Abstract
BCR-ABL tyrosine kinase plays a major role in the pathogenesis of chronic myeloid leukemia (CML) and is a proven target for drug development. Currently available drugs in the market are effective against CML; however, side-effects and drug-resistant mutations in BCR-ABL limit their full potential. Using high throughput virtual screening approach, we have screened several small molecule databases and docked against wild-type and drug resistant T315I mutant BCR-ABL. Drugs that are currently available, such as imatinib and ponatinib, were also docked against BCR-ABL protein to set a cutoff value for our screening. Selected lead compounds were further evaluated for chemical reactivity employing density functional theory approach, all selected ligands shows HLG value > 0.09900 and the binding free energy between protein-ligand complex interactions obtained was rescored using MM-GBSA. The selected compounds showed least ΔG score −71.53 KJ/mol to maximum −126.71 KJ/mol in both wild type and drug resistant T315I mutant BCR-ABL. Following which, the stability of the docking complexes were evaluated by molecular dynamics simulation (MD) using GROMACS4.5.5. Results uncovered seven lead molecules, designated with Drug-Bank and PubChem ids as DB07107, DB06977, ST013616, DB04200, ST007180 ST019342, and DB01172, which shows docking scores higher than imatinib and ponatinib.
Chronic myeloid leukemia (CML) arises due to the chromosomal aberration in which reciprocal translocation of the Abelson gene on chromosome 9 to break-point cluster region gene on chromosome 22 results in the creation of fusion oncogene, bcr-abl (9; 22) (q34; q11)1. The product of bcr-abl oncogene, BCR-ABL protein is a constitutively active tyrosine kinase that drives the disease CML through phosphorylation of many downstream effector molecules, including Grb2, RAK, ROS, PI3K, JNK, STAT5, AKT and Myc which consequently promoting cell proliferation2,3. Imatinib is the first FDA approved tyrosine kinase inhibitor (TKI) that has shown potent inhibitory effect on the progression of CML4. Despite imatinib's clinical success, resistance due to mutations and side effects are still a limitation of this drug5. To overcome resistance, second generation TKI's inhibitors, nilotinib6 and dasatinib7 were developed. However, neither compound effectively inhibits T315I mutant BCR-ABL, which constitutes 20% of all BCR-ABL mutations8,9,10.
Ponatinib is the only available drug that is designed to overcome T315I gatekeeper mutation11 and is efficient in inhibiting the mutant BCR-ABL12. Ponatinib exhibits triple carbon-carbon (ethynyl linkage) bond between the methyl phenyl and purine groups13,14. It holds the Isoleucine side chain without steric interference and without any loss of hydrogen bond (H_bond)11. It also showed similar binding pattern (DFG-out) as imatinib and nilotinib which interacts with Met318, Asp381 and the side chain of Glu286. In addition, the drug makes van der Waals contacts with Tyr253 and Phe382 because of squeezed conformation of the P-loop and DFG-out mode of activation loop respectively14. Despite potent inhibition capability, ponatinib shows severe side-effects like blood clotting in cardiac valves, chambers and cerebral vessels consequently leading to adverse conditions like myocardial infarction, cardiac stroke and cerebral infraction15. Therefore, a broad spectrum drug capable of inhibiting both wild-type and mutant BCR-ABL with fewer side effects is currently in demand. The availability of 3D-structure of the target protein and the structural details of ponatinib and T315I mutant BCR-ABL protein complex renders an opportunity to identify the most active drug candidate that can efficiently block the catalytic activity of BCR-ABL.
Using a virtual screening approach, we have screened and identified potent drug-like compounds, efficient against both wild-type and mutant BCR-ABL, from a large library of small molecules. Drug molecules exhibiting docking score higher than ponatinib were manually explored in molecular visualization tools in order to analyze their binding patterns. Highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) were calculated using DFT analysis to assess their chemical reactivity. Following which, protein-ligand complexes were incorporated in GROMACS for MD simulation to evaluate their structural stability throughout the trajectory period of simulation. Binding free energy for these complexes were then investigated to cross-check their binding affinity. Results of these studies uncovered seven lead molecules designated as DB07107, DB06977, ST013616, DB04200, ST007180 ST019342, and DB01172. Among these, DB07107, DB06977, ST013616, DB04200 and ST007180 were found to be more effective in blocking drug-resistant T315I mutant than the wild-type BCR-ABL. Interestingly ST019342, and DB01172 were effective only in mutant BCR-ABL.
Methods
Protein selection and preparation
The crystallographic co-ordinates for wild-type BCR-ABL (PDB ID: 3OXZ)16 and mutant T315I (PDB ID: 3QRJ)17 were retrieved from the Protein Data Bank (PDB). Prior to docking, protein structures were prepared by removing water molecules using Schrödinger software. Following which, bond orders were assigned, and hydrogen atoms were added to the crystal structures. Finally, a restrained minimization of the protein structure was performed using the default constraint of 0.30Å RMSD and the OPLS-2005 force field18.
Ligand preparation and grid generation
A total of 36,481 small molecules were retrieved in Structure Data Format (SDF) from various small molecule libraries which includes Ligand.info: Small-molecule Meta-Database19,20 (29,090), Drug Bank21,22 (6,825) and PubChem23 database (566). These small molecules were prepared using the LigPrep wizard of Schrödinger by assigning the bond orders and bond angles and then subjected to minimization using OPLS_2005 force field18. For accurate enumeration of ligand protonation states in biological condition we used Epik24,25. Then, Grid box of size 24 × 24 × 24 A3 was generated keeping ATP binding site of BCR-ABL (Glu286, Met318, Ile360, Ala380, Asp381) as centroid using the “Receptor Grid Generation” of Schrödinger Glide26.
Preparation of reference compounds
Reference compounds of FDA approved drugs such as ponatinib, bosutinib, bafetinib, dasatinib, nilotinib and imatinib were docked in the ATP binding site of both wild-type and mutant BCR-ABL. Ligands were prepared using the LigPrep module of Maestro followed by XP docking. The best docked complex score was set as the cutoff value for screening potential inhibitors of wild-type and mutant BCR-ABL.
Virtual screening
High throughput virtual screening (HTVS) was performed against the BCR-ABL using the Schrödinger software. Here, we used the virtual screening workflow of Glide Maestro for the docking. It performed the docking mainly in three phases namely, HTVS, SP (standard-precision) and XP (extra-precision). Using HTVS, we screened the large library of drug like molecules and reduced the intermediate conformations throughout the HTVS docking process. Successful compounds (10%) from HTVS were further assessed in SP docking for reliable docking of the screened compounds with high accuracy. To eliminate false-positive results, the best 10% of thriving compounds from SP docking were further incorporated for XP docking mode using advanced scoring.
Density functional theory analysis
Electronic effects of drug like molecules plays an important role in the pharmacological effects27. Therefore, drug candidates from the best binding poses were exported to Maestro 9.3 of Schrödinger 2012 version and geometry was optimized in the Jaguar panel using Becke's three-parameter exchange potential28 and Lee-Yang-Parr correlation functional (B3LYP) theory29,30 with 6-31G* basis set. Subsequently, surfaces (molecular orbital, density, potential) and atomic electrostatics potential charges (EPS) were monitored to compute the HOMO and LUMO. HOMO energy proposes the region of the small molecules, which can donate electron during the complex formation, while LUMO energy signifies the capacity of the molecule to accept the electrons from the partner protein. The difference in HOMO and LUMO energy, known as HOMO-LUMO gap energy, indicates the electronic excitation energy31 that is necessary to compute the molecular reactivity and stability of the compounds32.
