Abstract
Improving the effectiveness of adipose-tissue derived human mesenchymal stromal/stem cells (AMSCs) for skeletal therapies requires a detailed characterization of mechanisms supporting cell proliferation and multi-potency. We investigated the molecular phenotype of AMSCs that were either actively proliferating in platelet lysate or in a basal non-proliferative state. Flow cytometry combined with high-throughput RNA sequencing (RNASeq) and RT-qPCR analyses validate that AMSCs express classic mesenchymal cell surface markers (e.g., CD44, CD73/NT5E, CD90/THY1 and CD105/ENG). Expression of CD90 is selectively elevated at confluence. Self-renewing AMSCs express a standard cell cycle program that successively mediates DNA replication, chromatin packaging, cyto-architectural enlargement and mitotic division. Confluent AMSCs preferentially express genes involved in extracellular matrix (ECM) formation and cellular communication. For example, cell cycle-related biomarkers (e.g., cyclins E2 and B2, transcription factor E2F1) and histone-related genes (e.g., H4, HINFP, NPAT) are elevated in proliferating AMSCs, while ECM genes are strongly upregulated (>10 fold) in quiescent AMSCs. AMSCs also express pluripotency genes (e.g., POU5F1, NANOG, KLF4) and early mesenchymal markers (e.g., NES, ACTA2) consistent with their multipotent phenotype. Strikingly, AMSCs modulate expression of WNT signaling components and switch production of WNT ligands (from WNT5A/WNT5B/WNT7B to WNT2/WNT2B), while up-regulating WNT-related genes (WISP2, SFRP2 and SFRP4). Furthermore, post-proliferative AMSCs spontaneously express fibroblastic, osteogenic, chondrogenic and adipogenic biomarkers when maintained in confluent cultures. Our findings validate the biological properties of self-renewing and multi-potent AMSCs by providing high-resolution quality control data that support their clinical versatility.
Keywords: Mesenchymal stem cell, adipose-tissue derived stromal cells, pluripotent, multipotent, cell cycle, lineage-commitment, fibroblast, osteogenesis, chondrogenesis, adipogenesis, histone, cyclin, extracellular matrix, CD44, CD73, NT5E, CD90, THY1, CD105, ENG, NES, ACTA2, OCT4, POU5F1, NANOG, KLF4, CCND1, CCNB2, HIST1H3H, HIST1H4A, HIST2H4A, HIST2H4B, E2F1, E2F7, E2F8, HINFP, NPAT, ASPN, ECM2, FMOD, OGN, PODN, WISP2, SFRP2, SFRP4, WNT2, WNT2A, WNT5A, WNT5B, WNT7B, RARRES2, TNNT3, ADH1B, H19, CHI3L1
INTRODUCTION
Adipose-tissue derived human mesenchymal stromal/stem cells (AMSCs) grown in human platelet lysate (hPL) under good manufacturing practice (GMP) conditions provide a xenobiology-free source of cells that can be harvested in sufficient quantities to accommodate a range of therapeutic applications for tissue regeneration [Bieback et al., 2009; Crespo-Diaz et al., 2011; Jung et al., 2012; Mader et al., 2013]. GMP-hPL-AMSCs are derived from the stromal vascular fraction (SVF) of lipoaspirates during elective surgeries. The product of the harvesting procedure and subsequent proliferative expansion is a population of mesenchymal stromal cells that has high proliferative potential and is capable of multi-lineage differentiation [Crespo-Diaz et al., 2011]. Platelet lysate is a potent mixture of growth factors and extracellular ligands that has evolved to support tissue repair in vertebrates. Compared to bovine serum, hPL avoids issues with species-related biocompatibility and zoonotic transmission of infectious agents. Safe and effective application of GMP-hPL-AMSCs for treatment of non-lethal skeletal afflictions requires a thorough characterization of the molecular mechanisms that support their self-renewal and multi-lineage potential.
The biological potential of human AMSCs propagated in cell culture media with platelet derived products has been recognized for some time [Bieback et al., 2009; Blande et al., 2009; Crespo-Diaz et al., 2011; Iudicone et al., 2014; Siciliano et al., 2013]. Other studies have further corroborated this concept. For example, both single donor and pooled hPL are effective alternatives for fetal bovine serum or platelet-poor plasma for expansion and maintenance of AMSCs [Castegnaro et al., 2011; Shih et al., 2011]. In addition, hPL appears to stimulate cell proliferation more effectively than bovine products [Cholewa et al., 2011] and can be released from engineered nanoparticles to support surgical applications [Santo et al., 2012]. Platelet lysate is an effective and unique cocktail of mitogens and morphogens with potent effects on cellular phenotypes. For example, one potent mitogen of AMSCs is FGF2 [Rider et al., 2008]. We examined AMSCs at a high level of molecular resolution to understand their biological properties when cultured in hPL.
In this study, we comprehensively examined the molecular characteristics of AMSCs using a combination of high throughput RNASeq, real time RT-qPCR and flow cytometry that together interrogate mRNA and protein expression in proliferating and confluent cell populations. To understand the ‘stemness’ of AMSCs, we analyzed both pluripotency factors [Apostolou and Hochedlinger, 2013] and mesenchymal cell surface markers [Bourin et al., 2013]. To define the cell proliferative properties of AMSCs, we examined the expression of genes involved in cell cycle machinery, DNA replication and histones, similar to previous studies on human pluripotent embryonic stem cells [Becker et al., 2010; Becker et al., 2007]. To characterize the biosynthetic production capacity of AMSCs, we analyzed expression of extracellular matrix proteins that support production of skeletal tissues [Cawston and Young, 2010; Ma et al., 2013; Mafi et al., 2012]. Our data provide insight into the mechanistic underpinnings for the principal capabilities of AMSCs with respect to their (i) high proliferative potential, (ii) postulated stem cell properties, (iii) anabolic activities to produce extracellular matrix proteins for tissue repair, and (iv) developmental plasticity for spontaneous differentiation into distinct mesenchymal lineages.
MATERIALS AND METHODS
Culture conditions for AMSCs
Mesenchymal stromal cells were derived from lipo-aspirates obtained from consenting healthy donors with approval from the Mayo Clinic Institutional Review Board (IRB) as previously described [Crespo-Diaz et al., 2011; Mader et al., 2013]. Samples were obtained from three consenting normal patients (respectively, male/41 yr, female/32 yr, male/54 yr) that underwent elective removal of subcutaneous adipose tissue. Fat tissue was enzymatically digested using collagenase (Type I at 0.075%; Worthington Biochemicals) for 1.5 h at 37°C. Adipocytes were separated from the stromal vascular fraction by low speed centrifugation (400 g for 5 min). After the adipose supernatant was removed, the cell pellet was rinsed with PBS and passed through cell strainers (70µm followed by 40µm) (BD Biosciences). The resulting cell fraction was incubated in T-175 cm2 flasks at 37°C in 5% CO2 at a cell density of 1.0–2.5 ×103 cells/cm2 in standard culture medium (Advanced MEM) with 5% PLTMax (a clinical grade commercial platelet lysate product [MillCreekLifeSciences]), 2 U/ml heparin (hospital pharmacy), 2 mM L-glutamine (Invitrogen) and antibiotics (100 U/ml penicillin, 100 g/ml streptomycin). Cells were harvested while still actively proliferating or when they reached confluence (typically four days after plating).
IncuCyte Analysis
AMSCs were seeded in 24-well plates at 1,500 cells/cm2 (5 replicates) and cultures were monitored for confluence using the IncuCyte FLR live imaging system (Essen BioScience). Images from four defined fields in each well were captured every 3 hours for the duration of culture. The IncuCyte system determines confluence for a single field using internal image analysis software, and confluence at a given time point is then calculated as the average of all fields (20 fields) for the sample.
Flow Cytometry
Approximately 1×106 fresh AMSCs were added to two tubes and centrifuged at 486 × G for 5 minutes. The supernatant was removed and 50 µl of mouse serum was added to each cell pellet, vortexed, and incubated for 5 minutes at room temperature. The following antibodies were added to the control tube: CD3 FITC (BD Biosciences, clone SK7), CD3 PE (BD Biosciences, clone SK7), CD3 ECD (Coulter, clone UCHT1), CD3 PC5.5 (Coulter, clone UCHT1), CD3 PC7 (Coulter, clone UCHT1), CD3 APC (Coulter, clone UCHT1), CD3 APC-Alexa Fluor 750 (Coulter, clone UCHT1), CD3 Pacific Blue (Coulter, clone UCHT1), CD3 Krome Orange (Coulter, clone UCHT1). The following antibodies were used to determine AMSC phenotype: CD90 FITC (Coulter, clone F15-42-1-5), CD105 PE (Coulter, clone 1G2), CD14 ECD (Coulter, clone RMO52), CD73 PerCP-eFluor 710 (eBioscience, clone AD2), CD44 PC7 (eBioscience, clone IM7), HLA-ABC APC (eBioscience, clone W6/32), HLA-DR Pacific Blue (Coulter, clone Immu-357), CD45 Krome Orange (Coulter, clone J.33). The tubes were vortexed and incubated for 15 minutes at room temperature in the dark. A total of 3ml of PBSFE (PBS with 5mM NaEDTA (Sigma) and 1% Bovine Serum Albumin (Sigma) were added to each tube and centrifuged at 486 × G for 5 minutes. The supernatant was discarded and 200µl of 1% Paraformaldehyde (Electron Microscopy Sciences) was added prior to acquisition. The flow cytometric acquisition was performed on the Beckman Coulter Gallios (Coulter). The cells were acquired using forward and side scatter and gated to exclude debris. The cytometer acquired 50,000 AMSC events. The analysis was performed using Kaluza software (Coulter). For each donor unit, an overlay histogram with a logarithmic x-axis was created with three data sets: the Control tube from the proliferating cells, the AMSC tube from the proliferating cells, and the AMSC tube from the confluent cells. This overlay histogram was created for each marker. An overlay gate was added and set so the Control peak was approximately 1.0% positive. The proliferating and confluent peaks were then analyzed for percentage positive in the gate and also the overall arithmetic mean.
