Abstract
Introduction
Crohn’s disease (CD) and ulcerative colitis (UC) are chronic immunologically mediated diseases with a progressive relapsing remitting course. There is considerable heterogeneity in disease course and accurate prediction of natural history has been challenging. The phenotypic implication of increasing genetic predisposition to CD or UC is unknown.
Methods
The data source for our study was a prospective cohort of CD and UC patients recruited from a tertiary referral center. All patients underwent genotyping on the Illumina Immunochip. A genetic risk score (GRS) incorporating strength of association (log odds ratio) and allele dose for each of the 163 IBD-risk loci was calculated and phenotypic associations examined across GRS quartiles.
Results
Our study cohort included 1,105 patients (697 CD, 408 UC). Increasing genetic burden was associated with earlier age of diagnosis of CD (Ptrend=0.008). Patients in the highest GRS quartile were likely to develop disease 5 years earlier than those in the lowest quartile. Increasing genetic burden was also associated with ileal involvement in CD (Ptrend < 0.0001). The effect of genetic burden was independent of the NOD2 locus and was stronger among those with no NOD2 variants, and in never smokers. UC patients with an involved first degree relative had a higher genetic burden, but GRS was not associated with disease phenotype in UC.
Conclusion
Increasing genetic burden is associated with early age of diagnosis in CD but not UC. The expanded panel of IBD risk loci explains only a fraction of variance of disease phenotype suggesting limited clinical utility of genetics in predicting natural history.
Keywords: Genetic burden, phenotype, ileal disease, Crohn’s disease, age at diagnosis
INTRODUCTION
We are witnessing changes in the therapeutic paradigms for management of Crohn’s disease (CD) and ulcerative colitis (UC) which are complex diseases affecting an estimated 1.6 million Americans1, 2. Recognizing progressive bowel damage and the consequence of prolonged mucosal disruption, randomized trials have demonstrated that treating early on the course of the disease with efficacious anti-tumor necrosis factor (anti-TNF) biologic therapies may result in superior outcomes and prevent complications3-5. However, widespread adoption of such an approach is challenging owing to concerns regarding cost and safety3, 6. Genetic variants offer the promise of an unchanging, stable and readily available predictive tool if demonstrated to have strong prognostic implications for disease behavior and disease-related complications.
Advances in genetics have led to the identification of 163 distinct genetic risk loci for either CD or UC; the vast majority of loci are shared between the two diseases7. While these variants correlate well with disease risk, albeit with modest odds ratios, the implications of the genetic burden conferred by these loci on disease phenotype remains unexplored. The examination of genotype-phenotype correlations in IBD have been mostly through candidate single nucleotide polymorphism (SNP) analyses, using small cohorts of patients with significant associations not withstanding correction for multiple testing, or have preceded recent genetic discovery using fine mapping8, 9. Recent such analyses have suggested that rather than examining the association between single SNPs and disease behavior in CD and UC8, a more comprehensive approach examining overall genetic burden may yield more meaningful results. Indeed, other inflammatory diseases that share pathogenesis with IBD have demonstrating phenotypic implications to an increasing genetic burden10, 11. Chibnik et al. genotyped 542 Caucasian rheumatoid arthritis (RA) patients for 31 validated RA risk loci and found an association between increasing genetic burden, seropositivity, and erosive changes in RA10.
In this prospective study, we examined the phenotypic implications of a higher genetic risk burden for CD and UC using a novel genetic risk score method. We demonstrate contrasting effects of genetics on disease behavior across CD and UC suggesting differential contribution of genetics to disease phenotype for both diseases.
METHODS
Study cohort
We recruited adult patients, age > 18 years, with CD, UC, or indeterminate colitis in a prospective patient cohort, the prospective registry of IBD study at Massachusetts General Hospital (PRISM). All patients were interviewed by a study coordinator and provided detailed information regarding their disease which was verified by their treating physician. Disease location was classified according to the Montreal classification in CD as involving the ileum alone, colon alone, or ileocolonic with a modifier for upper gastrointestinal tract involvement and perianal disease. Disease behavior was stratified as inflammatory, stricturing, or penetrating disease. Disease extent in UC was according to the Montreal classification; patients with proctitis and left-sided colitis were collapsed into one stratum. Smoking status was classified as current, never, or former smoker at enrollment into the study. We also obtained information on medication use including use of immunomodulators (azathioprine, 6-mercaptopurine, and methotrexate) and anti-TNF agents (infliximab, adalimumab, certolizumab pegol). Disease phenotype and medication use was updated till August 2012. All patients provided informed consent and the study was approved by the Institutional Review Board of Partners Healthcare.
