Abstract
The binding of peptides to classical major histocompatibility complex (MHC) class I proteins is the single most selective step in antigen presentation. However, the peptide-binding specificity of cattle MHC (bovine leucocyte antigen, BoLA) class I (BoLA-I) molecules remains poorly characterized. Here, we demonstrate how a combination of high-throughput assays using positional scanning combinatorial peptide libraries, peptide dissociation, and peptide-binding affinity binding measurements can be combined with bioinformatics to effectively characterize the functionality of BoLA-I molecules. Using this strategy, we characterized eight BoLA-I molecules, and found the peptide specificity to resemble that of human MHC-I molecules with primary anchors most often at P2 and P9, and occasional auxiliary P1/P3/P5/P6 anchors. We analyzed nine reported CTL epitopes from Theileria parva, and in eight cases, stable and high affinity binding was confirmed. A set of peptides were tested for binding affinity to the eight BoLA proteins and used to refine the predictors of peptide–MHC binding NetMHC and NetMHCpan. The inclusion of BoLA-specific peptide-binding data led to a significant improvement in prediction accuracy for reported T. parva CTL epitopes. For reported CTL epitopes with weak or no predicted binding, these refined prediction methods suggested presence of nested minimal epitopes with high-predicted binding affinity. The enhanced affinity of the alternative peptides was in all cases confirmed experimentally. This study demonstrates how biochemical high-throughput assays combined with immunoinformatics can be used to characterize the peptide-binding motifs of BoLA-I molecules, boosting performance of MHC peptide-binding prediction methods, and empowering rational epitope discovery in cattle.
Keywords: Bovine leucocyte antigen, BoLA, Rational epitope discovery, CTL epitopes, Immunoinformatics
Introduction
Intracellular proteins are displayed for CD8+ T cells to survey as peptide fragments on the surface of all nucleated cells in the context of MHC-I molecules. The binding of peptides to MHC-I proteins is the most selective event in antigen presentation to CD8 T cells and stable interaction of the peptide– MHC complex is a prerequisite for effective surface presentation (Yewdell and Bennink 1999). This process has evolved to allow T cell surveillance of the intracellular environment of cells resulting in a critical aspect of the immune response to infection and oncogenesis.
The tick-borne parasite Theileria parva causes the deadly cattle disease East Coast fever (ECF) in cattle, which is of significant economic importance and a great threat to smallholder farmers in sub-Saharan Africa. Annually, about 1 million cattle die from ECF. Recently, it has been demonstrated that immunity against T. parva is dominated by a strong CD8+ T cell response that kills parasite infected lymphocytes (McKeever et al. 1994). We have previously proposed a reverse immunology strategy for effective and rational epitope discovery based on in silico prediction tools combined with experimental peptide-binding data from recombinant bovine MHCs (Nene et al. 2012). Our aim here is to extend this approach and improve the performance of the MHC peptide-binding prediction methods NetMHC (Lundegaard et al. 2008; Nielsen et al. 2003) and NetMHCpan (Hoof et al. 2009; Nielsen et al. 2007) by integrating peptide-binding affinity data for a limited set of BoLA-I (bovine leucocyte antigen) MHC class I molecules known to restrict T. parva CTL epitopes (Graham et al. 2006) and/or prevalent in breeds available to us (Codner et al. 2012). This will demonstrate how such an approach in a highly cost-effective manner can be used to guide the search for CTL epitopes in cattle.
Cattle have at least six MHC class I genes (Holmes et al. 2003). In each individual, only a subset of one to three loci are transcribed leading to a large set of variable haplotype combinations (Birch et al. 2006). Previous studies on peptide binding to cattle MHCs have mostly been small-scale using alanine scans and/or X-ray crystallography of one or two individual peptide sequences (De Groot et al. 2003; Hegde et al. 1999; Li et al. 2011; Macdonald et al. 2010; Sinnathamby et al. 2004), or pool sequencing of peptides eluted from bovine cells (Gaddum et al. 1996; Hegde et al. 1995) to characterize BoLA-I binding motifs.
Given this limited set of data, the performance of methods for predicting peptide binding to BoLA class I molecules remains suboptimal. In order to improve such methods and generate experimental peptide-binding data, we expressed recombinant bovine MHC class I proteins (BoLA-I) and determined their peptide-binding specificity. Our strategy was to use a nonameric Positional Scanning Combinatorial Peptide Library (PSCPL) in combination with a high-throughput peptide–MHC-I dissociation assay (Harndahl et al. 2011), and feed this data into peptide-binding prediction methods. We have earlier described how this approach can be used to characterize binding specificities of human MHC class I molecules (paper submitted for publication). We used the resulting data to identify discrete peptides from our peptide repository, which contains more than 9,000 different nonameric synthetic peptides. In parallel, we used the NetMHCpan method, prior to inclusion of BoLA binding data, to identify peptides from the same peptide repository that matches the predicted binding motif but did not match the PSCPL data. These individual experimental bovine peptide-binding data allowed us to construct refined versions of NetMHC and NetMHCpan. These methods demonstrated a significantly improved performance for prediction of known bovine CD8 T cell epitopes, leading to more accurate predictions of possible T cell epitopes from genomic data.
The work presented here is, to the best of our knowledge, the first study that provides detailed peptide-binding motifs and quantitative binding data for bovine MHC class I proteins. The resulting updated versions of NetMHCpan (version 2.8) and NetMHC (version 3.4) enable more reliable prediction of peptide binding to BoLA-I molecule and are publicly available via the CBS servers (www.cbs.dtu.dk/services).
Materials and methods
Peptides and peptide libraries
All peptides, except the known T. parva epitopes described below, were purchased from Schafer-N Copenhagen, Denmark. Briefly, peptides were synthesized using standard 9-fluorenylmethyloxycarbonyl (Fmoc) chemistry. The synthesized peptides were purified by reversed-phase high performance liquid chromatography (to at least >80 % purity frequently 95–99 %), validated by mass spectrometry and quantitated by weight. Peptides for the known T. parva CD8 T cell epitopes were purchased from Mimotopes (Victoria, Australia) purity=95 %. PSCPL peptides were synthesized using standard solid-phase Fmoc chemistry on 2-chlorotrityl chloride resins. Briefly, an equimolar mixture of 19 of the common Fmoc amino acids (excluding cysteine) was prepared for each synthesis and used for coupling in 8 positions, whereas a single type of Fmoc amino acid (including cysteine) was used in one position. This position was changed in each synthesis starting with the N-terminus and ending with the C terminus. In one synthesis, the amino acid pool was used in all nine positions. The library therefore consisted of 20×9+1 = 181 individual peptide libraries.
Twenty PSCPL sub-libraries describing position 1: AX8, CX8, DX8, …. YX8
Twenty PSCPL sub-libraries describing position 2: XAX7, XCX7, XDX7, …. XYX7
Etc.
Twenty PSCPL sub-libraries describing position 9: X8A, X8C, X8D, …. X8Y
A completely random peptide library: X9
X denotes the random incorporation of the amino acids from the mixture, whereas the single letter amino acid abbreviation is used to denote identity of the fixed amino acid.
The peptides in each synthesis were cleaved from the resin in trifluoroacetic acid–1,2-ethanedithiol–triisopropylsilane– water 95:2:1:3v/v/v/v, precipitated in cold diethyl ether, and extracted with water before desalting on C18-columns freeze drying and weighing.
Cloning, expression, and purification of biotinylated bovine MHC class I proteins
The recombinant biotinylated bovine MHC proteins were expressed the same way as recombinant human MHCs as previously described (Leisner et al. 2008; Ostergaard Pedersen et al. 2001). In short, the desired MHC sequence (residues 1–275) was back-translated and cloned into the IPTG inducible pET28a expression vector containing a biotin signal peptide (BSP) and a histidine affinity tag (HAT). The recombinant constructs were validated by DNA sequencing and transformed into the Escherichia coli strain BL21(DE3) previously transformed with the pACYC (Avidity, Denver) vector containing the BirA gene encoding biotin ligase. The transformed cells were grown in a 2-L fermentor (LabFors®), in media supplemented with kanamycin (50 µg/ml) and chloramphenicol (20 µg/ml) to maintain the pET28a derived expression plasmid and the BirA containing pACYC184 plasmid, respectively. When the culture reached an optical density of OD600=25 the media was supplemented with biotin (sigma #B4501, final concentration 125 µg/ml), and IPTG (1 mM) was added to induce expression of the MHC-I heavy chain gene and of the BirA gene; the induced culture was continued for 3 h. At the end of induction, protease inhibitor (PMSF 80 µg/L) was added and the cells were lysed in a cell-disrupter (basic Z, constant systems Ltd Daventry, UK) set at 2,300 bar. The released inclusion bodies containing biotinylated MHC-I heavy chain proteins were harvested by centrifugation (Sorwall RC6, 20 min, 17,000g).