Molecular dynamics simulations for protein ligand complexes
Two sets of Molecular Dynamics Simulations were performed using Gromacs 4.5.533. In the first set, we evaluated the stability of the mutant type of BCR-ABL and selected the best seven drug candidates from the docking study. In the second set, we evaluated drug candidates with wild-type BCR-ABL. We used MD for performing protein and ligand complexes as described34. The topology file for the selected small molecules were generated using the automated topology builder (ATB)35 in the framework of GROMOS 53A6 force-field36. The protein-ligand complexes were then solvated with TIP3P explicit water molecules and placed in the center of a cubic box of size 24 × 24 × 24 A3. Minimum 1.0 Å distance was maintained between the protein and the edge of the simulation box so that protein can fully immerse with water and rotate freely. Then, Particle Mesh Ewald (PME) method37 was used for the electrostatic energy calculation. It permits the use of the Ewald summation at a computational cost comparable to that of a simple truncation method of 10 Å or less, and the linear constraint solver (LINCS)38 algorithm was used for covalent bond constraints. Before minimization, the system was neutralized by adding 8 Na+ ions. The steepest descent approach (1000 ps) was used for each protein-ligand complex for energy minimization. Further NVT was performed for 100 ps to equilibrate the system with protein and ligand for constant volume, pressure (1 atm) and temperature (300 K). The final MD run was set to 10000 ps for each protein-ligand complex, and trajectories were saved for further analysis using Xmgrace and UCFC Chimera software39.
Rescoring of BCR-ABL and drug candidate complexes using interaction energy and MM-GBSA approach
Interaction energy and Gibbs free energy were calculated using Gromacs and Schrödinger software. Interaction energy for BCR-ABL and drug complexes was calculated by estimating the short range Lennard-Jones and short range Coulomb energy using the g_energy analysis tool of Gromacs software.
 |
Here, Eint stands for interaction energy, ELJ stands for short ranges Lennard-Jones and ECoul denotes short ranges for coulomb energy. It estimates the stability of proteins and drug candidate complexes. The binding free energy was calculated based on the following equation40.
Here, ΔEMM stands for the energy difference between ligands in complex and unliganded receptor.
 |
ΔGsolv is the difference in the GSA solvation energy of the complex and the sum of solvation energy for the unbound ligand protein complex. ΔGSolv is the difference in surface area energy for the complex and the sum of surface area energy for the protein and ligand.
Results and Discussion
Docking of reference compounds for HTVS
To set a cutoff value for docking studies, we docked all FDA approved drug such as ponatinib, bosutinib, bafetinib, dasatinib, nilotinib and imatinib using XP docking. Results showed that ponatinib has the highest binding affinity towards the T315I mutant BCR-ABL with a docking score of −11.050 kcal/mol while bosutinib, bafetinib, dasatinib, nilotinib and imatinib showed −5.513 kcal/mol, −4.689 kcal/mol, −4.702 kcal/mol, −3.772 kcal/mol, −3.593 kcal/mol and −3.78 kcal/mol, respectively (Table 1). Hence, for screening and identifying better lead molecules −11.00 kcal/mol was set as a cutoff value for XP docking.
Table 1. Glide XP results for the existing drug molecules with mutant and wild-type of BCR-ABL (T315I), by Schrödinger 9.3.
| Pubchem ID | Compound Name | Docking Score | HD:HA(Å) |
|---|---|---|---|
| Mutant type of BCR-ABL (T315I) | |||
| 24826799 | Ponatinib | −11.917 | Glu286:OE2::N3(2.865), Met318:N::N5 (2.815), Ile360:O::N2 (3.457), Asp381:N:: O1(2.867) |
| 5328940 | Bosutinib | −5.310 | Asp381:OD2::n2(2.737), Ile360:N::N5 (3.047) |
| 24853523 | Bafetinib | −4.644 | Asp381:O::N3(2.720), Arg362:nh2::n8 (3.223) |
| 3062316 | Dasatinib | −4.220 | Ile360:O:O1(2.827), Arg362:NH1::O1(2.959)Ile360:N::O1(3.095) |
| 644241 | Nilotinib | −3.617 | Arg362:NH1::O1(2.810), Asp381:OD2:N2 (3.029) |
| 5291 | Imatinib | −3.460 | Glu292:OE2::N2(3.092), Ile360:O::N4 (3.027) |
| Wild-type of BCR-ABL | |||
| 24826799 | Ponatinib | −10.285 | Met318: H::N5(1.9), Asp381: H:: O1 (2.1), Glu286: OE2::H17 (2.3), Asp 381: OD2:: H17 (2.3), Asp 381: OD2::H28 (2.5) |
| 5328940 | Bosutinib | −7.462 | Gly249: H::O3 (2.5), Asn322: HD21:N4 (2.0), Asn322: H:N4 (2.5) |
| 24853523 | Bafetinib | −10.188 | Asp381: OD2::H32 (1.9), ASP 381:H::O1 (2.1), GLU286: OE2::H19 (2.1), THR315: OG1::H23 (2.5), ASN322: H::N8 (2.7) |
| 3062316 | Dasatinib | −7.574 | GLU286: OE2::H15 (1.7) |
| 644241 | Nilotinib | −9.691 | Asp381: H:: O1 (2.3), Glu286: OE2::N2 (1.8) |
| 5291 | Imatinib | −10.047 | Asp381: OD2::N2(1.7), Asp381: H:: O1 (2.4), Glu 286: OE2::N3 (1.9) |
Binding pose analysis of the identified compounds
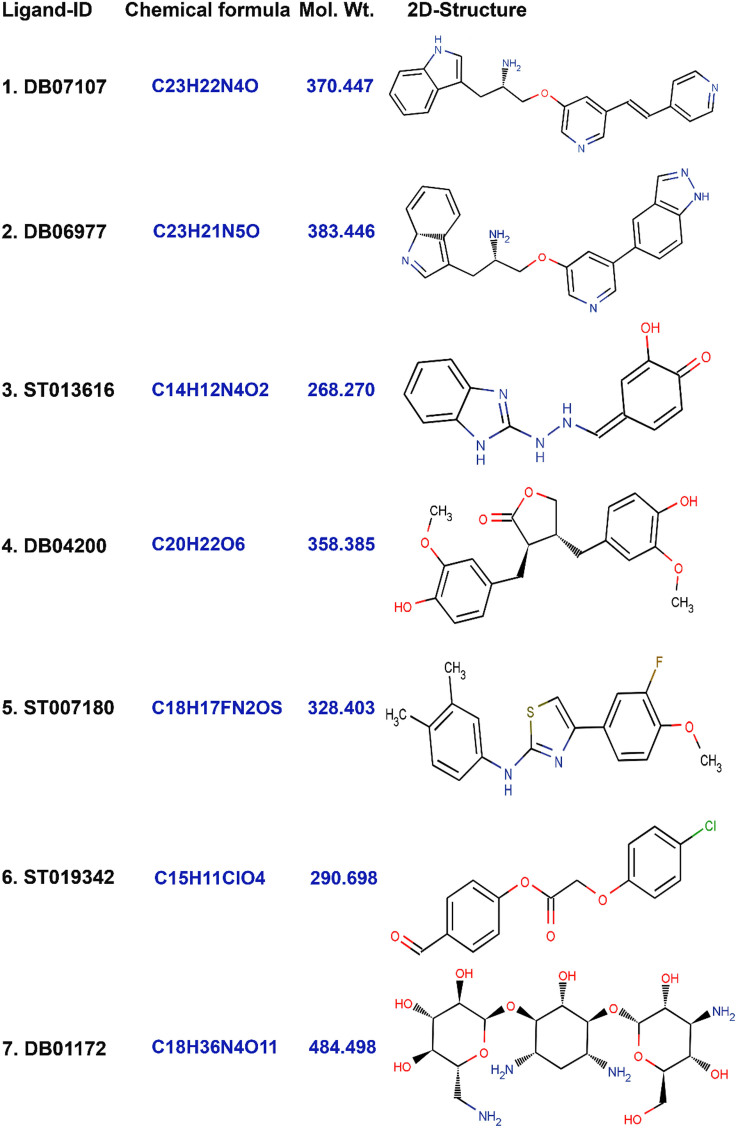
Results of binding pose analysis of seven lead molecules with better binding affinity and higher binding free energy than the reference compounds. Four of which, designated DB07107, DB06977, DB04200 and DB0117, were from DrugBank and ST007180, ST013616 and ST019342 were from the ligand.info database. The 2D conformations and drug details are given in (Figure 1).