Genotyping and genetic risk score calculation
Approximately 10mL of blood was drawn from all patients for extraction of the buffy coat. Genotyping was then performed on the Illumina Immunochip at the Broad Institute (Cambridge, MA). This is a custom-designed genetic chip with nearly 200,000 fine mapping loci that have previously described or have been putatively associated with a spectrum of inflammatory diseases12.
The weighted genetic risk score (GRS) was calculated by first extracting the 163 distinct IBD risk loci. For each risk allele, the genetic burden conferred by it was calculated by multiplying the log odds ratio of its association with disease from the recent expanded Immunochip analysis7 and allele dose (wild type, heterozygous, or homozygous). Wild type was assigned a weight of 0, with weights of 1 and 2 for hetero- and homozygosity. The cumulative GRS was then calculating by adding the contribution of each of the risk loci (Σ [log(odds ratio) × allele dose (0,1,2)]. The CD GRS was restricted to the 140 CD risk alleles while the UC-GRS included only the 133 UC risk alleles, with 120 SNPs shared across both diseases. The GRS was stratified into quartiles for examination of a dose-dependent effect. The continuous GRS was used to calculate the p-value for the trend across the strata.
Statistical Analysis
Genotypic extraction and calculation of the GRS was performed using Plink V1.0713. All SNPs met the Hardy-Weinberg equilibrium threshold of p > 0.001, genotyping call rate > 99%, and genotyping success rate > 80%. Continuous variables were summarized as medians and interquartile ranges (IQR) while categorical variables were expressed as proportions and compared using the chi-square test. Statistical analysis was performed using Stata 12.0 (StataCorp, College Station, TX). Linear regression analysis was performed with age at diagnosis as a dependent variable. For categorical outcomes of disease location, behavior, need for surgery, immunomodulator or anti-TNF therapy, logistic regression with genetic burden as a predictor variable was performed. We also performed analysis stratified by the NOD2 locus which has so far demonstrated the strongest phenotypic association14, as well as smoking status to examine potential gene-environment interactions. We also adjusted for the present of NOD2 variants in our multivariate model to demonstrate the effect of the GRS independent of the NOD2 locus. A p-value < 0.05 indicated statistical significance. To confirm the robustness of our results, we repeated the logistic regression models using bootstrapping with 1000 repetitions. We also divided our cohort into two equal random subsets of patients with CD and UC and compared the association between genetic burden and disease phenotype in both subcohorts.
RESULTS
Study Cohort
Our study included 1,105 patients (697 CD, 408 UC) genotyped on the Illumina Immunochip. The median age of diagnosis was 24 years among those with CD and 27 years for UC with median follow-up after diagnosis of 10 and 8 years respectively (Table 1). Approximately 27% of patients with CD and 21% of those with UC had a family history of IBD, with 5% having more than one first degree relative affected. A third of the CD patients had penetrating disease (31%); one-quarter had isolated colonic disease (26%), and a similar proportion had perianal disease (28%). Just over half the patients with ulcerative colitis (55%) had pancolitis. Immunomodulator use was frequent in both diseases, occurring in 72% of those with CD and 58% of those with UC. A total of 44% of patients with CD and 9% of those with UC underwent bowel resection. The median duration of disease was similar across all quartiles of genetic risk for both CD and UC (p > 0.10 for both comparisons). Out of a possible 280 risk SNPs (140 SNPs × 0, 1, or 2 for each allele) in CD, the median allele number was 146, corresponding to a median unweighted allele count of 0.54 (IQR 0.52 – 0.56). Among those with CD, 24% were heterozygous and 7% homozygous for a NOD2 polymorphism.