The inclusion bodies were washed in PBS, 0.5 % NP-40 (Sigma), 0.1% deoxycholic acid (DOC, Sigma) and dissolved at 4 °C overnight in 8 M urea-Tris buffer (8 M urea, 25 mM Tris pH 8) using 200 ml per 100 g wet cell paste, and cleared by centrifugation. The dissolved MHC-I proteins were then purified using Ni2+/IDA metal chelating affinity column chromatography, Hydrophobic ionic chromatography (phenyl sepharose high performance) and eventually by Superdex200 gel filtration column chromatography, driven by a Fast Protein Liquid Chromatography machine (P920 Amersham Biotech industries). Peak fractions were identified by A280 absorbance, SDS-PAGE analysis and pooled. Throughout purification the MHC proteins were dissolved in 8 M Urea to maintain denaturing conditions. At no time the MHCs were exposed to reducing conditions, which ensures purification of highly active pre-oxidized moieties as previously described (Ostergaard Pedersen et al. 2001). Protein concentrations were determined by BCA assay (Thermo Scientific Pierce®). The degree of biotinylation (usually >95 %) were determined by a gel-shift assay (Leisner et al. 2008) and the pre-oxidized, denatured proteins were stored at −24 °C until use.
Production of recombinant human β2m
Recombinant β2m was produced by overexpression in E. coli, refolded and purified as previously described (Ferre et al. 2005).
Production of recombinant bovine β2m
Recombinant bovine β2m was expressed in E. coli as described for the bovine MHC-I heavy chain. Inclusion bodies containing recombinant bovine β2m were dissolved in 8 M urea and purified by anion exchange chromatography (Q Sepharose Fast Flow, QSFF, Ge Healthcare Life Sciences 17-0510-01) under denaturing conditions. The protein was refolded overnight by drop-wise dilution into 25 mM Tris, 300 mM urea, pH 8, and concentrated by tangential flow ultrafiltration (Vivaflow). The concentrated product was buffer-exchanged into PBS by gel filtration (Superdex 200, Amersham Biosciences) and concentrated. As a result of E. coli expression, the recombinant bovine β2m protein includes an N-terminal methionine, and is hereafter named bovine met-β2m.
Radiolabeling of human and bovine met-β2m
Recombinant human β2m and bovine met-β2m were radiolabeled with iodine (125I) using the chloramine-T procedure (Hunter and Greenwood 1962). Twenty micrograms β2m was mixed with 1 mCi 125I (Perkin-Elmer, NEZ033A005MC) and 5 µl (1 mg/ml) chloramine-T (Sigma C9887) in a total volume of 65 µl. The reaction was stopped after 1 min by the addition of 5 µl metabisulfite (1 mg/ml). Free iodine was removed by size exclusion chromatography using a 1 ml Sephadex G10 column equilibrated in PBS/2% ethanol/0.1 % azide. The column was eluted with PBS/2 % ethanol/0.1 % azide and 200 µl fractions were collected. Column fractions were tested for gamma emission and labeled fractions were pooled. Radiolabeled β2m was stored at 4 °C for no more than 4 weeks.
Bovine peptide-binding motifs by scintillation proximity assay
A nonameric peptide-binding motif was determined for 8 recombinant bovine MHC proteins: BoLA-6*01301 (HD6), −2*01201 (T2A), −6*04101 (T2B), -T2C, −3*00101 (AW10), −3*00201 (JSP.1), −1*02301 (D18.4) and -T7. Here, the names in parenthesis (i.e., HD6) are the “old” allele names often used in the literature, and the BoLA names are from the IPD (Robinson et al. 2013). Three sequences are not available in the IPD. These are available from Genbank with the following GI’s; T2C −158516741, T5 −158516743, and T7 −158516745. The different MHC proteins are found in the following breed of cattle (data obtained from the IPD and Genbank): Holstein (BoLA-6*01301 (HD6), BoLA-2*01201 (T2A), BoLA-3*00201 (JSP.1), and BoLA-1*02301 (D18.4)), Boran (BoLA-3*00101 (AW10)), Jersey (BoLA-6*04101 (T2B)), Bos taurus×Bos indicus (BoLA-T2C and BoLA-T7), and Bos indicus (BoLA-T5). The nonameric PSCPL library (Stryhn et al. 1996) was evaluated in a recently developed MHC Class I peptide dissociation assay based on a scintillation proximity assay principle (SPA) (Harndahl et al. 2011). The recombinant bovine MHC-I proteins stored in 8 M urea was diluted at least 100 fold in a PBS/0.1 % Lutrol F68 buffer containing peptide and 125I radiolabeled recombinant bovine met-β2m to initiate pMHC complex formation. The reactions were carried out in streptavidin-coated scintillation 384-well HTS Flashplate® Plus microplates (Perkin-Elmer SMP410A001PK). The final concentration of the MHC-I heavy chain was between 12.5–100 nM depending on the specific activity of each protein. The mixture containing the recombinant bovine heavy chain, radiolabeled bovine met-β2m and the peptide mix with a specific amino acid in a given position was incubated overnight at 18 °C. After incubation, an excess amount (200 nM) of unlabeled bovine met-β2m was added to prevent re-association of dissociated complexes and the dissociation was initiated by transferring the plate to 37 °C. The plates were read continuously for 24 h at 37 °C in a Perkin-Elmer Topcount NXT Liquid Scintillation Counter.
Analysis of PSCPL data
The area under the dissociation curve (AUC) was calculated after background subtraction by summarizing the CPM from 0–24 h. The relative binding (RB), i.e., the relative contribution of each residue in the sub-library to peptide binding was calculated according to the equation:
AUC is the area under the dissociation curve from 0 to 24 h. The RB values of each amino acid in a given position were summarized and normalized so the sum equals 20. The anchor position (AP) values for a given position, defining the relative contribution of this position to peptide binding, were calculated as:
Generation of sequence logos
Sequence logos describing the amino acid binding specificity captured by the PSCPL were calculated using the Seq2Logo sequence logo generator (Thomsen and Nielsen 2012). In short, the RB values at each peptide position were normalized by the sum of the RB values at the given position. This normalized PSCPL matrix was submitted to the Seq2Logo server, and binding motif visualized as a p-weighted Kullback-Leibler logo generated using a flat amino acid background distribution (i.e. all amino acids are assumed to be found with an equal frequency). In this type of logo, a tall stack of letters characterizes anchor positions and amino acids presented on the positive y-axis are enriched in peptide binders and amino acids on the negative y-axis are depleted from peptide binders.
Stability of recombinant bovine pMHC complexes
The stability of the recombinant pBoLA-I complexes was determined using the aforementioned SPA-based peptide–MHC Class I dissociation assay (Harndahl et al. 2011).
