Abstract
Rationale
Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry combined with isotope labeling methods are effective for protein and peptide quantification, but limited in their multiplexing capacity, cost-effectiveness and dynamic range. This study investigates MALDI MS-based quantification of peptide phosphorylation without labeling, and aims to overcome the shot-to-shot variability of MALDI using a mathematical transformation and extended data acquisition times.
Methods
A linear relationship between the reciprocal of phosphopeptide mole fraction and the reciprocal of phosphorylated-to-unphosphorylated signal ratio is derived, and evaluated experimentally using three separate phosphopeptide systems containing phosphorylated serine, threonine and tyrosine residues: mixtures of phosphopeptide and its des-phospho-analog with known stoichiometry measured by vacuum MALDI-linear ion trap mass spectrometry and fit to the linear model. The model is validated for quantifying in vitro phosphorylation assays with inhibition studies on Cdk2/cyclinA.
Results
Dynamic range of picomoles to femtomoles, good accuracy (deviations of 1.5–3.0% from expected values) and reproducibility (RSD = 4.3–6.3%) are achieved. Inhibition of Cyclin-dependent Kinase phosphorylation by the classical inhibitors olomoucine and r-roscovitine was evaluated and IC50 values found to be in agreement with reported literature values. These results, achieved with single-point calibration, without isotope or chromatography, compare favorably to those arrived at using isotope dilution (p > 0.5 for accuracy).
Conclusion
The mathematical relationship derived here can be applied to a method that we term Double Reciprocal Isotope-free Phosphopeptide Quantification (DRIP-Q), as a strategy for quantification of in vitro phosphorylation assays, the first MALDI-based, isotope- and calibration curve-free method of its type. These results also pave the way for further systematic studies investigating the effect of peptide composition and experimental conditions on quantitative, label-free MALDI.
INTRODUCTION
Protein phosphorylation, the catalytic transfer of phosphate from ATP to a protein substrate by protein kinases, globally regulates cellular processes. Phosphorylation, one of the most important post-translational modifications, has been estimated to occur at 30–50% of the eukaryotic proteome as a switch of cellular functions [1]. Changes in phosphorylation of specific sites affect cellular signaling changes. Many methodologies have been used to identify and quantify phosphorylation, such as western blotting, radioactive phosphate labeling, flow cytometry, and mass spectrometry [2–4]. Mass spectrometry (MS)-based techniques have become an increasingly important tool in phosphorylation analysis, in which a signal corresponding to the phosphate moiety can be detected on the mass spectrum. MS is advantageous because it has the capability to readily determine site-specificity, and does not require the generation of sequence-specific antibodies.
Two ionization techniques, electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI), are generally used in protein mass spectrometry [5]. For most phosphorylation and phosphoproteomic analyses, ESI-MS must be coupled to liquid chromatography to simplify the complex spectra that arise from mixtures of peptides with multiple charge states. MALDI-MS has also been extensively used in phosphorylation analysis, and has complementary strengths to ESI. MALDI-MS requires only simple sample preparation, is generally fast and easy to use, and yields MS spectra that ordinarily do not require chromatographic separation. However, MALDI-MS is inherently less quantitative due to unpredictable differences in ionization efficiency. Heterogeneities in analyte/matrix co-crystallization, such as non-uniform analyte distribution within the crystals, variable thickness of the crystal layer, and differences in crystal size, can lead to significant changes in the absolute ion intensity, both within an individual sample crystal, and between different crystals [6].
To minimize these problems for phosphorylation quantification, isotope derivative methods are required in quantitative proteomic MALDI (as well as many ESI) approaches. Such methods take advantage of the fact that isotopomers respond identically under MS conditions, but are able to be separated by mass to charge ratios [7–12]. While effective, such approaches suffer from high cost, time-consuming analysis, requirement for specialized reagents, and dynamic range limitations [13–16].
Label-free strategies have been increasingly prominent as alternatives for quantitative proteomics due to their lower cost and convenience. Liquid chromatography MS (LC-MS) has been one of the fundamental techniques in the label-free relative-quantitative proteomics [17–19]. Chromatographic retention peaks can be integrated with external calibration to measure peptide abundance [20]. However, the chromatographic task is time-consuming and may suffer from interference problems. Mobile phase conditions must also be carefully controlled to avoid changing the solubility and ionization of the peptide analytes.
As an alternative to chromatography based label-free methods, label-free MALDI-MS methods are in rising demand for high-throughput proteomic analysis. The inherent limitations in reproducibility of MALDI-MS can at least in part be overcome by increasing the signal acquisition time. The signal from a single spot can be interrogated for several minutes to average the signal from thousands of laser shots, as compared to the smaller number of scans generally available when limited by a chromatographic time scale in ESI-LC-MS. Kinumi et al. has successfully measured phosphopeptides derived from the phosrestin protein by using a label-free MALDI approach with calibration construction by plotting signal ratios versus peptide amount ratios, quantifying the ratio of phosphopeptide to total peptide [21]. The calibration curve showed a linear correlation between the peak ratios and amount ratios. In other cases, the unphosphorylated and phosphorylated substrates have different ionization efficiencies, resulting from their differing physicochemical properties. For example, a study by Parker, et al. demonstrated non-linear relationships between signal ratios and amount ratios for seven different peptides using MALDI ionization [6]. This may be due to differential ionization effects between the different peptides.
In this study, we derive a double reciprocal transformation that accounts for differences in ionization efficiencies of analytes, allowing for the direct comparison of unphospho- and phosphopeptide signal in a linear fashion, and show experimentally that the linear relationship holds for three individual phosphopeptide systems containing phosphorylated serine, threonine and tyrosine in MALDI-linear ion trap mass spectrometry. Based on these results, we propose a MALDI-MS method for quantification of phosphorylation stoichiometry, termed Double Reciprocal Isotope-free Phosphopeptide Quantification (DRIP-Q). This method uses a single point calibration that does not require isotope labeling or chromatography and is shown to be robust and reproducible over 2 orders of magnitude in concentration levels for phosphorylation assays. The results obtained with this method are comparable to those obtained using traditional stable isotope labeling. Using simple mixtures of peptides to limit ion suppression effects, this study serves as proof of principle that MALDI-based label-free methods are feasible for quantitative phosphorylation analysis. To our knowledge, this is the first report of a calibration-curve-free MALDI-MS method for the determination of phosphorylation.
