Abstract
Background
Quisinostat (JNJ-26481585) is a second generation pyrimidyl-hydroxamic acid histone deacetylase (HDAC) inhibitor with high cellular potency towards class I and II HDACs. Quisinostat was selected for clinical development as it showed prolonged pharmacodynamic effects in vivo and demonstrated improved single agent antitumoral efficacy compared to other analogs.
Procedures
Quisinostat was tested against the PPTP in vitro panel at concentrations ranging from 1.0 nM to 10 μM and was tested against the PPTP in vivo panels at a dose of 5 mg/kg (solid tumors) or 2.5 mg/kg (ALL models) administered intraperitoneally daily x 21.
Results
In vitro quisinostat demonstrated potent cytotoxic activity, with T/C% values approaching 0% for all of the cell lines at the highest concentration tested. The median relative IC50 value for the PPTP cell lines was 2.2 nM, (range <1 nM to 19 nM). quisinostat induced significant differences in EFS distribution compared to control in 21 of 33 (64%) of the evaluable solid tumor xenografts and in 4 of 8 (50%) of the evaluable ALL xenografts. An objective response was observed in 1 of 33 solid tumor xenografts while for the ALL panel, two xenografts achieved complete response (CR) or maintained CR, and a third ALL xenograft achieved stable disease.
Conclusions
Quisinostat demonstrated broad activity in vitro, and retarded growth in the majority of solid tumor xenografts studied. The most consistent in vivo activity signals observed were for the glioblastoma xenografts and T-cell ALL xenografts.
Keywords: Preclinical Testing, Developmental Therapeutics, HDAC inhibitor
INTRODUCTION
Transcription of eukaryotic genes is tightly regulated by a complex interplay between DNA methylation and covalent histone modifications (acetylation, methylation, phosphorylation) [1]. The acetylation of lysine residues on histones H3 and H4 modifies the structure of nucleosomes leading to an open chromatin configuration, which facilitates gene transcription. The level of acetylation of histones is the result of dynamic interplay between histone acetyl transferases (HATs) and histone deacetylases (HDACs). Aberrant regulation of the acetylation/deacetylation equilibrium is considered to play a role in generating the abnormal gene expression patterns observed in many forms of cancer [2,3]. For example, the pathogenesis of acute promyelocytic leukemia (APL) and its responsiveness to treatment with all-trans retinoic acid is based on the recruitment by PML-RARα of an HDAC nuclear co-repressor complex to target genes [4].
There are 18 described HDACs distributed in four classes (including the sirtuins). Class I encompasses HDAC1, HDAC2, HDAC 3 and HDAC8. The activity of these HDACs is considered crucial for cell proliferation [5] and is frequently augmented in cancer [6]. Mice lacking Class I HDACs show a range of defects in embryogenesis, particularly of the vasculature, and are not viable [5]. Inhibition of HDAC6 leads to hyperacetylation of tubulin and HSP90, with the latter effect resulting in reduced activity and/or levels of HSP90 client proteins [7,8].
The effects of HDAC inhibition can be broadly grouped as follows: a) induction of apoptosis; b) modification of cell cycle (either G1 or G2/M phase arrest); c) differentiation induction; d) changes in angiogenesis and the tumor microenvironment; and e) production of reactive oxygen species [9]. HDAC inhibition induces a plethora of gene expression changes in cancer cells that may lead to these effects [10], with increases in levels of the cell cycle regulator p21waf being commonly reported as a consequence of HDAC inhibition [11,12]. Reports on the changes induced on gene expression upon histone deacetylase inhibitor (HDI) exposure range from 2% to more than 20% of genes [13,14]. Based on the relaxation of nucleosomes and increased transcription due to histone hyperacetylation, increases in gene expression would be the expected response to HDAC inhibition. There are, however, numerous examples of reduced expression of genes [15], reflecting the complex factors affecting the equilibrium established by HATs and HDACs on their gene targets. Furthermore, besides the lysine residues on histones there are a number of other proteins which can undergo structural and functional changes as a consequence of acetylation/deacetylation (reviewed in [16]), including transcription factors such as p53, Hif1a, Foxo proteins, p65, and STAT proteins.
HDIs are diverse in chemical structure including short chain fatty acid moieties, hydroxamic acid derived compounds, and cyclic oligopeptides. A large number of HDIs have been or are currently under investigation as therapeutic options for cancer [17]. The hydroxamic acid family of drugs (i.e. trichostatin A, vorinostat (SAHA), panobinostat, oxamflatin) is the larger group and many of its members are pan-HDIs (reviewed in [11]). Two HDIs, vorinostat and romidepsin, have received FDA approval for treatment of cutaneous T cell lymphoma [18,19].
Quisinostat is an orally available, second generation pyrimidyl-hydroxamic acid derivative HDAC inhibitor that has completed its phase 1 evaluation in adults with cancer [20]. Quisinostat was selected for development from among 140 candidate HDAC inhibitors based on its prolonged in vivo pharmacodynamic response against an ovarian cancer model [21]. JNJ-26481858 shows relative selectivity for HDAC1 and HDAC2 in enzymatic assays, but in cellular assays it shows broad spectrum activity against both Class I (e.g., HDAC1-3) and Class II (e.g., HDAC6) isozymes [21]. It is approximately 500-fold more potent than vorinostat at inhibiting HDAC1 [21]. Quisinostat potently induced apoptosis against leukemia cell lines [22], and it induced tumor growth inhibition as well as regressions against multiple adult cancer xenografts models [21,23]. Quisinostat was selected for systematic testing by the PPTP based on its improved preclinical efficacy relative to other HDIs and on the potential relevance of HDAC inhibition to the treatment of childhood cancers. This report describes testing of quisinostat as a single agent against the PPTP’s in vitro panel as well as against the in vivo tumor panel.