Bovine class I peptide-binding assay
Peptide affinity to bovine MHC-I molecules was determined using a peptide–MHC-I binding assay where pMHC-I complexes are detected via the luminescent oxygen channeling immunoassay (LOCI) principle (Harndahl et al. 2009). Briefly, lyophilized peptides were dissolved in PBS/0.1 % Lutrol F68 by sonication for 10 min. Five-fold dilution series of peptides were done in 384 well microplates using a Microlab® STAR liquid handling robot (Hamilton Robotics). Recombinant, denatured bovine heavy chains were diluted into PBS/0.1 % Lutrol® F 68/100 mM Tris/maleate (pH 6.6) containing pre-folded recombinant human β2m on ice. The HC/β2m mixture was added 1:1 to the peptide titrations and incubated for 48 h at 18 °C to ensure proper MHC-I complex folding. The final concentration of bovine heavy chain ranged from 0.5 to 10 nM and β2m concentrations ranged from 5 to 25 nM based on the optimal signal to background ratio for the specific protein obtained from dose-response experiments. Following complex folding, samples were transferred to 384 well Optiplates™ and mAb W6/32 conjugated acceptor beads and streptavidin coated donor beads (Perkin Elmer, 6762001 conjugated in-house with mAb W6/32 and 6760002) were added, giving a final bead concentration of 15 µg/ml of each. Plates were incubated overnight at 18 °C to obtain equilibrium and subsequently read in an Envision® multi label reader (Perkin Elmer) after equilibration to room temperature. All handling of LOCI reagents was done in dark or in green light. All data handling and non-linear regression was done using Microsoft® Office Excel® and GraphPad® Prism.
Binding affinity of recombinant human β2m and bovine met-β2m to BoLA-6*01301 (HD6)
The binding affinity of human β2m and bovine met-β2m to BoLA-6*01301 was measured in a competitive binding assay. In a 96-well HTS Flashplate® Plus microplate, a fixed concentration of 125I labeled bovine met-β2m, biotinylated BoLA 6*01301 (50 nM) and a binding peptide was incubated in the presence of increasing concentrations of unlabeled human β2m or bovine met-β2m (0.45 nM to 1 µM) for 24 h at 18 °C to allow pMHC-I complex folding. A parallel experiment was done using 125I labeled bovine. Plates were read in Perkin-Elmer Topcount NXT Liquid Scintillation Counter. Data was analyzed using Graphpad® Prism and IC50 values calculated using “Log agonist versus Inhibitor” model.
Peptide-binding specificity of BoLA 6*01301 (HD6) using recombinant bovine or human β2m
The peptide-binding specificity of BoLA-6*01301 was assessed using SPA driven PSCPL analysis (position 2 and 9 only) as described above using either 125I radiolabeled human β2m or bovine met-β2m. Dissociation was initiated by addition of excess of unlabeled β2m of the relevant species.
Results
Generation of BoLA-I molecules
We have previously generated recombinant MHC proteins from different species such as human, swine and others (Ferre et al. 2003; Ostergaard Pedersen et al. 2001; Pedersen et al. 2011). Here, we transfer the underlying approaches to the BoLA system. The focus of this study has been on the bovine alleles BoLA-6*01301 (HD6), BoLA-2*01201 (T2A), BoLA-6*04101 (T2B), BoLA-T2C, BoLA-3*00101 (AW10), BoLA-3*00201 (JSP.1), BoLA-1*02301 (D18.4), and BoLA-T7.
The recombinant biotinylated bovine MHC proteins were expressed and purified as described in “Materials and methods”. As seen by non-reducing SDS-PAGE analysis (Fig. 1a), we were able to isolate denatured, recombinant bovine MHC-I molecules of high purity. The degree of biotinylation was assessed in a biotin-avidin gel-shift assay and ranged between 90 and 100 % (Fig. 1a), thus providing fully biotinylated, recombinant BoLA-I molecules for biochemical assays.
Fig. 1.
Purification and in vivo biotinylation of recombinant bovine MHC molecules. Recombinant bovine MHC-I alpha chains were expressed in E. coli BL21(DE3) and inclusion bodies were isolated, dissolved and purified under denaturing conditions. In vivo biotinylation, via co-expression of BirA, was assessed in the purified protein product, shown here for BoLA-T2c (a), by a SDS-PAGE gel-shift assay where the biotinylated protein is incubated with increasing concentrations of avidin prior to SDS-PAGE analysis. Bovine met-β2m was expressed in E. coli BL21(DE3) and resulting inclusion bodies were isolated, dissolved in denaturing buffer, purified by ion-exchange chromatography and the protein refolded. After refolding the protein was purified size exclusion chromatography (b) resulting in a highly pure protein product, here visualized by non-reducing SDS-PAGE analysis of SEC peak fractions
Generation of BoLA met-β2m
Bovine met-β2m was likewise produced as described in the “Materials and methods” section. Following refolding, the protein was purified by size exclusion chromatography, resulting in a highly pure protein preparation (Fig. 1b).
Generation of peptide–BoLA-I binding and stability assays
We have reported two assays that enable high-throughput measurement of the stability of the pMHC-I complex (half-life, t1/2) and the peptide–MHC-I (pMHC-I) binding affinity, respectively (Harndahl et al. 2009, 2011). The former is based on the dissociation of radiolabeled β2m from pMHC-I complexes as an indirect measure of peptide dissociation, first shown by Parker et al. (1992), here modified to allow detection in a high-throughput setting via the SPA principle. The latter is based on detection of pMHC-I complexes from ligand saturation experiments via the luminescent oxygen channeling immunoassay principle (LOCI). Our aim was to use these two assays to measure the affinity and stability of pMHC complexes from bovine species. The LOCI-based assay depends on the mAb W6/32 epitope, that only recognizes folded pMHC-I complexes, while the dissociation assay is dependent on successful radiolabeling of β2m. As proof of concept, the immunodominant epitope VGYPKVKEEML restricted by the BoLA-6*01301 (HD6) allotype (Macdonald et al. 2010) was tested in the two assays. The VGYPKVKEEML-BoLA-6*01301 (HD6) complex dissociated at a rate that resembled rates observed for human epitopes (Harndahl et al. 2012) (t1/2 = 10-2 h, Fig. 2a). This shows that both the recombinant BoLA-I heavy chain and bovine met-β2m are functional and capable of forming stable pMHC-I complexes. Next, we tested whether the pBoLA-I complexes could be detected by the W6/32 monoclonal antibody in the LOCI assay. Dose-response titrations of recombinant BoLA-6*01301 (HD6) and bovine met-β2m in the presence and absence of the VGYPKVKEEML peptide yielded no peptide specific signal (data not shown). It has been reported that N-terminus of β2m is involved in the formation of the W6/32 epitope (Shields and Ribaudo 1998) and in the case presented here, the N-terminal methionine of the recombinant bovine met-β2m likely interferes with W6/32 detection. Alternatively, the failed W6/32 recognition may be explained by the lack of pan-specificity of the antibody to BoLA-I when associated with bovine met-β2m (Joosten et al. 1992). Substituting bovine met-β2m with human β2m, lacking the N-terminal methionine, however, proved highly suitable for the W6/32 based LOCI detection with BoLA-6*01301. As shown in Fig. 2b, human β2m support BoLA-I folding and W6/32 epitope formation and the binding affinity of the VGYPKVKEEML epitope to BoLA-6*01301 (HD6) was determined to 2.7 nM.
Fig. 2.
Development of BoLA-I ligand binding assays. Left panel Dissociation kinetics of the VGYPKVKEEML-BoLA-6*01301 (HD6) complex. Radiolabeled pBoLA-I complexes were formed by overnight incubation of recombinant, denatured, and biotinylated BoLA-6*01301 (HD6) with 125I-radiolabeled bovine met-β2m and excess of peptide. After addition of excess of unlabeled bovine met-β2m, the pBoLA-I complex decay was monitored continuously for 24 h. Right panel Binding of the immune dominant epitope VGYPKVKEEML to the restricting BoLA-6*01301(HD6) allotype. Peptide-BoLA-I complexes were generated by incubation of recombinant, denatured, and biotinylated BoLA-6*01301 (HD6) with excess of human β2m and increasing concentrations of peptide. pBoLA-I complexes were detected by luminescent oxygen channeling immunoassay (LOCI) via streptavidin conjugated donor beads and W6/32 conjugated acceptor beads, ensuring detection of correctly folded pBoLA-I complex only
We next assessed whether the use of human β2m in BoLA-I binding experiments affected the peptide-binding specificity of BoLA-I allotypes. To investigate this, we measured the binding affinity of human β2m versus bovine met-β2m to bovine MHC-I in a competitive binding assay. BoLA-6*01301 (HD6) and a binding peptide (VGYPKVKEEML) were incubated with either radiolabeled human β2m or bovine met-β2m and unlabeled human β2m or bovine met-β2m. The binding affinity of the bovine met-β2m to the recombinant bovine MHC-I heavy chain BoLA-6*01301 (HD6) was determined to be 6.7 nM while the affinity of the recombinant human β2m to BoLA-6*01301 (HD6) was 2.5 nM (data not shown). This suggests a very limited effect on the binding affinity of replacing bovine met-β2m with human β2m.