Real-time reverse transcriptase quantitative PCR (RT-qPCR) analysis
RNA was isolated one, four, and seven days after plating AMSCs (seeded at 3,000 cells/cm2) using the miRNeasy Mini Kit (Qiagen). Cells were actively proliferating for three days in culture and reached confluence on the fourth day of culture. RNA was then reverse transcribed into cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen). Gene expression was quantified using real-time PCR. Real-time qPCR reactions were performed with 10 ng cDNA per 10 µl with QuantiTect SYBR Green PCR Kit (Qiagen) and the CFX384 Real-Time System (BioRad). Transcript levels were normalized to a housekeeping gene, GAPDH. Gene expression levels were quantified using the 2^(−ΔΔ Ct) method. Gene specific primers are shown in Table 1.
Table 1.
Primer sequences used in PCR reactions.
| GENE NAME | FORWARD PRIMER SEQUENCE | REVERSE PRIMER SEQUENCE |
|---|---|---|
| GAPDH | ATGTTCGTCATGGGTGTGAA | TGTGGTCATGAGTCCTTCCA |
| HIST2H4A | AGCTGTCTATCGGGCTCCAG | CCTTTGCCTAAGCCTTTTCC |
| CCNB2 | CCGACGGTGTCCAGTGATTT | TGTTGTTTTGGTGGGTTGAACT |
| COL1A1 | GCTACCCAACTTGCCTTCATG | TGCAGTGGTAGGTGATGTTCTGA |
| COL2A1 | TGAAGGTTTTCTGCAACATGGA | TTGGGAACGTTTGCTGGATT |
| COL10A1 | AAGAATGGCACCCCTGTAATGT | ACTCCCTGAAGCCTGATCCA |
| POU5F1 | GCAATTTGCCAAGCTCCTGAA | AAGCTAAGCTGCAGAGCCTCAAAG |
| NANOG | CAACTGGCCGAAGAATAGCAATG | TGGTTGCTCCAGGTTGAATTGTT |
| KLF4 | AAGAGTTCCCATCTCAAGGCACA | GGGCGAATTTCCATCCACAG |
| RUNX2 | ATGTGTTTGTTTCAGCAGCA | TCCCTAAAGTCACTCGGTATGTGTA |
| SP7 | GCCATTCTGGGTTGGGTATC | GAAGCCGGAGTGCAGGTATCA |
| ATF4 | ATGACCGAAATGAGCTTCCTG | CTGGAGAACCCATGAGGTTTG |
| SOX5 | CAGCCAGAGTTAGCACAATAGG | CTGTTGTTCCCGTCGGAGTT |
| SOX9 | TGTATCACTGAGTCATTTGCAGTGT | AAGGTCTGTCAGTGGGCTGAT |
| PPARG | TGGAATTAGATGACAGCGACTTGG | CTGGAGCAGCTTGGCAAACA |
| KLF3 | TGTCTCAGTGTCATACCCATCT | ACCCCATACTTGTTAGGCTTCAT |
| THY1 (CD90) | ATGAAGGTCCTCTACTTATCCGC | GCACTGTGACGTTCTGGGA |
| NT5E (CD73) | AAGGACTGATCGAGCCACTC | GGAAGTGTATCCAACGATTCCCA |
| ENG (CD105) | TGCACTTGGCCTACAATTCCA | AGCTGCCCACTCAAGGATCT |
| PTPRC (CD45) | ACAGCCAGCACCTTTCCTAC | GTGCAGGTAAGGCAGCAGA |
| CD14 | CAACCTAGAGCCGTTTCTAAAGC | GCGCCTACCAGTAGCTGAG |
High Throughput RNA Sequencing & Bioinformatic analysis
RNA libraries were prepared according to the manufacturer’s instructions for the TruSeq RNA Sample Prep Kit v2 (Illumina). Briefly, poly-A mRNA was purified from total RNA using oligo dT magnetic beads. Purified mRNA was fragmented at 95°C for 8 minutes, eluted from the beads and primed for first strand cDNA synthesis. RNA fragments were copied into first strand cDNA using SuperScript III reverse transcriptase and random primers (Invitrogen). Next, second strand cDNA synthesis was performed using DNA polymerase I and RNase H. The double-stranded cDNA was purified using a single AMPure XP bead (Agencourt) clean-up step. The cDNA ends were repaired and phosphorylated using Klenow, T4 polymerase, and T4 polynucleotide kinase followed by a single AMPure XP bead clean-up. Blunt-ended cDNAs were modified to include a single 3’ adenylate (A) residue using Klenow exo- (3’ to 5’ exo minus). Paired-end DNA adaptors (Illumina) with a single “T” base overhang at the 3’ end were immediately ligated to the ‘A tailed’ cDNA population. Unique indexes, included in the standard TruSeq Kits (12-Set A and 12-Set B) were incorporated at the adaptor ligation step for multiplex sample loading on the flow cells. The resulting constructs were purified by two consecutive AMPure XP bead clean-up steps. The adapter-modified DNA fragments were enriched by 12 cycles of PCR using primers included in the Illumina Sample Prep Kit. The concentration and size distribution of the libraries was determined on an Agilent Bioanalyzer DNA 1000 chip. A final quantification, using Qubit fluorometry (Invitrogen), confirmed sample concentrations.
Libraries were loaded onto flow cells at concentrations of 8–10 pM to generate cluster densities of 700,000/mm2 following the standard protocol for the Illumina cBot and cBot Paired end cluster kit version 3. Flow cells were sequenced as 51 × 2 paired end reads on an Illumina HiSeq 2000 using TruSeq SBS sequencing kit version 3 and HCS v2.0.12 data collection software. Base-calling was performed using Illumina’s RTA version 1.17.21.3. The RNA-Seq data were analyzed using the MAPRSeq v.1.2.1 system for RNA-sequencing data analysis (http://bioinformaticstools.mayo.edu/research/maprseq/), the Bionformatics Core standard tool, which includes alignment with TopHat 2.0.6 [Kim et al., 2013] and gene counts with the HTSeq software (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html). Normalization and differential expression analysis were performed using edgeR 2.6.2 [Robinson et al., 2010]. Expression values for each gene were normalized to 1 million reads and corrected for gene length (reads per kilobasepair per million mapped reads, RPKM). RKPM values were determined from three different patients of similar age providing three independent biological replicates.
RESULTS
High throughput analysis of gene expression in AMSCs
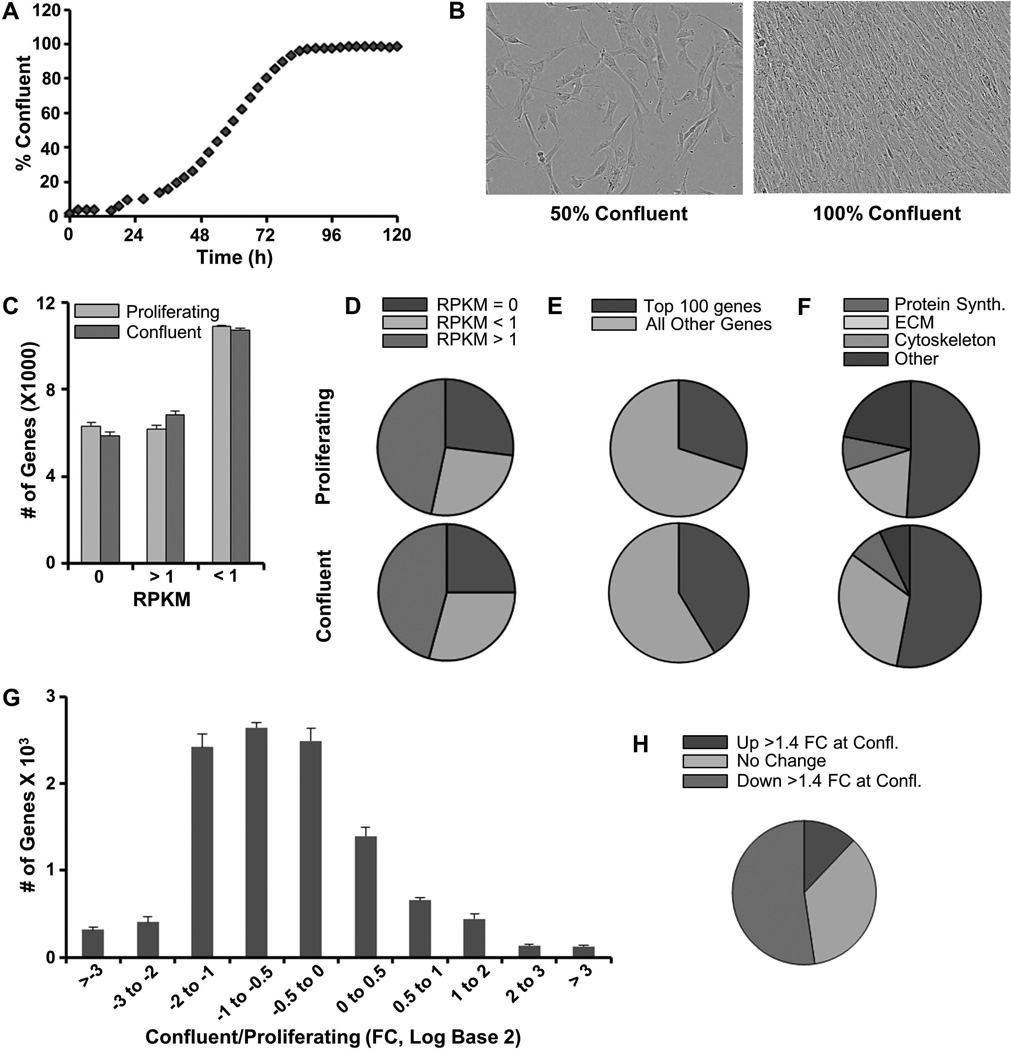