Figure 1. 2-D Structures of the finally selected seven lead molecules have been shown in the figure.
(1) DB07107, (2) DB06977, (3) ST013616, (4) DB04200, (5) ST007180, (6) ST019342, and (7) DB01172.
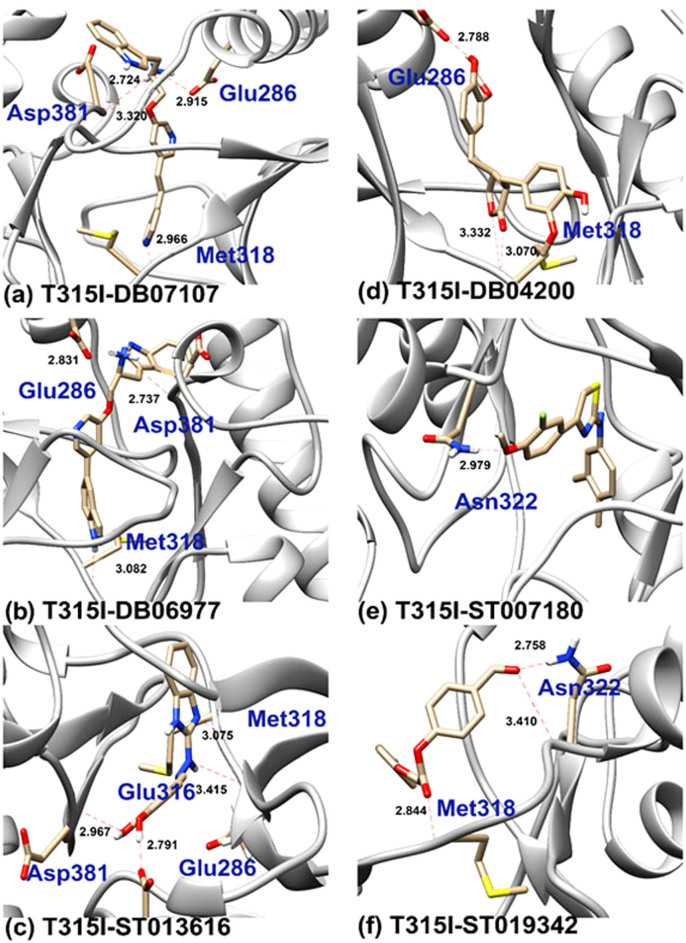
Interestingly, DB07107 (C23H22N4O) from DrugBank showed the highest binding energy with XP score of −14.045 kcal/mol (Figure 2a). To get an insight into their interacting pattern, we used UCFC Chimera molecular visualization tool and Glide for generating 2D interaction plots. Docking pose analysis revealed four hydrogen bonds (H_bonds) interactions with the ATP binding site residues of BCR-ABL. Here, we observed single H_bond with each Glu286 and Met318 residues with bond length of 2.92Å and 2.97Å while two H_bond formations were observed with Asp381 with the bond length of 3.32Å and 2.72Å.
Figure 2. Binding poses of the DB07107, DB06977, ST013616, DB04200, ST007180 and ST019342 lead molecules.
The proposed binding mode of the lead molecules has been shown in the stick format. Residues involved in Hydrogen bonding have been labeled with the Hydrogen bond in dotted red lines and bond length have been shown in Angstrom.
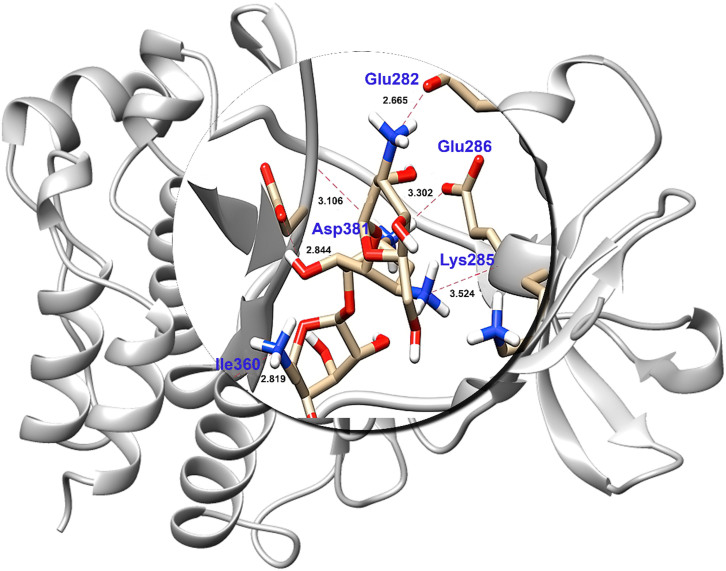
Other potent drugs, DB06977 (C23H21N5O), ST013616 (C14H12N4O2), DB04200 (C20H22O6), ST007180 (C18H17FN2OS), ST019342 (C15H11ClO4) (Figure 2b,2c,2d,2e,2f) and DB01172 (C18H36N4O11) (Figure 3) showed high binding affinity with XP scores of −13.163 kcal/mol, −12.065 kcal/mol, −12.041 kcal/mol, −11.555 kcal/mol, −11.033 kcal/mol and −8.433 kcal/mol, respectively (Table 2). Here, it is noticeable that the key residues of Glu286, Meth318 and Asp381 were involved in the H_bond interactions as has been observed with the marketed drug, ponatinib. Although DB01172 exhibits XP docking scored below the cuttoff value, we consider it as a potent inhibitor for BCR-ABL because it showed the highest number of H_bond with the hot spot residues of BCR-ABL. Moreover, it uncovered one side chain hydrogen bond with Glu282, Glu286, Asp381, Lys285 and one backbone hydrogen bond each with Asp381, Ile360. Therefore, we considered DB01172 for our further analysis.
Figure 3. Binding poses of the DB01172 lead molecule.
The proposed binding mode of the lead molecules has been shown in the stick format. Residues involved in Hydrogen bonding have been labeled with the Hydrogen bond in dotted red lines and bond length have been shown in Angstrom.