Table 1.
Cohort characteristics
| Crohn’s disease (n = 697) |
Ulcerative colitis (n = 408) |
|
|---|---|---|
| Median age at diagnosis (IQR) (in years) |
24 (18 – 33) | 27 (21 – 38) |
| Male sex | 324 (46) | 196 (48) |
| Median disease duration (IQR) (in years) |
10 (5 – 18) | 8 (4 – 16) |
| Family history of IBD | 188 (27) | 85 (21) |
| ≥ 2 first degree relatives with IBD |
36 (5) | 21 (5) |
| Disease behavior | ||
| Inflammatory | 319 (48) | |
| Stricturing | 150 (22) | |
| Penetrating | 207 (31) | |
| Disease location | ||
| Ileal | 151 (22) | |
| Colonic | 176 (26) | |
| Ileocolonic | 355 (52) | |
| Perianal disease | 194 (28) | |
| Disease extent | ||
| Proctitis | 44 (11) | |
| Limited colitis | 133 (34) | |
| Pancolitis | 214 (55) | |
| Immunomodulators | 503 (72) | 237 (58) |
| Biologic anti-TNFs | 392 (56) | 133 (33) |
| Bowel Resection | 304 (44) | 37 (9) |
| Smoking | ||
| Never | 421 (62) | 251 (64) |
| Former | 203 (30) | 125 (32) |
| Current | 53 (8) | 17 (4) |
TNF – tumor necrosis factor
GRS and phenotype in CD
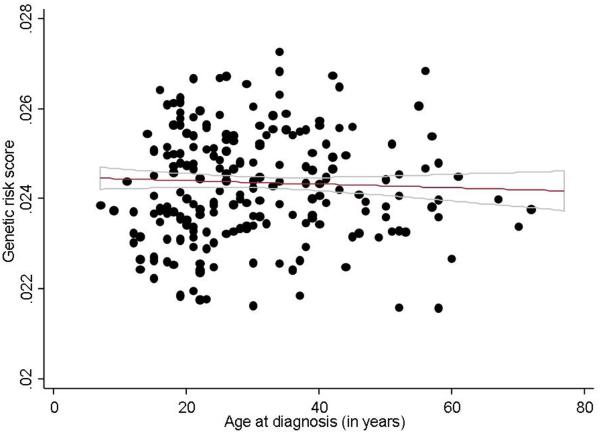
Table 2 describes the association between GRS quartile and disease phenotype in CD. There was a strong association between genetic burden and age at diagnosis. Compared to patients in the lowest quartile of GRS, those in the highest quartile had an age of onset nearly 5 years younger (25.4 years vs. 30.3 years, Regression co-efficient −4.9, 95% confidence interval (CI) −7.7 to −2.1; Ptrend 0.008) (Figure 1). Similarly, higher genetic burden also correlated with ileal disease location. Just over two-thirds of those in the lowest quartile of GRS had ileal involvement with their CD compared to 82% of those in the highest quartile (p=0.002). There was a highly significant linear trend across quartiles (p < 0.0001). We also observed a trend towards complicated CD (Ptrend 0.09), immunomodulator use (Ptrend 0.07) and bowel resection (Ptrend 0.05) with increasing GRS. However, the association with complicated disease and bowel resection were confounded by the association of the GRS with ileal disease location. Adjusting for disease location rendered those associations non-significant, and attenuated the trend towards immunomodulator use (Ptrend 0.12). Interestingly, there was no difference in GRS based on family history of IBD (Ptrend 0.49) or with history of IBD in multiple first degree relatives.
Table 2.
