Rheum. 2011;63:1707–17. doi: 10.1002/art.30312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei J, Fang F, Lam AP, et al. Wnt/beta-catenin signaling is hyperactivated in systemic sclerosis and induces Smad-dependent fibrotic responses in mesenchymal cells. Arthritis Rheum. 2012;64:2734–45. doi: 10.1002/art.34424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asano Y, Ihn H, Yamane K, et al. Increased expression of integrin alpha(v)beta3 contributes to the establishment of autocrine TGF-beta signaling in scleroderma fibroblasts. J Immunol. 2005;175:7708–18. doi: 10.4049/jimmunol.175.11.7708. [DOI] [PubMed] [Google Scholar]
- 25.Asano Y, Ihn H, Yamane K, et al. Increased expression of integrin alphavbeta5 induces the myofibroblastic differentiation of dermal fibroblasts. Am J Pathol. 2006;168:499–510. doi: 10.2353/ajpath.2006.041306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asano Y, Ihn H, Yamane K, et al. Increased expression levels of integrin alphavbeta5 on scleroderma fibroblasts. Am J Pathol. 2004;164:1275–92. doi: 10.1016/s0002-9440(10)63215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horan GS, Wood S, Ona V, et al. Partial inhibition of integrin alpha(v)beta6 prevents pulmonary fibrosis without exacerbating inflammation. Am J Respir Crit Care Med. 2008;177:56–65. doi: 10.1164/rccm.200706-805OC. [DOI] [PubMed] [Google Scholar]
- 28.Carracedo S, Lu N, Popova SN, et al. The fibroblast integrin alpha11beta1 is induced in a mechanosensitive manner involving activin A and regulates myofibroblast differentiation. J Biol Chem. 2010;285:10434–45. doi: 10.1074/jbc.M109.078766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munger JS, Huang X, Kawakatsu H, et al. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96:319–28. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 30.Margadant C, Sonnenberg A. Integrin-TGF-beta crosstalk in fibrosis, cancer and wound healing. EMBO Rep. 2010;11:97–105. doi: 10.1038/embor.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi-Wen X, Renzoni EA, Kennedy L, et al. Endogenous endothelin-1 signaling contributes to type I collagen and CCN2 overexpression in fibrotic fibroblasts. Matrix Biol. 2007;26:625–32. doi: 10.1016/j.matbio.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Atfi A, Dumont E, Colland F, et al. The disintegrin and metalloproteinase ADAM12 contributes to TGF-beta signaling through interaction with the type II receptor. J Cell Biol. 2007;178:201–8. doi: 10.1083/jcb.200612046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skubitz KM, Skubitz AP. Gene expression in aggressive fibromatosis. J Lab Clin Med. 2004;143:89–98. doi: 10.1016/j.lab.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Taniguchi T, Asano Y, Akamata K, et al. Serum levels of ADAM12-S: possible association with the initiation and progression of dermal fibrosis and interstitial lung disease in patients with systemic sclerosis. J Eur Acad Dermatol Venereol. 2013;27:747–53. doi: 10.1111/j.1468-3083.2012.04558.x. [DOI] [PubMed] [Google Scholar]
- 35.Dulauroy S, Di Carlo SE, Langa F, et al. Lineage tracing and genetic ablation of ADAM12(+) perivascular cells identify a major source of profibrotic cells during acute tissue injury. Nat Med. 2012;18:1262–70. doi: 10.1038/nm.2848. [DOI] [PubMed] [Google Scholar]
- 36.Egging D, van Vlijmen-Willems I, van Tongeren T, et al. Wound healing in tenascin-X deficient mice suggests that tenascin-X is involved in matrix maturation rather than matrix deposition. Connect Tissue Res. 2007;48:93–8. doi: 10.1080/03008200601166160. [DOI] [PubMed] [Google Scholar]