MATERIALS AND METHODS
In vitro testing
In vitro testing was performed using DIMSCAN, as previously described in a characterized panel of 23 cell lines [24]. Cells were incubated in the presence of quisinostat for 96 hours at concentrations from 1 nM to 10 μM and analyzed as previously described [24].
In vivo tumor growth inhibition studies
CB17SC scid−/− female mice (Taconic Farms, Germantown NY), were used to propagate subcutaneously implanted kidney/rhabdoid tumors, sarcomas (Ewing, osteosarcoma, rhabdomyosarcoma), neuroblastoma, and non-glioblastoma brain tumors, while BALB/c nu/nu mice were used for glioma models, as previously described [25,26]. Human leukemia cells were propagated by intravenous inoculation in female non-obese diabetic (NOD)/scid−/− mice as described previously [27]. Female mice were used irrespective of the patient gender from which the original tumor was derived. All mice were maintained under barrier conditions and experiments were conducted using protocols and conditions approved by the institutional animal care and use committee of the appropriate consortium member. Eight to ten mice were used in each control or treatment group. Tumor volumes (cm3) [solid tumor xenografts] or percentages of human CD45-positive [hCD45] cells [ALL xenografts] were determined as previously described [25]. Responses were determined using three activity measures as previously described [25]. An in-depth description of the analysis methods is included in the Supplemental Response Definitions section.
Statistical Methods
The exact log-rank test, as implemented using Proc StatXact for SAS®, was used to compare event-free survival distributions between treatment and control groups. P-values were two-sided and were not adjusted for multiple comparisons given the exploratory nature of the studies.
Pharmacodynamic studies
Rhabdomyosarcoma xenografts were harvested prior to treatment, or at 4, 8 and 24 hr after dose 1 (5 mg/kg). Additional tumors were harvested 24 hr post day 3 dosing and at 4 hr post day 4 dosing. Samples were prepared for immunoblotting as previously described [28]. Membranes were probed for acetylated Histone H3 and H4, p21 PARP and cleaved PARP. ALL-8 engrafted mice were treated with quisinostat at 2.5 mg/kg daily, and spleen cells (>90% huCD45) were harvested at days 0 (no treatment), 1, 3 and 7 and were analyzed for acetylated histone H4. Loading was normalized to GAPDH (solid tumors) or actin (ALL-8).
Drugs and Formulation
Quisinostat was provided to the Pediatric Preclinical Testing Program by Janssen Pharmaceutica, through the Cancer Therapy Evaluation Program (NCI). Powder was stored at room temperature, protected from light. Drug was formulated in 10% hydroxy-propyl-β-cyclodextrin, 25 mg/mL mannitol, in sterile water for injection, and made fresh prior to administration. Quisinostat was administered intraperitoneally (IP) to mice using a daily schedule. Based upon toxicity testing (non-tumored mice) a dose of 5 mg/kg was selected for the solid tumors, and at a reduced dose of 2.5 mg/kg was used for the ALL panel, for 21 days. Quisinostat was provided to each consortium investigator in coded vials for blinded testing.
RESULTS
In vitro testing
Quisinostat demonstrated potent cytotoxic activity, with T/C% values approaching 0% for all of the cell lines at the highest concentration tested. The median relative (Rel) IC50 value for the PPTP cell lines was 2.2 nM, with a range from <1 nM (MOLT-4, CHLA-9, and CHLA-258) to 19 nM (NB-EBc1), Table I. The ALL cell lines tended to be more sensitive to quisinostat (median Rel IC50 of 1.9 nM) and the rhabdomyosarcoma and neuroblastoma cell lines less sensitive (median Rel IC50 of 5.1 and 6.8 nM, respectively), but these differences were not significant.
Table I.
In vitro activity of JNJ-26481585 against PPTP cell lines.
| Cell Line | Histotype | Rel IC50 (nM) | Panel Rel IC50/Line Rel IC50 | Ymin% (Observed) | p53 Mutation Status |
|---|---|---|---|---|---|
| RD | Rhabdomyosarcoma | 7.2 | 0.30 | 0.02 | R248W |
| Rh41 | Rhabdomyosarcoma | 3.0 | 0.73 | 0.02 | Del1001–1013 |
| Rh18 | Rhabdomyosarcoma | 11.5 | 0.19 | 0.08 | WT |
| Rh30 | Rhabdomyosarcoma | 2.1 | 1.02 | 0.00 | R273C |
| BT-12 | Rhabdoid | 9.5 | 0.23 | 0.03 | A88T |
| CHLA-266 | Rhabdoid | 1.9 | 1.18 | 0.10 | A88T |
| TC-71 | Ewing sarcoma | 3.2 | 0.69 | 0.00 | R213(Stop) |
| CHLA-9 | Ewing sarcoma | <1.0 | >2.21 | 0.00 | ?? |
| CHLA-10 | Ewing sarcoma | 2.5 | 0.88 | 0.00 | ?? |
| CHLA-258 | Ewing sarcoma | <1.0 | >2.21 | 0.00 | WT |
| SJ-GBM2 | Glioblastoma | 1.6 | 1.40 | 0.11 | R273C |
| NB-1643 | Neuroblastoma | 3.8 | 0.57 | 0.11 | WT |
| NB-EBc1 | Neuroblastoma | 19.0 | 0.12 | 0.00 | WT |
| CHLA-90 | Neuroblastoma | 1.1 | 2.01 | 1.46 | E286K |
| CHLA-136 | Neuroblastoma | 9.8 | 0.22 | 0.56 | WT |
| NALM-6 | ALL | 2.2 | 0.98 | 0.00 | WT |
| COG-LL-317 | ALL | 1.3 | 1.75 | 0.00 | WT |
| RS4;11 | ALL | 3.2 | 0.69 | 0.00 | WT |
| MOLT-4 | ALL | <1.0 | >2.21 | 0.01 | WT |
| CCRF-CEM (1) | ALL | 3.1 | 0.71 | 0.00 | R248Q, R175H |
| CCRF-CEM (2) | ALL | 1.6 | 1.33 | 0.00 | R248Q, R175H |
| Kasumi-1 | AML | 1.1 | 2.02 | 0.00 | R248Q |
| Karpas-299 | ALCL | 1.9 | 1.16 | 0.09 | R273C |
| Ramos-RA1 | NHL | 1.4 | 1.57 | 0.00 | I254D |
| Median | 2.2 | 1.00 | 0.00 | ||
| Minimum | <1.0 | 0.12 | 0.00 | ||
| Maximum | 19.0 | >2.21 | 1.46 |
In vivo testing
Quisinostat was tested in vivo using a 5 mg/kg dose for solid tumors and 2.5 mg/kg for ALL xenografts, administered intraperitoneally daily for 3 weeks. Seventeen of 779 mice died during the study (2.2%), with 7 of 390 in the control arms (1.8%) and 10 of 389 (2.6%) in the quisinostat treatment arms. All 41 xenograft models studied were considered evaluable for efficacy. A complete summary of results is provided in Supplemental Table I, including total numbers of mice, number of mice that died (or were otherwise excluded), numbers of mice with events and average times to event, tumor growth delay, as well as numbers of responses and T/C values.