Genetic burden and disease phenotype in Crohn’s disease
| Phenotype | Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | P(linear trend) |
|---|---|---|---|---|---|
| Age at diagnosis | 0 (Reference) | −3.2 (−5.9 to −0.4) | −3.6 (−6.4 to −0.8) | −4.9 (−7.7 to −2.1) | 0.008 |
| Ileal disease | 1 (Reference) | 1.5 (0.9 - 2.4) | 1.9 (1.2 – 3.1) | 2.5 (1.5 – 4.1) | < 0.0001 |
| Perianal disease | 1 (Reference) | 1.5 (0.9 – 2.4) | 1.4 (0.9 – 2.2) | 1.2 (0.7 – 2.0) | 0.66 |
| Complicated CD | 1 (Reference) | 0.9 (0.6 – 1.4) | 0.9 (0.6 – 1.4) | 1.3 (0.8 – 2.0) | 0.09 |
| Bowel resection | 1 (Reference) | 1.4 (0.9 – 2.1) | 1.2 (0.8 – 1.8) | 1.5 (1.0 – 2.3) | 0.05 |
| IMM use | 1 (Reference) | 2.3 (1.4 – 3.7) | 1.4 (0.9 – 2.2) | 1.9 (1.2 – 3.1) | 0.07 |
| Anti-TNF use | 1 (Reference) | 1.9 (1.2 – 2.9) | 1.4 (0.9 – 2.2) | 1.4 (0.9 – 2.1) | 0.38 |
| Family history | 1 (Reference) | 1.3 (0.8 – 2.1) | 1.1 (0.7 – 1.7) | 1.2 (0.7 – 1.9) | 0.49 |
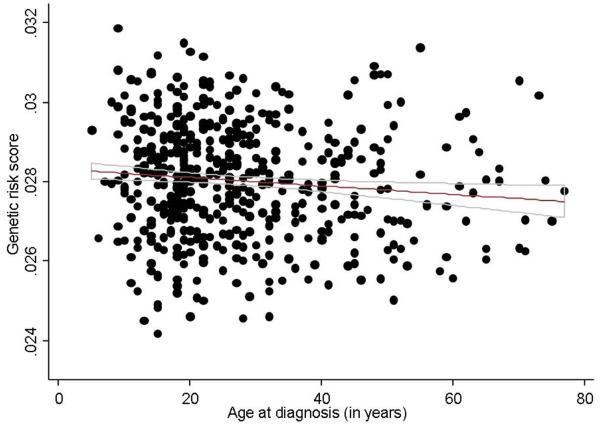
Figure 1. Association between genetic burden and age of diagnosis in Crohn’s disease and ulcerative colitis.
(a) Crohn’s disease
(b) Ulcerative colitis
We then examined the fraction of the variance in disease phenotypes predicted by the GRS. The area under the curve (AUC) for predicting ileal involvement in CD patients by the GRS was 0.60. However, the fraction of the disease variance (measured by the pseudo-R2) explained by the CD risk score was less than 5% for both age at diagnosis and ileal location suggesting that much of the variance in disease phenotype in CD remains unexplained by the IBD risk loci. The association (or lack thereof) between disease phenotypes and CD was similar after repeating the regression models by bootstrapping, as well after division of our cohort into two subsets.
Interactions with NOD2 and smoking status
Given the previously demonstrated phenotypic associations between the NOD2 locus and ileal fibrostenosing CD, we examined whether the GRS quartile interacts with any of the recently described six variants in the NOD2 locus15. We also examined gene-environment interactions by examining whether genetic burden differentially impacts phenotype based on smoking status. The GRS had similar influences on age of onset in individuals without any NOD2 variants (OR for Q4 vs. Q1: −4.3, 95% CI −7.7 to −0.8) as those with at least one NOD2 mutation (OR −4.1, 95% CI −8.5 to 0.4). The association between GRS and ileal involvement was also stronger for patients without a NOD2 mutation compared to those with at least one variant. Interestingly, GRS quartile had a stronger influence on age of onset in never smokers (−5.0, 95% CI −8.2 to −1.8) compared to ever smokers (−4.2, 95% CI −9.4 to 0.9).
