Abstract
Background
Although identification of the target mRNAs of micro RNAs (miRNAs) is essential to understanding their function, the low complementarity between miRNAs and their target mRNAs has complicated this process. In this study, we sought to identify miRNAs which reduce the expression of the transcription factor Zeb-2, a transcriptional repressor of E-cadherin which is known to be down regulated by members of the miR-200 family (miR-200a,b,c miR-429, and miR-141).
Findings
We first used a computational target predicting system to identify 82 candidate miRNAs which bound the 3′UTR region of the Zeb-2 mRNA. Of these 82 miRNAs, precursors for 51 were available in our miRNA precursor library. Pre-miR™ Precursor Molecules for these 51 miRNAs were co-transfected into NIH3T3 cells with a luciferase reporter vector containing the 3′UTR region of the Zeb-2 mRNA. Seven miRNAs (miR-141, mi-183, miR-200a, miR-200b, miR-200c, miR-429 and miR-666-5p) were shown to down-regulate luciferase activity and Western blotting analysis confirmed that Pre-miR™ Precursor Molecules for these seven miRNAs induced expression of E-cadherin and miScript target protector against miR-183 and miR-666-5p abrogated this effect. Moreover, an Anti-miR™ miRNA Inhibitor targeting miR-183 and miR-666-5p repressed expression of E-cadherin.
Conclusions
We have established a method to identify miRNAs that bind the 3′UTR region of the Zeb-2 mRNA and that induce expression of E-cadherin, possibly by down-regulating the expression of Zeb-2. Our method may be more widely applicable for identifying miRNAs that bind target mRNA 3′UTR regions and down-regulate the expression of proteins encoded by these mRNAs.
Background
MicroRNAs (miRNAs) are 22-nucleotide (nt) endogenous non-coding RNAs that exhibit a high degree of structural and functional conservation throughout different species. miRNAs are initially synthesized as long primary transcripts that are subsequently processed into -70-nt stem-loop pre-microRNAs by Drosha endonucleasae [1] and transported out of the nucleus by exportin 5 [2]. Dicer then processes these pre-microRNA in the cytoplasm to yield the -22-nt mature miRNAs [3]. Binding of miRNA to target mRNAs with perfect or near perfect complementarity has been shown to induce mRNA degradation, whereas imperfect complementarity reportedly induces translational regression. In this context, 7–8 nt sequence at the 5′ end of the miRNA sequence, known as the seed sequence, is known to be critical for efficient targeting.
miRNAs have been implicated in regulating complex physiological processes such as embryogenesis [4], organ development [5], and oncogenesis [6,7], and we have recently demonstrated their role in the pathology of kidney diseases [8]. To date over 2000 miRNAs have been identified in human, although the functional roles of the vast majority remain unknown.
Whereas identifying the target mRNAs of miRNAs is essential to understanding their biological roles, this has proven difficult due to the imperfect complementarily between miRNAs and their target mRNAs. cA recently described method for identifying miRNA target mRNAs using exogenously tagged Ago2 [9] has proven effective, but has a complicated protocol and carries a high rate of identification of artifactual mRNAs such as mitochondrial mRNAs. Other approaches have used microarray analysis to identify miRNAs targeting disease-related proteins. Global expression profiling of cancer cell lines overexpressing miR-16, for example, identified 27 candidate targets of this miRNA, of which three (MAP7, PRDM44 and CDS2) were experimentally validated [10]. In general however, such methods have proven unreliable for detection of disease-related miRNA target genes. Here, we set out to establish a reliable method for identifying miRNAs which down-regulate the expression of a specific target protein.
Many miRNAs have been shown to bind to the 3′UTR region of their target mRNA. Previous studies have demonstrated miRNA-mediated repression of the translation of a luciferase reporter gene to which the 3′UTR coding region of the target mRNA has been spliced [11]. In this proof-of-principle study, we used a similar method to identify miRNAs which down-regulated expression of the E-box-binding zinc-finger transcription factor Zeb-2, which has previously been shown to be repressed by members of the miR-200 family (miR-200a,b,c miR-429, and miR-141) [11-13].