AMSCs perform cell autonomous functions related to cell proliferation, phenotype maintenance and cellular metabolism, as well as functions linked to its extracellular environment and cell/cell communication. To understand what cellular, molecular and biochemical functions are executed by AMSCs during proliferative and non-proliferative phases, we examined gene expression profiles in pre-confluent and confluent AMSCs using high-throughput RNASeq analysis of cells derived from three healthy patients. AMSCs in culture undergo proliferative expansion and reach confluence after about three to five days depending on seeding density, as evidenced by IncuCyte cell density analysis and microscopic visualization (Fig. 1A & B). In both pre-confluent and confluent cultures, the 100 most highly expressed genes account for 30–40% of all mapped transcripts (Fig. 1A–C). These very most abundant mRNAs (expressed at >300 RPKM) encode mostly proteins involved in translation (e.g., ribosomal proteins; ~50% of the ‘Top 100’ most abundant mRNAs), extracellular matrix proteins (ECM) and cytoskeletal proteins (Fig. 1F). Annotation analysis of this set of highly abundant mRNAs indicates that quiescent cells express a larger number of mRNAs for ECM proteins than actively dividing AMSCs (Fig. 1F). Of all annotated genes (n = 23,338) mapping of RNA reads from pre-confluent and confluent AMSC cultures reveals that nearly half (n = 11,044; ~47%) are expressed at levels greater than 1 RPKM (Fig. 1C & D). About 9% of these genes change by more than 4 fold when comparing proliferative and quiescent states (n= 241 up regulated and n= 716 down regulated) (Fig. 1G & H). A smaller gene subset (n=334 of 11,044; ~3%) is modulated by at least an order of magnitude, reflected by a greater than 10 fold-change in mRNA expression when cells transition from a proliferative to post-proliferative phase. There are 251 genes that are highly down-regulated (>10 fold) and 83 genes that are up-regulated at confluence. Functional annotation analysis using DAVID6.7 indicates that the primary gene ontology (GO) category for the down-regulated genes is cell cycle control (n=107 of 251 total; enrichment score 41.51). The majority of these genes are involved in mitosis and S phase related functions associated with DNA replication and chromosome segregation. In essence and as expected, self-renewing AMSCs express a standard cell cycle program that successively mediates DNA replication, chromatin packaging, cyto-architectural enlargement and mitotic division. The up-regulated genes are primarily secreted proteins including cell signaling ligands and extracellular matrix proteins (n=43 of 83 total; enrichment score 11.48). Taken together, the gene expression results show that proliferating AMSCs mostly produce new intracellular components, while post-proliferative cells produce a tissue-like extracellular matrix and proteins that mediate intercellular communication.
FIGURE 1. High throughput analysis of gene expression in proliferating and quiescent confluent AMSCs.
Incucyte cell density analysis showing cell confluence over time (A). Representative microscopy images of cell cultures at different levels of confluence (B). Number of genes not expressed, expressed at low levels, and expressed at high levels in proliferating and confluent AMSCs (C). Pie chart of data in panel A (D). Expression (RPKM) of the 100 most abundant genes relative to all other genes in proliferating and confluent AMSCs (E). The top 100 expressing genes divided by class in proliferating and confluent AMSCs (F). Differential expression of genes in proliferating and confluent AMSCs (G). Significant (FC > 1.4) gene expression differences between proliferating and confluent AMSCs (H). Reads per kilobasepair per million mapped reads = RPKM, Fold Change = FC. RNASeq data were analyzed as averages (and standard deviations) for three consenting normal patients (n= 3).
Proliferation-related expression of cell cycle regulatory genes supporting AMSC self-renewal
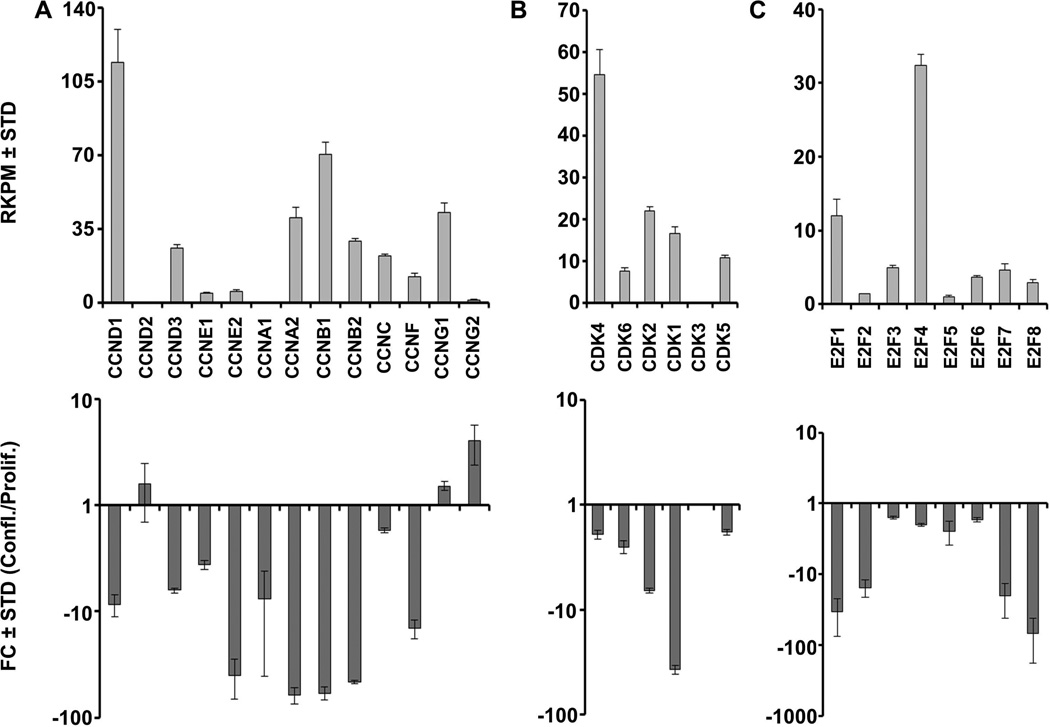
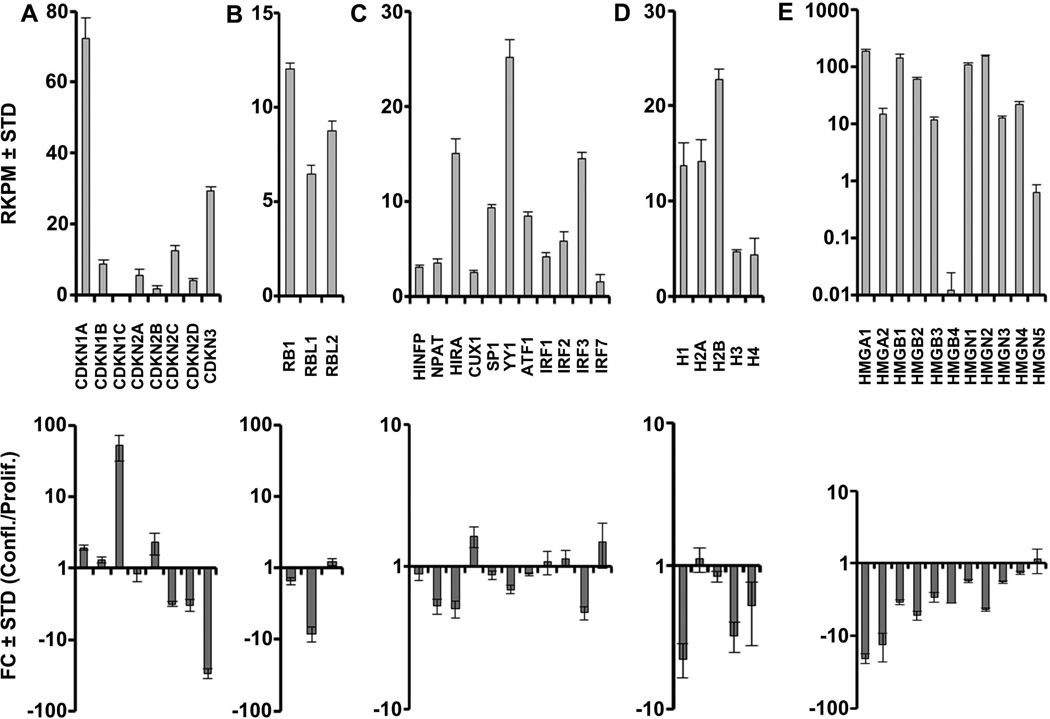
Self-renewal of pluripotent human stem cells is controlled by the general cell cycle regulatory machinery involving cyclins, cyclin-dependent kinases, retinoblastoma-related pocket proteins and E2F proteins [Becker et al., 2010; Becker et al., 2007]. To assess whether similar mechanisms are operative in multipotent somatic AMSCs, we collated RNASeq data on canonical cell cycle regulators. As discussed above (see Fig. 1), more than 40% (107 of 251) of genes that are strongly down-regulated at confluence are involved in cell cycle control. Proliferating, but not confluent AMSCs, prominently express two G1 phase-related D-type cyclins (CCND1 and CCND3) that transduce growth factor responses, two G1/S phase-related E-type cyclins (CCNE1 and CCNE2) that are key targets of E2Fs, and three A/B type cyclins (e.g., CCNA2, CCNB1 and CCNB2) that control progression through G2 and M phases (Fig. 2A). Paralleling these changes in cyclin expression are reductions in the expression of the cognate CDKs (CDK4, CDK6, CDK2 and CDK1) (Fig. 2B), as well as two activating members of the E2F family (E2F1 and E2F2) at confluence (Fig. 2C). Apart from the suppression of cell cycle stimulatory proteins, post-proliferative AMSCs express higher levels of the cell cycle inhibitors CDKN1A, CDKN1C and CDKN2B, consistent with their roles in arresting cell proliferation (Fig. 3A). Also of note are the down-regulation of Cyclin F (CCNF), which is involved in maintenance of the pluripotent cell cycle, and the up-regulation of the cell cycle inhibitory G type cyclin G2 (CCNG2) (Fig 2A).