EXPERIMENTAL
Materials
HPLC grade water, acetonitrile, and formic acid were purchased from Fisher Scientific (Pittsburg, PA, USA). Synthetic peptides, AAAAYRAAR, LRWGFTTPDKKHQKEPPF, and RQSVELHSPQSLPR, were used for the quantitative method development. The peptide, AAAAYRAAR, and its phosphorylated form, AAAApYRAA, and their [13C3]-alanine (labels on the third alanine residue) labeled forms, were synthesized and purified by University of Utah DNA/peptide facility and analyzed by quantitative amino acid analysis. The HIV-RT peptides, LRWGFTTPDKKHQKEPPF, containing a CDK phosphorylation consensus sequence (S/T)PXR/K, and its phosphorylated form, LRWGFTpTPDKKHQKEPPF, were synthesized in house using standard fmoc-based solid-phase peptide chemistry. The Aquaporin peptide, RQSVELHSPQSLPR, and its two phosphorylated forms at S256 and S261, RQpSVELHSPQSLPR, and RQSVELHpSPQSLPR, were purchased from Anaspec (Fremont, CA).
MALDI-MS quantitative analysis
Peptide standards, AAAAYRAAR (peptide 1), LRWGFTTPDKKHQKEPPF (peptide 2), and RQSVELHSPQSLPR (peptide 3), and their phosphopeptides were selected as three different model systems to establish the label-free MALDI-MS quantitative method. 1 µL of the sample mixture of the unphospho- and phosphopeptide standards with various phosphorylation fractions was mixed with 9 µL of matrix solution. Then, 1 µL of the peptide mixture with the matrix was spotted on a MALDI plate prior to MALDI-MS analysis. MALDI-MS experiments were carried out on a Thermo LTQ XL linear ion trap mass spectrometer (Thermo Scientific) was equipped with a MALDI ion source utilizing a nitrogen laser (337 nm) firing at 60 Hz. α-Cyano-4-hydroxycinnamic acid (CHCA) was purchased from Sigma-Aldrich (St. Louis, MO, USA) and 2,5-Dihydroxybenzoic acid (DHB) was purchased from Acros Organics (Morris Plains, NJ, USA) and used without further purification. A saturated aqueous solution of a-Cyano-4-hydroxycinnamic acid (CHCA) (Sigma-Aldrich, St. Louis, MO, USA) in 0.1 % trifluoroacetic acid/acentonitrol (30/70, v/v) or 2,5-Dihydroxybenzoic acid (DHB) (Acros Organics Morris Plains, NJ, USA) mixed with water / acetonitrile (50/50, v/v) in a 1:4 ratio (v/v) was used as a matrix solution. CHCA was utilized for peptide 1, and DHB for all other experiments. Full scan mass spectra were acquired in positive mode by 400 scans with automated gain control (AGC) set at 30,000 counts with automated spectrum filter (ASF) off, 3 microscan/step, and processed by XcaliburTM 2.0.7 software. Laser energies between 7.6 and 13 µJ were utilized according to instrument settings. Quantitative data was processed manually for the most intense isotope of each peptide; similar results were obtained using either peak heights or integrated peak areas.
In vitro Phosphatase reaction
Dephosphorylated peptide AAAAYRAAR was generated enzymatically.The phosphopeptide (AAAApYRAAR) was applied to the phosphatase reaction, which was performed in a final volume of 50 µL, containing 25mM of Ammonium bicarbonate buffer, 4 Units of Alkaline Phosphatase (Roche), and 1.56 nmol of the phosphopeptide substrate at 30 °C for 1 h. The reaction was quenched by adding 20% acetic acid into the reaction sample, and heating it for at 90 °C for 10 min. A control (inactivated phosphatase) experiment was carried out by adding 20 µL of 20% acetic acid and heating at 90 °C for 10 min before adding the substrate, so that the phosphopeptide remained intact. This procedure allowed us to generate solutions of phosphopeptide and corresponding des-phosphopeptide of equal concentration in identical chemical milieus. The reaction and control samples were diluted and mixed in different phosphorylation fractions and subjected to MALDI-MS analysis.
Isotope dilution MALDI-MS
[13C3]-alanine labeled unphosphopeptide U* (AAA*AYRAAR) and labeled phosphopeptide P* (AAA*ApYRAAR) phosphorylated forms served as internal standards. The labeled standards were mixed with the unlabeled standards (U and P) in 1:1 ratio with the amount ranging from 0.1 –20 pmol/µL and analyzed by MALDI-LTQ. The amount of P and U in unknown samples was determined by using calibration methodology. Calibration curves were constructed for each labeled pair by plotting signal ratios of labeled to unlabeled peptides versus their amount ratios (i.e. the signal ratios of P* to P versus their amount ratios; the signal ratios of U* to U versus their amount ratios) [5].
In vitro kinase assays
To investigate CDK inhibitor activity, CDK2/CyclinA was assayed using 1.4 µM HIV-1 RT peptide LRWGFTTPDKKHQKEPPF in the presence of 2 mM ATP and 1× NEBuffer (50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, and 1 mM DTT, New England BioLabs Inc.) at 30°C for 15 min with the addition of CDK inhibitors, Roscovitine at a concentration of 0.1 µM to 5 µM or Olomoucine, at a concentration of 1 µM to 50 µM. A control experiment was performed with no inhibitor addition. Following the incubation, 1 µL of each sample was added into 9 µL of the matrix solution for MALDI analysis. IC50 values are calculated by nonlinear regression analysis using GraphPad Prism (version 6 Prism Software Inc.).
Results and Discussion
MS is a fast, high-throughput, and probe-free method for phosphorylation quantification. Both HPLC-ESI-MS and MALDI-MS can be used for phosphorylation quantification. HPLC-ESI-MS is considered to be more reproducible for phosphorylation quantification than MALDI-MS, even though MALDI is advantageous due to its analytical speed and accommodation of more complex samples in the absence of additional separations. When used with isotope dilution quantitative strategy, MALDI-MS shows sufficient precision and reproducibility for peptide quantification, whereas isotope-free MALDI methods still face some challenges. For instance, phosphorylation may cause a change of ionization efficiency, which will result in non-linearity between the signal ratios and actual mole ratios. This condition has been previously observed in calibration curves for isotope-free MALDI-MS [6]. To resolve this problem requires that differences in ionization efficiency between the unphosphorylated and phosphorylated peptide be accounted for. Here, we propose a new mathematic approach to find a linear correlation between signals and actual amounts for phosphorylation quantification. This method factors in ionization efficiency differences, and allows for the quantification of phosphorylation without calibration curves, stable isotopes, or chromatography.
