Abstract
The general control nonderepressible 2 (GCN2) kinase is a nutrient-sensing pathway that responds to amino acids deficiency and induces a genetic program to effectively maintain cellular homeostasis. Here we established the conserved role of Caenorhabditis elegans GCN-2 under amino acid limitation as a translation initiation factor 2 (eIF2) kinase. Using a combination of genetic and molecular approaches, we showed that GCN-2 kinase activity plays a central role in survival under nutrient stress and mediates lifespan extension conferred by dietary restriction (DR) or inhibition of the major nutrient-sensing pathway, the target of rapamycin (TOR). We also demonstrated that the GCN-2 and TOR signaling pathways converge on the PHA-4/FoxA transcription factor and its downstream target genes to ensure survival of the whole organism under a multitude of stress conditions, such as nutrient scarcity or environmental stresses. This is one step forward in the understanding of evolutionary conserved mechanisms that confer longevity and healthspan.
Keywords: general control nonderepressible 2, aging, target of rapamycin, Caenorhabditis elegans, PHA-4
Introduction
The ability of most organisms to survive relies on their capability to rapidly trigger a coordinated systemic response upon nutrient or environmental stresses. In eukaryotes, such a stress response involves the inhibition of global protein synthesis, with a concomitant reprogramming of gene expression, which allow cells to conserve resources, maintain cellular homeostasis, and effectively deal with stress (Spriggs et al., 2010). Inhibition of protein synthesis is attained through phosphorylation of the alpha subunit of the translation initiation factor 2 (eIF2α) by specific protein kinases, each activated by different stress signals (Wek et al., 2006). General control nonderepressible 2 is the only eIF2α kinase conserved from yeast to mammals that regulates amino acid transport and metabolism in response to nutrient depletion (Hinnebusch, 2005). GCN2 can also be activated by other stresses, such as UV irradiation, and has been shown to regulate many other vital cellular processes in mammals, such as lipid metabolism, oxidative stress resistance, feeding behavior, NF-kB signaling upon UV radiation, synaptic plasticity, and memory (Harding et al., 2003; Costa-Mattioli et al., 2009).
Phosphorylation of eIF2α under stress results in inhibition of global protein synthesis, which is accompanied by favored translation of specific mRNAs that adapt the organism to stress. These mRNAs include potent transcription factors such as GCN4 in yeast (Natarajan et al., 2001) and ATF4 in mammals (Harding et al., 2003). The proposed model for the translation of these mRNAs, under amino acid limitation, involves the upstream open reading frames (uORFs) that are located in their 5′-UTR (Tzamarias & Thireos, 1988; Vattem & Wek, 2004). These uORFs are preferentially translated in the nonstressed condition, leading to synthesis of incorrect peptides and precluding translation of the authentic gene initiation site (Hinnebusch, 2005). Under stress, the inhibitory effect of eIF2α phosphorylation to the levels of the active ternary complex increases the probability that ribosomal scanning will bypass the uORFs, and translation re-initiation will occur at the canonical gcn4 or atf4 initiation site.
The mechanisms of translational and transcriptional reprogramming under stress have emerged as important mediators of lifespan extension and stress resistance in Caenorhabditis elegans (Curran & Ruvkun, 2007; Hansen et al., 2007; Syntichaki et al., 2007; Rogers et al., 2011). Consistent with this, aging in many organisms is modulated by conserved signaling pathways that affect numerous cellular processes, involving regulation of translation. One such pathway is that of the target of rapamycin (TOR) kinase, which responds to hormonal, nutrient, and environmental stress signals to regulate growth, differentiation, and metabolism of eukaryotes (Cornu et al., 2013). The TOR pathway has been identified as a mediator of DR-induced longevity in yeast, worms, flies, and mice (Kapahi et al., 2010), whereas differential expression of TOR signaling has been recently associated with advancing age in human populations (Harries et al., 2012; Passtoors et al., 2013). The downstream targets of TOR signaling involve mRNA translation, ribosome synthesis, transcription, stress response, metabolism, and autophagy. All these cellular processes have been linked to the longevity effects of TOR disruption in multiple species.
Despite the well-established link between nutrient limitation and lifespan in many eukaryotes, the interplay between the two major nutrient-sensing pathways, GCN2 and TOR, in longevity still remains unclear. Reduced availability of nutrients such as nitrogen, carbon, or amino acids inhibits TOR and induces autophagy in yeast and mammals (Jung et al., 2010). Also in mice, the impaired amino acid uptake in intestinal cells leads to increased phosphorylation of eIF2α, suggesting activation of the GCN2 pathway and reduced TOR signaling (Broer et al., 2011). Rapamycin (a TOR-specific inhibitor) has been shown to derepress translation of yeast GCN4 through activation of GCN2 (Cherkasova & Hinnebusch, 2003; Kubota et al., 2003). Genetic evidence in yeast suggests that DR, TOR inhibition or depletion of the 60S ribosomal subunits mediate replicative lifespan partially through the activation of the GCN4 transcription factor (Steffen et al., 2008). Recently, a role of GCN2 in the aging process was suggested in mice, where the absence of GCN2 affects macronutrient selection changes during aging (Maurin et al., 2012). Herein, we have established the conserved function of the GCN-2 kinase in C. elegans under amino acid limitation, and we showed that loss of GCN-2 activity is not required for normal lifespan, but affects the lifespan of nutrient-sensitized worms. We revealed that GCN-2 signaling positively regulates the induction of PHA-4/FoxA transcription factor under nutrient or oxidative stress, as part of the adaptive response that ensures stress survival and longevity.
Results
A GCN-2-dependent phosphorylation of eIF2α under amino acid limitation
In C. elegans, the sole homolog of yeast/mammalian GCN2 is encoded by the gene Y81G3A.3 (gcn-2) and phosphorylates the eIF2α subunit at the putative phosphorylation site Ser49 (Nukazuka et al., 2008). Lately, it was shown that GCN-2 is required for the induced phospho-eIF2α levels during mitochondrial or osmotic stress (Baker et al., 2012; Lee & Strange, 2012), but its function during nutrient deprivation or other stresses has not been established. The predicted GCN-2 protein of 1696 amino acids shares 24.2% identity (40.7% similarity) with human HsGCN2 and 21% identity (35.3% similarity) with yeast ScGCN2 (EMBOSS Align-EMBL/EBI), having all the functional domains that characterize the kinase across species (Fig. 1A). To investigate the function of GCN-2, we used the existing gcn-2 mutants (ok871 and ok886), both of which have an in-frame deletion (Wormbase WS230), lacking part of the internal coding sequence (Fig. 1A). In both gcn-2 alleles, we detected a truncated mRNA transcript (lanes 1 and 2 in Fig. 1B, left pane) expressed at the same levels as the wild-type (N2) transcript (Fig. 1B, right pane). By measuring the basal levels of eIF2α phosphorylation in whole protein extracts of N2 worms subjected to gcn-2(RNAi) (lane 2 in Fig. 1C) or of each gcn-2 mutant (lanes 3–4 in Fig. 1C), compared to untreated animals (lane 1 in Fig. 1C), we verified that both ok871 and ok886 are loss-of-function alleles of gcn-2.
Figure 1.