We first established dual luciferase reporter vector containing the 3′UTR coding region of Zeb-2 mRNA spliced to the 3′ end of the luciferase coding region. We then transfected cultured cells with this reporter vector in 96-well plates and 48 hours later transfected cells with a series of miRNA precursors. Luciferase activity was assayed after 72 hours later to identify miRNAs which bound the 3′UTR of Zeb-2 and subsequently reduced luciferase expression. This method proved cumbersome however, prompting us to use a computational target predicting system to narrow down the number of screened miRNAs to a figure (<100 miRNAs) that was manageable, which a previous study had shown was likely to yield true functional miRNAs (data not shown). We anticipate that our new method will be valuable for future characterization of miRNA function.
Materials and methods
Prediction of miRNAs by computational target predicting system
To detect candidate miRNAs targeting Zeb2, we first evaluated a series of miRNA precursors. To narrow the screened miRNAs to below 100 miRNAs, we used a computational target predicting system (miRanda) containing updated sequences for all known miRNAs [14-16]. Cutoff scores for selection of candidate miRNAs were < -20.0 for energy and >120 for binding.
ZEB2 3′-UTR luciferase reporter vector
We designed a dual luciferase reporter vector containing the 3′UTR coding region of Zeb-2 mRNA spliced to the 3′end of the luciferase coding region. Firstly, the 3′-UTR for ZEB2 was PCR-amplified from mouse genomic DNA. The PCR primers used to amplify the Zeb-2 3′-UTR were 5′-CAGTTCAGCCAAGACAGAGT-3′ (forward) and 5′-TTCGAGCATGGTCATTTTC-3′UTR (reverse). The amplified 3′-UTRs was cloned downstream of the firefly luciferase coding region in the pGL3-promoter luciferase reporter vector (Clontech). Sequence analysis confirmed the accuracy of the PCR procedure.
miRNA precursor library
In this study we used the Pre-miR™ miRNA Precursor Library - Mouse V3 (Ambion co.).
Pre-miR™ miRNA Precursor Molecules are small, chemically modified double-stranded RNA molecules designed to mimic endogenous mature miRNAs. The Pre-miR™ miRNA Precursor Library–Mouse V3 consists of 379 miRNA mimics corresponding to 379 mouse mature miRNAs cataloged in version 9.2 of the miRBase Sequence Database.
Cell culture
NIH3T3 and HK-2 cell lines were provided by the RIKEN BRC through the National Bio-Resource Project of MEXT, Japan. NIH3T3 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% bovine serum. HK-2 cells were grown in DMEM supplemented with 5% fetal bovine serum. Cells were routinely cultured at 37°C in a humidified atmosphere of 95% air-5% CO2.
Transfection and luciferase assays
NIH3T3 cells were seeded in 96 well plate (Sterilin UK). 24 hr later, 0.1 μg of reporter plasmid was transfected using the Lipofectamine LTX system (Invitrogen), according to the manufacturer’s protocols. To assess the effect of miRNAs on reporter activity, 2.5 pmol of synthetic Pre-miR™ miRNA Precursor Molecules and Pre-miR™ miRNA Precursor Molecules-Negative Control (Ambion co.) were transfected 48 hours later using the Lipofectamine RNAiMax system (Invitrogen), according to the manufacturer’s protocols. Cells were lysed after 24 hours in 40 μl passive lysis buffer (Promega). Measurements were performed using the Promega luciferase assay system and the GloMax 96 Microplate Luminometer (Promega). Each sample was measured in four replicates. The luciferase activity of each lysate was normalized to the luciferase activity of the Pre-miR™ Precursor Molecules-Negative control.
Western blotting of E-cadherin
Western blotting analysis of E-cadherin was performed in NIH3T3 cells. To investigate the effect of miRNAs on Zeb-2 levels, NIH3T3 cells were transfected with 20 pmol/ml Pre-miR™ Precursor Molecules and Anti-miR™ miRNA Inhibitor (Ambion) using the Lipofectamine RNAiMax system (Invitrogen). To show a direct miRNA-mRNA interaction at the predicted target sites, NIH3T3 cells were transfected with 20 pmol/ml miRNA precursors and 300 pmol miScript target protector (Qiagen) against miR-183 or miR-666-5p simultaneously. After 24 hours the cell lysate was extracted and analysed by Western blotting using a mouse monoclonal anti-E-cadherin antibody (BD Bioscience). as previously described [8]. To confirm that the same amount of protein was investigated, the expression of beta-actin was also investigated.