GRS and phenotype in UC
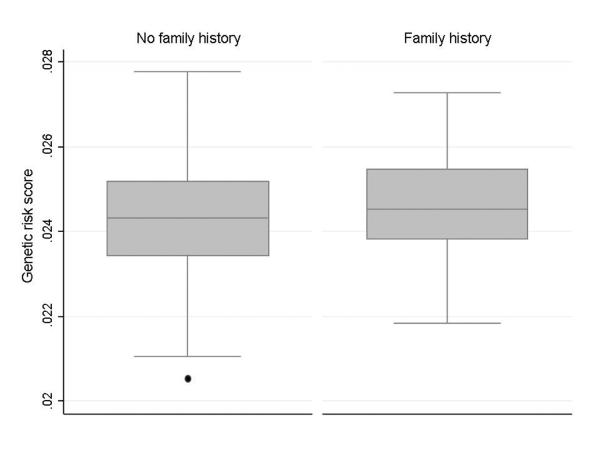
In contrast to CD, the GRS did not predict age of diagnosis, extent of disease or severity defined as the need for immunomodulator, anti-TNF biologics, or colectomy in UC (Ptrend for all > 0.20) (Table 3). We observed a direct association between family history of IBD and genetic burden of UC. Patients in the highest quartile of GRS were twice as likely to have a first degree relative with a history of IBD (Odds ratio (OR) 1.9, 95% CI 1.0 – 3.9; Ptrend 0.03) (Figure 2) compared to those with the lowest quartile of genetic burden. Patients in the highest GRS quartiles were also twice as likely to have two or more first degree relatives (OR 2.29, p=0.18) with a non-significant association due to small number of patients in this category.
Table 3.
Genetic burden and disease phenotype in Ulcerative colitis
| Phenotype | Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | P(linear trend) |
|---|---|---|---|---|---|
| Age at diagnosis | 0 (Reference) | 0.2 (−3.4 to 3.8) | −1.6 (−5.2 to 2.0) | −0.8 (−4.5 to 2.7) | 0.39 |
| Pancolitis | 1 (Reference) | 1.4 (0.8 – 2.5) | 1.3 (0.7 – 2.2) | 1.3 (0.8 – 2.3) | 0.76 |
| Colectomy | 1 (Reference) | 3.0 (1.1 – 7.8) | 1.4 (0.5 – 4.1) | 1.2 (0.4 – 3.6) | 0.92 |
| IMM use | 1 (Reference) | 0.8 (0.5 – 1.4) | 1 (0.6 – 1.8) | 0.8 (0.5 – 1.4) | 0.70 |
| Anti-TNF use | 1 (Reference) | 0.7 (0.4 – 1.4) | 0.9 (0.5 – 1.6) | 0.9 (0.5 – 1.6) | 0.63 |
| Family history | 1 (Reference) | 1.5 (0.7 – 3.0) | 1.3 (0.6 – 2.7) | 1.9 (1.0 – 3.9) | 0.03 |
Figure 2. Association between family history and genetic burden in ulcerative colitis.
DISCUSSION
In a large, prospectively followed and genotyped cohort of 1,105 patients with IBD, we demonstrate that increasing genetic burden of disease predisposes towards ileal disease location and early age of diagnosis in CD, with a weaker trend towards severe manifestations of disease influenced by the tendency for ileal involvement. In contrast, increasing genetic burden was more common in UC patients with an involved first degree relative, but had had no implications on disease phenotype or severity.
A recent study by Jung et al. examined the phenotypic implications of 53 CD-risk alleles in a French cohort8. The presence of NOD2 risk alleles was associated with early age of onset and ileal involvement. While other phenotypic associations between the interleukin-23 receptor (IL23R) polymorphisms and colonic disease as well as autophagy mutations at the ATG16L1 and IRGM loci on non-inflammatory disease, such associations were not robust to replication in an independent cohort8. Weersma et al. genotyped a large Dutch cohort for five disease risk alleles, namely NOD2, IBD5, DLG5, ATG16L1 and IL23R16. Increasing genetic burden quantified in their study by assigning an equal weight to polymorphisms at each of the loci revealed an association with penetrating or stricturing behavior and need for surgery. However, their study pre-dated discovery of several newly identified loci and also did not account for the potential differential strength of association conferred by the loci owing to varying effects on CD risk. Furthermore, associations with complicated disease in their study did not adjust for pre-disposition to ileal disease16. In our study, as was noted by Jung et al., adjusting for ileal location of CD neutralizes the associations observed with penetrating, fibro-stenosing or surgical CD, suggesting little added clinical utility to genotyping.