FIGURE 2. Expression of cell cycle regulatory genes in proliferating and confluent AMSCs by RNA-Seq.
Expression of cyclins (A), cyclin-dependent kinases (B), and E2F family members (C) in proliferating AMSCs (top) and fold change expression values comparing confluent and proliferating AMSCs (bottom). Reads per kilobasepair per million mapped reads = RPKM, Fold Change = FC, n = 3.
FIGURE 3. Expression of cell cycle inhibitors, histone regulators, histones, and chromatin-associated genes in proliferating and confluent AMSCs by RNA-Seq.
Expression of cyclin-dependent kinase inhibitors (A), Rb family members (B), histone regulators (C), histones (D), and chromatin-associated genes (E) in proliferating AMSCs (top) and fold change expression values comparing confluent and proliferating AMSCs (bottom). Histone mRNAs are derived from multiple distinct gene copies that essentially express the same protein. Because individual mRNA species are not efficiently detected in RNASeq due to the lack of a polyA tail, expression values are represented as the aggregate values of multiple different mRNAs for each histone gene subclass. Reads per kilobasepair per million mapped reads = RPKM, Fold Change = FC, n = 3.
Beyond E2F dependent mechanisms that govern cell cycle progression, actively proliferating AMSCs express high levels of DNA replication dependent histone mRNAs (e.g., H3 and H4) (Fig. 3D), which are functionally required for the packaging of DNA at the G1/S phase transition, as well as select non-histone proteins (e.g., HMGA1, HMGB1, HMGB1) (Fig. 3E). Expression of histone and HMG mRNAs is down-regulated in quiescent cells, which no longer replicate DNA, but AMSCs continue to express critical histone gene transcription factors (e.g., HINFP/HiNF-P, CUX1/HiNF-D) (Fig. 3C). Taken together, the down-regulation of mRNAs encoding principal cell cycle regulators and histone genes is consistent with retention of normal cell growth mechanisms in AMSCs that support both self-renewal through mitosis and the cessation of proliferation during quiescence.
Maintenance of the multipotent state in AMSCs
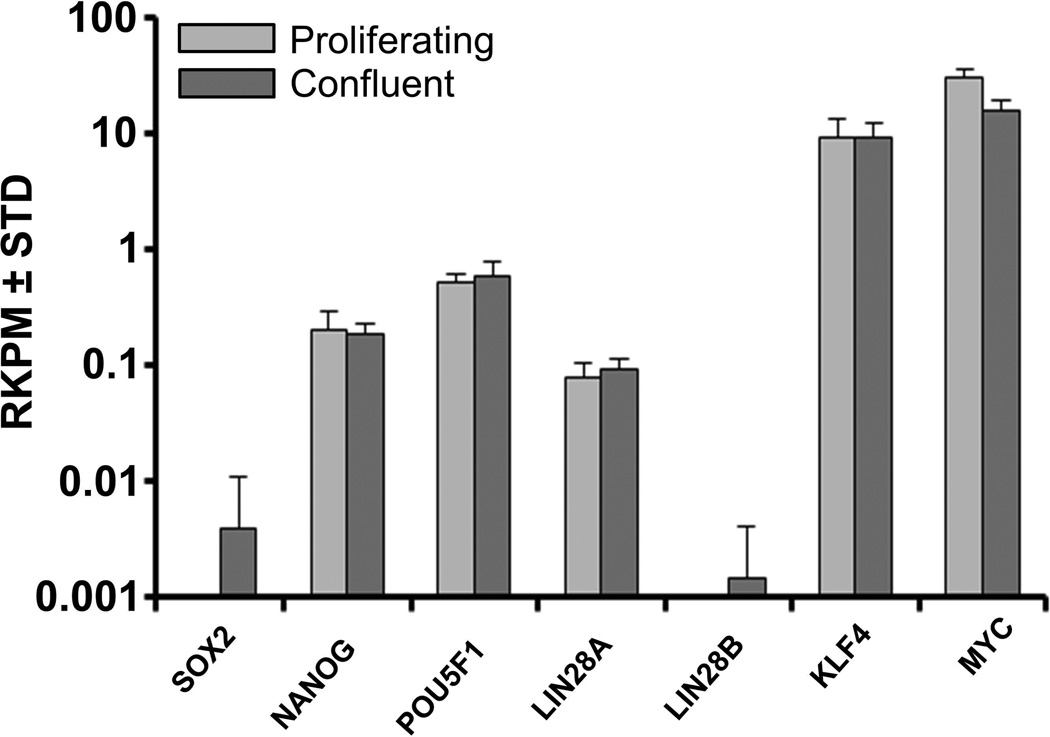
Pluripotent embryonic stem cells derived from the inner cell mass of blastocysts have a unique phenotype that permits phenotypic conversion into any other cell type during development. However, somatic cells derived from adult tissues have less developmental plasticity. Furthermore, the diversity of cells that can reside in tissues derived from human organs provides constraints on the purity of adult cell populations that can be isolated. AMSCs are purified from the stromal vascular fraction of adipose tissue, which enriches for mesenchymal stromal cells. To qualify their “stemness”, we investigated whether AMSCs express the classical regulatory factors present in embryonic stem cells. AMSCs express robust levels of the programming factors MYC and KLF4 (>10 RKPM) based on RNASeq analysis (Fig. 4). In addition, they constitutively express the classical pluripotency related genes NANOG, POU5F1 and LIN28A at relatively low levels (between 0.1 and 1 RKPM), regardless of whether the cells are proliferating or confluent. In addition, MSCs have low but detectable levels of TDGF1, NODAL and ZFP42/REX1 (<0.1 RKPM), but do not express appreciable levels of SOX2 or LIN28B (data not shown). The selective expression of only a subset of the pluripotency markers is consistent with the concept that AMSCs are not pluripotent cells, but immature somatic mesenchymal cells with multi-lineage potential.
FIGURE 4. Expression of pluripotent genes in proliferating and confluent AMSCs by RNA-Seq.
Reads per kilobasepair per million mapped reads = RPKM, n = 3.