Effect of miRNAs on epithelial and mesenchymal transition
To examine the effect of microRNAs to epithelial mesenchymal transition, we performed morphological observation and fluorescent staining of filamentous actin (F-actin) in transfected cells. Normal human kidney HK-2 cells (purchased from RIKEN) were transfected with Pre-miR™ miRNA Precursor Molecules for miR-183or miR-666-5p. 24 hours later 4 ng/ml TGF-beta was added and another 24 hours later, cells were observed under phase contrast microscope. Cells were then fixed with 4% paraformaldehyde and incubated with Rhodamine-conjugated Phalloidin (CHEMICON) for 30 min, before observation under a laser microscope.
Statistical analysis
All data are reported as mean ± SD. When comparisons were made between two different groups, statistical significance was determined using the Student’s t-test.
Findings
Results
Prediction of Zeb-2 mRNA 3’UTR-binding miRNAs
We identified in silico 82 candidate miRNAs (Table 1) which were predicted to directly bind the 3′UTR of the Zeb-2 mRNA and regulate expression of Zeb-2. These candidates included all miRNAs, namely miR-141, miR-200a,b,c and miR-429, previously shown to directly regulate Zeb-2.
Table 1.
Candidate miRNAs which can bind 3′UTR of Zeb-2 mRNA
| mmu-miR-1187 |
mmu-miR-2142 |
mmu-miR-467d |
| mmu-miR-1188 |
mmu-miR-22 |
mmu-miR-469 |
| mmu-miR-124 |
mmu-miR-23a |
mmu-miR-470 |
| mmu-miR-125a-3p |
mmu-miR-23b |
mmu-miR-471 |
| mmu-miR-136 |
mmu-miR-27a |
mmu-miR-540-5p |
| mmu-miR-138 |
mmu-miR-28 |
mmu-miR-541 |
| mmu-miR-141 |
mmu-miR-298 |
mmu-miR-547 |
| mmu-miR-152 |
mmu-miR-301b |
mmu-miR-574-5p |
| mmu-miR-155 |
mmu-miR-320 |
mmu-miR-666-5p |
| mmu-miR-183 |
mmu-miR-324-3p |
mmu-miR-669c |
| mmu-miR-1896 |
mmu-miR-328 |
mmu-miR-669 g |
| mmu-miR-1906 |
mmu-miR-33 |
mmu-miR-674 |
| mmu-miR-1927 |
mmu-miR-342-3p |
mmu-miR-680 |
| mmu-miR-1931 |
mmu-miR-34a |
mmu-miR-686 |
| mmu-miR-1933-5p |
mmu-miR-379 |
mmu-miR-691 |
| mmu-miR-1935 |
mmu-miR-380-3p |
mmu-miR-693-3p |
| mmu-miR-1941-5p |
mmu-miR-409-5p |
mmu-miR-697 |
| mmu-miR-1948 |
mmu-miR-429 |
mmu-miR-698 |
| mmu-miR-1958 |
mmu-miR-448 |
mmu-miR-706 |
| mmu-miR-1960 |
mmu-miR-449a |
mmu-miR-708 |
| mmu-miR-1962 |
mmu-miR-449b |
mmu-miR-712 |
| mmu-miR-1966 |
mmu-miR-449c |
mmu-miR-742 |
| mmu-miR-200a |
mmu-miR-452 |
mmu-miR-872 |
| mmu-miR-200b |
mmu-miR-466b-5p |
mmu-miR-873 |
| mmu-miR-200c |
mmu-miR-466c-5p |
mmu-miR-880 |
| mmu-miR-205 |
mmu-miR-466e-5p |
mmu-miR-881 |
| mmu-miR-20a |
mmu-miR-467c |
mmu-miR-96 |
| mmu-miR-2138 |
Screening of miRNAs which can down-regulate the expression of Zeb-2
Of these 82 miRNAs, Pre-miR™ miRNA Precursor Molecules for 51which were available in our Pre-miR™ miRNA Precursor Library, were co-transfected into NIH3T3 cells along with a luciferase reporter vector containing the 3′UTR coding region of Zeb-2. Pre-miR™ miRNA Precursor Molecules for miR-141, miR-183, miR-200a, miR-200b, miR-200c, miR-429 and miR-666-5p down-regulated luciferase activity below 80% compared to control, (miR-141: 0.6 ± 0.106, miR183: 0.6 ± 0.106, miR-200a: 0.645 ± 0.031, miR-200b: 0.752 ± 0.163, miR-200c: 0.603 ± 0.03, miR-429: 0.633 ± 0.076) (Table 2).
Table 2.