We next investigated if the use of human β2m altered the specificity of the bovine class I peptide-binding groove. Using a nonameric positional scanning combinatorial peptide library (PSCPL) in combination with a scintillation proximity assay (SPA)-based pMHC-I dissociation assay, we analyzed the specificity of peptide positions 2 and 9 in BoLA-6*01301 (HD6) with both radiolabeled human β2m and bovine met-β2m. We observed very limited change in the specificity of these two key positions (see Supplementary file 1). From these results, we concluded that the use of human β2m is a valid substitution for bovine β2m in pBoLA-I binding assays, and for the remainder of this study used recombinant human β2m for LOCI based peptide-binding experiments for all recombinant bovine heavy chains.
Determining MHC-I peptide-binding motifs by positional scanning combinatorial peptide libraries
We used the approach described earlier (paper submitted for publication), combining the dissociation assay and the Positional Scanning Combinatorial Peptide Library (PSCPL), to characterize the peptide-binding motifs of eight BoLA-I molecules. A prerequisite to a PSCPL analysis is that a given recombinant MHC protein can bind the reference X9 peptide library, and thereby generate a specific signal compared to a negative control without peptide. Figure 3 shows an example of BoLA-6*01301 (HD6) bound to three different sub-libraries; XXXXXXXXX (X9), XXXXXXXXL (X8L) and XXXXXXXXN (X8N). A large increase in binding and stability of the X8L library compared to X9, and a large decrease for the X8N library is seen, showing that leucine (L) is a highly favored and asparagine (N) a highly disfavored amino acid at the ninth position of a nonameric peptide. The ratio between the area under the dissociation curve (AUC) of a sub-library and the AUC of the reference library X9 is the relative binding (RB). RB values below 0.5 are considered as disfavored for binding, values 0.5<RB<2 are accepted and residues with values above 2 are considered favored for binding.
Fig. 3.
Positional Scanning Combinatorial Peptide Library (PSCPL) analysis of the binding specificity of BoLA 6*01301 (HD6) measured by pMHC-I dissociation. The pBoLA-I dissociation curves are shown for two sub-libraries (X8L and X8N) and the reference library (X9). The relative binding (RB) values are calculated for the dissociation data as the ratio of the area under the dissociation curve (AUC) of the sub-library and the AUC for the reference library (X9)
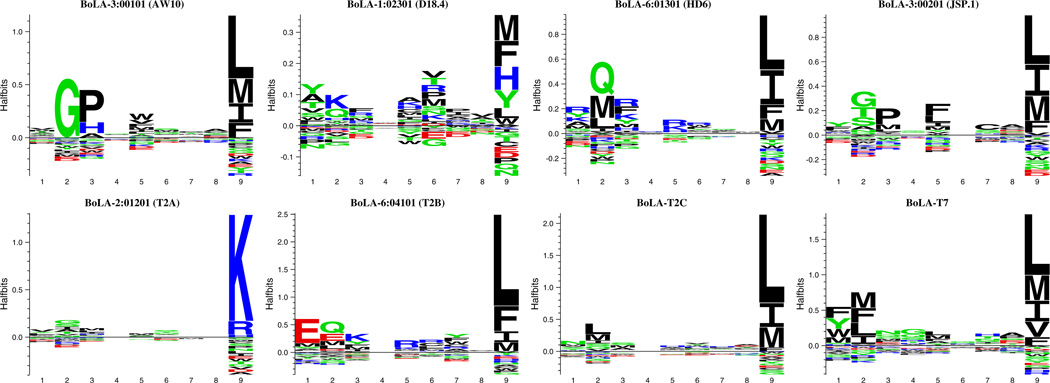
Measurement of the pMHC-I complex stability of all 180 nonameric sub-libraries of the PSCPL can be summarized as a detailed binding matrix, which identifies favored and disfavored amino acid residues for each amino acid position. The PSCPL-derived binding matrices represent BoLA-I binding motifs and can be shown as position specific weight matrices of RB values or visualized as logos using the Seq2Logo web-server (Thomsen and Nielsen 2012). An example of a PSCPL derived peptide-binding motif is shown in Fig. 4 for the molecule BoLA-6*01301 (HD6). This molecule was found to have a well-defined peptide-binding motif characterized by two anchor positions: one anchor at the C-terminal (P9) with preference for hydrophobic amino acids and one anchor at P2 with preference for neutral (Q) and hydrophobic (M, L) amino acids. The C-terminal and P2 anchor positions is in agreement with the motif suggested earlier obtained using alanine scan (Macdonald et al. 2010). Of note is that Macdonald et al. showed that for the given peptide analyzed in their study, P5 had greater involvement than the P2 residue in MHC binding. This observation is not reproduced in our findings.
Fig. 4.
PSCPL derived peptide-binding motif of BoLA-6*01301 (HD6). Left panel Peptide-binding matrix obtained by peptide-MHC-I dissociation assay based PSCPL analysis. Relative binding values >2 are considered favored amino acids, values 0.5–1.5 are considered tolerated and values below 0.5 are considered disfavored. AP values >10 are considered anchor positions. Right panel Sequence logo representation of the binding motif. The sequence logo was generated using the Seq2Logo server (Thomsen and Nielsen 2012) as described in the text. Hydrophobic amino acids F, I, L, and M are favored at the anchor position 9. At the anchor position 2 amino acids Q, M, and L are favored
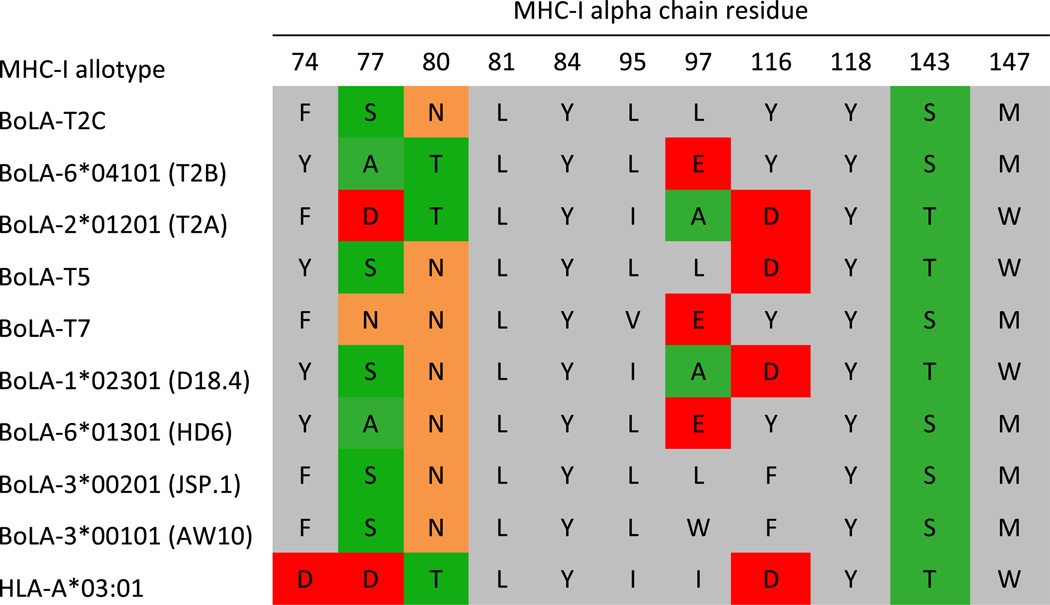
Using this SPA-based PSCPL analysis, we were able to characterize the binding motif of 8 of the 9 recombinant BoLA-I allotypes produced (BoLA-6*01301 (HD6), BoLA-2*01201 (T2A), BoLA-6*04101 (T2B), BoLA-T2C, BoLA-3*00101 (AW10), BoLA-3*00201 (JSP.1), BoLA-l*02301 (D18.4), and BoLA-T7). For the BoLA-T5 molecule, we were unable to obtain a peptide-specific signal in the SPA driven pMHC-I dissociation hindering PSCPL analysis, and this molecule was not analyzed further. A sequence logo representation of the binding motifs is shown in Fig. 5 (the individual weight matrices are shown in Supplementary file 2). All BoLA-I proteins except BoLA-2*01201 (T2A) showed a strong preference for amino acids with hydrophobic properties at the C-terminal position of the peptide; in particular favoring the amino acids F, I, L, and M. The F pocket of the MHC-I proteins is formed by the following polymorphic residues 74, 77, 80, 81, 84, 95, 97, 116, 118, 143, and 147 of the MHC-I alpha chain (Nielsen et al. 2007). Of these, positions 77, 80, 81, and 116 are considered key residues (Sidney et al. 2008). Figure 6 shows the amino acids forming the F pocket of the different MHC proteins. It appears that the majority of the pocket residues belong to functional groups that favor interactions with hydrophobic residues. One outlier in the pocket analysis is BoLA-2*01201 (T2A). This molecule has a strong C-terminal preference for positively charged residues arginine and lysine (see Fig. 5). A similar C-terminal preference is observed for the human HLA-A*03:01 molecule (McMahon et al. 2011). Comparing the residues lining the F pocket of the HLA-A*03:01 molecule with the BoLA molecules offers a potential explanation of the observed binding motifs preferences. In contrast to the other BoLA molecules, both the BoLA-2*01201 (T2A) and the HLA-A*03:01 molecules have a D in positions 77 and 116, resulting in an acidic C-terminal pocket suggesting that these amino acids are critical in determining the F pocket preference for basic amino acids.