Expression profiling of cell surface markers as a proxy for immuno-phenotyping of hPL-AMSCs
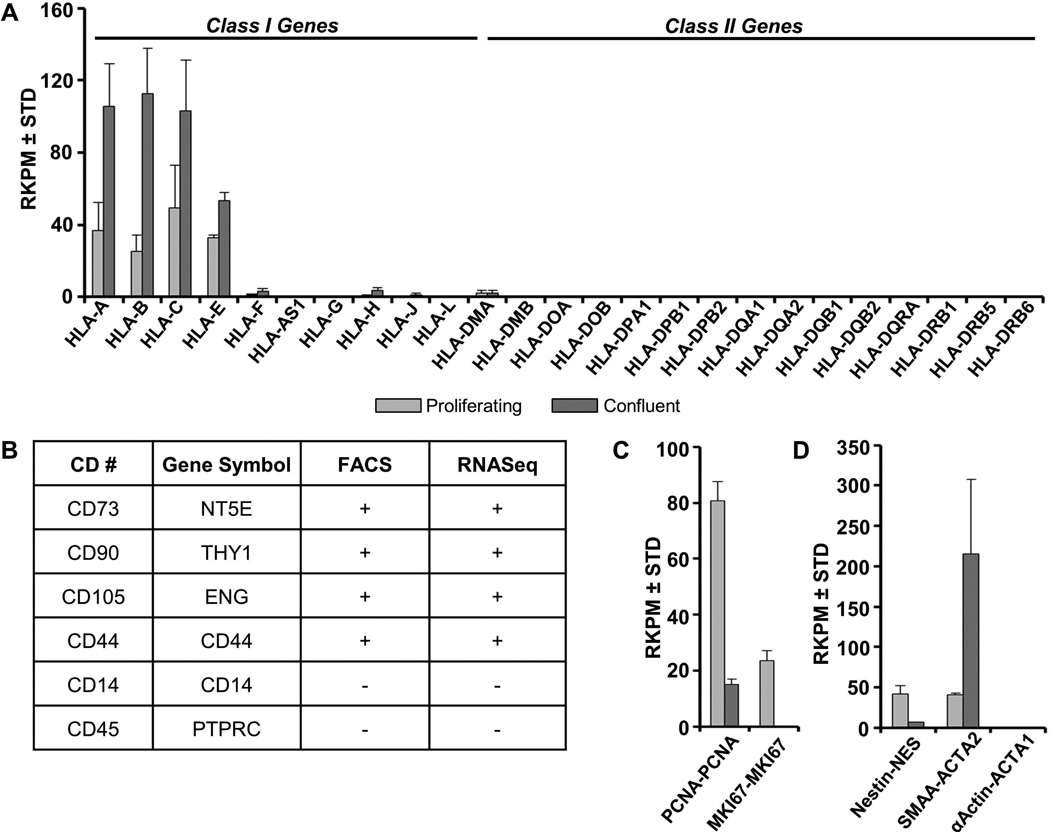
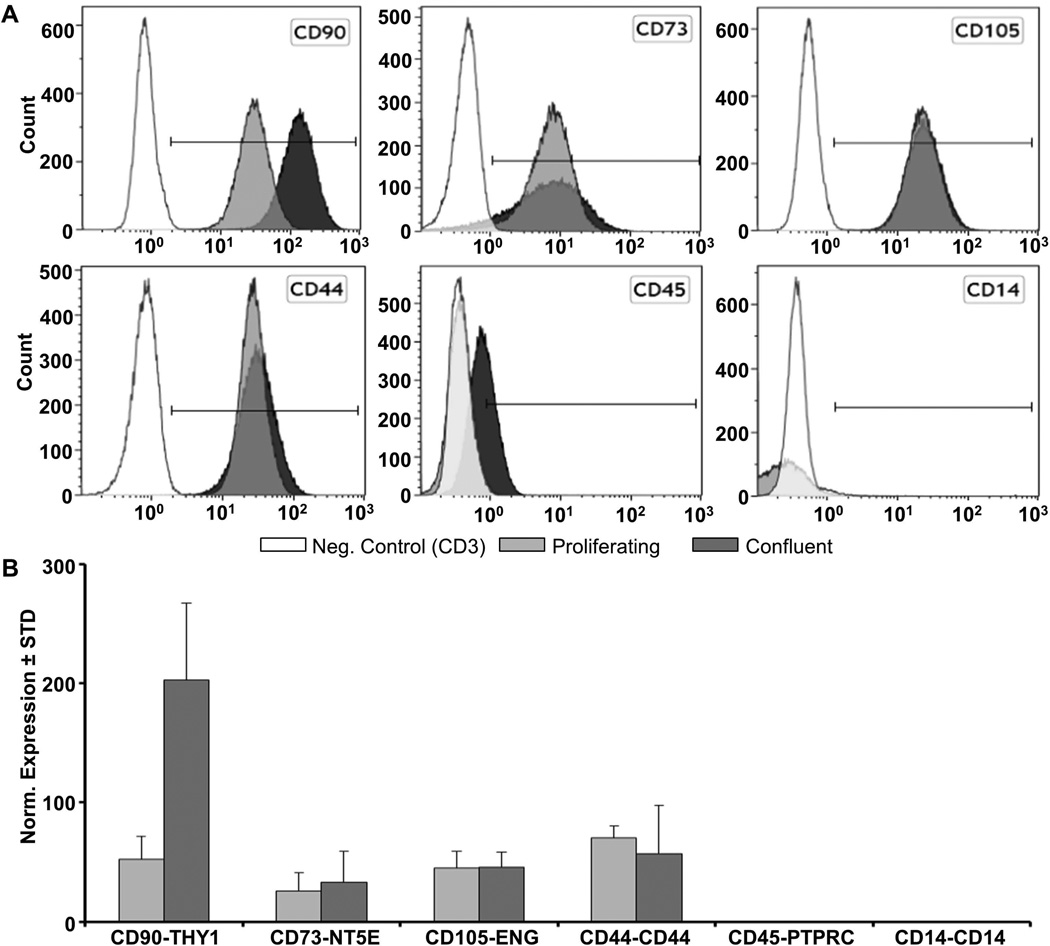
GMP-hPL-AMSCs prepared for use in clinical trials are routinely characterized for the presence of standard cell surface markers by flow cytometry [Crespo-Diaz et al., 2011; Mader et al., 2013]. AMSCs are immuno-positive at the cell surface for typical mesenchymal stromal markers CD44, CD73/NT5E, CD90/THY1 and CD105/ENG, but negative for CD14, CD45/PTPRC (Fig. 5B, data not shown). Corroborating these data, mRNAs for these and other cell surface markers are detected by high throughput RNA Sequencing analysis and RT-qPCR (Fig. 5A & B, data not shown). Interestingly, levels of CD90/THY1 increase in confluent AMSCs based on both flow cytometry and RT-qPCR (Fig. 6). In contrast, CD105/ENG does not change, while CD44 and CD73 each exhibit subtle changes in cell surface expression by flow cytometry, but differences in the corresponding mRNA levels as measured by RT-qPCR do not reach statistical significance (Fig. 6).
FIGURE 5. Gene expression analysis as proxy biomarkers for cell surface expression of proliferating and confluent AMSCs by RNA-Seq.
Expression of class I and II CD markers in proliferating and confluent AMSCs (A). Comparison of CD marker detection by flow cytometry and RNA-Seq in proliferating AMSCs ([+] represents >90% positive cells and >10 RPKM awhile [−] represents <5% positive cell or <1 RPKM) (B). Expression of proliferation and viability markers in proliferating and confluent AMSCs (C). Expression of additional molecular biomarkers for MSC identity in proliferating and confluent AMSCs (D). Reads per kilobasepair per million mapped reads = RPKM, n = 3.
FIGURE 6. Comparison of CD marker expression by flow cytometry and RT-qPCR.
Flow cytometry and RT-qPCR analysis with proliferating and confluent AMSCs was performed as described in methods. Flow cytometry analysis of CD markers in proliferating and confluent AMSCs (n=4, representative data shown) (A). RT-qPCR analysis of CD markers in proliferating and confluent AMSCs (B). Gene expression was normalized to GAPDH (set at 100), n=4.
In addition, AMSCs are negative for class I Major Histocompatibility (MHC-I) antigens and negative for class II MHC and HLA-DR antigens by flow cytometry [Crespo-Diaz et al., 2011] and RNASeq (Fig. 5A). The International Federation of Adipose Therapeutics and Science (IFATS) and International Society for Cellular Therapy (ISCT) also recommend additional definitions for SVF-derived adherent mesenchymal stromal cells in culture, including the negative markers CD235A/GYPA, CD31/PECAM1 and CD106/VCAM1, as well as the positive markers CD13/ANPEP and CD36 [Bourin et al., 2013]. CD106/VCAM1 and CD36 discriminate between AMSCs and bone marrow derived mesenchymal stromal cells (BMSCs) [Bourin et al., 2013]. Based on RNASeq results, AMSCs express relatively high mRNA levels for CD13/ANPEP, but relatively low amounts of CD36 mRNA, while expression of CD106/VCAM1 mRNA is at or below the level of detection (data not shown). Hence, AMSCs express mRNAs for all the recommended cell surface markers and do not express negative markers at appreciable levels.
The IFATS/ISCT panel also recommended tests for viability and fibroblastoid colony-forming units (CFU-Fs). In this study, we monitored MKI67 (encoding the proliferation specific antigen Ki67) and PCNA (encoding an S phase specific DNA polymerase subunit) as biomarkers for proliferative potential and cell viability. The well-established classical markers nestin (NES) and smooth muscle alpha actin (ACTA2/SMAA) were analyzed as additional molecular biomarkers for mesenchymal stem cell identity. Proliferating but not confluent AMSCs express high levels of MKI67 and PCNA (Fig. 5C). AMSCs also express robust levels of both NES and ACTA2/SMAA (Fig. 5D), albeit that expression for both markers changes in opposite directions as AMSCs become post-proliferative.
In conclusion, the RNA expression analysis presented here indicates that AMSCs express all expected markers for mesenchymal stromal cells with multi-lineage potential. Furthermore, the consistency between immuno-phenotyping data and mRNA levels of CD markers and other membrane-associated proteins indicates that gene expression data can be used effectively to complement cell surface expression analysis by flow cytometry.
Post-proliferative activation of ECM-related genes proteins, imprinted genes and genes for WNT signaling components in confluent AMSCs
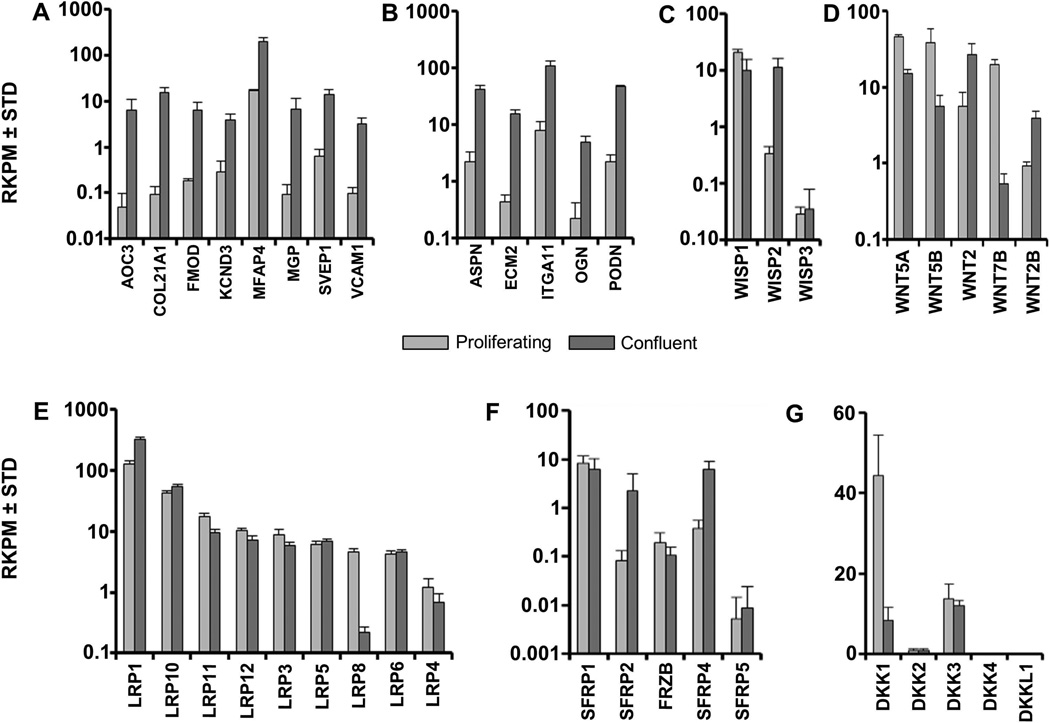
Global gene expression analysis reveals that confluent AMSCs cease high level production of proteins required for cytoskeletal and genomic proteins and instead switch to production of proteins that form an extracellular matrix and/or initiate cell/cell communication. We established which specific genes are both most prominently up-regulated and robustly expressed by filtering our RNAseq dataset for genes exhibiting, respectively, at least a 10 fold difference in gene expression and a relative expression in confluent cells of at least 1 RKPM. Among this set of genes were different types of ECM genes including Collagen Type XXI (COL21A1), fibromodulin (FMOD) and matrix gla protein (MGP), and several leucine-rich repeat proteins such as osteoglycin (OGN), podocan (PODN) (Fig. 7A & B).