Relative luciferase activity of candidate miRNA
| Control | 1 ± 0.06 |
|---|---|
| miR-124 |
1.15 ± 0.025 |
| miR-136 |
1.17 ± 0.018 |
| miR-138 |
0.952 ± 0.16 |
| miR-141 |
0.6 ± 0.106 |
| miR-152 |
0.919 ± 0.044 |
| miR-155 |
0.934 ± 0.183 |
| miR-183 |
0.769 ± 0.022 |
| miR-200a |
0.645 ± 0.031 |
| miR-200b |
0.752 ± 0.163 |
| miR-200c |
0.603 ± 0.03 |
| miR-205 |
0.927 ± 0.017 |
| miR-20a |
0.963 ± 0.017 |
| miR-22 |
0.954 ± 0.157 |
| miR-23a |
0.896 ± 0.038 |
| miR-23b |
0.904 ± 0.051 |
| miR-27a |
0.984 ± 0.156 |
| miR-28 |
1.071 ± 0.038 |
| miR-298 |
0.874 ± 0.08 |
| miR-301b |
1.032 ± 0.119 |
| miR-320 |
0.964 ± 0.007 |
| miR-324-3p |
1.194 ± 0.031 |
| miR-328 |
1.202 ± 0.016 |
| miR-33 |
1.051 ± 0.125 |
| miR-342-3p |
0.854 ± 0.086 |
| miR-34a |
0.917 ± 0.043 |
| miR-379 |
1.062 ± 0.140 |
| miR-380-3p |
0.958 ± 0.016 |
| miR-429 |
0.633 ± 0.076 |
| miR-448 |
0.931 ± 0.133 |
| miR-449a |
1.027 ± 0.023 |
| miR-449b |
1.022 ± 0.020 |
| miR-452 |
0.826 ± 0.053 |
| miR-469 |
1.096 ± 0.112 |
| miR-470 |
0.948 ± 0.062 |
| miR-471 |
0.814 ± 0.059 |
| miR-541 |
0.985 ± 0.124 |
| miR-547 |
0.934 ± 0.04 |
| miR-666-5p |
0.762 ± 0.022 |
| miR-669c |
0.965 ± 0.117 |
| miR-674 |
0.993 ± 0.015 |
| miR-680 |
1.148 ± 0.025 |
| miR-686 |
1.176 ± 0.009 |
| miR-691 |
1.018 ± 0.202 |
| miR-693-3p |
0.889 ± 0.005 |
| miR-697 |
0.979 ± 0.055 |
| miR-698 |
1.002 ± 0.213 |
| miR-706 |
1.037 ± 0.037 |
| miR-708 |
0.983 ± 0.042 |
| miR-712 |
0.821 ± 0.216 |
| miR-742 |
0.976 ± 0.047 |
| miR-96 | 0.927 ± 0.033 |
Prediction of target sites of the identified miRNAs in the Zeb-2 3′UTR
The miRanda computational target predicting system identified potential target sites for the candidate miRNAs in the Zeb-2 3′UTR (Figure 1). miR-183, miR-200a, miR-200b and miR-666-5p have at least one potential target site, while miR-141 has two, miR-429 has three and miR-200c has six.
Figure 1.
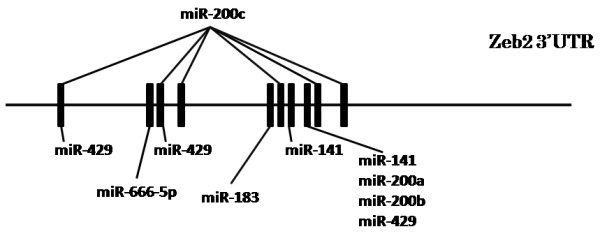
Schematic of putative detected miRNAs target sites for Zeb-2 3′UTR. Black boxes represent target sites for each miRNAs.
Western blotting of E-cadherin
Zeb-2 has been shown to be a transcriptional repressor of E-cadherin. Since we were unable to obtain an appropriate antibody for Zeb-2, we performed western blotting of E-cadherin as a surrogate marker for Zeb-2 expression. Western blotting analysis showed that Pre-miR™ Precursor Molecules for the seven miRNAs referred to above up-regulated expression of E-cadherin (Figure 2a), most likely due to their down-regulation of the expression of Zeb-2. Focusing on miR-183 and miR-666-5p, we next examined whether Anti-miR™ miRNA Inhibitor against miR-183 and miR-666-5p could down-regulate the expression of E-cadherin. Western blotting analysis confirmed that this was the case (Figure 2b). miScript target protectors are modified RNA oligonucleotides complementary to specific target sites and which do not bind other sequences [17]. Transfection of custum-desigend miScript target protectors against the predicted miR-183 and miR-666-5p target sites in the Zeb-2 3′UTR abrogated the effect of the Pre-miR™ Precursor Molecules for miR-183 and miR-666-5p (Figure 2c).