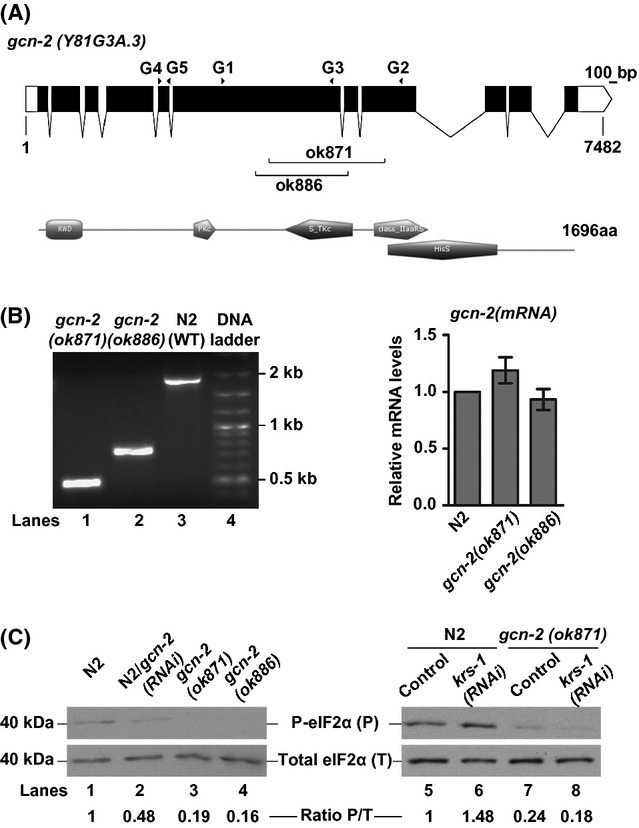
Conservation in gene structure and function of Caenorhabditis elegans gcn-2. (A) Gene structure and predicted protein domains of GCN-2, designed using the Prosite MyDomains (http://prosite.expasy.org/mydomains/): Black boxes represent exons linked by lines corresponding to introns, and white boxes indicate the 5′ and 3′ UTR found in a second alternative isoform (http://www.wormbase.org). The graphic was created using the Exon-Intron Graphic Maker (http://wormweb.org/exonintron). Branches point to the sequences deleted in the two alleles ok871 and ok886. Black arrowheads indicate the position of primers used in this study (Table S2). (B) Reverse transcription (RT)–PCR analysis with primers G1/G2 (shown in A pane) and qRT-PCR analysis with primers G4/G5 (shown in A pane). (C) Western blot analysis showing the levels of phosphorylated (P-eIF2α) normalized by the total amount of eIF2α, in whole worm extracts. Basal levels of P-eIF2α under well-fed conditions in untreated or gcn-2(RNAi)-treated N2 and gcn-2 mutants (left pane). Induced levels of P-eIF2α under amino acid limitation in krs-1(RNAi) treated worms (right pane).
To determine whether worm GCN-2 kinase responds to amino acid limitation, we raised worms on plates seeded with bacteria expressing dsRNA against krs-1 gene, the worm lysil-tRNA synthetase. Aminoacyl-tRNA synthetases (AARSs) catalyze the ligation of specific amino acids to their cognate tRNAs and are important for cellular protein synthesis. Changes in the levels of AARSs affect the levels of uncharged tRNAs, and this consists the major signal for GCN2 activation and eIF2α phosphorylation in other organisms. We found that worms grown on bacteria expressing dsRNA for krs-1 either arrested in early larval stages or became adults with low brood size (∼20% of the normal), depending on the starting time of RNAi treatment (eggs or L3–L4 stage, respectively). In such krs-1(RNAi)-fed adults, we monitored an increase (∼50%) of the phospho-eIF2α levels (lane 6 in Fig. 1C), compared to untreated control animals (lane 5 in Fig. 1C). We observed the same level of induction using RNAi for lrs-1, encoding a leucyl-tRNA synthetase (lanes 1–2 in Fig. S1). Moreover, this increase depends on GCN-2 activity (lanes 7–8 in Fig. 1C and lanes 3–4 in Fig. S1). All these are consistent with the conserved role of worm GCN-2 as an eIF2α kinase under amino acid limitation conditions.
Favored translation of atf-5 during amino acid limitation
Phosphorylation of eIF2α in yeast and mammals has two consequences: inhibition of global protein synthesis and induction of specific mRNA translation. In C. elegans, it has been shown that knockdown of several genes encoding AARSs reduces [35S]methionine incorporation and protein synthesis rate (Anderson et al., 2009), in agreement with the increased phospho-eIF2α levels shown in Fig. 1C (compare lanes 5 and 6). Therefore, we tested whether phosphorylation of eIF2α under amino acid deprivation could also induce translation of specific mRNAs. The worm homolog of yeast GCN4 and the related mammalian ATF4 transcription factor is encoded by the atf-5 (T04C10.4) gene, bearing two upstream ORFs (Fig. 2A). To evaluate the role of uORFs in atf-5 mRNA translation, we created transgenic animals carrying either the intact atf-5 gene including the two uORFs (BRF140) or the atf-5 coding sequence lacking both uORFs (BRF152). In both cases, the transgenes were fused at C′-terminus with gfp, and their expression was driven by the atf-5 promoter. BRF140 worms showed only a weak fluorescent signal with atf-5::gfp expression in the nucleus of few cells, varying between individuals (Fig. 2B). In contrast, BRF152 worms displayed a bright GFP signal in the nucleus of all cells (Fig. 2B). As the expression levels of atf-5 mRNA are increased ∼10-fold in BRF140 and ∼twofold in BRF152 young adults, compared to endogenous mRNA (Fig. 2B), it becomes evident that the presence of the uORFs in the 5′ leader of atf-5mRNA is inhibitory for its translation.
Figure 2.
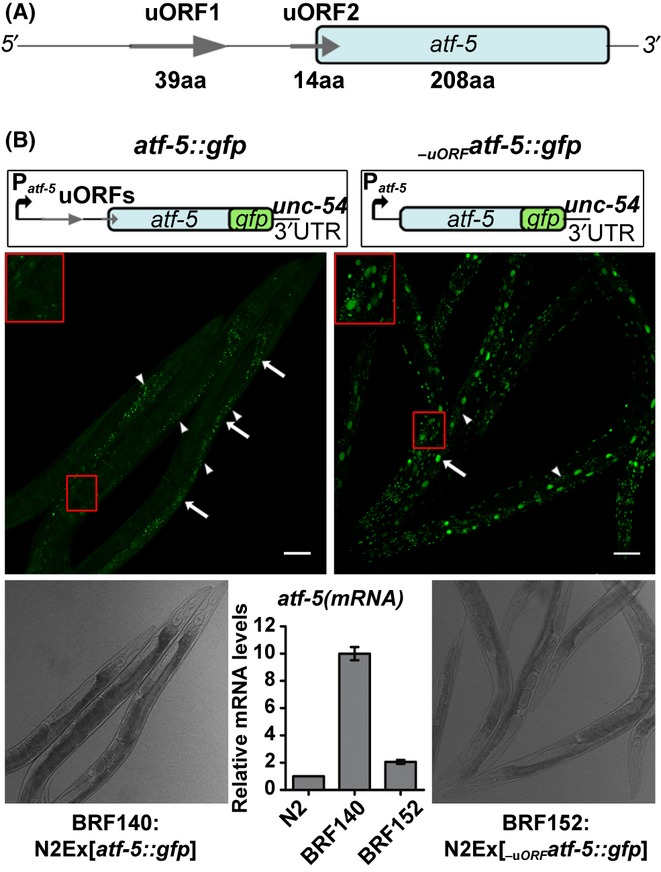
Translational control of atf-5 gene expression. (A) Schema of uORFs within the atf-5 5′-UTR. The amino acid length of the predicted translated uORF and the coding sequence of atf-5 gene are shown. (B) Confocal and bright field images of 1-day-old transgenic worms expressing the translational fusion of the intact atf-5 (left box) or the uORF-less atf-5 transgene (right box) under normal feeding conditions. A larger magnification of the area in the red box is shown on the top left. White arrows indicate fluorescent nuclei, and white arrowheads show regions of autofluorescence. All images were taken at 20× magnification under the same microscopy settings (scale bar: 50 μm). The levels of atf-5 mRNA in N2, BRF140, and BRF152 worms, normalized to ama-1(mRNA), were quantified using qRT–PCR.
The inhibitory effect of uORFs in the translation of the intact atf-5 mRNA was also greatly ameliorated in response to amino acid limitation, as this was recapitulated through RNAi-mediated silencing of AARSs genes. Transgenic BRF140 worms subjected to krs-1(RNAi) showed enhanced fluorescent signal in neurons, hypodermis, muscles, and intestine (Fig. 3). The fluorescence intensity in the few L3-arrested progeny (F1) of the RNAi-treated animals (P0) almost attained the brightness of BRF152 worms. This enhancement was dependent on GCN-2 function as we showed by expressing the same intact atf-5 transgene in the gcn-2(ok871) background (BRF144 in Fig. 3). Only the basal signal in varying cells and the autofluorescence of the intestine were observed. Similar results (Fig. S2) were obtained by inactivating an arginyl-tRNA synthetase (rrt-1) gene. The gcn-2-dependent induction of atf-5::gfp mirrored the gcn-2-dependent phosphorylation of eIF2α upon amino acid limitation. Consequently, direct inactivation of eIF2α by RNAi, instead of phosphorylation, bypasses the requirement for gcn-2 resulting in upregulation of atf-5::gfp transgene in both N2 and gcn-2 worms (Fig. S2). We also observed a gcn-2-independent induction of atf-5::gfp transgene after treatment of worms with a potent inducer of ER stress, tunicamycin (Fig. S2). Thus, the two uORFs direct the translational activation of the C. elegans atf-5 gene under nutrient or other stresses, in agreement with the yeast gcn4 and mammalian atf4 homologs.