Mathematic Approach for phosphorylation quantification
The approach proposed here accounts for possible differences in ionization efficiencies for a phosphopeptide (P) and its unphosphorylated analog (U). It requires the assumption (which we later confirm experimentally) that signal intensity is directly proportional to sample concentration, where the proportionality constant is defined as the ionization efficiency, I. Therefore,
| (Eq. 1a) |
| (Eq. 1b) |
where S, I and M represent observed mass spectral signal intensity, ionization efficiency and amount in moles, and the subscript P or U indicates the phosphopeptide or its unphosphorylated analog.
Eqs. 1a and 1b can be combined to give:
| (Eq. 2a) |
where (m) is the mole fraction of phosphopeptide P, and (1−m) is the mole fraction of U. For example, if m =0.1, 10% of the total peptide is phosphorylated.
Letting the signal ratio, , and the instrumental response factor ratio , the equation becomes:
| (Eq. 3b) |
Converting S = f(m) into m = f(S), yields the hyperbolic relationship:
| (Eq. 3c) |
Taking the reciprocal of both sides of equation gives
| (Eq. 3d) |
A plot of versus yields a straight line with a y-intercept of 1 and a slope of a. Therefore, once the response factor ratio, a, has been determined, the phosphorylation fraction m can be calculated by using the observed signal intensity ratio S of phospho- to unphospho-peptide. This method only requires determination of a and S, and therefore avoids time-consuming calibration curve construction.
Consistency of instrument response factor, a
For Eq. 3d to hold, the response factor ratio, a, must be constant. To investigate the linear correlation between the reciprocals of the phosphorylation percentages and signal ratios in Eq. 3d, the response factor ratio was tested for three peptide models over a range of phosphopeptide to unphosphosphopeptide concentration ratios. The three models (sequences given in Table 1) include peptides that may be phosphorylated at tyrosine, threonine or serine.
Table 1.
Instrument response factor (a) at different phosphorylation fractions(m)
| Instrumental Response Factor Ratio, α* | |||||
|---|---|---|---|---|---|
| ma | 1b | 2c | ma | 3ad | 3bd |
| 0.10 | 0.670 ± 0.027 | 0.189 ± 0.016 | 0.32 | 0.723 ± 0.034 | 1.032 ± 0.073 |
| 0.20 | 0.671 ± 0.029 | 0.176 ± 0.014 | 0.38 | 0.716 ± 0.015 | 1.017 ± 0.074 |
| 0.30 | 0.672 ± 0.037 | 0.177 ± 0.014 | 0.43 | 0.691 ± 0.038 | 1.062 ± 0.105 |
| 0.40 | 0.634 ± 0.009 | 0.180 ± 0.006 | 0.48 | 0.731 ± 0.042 | 1.022 ± 0.045 |
| 0.50 | 0.669 ± 0.005 | 0.189 ± 0.014 | 0.54 | 0.763 ± 0.047 | 1.006 ± 0.095 |
| 0.60 | 0.639 ± 0.032 | 0.186 ± 0.017 | 0.63 | 0.732 ± 0.024 | 1.023 ± 0.086 |
| 0.70 | 0.658 ± 0.056 | 0.182 ± 0.008 | 0.74 | 0.711 ± 0.038 | 1.107 ± 0.034 |
| 0.80 | 0.600 ± 0.023 | 0.199 ± 0.017 | 0.91 | 0.787 ± 0.086 | 1.107 ± 0.131 |
| Average | 0.652 | 0.184 | 0.733 | 1.047 | |
| R.S.D.e | 3.94% | 4.32% | 4.15% | 3.86% | |
phosphorylation fraction
peptide 1 and phosphopep. 1 (AAAAYRAAR, AAAApYRAA) with total amount of peptide = 3.1 pmol.
peptide 2 and phosphopep.2 (LRWGFTTPDKKHQKEPPF, RWGFTpTPDKKHQKEPPF) with total amount of peptide = 1.4 pmol
peptide 3, phosphopeptide 3a, phosphopeptide 3b (RQSVELHSPQSLPR, RQpSVELHSPQSLPR, RQSVELHpSPQSLPR) with total amount of peptide = 0.1 pmol
n = 3
peptide 1 analyzed using CHCA matrix; peptides 2 and 3 analyzed using DHB matrix. Laser energy for CHCA = 7.6 µJ, and for DHB = 11 µJ
For each model, the phosphopeptide and unphosphopeptide were mixed at nine different mole fractions of phosphopeptide (m), and the response factor ratio a was determined for each mixture from the observed mass spectrometric signal intensity ratio S. The response factor ratio is found to be consistent across the range of concentrations for each model (Table 1, data shown in supplemental figure S1; similar results are obtained when peak heights or integrated peak areas are used), with RSDs for a varying from 3.86 % to 4.32 % for the three models, confirming the linear correlation between and in Eq. 3d. These results demonstrate the potential for applying Eq. 3d to quantification of phosphorylation by isotope-free MALDI-MS.
Effect of laser energy on phosphorylation quantification
MALDI mass spectrometric signal response may vary with laser intensity. Changes in the signal response might further affect the signal ratios (S) and the consistency of a values in Eq. 3d. For example, low laser intensity may lead to low signal response due to the deficiency in sample ionization, whereas excessively high laser intensity may produce a high signal background, lowering the signal to noise ratio. To investigate the effect of laser intensity on the constant a in Eq. 3d, peptide 2 in its unphospho- and phosphoforms were mixed at various phosphorylation fractions (m), and were analyzed by MALDI with laser energies of 8 and 13 µJ. For each mixture, the response factor ratio a value was calculated from the observed signal ratio (S) and the known phosphorylation fraction (m). Average a values were 0.231 ± 5.70% RSD and 0.158 ± 4.44% RSD for 8 and 13 µJ, respectively (Table 2). These results indicate that changes in laser intensity can differentially affect the ionization efficiencies of the unphospho- and phosphopeptides, but the ionization efficiency ratio a is consistent with a small RSD at a constant laser energy across a wide range of phosphorylation fractions. As a result, care should be taken to ensure that the same laser intensity is applied to all samples in an experiment; over longer periods of time, from several weeks to months, actual laser output may drift and re-calibration may become important.