Table 2. Glide XP results for the seven lead molecules with the mutant (T315I) and wild-type of BCR-ABL, by Schrödinger 9.3.
| Drug Bank ID | Docking Score | HD:HA(Å) | |
|---|---|---|---|
| T315I mutant | DB07107 | −14.031 | Glu286:OE2:: N4(2.92), Met318:N:: N1 (2.97), Asp381:O1:: N(3.32), Asp381:N4:: O (2.72) |
| DB06977 | −13.163 | Glu286:Oe2:: N5(2.81), Glu381:N5:: O (2.73), Met318:O:: N3(3.08) | |
| ST013616 | −12.106 | Glu286:OE2:: O1(2.79), Met381:O2:: O(2.96), Glu316:O: N2 (3.41), Met318:N:: N3(3.07) | |
| DB04200 | −12.065 | Glu286:OE2::O5(2.78), Met318: N:: O2 (3.07), Met318:N::O1(3.33) | |
| ST007180 | −11.555 | Asn322 ND2: O1 (2.79) | |
| ST019342 | −11.033 | Met318 N1: N (2.84), Asn322 N: O4 (3.41)Asn322 Nd2: O4 (2.75) | |
| DB01172 | −8.603 | Glu282 OE1: N4 (2.66), Lys285 O: N3 (3.52), Glu286 OE2: N2 (3.03), Ile360 O: N1 (2.82), Asp381 OD2: O4 (2.84), Asp381 O: N2 (3.10) | |
| Wild-type | DB07107 | −10.22 | Glu286:Oe2::N2(2.658), Asp381:O::N2 (2.600), Asp381:N::O1(3.497), Met318:N:: N5(3.043) |
| DB06977 | −10.94 | Asp381:O::N2(2.599), Glu286:Oe2::N2 (2.727) | |
| ST013616 | −8.17 | Lys 271:N2::O1(3.031), Glu286:Oe2::O2 (2.784), Glu286:Oe2::O1(2.627), Glu316: O::N2(3.228), Met318:N::N3(3.020) | |
| DB04200 | −9.76 | Met318:N::O1(3.177), Asn322:O3:: Nd2(2.972) Asp381:O::O6(2.915) | |
| ST007180 | −9.62 | Met318:O::N1(3.215), Met318:O:: S1(3.907) | |
| ST019342 | −8.33 | Met318:N::O2(3.016), Asn322:Nd2::O4 (3.129) | |
| DB01172 | −9.72 | Ile360:O::O8(2.900), Glu286:Oe2::N2(3.136), Asp381:N::O10 (2.787), Asp381: O::O10 (3.227), Asp381:Od2::O7(3.486), Glu286:Oe2::O3(2.963), His361:O::N4 (2.800), Asp381:Od2::Od2(2.647), Arg362:Nh1::O8 (3.365) |
Assessment of identified drugs in Wild-type of BCR-ABL
To determine the efficacy of the selected drug in binding to and inhibiting wild-type of BCR-ABL we performed docking studies as described in the methods section. To compare their efficacy and binding pattern with our screened compounds we again docked FDA approved drugs with wild-type of BCR-ABL.
As shown in table 2, DB07107 and DB06977 showed better docking scores than existing drugs while others showed results comparable to those of marketed drugs like ponatinib, bosutinib, bafetinib, dasatinib, nilotinib and imatinib. In the case of DB06977, although one H_bond is missing (Met318) with the wild-type of BCR-ABL complex compared to T315I mutant type, it still exhibits a favorable binding score of −10.94 kcal/mol. While, in the case of ST013616, one extra H_bond interaction is found to interact with Lys271. However, it did not show H-bond interaction with Asp381. Most of the selected drugs in these studies showed slightly less efficacy against wild-type compared to mutant BCR-ABL but better when compared to ponatinib. These investigations lead us to conclude that DB07107, DB06977, ST013616, DB04200 and ST007180 are more effective against the mutant than the wild-type BCR-ABL. ST019342, and DB01172 are effective only against mutant BCR-ABL.
DFT analysis
HOMO-LUMO plays an important role in stabilizing the interactions between drug and receptor protein32. Hence, the orbital energy of both HOMO and LUMO and the gap between them was calculated to estimate the chemical reactivity of the selected compounds using DFT (Table 3). HOMO-LUMO plots were generated to analyze the atomic contribution for these orbitals. The plots of HOMO and LUMO show the positive electron density in red color and negative electron density in blue (Figures 4 and 5). All the selected drug candidates showed minimal HOMO-LUMO gap with the average energy difference of 0.13 eV, signifying molecular reactivity. The lowest energy gap was exhibited by DB06977 with the HOMO-LUMO gap energy of 0.09911 eV and the highest energy gap was observed in DB04200 with the gap energy of 0.18442 eV (Table 3). HOMO energy was higher for DB07107 (Figure 4) and DB01172 (Figure 5), when compared to LUMO energy indicating an ability to donate the electrons rather than accept electrons with their partner receptor-binding site region.
Table 3. Frontier orbital energies of best seven lead compounds.
| Sl.No | Compound | HOMO (eV) | LUMO (eV) | HLG (eV)* |
|---|---|---|---|---|
| 1. | DB07107 | −0.29572 | −0.14704 | 0.14868 |
| 2. | DB06977 | −0.28270 | −0.18359 | 0.09911 |
| 3. | ST013616 | −0.18235 | −0.06262 | 0.11973 |
| 4. | DB04200 | −0.19362 | −0.00920 | 0.18442 |
| 5. | ST007180 | −0.18628 | −0.03046 | 0.15582 |
| 6. | ST019342 | −0.23036 | −0.07439 | 0.15597 |
| 7. | DB01172 | −0.57549 | −0.40945 | 0.16604 |
Figure 4. Plots of highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) of DB07107, DB06977, ST013616 and DB04200.
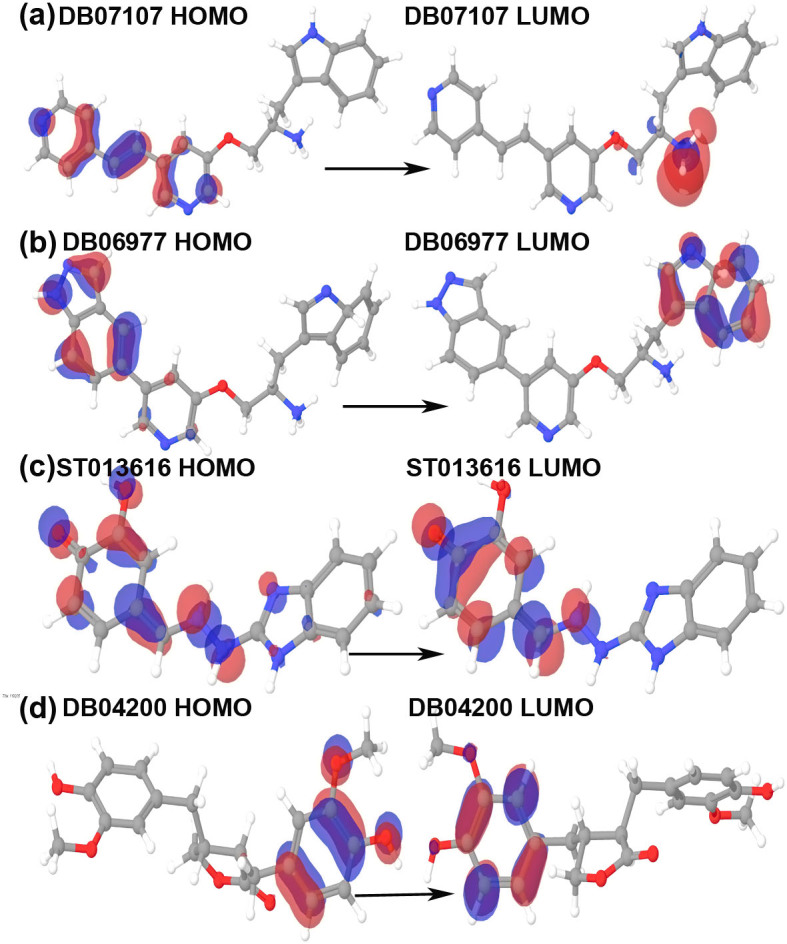
The positive electron density has been shown in red color while negative have been shown in blue.