Figure 2.
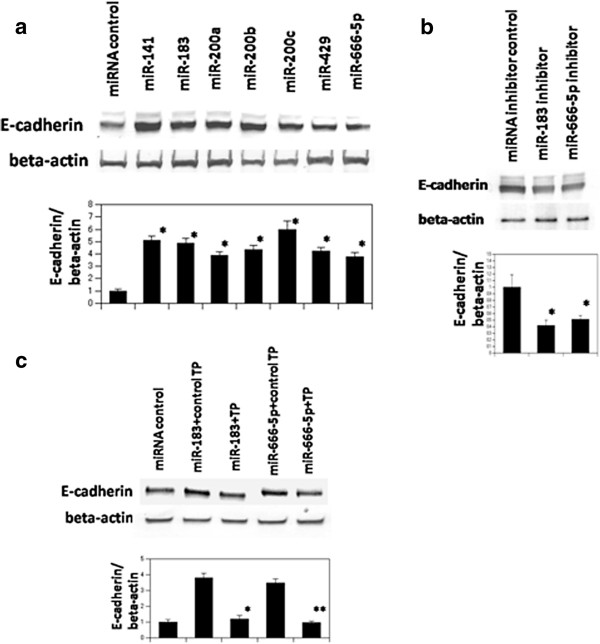
Western blotting of E-cadherin. (a) Western blotting of E-cadherin in NIH3T3 cells transfected with control miR or Pre-miR™. Precursor Molecules individually. *p < 0.05, compared to cells transfected with Pre-miR™ miRNA Precursor Molecules-Negative Control. (b) Western blotting of E-cadherin in NIH3T3 cells transfected with Anti-miR™ miRNA Inhibitor against miR-183 and miR-666-5p. *p < 0.05, compared to cells transfected with Anti-miR™ miRNA Inhibitor-negative control. (c) Western blotting of E-cadherin in NIH3T3 cells transfected with Pre-miR™ Precursor Molecules for miR-183 or miR-666-5p precursors and each Script Target Protector. *p < 0.05, compared to cells transfected with Pre-miR™ Precursor Molecules for miR-183 and miScript target protector-negative control, **p < 0.05, compared to cells transfected with re-miR™ Precursor Molecules for miR-666-5p and miScript target protector-negative control. The data are mean ± S.D. of triplicates and expression levels of E-cadherin were normalized to beta-actin with further normalization to the negative control.
Cluster analysis of seven miRNAs
The miR-200 family of miRNAs has been mapped to two separate clusters of less than 2000 bp each in the mouse genome (Figure 3 upper panel) [3]. The first cluster (Cluster 1) contains miR-200a, miR-200b and miR-429 and is located in chromosome 4. The second cluster (Cluster 2), containing miR-200c and miR-141, is located in a 500-bp region of chromosome 6. The two remaining miRNAs we identified, miR-183 and miR-666-5p, are located on chromosome 6 (30119 K) and chromosome 12 (110955 K) respectively. The five miR-200 family miRNAs located in the chromosome 4 and 6 clusters contain very similar seed sequences (Figure 3 lower panel) which are not shared with miR-183 and miR-666-5p.
Figure 3.
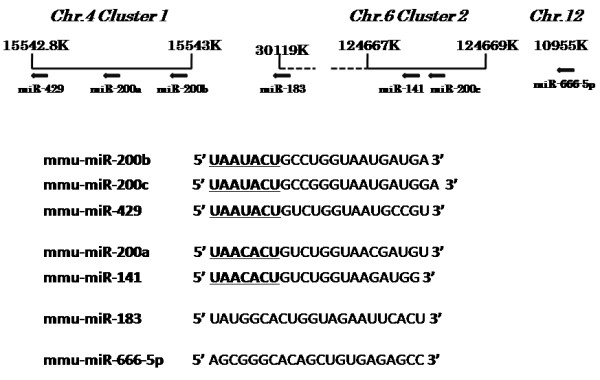
Upper panel, schematic of chromosomal location of the detected miRNAs in the mouse genome. Lower panel, sequence alignment of the detected miRNAs. Nucleotides 2–7, representing their seed sequences, are underlined.