Table 2.
Effect of MALDI laser energy on instrument response factor (a) and accuracy
| 8 µJ | 13 µJ | |||||
|---|---|---|---|---|---|---|
| ma | Response factor ratio, a |
mc (observed) |
% error | Response factor ratio, a |
mc (observed) |
% error |
| 0.090 | 0.226 ± 0.019b | 0.088 | −2.62 | 0.157 ± 0.008 | 0.089 | −1.04 |
| 0.110 | 0.215 ± 0.015 | 0.102 | −7.14 | 0.154 ± 0.004 | 0.107 | −3.17 |
| 0.140 | 0.217 ± 0.013 | 0.131 | −6.15 | 0.157 ± 0.014 | 0.139 | −0.74 |
| 0.200 | 0.231 ± 0.010 | 0.200 | −0.23 | 0.148 ± 0.005 | 0.188 | −6.16 |
| 0.330 | 0.231 ± 0.012 | 0.330 | −0.08 | 0.157 ± 0.010 | 0.328 | −0.60 |
| 0.670 | 0.244 ± 0.016 | 0.684 | 2.10 | 0.164 ± 0.002 | 0.680 | 1.45 |
| 0.800 | 0.255 ± 0.011 | 0.818 | 2.21 | 0.172 ± 0.007 | 0.815 | 1.82 |
| Average 0.231 | RMS (%) | Average 0.158 | RMS (%) | |||
| R.S.D. 5.70% | 3.87 | R.S.D. 4.44% | 2.81 | |||
phosphorylation fraction of Pep. 2 mixture with total peptide = 1.25 pmol
standard deviation, n = 5
for each mixture with a known m, the m (observed) is calculated using the response factor a derived as an mean of the a values from the other six mixtures
To verify that phosphorylation fraction in a sample can be readily calculated from data with the proposed mathematical strategy, the phosphorylation fraction (m) calculation (Eq 3d) was assessed using the data already acquired as a test case. To do this, an experimental phosphorylation fraction was calculated for each sample by applying the observed peak ratio S and the average a value derived from the other 6 measurements Eq. 3d. This was done for both the 8 and 13 µJ laser energy samples. The experimental phosphorylation fractions were then compared to their known values, and accuracy determined as % error (Table 2), obtained on the basis of five experimental replicates per sample. The % errors for the experimental m values fall in the range of 0.08 % – 7.14 % (overall RMS error of 3.87%), and 0.60 % – 6.16 % (RMS error of 2.81%), for 8 and 13 µJ laser energy, respectively. These error values suggest that the ionization efficiency ratio a can indeed be applied to the Eq. 3d for accurate isotope-free MALDI quantification of phosphorylation.
Linearity, dynamic range and reproducibility
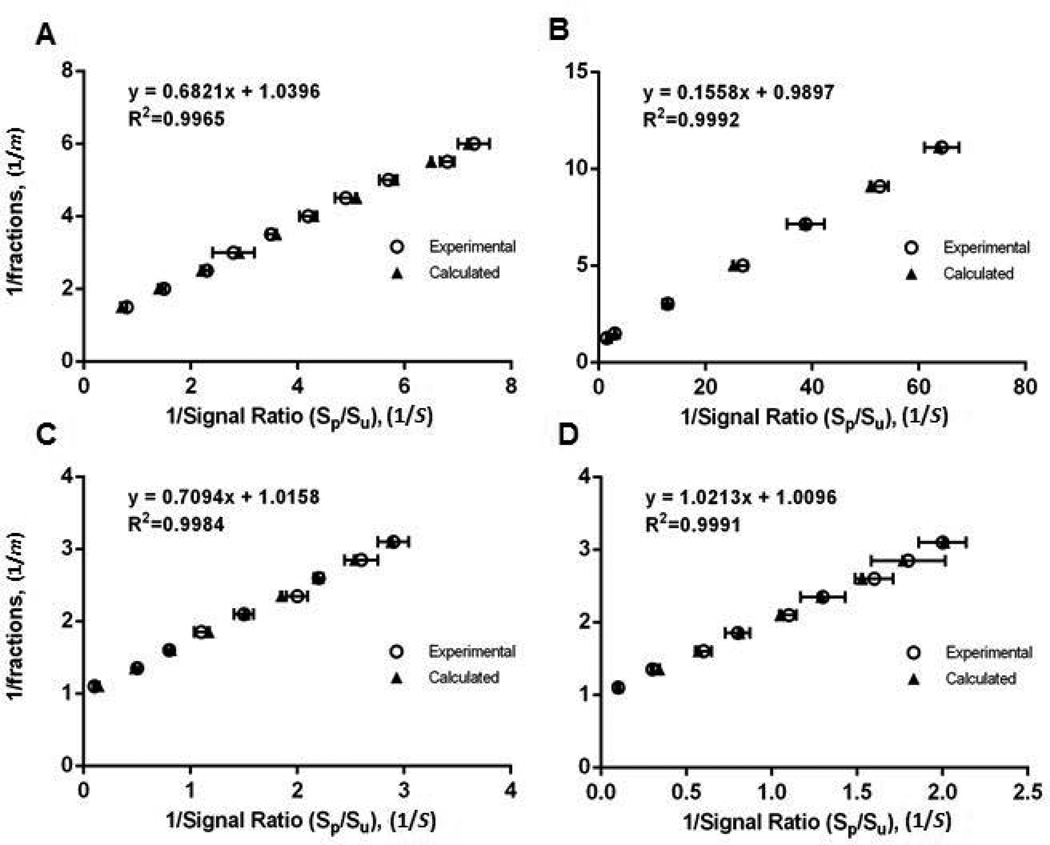
After establishing consistency of the response factor a, across a wide range of phosphorylation fractions, in three different peptide models (Table 1), the same model peptides are further subjected to MALDI-MS analysis to investigate the linearity of the equation. The reciprocals of observed signal ratios (1/S) and the known phosphorylation fractions (1/m) are plotted in Fig. 1, and good linearity of Eq. 3d is shown among all peptide models with R2 values above 0.996.
Fig. 1. Linearity of double reciprocal equation.