Several studies have examined genetic predictors of age of onset of IBD and have yielded inconsistent results. The genetic risk allele with one of the strongest effect sizes on CD risk is the NOD2 locus. However, two studies by Abreu et al. and Ahmad et al. found no association between NOD2 carriage and age of onset of CD17, 18. In contrast, Annese et al. found a dose-dependent effect of NOD2 with mean age of diagnosis of 33, 29, and 24 years among those with 0, 1, or 2 NOD2 mutations19. In a Dutch cohort with genotyping restricted to five disease-associated loci, increasing numbers of risk alleles was associated with diagnosis younger than 40 years of age16. Our study extends these previous findings by confirming that genetic burden predisposes to early age of diagnosis in CD, and that this effect is not influenced solely by the presence of the NOD2 locus as those with no NOD2 variants experienced a more robust effect of the GRS. Interestingly, the effect was also stronger among non-smokers, suggesting a possible complementary effect between genetics and environment on disease onset. There is limited data on genetic determinants of age of onset in UC; our findings suggest that genetic burden of UC-risk loci may not predispose to early onset in UC.
Our study has several implications. To our knowledge, ours is the first large study using the expanded Immunochip risk loci as well as the genetic risk score approach to examining genotype-phenotype relationships in CD and UC. With recent strategies for the management of CD suggesting an early, aggressive approach with “top-down” therapy to prevent progressive bowel damage, there is increasing interest in prediction tools that may identify patients at high risk for adverse disease outcomes. Indeed, genotype promises to be a stable and readily available predictive tool. However, the low degree of variance explained by the burden of IBD genetic risk-alleles on age of diagnosis and ileal location of CD suggests that known IBD risk-alleles may be only weak determinants of phenotype of disease. Furthermore, the association between genetic burden and complicated disease, need for bowel surgery, immunomodulator or anti-TNF biologic therapies was influenced by the predisposition towards ileal involvement, and were no longer significant on adjusting for disease location. As ileal disease is a clinical parameter that can be readily ascertained, there appears to be limited added utility of genotyping for known risk loci with intent of predicting disease course. Future studies examining potential genetic predictors of adverse disease outcomes should consider expanding analyses to additional loci not yet implicated as being IBD-risk alleles.
The dichotomy in effect between CD and UC is interesting. Despite the vast majority of the IBD risk loci being shared between the two diseases, known genetic risk alleles explain a smaller fraction of the disease variance in UC than CD7, 20, 21. Furthermore, the influence of genetics on disease phenotype in UC has yielded less consistent and weaker associations, similar to our analysis9. The susceptibility to environmental influences are also different between the two diseases, suggesting that despite involvement of common pathways, CD and UC remain at least two distinct entities and require separate approaches to prediction of risk and modification of the environmental influences. Finally, epigenetic alterations have been described in IBD, with enrichment of methylation sites near genes involved in the Th17 pathway22. While one small study did not identify an association between cross-sectional disease activity or treatment and epigenetic alterations, larger prospective studies of the implications of epigenetic changes in CD and UC may shed further light on determinants of disease phenotypes and clinical course22, 23.
There are a few limitations of our study. First, it was conducted at a single tertiary academic center and is thus likely skewed towards a cohort of patients with more severe disease as evidenced by a high frequency of use of immunomodulator or biologic therapy. Nevertheless, the lack of strong genetic predictors of disease phenotype even in a cohort enriched for adverse outcomes suggests that the contribution of the IBD-risk alleles to influencing disease behavior is weak.
In conclusion, we demonstrate an association between increasing genetic burden and early age of onset in CD but not UC. A higher complement of disease risk loci also predisposes towards ileal involvement in CD independent of the NOD2 locus, but the variance explained by genetics on disease phenotype is weak. Further studies should expand variants examined to outside the IBD-risk alleles as well as examine the influence of the dynamic external and internal environment (microbiome) and epigenetic changes on disease phenotype and behavior. This requires continued international collaborations and accrual of large cohorts of patients with genetic and detailed and longitudinal phenotypic data in order to develop accurate and clinically useful prediction tools.
What is current knowledge?
A total of 163 genetic polymorphisms have been described in association with Crohn’s disease (CD)or ulcerative colitis (UC)
There is substantial heterogeneity in the natural history of CD and CD.