Figure 3.
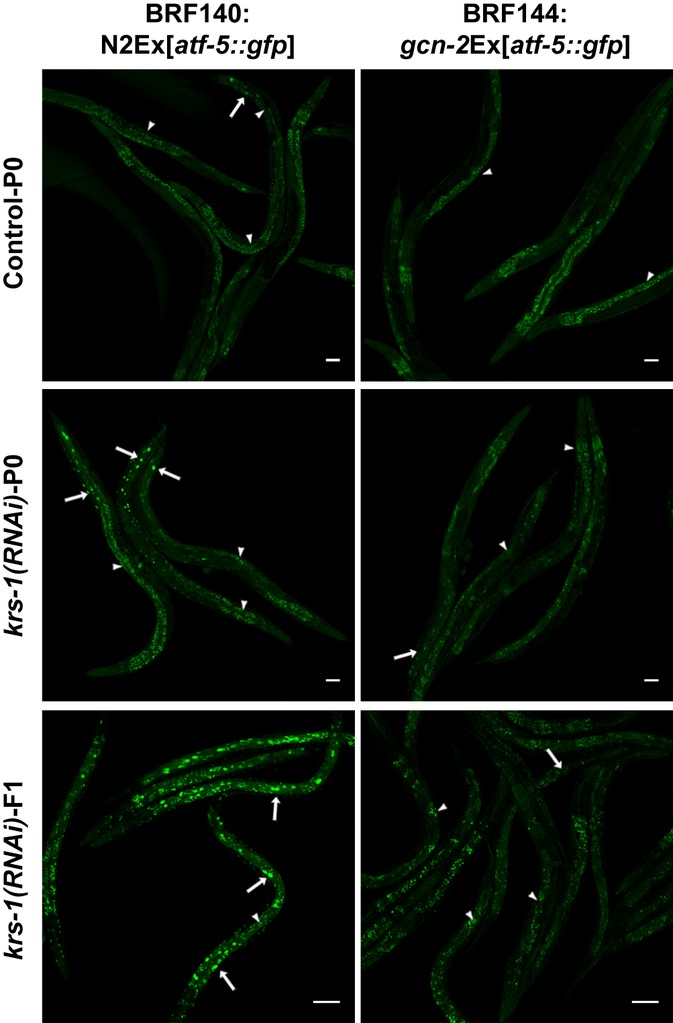
General control nonderepressible-2-dependent translational control of atf-5 under amino acid limitation. Confocal images of N2 or gcn-2(ok871) worms, both carrying a translational fusion of atf-5::gfp, fed Control or krs-1(RNAi) expressing bacteria. The images show 1-day adults fed with each RNAi from eggs (P0) or their L3-arrested progeny (F1) in krs-1(RNAi) worms. White arrows indicate fluorescent nuclei; white arrowheads show regions of autofluorescence. All images were taken at 20× magnification under the same microscopy settings (scale bar: 50 μm).
General control nonderepressible-2 signaling influences the longevity of nutrient-sensitized worms
The conservation of GCN-2 signaling as a nutrient-sensing pathway in C. elegans offers the opportunity to address its impact in the aging process, which is strongly affected by the nutrient status of the organism. We performed phenotypic and lifespan analysis in both gcn-2 (ok871 and ok886) worms as well as in the atf-5(ok576) null mutant (Wormbase WS230). All three mutants behaved indistinguishably from N2 strain under normal conditions in growth, development, movement, fecundity (Fig. S3A), and lifespan (Fig. 4A and Table 1). Similarly, postdevelopmental RNAi of gcn-2 or atf-5 had no significant effect on the lifespan of N2 worms (Fig. 4B and Table 2). As GCN2 is activated under amino acid limitation, we monitored the lifespan of N2 and gcn-2 mutants subjected to rrt-1(RNAi) or krs-1(RNAi), during adulthood only. In all cases, knockdown of either tRNA synthetase gene remarkably shortened lifespan of worms (Fig. S4A and 1 mm isopropylb-D-thiogalactopyranoside (IPTG) in Table 2; Table S3). However, when we applied weaker RNAi conditions to partially inactivate rrt-1 or krs-1 gene, we revealed significantly increased sensitivity of both gcn-2 mutants, compared to N2 (Fig. S4B and 0.25 mm IPTG in Table 2). Worms deficient in GCN-2 kinase activity had a mean lifespan ∼10–15% shorter than the mean lifespan of wild-type. Surprisingly, atf-5 mutants were not more sensitive than N2 worms under these conditions (Table S3).
Figure 4.
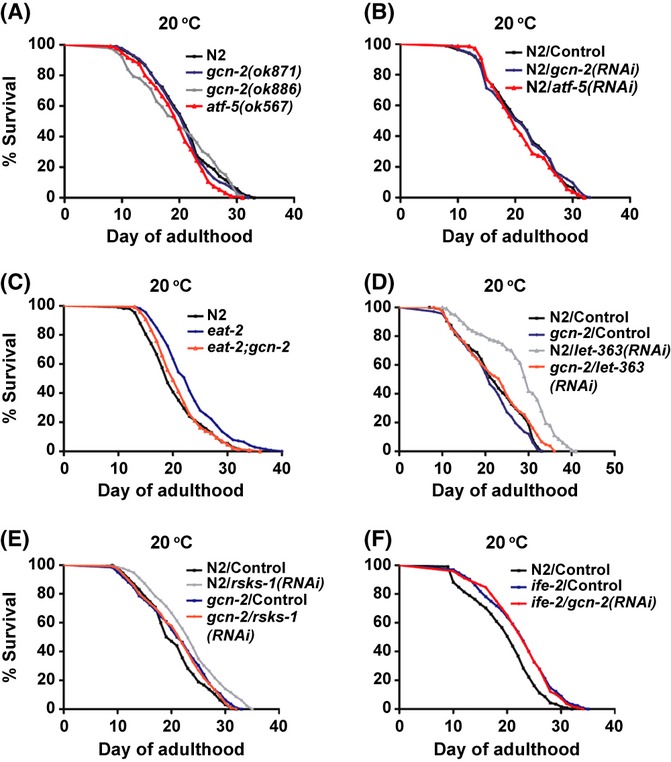
Loss of GCN-2 function affects lifespan only under nutrient stress. Survival curves of (A) N2, gcn-2, and atf-5 mutants fed OP50 bacteria (B) N2 worms treated with gcn-2(RNAi) or atf-5(RNAi) from L4s (C) eat-2(ad465) and eat-2(ad465);gcn-2(ok871) fed OP50 bacteria (D) N2 and gcn-2(ok871) treated with let-363(RNAi) from their first day of adulthood (E) N2 and gcn-2(ok871) treated with rsks-1(RNAi) from L4s (F) N2 and ife-2(ok306) treated with gcn-2(RNAi) from L4s.
Table 1.
Lifespan experiments in OP-50 plates*
| Strain | Treatment | Median/Max lifespan (days)† | Mean lifespan ± SEM (days)‡ | Number (T/C)§ | P-value against N2¶ | P-value against specific strain** | |
|---|---|---|---|---|---|---|---|
| Fig. 4A | N2 | 20 °C | 21/30.5 | 21.33 ± 0.33 | 99/6 | ||
| gcn-2(ok871) | ≫ | 21/29.4 | 20.67 ± 0.33 | 80/6 | 0.4864 | ||
| gcn-2(ok886) | ≫ | 20/30 | 19.33 ± 0.67 | 88/3 | 0.7357 | ||
| atf-5(ok576) | ≫ | 20/27.7 | 19.67 ± 0.33 | 98/3 | 0.0347 | ||
| Fig. 4C | N2 | 20 °C | 19/31 | 19.4 ± 0.55 | 110/2 | ||
| eat-2(ad465) | ≫ | 23/35.6 | 23.25 ± 0.25 | 160/17 | 0.0001 | ||
| eat-2(ad465);gcn-2(ok871) | ≫ | 20/31.4 | 20.5 ± 0.87 | 145/14 | 0.4172 | 0.0009 |
Data from representative experiments are shown. Data from independent repeats of each longevity assay are shown in Table S3 (Supporting information).