The linearity of the double reciprocal procedure is assessed by comparing the experimentally determined and calculated double reciprocal plots using phosphorylated and unphosphorylated analogs of (A) peptide 1 (B) peptide 2 (C) peptide 3A and (D) peptide 3B
The dynamic range of the method is further determined by evaluating the consistency of the response factors (a) among pep. 2 mixtures of unphospho/ phosphopeptides (m = 0.5) with a serial dilution. A consistent signal ratio (1/S) was observed within a range of sample amount from 7.0 to 0.04 pmol with a RMS error being 0.62 (± 8.6 % of mean), shown in Table 3 (spectra shown in supplemental figure S2). This suggests that the proposed mathematical method is feasible for phosphorylation quantification with a dynamic range of at least two orders of magnitude, down to 40 fmol of sample on the plate. At levels below this, the spot-to-spot variability in a increases to less reliable levels (not shown).
Table 3.
Effect of sample concentration on reponse factor ratio (a)
| Amounta (pmol) | Response factor ratio, a |
|---|---|
| 7.0 | 0.144 ± 0.007b |
| 3.5 | 0.140 ± 0.006 |
| 1.7 | 0.133 ± 0.016 |
| 0.8 | 0.144 ± 0.013 |
| 0.4 | 0.139 ± 0.013 |
| 0.04 | 0.140 ± 0.011 |
| Average 0.140 | |
| R.S.D. (%) 2.84 | |
the mixture of LRWGFTTPDKKHQKEPPF and LRWGFTpTPDKKHQKEPPF (m=0.5; 1:1, w/w) by using13 µJ laser energy
standard deviation, n=3
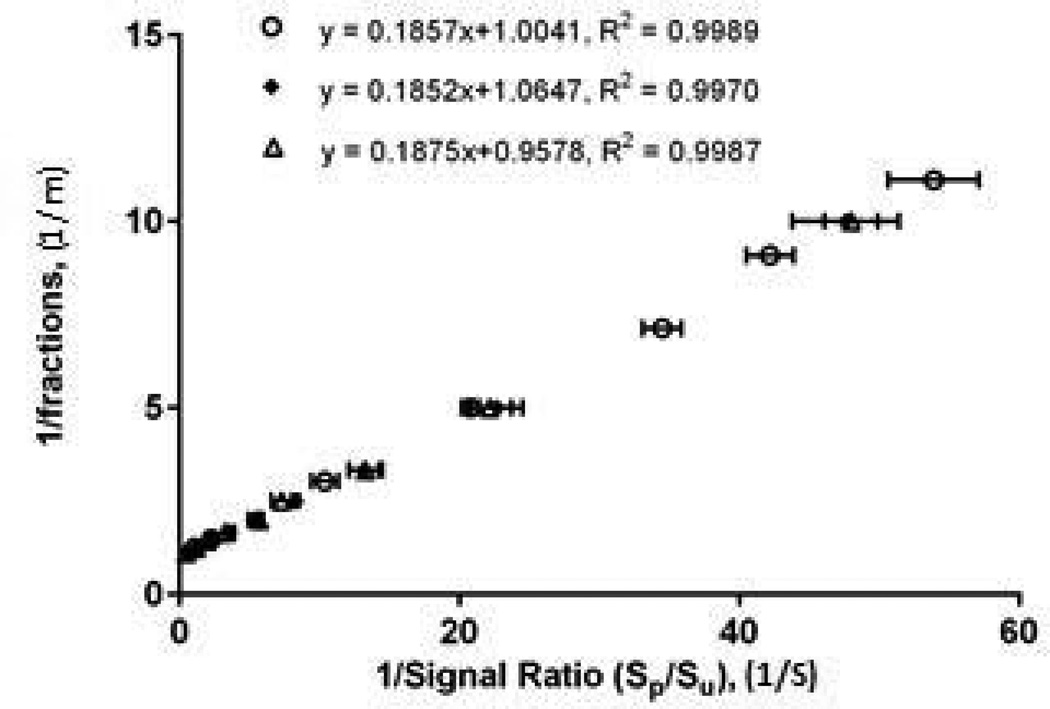
To investigate reproducibility, we conducted three experiments in separate days within a two week period under the same instrumental conditions. Fig. 2 shows the plots of the concentration curves for analysis of the peptide 2 mixtures with different phosphorylation fractions across the three experiments. Each of the curves was assigned a random range of phosphorylation fractions with R2 values > 0.99, showing linearity. In addition, a p-value of 0.88 was calculated for homogeneity of regression using ANCOVA, demonstrating good agreement among the three curves, with the mean slope of 0.186, and inter-day CV of 0.65% (n=3).
Fig. 2. Inter-day reproducibility.
Double reciprocal plots for peptide 2 obtained from three individual experiments performed on different days in a 2 week period
Validation of phosphorylation quantification by isotope dilution MALDI-MS
The previous studies on model peptide systems demonstrate the ability to monitor the amount of phosphorylation in a sample using Eq. 3d. Because consistent a values can be obtained under a variety of conditions, the mathematical procedure provides a rapid and low-cost strategy for phosphorylation quantification, that we term Double Reciprocal Isotope-free Phosphorylation Quantification (DRIP-Q) directly from the observed signal ratios and the a values without isotopes or chromatography.
In order to validate the proposed DRIP-Q method, it was compared with isotope dilution MS, an established method previously shown to be a robust and precise quantitative MS platform for peptide analysis [5]. The isotope dilution strategy utilizes heavy stable isotope-labeled internal standards that are otherwise identical to the analyte; because the analyte and isotopomeric standard have identical chemical structures, they do not suffer from the typical MALDI-related difficulties of heterogeneous crystal distribution and differential ionization. However, its effectiveness as a quantitative method is tempered by the need for cost of producing isotopically labeled standards.
To compare the two methods, mixtures of a peptide 1, AAAAYRAAR and its phosphoform, at 30% (w/w) and 40% (w/w) phosphorylation concentrations (m = 0.3 and 0.4), were subjected to both isotope dilution and DRIP-Q analysis. To perform the isotope dilution analysis, phosphorylated and unphosphorylated versions of peptide 1 were both synthesized with unlabeled or [13C3]-Alanine at the first position. The labeled and unlabeled peptides were mixed in defined amounts and subjected to MALDI analysis. The results of DRIP-Q method and isotope dilution MALDI-MS method are compared in Table 4.
Table 4.