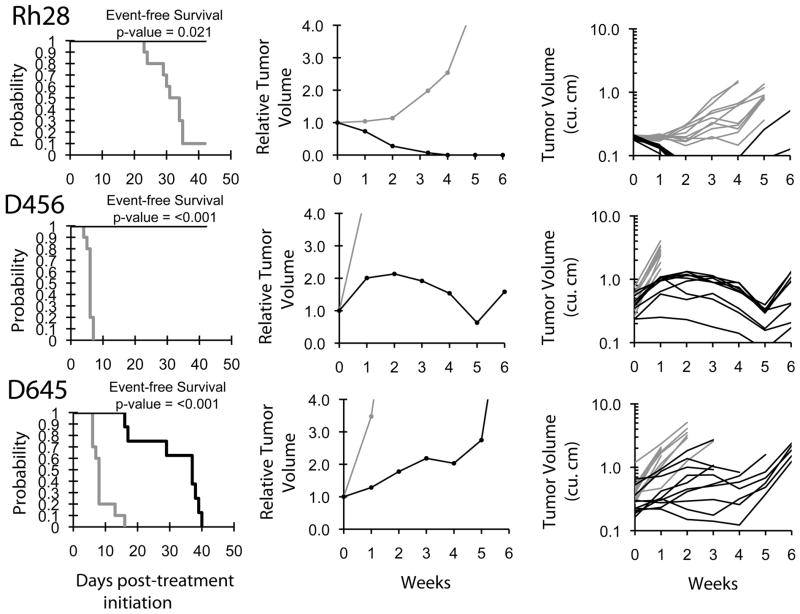
Quisinostat induced significant differences in EFS distribution compared to control in 21 of 33 (64%) evaluable solid tumor xenografts, and in 4 of 8 (50%) of the evaluable ALL xenografts (Table II). Quisinostat induced tumor growth inhibition meeting criteria for intermediate EFS T/C activity (EFS T/C > 2) in 6 of 32 (19%) evaluable solid tumor xenografts. The only solid tumor panel with two xenografts with EFS T/C > 2 was the glioblastoma panel, with both D645 and D456 showing substantial delay in time to event (EFS T/C of 4.5 and >6.7, respectively). For the ALL panel, 3 of 8 (38%) xenografts met criteria for intermediate activity, while one T lineage xenograft (ALL-16) met criteria for high activity.
Table II.
Summary of in Vivo Activity of JNJ-26481585
| Line | Tumor Type | Time to Event | P-value | EFS T/C | Median Final RTV | Tumor Volume T/C | T/C Volume Activity | EFS Activity | Response Activity |
|---|---|---|---|---|---|---|---|---|---|
| BT-29 | Rhabdoid | 31.4 | <0.001 | 2.3 | >4 | 0.47 | Low | Int | Int |
| KT-11 | Wilms | 12.9 | 0.007 | 1.4 | >4 | 0.64 | Low | Low | Low |
| KT-13 | Wilms | 18.1 | <0.001 | 2.1 | >4 | 0.32 | Int | Int | Int |
| SK-NEP-1 | Ewing | 12.9 | <0.001 | 2.1 | >4 | 0.55 | Low | Int | Int |
| EW5 | Ewing | 14.5 | 0.004 | 1.2 | >4 | 0.73 | Low | Low | Low |
| TC-71 | Ewing | 6.3 | 0.994 | 1.1 | >4 | 0.81 | Low | Low | Low |
| CHLA258 | Ewing | 10.1 | 0.807 | 1.2 | >4 | 0.91 | Low | Low | Low |
| Rh28 | ALV RMS | > EP | <0.001 | > 1.3 | 0.0 | 0.22 | Int | NE | High |
| Rh30 | ALV RMS | 12.7 | 0.232 | 1.3 | >4 | 0.62 | Low | Low | Low |
| Rh30R | ALV RMS | 23.4 | <0.001 | 1.8 | >4 | 0.43 | Int | Low | Int |
| Rh41 | ALV RMS | 13.8 | 0.071 | 0.8 | >4 | 1.39 | Low | Low | Low |
| Rh18 | EMB RMS | 15.9 | 0.008 | 2.1 | >4 | 0.60 | Low | Int | Int |
| BT-28 | Medulloblastoma | 11.6 | 0.021 | 1.6 | >4 | 0.68 | Low | Low | Int |
| BT-45 | Medulloblastoma | 6.9 | 0.207 | 1.1 | >4 | 0.78 | Low | Low | Low |
| BT-44 | Ependymoma | 11.2 | <0.001 | 1.3 | >4 | 0.26 | Int | Low | Low |
| BT-35 | Astrocytoma | 6.1 | <0.001 | 1.7 | >4 | 0.40 | Int | Low | Int |
| BT-40 | Astrocytoma | 6.0 | 0.281 | 1.0 | >4 | 1.06 | Low | Low | Low |
| GBM2 | Glioblastoma | 15.1 | 0.355 | 1.8 | >4 | 0.84 | Low | Low | Int |
| BT-39 | Glioblastoma | 10.8 | 0.632 | 1.1 | >4 | 0.84 | Low | Low | Low |
| D645 | Glioblastoma | 37.0 | <0.001 | 4.5 | >4 | 0.40 | Int | Int | Int |
| D456 | Glioblastoma | > EP | <0.001 | > 6.7 | 1.7 | 0.40 | Int | Int | Int |
| NB-SD | Neuroblastoma | 10.4 | 0.031 | 1.2 | >4 | 0.76 | Low | Low | Low |
| NB-1771 | Neuroblastoma | 19.8 | 0.055 | 1.5 | >4 | 0.78 | Low | Low | Low |
| NB-1691 | Neuroblastoma | 10.9 | 0.017 | 1.4 | >4 | 0.81 | Low | Low | Low |
| NB-EBc1 | Neuroblastoma | 15.7 | <0.001 | 1.3 | >4 | 0.53 | Low | Low | Low |
| CHLA-79 | Neuroblastoma | 9.6 | 0.801 | 1.0 | >4 | 1.02 | Low | Low | Low |
| NB-1643 | Neuroblastoma | 6.6 | 0.893 | 1.1 | >4 | 1.01 | Low | Low | Low |
| OS-1 | Osteosarcoma | 32.5 | 0.054 | 1.3 | >4 | 0.88 | Low | Low | Low |
| OS-2 | Osteosarcoma | 32.7 | <0.001 | 1.3 | >4 | 0.53 | Low | Low | Low |
| OS-17 | Osteosarcoma | 20.2 | 0.001 | 1.1 | >4 | 0.78 | Low | Low | Low |