Fig. 5.
Sequence logo representation of the binding motif of the 8 BoLA-I molecules. Logos were generated using the Seq2Logo server (Thomsen and Nielsen 2012) as described in the text
Fig. 6.
Amino acids forming the F pocket of the different BoLA-I proteins and the human allomorph HLA-A*03:01. The following color scheme was used: green small non-polar amino acids, gray hydrophobic amino acids, red negative charged amino acids, orange polar. F pocket contact residues were identified as described in (Nielsen et al. 2007). Numbers in the first row correspond to residue positions in the mature MHC protein sequence (according to the IMGT nomenclature)
Biochemical characterization of known BoLA-I restricted epitopes
A small set of known T. parva CD8 restricted epitopes restricted by the BoLA-I molecules included in this study has been reported (Graham et al. 2008). To further validate the functionality of the peptide-binding and stability assays described here, we measured the binding affinity and stability of nine T. parva epitopes with their cognate BoLA-I molecules using the LOCI based binding assay to determine the equilibrium dissociation constant (binding affinity, KD in nM) and the SPA-based dissociation assay to determine the stability of the pMHC complexes (half-life, t1/2 in hours).
The results, shown in Table 1 confirm that the epitope sequences form high affinity, stable complexes with the corresponding BoLA-I restriction element. For example, the BoLA-6*01301 (HD6) restricted 11mer epitope VGYPKVKEEML was found to have an affinity of 2.7 nM and a half-life (t1/2) of 11 h The BoLA-6*04101 (T2B) restricted epitope SHEELKKLGML, however, could not be detected in the stability assay, and the affinity was determined to be 4,558 nM. This strongly suggested that this peptide sequence is not a minimal epitope, and that the epitope is nested within this sequence. This has been recently confirmed (Svitek et al. 2014).
Table 1.
Peptide–BoLA-I binding affinity and complex stability of known T. parva CTL epitopes
| Epitope name | Sequence | MHC-I restriction |
pBoLA-I binding affinity (EC50, nM) |
pBoLA-I complex stability (half-life, h) |
Alternative restriction |
pBoLA-I binding affinity (EC50, nM) |
pBoLA-I complex stability (half-life, h) |
|---|---|---|---|---|---|---|---|
| Tp1214–224 | VGYPKVKEEML | HD6 (6*01301) | 3 | 11 | T2B (6*04101) | 12 | 14 |
| Tp227–37 | SHEELKKLGML | T2B (6*04101) | 4558 | Not detectable | |||
| Tp249–59 | KSSHGMGKVGK | T2A (2*01201) | 364 | 2 | |||
| Tp296–104 | FAQSLVCVL | T2C | 144 | 57 | JSP.1 (3*00201) | 767 | 4 |
| Tp298–106 | QSLVCVLMK | T2A (2*01201) | 18 | 3 | |||
| Tp4328–336 | TGASIQTTL | AW10 (3*00101) | 40 | 23 | JSP.1 (3*00201) | 564 | 2 |
| Tp587–95 | SKADVIAKY | T5 | D18.4 (1*02301) | 15 | 18 | ||
| Tp7 | EFISFPISL | T7 | 252 | 14 | |||
| Tp 8379–387 | CGAELNHFL | AW10 (3*00101) | 30 | 14 | JSP.1 (3*00201) | 431 | 2 |
| Tp9199–228 | AKFPGMKKSK | D18.4 (1*02301) | 100 | 4 |
The peptide-BoLA-I binding affinity (EC50) and complex stability (t1/2) of known T. parva epitopes was measured for published ((Nene et al. 2012)) and proposed alternative restriction elements
Cross reactivity of known BoLA-I restricted epitopes
In order to investigate the cross reactivity between epitopes and MHC molecules, we measured the binding affinity and complex stability of the 9 T. parva epitopes described above and the T5 epitope SKADVIAKY to the eight BoLA-I molecules. Note, the original T5 restriction of SKADVIAKY was not measured, as we were not able to develop functional assays for this molecule. The result of this analysis is shown in Table 1. Only a limited degree of cross binding was observed between the different molecules. One striking example of cross binding is the suggested restriction of VGYPKVKEEML to BoLA-6*04101 (T2B) with an affinity of 12 nM and a half-life time of 12 h. BoLA-6*01301 (HD6) and BoLA-6*04101 (T2B) are very similar in sequence, and only five amino acids differ between the two full protein sequence with four of these forming part of the pseudo sequence lining the MHC binding cleft (Nielsen et al. 2007). Further, the BoLA-T2C restricted epitope FAQSLVCVL and the BoLA-3*00101 (AW10) restricted epitope TGASIQTTL were both found to cross-bind to BoLA-3*00201 (JSP.1) and the BoLA-T5 restricted epitope SKADVIAKY was found to bind with an affinity of 15 nM and a stability of 18 h to BoLA-1*02301 (D18.4). The latter observation confirms what has been reported earlier (MacHugh et al. 2011; Svitek et al. 2014).
Testing discrete peptides
The PSCPL-driven analysis is ideally suited to give a first representation of the peptide-binding motif. However, it is not well suited to capture correlated effects where an amino acid in one position may affect the specificity of another position. The latter is most efficiently captured when investigating the binding of a large number of discrete peptides. Approximately 90 discrete high-scoring peptides for each BoLA-I molecule were selected from our peptide repository containing more than 9,000 distinct nonamer peptides according to each of the PSCPL matrices described above multiplying the relative binding (RB) values for each amino acid in each position. To complement this PSCPL matrix-driven selection, an additional set of 90 discrete peptides was selected for each MHC molecule with high-predicted binding affinity according to the NetMHCpan prediction method but low predicted affinity according to the PSCPL matrix prediction. This second selection complements the PSCPL selection by choosing peptides that do not fit the motif captured by the PSCPL but are predicted to bind by the NetMHCpan method (Hoof et al. 2009; Nielsen et al. 2007) as trained without prior explicit knowledge about the specificity of the given BoLA-I molecule. We have earlier demonstrated that such a complementary selection strategy is a highly cost-effective approach, which allows the identification of individual peptides highly enriched in MHC binders and rich in information critical for the subsequent training of improved algorithms (paper submitted for publication).