Comparison of phosphorylation quantification by DRIP-Q method and isotope dilution method
| fraction (m)a | DRIP-Q method | Isotope dilution method | p valueb |
|---|---|---|---|
| 0.3 | 0.291 ± 0.018 | 0.295 ± 0.005 | 0.58 |
| 0.4 | 0.394 ± 0.017 | 0.391 ± 0.005 | 0.63 |
mixtures of AAAAYRAAR/AAAApYRAAR with phosphopeptide fraction (0.3, 0.4) in an amount range of 0.9– 2.0 pmol
t-test (two tails, n=5)
The phosphorylation fraction of each peptide mixture samples was determined at 0.291 and 0.394 in agreement with the results (0.295 and 0.391, p > 0.60) obtained from isotope dilution method. Quantification by isotope dilution yields a smaller standard deviation than the DRIP-Q method (1.7–1.8% RSD vs 4.3–6.3% RSD, p < 0.01), a result that could be expected since the use of isotopic internal standards eliminates some of the intrinsic uncertainty in MALDI quantification. Furthermore, other groups have reported average RSDs of 16.6% and 7.9 % for label-free ESI LC-MS method and Isobaric Tags for Relative and Absolute Quantification (iTRAQ) method, respectively [22]. While ESI is thought to be more quantitatively reproducible than MALDI, the DRIP-Q method performs comparably to or better than these reported methods, possibly because the MALDI technique allows for longer acquisition time. In this analysis, spectra from approximately 18,000 laser shots were acquired over 5 minutes (laser firing at a repetition rate of 60 Hz), allowing for the averaging of much more data than typically available over the elution time of a peak in LC-MS; indeed MALDI shows substantial improvement in reproducibility as the number of laser shots increases (supplemental figure S3). A similar phenomenon is observed by Johnson et al. when evaluating the QconCAT isotope label-based phosphorylation stoichiometry methodology [23]. For peptides analyzed by both MALDI and ESI, they observe RSDs of 1.1% – 16.2% for MALDI and 2.0% – 30.5% for ESI, with MALDI ionization giving consistently more reproducible results. Together, these data suggest the variations between DRIP-Q method and isotope dilution MALDI-MS method are comparable, with excellent accuracy, in spite of no isotope labeling or calibration curve needed in the proposed DRIP-Q method. The somewhat better precision of isotope labeling does not lead to an obvious increase in accuracy, and is offset by the lower cost of the isotope-free DRIP-Q method.
Applications to Cyclin-dependent kinase inhibition
This study provides a simple and accurate quantitative method to determine phosphorylation by MS, with potential applications for assessing kinase activity and inhibitor potency. Cyclin-dependent kinases (CDKs) are proline-directed serine/threonine kinases that regulate eukaryotic cell cycle. Inhibitors of CDKs are currently under investigation as potential therapeutics for cancer and viral infection [24–25]. For example, p21 inhibition of CDK2-dependent phosphorylation of HIV-reverse transcriptase significantly reduced the efficacy of HIV-1 transcription [26]. Here, the DRIP-Q method is evaluated as a method for assessing the in vitro inhibition of CDK.
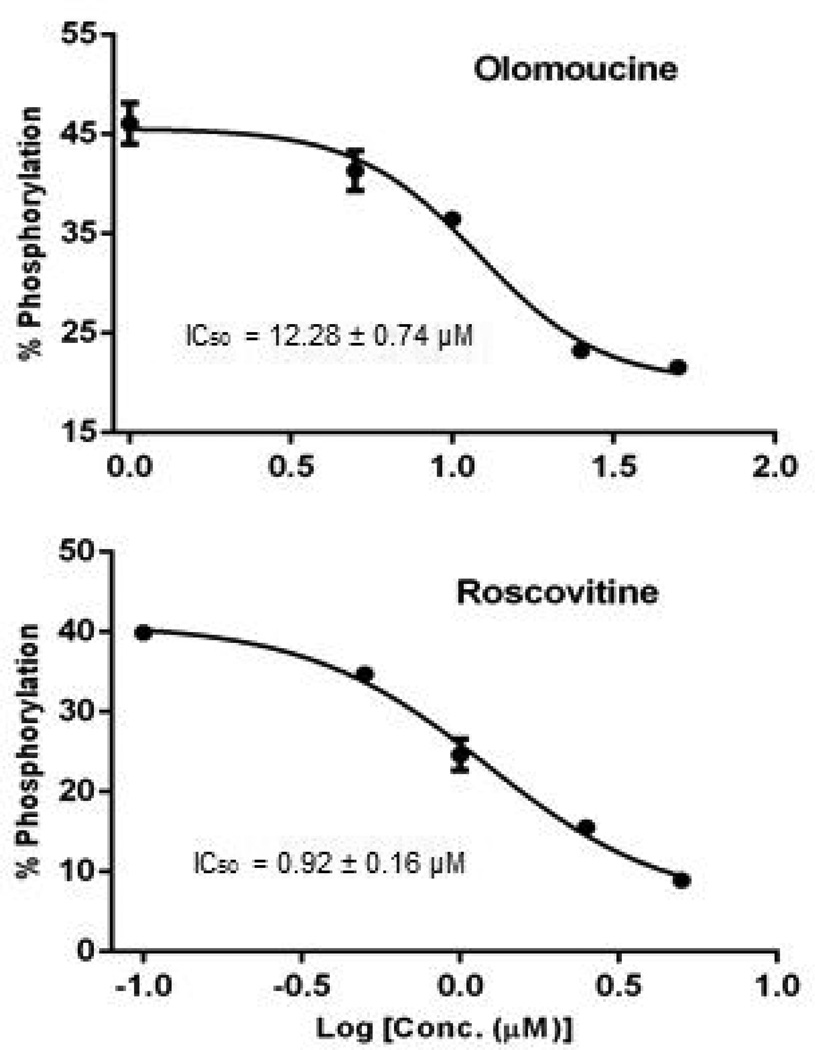
r-Roscovitine and Olomoucine are well known pharmacological ATP-competitive inhibitors of CDKs. In a recent paper, r-Roscovitine has also been reported as a potential pharmacological CDK inhibitor of HIV-1 replication [27]. Peptide 2 (LRWGFTTPDKKHQKEPPF) derived from the sequence of HIV reverse transcriptase was subjected to in vitro phosphorylation by Cyclin A/CDK2. After incubation of kinase, peptide and ATP, the reaction mixture was subjected to DRIP-Q analysis to determine the IC50 (50% inhibition of the control) values of r-Roscovitine and Olomoucine. The mathematical a value for the system was determined using a single point measurement of phosphorylated and unphosphorylated standards (supplemental figure S4), and found to have a value of a = 0.184 ± 0.017. This a value was then applied to peptide/phosphopeptide signal intensity ratios according to Eq. 3d without the construction of additional calibration curves. Inhibition plots were generated, and IC50 s of r-Roscovitine and Olomoucine were found to be 0.92 ± 0.16 µM and 12.28 ± 0.74 µM, respectively (Fig 3).