- 37.Kimura A, Inose H, Yano F, et al. Runx1 and Runx2 cooperate during sternal morphogenesis. Development. 2010;137:1159–67. doi: 10.1242/dev.045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Q, Eberspaecher H, Lefebvre V, et al. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997;209:377–86. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida CA, Yamamoto H, Fujita T, et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952–63. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam AP, Flozak AS, Russell S, et al. Nuclear beta-catenin is increased in systemic sclerosis pulmonary fibrosis and promotes lung fibroblast migration and proliferation. Am J Respir Cell Mol Biol. 2011;45:915–22. doi: 10.1165/rcmb.2010-0113OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carthy JM, Garmaroudi FS, Luo Z, et al. Wnt3a induces myofibroblast differentiation by upregulating TGF-beta signaling through SMAD2 in a beta-catenin-dependent manner. PLoS ONE. 2011;6:e19809. doi: 10.1371/journal.pone.0019809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang W, Wei K, Jacobs SS, et al. SPARC suppresses apoptosis of idiopathic pulmonary fibrosis fibroblasts through constitutive activation of beta-catenin. J Biol Chem. 2010;285:8196–206. doi: 10.1074/jbc.M109.025684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhattacharyya S, Wei J, Varga J. Understanding fibrosis in systemic sclerosis: shifting paradigms, emerging opportunities. Nat Rev Rheumatol. 2012;8:42–54. doi: 10.1038/nrrheum.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinz B, Pittet P, Smith-Clerc J, et al. Myofibroblast development is characterized by specific cell-cell adherens junctions. Mol Biol Cell. 2004;15:4310–20. doi: 10.1091/mbc.E04-05-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider DJ, Wu M, Le TT, et al. Cadherin-11 contributes to pulmonary fibrosis: potential role in TGF-beta production and epithelial to mesenchymal transition. FASEB J. 2012;26:503–12. doi: 10.1096/fj.11-186098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wynn TA. Common and unique mechanisms regulate fibrosis in various fibroproliferative diseases. J Clin Invest. 2007;117:524–9. doi: 10.1172/JCI31487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gardner H, Shearstone JR, Bandaru R, et al. Gene profiling of scleroderma skin reveals robust signatures of disease that are imperfectly reflected in the transcript profiles of explanted fibroblasts. Arthritis Rheum. 2006;54:1961–73. doi: 10.1002/art.21894. [DOI] [PubMed] [Google Scholar]
- 48.Whitfield ML, Finlay DR, Murray JI, et al. Systemic and cell type-specific gene expression patterns in scleroderma skin. Proc Natl Acad Sci U S A. 2003;100:12319–24. doi: 10.1073/pnas.1635114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hughes T, Ture-Ozdemir F, Alibaz-Oner F, et al. Epigenome-wide scan identifies a treatment-responsive pattern of altered DNA methylation among cytoskeletal remodeling genes in monocytes and CD4+ T cells in Behcet’s disease. Arthritis Rheumatol. 2014 Feb 19; doi: 10.1002/art.38409. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano K, Whitaker JW, Boyle DL, et al. DNA methylome signature in rheumatoid arthritis. Ann Rheum Dis. 2013;72:110–17. doi: 10.1136/annrheumdis-2012-201526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altorok N, Coit P, Hughes T, et al. Genome-Wide DNA Methylation Patterns in Naive CD4+ T Cells From Patients With Primary Sjogren’s Syndrome. Arthritis Rheumatol. 2014;66:731–9. doi: 10.1002/art.38264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeffries MA, Dozmorov M, Tang Y, et al. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics. 2011;6:593–601. doi: 10.4161/epi.6.5.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanders YY, Ambalavanan N, Halloran B, et al. Altered DNA methylation profile in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;186:525–35. doi: 10.1164/rccm.201201-0077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rabinovich EI, Kapetanaki MG, Steinfeld I, et al. Global methylation patterns in idiopathic pulmonary fibrosis. PloS ONE. 2012;7:e33770. doi: 10.1371/journal.pone.0033770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de la Rica L, Urquiza JM, Gomez-Cabrero D, et al. Identification of novel markers in rheumatoid arthritis through integrated analysis of DNA methylation and microRNA expression. J Autoimmun. 2013;41:6–16. doi: 10.1016/j.jaut.2012.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.