| OS-9 | Osteosarcoma | 34.4 | <0.001 | 1.5 | >4 | 0.56 | Low | Low | Low |
| OS-33 | Osteosarcoma | 19.5 | 0.029 | 1.3 | >4 | 0.83 | Low | Low | Low |
| OS-31 | Osteosarcoma | 26.2 | 0.004 | 1.3 | >4 | 0.65 | Low | Low | Low |
| ALL-2 | ALL B-precursor | 32.9 | 0.004 | 1.9 | >25 | . | Low | Int | |
| ALL-3 | ALL B-precursor | 38.5 | <0.001 | 4.0 | >25 | . | Int | Int | |
| ALL-4 | ALL B-precursor | 4.5 | 0.612 | 0.9 | >25 | . | Low | Low | |
| ALL-7 | ALL B-precursor | 4.8 | 0.029 | 0.7 | >25 | . | Low | Low | |
| ALL-8 | ALL T-cell | > EP | 0.019 | > 4.6 | 23.6 | . | Int | High | |
| ALL-16 | ALL T-cell | > EP | 0.103 | > 2.5 | 0.2 | . | High* | High | |
| ALL-17 | ALL B-precursor | 19.1 | 0.025 | 2.4 | >25 | . | Int | Int | |
| ALL-19 | ALL B-precursor | 3.0 | 0.467 | 1.0 | >25 | . | Low | Low |
High EFS T/C is called despite p>0.05 resulting from irregular growth in control animals.
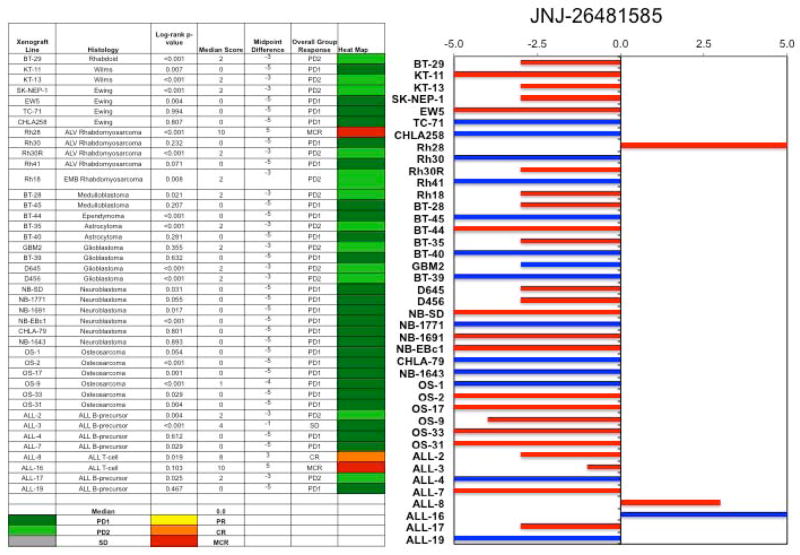
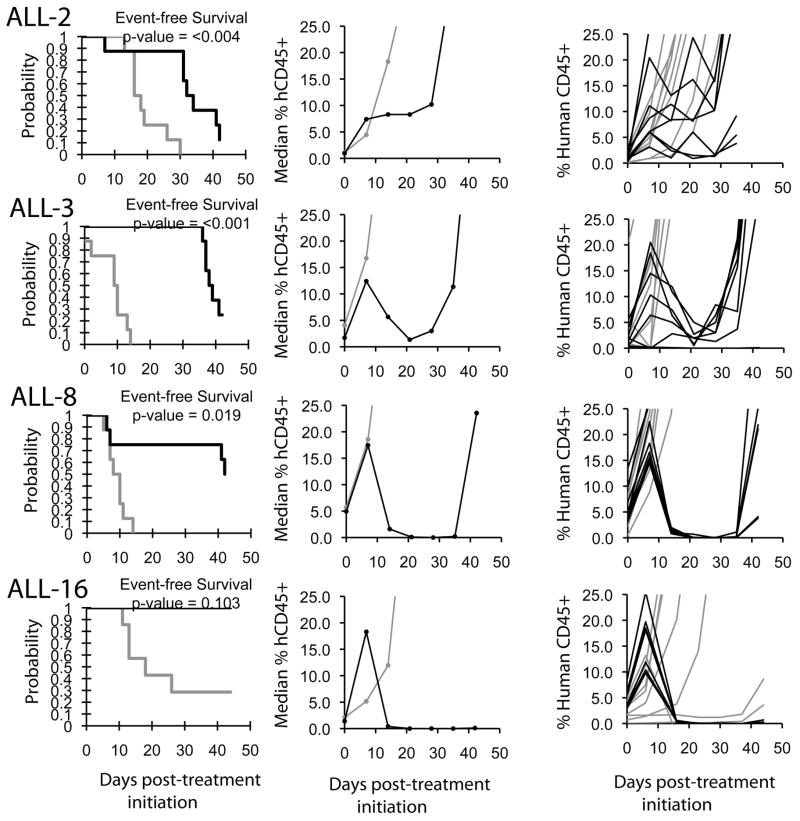
An objective response was observed in 1 of 33 solid tumor xenografts: Rh28 in the rhabdomyosarcoma panel achieved a maintained complete remission (MCR). For the ALL panel, two xenografts achieved CR and MCR, respectively, and a third xenograft achieved stable disease. The two ALL xenografts achieving complete responses (ALL-8 and ALL-16) are both T-cell immunophenotype, and the B-precursor ALL xenograft achieving stable disease (ALL-3) has an MLL gene rearrangement. The in vivo testing results for the objective response measure of activity are presented in Figure 1 in a ‘heat-map’ format as well as a ‘COMPARE’-like format, based on the scoring criteria described in the Supplemental Response Definitions section. The latter analysis demonstrates relative tumor sensitivities around the midpoint score of 5 (stable disease). Examples of the solid tumor xenografts showing either regression (Rh28) or extended tumor growth delay (D456 and D645) are shown in Figure 2. Examples of relative leukemia growth curves for ALL xenografts meeting criteria for intermediate or high activity are shown in Figure 3.
Figure 1.
Quisinostat in vivo objective response activity, left: The colored heat map depicts group response scores. A high level of activity is indicated by a score of 6 or more, intermediate activity by a score of ≥2 but <6, and low activity by a score of <2. Right: representation of tumor sensitivity based on the difference of individual tumor lines from the midpoint response (stable disease). Bars to the right of the median represent lines that are more sensitive, and to the left are tumor models that are less sensitive. Red bars indicate lines with a significant difference in EFS distribution between treatment and control groups, while blue bars indicate lines for which the EFS distributions were not significantly different.
Figure 2.
Quisinostat activity in vivo against individual solid tumor xenografts. Kaplan-Meier curves showing the probability for EFS (left), relative tumor volume (center), and individual tumor volumes (right) graphs are shown for selected xenografts (Rh28, D456, and D645). Controls (gray lines); Treated (black lines), significances of the difference between treated and control groups are included.