FIGURE 7. Expression of extra-cellular matrix and WNT-related genes in proliferating and confluent AMSCs by RNA-Seq.
Expression of cell adhesion genes (A), leucine rich repeat genes (B), WISPs (C), WNTs (D), LRPs (E) SFRPs (F), and DKKs (G) in proliferating and confluent AMSCs. Reads per kilobasepair per million mapped reads = RPKM, n = 3.
Confluent AMSCs selectively up-regulate the WNT-inducible stimulatory protein WISP2 by almost one order of magnitude (Fig. 7C), suggesting that WNT signaling may be more active in confluent cells. Therefore, we examined the expression levels of WNT signaling components. Levels of LRP and FZD proteins that function as WNT receptors/co-receptors remain fairly constant in AMSCs (Fig. 7E, data not shown), except in the case of osteogenic receptor LRP8 [Zhang et al., 2012], which is down regulated. AMSCs change the expression level of distinct WNT ligands that mediate canonical or non-canonical WNT signaling. While proliferating AMSCs express WNT5A, WNT5B and WNT7B, confluent cells express the non-canonical WNT2 and WNT2B ligands (Fig. 7D). Furthermore, AMSCs up-regulate expression of two of the five SFRP/FRZB decoy receptors (SFRP2 and SFRP4) that interact with WNT ligands (Fig 7F). Additionally, post-proliferative AMSCs exhibit a selectively down-regulation of the WNT inhibitor DKK1 (Fig. 7G). Taken together, these results show that AMSCs modify WNT signaling components at confluence.
Spontaneous differentiation of AMSCs
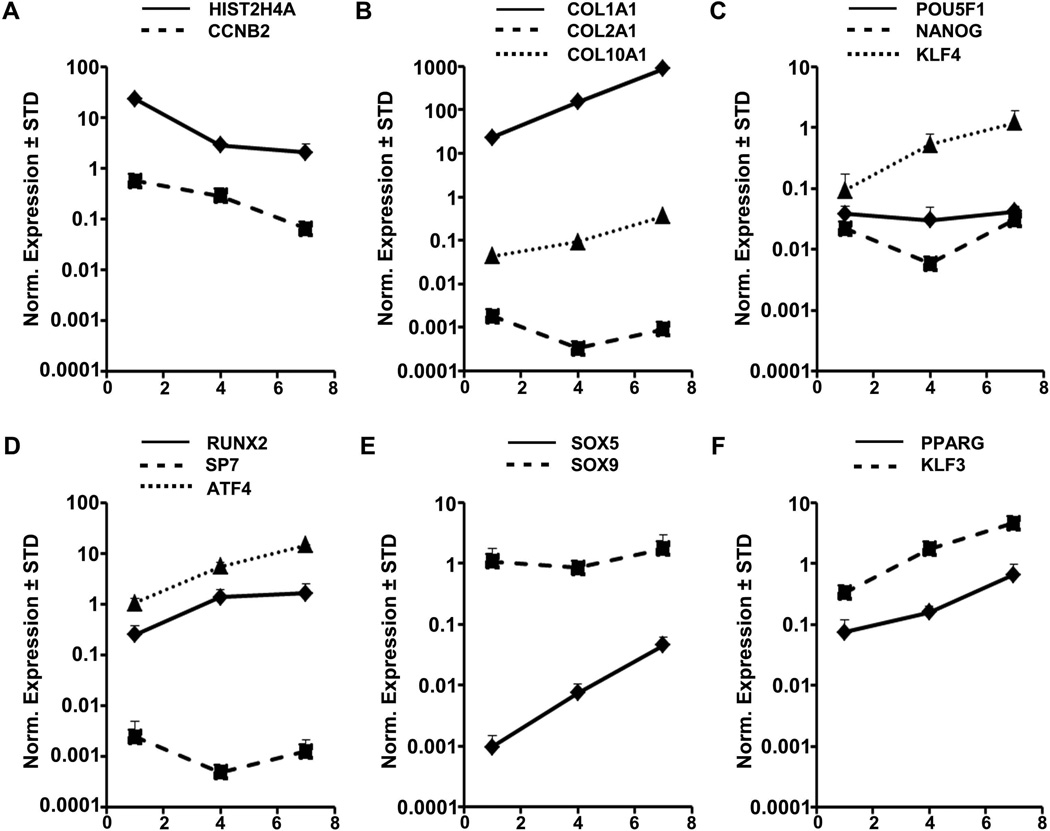
Cessation of proliferation results in a pronounced switch from cytoskeletal and nuclear components to extra-cellular matrix proteins. We next addressed whether continuous culture of AMSCs in base medium with hPL may change their phenotype. RT-qPCR analysis shows that the expression response of AMSCs is rather heterogeneous from a mesenchymal lineage-commitment perspective. Maintenance of cells for 7 days decreases cell proliferation as expected (Fig. 8A). Subsequently, cultures exhibit up-regulation of the chondrogenic marker SOX5 but not SOX9 or COL2A1 (Fig. 8B & E), the osteogenic genes RUNX2 and ATF4, but not SP7/Osterix (Fig. 8D), the adipogenic markers PPARG and KLF3 (Fig. 8F), as well as the reprogramming gene KLF4, but not POU5F1/OCT4 or NANOG (Fig. 8C). While our data do not discriminate between gene expression effects in individual cells versus cell population effects, our data indicate that non-proliferating AMSCs begin to adopt more specialized cellular phenotypes without committing to well-characterized chondrogenic or osteogenic cell fates.
FIGURE 8. Expression of mesenchymal lineage genes in AMSCs by RT-qPCR.
Cells were plated and RNA isolated and processed as described in methods. Expression of cell cycle genes (A), extra-cellular matrix genes (B), pluripotency genes (C), osteogenic transcription factors (D), chondrogenic transcription factors (E), and adipogenic transcription factors (F). X-Axis indicates day of culture where day 1 are actively dividing AMSCs, day 4 are confluent AMSCs, and day 7 are post-confluent AMSCs. Gene expression was normalized to GAPDH (set at 100), n = 3.
DISCUSSION
This study examined the biological phenotype of AMSCs expanded in hPL to understand the therapeutic efficacy of this clinically versatile cell source for tissue repair. We comprehensively examined AMSCs for these and other molecular characteristics using a combination of RNASeq, RT-qPCR and flow cytometry to interrogate mRNA and protein expression in proliferating and confluent cell populations. Our data provide insight into the mechanistic underpinnings for the principal capabilities of AMSCs with respect to their (i) postulated stem cells properties, (ii) developmental plasticity for spontaneous differentiation into distinct mesenchymal lineages, (iii) high proliferative potential and ability for self-renewal, and (iv) anabolic activities to produce extracellular matrix proteins for musculoskeletal tissue repair.
Our study provides a molecular framework for biomedical applications of clinical-grade AMSCs grown in hPL. Since our initial published studies on the growth of AMSCs in this medium [Crespo-Diaz et al., 2011], a number of other studies have examined distinct biological properties of hPL-AMSCs. Similar to our previous work, several reports have shown that pooled hPL increases proliferation rates of AMSCs compared to fetal bovine serum, while not compromising genomic integrity, differentiation potential or cell surface expression of selected CD markers [Ben Azouna et al., 2012; Jung et al., 2012; Kinzebach and Bieback, 2013; Pawitan, 2012; Trojahn Kolle et al., 2013; Warnke et al., 2013; Witzeneder et al., 2013]. Lohman and colleagues noted that the beneficial properties of hPL are age-dependent [Lohmann et al., 2012]. The versatility of hPL is reflected by its ability to regulate cell migration and chemotaxis, as well as promote angiogenesis and extracellular matrix mineralization through chemokine production [Fekete et al., 2012; Leotot et al., 2013]. Another advantage of hPL is that it can be used as a solid surface [Walenda et al., 2012]. Furthermore, hPL is more effective than bovine-based growth media in enhancing the immunomodulatory properties of AMSCs [Menard et al., 2013; Schallmoser and Strunk, 2013]. Human platelet lysate permits chondrogenic differentiation [Hildner et al., 2013] and suppresses adipogenic differentiation [Lange et al., 2012], which is an undesirable cell fate for musculoskeletal tissue regeneration. Taken together, hPL as a liquid or gel has many positive biological effects on AMSCs, and is superior in promoting cell growth.