The effect to epithelial and mesenchymal transition
Observation under phase contrast microscope revealed that the addition of Transforming growth factor (TGF)-β resulted in the appearance of the hallmarks of epithelial to mesenchymal transition, including a change in morphology from round compact shape to spindle shaped. This was accompanied by a rearrangement of actin filaments from a cortical to a stress-fiber pattern (Figure 3) [10]. Transfection of Pre-miR™ miRNA Precursor Molecules for miR-183 and miR-666-5p inhibited the appearance of these changes (Figure 4).
Figure 4.

TGF-beta induced epithelial mesenchymal transition to HK-2 cells which was inhibited by Pre-miR™ Precursor Molecules for miR-183 and miR-666-5p. Phase contrast and Immunofluorescence staining of F-actin of HK-2 cells which was stimulated with 4 ng/ml TGF-beta and, Pre-miR™ Precursor Molecules for miR-183 and miR-666-5p.
Discussion
We report here a novel method of identifying miRNAs which bind to the 3′UTR region of the Zeb 2 mRNA and that up-regulate expression of E-cadherin. Several reports have shown that members of the miR-200 family (miR-200a,b,c, miR-141 and miR-429) inhibit epithelial mesenchymal transition (EMT) through direct targeting of ZEB1 and ZEB2, which encode transcriptional repressors of E-cadherin in kidney tubular cells [4], breast cancer cells [5], and mammary epithelial cells [3]. Recent reports have indicated that a double-negative feedback loop between ZEB1, ZEB2 and miRNA-200 family members regulates EMT in kidney tubular cells [18].
In this report, besides confirming down-regulation of Zeb-2 by member of the miR-200 family, we have shown that miR-183 and miR-666-5p can also down-regulate Zeb-2. Using a novel screening method, we demonstrated that while miR-23a, 298, 342-3p,452, 471, 693-3p, 712 down-regulated luciferase activity (above 80% compared to control), they had no effect on E-cadherin levels, and excluded them from further analysis.
While no target of miR-666-5p has been previously reported, several studies have shown that miR-183 is up-regulated in prostate carcinoma and breast cancer [19,20]. Given that EMT has been known to be associated with tumorigenesis, it may be that up-regulation of miR-183 inhibits EMT in tumor tissue.
Our study contains another important new finding. Many computational target prediction systems generate large numbers of candidate miRNAs that potentially down-regulate the expression of the proteins encoded by these mRNAs. While we can not know in fact how many miRNAs down-regulate the expression of a given protein, based on our results, only about seven miRNAs can up-regulate expression of E-cadherin, most likely due to their down-regulation of the expression of Zeb-2. We have not investigated all currently known miRNAs, however we can infer that the number of miRNAs that can down-regulate a single protein is limited.
Identifying the target mRNAs of miRNAs is essential to understanding the cellular regulatory networks in which miRNAs are involved, but due to the low complementarity between miRNAs and their target mRNAs, only a few mammalian target mRNAs have been identified. To ameliorate this situation, a variety of prediction algorithms, such as miRanda [15,21], TargetScan [14,22,23], Pic Tar [24], RNA22 [25], RNA hybrid [26], PITA [27], EiM Mo [28] and DIANA [29], have been developed. These algorithms use a spectrum of parameters, including binding energy of the duplex structure, evolutionary conservation of the target site and secondary structure of 3′UTR. Despite this, they generate multiple false positive candidates and, accordingly, experimental verification of predicted miRNA-mRNA interactions must be performed. High-throughput methods successfully used in validation of miRNA target sites, including microarray and proteomic analysis, are based on measuring changes in the mRNA and protein level in response to miRNA introduction [10]. However, they cannot identify microRNAs which down-regulate a specific target protein and reporter assay such as that used in this study are required to validate candidate miRNAs routinely and with high specificity.
Our study contains one more important new finding. Until now, we have been unable to determine the number of seed sequence matches needed for the binding of miRNAs to target mRNA 3′UTR. In this reports, we identified seven miRNAs which can directly regulate the expression of Zeb-2. The number of seed sequence matches for these miRNAs were eight (miR-200a, miR-200c, miR-183, miR-429 and miR-666-5p) and seven (miR-141 and miR-200b) (Figure 5), indicating that at least seven seed sequence matches are desirable for a miRNA to bind to a target mRNA 3′UTR. This information is useful for the identification of miRNAs which can directly regulate disease related proteins.