Figure 3.
Quisinostat activity in vivo against individual ALL xenografts. Kaplan-Meier curves showing the probability for EFS (left), median leukemia engraftment (center) as detected in peripheral blood (see Materials and Methods), and individual leukemia engraftment (right) graphs are shown for selected xenografts (ALL-2, ALL-3, ALL-8, and ALL19). Controls (gray lines); Treated (black lines), significances of the difference between treated and control groups are included.
Pharmacodynamic studies
To determine whether at the doses and schedule used quisinostat achieved target inhibition, we examined tumor tissue from 3 rhabdomyosarcoma models with differing responses and ALL-8 that had a complete response to treatment (Supplemental Figure 1). Three tumors were assayed at each time point to determine whether there was a drug induced increase in acetylated histones H3 and H4, and p21CIP1. Apoptosis was monitored by assessing cleaved and total PARP. In the most sensitive model, Rh28, there was a marked increase in acetylated H3 4 and 8 hr after dose 1. H3 and H4 acetylation was elevated compared to controls 24 hr after daily dose 3. p21CIP increase paralleled that of acetylated histones, and cleaved PARP was detected in each Rh28 tumor 4 hr after the fourth dose. In the non-responsive Rh30 model increases in H3 and H4 acetylation were similar to those determined in Rh28 xenograft, although p21 induction was seen only after 8 hr, and cleaved PARP was not detected. For the other non-responsive model, Rh41, there was a robust increase in H3 and H4 acetylation at 4 and 8 hr after dose 1 but was deminshed in the 96 and 100 hr samples. Levels of p21CIP1 increased marginally, if at all. For the ALL-8 model, tumor from spleen was analyzed 4 hr after dosing on days 1, 3, and 7. As shown in Supplemental Figure 1D, there was a cumulative increase in acetylated H4 histone.
DISCUSSION
Interest in HDIs and their potential use for cancer treatment has grown substantially in the recent years. In addition to their single agent effects, HDIs have demonstrated sensitization of cancer cells both to radiation and chemotherapeutic treatments [29,30]. A number of reports have described the preclinical activity of HDIs against pediatric cancers, including reports for romidepsin [28,31,32], vorinostat [33,34], and etinostat [35]. Pediatric phase 1 trials of vorinostat and romidepsin have been conducted [36,37], and frontline studies of vorinostat given with radiation therapy have been developed for children with high grade gliomas and diffuse intrinsic pontine glioma. The emerging themes from the prior preclinical reports are that HDIs are strong inducers of apoptosis when tested in vitro, with potency ranging from low micromolar IC50 values for vorinostat to low nanomolar IC50 values for romidepsin. However, in vivo activity for HDIs against pediatric xenografts has been limited. For example, vorinostat induced no objective responses among 30 evaluable solid tumor and 8 evaluable ALL xenografts [33]. Romidepsin at clinically relevant doses induced regressions in 3 of 39 pediatric solid tumor models, with several other xenografts showing prolonged stable disease [28].