Max lifespan is the mean of the last 10% surviving worms.
Mean lifespan and standard error of the mean (SEM) of 2–4 plates.
Total number (T) of dead and censored (C) worms/censored (C).
P-value from log-rank test comparing a mutant strain to N2 wild-type strain (<0.05 is considered statistically significant).
P-value from log-rank test comparing a double mutant to a specific single mutant, for example eat-2;gcn-2 vs. eat-2 etc.
Table 2.
Lifespan experiments in RNAi plates*
| Strain/RNAi | Treatment/ IPTG† | Median/Max lifespan (days)‡ | Mean lifespan ± SEM (days)§ | Number (T/C)¶ | P-value against control** | P-value against specific control†† | |
|---|---|---|---|---|---|---|---|
| Fig. 4B | N2/Control | 20 °C/1 mm | 21/30 | 20.83 ± 0.44 | 83/3 | ||
| N2/gcn-2(RNAi) | ≫ | 21/31.3 | 20.67 ± 0.33 | 88/3 | 0.6489 | ||
| N2/atf-5(RNAi) | ≫ | 20/29 | 19.5 ± 0.29 | 70/5 | 0.5680 | ||
| Fig. S4 A | N2/Control | 20 °C/1 mm | 21/30.1 | 21 ± 0.58 | 124/5 | ||
| N2/rrt-1(RNAi) | ≫ | 10/16.1 | 10 ± 0.58 | 119/10 | <0.0001 | ||
| N2/krs-1(RNAi) | ≫ | 8/11.5 | 7.83 ± 0.16 | 124/6 | <0.0001 | ||
| Fig. S4 A | N2/Control | 20 °C/1 mm | 22/30.4 | 22 ± 1 | 118/9 | ||
| N2/rrt-1(RNAi) | ≫ | 13/20.6 | 13 ± 0.57 | 125/1 | <0.0001 | ||
| gcn-2(ok871)/Control | ≫ | 21/30.4 | 22.17 ± 1.69 | 117/5 | 0.0978 | ||
| gcn-2(ok871)/rrt-1(RNAi) | ≫ | 13/19.1 | 12.67 ± 0.67 | 108/6 | <0.0001 | 0.4078 | |
| Fig. S4 B | N2/Control | 20 °C/0.25 mm | 22/30.6 | 22.50 ± 0.29 | 80/5 | ||
| N2/rrt-1(RNAi) | ≫ | 19/28.3 | 18.83 ± 0.44 | 140/5 | <0.0001 | ||
| gcn-2(ok871)/Control | ≫ | 22/30.7 | 21.67 ± 0.67 | 76/8 | 0.6962 | ||
| gcn-2(ok871)/rrt-1(RNAi) | ≫ | 16/27.2 | 16.33 ± 0.33 | 138/2 | <0.0001 | <0.0001 | |
| Fig. S4 B | N2/Control | 20 °C/0.25 mm | 20/28.6 | 19.67 ± 0.67 | 91/5 | ||
| N2/rrt-1(RNAi) | ≫ | 17/25.8 | 16.33 ± 0.33 | 185/8 | <0.0001 | ||
| gcn-2(ok880)/Control | ≫ | 19/26 | 18.67 ± 0.33 | 92/2 | 0.1081 | ||
| gcn-2(ok880)/rrt-1(RNAi) | ≫ | 15/22.9 | 14.83 ± 0.16 | 177/6 | <0.0001 | <0.0001 | |
| Fig. S4 B | N2/Control | 20 °C/0.25 mm | 21/28.6 | 21.33 ± 0.67 | 82/12 | ||
| N2/krs-1(RNAi) | ≫ | 12/14.4 | 12 ± 0.0 | 96/9 | <0.0001 | ||
| gcn-2(ok871)/Control | ≫ | 22/27.9 | 22 ± 1 | 83/13 | 0.9383 | ||
| gcn-2(ok871)/krs-1(RNAi) | ≫ | 11/12.3 | 11 ± 0.0 | 79/9 | <0.0001 | <0.0001 | |
| Fig. 4D | N2/Control | 20 °C/0.25 mm | 22/31.7 | 21.5 ± 0.5 | 66/2 | ||
| N2/let-363(RNAi) | ≫ | 30/39 | 29.63 ± 1.14 | 120/3 | <0.0001 | ||
| gcn-2(ok871)/Control | ≫ | 21/31.2 | 21.25 ± 0.75 | 69/2 | 0.2995 | ||
| gcn-2(ok871)/let-363(RNAi) | ≫ | 24/34.4 | 24 ± 1.08 | 113/4 | 0.0279 | ||
| atf-5(ok576)/Control | ≫ | 21/30.4 | 21.5 ± 0.5 | 97/1 | 0.0456 | ||
| atf-5(ok576)/let-363(RNAi) | ≫ | 27/35.3 | 27.13 ± 0.77 | 111/3 | <0.0001 | ||
| Fig. 4F | N2/Control | 20 °C/1 mm | 21/29.2 | 21 ± 0.58 | 119/2 | ||
| ife-2(ok306)/Control | ≫ | 23/32.4 | 23 ± 0.41 | 133/7 | 0.0002 | ||
| ife-2(ok306)/gcn-2(RNAi) | ≫ | 23/31.5 | 23.25 ± 0.25 | 124/5 | 0.1072 | ||
| ife-2(ok306)/atf-5(RNAi) | ≫ | 23/30.3 | 22.83 ± 0.44 | 123/4 | 0.1696 | ||
| Fig. 4E | N2/Control | 20 °C/1 mm | 20/30 | 19.75 ± 0.48 | 112/12 | ||
| N2/rsks-1(RNAi) | ≫ | 23/34.1 | 23.25 ± 0.75 | 135/5 | <0.0001 | ||
| gcn-2(ok871)/Control | ≫ | 22/30.8 | 21.88 ± 0.87 | 131/18 | 0.1485 | ||
| gcn-2(ok871)/rsks-1(RNAi) | ≫ | 22/30.2 | 22.13 ± 0.72 | 130/8 | 0.6481 | ||
| atf-5(ok576)/Control | ≫ | 19/28 | 19 ± 0.41 | 101/15 | 0.0495 | ||
| atf-5(ok576)/rsks-1(RNAi) | ≫ | 20/30.7 | 20.75 ± 0.75 | 134/18 | 0.0162 |
IPTG, isopropylb-D-thiogalactopyranoside.
Data from representative experiments are shown. Data from independent repeats of each longevity assay are shown in Table S3 (Supporting information).
IPTG final concentration in bacterial cultures.
Max lifespan is the mean of the last 10% surviving worms.
Mean lifespan and standard error of the mean (SEM) of 2–4 plates.
Total number (T) of dead and censored (C) worms/censored (C).
P-value from log-rank test comparing a RNAi-treated strain to isogenic Control strain. P < 0.05 is considered statistically significant.
P-value from log-rank test comparing a mutant strain to the equal treated N2 population, for example gcn-2/Control vs. N2/Control or gcn-2/RNAi vs. N2/RNAi.
Dietary restriction slows aging across animal species and, in C. elegans, a well-studied feeding defective mutant that has decreased food uptake and increased lifespan is the eat-2(ad465) (Avery, 1993; Lakowski & Hekimi, 1998). We examined the effect of gcn-2 deletion in the lifespan of eat-2 worms by creating the double mutant eat-2(ad465);gcn-2(ok871). Loss of GCN-2 activity suppressed the longevity of eat-2 worms, making them live as wild-type (Fig. 4C and Table 1). This is not due to higher pumping rate of eat-2;gcn-2, compared to eat-2 (data not shown) and eat-2;gcn-2 worms had similar reduced fecundity as eat-2 worms (Fig. S3B). In addition, the double mutant exhibited a further delay in development than the single eat-2 mutant (at 90 h posthatching, ∼30% of eat-2;gcn-2 animals became adults vs. ∼90% in eat-2 population), suggesting that gcn-2 activity is required for both normal growth and longevity of eat-2 worms.