One concern with propagating AMSCs in culture using hPL is that this rich source of cytokines and growth factors may render cells hyper-proliferative and thus may favor cells that are best at at self-propagation (e.g., cancer stem cells). One counter-argument is that the growth factors in hPL are in such abundance that there should be no selective advantage to serum independence, which is a hallmark of tumor cells. Our studies show that AMSCs become effectively post-proliferative and quiescent when they become contact inhibited in cell culture, as reflected by down regulation of diagnostic cell cycle markers and up regulation of ECM proteins. AMSCs do express high mRNA levels of the immortalizing MYC gene but mRNA levels of telomerase/TERT are below the detection level of RNASeq. Hence, AMSCs exhibit properties of normal contact-inhibited human cells.
Our current findings validate PL-AMSCs as multipotent stromal cells that express cell surface proteins characteristic of mesenchymal stromal cells: CD44, CD73/NT5E, CD90/THY1 and CD105/ENG, but negative for CD14, CD45/PTPRC [Bourin et al., 2013; Crespo-Diaz et al., 2011]. The present work extends these earlier findings by showing that cell surface expression of these proteins is modulated when cells reach confluence and that their detection is corroborated by selective detection of their respective mRNAs. The ‘stemness’ of AMSCs is also reflected by selective expression of factors that are known to control pluripotency (NANOG, POU5F1 and LIN28A) [Apostolou and Hochedlinger, 2013]. We note that the respective mRNA levels for these genes in AMSCs are at least one to two orders of magnitude lower than that for pluripotent embryonic stem cells (data not shown). Consistent with their mesenchymal rather than pluripotent phenotype, AMSCs express the mesenchymal markers Nestin/NES and SMAA/ACTA2, as well as classical transcription factors characteristic for mesenchymal lineages (e.g., RUNX2, SOX9 and PPARG). PL-AMSCs have tri-lineage potential [Crespo-Diaz et al., 2011]. Yet, our current study shows that these cells will spontaneously adopt more mature but less defined mesenchymal phenotypes when they are maintained in a post-proliferative confluent state. Thus, AMSCs have a dynamic phenotype and retention of multi-potency may require an active gene regulatory program that sustains a self-renewing multi-potent state.
Proliferating AMSCs specifically express a standard cell cycle program of cyclins and CDKs, E2Fs and RB related pocket proteins to drive cell cycle progression, as well as histone genes and cognate regulators to support packaging of newly replicated DNA, similar to results shown in previous studies on human pluripotent embryonic stem cells (ESCs) [Becker et al., 2010; Becker et al., 2006; Becker et al., 2007]. Human ES cells have an abbreviated G1 and use a CDK4/cyclin D2 (CCND2) related mechanisms to stimulate cell cycle progression [Becker et al., 2006]. AMSCs, which represent multipotent somatic cells, express high levels of CDK4, but in contrast to ESCs they express CCND1 and to a lesser degree CCND3 instead of CCND2. Also, while E2F5 is the most prominent E2F member expressed in ESCs, E2F1 is highly expressed in multipotent AMSCs. Thus, commitment of ESCs and AMSCs for self-renewal is mediated by different cyclin and E2F subtypes, consistent with fundamental requirements for maintaining pluripotent versus multipotent cellular states.
Our study provides a comprehensive analysis of the molecular phenotype by defining the transcriptomes of proliferating and confluent AMSCs using RNASeq. Jääger and co-workers used RNA-seq analysis to compare AMSCs with dermal fibroblasts and showed that AMSCs have a fundamentally distinct cellular phenotype compared to dermal fibroblasts, reflecting their different developmental origins [Jaager et al., 2012] although both AMSCs and dermal fibroblasts have multi-lineage potential. Anabolic properties of AMSCs in generating skeletal tissues are linked to extracellular matrix protein production [Cawston and Young, 2010; Ma et al., 2013; Mafi et al., 2012; Wu et al., 2013; Wu et al., 2012]. Our work reveals that AMSCs modulate the expression of a number of ECM proteins as cells reach confluence, suggesting that non-proliferating AMSCs are competent at forming and/or remodeling musculoskeletal tissue matrices. Corroborating this idea, Tokunaga and co-workers have presented gene expression data indicating stromal cell type specific differences in the expression of ECM proteins, adhesion proteins and matrix metalloproteases [Tokunaga et al., 2014]. Another interesting observation from our study is that AMSCs robustly and selectively express distinct WNT signaling components, and expression of these WNT related proteins is altered when cells reach confluence. These results complement and extend studies by Torensma and colleagues who profiled bone marrow stromal cells and AMSCs using an Affymetrix exon microarray platform. They noted changes in the expression of proteins within the WNT pathway between BMSCs and AMSCs [Torensma et al., 2013]. Further studies will be required to establish the significance of difference in WNT signaling factors and ECM proteins, both between different mesenchymal cell types and in relation to their proliferative potential.
In conclusion, this study provides a comprehensive characterization of expression programs in AMSCs and shows that distinct cell cycle regulatory pathways are operative during AMSC self-renewal and that tissue-anabolic programs are activated in post-proliferative AMSCs. Cessation of AMSC proliferation occurs concomitant with changes in the expression of WNT signaling components, and spontaneous expression of different mesenchymal phenotypes (e.g., fibroblastic, osteogenic, chondrogenic and adipogenic phenotypes). This work validates the clinical use of AMSCs grown in hPL and provides a roadmap for strategies that engineer and tailor their biological properties for different applications in regenerative medicine.
Acknowledgments
We thank the members of our research groups (Fuhua Xu, Xiaodong Li, Grant Meeker, Catalina Galeano, Meghan McGee-Lawrence, Bridget Stensgard, David Razidlo), as well as our University of Twente colleagues Janine Post and Marcel Karperien for stimulating discussions, and sharing of expertise and biological reagents. We also thank Chris Kolbert and Bruce Eckloff for assistance with RNASeq, as well as Jeff Nie, Jaime Davila and Jean-Pierre Kocher for providing bioinformatics support.
Funding: This work was supported by intramural funds from the Center of Regenerative Medicine at Mayo Clinic and NIH R01 grants AR049069 (to AvW), R01 DE020194 (to JJW) and R01 CA139322 (to GSS & JLS), and a Patrick J. Kelly Research Fellowship Kelly Fellowship award (to SMR). This publication was also made possible in part by a career development award from the American Hand Society and the National Center for Advancing Translational Sciences (UL1 TR000135) (to S Kakar).
Footnotes
Conflict of Interest: Dr. Allan Dietz has a commercial interest in Mill Creek Life Sciences which manufactures the clinical grade commercial platelet lysate product used for maintaining adipose-tissue derived mesenchymal stem cells.
LITERATURE CITED
- Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KA, Ghule PN, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Cyclin D2 and the CDK substrate p220(NPAT) are required for self-renewal of human embryonic stem cells. Journal of cellular physiology. 2010;222:456–464. doi: 10.1002/jcp.21967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. Journal of cellular physiology. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. Journal of cellular physiology. 2007;210:517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- Ben Azouna N, Jenhani F, Regaya Z, Berraeis L, Ben Othman T, Ducrocq E, Domenech J. Phenotypical and functional characteristics of mesenchymal stem cells from bone marrow: comparison of culture using different media supplemented with human platelet lysate or fetal bovine serum. Stem cell research & therapy. 2012;3:6. doi: 10.1186/scrt97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieback K, Hecker A, Kocaomer A, Lannert H, Schallmoser K, Strunk D, Kluter H. Human alternatives to fetal bovine serum for the expansion of mesenchymal stromal cells from bone marrow. Stem cells. 2009;27:2331–2341. doi: 10.1002/stem.139. [DOI] [PubMed] [Google Scholar]
- Blande IS, Bassaneze V, Lavini-Ramos C, Fae KC, Kalil J, Miyakawa AA, Schettert IT, Krieger JE. Adipose tissue mesenchymal stem cell expansion in animal serum-free medium supplemented with autologous human platelet lysate. Transfusion. 2009;49:2680–2685. doi: 10.1111/j.1537-2995.2009.02346.x. [DOI] [PubMed] [Google Scholar]
- Bourin P, Bunnell BA, Casteilla L, Dominici M, Katz AJ, March KL, Redl H, Rubin JP, Yoshimura K, Gimble JM. Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: a joint statement of the International Federation for Adipose Therapeutics and Science (IFATS) and the International Society for Cellular Therapy (ISCT) Cytotherapy. 2013;15:641–648. doi: 10.1016/j.jcyt.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castegnaro S, Chieregato K, Maddalena M, Albiero E, Visco C, Madeo D, Pegoraro M, Rodeghiero F. Effect of platelet lysate on the functional and molecular characteristics of mesenchymal stem cells isolated from adipose tissue. Current stem cell research & therapy. 2011;6:105–114. doi: 10.2174/157488811795495440. [DOI] [PubMed] [Google Scholar]
- Cawston TE, Young DA. Proteinases involved in matrix turnover during cartilage and bone breakdown. Cell and tissue research. 2010;339:221–235. doi: 10.1007/s00441-009-0887-6. [DOI] [PubMed] [Google Scholar]