Quisinostat demonstrated potent cytotoxic activity in vitro, with T/C% values approaching 0% for all of the cell lines at the highest concentration tested. The 2.2 nM median IC50 value for quisinostat was far lower than the IC50 value observed for the PPTP in vitro cell line panel for vorinostat (1.4 μM) [33]. One of the most sensitive cell lines was MOLT-4, a T-cell ALL cell line, which is consistent with the in vivo testing finding of greatest sensitivity for T-cell ALL xenografts. However, there were only modest differences in drug sensitivity in vitro among the cell lines tested, which represent a range of cancer types. The p53 mutation status showed no relationship with sensitivity to quisinostat, as approximately equal numbers of mutant p53 cell lines had IC50 values above and below the median for the entire panel. These results are consistent with recent reports showing no clear relationship between p53 mutation status and response of pediatric preclinical models to HDIs [28,31].
The treatment of engrafted mice, at the selected doses was well tolerated and toxicity levels were low (no models were excluded because of toxicity). The ALL panel was treated at a reduced dose with respect to the solid tumor panel, as the dose of 5 mg/kg for 21 days exceeded the MTD for non-engrafted NOD/SCID mice. This dose reduction does not seem to have limited the efficacy of the treatments with respect to that observed for the solid tumors.
Promising results obtained with quisinostat for cell line in vitro testing were only partially confirmed when efficacy was tested in vivo against the pediatric cancer xenograft panel, with only one objective response among 33 solid tumor xenografts and 2 objective responses among 8 ALL xenografts observed. We have reported similar discrepancies between in vitro and in vivo findings for other single agents [33,38,39], with the explanation likely reflecting in part the inability in the in vivo setting to achieve the depth and duration of target inhibition produced in vitro. The fact that quisinostat was specifically selected based on its in vivo pharmacodynamic activity suggested that it would overcome this limitation [21], which we have confirmed to some extent. In the case of vorinostat tested as a single agent, there were no xenografts with delays in time to event of two-fold or greater [33], whereas for quisinostat this level of activity was observed in approximately 20% of the solid tumor xenografts. Additionally, objective responses were observed for quisinostat, but not for vorinostat.
The most consistent solid tumor activity signals observed were for the glioblastoma xenograft panel, with both D456 and D645 showing prolonged time to event. These xenografts could be valuable in identifying markers of sensitivity for this histotype. The results for the glioblastoma xenograft D456 are impressive, with control of tumor growth during the 21 days of treatment as well as for an additional 21 days of follow-up. Thus, further preclinical evaluation of the utility of quisinostat for glioblastoma is indicated. The response in the rhabdomyosarcoma xenograft Rh28 was also impressive, but 4 additional rhabdomyosarcoma xenografts showed only limited responses to quisinostat. These results are reminiscent of those obtained for romidepsin, which also induced regressions in a single xenograft (Rh66) from among 10 rhabdomyosarcoma xenografts tested [28]. Of note, Rh28 showed tumor growth delay, but not regression, in response to treatment with romidepsin.
The T-cell ALL responses are particularly interesting, as leukemia burden seemed to increase (as detected in circulation) for a week following initiation of treatment, but thereafter the leukemia cells quickly disappeared and remission was attained. A similar pattern of response was observed for the B-precursor ALL xenograft ALL-3, which showed a clear treatment effect to quisinostat, but failed to meet criteria for objective response. These patterns of blast presence in blood could be indicators of differentiation induction or modification of the expression of homing/attachment molecules which prompt them to exit the bone marrow into peripheral blood. Alteration in the chemokine CXCR4 has been reported by the HDI vorinostat in ALL [40], and a similar effect could help explain our findings. Efficacy testing in vivo on a larger panel of ALL, particularly of T lineage, is required to address the question about a subtype-specific effect that might justify clinical development of JNJ-26481585 for this ALL subtype. Regarding the anti-leukemia effect of quisinostat against the MLL-rearranged (t11;19) ALL-3 xenograft, connectivity map gene expression analyses predicted that HDIs would reverse the leukemia-specific gene signature associated with MLL-AF4 infant ALL [41]. Further evaluation of quisinostat against ALL xenografts with MLL-rearrangement is warranted to develop a more robust dataset testing the role of HDAC inhibition for this high-risk ALL subtype.
At the doses used (5 mg/kg for solid tumors and 2.5 mg/kg for leukemias) quisinostat increased acetylation of histones H3 and H4 in each solid tumor and leukemia model. However, for the rhabdomyosarcoma models induction of acetylated histones was equally robust in responsive as non-responsive models. Similar induction of p21CIP1 was also seen in responsive Rh28 xenografts as in non-responsive Rh30. The only characteristic that distinguished Rh28 xenografts was induction of cleaved PARP after the fourth daily dose. For ALL-8 there was a significant and cumulative increase in acetylated H4 histone with daily dosing. Thus, similar to other studies [28], target inhibition resulting in increased histone acetylation did not correlate with tumor responses.
Single agent activity for quisinostat exceeded that observed for another HDI [33], but nonetheless was restricted to a minority of models. This suggests that identifying biological predictors of response could be a clinically relevant line of future research. Further testing of ALL subtypes such as T-cell ALL and MLL-rearranged ALL is a priority. Additional opportunities arise from the evidence for synergy that this family of drugs has shown with established anticancer agents [30]. While single agent activity tends to be a positive indicator of potential efficacy in combination therapy, the inclusion of this HDI into drug combinations is appealing based on its improved pharmacodynamic profile and the observed effects in vivo. Particularly interesting novel agents that may prove effective in combination with quisinostat in the pediatric setting include hypomethylating agents [42], proteasome inhibitors [43], and pro-apoptotic drugs such as ABT-263, based on the reported changes induced by quisinostat on pro- and anti-apoptotic molecules, and in particular on MCL1 levels [44].