Several studies in diverse species have linked the DR effect on longevity with the inhibition of the TOR kinase pathway (Kapahi et al., 2004; Kaeberlein et al., 2005; Hansen et al., 2008). We examined whether loss of GCN-2 signaling affects the long life of worms with reduced TOR (LET-363) kinase activity. By subjecting N2, gcn-2, and atf-5 worms in let-363(RNAi) from their first day of adulthood, we observed that TOR inhibition failed to increase lifespan in gcn-2 mutants (Fig. 4D and Table 2). We verified by quantitative reverse transcription PCR (qRT–PCR) that gcn-2 mutants were responsive to feeding RNAi at the same level as N2 (Fig. S5). Two downstream effectors of the TOR pathway are the rsks-1/S6 kinase and ife-2/eIF4E translation initiation factor, which when inactivated extends worm’s lifespan (Hansen et al., 2007; Pan et al., 2007; Syntichaki et al., 2007). We found that inactivation of rsks-1 failed to increase the lifespan in gcn-2 mutants (Fig. 4E and Table 2). However, inactivation of gcn-2 gene in ife-2(ok306) worms did not alter their long life (Fig. 4F and Table 2). Finally, loss of atf-5 did not alter the longevity induced by inactivation of TOR and its effectors (Table 2), suggesting that loss of ATF-5, in contrast to GCN-2, is not sufficient to alter lifespan of nutrient-sensitized worms.
General control nonderepressible-2 regulates the activation of PHA-4 in response to TOR disruption
In C. elegans, the transcription factor PHA-4/FoxA is selectively required for lifespan extension by DR or reduced TOR signaling (Panowski et al., 2007; Steffen et al., 2008). It was also shown that inactivation of let-363 and rsks-1, but not ife-2, promotes PHA-4 activity to increase lifespan, indicating that the TOR and rsks-1/S6K signaling antagonizes PHA-4 (Steffen et al., 2008). The expression levels of pha-4 are increased in TOR-deficient and eat-2(ad1116) animals (Panowski et al., 2007; Lapierre et al., 2011). We observed similar transcriptional induction of pha-4 in eat-2(ad465), N2/let-363(RNAi), and N2/rrt-1(RNAi) worms, but this induction was impaired in the absence of GCN-2, at least for the RNAi-treated animals (Fig. 5A). Thus, GCN-2 signaling positively regulates pha-4 transcript levels in response to reduced TOR pathway.
Figure 5.
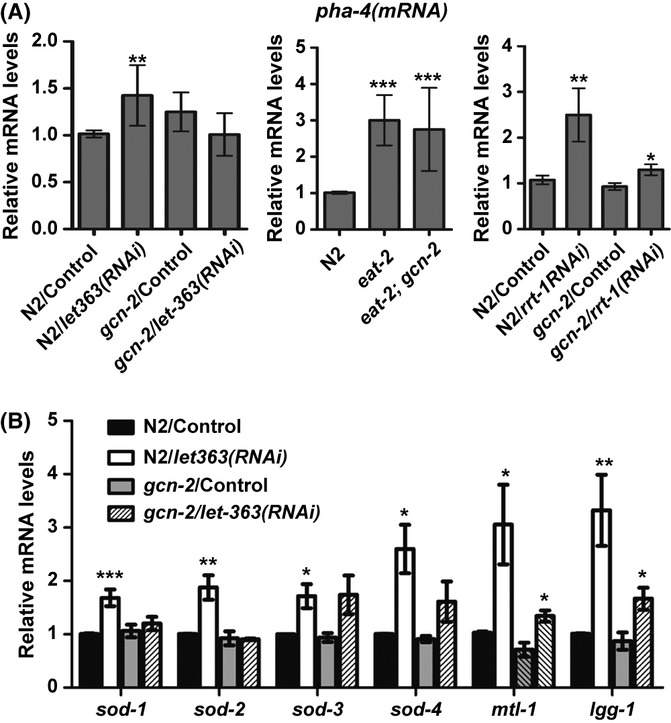
General control nonderepressible-2 influences the induction of pha-4 and its downstream targets in response to TOR inactivation. qRT–PCR of pha-4(mRNA) in (A) 1-day adults of N2 and gcn-2(ok871) treated with let-363(RNAi) from L3 stage; N2, eat-2, and eat-2;gcn-2 young adults raised on OP-50 bacteria at 20 °C; and in the F1 progenies (L2–L3 stage) of N2 and gcn-2(ok871) treated with rrt-1(RNAi) (B) qRT–PCR of sod-1, sod-2, sod-3, sod-4, mtl-1, and lgg-1 on 1-day adults of N2 and gcn-2(ok871) treated with let-363(RNAi) from L3 stage. Quantification of each mRNA level, relative to ama-1 mRNA and the mean ± SD of biological triplicates are shown. The asterisks represent statistical significant difference from N2/Control or gcn-2/Control (*P < 0.05, ** P < 0.01, ***P < 0.001 in unpaired t-test).
It has been suggested that increased expression of pha-4 during DR or TOR inhibition facilitates its binding to DR-specific genes, in an analogous manner to its expression and binding specificity during embryogenesis (Panowski et al., 2007). Such PHA-4 targets are genes mostly involved in metabolic processes and defense responses (Panowski et al., 2007; Zhong et al., 2010). The superoxide dismutase (sod) gene family, responsible for scavenging ROS, includes members that are differentially regulated by PHA-4 and DAF-16/FoxO transcription factors in response to DR or reduced insulin/IGF-1 signaling (ISS), respectively. PHA-4 regulates the expression of sod-1, sod-2, sod-4, and sod-5, but not sod-3, in eat-2 mutants, while DAF-16 regulates the expression of sod-1, sod-3, and sod-5 in ISS mutants (Panowski et al., 2007). By measuring the mRNA levels of sod genes in let-363(RNAi)-fed worms, we found a clear induction of PHA-4-regulated sod-1, sod-2, and sod-4 (sod-5 was not tested) (Fig. 5B), similarly to eat-2 mutants. Moreover, this induction was GCN-2-dependent (Fig. 5B). Curiously, we observed an induction of DAF-16-regulated sod-3 in let-363(RNAi)-fed worms, which was totally GCN-2-independent (Fig. 5B). Although TOR disruption extends lifespan independently of DAF-16, the induction of sod-3 in TOR-deficient worms might be due to reduced ISS and activation of DAF-16 or alternatively, a mild stress response to elevated mitochondrial ROS under these conditions (Ristow & Zarse, 2010). Evidence against the first hypothesis comes from the lower induction of sod-3 here than in response to lower ISS and that the expression of another DAF-16 target, hsp-16.2, did not change in let-363(RNAi)-treated worms (data not shown).
We also tested the expression of mtl-1 gene, encoding for a metallothionein involved in detoxification/stress adaptation. This is a DAF-16 target under reduced ISS, but is a candidate PHA-4 target in starved L1s (Zhong et al., 2010). We observed an upregulation of mtl-1 transcript in let-363(RNAi)-treated animals, which was partially GCN-2-dependent (Fig. 5B). PHA-4 is also directly involved in the induction of autophagy-related genes, and autophagy is required for lifespan extension in response to DR and TOR disruption (Jia & Levine, 2007; Hansen et al., 2008; Zhong et al., 2010; Lapierre et al., 2011). Again, we measured less induction of the autophagic gene lgg-1 in let-363(RNAi)-fed worms when GCN-2 activity was missing, compared to controls (Fig. 5B). Similarly, the induction of mtl-1 and lgg-1 was diminished in eat-2;gcn-2 compared to eat-2 worms (Fig. S6). Taken together, loss of GCN-2 impairs the induction of pha-4 and specific downstream targets, under conditions of DR or TOR inactivation.