Each peptide within these data sets was next assayed for binding affinity to the respective BoLA-I molecules using the LOCI based pMHC-I binding assay. The results of these screenings are summarized in Table 2. Several observations can be made from the data presented in Table 2. First of all, it is clear that the PSCPL matrices in most cases have a very high predictive capacity for identification of binders to the BoLA-I molecules. Only one molecule, BoLA-3*00101 (AW10), had a very low performance with only 9 % of the selected peptides being binders. For the remaining seven molecules on average 55%[26–93 %] of the selected peptides were found to bind the respective molecule (KD<500 nM). A second important observation is that the NetMHCpan selected peptides also to some degree [9–70 %, excluding BoLA-3*00101 (AW10)] bound the respective BoLA-I molecules. This is somewhat surprising since the NetMHCpan peptides were explicitly selected to not match the binding motif described by the PSCPL’s. These observations thus suggest that the PSCPL matrix approach in general only captures a subset of the overall binding specificity of an MHC molecule, and that the characterization of the specificity can be effectively complemented using predictions of the NetMHCpan method. That the two approaches are complementary is further underlined by the fact that only between 20 and 60 % of the identified BoLA binding peptides would have been predicted using the NetMHCpan method trained without any prior knowledge about the specificity of the BoLA molecules (data not shown).
Table 2.
Peptide binding to BoLA-I molecules
| MHC allotype | N | KD <50 nM | 50 nM<KD<500 nM | 500 nM<KD<5000 nM | KD>5000 nM |
|---|---|---|---|---|---|
| PSCPL selected peptides | |||||
| 3*00101 (AW10) | 83 | 1 (1 %) | 7 (8 %) | 18 (22 %) | 57 (69 %) |
| 1*02301 (D18.4) | 83 | 8 (10 %) | 42(51 %) | 12 (14 %) | 21 (25 %) |
| 6*01301 (HD6) | 87 | 50 (57 %) | 27(31 %) | 6 (7 %) | 3 (3 %) |
| 3*00201 (JSP.1) | 84 | 6 (7 %) | 16 (19 %) | 18 (21 %) | 44 (52 %) |
| 2*01201 (T2A) | 80 | 0 (0 %) | 39 (49 %) | 35 (44 %) | 6 (8 %) |
| 6*04101 (T2B) | 84 | 10(12%) | 17 (20 %) | 14 (17 %) | 43 (51 %) |
| T2C | 90 | 65 (72 %) | 19 (21 %) | 4 (4 %) | 2 (2 %) |
| T7 | 83 | 0 (0 %) | 29 (35 %) | 37 (45 %) | 17 (20 %) |
| NetMHCpan selected peptides (not matching PSCPL) | |||||
| 3*00101 (AW10) | 84 | 0 (0 %) | 0 (0 %) | 1 (1 %) | 83 (99 %) |
| 1*02301 (D18.4) | 84 | 5 (6 %) | 54 (64 %) | 15 (18 %) | 10 (12 %) |
| 6*01301 (HD6) | 85 | 12(14%) | 39 (46 %) | 29 (34 %) | 4 (5 %) |
| 3*00201 (JSP.1) | 74 | 1 (1 %) | 9 (12 %) | 22 (30 %) | 42 (57 %) |
| 2*01201 (T2A) | 87 | 0 (0 %) | 8 (9 %) | 33 (38 %) | 46 (53 %) |
| 6*04101 (T2B) | 73 | 0 (0 %) | 11 (15 %) | 22 (30 %) | 40 (55 %) |
| T2C | NA | NA | NA | NA | NA |
| T7 | 66 | 2 (3 %) | 8 (12 %) | 32 (48 %) | 24 (36 %) |
Peptide binding to BoLA-I molecules was measured for two sets of peptides; one consisting of peptides predicted to bind according the PSCPL derived binding motif (upper half) and one set of peptides predicted to bind according to the NetMHCpan prediction method but not by the PSCPL binding motif
In silico validation of known BoLA-I epitopes
The discrete peptide-binding affinity data were next used to retrain the NetMHCpan and NetMHC methods. The updated versions are available at www.cbs.dtu.dk/services/NetMHCpan and www.cbs.dtu.dk/services/NetMHC. Using the updated prediction methods, we next used a set of BoLA-I epitopes earlier described in the literature (MacHugh et al. 2011; Nene et al. 2012) to investigate to what degree a consensus method defined as a simple average of the NetMHCpan and NetMHC prediction values was able to predict the suggested restrictions. The analysis was performed as described previously (Nene et al. 2012). In short, for each epitope, the evaluation was made by predicting the binding affinity for all 8 to 11mer peptides contained within the source protein, and next calculating the fraction of peptides with a predicted binding affinity stronger than the predicted binding affinity of the known CTL epitope (the false-positive prediction rate, FR). In situations where the BoLA-I molecule was not covered by the NetMHC method, only the NetMHCpan predictions were used. The result of this analysis is shown in Table 3.
Table 3.
In silico prediction of known T. parva and T. annulata CTL epitopes
| Pathogen | Epitope | BoLA | Protein | #pep | #FP | FR | Alternative | FR |
|---|---|---|---|---|---|---|---|---|
| T. parva | VGYPKVKEEML | 6*01301 (HD6) | Tp1 | 2138 | 34 | 0.016 | ||
| SHEELKKLGML | 6*04101 (T2B) | Tp2 | 662 | 14 | 0.021 | EELKKLGML | 0.000 | |
| DGFDRDALF | 6*04101 (T2B) | Tp2 | 662 | 143 | 0.216 | GFDRDALF | 0.041 | |
| KSSHGMGKVGK | 2*01201 (T2A) | Tp2 | 662 | 3 | 0.005 | |||
| FAQSLVCVL | T2C | Tp2 | 662 | 15 | 0.023 | |||
| QSLVCVLMK | 2*01201 (T2A) | Tp2 | 662 | 5 | 0.008 | |||
| KTSIPNPCKW | 2*01201 (T2A) | Tp2 | 662 | 91 | 0.137 | KTSIPNPCK | 0.015 | |
| TGASIQTTL | 3*00101 (AW10) | Tp4 | 2282 | 7 | 0.003 | |||
| SKADVIAKY | T5 | Tp5 | 586 | 2 | 0.005 | |||
| EFISFPISL | T7 | Tp7 | 2850 | 184 | 0.065 | FISFPISL | 0.001 | |
| CGAELNHFL | 3*00101 (AW10) | Tp8 | 1726 | 12 | 0.007 | |||
| AKFPGMKKSK | 1*02301 (D18.4) | Tp9 | 1302 | 68 | 0.052 | |||
| Ave | 0.046 | 0.015 | ||||||
| T. annulata | SKADVIAKY | 1:02301 | Ta5 | 586 | 4 | 0.007 | ||
| QRSPMFEGTL | 3:00201 | Ta9 | 1306 | 15 | 0.011 | RSPMFEGTL | 0.000 | |
| ERSPTFGGPLa | 3:00201 | Ta9 | 1306 | 36 | 0.028 | RSPTFGGPL | 0.000 | |
| SKFPKMRMG | 1:02301 | Ta9 | 1306 | 96 | 0.074 | SKFPKMRM | 0.007 | |
| KRKTEGYVF | 2:02201 | Ta11 | 1630 | 23 | 0.014 | |||
| Ave | 0.027 | 0.006 |
The columns are: Epitope the CTL epitope sequence from the literature; MHC the corresponding restriction element; Protein the source protein ID; #pep the number of unique 8-11mer peptides contained within the source protein; #FP the number of false-positive predictions (peptides with a predicted binding affinity value stronger than the known epitope); FR the false-positive ratio (#FP/#pep); Alternative the predicted alternative epitope, and FR the false-positive ratio for the alternative epitope. If no alternative epitope was predicted, the last column is left empty. (Ave) gives the average value for the peptide subset. Data from (Nene et al. 2012) and (MacHugh et al. 2011)
Allelic variant of QRSPMFEGTL (Ivan Morrison, personal communication)
Several important conclusions can be drawn from the analysis. First and foremost, it is evident that the NetMHC consensus method in the majority of cases was able to accurately identify the known BoLA-I epitope within the given source protein sequence. However, in some cases, the epitope sequence obtained from the literature matched the predicted binding motif of the restricting BoLA-I molecule very poorly, as seen with the T. parva epitopes SHEELKKLGML and EFISFPISL restricted to BoLA-2*01201 (T2A) and BoLA-T7, respectively. These peptides are predicted with false-positive rates of 2 and 6.5 % having 14 and 184, alternative peptides, respectively, with a stronger predicted binding affinity value than the reported epitope within the source protein sequence. In contrast to this, the alternative nested peptides EELKKLGML and FISFPISL have very low false-positive rates (0 and 0.1 %). A similar argument can be made for most other epitopes with a FR above 2.5 %. In all these cases, an optimal epitope nested within the original epitope can be identified with corresponding FR values close to or even equal to 0. Only one epitope, the Tp9 epitope AKFPGMKKSK restricted to BoLA-1*02301 (D18.4), remains poorly described following these arguments. This epitope is predicted with a false position rate of 5 %, and no nested alternative peptides are found with a lower FR value.