Supplementary Material
Acknowledgments
This work was supported by NO1-CM-42216, CA21765, and CA108786 from the National Cancer Institute and used quisinostat supplied by Janssen Pharmaceutica (Johnson & Johnson Pharmaceutical Research and Development), Beerse, Belgium. In addition to the authors represents work contributed by the following: Sherry Ansher, Catherine A. Billups, Ingrid Boehm, Joshua Courtright, Edward Favours, Henry S. Friedman, Danuta Gasinski, Debbie Payne-Turner, Chandra Tucker, Joe Zeidner, Jianrong Wu, Ellen Zhang, and Jian Zhang. Children’s Cancer Institute Australia for Medical Research is affiliated with the University of New South Wales and Sydney Children’s Hospital.
Footnotes
Conflict of interest statement: The authors consider that there are no actual or perceived conflicts of interest.
References
- 1.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 2.Cress WD, Seto E. Histone deacetylases, transcriptional control, and cancer. Vol. 184. John Wiley & Sons, Inc; 2000. pp. 1–16. [DOI] [PubMed] [Google Scholar]
- 3.Timmermann S, Lehrmann H, Polesskaya A, et al. Histone acetylation and disease. Cellular and Molecular Life Sciences. 2001;58(5):728–736. doi: 10.1007/PL00000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin RJ, Nagy L, Inoue S, et al. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391(6669):811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 5.Lagger G, O’Carroll D, Rembold M, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21(11):2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weichert W. HDAC expression and clinical prognosis in human malignancies. Cancer Letters. 2009;280(2):168–176. doi: 10.1016/j.canlet.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 7.Kekatpure VD, Dannenberg AJ, Subbaramaiah K. HDAC6 modulates Hsp90 chaperone activity and regulates activation of aryl hydrocarbon receptor signaling. The Journal of biological chemistry. 2009;284(12):7436–7445. doi: 10.1074/jbc.M808999200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Kovacs JJ, Murphy PJ, Gaillard S, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Molecular cell. 2005;18(5):601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 9.Qian DZ, Kato Y, Shabbeer S, et al. Targeting Tumor Angiogenesis with Histone Deacetylase Inhibitors: the Hydroxamic Acid Derivative LBH589. Clinical Cancer Research. 2006;12(2):634–642. doi: 10.1158/1078-0432.CCR-05-1132. [DOI] [PubMed] [Google Scholar]
- 10.Sambucetti LC, Fischer DD, Zabludoff S, et al. Histone Deacetylase Inhibition Selectively Alters the Activity and Expression of Cell Cycle Proteins Leading to Specific Chromatin Acetylation and Antiproliferative Effects. Journal of Biological Chemistry. 1999;274(49):34940–34947. doi: 10.1074/jbc.274.49.34940. [DOI] [PubMed] [Google Scholar]
- 11.Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov. 2002;1(4):287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 12.Richon VM, Sandhoff TW, Rifkind RA, et al. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(18):10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peart MJ, Smyth GK, van Laar RK, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(10):3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser KB, Staver MJ, Waring JF, et al. Gene Expression Profiling of Multiple Histone Deacetylase (HDAC) Inhibitors: Defining a Common Gene Set Produced by HDAC Inhibition in T24 and MDA Carcinoma Cell Lines. Mol Cancer Ther. 2003;2(2):151–163. [PubMed] [Google Scholar]
- 15.Zupkovitz G, Tischler J, Posch M, et al. Negative and Positive Regulation of Gene Expression by Mouse Histone Deacetylase 1. Mol Cell Biol. 2006;26(21):7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spange S, Wagner T, Heinzel T, et al. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. The International Journal of Biochemistry & Cell Biology. 2009;41(1):185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 17.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6(1):38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 18.Duvic M, Talpur R, Ni X, et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109(1):31–39. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mann BS, Johnson JR, Cohen MH, et al. FDA Approval Summary: Vorinostat for Treatment of Advanced Primary Cutaneous T-Cell Lymphoma. Oncologist. 2007;12(10):1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 20.Venugopal B, Baird R, Kristeleit R, et al. A Phase I study of quisinostat (JNJ-26481585), an oral hydroxamate histone deacetylase inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-13-0312. [DOI] [PubMed] [Google Scholar]
- 21.Arts J, King P, Marien A, et al. JNJ-26481585, a novel “second-generation” oral histone deacetylase inhibitor, shows broad-spectrum preclinical antitumoral activity. Clin Cancer Res. 2009;15(22):6841–6851. doi: 10.1158/1078-0432.CCR-09-0547. [DOI] [PubMed] [Google Scholar]
- 22.Tong W-G, Wei Y, Stevenson W, et al. Preclinical antileukemia activity of JNJ-26481585, a potent second-generation histone deacetylase inhibitor. Leukemia Research. 2010;34(2):221–228. doi: 10.1016/j.leukres.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 23.Arts J, Marien A, King P, et al. A novel “second-generation” oral pan-Histone Deacetylase (HDAC) inhibitor showing broad-spectrum preclinical antitumor activity against solid and haematological malignancies. Proceedings of the 99th Annual Meeting of the American Association for Cancer Research; 2008. p. Abstr #2444. [Google Scholar]