Figure 5. Plots of highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) ST007180, ST013942 and DB01172.
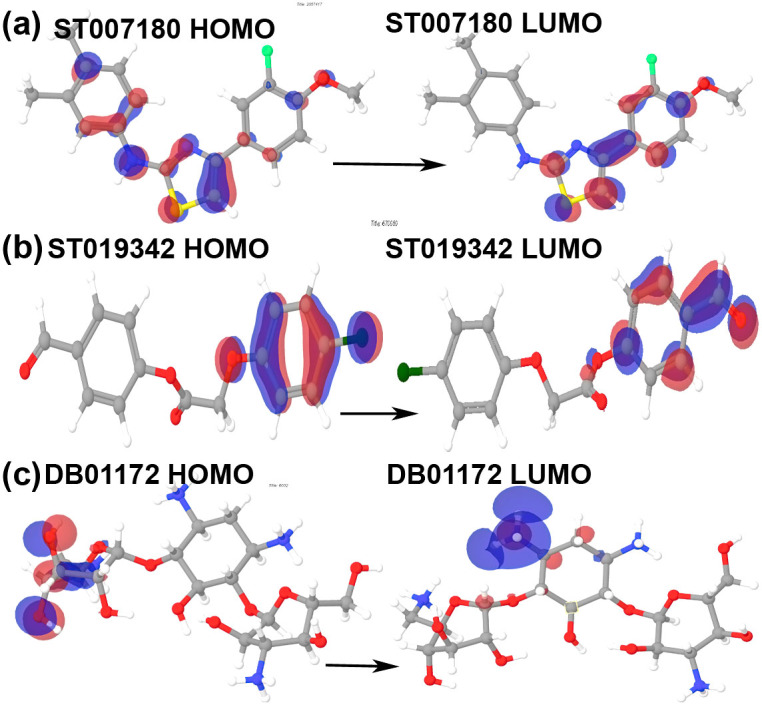
The positive electron density has been shown in red color while negative have been shown in blue.
Molecular dynamics simulations
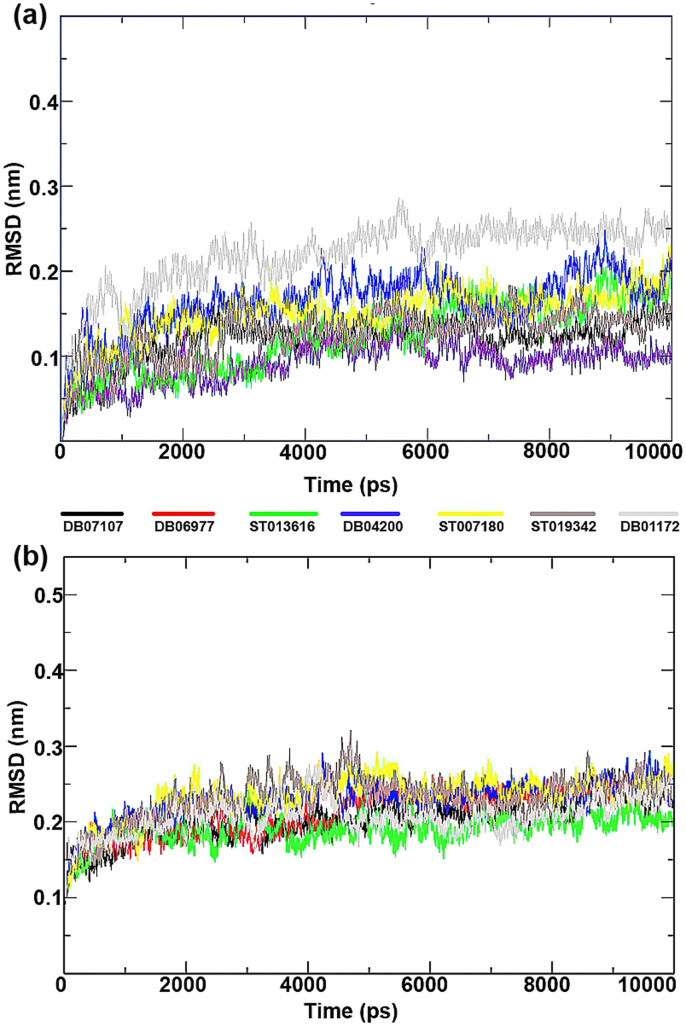
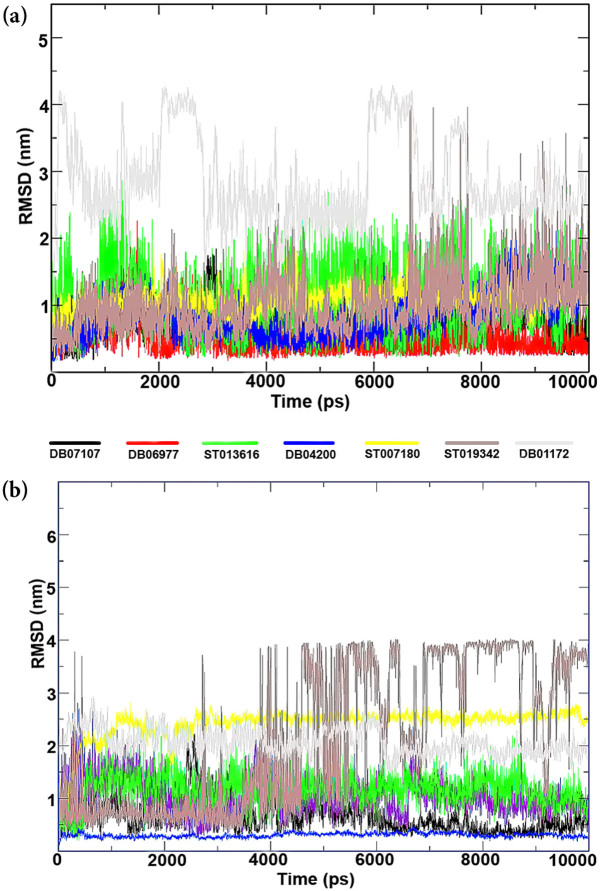
To compare the structural behavior and flexibility of the wild-type and mutant BCR-ABL, all lead compounds were incorporated in Gromacs4.5.5 and MD was performed for 10 ns of each complexes. Root mean square deviations (RMSD) of the wild-type and mutant BCR-ABL were calculated against their initial structure in the protein-ligand complexes and graphs were generated to compare the flexibility of the backbone of the proteins using the Xmgrace software. Throughout the simulation period, no significant fluctuations were observed in the backbone of the wild-type and mutant T315I BCR-ABL, implying that the binding of drug candidates at the active site of the proteins is not only stable and strong but also does not disturb the protein backbone stability (Figure 6).
Figure 6. Time dependence of root mean square deviations (RMSDs) of the Backbone of mutant (T3I51)and wild type of BCR-ABL have been shown in figure against the initial structure during 10,000 ps molecular dynamics (MD) simulation.