Fig. 3. In vitro inhibition of CDK2 measured by DRIP-Q.
Inhibitory activities of Roscovitine and Olomoucine against CDK2/cyclin A. Peptide 2 is used as substrate in invitro kinase assays
This result suggests that both r-Roscovitine and Ololomoucine have low micromolar inhibitory activity against the substrate peptide2 in the presence of CDK2/CyclinA. The inhibitory activity of r-Roscovitine is about 10 times higher than Olomoucine for CDK phosphorylation of peptide 2. Prior published studies report similar IC50 range of r-Roscovitine (0.65–1.8 µM) [28–29] and Olomoucine (7–10 µM) [30–31] for CDK activity, consistent with the results that r-Roscovitine has also an order of magnitude more inhibitory activity than Olomoucine. The consistency indicates that the proposed DRIP-Q method is feasible to provide a reference method for simply, quickly and accurately quantifying phosphorylation in kinase assays.
Conclusion
This paper describes a novel quantitative strategy combined with isotope-free MALDI-MS to quantify phosphorylation in in-vitro assays. Only a single point calibration is required, using a mix of phosphorylated and unphosphorylated standards. Based on the proposed DRIP-Q method, phosphorylation concentration in an assay sample was simply calculated by using its signal ratios on the mass spectrum with a consistent instrument response factor. Excellent reproducibility, precision, dynamic range and accuracy have been determined by using three different peptides, containing different phosphorylation sites (Serine, Threonine, and Tyrosine respectively), and issues with MALDI crystal heterogeneities are adequately overcome by sufficient signal averaging. Method validation has been carried out by comparing DRIP-Q method to an isotope-dilution method. In addition, the method has been successfully applied to an in vitro assay to monitor the inhibition activity of CDK inhibitors by determining IC50 values consistent with previous measurements.
This study suggests that MALDI-linear ion trap mass spectrometry holds the potential for label-free, chromatography-free and calibration curve-free methods development and application. However, it does require careful assessment of experimental conditions, particularly laser energy. Additionally, further studies of how DRIP-Q responds to mixtures with complex backgrounds such as proteomic samples are being undertaken. Nonetheless, the work here demonstrates the feasibility of MALDI-linear ion trap MS for quantitative work such as drug/inhibitor screening by in vitro kinase assay, and lays the foundation for further analysis of systematics, experimental conditions including choice of matrix, and more generalized applications.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Rinku Prithiani for technical assistance and the University of Utah DNA/peptide facility for peptide synthesis. ML is supported by the National Institutes of Health (AI093203 and AI098487), Clinical Scientist Development Award from the Doris Duke Charitable Foundation (grant 2009034), and American Foundation for AIDS Research (grant 108302-51-RGRL). EJC is supported by the National Institutes of Health (SC2GM086343) and Professional Staff Congress – City University of New York.
References
- 1.Kreegipuu A, Blom N, Brunak S. Phospho Base, a database of phosphorylation sites: release 2.0. Nucleic Acids Res. 1999;27:237. doi: 10.1093/nar/27.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, Polakiewicz RD, Comb MJ. Phosphoptrotein Analysis Using Antibodies Broadly Reactive against Phosphorylated Motifs. J. Biol. Chem. 2002;277:39379. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 3.Ma H, Deacon S, Horiuchi K. The challenge of selecting protein kinase assays for lead discovery optimization. Expert Opin. Drug Discovery. 2008;3:607. doi: 10.1517/17460441.3.6.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia Y, Quinn CM, Kwak S, Talanian RV. Current in vitro kinase assay technologies: the quest for a universal format. Curr. Drug Discovery Technol. 2008;5:59. doi: 10.2174/157016308783769414. [DOI] [PubMed] [Google Scholar]
- 5.Kuklenyik Z, Boyer AE, Lins R, Quinn CP, Gallegos-Candela M, Woolfitt A, Pirkle JL, Barr JR. Comparison of MALDI-TOF-MS and HPLC-ESI-MS/MS for Endopeptidase Activity-Based Quantification of Anthrax Lethal Factor in Serum. Anal. Chem. 2011;83:1760. doi: 10.1021/ac1030144. [DOI] [PubMed] [Google Scholar]
- 6.Parker L, Engel-Hall A, Drew K, Steinhardt G, Helseth DL, Jabon D, McMurry T, Angulo DS, Kron SJ. Investigating quantitation of phosphorylation using MALDI-TOF mass spectrometry. J. Mass Spectrom. 2008;43:518. doi: 10.1002/jms.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunasekera K, Wüthrich D, Braga-Lagache S, Heller M, Ochsenreiter T. Proteome remodeling during development from blood to insect-form Trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genomics. 2012;13:556. doi: 10.1186/1471-2164-13-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong J, Kim M, Chaerkady R, Wu X, Huang T, Getnet D, Mitchell CJ, Palapetta SM, Sharma J, O'Meally RN, Cole RN, Yoda A, Moritz A, Loriaux MM, Rush J, Weinstock DM, Tyner JW, Pandey A. A. TSLP Signaling Network Revealed by SILAC-Based Phosphoproteomics. Mol. Cell. Proteomics. 2012;11:6. doi: 10.1074/mcp.M112.017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer L, Chen ZA, Rappsilber J. Quantitative cross-linking/mass spectrometry using isotope-labelled crosslinkers. J Proteomics. 2013;88:120. doi: 10.1016/j.jprot.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye X, Luke B, Andresson T, Blonder J. 18O stable isotope labeling in MS-based proteomics. Brief Funct. Genomic Proteomics. 