- 24.Kang MH, Smith MA, Morton CL, et al. National Cancer Institute pediatric preclinical testing program: Model description for in vitro cytotoxicity testing. Pediatr Blood Cancer. 2011;56:239–249. doi: 10.1002/pbc.22801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houghton PJ, Morton CL, Tucker C, et al. The Pediatric Preclinical Testing Program: description of models and early testing results. Pediatr Blood Cancer. 2007;49(7):928–940. doi: 10.1002/pbc.21078. [DOI] [PubMed] [Google Scholar]
- 26.Houghton PJ, Morton CL, Kolb EA, et al. Initial testing (stage 1) of the proteasome inhibitor bortezomib by the Pediatric Preclinical Testing Program. Pediatr Blood Cancer. 2008;50(1):37–45. doi: 10.1002/pbc.21214. [DOI] [PubMed] [Google Scholar]
- 27.Liem NL, Papa RA, Milross CG, et al. Characterization of childhood acute lymphoblastic leukemia xenograft models for the preclinical evaluation of new therapies. Blood. 2004;103(10):3905–3914. doi: 10.1182/blood-2003-08-2911. [DOI] [PubMed] [Google Scholar]
- 28.Graham C, Tucker C, Creech J, et al. Evaluation of the antitumor efficacy, pharmacokinetics, and pharmacodynamics of the histone deacetylase inhibitor depsipeptide in childhood cancer models in vivo. Clin Cancer Res. 2006;12(1):223–234. doi: 10.1158/1078-0432.CCR-05-1225. [DOI] [PubMed] [Google Scholar]
- 29.Camphausen K, Tofilon PJ. Inhibition of Histone Deacetylation: A Strategy for Tumor Radiosensitization. Journal of Clinical Oncology. 2007;25(26):4051–4056. doi: 10.1200/JCO.2007.11.6202. [DOI] [PubMed] [Google Scholar]
- 30.Bots M, Johnstone RW. Rational Combinations Using HDAC Inhibitors. Clinical Cancer Research. 2009;15(12):3970–3977. doi: 10.1158/1078-0432.CCR-08-2786. [DOI] [PubMed] [Google Scholar]
- 31.Panicker J, Li Z, McMahon C, et al. Romidepsin (FK228/depsipeptide) controls growth and induces apoptosis in neuroblastoma tumor cells. Cell cycle (Georgetown, Tex) 2010;9(9):1830–1838. doi: 10.4161/cc.9.9.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakimura R, Tanaka K, Nakatani F, et al. Antitumor effects of histone deacetylase inhibitor on Ewing’s family tumors. International journal of cancer. 2005;116(5):784–792. doi: 10.1002/ijc.21069. [DOI] [PubMed] [Google Scholar]
- 33.Keshelava N, Houghton PJ, Morton CL, et al. Initial testing (stage 1) of vorinostat (SAHA) by the Pediatric Preclinical Testing Program. Pediatr Blood Cancer. 2009;53(3):505–508. doi: 10.1002/pbc.21988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.More SS, Itsara M, Yang X, et al. Vorinostat increases expression of functional norepinephrine transporter in neuroblastoma in vitro and in vivo model systems. Clin Cancer Res. 2011;17(8):2339–2349. doi: 10.1158/1078-0432.CCR-10-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaboin J, Wild J, Hamidi H, et al. MS-27-275, an Inhibitor of Histone Deacetylase, Has Marked in Vitro and in Vivo Antitumor Activity against Pediatric Solid Tumors. Cancer Res. 2002;62(21):6108–6115. [PubMed] [Google Scholar]
- 36.Fouladi M, Furman WL, Chin T, et al. Phase I study of depsipeptide in pediatric patients with refractory solid tumors: a Children’s Oncology Group report. J Clin Oncol. 2006;24(22):3678–3685. doi: 10.1200/JCO.2006.06.4964. [DOI] [PubMed] [Google Scholar]
- 37.Fouladi M, Park JR, Stewart CF, et al. Pediatric phase I trial and pharmacokinetic study of vorinostat: a Children’s Oncology Group phase I consortium report. J Clin Oncol. 2010;28(22):3623–3629. doi: 10.1200/JCO.2009.25.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith MA, Morton CL, Phelps DA, et al. Stage 1 testing and pharmacodynamic evaluation of the HSP90 inhibitor alvespimycin (17-DMAG, KOS-1022) by the pediatric preclinical testing program. Pediatr Blood Cancer. 2008;51(1):34–41. doi: 10.1002/pbc.21508. [DOI] [PubMed] [Google Scholar]
- 39.Morton CL, Houghton PJ, Gorlick R, et al. Initial testing of aplidin by the pediatric pre-clinical testing program. Pediatr Blood Cancer. 2009;53(3):509–512. doi: 10.1002/pbc.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crazzolara R, Jöhrer K, Johnstone RW, et al. Histone deacetylase inhibitors potently repress CXCR4 chemokine receptor expression and function in acute lymphoblastic leukaemia. British Journal of Haematology. 2002;119(4):965–969. doi: 10.1046/j.1365-2141.2002.03955.x. [DOI] [PubMed] [Google Scholar]
- 41.Stumpel D, Schneider P, Seslija L, et al. Connectivity Mapping Identifies HDAC Inhibitors as Suitable Candidates for the Treatment of t(4;11)-Positive Acute Lymphoblastic Leukemia In Infants. Blood (ASH Annual Meeting Abstracts) 2010;116:Abstr #3222. [Google Scholar]
- 42.Cameron EE, Bachman KE, Myohanen S, et al. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21(1):103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 43.Gao N, Dai Y, Rahmani M, et al. Contribution of Disruption of the Nuclear Factor-κB Pathway to Induction of Apoptosis in Human Leukemia Cells by Histone Deacetylase Inhibitors and Flavopiridol. Molecular Pharmacology. 2004;66(4):956–963. doi: 10.1124/mol.104.002014. [DOI] [PubMed] [Google Scholar]
- 44.Stühmer T, Arts J, Chatterjee M, et al. Preclinical anti-myeloma activity of the novel HDAC-inhibitor JNJ-26481585. British Journal of Haematology. 149(4):529–536. doi: 10.1111/j.1365-2141.2010.08126.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.