To investigate further if the suggested alternative epitopes were indeed stronger binders compared to the original epitopes, we measured binding affinity values of six of the eight peptides from Table 3 where the in silico predictions suggested alternative truncated optimal minimal epitopes. In all cases, the experimental validations confirmed a stronger binding affinity for the suggested alternative minimal epitopes (see Table 4).
Table 4.
Experimental validation of binding affinity of 5 known epitopes and the alternative minimal epitopes suggested by in silico predictions
| BoLA | Protein | Epitope | KD (nM) | Alternative | KD (nM) |
|---|---|---|---|---|---|
| 6*04101 (T2B) | Tp2 | SHEELKKLGML | 4,558 | EELKKLGML | 2 |
| T7 | Tp7 | EFISFPISL | 252 | FISFPISL | 142 |
| BoLA-3:00201 | Ta9 | QRSPMFEGTL | 309 | RSPMFEGTL | 8 |
| BoLA-1:02301 | Ta9 | SKFPKMRMG | 74 | SKFPKMRM | 40 |
| BoLA-3:00201 | Ta9 | ERSPTFGGPL | 799 | RSPTFGGPL | 5 |
Additional amino acids flanking the minimal epitope are underlined
Discussion
In this work, we have presented a strategy to analyze the peptide-binding motif of recombinant MHC-I proteins from cattle by measurement of the peptide–MHC-I complex stability of positional scanning combinatorial peptide libraries in a scintillation proximity assay (Harndahl et al. 2011). Our work demonstrates that the strategy published earlier for the human HLA system can be implemented in the analysis of MHCs from other species such as cattle.
By assaying a positional scanning combinatorial peptide library (PSCPL) in a high-throughput scintillation proximity dissociation assay, we obtained peptide-binding motifs of eight recombinant bovine MHC’s, all showing a dominant anchor at the C terminus of the peptide. The anchor in the amino terminal end of the peptide was at P2 in most cases, with some peptides having additional anchors at positions 1, 3, 5, or 6.
We confirmed these data by analyzing the binding properties of 9 published epitopes from T. parva to the proposed restricting MHC allotypes and eight of these were found to bind and form stable complexes with restricting BoLA-I molecule. We could not detect binding of the BoLA-6*04101 (T2B) restricted SHEELKKLGML epitope in the dissociation assay and the affinity was measured to be 4,558 nM, which makes it an unlikely epitope. In contrast, the nested peptide EELKKLGML bound with high affinity and stability to BoLA-6*04101, suggesting that this truncated peptide could be the minimal epitope. This has recently been experimentally confirmed by comparing the fraction of T cells stained by tetramers formed with the two peptide sequences (Svitek et al. 2014). When expanding the analysis to other BoLA-I restricted CTL epitopes, similar arguments could be put forward in cases of weak predicted binding affinity. In all cases, the alternative epitopes were predicted to be nested within a longer peptide, and in all cases the alternative epitope was found to have improved binding to the corresponding BoLA-I restriction element. These findings strongly suggest that, in such cases, the optimal epitope is nested within the previously defined epitope. Therefore, the above findings suggest that the identification of the original epitopes, in most cases performed using IFN-γ ELISpot assays (Graham et al. 2008), was dependent on the additional amino acids flanking the minimal epitope being trimmed off by peptidases prior to binding to BoLA-I. The actual epitope is defined by the nested peptide that has improved binding affinity and stability to the BoLA-I molecule.
Using PSCPL-derived binding motifs and the immunoinformatics prediction method NetMHCpan, we selected a large set of discrete peptides that were experimentally assayed for binding to the BoLA-I molecules. Incorporating these binding data in the training of the NetMHC and NetMHCpan methods led to improved prediction accuracy of known T. parva CTL epitopes. We then tested the nine epitopes for cross binding to each of the eight MHCs, and observed cross binding between a series of MHCs; BoLA-6*04101 (T2B) and BoLA-6*01301 (HD6), BoLA-T2C and BoLA-3*00201 (JSP.1), and BoLA-3*00101 (AW10) and BoLA-3*00201 (JSP.1). In all cases, cross binding was found to be consistent with predicted functional (i.e., binding motif) similarities between the different molecules.
We have outlined a highly cost-effective procedure for characterization of the peptide-binding motifs and functionality for bovine MHC class I proteins. The direct outcome of the procedure is a position specific scoring matrix characterizing the MHC binding motif, and improved prediction methods enabling reliable prediction of peptide binding to BoLA-I. The resulting improved versions of NetMHCpan (version 2.8) and NetMHC (version 3.4) are both publicly available. The described procedure is not limited to the BoLA MHC molecules described here and could readily be expandable (and has been (Follin et al. 2013; Pedersen et al. 2011)) to other BoLA molecules and other species of interest, given that the MHC protein sequences are available.
In conclusion, we have demonstrated how the use of high-throughput biochemical peptide-binding assays combined with state-of-the art immunoinformatics can be effectively used to functionally characterize the binding motif of BoLA class I molecules. This has resulted in the enhanced performance of BoLA-I peptide-binding prediction methods, empowering rational epitope discovery and facilitating the identification and understanding of the cellular immune responses to infection and during vaccine trials in cattle.
Supplementary Material
Acknowledgments
This research was funded through an award (No. 0965346) from the BREAD program of the National Science Foundation (USA); MN is a researcher at the Argentinean national research council (CONICET).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00251-014-0802-5) contains supplementary material, which is available to authorized users.
Competing interests No competing interests
Contributor Information
Andreas M. Hansen, Laboratory of Experimental Immunology, Department of International Health, Immunology and Microbiology, Faculty of Health Sciences, University of Copenhagen, Copenhagen, Denmark
Michael Rasmussen, Laboratory of Experimental Immunology, Department of International Health, Immunology and Microbiology, Faculty of Health Sciences, University of Copenhagen, Copenhagen, Denmark.
Nicholas Svitek, International Livestock Research Institute (ILRI), P.O. Box 30709, Nairobi 00100, Kenya.
Mikkel Harndahl, Laboratory of Experimental Immunology, Department of International Health, Immunology and Microbiology, Faculty of Health Sciences, University of Copenhagen, Copenhagen, Denmark.
William T. Golde, Plum Island Animal Disease Center, Agricultural Research Service, USDA, Greenport, NY, USA
John Barlow, Department of Animal Sciences, University of Vermont, Burlington, VT, USA.
Vishvanath Nene, International Livestock Research Institute (ILRI), P.O. Box 30709, Nairobi 00100, Kenya.
Søren Buus, Laboratory of Experimental Immunology, Department of International Health, Immunology and Microbiology, Faculty of Health Sciences, University of Copenhagen, Copenhagen, Denmark.
Morten Nielsen, Center for Biological Sequence Analysis, Technical University of Denmark, Kongens Lyngby, Denmark, mniel@cbs.dtu.dk; Instituto de Investigaciones Biotecnológicas, Universidad Nacional de San Martín, San Martín, Buenos Aires, Argentina.