Figure 5.

Sequence alignment of microRNAs. Their seed sequences are underlined.
Immunoprecipitation of RNA induced silencing complex (RISC)-associated mRNA requires a large amount of starting material. Using a modification of this protocol, Hayashida et al. constructed an efficient and convenient system for analyzing the mRNA content of RISC [2], and identified mRNA targets of individual miRNAs. The RISC complex however, contains multiple RNA species. For example, Hayashida et al. showed that while almost all of the cDNA recovered from RISC was miRNA (94%), the remainder contained rRNA and tRNA clones (3% and 2%, respectively) and cDNA clones (1%) with hits in genomic DNA sequences. These facts indicate the difficulty of identifying target mRNAs in the RISC complex.
We set out in this study to develop a new method for identifying target mRNAs for miRNAs. The originality of our method is that it is based upon a functional endpoint, namely down-regulation of a target protein. Using our method we will be in a position to identify miRNAs which can bind mRNA 3′UTR regions and down-regulate the expression of the encoded proteins. In future, we hope that the joint efforts of researchers all over the world will enable us to establish a database of miRNAs which can down-regulate the expression of specific protein targets.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SO: performed and analysed experiments, and wrote the manuscript. TM: performed prediction of miRNAs by computational target predicting system. YO: performed experiments. ES, HN, MT, KK, YH, TF: coordinated and oversaw the project. All authors read and approved the final manuscript.
Contributor Information
Shigeyoshi Oba, Email: sooba-npr@umin.ac.jp.
Takayuki Mizutani, Email: mirtmizutan@gmail.com.
Etsu Suzuki, Email: esuzuki-tky@umin.ac.jp.
Hiroaki Nishimatsu, Email: nishimatsu-uro@h.u-tokyo.ac.jp.
Masao Takahashi, Email: masaotakahashi-gi@umin.ac.jp.
Yousuke Ogawa, Email: Oga-fukuryu-union72722@docomo.ne.jp.
Kenjiro Kimura, Email: kimura@marianna-u.ac.jp.
Yasunobu Hirata, Email: hiratay-tky@umin.ac.jp.
Toshiro Fujita, Email: Toshiro.FUJITA@rcast.u-tokyo.ac.jp.
References
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science. 2011;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Yi R, O’Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- Nat Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Oba S, Kumano S, Suzuki E, Nishimatsu H, Takahashi M, Takamori H, Kasuya M, Ogawa Y, Sato K, Kimura K, Homma Y, Hirata Y, Fujita T. miR-200b precursor can ameliorate renal tubulointerstitial fibrosis. PLoS One. 2010;5:e13614. doi: 10.1371/journal.pone.0013614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashida Y, Nishibu T, Inoue K, Kurokawa T. A useful approach to total analysis of RISC-associated RNA. BMC Research Notes. 2009;2:169. doi: 10.1186/1756-0500-2-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Liang H, Deng T, Zhu K, Zhang S, Wang N, Jiang X, Wang X, Liu R, Zen K, Zhang CY, Ba Y, Chen X. The identification of novel targets of miR-16 and characterization of their biological functions in cancer cells. Mol Cancer. 2013;12:92. doi: 10.1186/1476-4598-12-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1and SIP1. Nature cell biology. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. MiRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JM, Lahiri DKML. MicroRNA-101 downregulates Alzheimer’s amyloid-beta protein levels in human cell cultures and is differentially expressed. Biochem Biophs Res Commun. 2011;404:889–895. doi: 10.1016/j.bbrc.2010.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implication of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- Lehmann U, Streichert T, Otto B, Albat C, Hasemeier B, Christgen H, Schipper E, Hille U, Kreipe HH, Länger F. Identification of differentially expressed microRNAs in human male breast cancer. BMC Cancer. 2010;10:109. doi: 10.1186/1471-2407-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Snader C. The microRNA.org resource targets and exression. 2008. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farth KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S, Grün D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, Kao HL, Gunsalus KC, Pachter L, Piano F, Rajewsky N. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M. RNA hybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of the site accessibility in microRNAs target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinforma. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Simossis VA, Sethupathy P, Vergoulis T, Koziris N, Sellis T, Tsanakas P, Hatzigeorgiou AG. Accurate microRNAs target prediction correlates with protein repression levels. BMC Bioinforma. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]