To ensure the binding stability of the drug candidates in the active site of proteins, ligand positional RMSD of each lead molecule were generated and analyzed as described study41. ST019342 showed more and continues fluctuations in the noticeable window size of 0.2–2.5 nm (Figure 7a) for wild-type and 0.2–4 nm for the mutant BCR-ABL complex (Figure 7b). Co-ordinates of ST019342-BCR-ABL (wild-type and mutant) complex were downloaded from the trajectory in the interval of 1000 ps and investigated in PyMOL for protein-ligand interactions. Our investigation uncovered that ST019342 has an unstable binding affinity towards BCR-ABL. It exhibits weak hydrogen bond interactions with the receptor binding site, which leads to the inefficient inhibition of BCR-ABL. Upon investigation of downloaded timeframe of MD, we observed that after 2 to 5 ns ST019342 was not bound to the binding pocket. It suggests its inability to inhibit the protein target efficiently while other drug candidates have shown stable and strong binding affinity.
Figure 7. Backbone RMSD values of drug candidates from both the types (wild and mutant) of protein ligand complexes were generated against the initial structures of protein-ligand complexes during 10,000 ps of molecular dynamics (MD) simulation period.
Graphs were plotted using Xmgrace, a 3-D plotting tool.
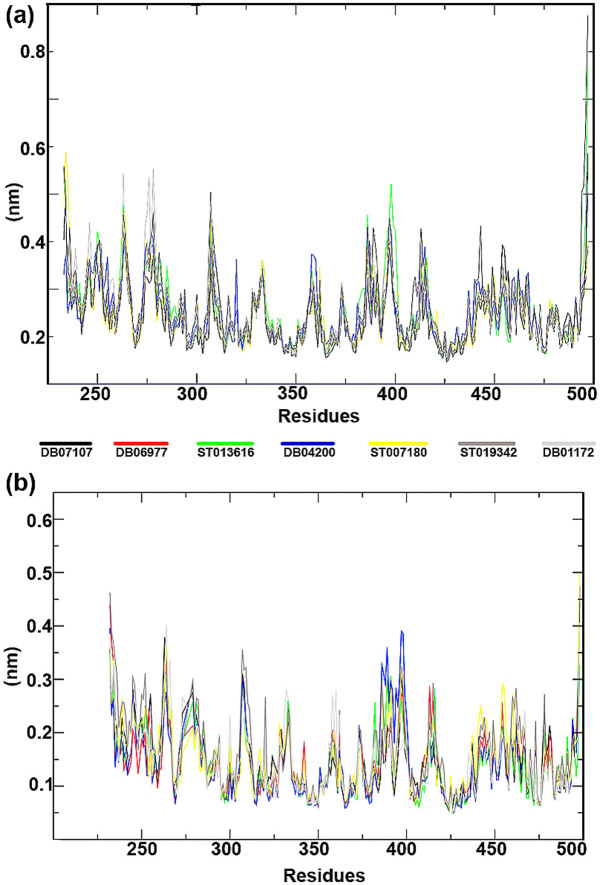
In order to calculate the residual mobility of each lead molecules in BCR-ABL-ligand complexes (wild-type and mutant), Root Mean Square Fluctuation (RMSF) was calculated in each complexes and the graph was plotted against the residue number based on the trajectory period of MD simulation. None of the drug candidates showed noticeable changes in their residual level as the general profile of residual fluctuation of wild-type and T315I is minimal in each complex without any abnormal fluctuation (Figure 8).
Figure 8. Root mean square fluctuations (RMSF) from the initial structures of protein-ligand complexes during the trajectory period of simulation.
Hydrogen bond analysis
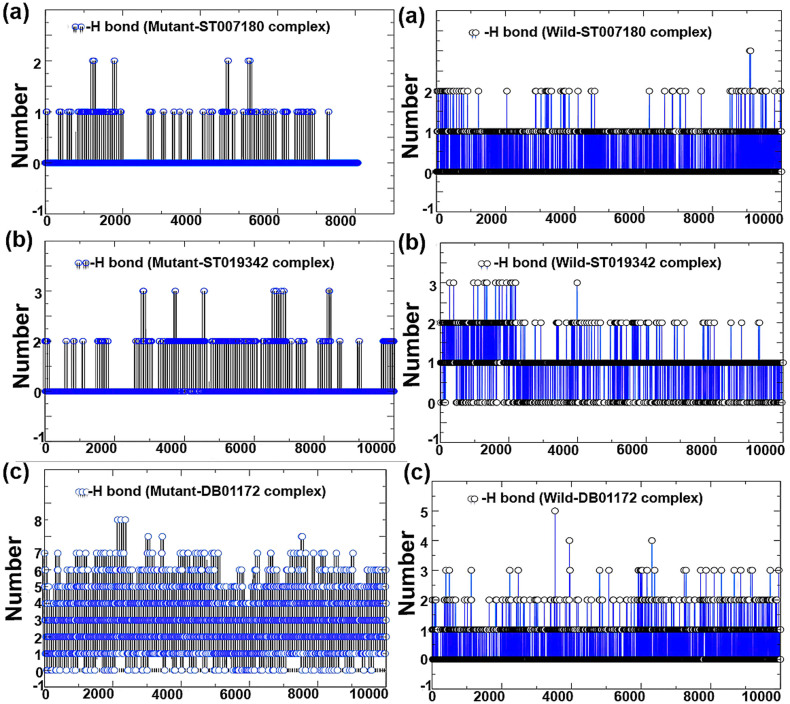
To determine the stability of hydrogen bonds with the ATP binding site of protein, MD analysis of BCR-ABL and the selected drug candidate's complex stability were monitored during the trajectory period. Hydrogen bond profiles between the selected drugs and BCR-ABL (T315I and wild-type) were calculated using the g_hbond utility of GROMACS. This analysis revealed that DB01172 comprises 6–7 (highest) average H_bonds during the simulation period sharing four H_bonds with Glu282, Lys285, Glu286 and Ile360, and two H_bonds with Asp381 (Figures 9 and 10) in T315I (mutant) while Wild-type showed poor H_bond interactions with 1–2 H_bonds on average during the trajectory period. The same pattern was also observed in the case of ST007180 and ST019342. ST007180 and ST019342 showed less number of hydrogen bond plots in the mutant BCR-ABL compared to wild type. To gain insight into the hydrogen bond instability, we downloaded the drug-receptor complexes in the interval of one ns for DB01172, ST007180 and ST019342. Following which, the structures were superimposed to monitor the binding pose of the drug candidates throughout the trajectories. Our investigation revealed that DB01172 is moving away from the ATP binding site of BCR-ABL in both mutant and wild-type towards the north-west (130°) and east direction (180°) (Figure 11). Compound ST007180 showed satisfactory results, while ST019342 showed slight movement in the binding site.
Figure 9. Total number of inter-molecular H_bond interactions between the drug compounds (DB07107, DB06977, ST013616 and DB04200) and BCR-ABL (mutant and wild).
Figure 10. Total number of inter-molecular H_bond interactions between BCR-ABL (mutant and wild) and ST007180, ST019342 and DB01172.
Figure 11. Spnapshot of the superimposed structures of mutant and wild type of DB01172 and ST007180 and ST019342.

Stuctures were downloaded from the trajectory file in the interval of 1 ns.
Other complexes, DB07107, DB06977, ST013616 and DB04200 exposed an average number of hydrogen bonds 2–3 (Glu286, Met318 and Asp381), 3 (Glu286, Met318 and Asp381), 3–4 (Glu286, Asp381, Glu316, Met318), 2–3 (Glu286 and Met318), 1 (322) and 1–2 (Met318 and Asn322), respectively in both wild-type and mutant BCR-ABL. The average number of H_bonds throughout the MD period signifies their continuous contribution in H_bond interactions, which provides stability to the complexes in holding it to the ATP binding position. This suggests that the functionality and ability of these compounds can efficiently inhibit BCR-ABL.