General control nonderepressible-2 signaling has a protective role against a multitude of stresses
Inactivation of various components of the TOR pathway in yeast and C. elegans (Powers et al., 2006; Hansen et al., 2007; Pan et al., 2007; Syntichaki et al., 2007) leads to increased stress resistance, but the underlying mechanisms remain elusive. The transcriptional activity of PHA-4 under DR or reduced TOR should be part of an adaptive mechanism of cells to cope with environmental stresses and live longer. Based on our findings that GCN-2 activity positively regulates expression of PHA-4 and some of its targets, which are genes involved in stress defense, we assessed sensitivity of TOR-deficient worms to oxidative stress, in the presence or absence of GCN-2. Whereas let-363(RNAi)-treated animals were more resistant to sodium arsenite than untreated controls, the deletion of gcn-2 partially suppressed this resistance (Fig. 6A). Likewise, eat-2 worms were more resistant to sodium arsenite than the eat-2;gcn-2 (Fig. 6B). We also observed that knockdown of pha-4 decreased the percentage of N2 that survived this stress but had smaller effect on gcn-2 worms, which were more sensitive than N2 (Fig. 6C). As our data suggest that oxidative stress can induce pha-4 in a GCN-2-dependent manner, we confirmed this by quantification of the fluorescent signal of a pha-4 promoter-driven GFP reporter in N2 and gcn-2(RNAi) worms, under sodium arsenite (Figs 6D and S7). This was also verified by measuring the mRNA levels of the endogenous pha-4 gene in N2 and gcn-2, treated with sodium arsenite (Fig. 6E). Finally, we observed increased sensitivity of gcn-2 worms after heat shock or UV irradiation, compared to N2 (Fig. S8). Combined these results suggest a broad role of GCN-2 on stress response and survival, through its function on translation regulation and/or transcriptional induction of specific programs that adapt cells to each stress (Fig. 6F).
Figure 6.
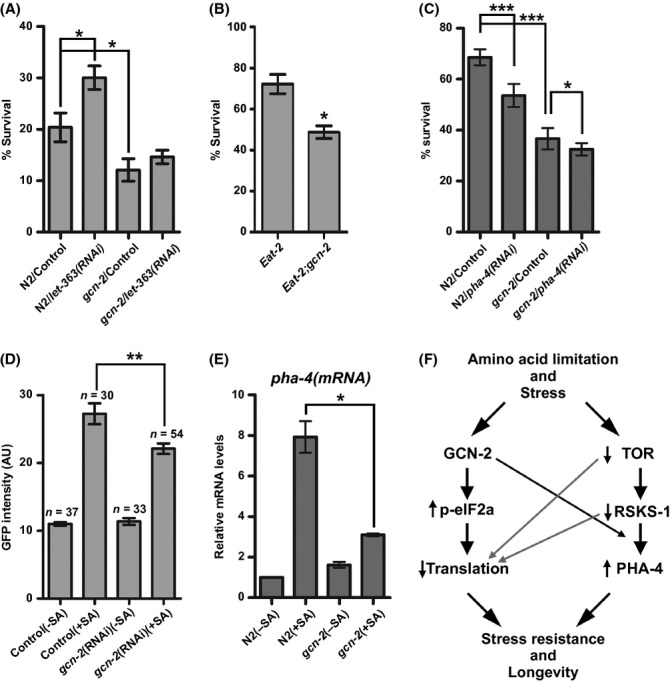
GCN-2 affects pha-4 induction and survival of TOR-deficient and eat-2 worms under oxidative stress. Survival to sodium arsenite (SA) of 1-day adults of (A) N2 and gcn-2(ok871) fed with let-363(RNAi) from L3 stage (B) eat-2(ad465) and eat-2(ad465);gcn-2(ok871) (C) N2 and gcn-2(ok871) fed with pha-4(RNAi) from eggs (D) Quantification in arbitrary units (AU) of GFP signal in the intestine of 1-day adults expressing a membrane-bound GFP under the pha-4 promoter, fed either Control or gcn-2(RNAi) expressing bacteria and treated or not with SA (15 mm for 3 h before observation). Fluorescence intensity was measured from several confocal images using ImageJ. The total number (n) of areas counted and the mean ± SD are shown (E) qRT–PCR of pha-4(mRNA) in 1-day N2 or gcn-2 worms treated or not with SA (15 mm for 1.5 h). Quantification of each mRNA level, relative to ama-1 mRNA, and the mean ± SD of biological triplicates are shown (*P < 0.05, **P < 0.01, ***P < 0.001 in unpaired t-test) (F) A model illustrating the function of GCN-2 in response to nutrient and other stresses.
Discussion
Aging is a complex biological process critically influenced by endogenous or exogenous signals that affect basic mechanisms and pathways, related to metabolism and stress response. Reduced nutrient signals can effectively alter the lifespan in many organisms, and nutrient-sensing pathways have acquired central role in the longevity determination. One such is the TOR kinase pathway that regulates both anabolic and catabolic processes, important for stress management and long-term survival. Another nutrient-sensing pathway is that of GCN2 kinase, which phosphorylates eIF2α translation factor in response to nutrient or other stresses. However, the impact of GCN2 function and its possible connection to TOR pathway in lifespan determination have not been investigated. C. elegans is a primary model organism for aging studies and has profoundly contributed to the determination of genetic and environmental factors that affect aging in organismal level.
We established the conserved role of C. elegans GCN-2 as an eIF2α kinase under nutrient stress and showed that the mRNA of atf-5, encoding a worm homolog of yeast GCN4 and mammalian ATF4 transcription factors, is under translational control by GCN-2. In normal culture conditions, deletion of gcn-2 was dispensable for growth, fertility, and lifespan of worms. However, under amino acid limitation, loss of gcn-2 shortened lifespan, supporting its function under nutrient deprivation. Furthermore, gcn-2 deletion decreased the long lifespan of nutrient-responsive worms, such as eat-2 mutants, a genetic model of DR (Lakowski & Hekimi, 1998), or RNAi-treated worms for let-363/TOR, and its downstream target rsks-1/S6K. We have revealed a novel role of GCN-2 in modulating pha-4/FoxA expression under conditions of TOR inactivation or amino acid deprivation. PHA-4/FoxA transcription factor is a master regulator of organ development, but is also required for survival of larvae under starvation and DR-induced longevity in adults (Hansen et al., 2007; Panowski et al., 2007; Sheaffer et al., 2008; Zhong et al., 2010). Thousands of genes are candidate PHA-4 targets, with diverse roles in many biological processes and preferentially bound in specific conditions (Zhong et al., 2010).
Under DR, PHA-4 activity regulates the expression of many autophagy-related genes or certain sod genes, encoding superoxide dismutase isoforms (Morck & Pilon, 2006; Panowski et al., 2007; Hansen et al., 2008). Here we showed that the induction of such stress-responsive genes in let-363(RNAi)-treated worms was dependent on GCN-2, consistent with the lower induction of pha-4 in gcn-2 mutants, relative to N2. Accordingly, gcn-2 deletion rendered wild-type, eat-2, or TOR-deficient worms more sensitive to sodium arsenite-induced oxidative stress, compared to the GCN-2 proficient animals. Also gcn-2 worms were more susceptible to other stresses, such as heat shock and UV irradiation. Lately, it was shown that activation of GCN-2 signaling protects cells during mitochondrial and hypertonic stress (Baker et al., 2012; Lee & Strange, 2012). A common cellular response to stress is the inhibition of global translation and induction of specific translational/transcriptional programs to maintain their intracellular homeostasis. GCN-2 has a central role in this response and participates in stress management by activating key transcription factors, such as ATF4 and NF-kB in mammals (Wek et al., 2006). This stress-induced reprogramming would also determine lifespan. A role of the yeast GCN4 in the lifespan extension by mutations in TOR, S6K, and 60S subunits has been reported (Steffen et al., 2008). Although GCN-2 increases atf-5 synthesis under amino acid limitation, loss of atf-5 did not recapitulate the effect of gcn-2 deletion in stress survival and lifespan. We speculate that ATF-5 has either more specific roles not related to the longevity mechanisms in worms or redundant function(s) with other transcription factors (e.g., PHA-4), so worms can compensate for its loss.
Overall, we demonstrated the central role of worm GCN-2 in stress response and revealed that the PHA-4 transcription factor is part of the GCN-2 signaling in response to nutrient and oxidative stress. Although longevity is not always linked to increased stress resistance and overexpression of antioxidants genes does not necessary lead to longevity, such an association could rely on specific genetic or environmental backgrounds. Studies in yeast, worms, flies, and mice suggest that an induction of oxidative stress and mitochondrial respiration may be a mechanism of DR-induced longevity (Ristow & Zarse, 2010; Pan et al., 2011). In Drosophila, SOD1 is required for lifespan extension by protein restriction only when sugar level is high (Sun et al., 2012). Conversely, genetic data in C. elegans support that oxidative stress is uncoupled from aging, and deletions of individual or combinations of antioxidant genes did not reduce the lifespan of N2, eat-2, or ISS mutants (Van Raamsdonk & Hekimi, 2010). This might be due to their small contribution on the whole adaptive response that determines longevity. This adaptive response should involve a coordinated function of several genes and pathways, regulated by transcription factors with a broad cellular role.