Genetics may influence disease phenotype across both diseases
What is new here?
Increasing genetic burden conferred by CD-specific risk loci is associated with earlier age of diagnosis, and ileal localization in CD. Association with disease complications is likely mediated through this tendency for ileal disease.
Genetic burden does not influence disease extent or severity in UC. However a higher genetic burden may be seen in individuals with a family history of IBD in a first degree relative.
Acknowledgements
We acknowledge the valuable contributions of the participants and the research staff of our registry. The work was presented as an oral presentation at Digestive Disease Week 2013, Orlando, FL, USA.
Source of funding: This work is supported by the National Institutes of Health (NIH) (P30 DK043351) to the Center for Study of Inflammatory Bowel Diseases. Ananthakrishnan is supported in part by a grant from the National Institutes of Health (K23 DK097142). Xavier is supported by grants U01 DK062432 & R01 DK064869 from the NIH.
Footnotes
Financial Conflicts of Interest: None
REFERENCES
- 1.Loftus EV., Jr. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–17. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 2.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, Kaplan GG. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54. doi: 10.1053/j.gastro.2011.10.001. e42; quiz e30. [DOI] [PubMed] [Google Scholar]
- 3.Van Assche G, Lewis JD, Lichtenstein GR, Loftus EV, Ouyang Q, Panes J, Siegel CA, Sandborn WJ, Travis SP, Colombel JF. The London position statement of the World Congress of Gastroenterology on Biological Therapy for IBD with the European Crohn's and Colitis Organisation: safety. Am J Gastroenterol. 106:1594–602. doi: 10.1038/ajg.2011.211. quiz 1593, 1603. [DOI] [PubMed] [Google Scholar]
- 4.D'Haens G, Baert F, van Assche G, Caenepeel P, Vergauwe P, Tuynman H, De Vos M, van Deventer S, Stitt L, Donner A, Vermeire S, Van de Mierop FJ, Coche JC, van der Woude J, Ochsenkuhn T, van Bodegraven AA, Van Hootegem PP, Lambrecht GL, Mana F, Rutgeerts P, Feagan BG, Hommes D. Early combined immunosuppression or conventional management in patients with newly diagnosed Crohn's disease: an open randomised trial. Lancet. 2008;371:660–7. doi: 10.1016/S0140-6736(08)60304-9. [DOI] [PubMed] [Google Scholar]
- 5.Colombel JF, Sandborn WJ, Reinisch W, Mantzaris GJ, Kornbluth A, Rachmilewitz D, Lichtiger S, D'Haens G, Diamond RH, Broussard DL, Tang KL, van der Woude CJ, Rutgeerts P. Infliximab, azathioprine, or combination therapy for Crohn's disease. N Engl J Med. 2010;362:1383–95. doi: 10.1056/NEJMoa0904492. [DOI] [PubMed] [Google Scholar]
- 6.Afif W, Loftus EV., Jr. Safety profile of IBD therapeutics: infectious risks. Gastroenterol Clin North Am. 2009;38:691–709. doi: 10.1016/j.gtc.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern D, Hun KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amineinejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Cohain A, Cichon S, D'Amato M, De Jong DJ, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Gieger C, Karlsen J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro MD, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor K, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zhang B, Zhang CK, Zhao H, Consortium TIIG, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly M, Franke A, Parkes M, Vermeire S, Barret JC, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012 doi: 10.1038/nature11582. S. Z. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung C, Colombel JF, Lemann M, Beaugerie L, Allez M, Cosnes J, Vernier-Massouille G, Gornet JM, Gendre JP, Cezard JP, Ruemmele FM, Turck D, Merlin F, Zouali H, Libersa C, Dieude P, Soufir N, Thomas G, Hugot JP. Genotype/phenotype analyses for 53 Crohn's disease associated genetic polymorphisms. PLoS One. 2012;7:e52223. doi: 10.1371/journal.pone.0052223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ananthakrishnan AN, Xavier RJ. How Does Genotype Influence Disease Phenotype in Inflammatory Bowel Disease? Inflamm Bowel Dis. 2013 doi: 10.1097/MIB.0b013e318281f5c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chibnik LB, Keenan BT, Cui J, Liao KP, Costenbader KH, Plenge RM, Karlson EW. Genetic risk score predicting risk of rheumatoid arthritis phenotypes and age of symptom onset. PLoS One. 2011;6:e24380. doi: 10.1371/journal.pone.0024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurreeman F, Liao K, Chibnik L, Hickey B, Stahl E, Gainer V, Li G, Bry L, Mahan S, Ardlie K, Thomson B, Szolovits P, Churchill S, Murphy SN, Cai T, Raychaudhuri S, Kohane I, Karlson E, Plenge RM. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet. 88:57–69. doi: 10.1016/j.ajhg.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adler J, Rangwalla SC, Dwamena BA, Higgins PD. The prognostic power of the NOD2 genotype for complicated Crohn's disease: a meta-analysis. Am J Gastroenterol. 2011;106:699–712. doi: 10.1038/ajg.2011.19. [DOI] [PubMed] [Google Scholar]
- 15.Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, Fennell T, Kirby A, Latiano A, Goyette P, Green T, Halfvarson J, Haritunians T, Korn JM, Kuruvilla F, Lagace C, Neale B, Lo KS, Schumm P, Torkvist L, Dubinsky MC, Brant SR, Silverberg MS, Duerr RH, Altshuler D, Gabriel S, Lettre G, Franke A, D'Amato M, McGovern DP, Cho JH, Rioux JD, Xavier RJ, Daly MJ. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–73. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weersma RK, Stokkers PC, van Bodegraven AA, van Hogezand RA, Verspaget HW, de Jong DJ, van der Woude CJ, Oldenburg B, Linskens RK, Festen EA, van der Steege G, Hommes DW, Crusius JB, Wijmenga C, Nolte IM, Dijkstra G. Molecular prediction of disease risk and severity in a large Dutch Crohn's disease cohort. Gut. 2009;58:388–95. doi: 10.1136/gut.2007.144865. [DOI] [PubMed] [Google Scholar]
- 17.Abreu MT, Taylor KD, Lin YC, Hang T, Gaiennie J, Landers CJ, Vasiliauskas EA, Kam LY, Rojany M, Papadakis KA, Rotter JI, Targan SR, Yang H. Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn's disease. Gastroenterology. 2002;123:679–88. doi: 10.1053/gast.2002.35393. [DOI] [PubMed] [Google Scholar]
- 18.Ahmad T, Armuzzi A, Bunce M, Mulcahy-Hawes K, Marshall SE, Orchard TR, Crawshaw J, Large O, de Silva A, Cook JT, Barnardo M, Cullen S, Welsh KI, Jewell DP. The molecular classification of the clinical manifestations of Crohn's disease. Gastroenterology. 2002;122:854–66. doi: 10.1053/gast.2002.32413. [DOI] [PubMed] [Google Scholar]
- 19.Annese V, Lombardi G, Perri F, D'Inca R, Ardizzone S, Riegler G, Giaccari S, Vecchi M, Castiglione F, Gionchetti P, Cocchiara E, Vigneri S, Latiano A, Palmieri O, Andriulli A. Variants of CARD15 are associated with an aggressive clinical course of Crohn's disease--an IG-IBD study. Am J Gastroenterol. 2005;100:84–92. doi: 10.1111/j.1572-0241.2005.40705.x. [DOI] [PubMed] [Google Scholar]
- 20.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–78. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nimmo ER, Prendergast JG, Aldhous MC, Kennedy NA, Henderson P, Drummond HE, Ramsahoye BH, Wilson DC, Semple CA, Satsangi J. Genome-wide methylation profiling in Crohn's disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm Bowel Dis. 2012;18:889–99. doi: 10.1002/ibd.21912. [DOI] [PubMed] [Google Scholar]
- 23.Kellermayer R. Epigenetics and the developmental origins of inflammatory bowel diseases. Can J Gastroenterol. 2012;26:909–15. doi: 10.1155/2012/526408. [DOI] [PMC free article] [PubMed] [Google Scholar]