Interaction energy and binding free energy
To estimate the binding association between wild-type and mutant of BCR-ABL, two different approaches were carried out. Firstly, crude interaction energy was estimated based on the short-range energy of the system. Next, to determine the binding affinity between BCR-ABL and drug complexes we scored the MM-GBSA energy of the complexes. Interaction energy data suggests that all the selected drug compounds have higher interaction energy of 203444.3 kcal/mol, −203449.57 kcal/mol, −292066.20 kcal/mol, −310571.94 kcal/mol, −295967.25 kcal/mol, −292055.68 kcal/mol, −292134.56 kcal/mol in DB07107, DB06977, ST013616, DB04200, ST007180, ST019342, and DB01172 respectively, for the mutant than wild-type which showed −232088.91 kcal/mol, −232068.12 kcal/mol, −232062.86 kcal/mol, −232033.94 kcal/mol, −232030.35 kcal/mol, −232091.54 kcal/mol, and −232062.86 kcal/mol for DB07107, DB06977, ST013616, DB04200, ST007180, ST019342, and DB01172 respectively (Table 4).
Table 4. Interactions energy and binding free energy of mutant and wild-type of BCR-ABL-ligand complexes.
| Ligand | Averagea | Average (wild-type) | Int. energya (Kcal/mol) | Int. energyb (Kcal/mol) | MM-GBSAa | MM-GBSAb |
|---|---|---|---|---|---|---|
| DB07107 | 159619 LJ (SR)−1.01083e + 06 Q(SR) | 184840 LJ (SR)−1.1559e + 06 Q (SR) | −203444.31 | −232088.91 | −112.76 | −112.25 |
| DB06977 | 159597 LJ (SR)−1.01083e + 06 Q(SR) | 184807 LJ (SR)−1.15578e + 06 Q (SR) | −203449.57 | −232068.12 | −126.71 | −124.78 |
| ST013616 | 237095 LJ(SR)−1.4591e + 06 Q(SR) | 184949 LJ (SR)−1.1559e + 06 Q (SR) | −292066.20 | −232062.86 | −71.53 | −82.32 |
| DB04200 | 159667 LJ (SR)−1.4591e + 06 Q(SR) | 184780 LJ (SR) -−1.15561e + 06 Q (SR) | −310571.94 | −232033.94 | −103.15 | −103.08 |
| ST007180 | 243003 LJ(SR)−1.48133e + 06 Q(SR) | 184855 LJ (SR)−1.15567e + 06 Q (SR) | −295967.25 | −232030.35 | −93.59 | −81.26 |
| ST019342 | 237089 LJ (SR)−1.45905e + 06 Q(SR) | 184879 LJ (SR)−1.15595e + 06 Q (SR) | −292055.68 | −232091.54 | −73.73 | −75.83 |
| DB01172 | 237089 LJ(SR)−1.45938e + 06 Q(SR) | 184949 LJ (SR)−1.1559e + 06 Q (SR) | −292134.56 | −232062.86 | −83.82 | −72.25 |
a: mutant.
b: wild-type.
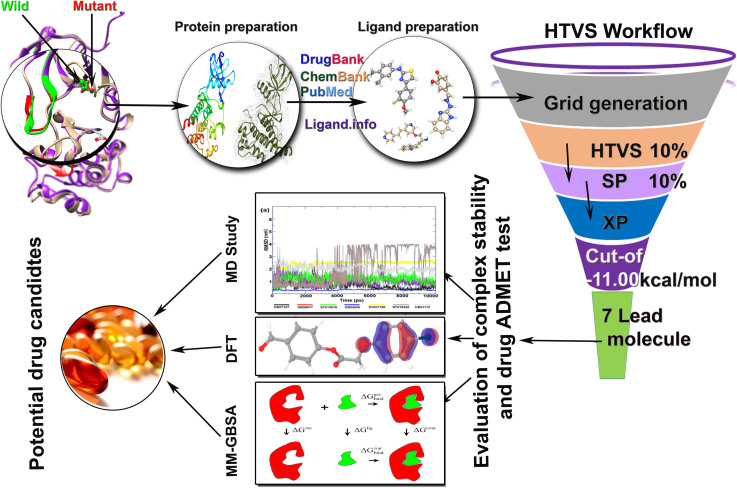
Re-scoring of complexes using Prime MM-GBSA results indicated that all the selected drug candidates displayed higher binding free energy (Table 4). Moreover, all of them displayed results ideal for being a drug candidate in the treatment of CML bearing wild-type or mutant BCR-ABL. Our studies show that these drugs can efficiently inhibit BCR-ABL by binding to DFG out conformation of BCR-ABL protein and can obstruct its catalytic activity. Hence, we recommend DB07107, DB06977, ST013616, DB04200 and ST007180 to be experimentally tested and further authenticated. Since ST019342 and DB01172 are not recommended in the broad-spectrum purpose due to their unbalanced backbone conformation in wild-type during the MD study, we recommend for further experimentation and use against T315I. The entire work flow chart of this study is shown in Figure 12.
Figure 12. Graphical abstract.
Conclusions
Using a virtual screening approach, we performed a molecular docking study for both wild-type and mutant (T315I) of BCR-ABL. The study yielded seven lead molecules which showed promising results against both wild-type and T315I mutant BCR-ABL. The bioactivity of these selected lead compounds were determined using frontier orbital study and the results showed that the selected drug candidates are chemically reactive. The stability of the protein-ligand complexes were investigated through 10 ns of MD simulation of each complex for both types. MD simulation studies illustrated the dynamic behavior of protein-ligand complexes. Results showed that the protein backbone and ligand backbone of DB07107, DB06977, ST013616, DB04200, ST007180 and DB01172 compounds are stable throughout the simulation period without any significant fluctuation within this period. In contrast, backbone of ST019342 showed anomalous fluctuations throughout the simulation period while DB01172 showed instability in RMSD graph with the wild-type BCR-ABL. Hydrogen bond analysis revealed that DB01172 has six hydrogens on average, while in wild-type it has 2–3 on average, implying low efficacy towards the wild-type of target. Therefore, in terms of broad spectrum therapy for CML ST019342 and DB01172 drugs are not recommended while DB07107, DB06977, ST013616, DB04200 and ST007180 showed remarkable results during the trajectory period. These drugs promise a new gateway for the further development of anti CML therapeutics targeting BCR-ABL. In summary, DB07107, DB06977, ST013616, DB04200 and ST007180 are more effective in inhibiting mutant than wild-type of BCR-ABL. ST019342 and DB01172 are effective only in mutant BCR-ABL.
Author Contributions
H.N., O.P. and R.B. conceived the study. H.N. and O.P. performed the model output analysis and wrote the initial draft of the paper. H.N., O.P., M.S.K. and R.B. contributed to interpreting results, discussion of the associated dynamics, and improvement of this paper.
Acknowledgments
The research work was carried out in the laboratory of the Centre for Excellence in Bioinformatics, and Department of Biochemistry & Molecular Biology, Pondicherry University-India. Hemanth Naick thanks Pondicherry University and UGC-BSR Research fellowship for the financial assistant. Om Prakash Sharma is grateful to the Council of Scientific & Industrial Research (CSIR), India for the Senior Research Fellowship (09/559/(0085)/2012/EMR-I) to pursue their Ph.D. degree.