The function of PHA-4 in promoting survival of L1 larvae under starvation is dictated by the expression of several targets, mostly related to stress defense and metabolic processes (Zhong et al., 2010). It is likely that the function of PHA-4 in the modulation of adult lifespan extension under DR entails similar targets and cellular processes. Increase in antioxidant defense and catabolic processes such as autophagy protect cells from the accumulation of damaged proteins or organelles. On the other hand, metabolic alterations induced by mitochondrial function or regulators of fatty acid metabolism could significantly modulate longevity. In yeast, activation of Rim15 kinase by nutrient depletion positively regulates the stress-responsive transcription factors Msn2/4 and Gis1 to protect cells and increase lifespan under DR and TOR inhibition (Powers et al., 2006; Wei et al., 2009). This is accomplished through the induction of stress defense genes and by switching metabolism from respiration to glycolysis and glycerol synthesis, generating a DR-like environment that enhances stress survival and lifespan (Wei et al., 2009). Our work provides the first genetic evidence that GCN-2 activity can influence the outcome of DR effect on lifespan by modulating the TOR/S6K signaling and its downstream transcription factor PHA-4. GCN-2 signaling positively regulates the induction of PHA-4 under nutrient or oxidative stress, by a yet unidentified mechanism. The next challenge will be to identify the downstream targets of TOR pathway that are controlled by GCN-2 and contribute to stress resistance and longevity.
Experimental procedures
Caenorhabditis elegans strains and culture
Standard methods of culturing and handling worms were used. Worms were raised on NGM plates seeded with E. coli OP50 as food. For tunicamycin treatments, L4 larvae were transferred on NGM plates with tunicamycin (5 μg/mL) for 24 h. See Table S1 (Supporting information) for all strains used in this study. Wild-type Bristol N2 and single mutant strains were provided by the Caenorhabditis Genetics Center (CGC, University of Minnesota, Minneapolis, MN, USA). The gcn-2(ok871), gcn-2(ok886), and atf-5(ok576) were outcrossed four times with the N2, and the relevant mutations were tracked in F2 progeny by PCR. For genotyping the mutants, primers G1/G2 and G1/G3 (Table S2) were used for both gcn-2 alleles vs. N2, and for atf-5(ok576), the set of primers A1/A2 (Table S2). The double mutant eat-2(ad465);gcn-2(ok871) was made by crossing the desired strains and selecting F2 progeny by PCR for carrying both mutations. To track the ad465 point mutation in eat-2, a PCR fragment using primers E1/E2 (Table S2) was digested with the restriction enzymes BamHI-SfcI. Transgenic animals were generated by microinjection of plasmid DNAs into the gonad of young adult N2 worms, using rol-6(su1006) as cotransformation marker. BRF144 was obtained by genetic cross of BRF140 with BRF162 males (Table S1).
Constructs
RNAi plasmids were constructed by inserting gene-specific PCR product, amplified from genomic DNA using the relevant primers (Table S2), into the RNAi feeding vector pL4440 (Andy Fire Kit 1999, Addgene plasmid 1654, Cambridge, MA, USA). For eIF2α(RNAi) plasmid, a KpnI digest fragment from pHSG399-eIF2α plasmid (provided by Dr. Shin Takagi (Nukazuka et al., 2008)) was subcloned to pL4440. RNAi clone for let-363 and rsks-1 was previously described (Syntichaki et al., 2007). For the intact atf-5::gfp transgene, a genomic PCR fragment with the A1/A4 primers (Table S2) containing the 1612 bp sequence upstream of the atf-5 coding region, and the 1413 bp atf-5 coding region (including uORFs) was cloned into the pPD95.77 vector (Andy Fire Kit 1995, Addgene plasmid 1494, Cambridge, MA, USA) in fusion with gfp at the C’-terminal. For the uORF-less atf-5::gfp, we first cloned the promoter region of atf-5, amplified with the set of primers A1/A2 (Table S2), into the pPD95.77 vector, and in this plasmid (Patf-5::GFP), we added the coding region of atf-5 without uORFs, amplified with the primers A5/A4 (Table S2).
Western blot analysis
Worms of each strain, grown on OP50 or RNAi plates at 20 °C, were collected in M9 buffer when just reaching adulthood. After 2–3 washes to remove bacteria, they were frozen in ethanol dry ice and, before loading onto SDS-PAGE, worm pellets were boiled in 50 μL 2X SDS-sample buffer for 10 min. Primary antibodies were a polyclonal antibody raised against worm eIF2α, a kind gift from Dr. Shin Takagi (Nukazuka et al., 2008), for total (T)-eIF2α and an anti-phospho-eIF2α antibody (Santa Cruz Biotechnology, sc-101670) for phosphorylated (P)-eIF2α levels. A secondary anti-rabbit IgG antibody (HRP) was used for immunoblot signal detection with ECL (Thermo Fisher Scientific Inc., Waltham, MA, USA). Quantification of immunoblot signals was performed using ImageJ software (ImageJ, U. S. National Institutes of Health, Bethesda, MD, USA). Ratio of P- to T-eIF2α levels was measured in two independent experiments.
RNA interference
The RNAi clones, expressing dsRNA from the indicated genes in HT115(DE3) E. coli bacteria, were grown with ampicillin (50 μg/mL) and tetracycline (10 μg/mL) in LB medium. On the following days, fresh cultures with ampicillin were induced with 0.25 or 1 mm isopropylb-D-thiogalactopyranoside (IPTG) and seeded on RNAi plates. Bacteria carrying the empty vector (pL4440) and treated likewise were used as control cultures.
Microscopy
The expression pattern of transgenic worms was monitored by mounting sodium azide- or levamizole-treated animals on 2% agarose pads, on glass microscope slides. Animals were imaged either under fluorescence microscope or using a Leica TCS SP5 confocal imaging system. Images shown from confocal are 2D maximal projections of z-stacks.
RNA isolation and quantitative reverse transcription PCR
Total RNA was prepared from frozen worm pellets, of the indicated genetic backgrounds and developmental stages, using a NucleoSpin RNA XS kit (Macherey–Nagel, Dueren, Germany) and measured by Quant-iT RNA assay kit (Invitrogen, Molecular Probes, Eugene, OR, USA). Total RNA was reverse transcribed with iScript™ cDNA synthesis kit (Biorad, Hercules, CA, USA), and quantitative PCR was performed using the SsoFast™ EvaGreen supermix (BioRad) in the MJ MiniOpticon system (BioRad). The relative amounts of mRNA were determined using the comparative Ct method for quantification, and the gene expression data are presented as the fold change in each strain relative to N2. qRT–PCR was performed in at least two independent samples in triplicates, and each sample was independently normalized to endogenous reference ama-1. The mean ± the standard deviation (SD) of at least two independent experiments is presented. The sequence of primers used for qRT–PCR is available upon request.
Lifespan assays
Lifespan assays were conducted as described previously (Syntichaki et al., 2007). Briefly, L4 larvae of each strain were transferred to NGM plates or RNAi plates, at the assay temperature. Animals were transferred every two days to fresh plates and were daily scored for surviving worms. Animals that failed to respond to stimulation by touch were referred as dead, whereas that bagged, exploded, or crawled off the plates were referred as censored in the analysis. Day 0 of adulthood was defined the day that the L4s were transferred to plates. Lifespan and statistical analysis were performed using GraphPad Prism, version 5 (GraphPad Software, San Diego, CA, USA). Each population is compared with the appropriate control population using the log-rank test.
Fertility assay
Worms of each genotype were grown at 20 °C, and 5–10 L4 hermaphrodites were placed on individual NGM plates to lay eggs. Animals were transferred daily to fresh plates until egg-laying ceased and the hatched progeny were counted in each plate. The total number of progeny per worm (brood size) was counted for each genotype, and the average brood size (mean ± SD) of each strain was plotted. Unpaired t-test was used to calculate P-values in GraphPad Prism 5.