- Cholewa D, Stiehl T, Schellenberg A, Bokermann G, Joussen S, Koch C, Walenda T, Pallua N, Marciniak-Czochra A, Suschek CV, Wagner W. Expansion of adipose mesenchymal stromal cells is affected by human platelet lysate and plating density. Cell transplantation. 2011;20:1409–1422. doi: 10.3727/096368910X557218. [DOI] [PubMed] [Google Scholar]
- Crespo-Diaz R, Behfar A, Butler GW, Padley DJ, Sarr MG, Bartunek J, Dietz AB, Terzic A. Platelet lysate consisting of a natural repair proteome supports human mesenchymal stem cell proliferation and chromosomal stability. Cell transplantation. 2011;20:797–811. doi: 10.3727/096368910X543376. [DOI] [PubMed] [Google Scholar]
- Fekete N, Rojewski MT, Furst D, Kreja L, Ignatius A, Dausend J, Schrezenmeier H. GMP-compliant isolation and large-scale expansion of bone marrow-derived MSC. PloS one. 2012;7:e43255. doi: 10.1371/journal.pone.0043255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner F, Eder MJ, Hofer K, Aberl J, Redl H, van Griensven M, Gabriel C, Peterbauer-Scherb A. Human platelet lysate successfully promotes proliferation and subsequent chondrogenic differentiation of adipose-derived stem cells: a comparison with articular chondrocytes. Journal of tissue engineering and regenerative medicine. 2013 doi: 10.1002/term.1649. [DOI] [PubMed] [Google Scholar]
- Iudicone P, Fioravanti D, Bonanno G, Miceli M, Lavorino C, Totta P, Frati L, Nuti M, Pierelli L. Pathogen-free, plasma-poor platelet lysate and expansion of human mesenchymal stem cells. Journal of translational medicine. 2014;12:28. doi: 10.1186/1479-5876-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaager K, Islam S, Zajac P, Linnarsson S, Neuman T. RNA-seq analysis reveals different dynamics of differentiation of human dermis- and adipose-derived stromal stem cells. PloS one. 2012;7:e38833. doi: 10.1371/journal.pone.0038833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Panchalingam KM, Wuerth RD, Rosenberg L, Behie LA. Large-scale production of human mesenchymal stem cells for clinical applications. Biotechnology and applied biochemistry. 2012;59:106–120. doi: 10.1002/bab.1006. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzebach S, Bieback K. Expansion of Mesenchymal Stem/Stromal Cells under Xenogenic-Free Culture Conditions. Advances in biochemical engineering/biotechnology. 2013;129:33–57. doi: 10.1007/10_2012_134. [DOI] [PubMed] [Google Scholar]
- Lange C, Brunswig-Spickenheier B, Eissing L, Scheja L. Platelet lysate suppresses the expression of lipocalin-type prostaglandin D2 synthase that positively controls adipogenic differentiation of human mesenchymal stromal cells. Experimental cell research. 2012;318:2284–2296. doi: 10.1016/j.yexcr.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Leotot J, Coquelin L, Bodivit G, Bierling P, Hernigou P, Rouard H, Chevallier N. Platelet lysate coating on scaffolds directly and indirectly enhances cell migration, improving bone and blood vessel formation. Acta biomaterialia. 2013;9:6630–6640. doi: 10.1016/j.actbio.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Lohmann M, Walenda G, Hemeda H, Joussen S, Drescher W, Jockenhoevel S, Hutschenreuter G, Zenke M, Wagner W. Donor age of human platelet lysate affects proliferation and differentiation of mesenchymal stem cells. PloS one. 2012;7:e37839. doi: 10.1371/journal.pone.0037839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Landman EB, Miclea RL, Wit JM, Robanus-Maandag EC, Post JN, Karperien M. WNT signaling and cartilage: of mice and men. Calcified tissue international. 2013;92:399–411. doi: 10.1007/s00223-012-9675-5. [DOI] [PubMed] [Google Scholar]
- Mader EK, Butler G, Dowdy SC, Mariani A, Knutson KL, Federspiel MJ, Russell SJ, Galanis E, Dietz AB, Peng KW. Optimizing patient derived mesenchymal stem cells as virus carriers for a phase I clinical trial in ovarian cancer. Journal of translational medicine. 2013;11:20. doi: 10.1186/1479-5876-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafi P, Hindocha S, Mafi R, Khan WS. Evaluation of biological protein-based collagen scaffolds in cartilage and musculoskeletal tissue engineering--a systematic review of the literature. Current stem cell research & therapy. 2012;7:302–309. doi: 10.2174/157488812800793045. [DOI] [PubMed] [Google Scholar]
- Menard C, Pacelli L, Bassi G, Dulong J, Bifari F, Bezier I, Zanoncello J, Ricciardi M, Latour M, Bourin P, Schrezenmeier H, Sensebe L, Tarte K, Krampera M. Clinical-grade mesenchymal stromal cells produced under various good manufacturing practice processes differ in their immunomodulatory properties: standardization of immune quality controls. Stem cells and development. 2013;22:1789–1801. doi: 10.1089/scd.2012.0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawitan JA. Platelet rich plasma in xeno-free stem cell culture: the impact of platelet count and processing method. Current stem cell research & therapy. 2012;7:329–335. doi: 10.2174/157488812802481508. [DOI] [PubMed] [Google Scholar]
- Rider DA, Dombrowski C, Sawyer AA, Ng GH, Leong D, Hutmacher DW, Nurcombe V, Cool SM. Autocrine fibroblast growth factor 2 increases the multipotentiality of human adipose-derived mesenchymal stem cells. Stem cells. 2008;26:1598–1608. doi: 10.1634/stemcells.2007-0480. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo VE, Duarte AR, Popa EG, Gomes ME, Mano JF, Reis RL. Enhancement of osteogenic differentiation of human adipose derived stem cells by the controlled release of platelet lysates from hybrid scaffolds produced by supercritical fluid foaming. Journal of controlled release : official journal of the Controlled Release Society. 2012;162:19–27. doi: 10.1016/j.jconrel.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Schallmoser K, Strunk D. Generation of a pool of human platelet lysate and efficient use in cell culture. Methods in molecular biology. 2013;946:349–362. doi: 10.1007/978-1-62703-128-8_22. [DOI] [PubMed] [Google Scholar]
- Shih DT, Chen JC, Chen WY, Kuo YP, Su CY, Burnouf T. Expansion of adipose tissue mesenchymal stromal progenitors in serum-free medium supplemented with virally inactivated allogeneic human platelet lysate. Transfusion. 2011;51:770–778. doi: 10.1111/j.1537-2995.2010.02915.x. [DOI] [PubMed] [Google Scholar]
- Siciliano C, Ibrahim M, Scafetta G, Napoletano C, Mangino G, Pierelli L, Frati G, De Falco E. Optimization of the isolation and expansion method of human mediastinal-adipose tissue derived mesenchymal stem cells with virally inactivated GMP-grade platelet lysate. Cytotechnology. 2013 doi: 10.1007/s10616-013-9667-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M, Inoue M, Jiang Y, Barnes RH, 2nd, Buchner DA, Chun TH. Fat depot-specific gene signature and ECM remodeling of Sca1 adipose-derived stem cells. Matrix biology : journal of the International Society for Matrix Biology. 2014 doi: 10.1016/j.matbio.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torensma R, Prins HJ, Schrama E, Verwiel ET, Martens AC, Roelofs H, Jansen BJ. The impact of cell source, culture methodology, culture location, and individual donors on gene expression profiles of bone marrow-derived and adipose-derived stromal cells. Stem cells and development. 2013;22:1086–1096. doi: 10.1089/scd.2012.0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojahn Kolle SF, Oliveri RS, Glovinski PV, Kirchhoff M, Mathiasen AB, Elberg JJ, Andersen PS, Drzewiecki KT, Fischer-Nielsen A. Pooled human platelet lysate versus fetal bovine serum-investigating the proliferation rate, chromosome stability and angiogenic potential of human adipose tissue-derived stem cells intended for clinical use. Cytotherapy. 2013;15:1086–1097. doi: 10.1016/j.jcyt.2013.01.217. [DOI] [PubMed] [Google Scholar]
- Walenda G, Hemeda H, Schneider RK, Merkel R, Hoffmann B, Wagner W. Human platelet lysate gel provides a novel three dimensional-matrix for enhanced culture expansion of mesenchymal stromal cells. Tissue engineering. Part C, Methods. 2012;18:924–934. doi: 10.1089/ten.TEC.2011.0541. [DOI] [PubMed] [Google Scholar]
- Warnke PH, Humpe A, Strunk D, Stephens S, Warnke F, Wiltfang J, Schallmoser K, Alamein M, Bourke R, Heiner P, Liu Q. A clinically-feasible protocol for using human platelet lysate and mesenchymal stem cells in regenerative therapies. Journal of cranio-maxillo-facial surgery : official publication of the European Association for Cranio-Maxillo-Facial Surgery. 2013;41:153–161. doi: 10.1016/j.jcms.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Witzeneder K, Lindenmair A, Gabriel C, Holler K, Theiss D, Redl H, Hennerbichler S. Human-derived alternatives to fetal bovine serum in cell culture. Transfusion medicine and hemotherapy : offizielles Organ der Deutschen Gesellschaft fur Transfusionsmedizin und Immunhamatologie. 2013;40:417–423. doi: 10.1159/000356236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Cai X, Zhang S, Karperien M, Lin Y. Regeneration of articular cartilage by adipose tissue derived mesenchymal stem cells: perspectives from stem cell biology and molecular medicine. Journal of cellular physiology. 2013;228:938–944. doi: 10.1002/jcp.24255. [DOI] [PubMed] [Google Scholar]
- Wu L, Prins HJ, Helder MN, van Blitterswijk CA, Karperien M. Trophic effects of mesenchymal stem cells in chondrocyte co-cultures are independent of culture conditions and cell sources. Tissue engineering. Part A. 2012;18:1542–1551. doi: 10.1089/ten.TEA.2011.0715. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang X, Zhang L, Zhou F, van Dinther M, Ten Dijke P. LRP8 mediates Wnt/beta-catenin signaling and controls osteoblast differentiation. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2012;27:2065–2074. doi: 10.1002/jbmr.1661. [DOI] [PubMed] [Google Scholar]