2009;8:136. doi: 10.1093/bfgp/eln055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouw JW, Krijgsveld J, Heck AJR. Quantitative proteomics by metabolic labeling of model organisms. Mol. Cell. Proteomics. 2010;9:11. doi: 10.1074/mcp.R900001-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boersema PJ, Foong LY, Ding VMY, Lemeer S, Breukelen BV, Philp R, Boekhorst J, Snel B, Hertog JD, Choo ABH, Heck AJR. In-depth Quanlitative and Quantitative Profiling of Tyrosine Phosphorylation Using a Combination of Phosphopeptide Immunoaffinity Purification and Stable Isotope Dimethyl Labeling. Mol. Cell. Proteomics. 2010;9:84. doi: 10.1074/mcp.M900291-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turck CW, Falick AM, Kowalak JA, Lane WS. The Association of Biomolecular Resource Facilities Proteomics Research Group 2006 Study: Reletive protein quantitation. Mol. Cell. Proteomics. 2007;6:1291. doi: 10.1074/mcp.M700165-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Liu Q, Zimmerman LJ, Ham AL, Slebos RJC, Rahman J, Kikuchi T, Massion PP, Carbone DP, Billheimer D, Liebler DC. Methods for peptide and protein quantitation by liquid chromatography-multiple reaction monitoring mass spectrometry. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.006593. M110.006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman HM, Schutt KL, Dieter EM, Lamos SM. Relative quantification of biomarkers using mixed-isotope labeling coupled with MS. Bioanalysis. 2012;4:2525. doi: 10.4155/bio.12.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Wen B, Zhou B, Yang L, Cha C, Xu S, Qiu X, Wang Q, Sun H, Lou X, Zi J, Zhang Y, Lin L, Liu S. Correction to quantitative analysis of the human AKR family members in cancer cell lines using the mTRAQ/MRM approach. J. Proteome Res. 2013;12:2022. doi: 10.1021/pr301153z. [DOI] [PubMed] [Google Scholar]
- 17.Rao PV, Reddy AP, Lu X. Proteomic identification of salivary biomarkers of type-2 diabetes. J. Proteome Res. 2009;8:239. doi: 10.1021/pr8003776. [DOI] [PubMed] [Google Scholar]
- 18.Levin Y, Schwarz E, Wang L, Leweke FM, Bahn S. Label-free LC-MS/MS quantitative proteomics for large scale biomarker discovery in complex samples. J. Sep. Sci. 2007;30:2198. doi: 10.1002/jssc.200700189. [DOI] [PubMed] [Google Scholar]
- 19.Asara JM, Christofk HR, Freimark LM, Cantley LC. A label-free quantification method by MS/MS TIC compared to SILAC and spectral counting in a proteomics screen. Proteomics. 2008;8:994. doi: 10.1002/pmic.200700426. [DOI] [PubMed] [Google Scholar]
- 20.Zhu W, Smith JW, Huang CM. Mass spectrometry-based label free quantitative proteomics. J. Biomed. Biotechnol. 2010;2010:840518. doi: 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinumi T, Niki E, Shigeri Y, Matsumoto H. Affinity-Tagged Phosphorylation Assay by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (ATPA-MALDI): Application to calcium/Calmodulin-dependent Protein Kinase. J. Biochem. 2005;138:791. doi: 10.1093/jb/mvi178. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Alvarez S, Hicks LM. Comprehensive comparison of iTRAQ and Label-free LC-Based Quantitative Proteomics Approaches Using Two Chlamydomonas reinhardtii Strains of Interest for Biofuels Engineering. J. Proteome Res. 2012;11:487. doi: 10.1021/pr2008225. [DOI] [PubMed] [Google Scholar]
- 23.Johnson H, Eyers CE, Eyers PA, Benyon RJ, Gaskell SJ. Rigorous determination of the stoichiometry of protein phosphorylation using mass spectrometry. J Am. Soc Mass. Spectrometry. 2009;20:2211. doi: 10.1016/j.jasms.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Holcakova J, Muller P, Tomasec P, Hrstka R, Nekulova M. Inhibition of Post-Transcriptional RNA Processing by CDK Inhibitors and Its Implication in Anti-Viral Therapy. PLoS ONE. 2014;9:e89228. doi: 10.1371/journal.pone.0089228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cicenas J, Valius M. The CDK inhibitors in cancer research and therapy. J. Cancer Res. Clin. Oncol. 2011;137:10. doi: 10.1007/s00432-011-1039-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leng J, Ho H-P, Buzon M, Pereyra F, Walker BD, Yu XG, Chang EJ, Lichterfeld M. A cell-intrinsic inhibitor of HIV-1 reverse transcription in CD4+ T cells from elite controllers. Cell Host & Microbe. 2014;15:717. doi: 10.1016/j.chom.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guendel I, Agbottah ET, Kehn-Hall K, Kashanchi F. Inhibition of human immunodeficiency virus type-1 by Cdk inhibitors. AIDS Research and Therapy. 2010;7:7. doi: 10.1186/1742-6405-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meijer L, Kim SH. Chemical inhibitors of cyclin-dependent kinases. Methods Enzymol. 1997;283:113. doi: 10.1016/s0076-6879(97)83011-x. [DOI] [PubMed] [Google Scholar]
- 29.Bach S, Knockaert M, Reinhardt J, Lozach O, Schmitt S, Baratte B, Koken M, Coburn LT, Jiang T, Liang D, Galons H, Dierick J-F, Pinna LA, Meggio F, Totzke F, Schächtele C, Lerman AS, Carnero A, Wan Y, Gray N, Meijer L. Roscovitine targets, protein kinases and pyridoxal kinase. J. Biol. Chem. 2005;280:31208. doi: 10.1074/jbc.M500806200. [DOI] [PubMed] [Google Scholar]
- 30.Vesely J, Havlicek L, Strnad M, Blow JJ, Donella-Deana A, Pinna L, Letham DS, Kato J, Detivaud L, Leclerc S, Meijer L. Inhibition of cyclin-dependent kinases by purine analogues. Eur. J. Biochem. 1994;224:771. doi: 10.1111/j.1432-1033.1994.00771.x. [DOI] [PubMed] [Google Scholar]
- 31.Raynaud FI, Whittaker SR, Fischer PM. In vitro and In vivo Pharmacokinetic-Pharmacodynamic Relationships for the Trisubstituted Aminopurine Cyclin-Dependent Kinase Inhibitors Olomoucine, Bohemine and CYC202. Clin. Cancer Res. 2005;11:4875. doi: 10.1158/1078-0432.CCR-04-2264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.