Stress resistance assays
For heat-shock assays, 1-day-old worms were shifted at 35 °C for 6 h. After one night of recovery at 20 °C, the percentage of worms surviving was determined. For UV resistance assays, 5-day adults were irradiated to NGM plates without bacteria at 0.2J/cm2 and then were transferred to NGM plates with food at 20 °C. Three days later, the percentage of worms surviving was determined. For oxidative stress, 1-day adults of each strain were transferred on NGM plates with 3 mm (for eat-2 and eat-2;gcn-2) or 5 mm (for N2 and gcn-2) sodium arsenite, and the percentage of worms surviving was determined after 20 h or 48–58 h at 20 °C, respectively. The average (mean ± SD) of at least three independent experiments with ∼100 individual for each strain per experiment was plotted. Unpaired t-test was used to calculate P-values in GraphPad Prism 5.
Acknowledgments
P.S. dedicates this work to the memory of her mentor and life companion George Thireos, for his continuous encouragement, inspiration, and support all these years. We are grateful to Dr. Shin Takagi of Nagoya University for providing the anti-eIF2α antibody and pHSG399-eIF2α construct. We thank the BIU of BRFAA for using the confocal system and all the members of the P.S. laboratory for technical assistance and helpful discussion. We would like to acknowledge Irene Topalidou, Dimitris Stravopodis, Antony Gavalas, George Diallinas, and Dimitris Thanos for their critical reading and suggestions on this manuscript. Some strains were provided by the Caenorhabditis Genetic Center, which is funded by the National Institutes for Health National Center for Research Resources (Minneapolis). The research leading to these results has received funding from the European Research Council under the European Union’s Seventh Framework Program (FP/2007-2013)/ERC Grant Agreement n. [201975].
Author contributions
PS and GT conceived and designed the experiments; AR, ArV, AnV, SP, and PS designed/performed experiments and analyzed the data; PS wrote the manuscript.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site.
Fig. S1 Inactivation of leucyl-tRNA synthetase lrs-1 induces phospho-eIF2α levels in a gcn-2-dependent manner.
Fig. S2 Induction of atf-5::gfp transgene by eIF2α(RNAi) or tunicamycin does not require GCN-2 activity.
Fig S3 gcn-2 deletion does not alter fertility in wild-type or eat-2 mutants.
Fig. S4 Loss of GCN-2 activity sensitizes animals to amino acid limitation.
Fig. S5. RNAi efficiency is not affected in gcn-2 mutant worms.
Fig. S6 GCN-2 regulates the induction of PHA-4 target genes in eat-2 mutants.
Fig. S7 Inactivation of gcn-2 affects the induction of a Ppha-4::gfp reporter under oxidative stress.
Fig. S8 Loss of gcn-2 but not atf-5 increases the stress sensitivity of worms.
Table S1 List of the strains used in this study.
Table S2 List of primers used for plasmid construction in this study.
Table S3 Summary of data from independent repeats of lifespan experiments.
References
- Anderson LL, Mao X, Scott BA, Crowder CM. Survival from hypoxia in C. elegans by inactivation of aminoacyl-tRNA synthetases. Science. 2009;323:630–633. doi: 10.1126/science.1166175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993;133:897–917. doi: 10.1093/genetics/133.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha Kinase GCN-2. PLoS Genet. 2012;8:e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer A, Juelich T, Vanslambrouck JM, Tietze N, Solomon PS, Holst J, Bailey CG, Rasko JE, Broer S. Impaired nutrient signaling and body weight control in a Na+ neutral amino acid cotransporter (Slc6a19)-deficient mouse. J. Biol. Chem. 2011;286:26638–26651. doi: 10.1074/jbc.M111.241323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003;17:859–872. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu M, Albert V, Hall MN. mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 2013;23:1–10. doi: 10.1016/j.gde.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Harries LW, Fellows AD, Pilling LC, Hernandez D, Singleton A, Bandinelli S, Guralnik J, Powell J, Ferrucci L, Melzer D. Advancing age is associated with gene expression changes resembling mTOR inhibition: evidence from two human populations. Mech. Ageing Dev. 2012;133:556–562. doi: 10.1016/j.mad.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Obata T, Ota K, Sasaki T, Ito T. Rapamycin-induced translational derepression of GCN4 mRNA involves a novel mechanism for activation of the eIF2 alpha kinase GCN2. J. Biol. Chem. 2003;278:20457–20460. doi: 10.1074/jbc.C300133200. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl Acad. Sci. U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, Gelino S, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Strange K. GCN-2 dependent inhibition of protein synthesis activates osmosensitive gene transcription via WNK and Ste20 kinase signaling. Am. J. Physiol. Cell Physiol. 2012;303:C1269–C1277. doi: 10.1152/ajpcell.00294.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurin AC, Chaveroux C, Lambert-Langlais S, Carraro V, Jousse C, Bruhat A, Averous J, Parry L, Ron D, Alliot J, Fafournoux P. The amino acid sensor GCN2 biases macronutrient selection during aging. Eur. J. Nutr. 2012;51:119–126. doi: 10.1007/s00394-011-0205-4. [DOI] [PubMed] [Google Scholar]
- Morck C, Pilon M. C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev. Biol. 2006;6:39. doi: 10.1186/1471-213X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukazuka A, Fujisawa H, Inada T, Oda Y, Takagi S. Semaphorin controls epidermal morphogenesis by stimulating mRNA translation via eIF2alpha in Caenorhabditis elegans. Genes Dev. 2008;22:1025–1036. doi: 10.1101/gad.1644008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Passtoors WM, Beekman M, Deelen J, van der Breggen R, Maier AB, Guigas B, Derhovanessian E, van Heemst D, de Craen AJ, Gunn DA, Pawelec G, Slagboom PE. Gene expression analysis of mTOR pathway: association with human longevity. Aging Cell. 2013;12:24–31. doi: 10.1111/acel.12015. [DOI] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: the concept of mitochondrial hormesis (mitohormesis) Exp. Gerontol. 2010;45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Rogers AN, Chen D, McColl G, Czerwieniec G, Felkey K, Gibson BW, Hubbard A, Melov S, Lithgow GJ, Kapahi P. Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab. 2011;14:55–66. doi: 10.1016/j.cmet.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr. Biol. 2008;18:1355–1364. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol. Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Komatsu T, Lim J, Laslo M, Yolitz J, Wang C, Poirier L, Alberico T, Zou S. Nutrient-dependent requirement for SOD1 in lifespan extension by protein restriction in Drosophila melanogaster. Aging Cell. 2012;11:783–793. doi: 10.1111/j.1474-9726.2012.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Thireos G. Evidence that the GCN2 protein kinase regulates reinitiation by yeast ribosomes. EMBO J. 1988;7:3547–3551. doi: 10.1002/j.1460-2075.1988.tb03231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S. Reactive oxygen species and aging in Caenorhabditis elegans: causal or casual relationship? Antioxid. Redox Signal. 2010;13:1911–1953. doi: 10.1089/ars.2010.3215. [DOI] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl Acad. Sci. U S A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Fabrizio P, Madia F, Hu J, Ge H, Li LM, Longo VD. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 2009;5:e1000467. doi: 10.1371/journal.pgen.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HY, Preston E, Slightham C, Hillier LW, Brock T, Agarwal A, Auerbach R, Hyman A, Gerstein M, Mango SE, Kim SK, Waterston RH, Reinke V, Snyder M. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 2010;6:e1000848. doi: 10.1371/journal.pgen.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Inactivation of leucyl-tRNA synthetase lrs-1 induces phospho-eIF2α levels in a gcn-2-dependent manner.
Fig. S2 Induction of atf-5::gfp transgene by eIF2α(RNAi) or tunicamycin does not require GCN-2 activity.
Fig S3 gcn-2 deletion does not alter fertility in wild-type or eat-2 mutants.
Fig. S4 Loss of GCN-2 activity sensitizes animals to amino acid limitation.
Fig. S5. RNAi efficiency is not affected in gcn-2 mutant worms.
Fig. S6 GCN-2 regulates the induction of PHA-4 target genes in eat-2 mutants.
Fig. S7 Inactivation of gcn-2 affects the induction of a Ppha-4::gfp reporter under oxidative stress.
Fig. S8 Loss of gcn-2 but not atf-5 increases the stress sensitivity of worms.
Table S1 List of the strains used in this study.
Table S2 List of primers used for plasmid construction in this study.
Table S3 Summary of data from independent repeats of lifespan experiments.