References
- Quintas-Cardama A. & Cortes J. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood. 113, 1619–1630 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergast A. M. et al. BCR-ABL-induced oncogenesis is mediated by direct interaction with the SH2 domain of the GRB-2 adaptor protein. Cell. 75, 175–185 (1993). [PubMed] [Google Scholar]
- Deininger M. W., Goldman J. M. & Melo J. V. The molecular biology of chronic myeloid leukemia. Blood. 96, 3343–3356 (2000). [PubMed] [Google Scholar]
- Capdeville R., Buchdunger E., Zimmermann J. & Matter A. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat Rev Drug Discov. 1, 493–502 (2002). [DOI] [PubMed] [Google Scholar]
- O'Hare T., Eide C. A. & Deininger M. W. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood. 110, 2242–2249 (2007). [DOI] [PubMed] [Google Scholar]
- Weisberg E. et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 7, 129–141 (2005). [DOI] [PubMed] [Google Scholar]
- Sawyers C. L. Even better kinase inhibitors for chronic myeloid leukemia. N. Engl. J. Med. 362, 2314–2315 (2010). [DOI] [PubMed] [Google Scholar]
- O'Hare T. et al. In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer Res. 65, 4500–4505 (2005). [DOI] [PubMed] [Google Scholar]
- Deguchi Y. et al. Comparison of imatinib, dasatinib, nilotinib and INNO-406 in imatinib-resistant cell lines. Leukemia Res. 32, 980–983 (2008). [DOI] [PubMed] [Google Scholar]
- Redaelli S. et al. Activity of bosutinib, dasatinib, and nilotinib against 18 imatinib-resistant BCR/ABL mutants. J. Clin. Oncol. 27, 469–471 (2009). [DOI] [PubMed] [Google Scholar]
- O'Hare T. et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 16, 401–412 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare T. et al. Inhibition of wild-type and mutant Bcr-Abl by AP23464, a potent ATP-based oncogenic protein kinase inhibitor: implications for CML. Blood. 104, 2532–2539 (2004). [DOI] [PubMed] [Google Scholar]
- Frankfurt O. & Licht J. D. Ponatinib--a step forward in overcoming resistance in chronic myeloid leukemia. Clin Cancer Res. 19, 5828–5834 (2013). [DOI] [PubMed] [Google Scholar]
- Reddy E. P. & Aggarwal A. K. The ins and outs of bcr-abl inhibition. Genes & cancer. 3, 447–454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariad suspends ponatinib sales. Cancer Discov. 4, 6–7 (2014). [DOI] [PubMed] [Google Scholar]
- Zhou T. et al. Structural mechanism of the Pan-BCR-ABL inhibitor ponatinib (AP24534): lessons for overcoming kinase inhibitor resistance. Chem Biol Drug Des. 77, 1–11 (2011). [DOI] [PubMed] [Google Scholar]
- Chan W. W. et al. Conformational control inhibition of the BCR-ABL1 tyrosine kinase, including the gatekeeper T315I mutant, by the switch-control inhibitor DCC-2036. Cancer Cell. 19, 556–568 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks J. L. et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 26, 1752–1780 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Grotthuss M., Pas J. & Rychlewski L. Ligand-Info, searching for similar small compounds using index profiles. Bioinformatics. 19, 1041–1042 (2003). [DOI] [PubMed] [Google Scholar]
- von Grotthuss M., Koczyk G., Pas J., Wyrwicz L. S. & Rychlewski L. Ligand.Info small-molecule Meta-Database. Comb Chem High Throughput Screen. 7, 757–761 (2004). [DOI] [PubMed] [Google Scholar]
- Knox C. et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 39, D1035–1041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S. et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36, D901–906 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan E., Y W., Paul A. & Stephen H. PubChem: Integrated Platform of Small Molecules and Biological Activities. Annu Rep Comput Chem. 4, 217–240 (2008). [Google Scholar]
- Greenwood J. R., Calkins D., Sullivan A. P. & Shelley J. C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput. Aided Mol. Des. 24, 591–604 (2010). [DOI] [PubMed] [Google Scholar]
- Shelley J. C. et al. Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 21, 681–691 (2007). [DOI] [PubMed] [Google Scholar]
- Friesner R. A. et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 49, 6177–6196 (2006). [DOI] [PubMed] [Google Scholar]
- Matysiak J. Evaluation of electronic, lipophilic and membrane affinity effects on antiproliferative activity of 5-substituted-2-(2,4-dihydroxyphenyl)-1,3,4-thiadiazoles against various human cancer cells. Eur. J. Med. Chem. 42, 940–947 (2007). [DOI] [PubMed] [Google Scholar]
- Becke A. D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652 (1993). [Google Scholar]
- Gill P. M. W., Johnson B. G., Pople J. A. & Frisch M. J. The performance of the Becke—Lee—Yang—Parr (B—LYP) density functional theory with various basis sets. Chem. Phys. Lett. 197, 499–505 (1992). [Google Scholar]
- Stephens P. J., Devlin F. J., Chabalowski C. F. & Frisch M. J. Ab Initio Calculation of Vibrational Absorption and Circular Dichroism Spectra Using Density Functional Force Fields. J. Phys. Chem. 98, 11623–11627 (1994). [Google Scholar]
- Zhan C.-G., Nichols J. A. & Dixon D. A. Ionization Potential, Electron Affinity, Electronegativity, Hardness, and Electron Excitation Energy: Molecular Properties from Density Functional Theory Orbital Energies. J. Phys. Chem. A. 107, 4184–4195 (2003). [Google Scholar]
- Zheng Y. et al. Design, synthesis, quantum chemical studies and biological activity evaluation of pyrazole-benzimidazole derivatives as potent Aurora A/B kinase inhibitors. Bioorg. Med. Chem. Lett.s. 23, 3523–3530 (2013). [DOI] [PubMed] [Google Scholar]
- Pronk S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 29, 845–854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma O. P., Vadlamudi Y., Liao Q. H., Strodel B. & Kumar M. S. Molecular modeling, dynamics, and an insight into the structural inhibition of cofactor independent phosphoglycerate mutase isoform 1 from Wuchereria bancrofti using cheminformatics and mutational studies. J Biomol Struct Dyn. 31, 765–778 (2013). [DOI] [PubMed] [Google Scholar]
- Malde A. K. et al. An Automated Force Field Topology Builder (ATB) and Repository: Version 1.0. J. Chem. Theory Comput. 7, 4026–4037 (2011). [DOI] [PubMed] [Google Scholar]
- Oostenbrink C., Villa A., Mark A. E. & van Gunsteren W. F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 25, 1656–1676 (2004). [DOI] [PubMed] [Google Scholar]
- Wang H., Dommert F. & Holm C. Optimizing working parameters of the smooth particle mesh Ewald algorithm in terms of accuracy and efficiency. J Chem Phys. 133, 034117 (2010). [DOI] [PubMed] [Google Scholar]
- Amiri S., Sansom M. S. & Biggin P. C. Molecular dynamics studies of AChBP with nicotine and carbamylcholine: the role of water in the binding pocket. Protein Eng Des Sel. 20, 353–359 (2007). [DOI] [PubMed] [Google Scholar]
- Pettersen E. F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- Lyne P. D., Lamb M. L. & Saeh J. C. Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. J Med Chem. 49, 4805–4808 (2006). [DOI] [PubMed] [Google Scholar]
- Sharma O. P. et al. Modeling, docking, simulation, and inhibitory activity of the benzimidazole analogue against beta-tubulin protein from Brugia malayi for treating lymphatic filariasis. Med Chem Res. 21, 2415–2427 (2012). [Google Scholar]