References
- Birch J, Murphy L, MacHugh ND, Ellis SA. Generation and maintenance of diversity in the cattle MHC class I region. Immunogenetics. 2006;58:670–679. doi: 10.1007/s00251-006-0137-y. [DOI] [PubMed] [Google Scholar]
- Codner GF, Stear MJ, Reeve R, Matthews L, Ellis SA. Selective forces shaping diversity in the class I region of the major histocompatibility complex in dairy cattle. Anim Genet. 2012;43:239–249. doi: 10.1111/j.1365-2052.2011.02239.x. [DOI] [PubMed] [Google Scholar]
- De Groot AS, Nene V, Hegde NR, Srikumaran S, Rayner J, Martin W. T cell epitope identification for bovine vaccines: an epitope mapping method for BoLA A-11. Int J Parasitol. 2003;33:641–653. doi: 10.1016/s0020-7519(03)00051-1. [DOI] [PubMed] [Google Scholar]
- Ferre H, et al. Purification of correctly oxidized MHC class I heavy-chain molecules under denaturing conditions: a novel strategy exploiting disulfide assisted protein folding. Protein Sci. 2003;12:551–559. doi: 10.1110/ps.0233003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre H, Ruffet E, Nielsen LL, Nissen MH, Hobley TJ, Thomas OR, Buus S. A novel system for continuous protein refolding and on-line capture by expanded bed adsorption. Protein Sci. 2005;14:2141–2153. doi: 10.1110/ps.051396105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follin E, Karlsson M, Lundegaard C, Nielsen M, Wallin S, Paulsson K, Westerdahl H. In silico peptide-binding predictions of passerine MHC class I reveal similarities across distantly related species, suggesting convergence on the level of protein function. Immunogenetics. 2013;65:299–311. doi: 10.1007/s00251-012-0676-3. [DOI] [PubMed] [Google Scholar]
- Gaddum RM, Willis AC, Ellis SA. Peptide motifs from three cattle MHC (BoLA) class I antigens. Immunogenetics. 1996;43:238–239. doi: 10.1007/BF00587307. [DOI] [PubMed] [Google Scholar]
- Graham SP, et al. Theileria parva candidate vaccine antigens recognized by immune bovine cytotoxic T lymphocytes. Proc Natl Acad Sci U S A. 2006;103:3286–3291. doi: 10.1073/pnas.0511273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SP, et al. Characterization of the fine specificity of bovine CD8 T-cell responses to defined antigens from the protozoan parasite Theileria parva. Infect Immun. 2008;76:685–694. doi: 10.1128/IAI.01244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harndahl M, Justesen S, Lamberth K, Roder G, Nielsen M, Buus S. Peptide binding to HLA class I molecules: homogenous, high-throughput screening, and affinity assays. J Biomol Screen. 2009;14:173–180. doi: 10.1177/1087057108329453. [DOI] [PubMed] [Google Scholar]
- Harndahl M, Rasmussen M, Roder G, Buus S. Real-time, high-throughput measurements of peptide-MHC-I dissociation using a scintillation proximity assay. J Immunol Methods. 2011;374:5–12. doi: 10.1016/j.jim.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harndahl M, Rasmussen M, Roder G, Dalgaard Pedersen I, Sorensen M, Nielsen M, Buus S. Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur J Immunol. 2012;42:1405–1416. doi: 10.1002/eji.201141774. [DOI] [PubMed] [Google Scholar]
- Hegde NR, Ellis SA, Gaddum RM, Tregaskes CA, Sarath G, Srikumaran S. Peptide motif of the cattle MHC class I antigen BoLA-A11. Immunogenetics. 1995;42:302–303. doi: 10.1007/BF00176450. [DOI] [PubMed] [Google Scholar]
- Hegde NR, Deshpande MS, Godson DL, Babiuk LA, Srikumaran S. Bovine lymphocyte antigen-A11-specific peptide motif as a means to identify cytotoxic T-lymphocyte epitopes of bovine herpesvirus 1. Viral Immunol. 1999;12:149–161. doi: 10.1089/vim.1999.12.149. [DOI] [PubMed] [Google Scholar]
- Holmes EC, Roberts AF, Staines KA, Ellis SA. Evolution of major histocompatibility complex class I genes in Cetartiodactyls. Immunogenetics. 2003;55:193–202. doi: 10.1007/s00251-003-0560-2. [DOI] [PubMed] [Google Scholar]
- Hoof I, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter WM, Greenwood FC. Preparation of iodine-131 labelled human growth hormone of high specific activity. Nature. 1962;194:495–496. doi: 10.1038/194495a0. [DOI] [PubMed] [Google Scholar]
- Joosten I, Teale AJ, van der Poel A, Hensen EJ. Biochemical evidence of the expression of two major histocompatibility complex class I genes on bovine peripheral blood mononuclear cells. Anim Genet. 1992;23:113–123. doi: 10.1111/j.1365-2052.1992.tb00030.x. [DOI] [PubMed] [Google Scholar]
- Leisner C, et al. One-pot, mix-and-read peptide-MHC tetramers. PLoS One. 2008;3:el678. doi: 10.1371/journal.pone.0001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. Two distinct conformations of a rinderpest virus epitope presented by bovine major histocompatibility complex class I N*01801: a host strategy to present featured peptides. J Virol. 2011;85:6038–6048. doi: 10.1128/JVI.00030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald IK, et al. MHC class I bound to an immunodominant Theileria parva epitope demonstrates unconventional presentation to T cell receptors. PLoS Pathog. 2010;6:e1001149. doi: 10.1371/journal.ppat.1001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacHugh ND, et al. Extensive polymorphism and evidence of immune selection in a highly dominant antigen recognized by bovine CD8 T cells specific for Theileria annulata. Infect Immun. 2011;79:2059–2069. doi: 10.1128/IAI.01285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeever DJ, Taracha EL, Innes EL, MacHugh ND, Awino E, Goddeeris BM, Morrison WI. Adoptive transfer of immunity to Theileria parva in the CD8+ fraction of responding efferent lymph. Proc Natl Acad Sci U S A. 1994;91:1959–1963. doi: 10.1073/pnas.91.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon RM, Friis L, Siebold C, Friese MA, Fugger L, Jones EY. Structure of HLA-A*0301 in complex with a peptide of proteolipid protein: insights into the role of HLA-A alleles in susceptibility to multiple sclerosis Acta crystallographica Section D. Biol Crystallogr. 2011;67:447–454. doi: 10.1107/S0907444911007888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, et al. Designing bovine T cell vaccines via reverse immunology. Ticks Tick-Borne Dis. 2012;3:188–192. doi: 10.1016/j.ttbdis.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Nielsen M, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard Pedersen L, et al. Efficient assembly of recombinant major histocompatibility complex class I molecules with preformed disulfide bonds. Eur J Immunol. 2001;31:2986–2996. doi: 10.1002/1521-4141(2001010)31:10<2986::aid-immu2986>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Parker KC, et al. Sequence motifs important for peptide binding to the human MHC class I molecule, HLA-A2. J Immunol. 1992;149:3580–3587. [PubMed] [Google Scholar]
- Pedersen LE, et al. Porcine major histocompatibility complex (MHC) class I molecules and analysis of their peptide-binding specificities. Immunogenetics. 2011;63:821–834. doi: 10.1007/s00251-011-0555-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Halliwell JA, McWilliam H, Lopez R, Marsh SG. IPD—the immuno polymorphism database. Nucleic Acids Res. 2013;41:D1234–D1240. doi: 10.1093/nar/gks1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields MJ, Ribaudo RK. Mapping of the monoclonal antibody W6/32: sensitivity to the amino terminus of beta2-microglobulin. Tissue Antigens. 1998;51:567–570. doi: 10.1111/j.1399-0039.1998.tb02994.x. [DOI] [PubMed] [Google Scholar]
- Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnathamby G, Seth S, Nayak R, Shaila MS. Cytotoxic T cell epitope in cattle from the attachment glycoproteins of rinderpest and peste des petits ruminants viruses. Viral Immunol. 2004;17:401–410. doi: 10.1089/vim.2004.17.401. [DOI] [PubMed] [Google Scholar]
- Stryhn A, Pedersen LO, Romme T, Holm CB, Holm A, Buus S. Peptide binding specificity of major histocompatibility complex class I resolved into an array of apparently independent subspecificities: quantitation by peptide libraries and improved prediction of binding. Eur J Immunol. 1996;26:1911–1918. doi: 10.1002/eji.1830260836. [DOI] [PubMed] [Google Scholar]
- Svitek N, et al. Use of “one-pot, mix-and-read” peptide-MHC class I tetramers and predictive algorithms to improve detection of cytotoxic T lymphocyte responses in cattle. Vet Res. 2014;45:50. doi: 10.1186/1297-9716-45-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen MC, Nielsen M. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 2012;40:W281–W287. doi: 10.1093/nar/gks469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.