Abstract
Within two years of the re-discovery of Mendelism, Bateson and Saunders had described six traits in non-laboratory animals (five in chickens and one in cattle) that show single-locus (Mendelian) inheritance. In the ensuing decades, much progress was made in documenting an ever-increasing number of such traits. In 1987 came the first discovery of a causal mutation for a Mendelian trait in non-laboratory animals: a non-sense mutation in the thyroglobulin gene (TG), causing familial goitre in cattle. In the years that followed, the rate of discovery of causal mutations increased, aided mightily by the creation of genome-wide microsatellite maps in the 1990s and even more mightily by genome assemblies and single-nucleotide polymorphism (SNP) chips in the 2000s. With sequencing costs decreasing rapidly, by 2012 causal mutations were being discovered in non-laboratory animals at a rate of more than one per week. By the end of 2012, the total number of Mendelian traits in non-laboratory animals with known causal mutations had reached 499, which was half the number of published single-locus (Mendelian) traits in those species. The distribution of types of mutations documented in non-laboratory animals is fairly similar to that in humans, with almost half being missense or non-sense mutations. The ratio of missense to non-sense mutations in non-laboratory animals to the end of 2012 was 193:78. The fraction of non-sense mutations (78/271 = 0.29) was not very different from the fraction of non-stop codons that are just one base substitution away from a stop codon (21/61 = 0.34).
Keywords: genome assembly, microsatellite, missense, non-laboratory animals, non-sense, Online Mendelian Inheritance in Animals (OMIA), single nucleotide polymorphisms (SNPs), SNP chip
Introduction
Mendelian traits in non-laboratory animals
As soon as Mendelism was rediscovered in 1900, enormous scientific effort was devoted to investigating the extent to which Mendelian inheritance occurs across the plant and animal kingdoms. Progress was spectacular. Just two years later, in his first report to the Evolution Committee of the Royal Society, Bateson (1902) described five Mendelian animal traits (all in chickens), namely pea comb (OMIA 000782-9031), rose comb (OMIA 000884-9031), polydactyly (OMIA 000810-9031), shank (skin) colour (OMIA 001449-9031) and white plumage (dominant white; OMIA 000373-9031). In an adjacent paper in the same report, Bateson & Saunders (1902) added polled in cattle (OMIA 000483-9913). Interestingly, they also speculated (correctly, as it turns out) that the characteristic plumage colour of Blue Andalusian chickens (OMIA 001154-9031) is due to heterozygosity. In the second report to the Royal Society's Evolution Committee, Bateson & Punnett (1905) presented an early example of a classic 9:3:3:1 Mendelian ratio, for crosses involving the pea comb locus and the rose comb locus. In reporting that the double-dominant genotype in these results is the ‘walnut’ comb, they also reported the first-ever example of interaction between alleles at different loci, for which Bateson (1907) a few years later coined the term ‘epistatic’. The second report also included a lengthy account by Hurst (1905) of extensive crossing experiments in chickens. In addition to the traits included in the 1902 reports, Hurst added feathered shanks (OMIA 000839-9031) and crested head (OMIA 000240-9031), plus a few others for which the evidence was probably insufficient. The strong emphasis on chickens in these early reports is a reflection of the relative ease and speed of breeding this species, compared with other domesticated animal species.
Whole books on Mendelism soon appeared. Punnett's (1905) Mendelism included ‘unpigmented’ (albinism; OMIA 000202-9986) and long hair (angora; OMIA 000439-9986) in rabbits and pea comb, rose comb, dominant white and Blue Andalusian in chickens. Bateson's (1909) Mendel's Principles of Heredity included Manx tail (OMIA 000975-9685) and polydactyly (OMIA 000810-9685) in cats; frizzle (OMIA 000394-9031) and rumplessness (OMIA 001633-9031) in chickens; and various coat colours in cats, cattle and horses. In 1911, Castle's Heredity in Relation to Evolution and Animal Breeding included polled in sheep (OMIA 000483-9940).
In the ensuing decades, catalogues of ever-increasing numbers of Mendelian traits were published for all the major domesticated animal species. Notable among the cataloguers are O.N. Eaton, F.B. Hutt, I.M. Lerner, K. Huston, R. Robinson, D.F. Patterson, J.J. Lauvergne, L. Ollivier, S.M. Dennis, H.W. Leipold, D. Hamori, W. Wegner, R.G. Somes, R.D. Jolly and P. Sellier. The reviews by these authors are too numerous to list here. They are listed in the ‘Reviews’ section of the ‘Landmarks, Reviews, Maps’ tab of the Online Mendelian Inheritance in Animals (OMIA) web site at http://omia.angis.org.au.
The state of knowledge of Mendelian traits in non-laboratory animals up to 2012 is summarised in Table1. Also shown in Table1 are the numbers of Mendelian traits for which causal mutations were known up to 2012. (Here, the word ‘causal’ is used broadly in the sense of causal or key; similarly, the word ‘mutations’ is used generically in the sense of ‘variants’.) The aim of this review is to summarise the history of the discovery of these causal mutations, with a particular emphasis on the methods of discovery. The review also provides some analyses of the types of mutations that have been recorded.
Table 1.
The numbers of single-locus (Mendelian) traits published in non-laboratory animals and the numbers of such traits for which causal mutations have been discovered, up to the end of 2012. Hyperlinks to lists, and thence to details, of the actual traits summarised in this table are available from the home page of Online Mendelian Inheritance in Animals (OMIA): http://omia.angis.org.au. The last two rows indicate the number of missense and non-sense causal mutations reported for the seven species with the greatest number of published causal mutations.
| Animal species | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dog | Cattle | Chicken | Sheep | Cat | Pig | Horse | Goat | Other | Total | |
| Mendelian trait | 221 | 146 | 125 | 88 | 77 | 49 | 40 | 13 | 239 | 998 |
| Mendelian trait; causal mutation known | 155 | 82 | 37 | 40 | 44 | 23 | 29 | 8 | 81 | 499 |
| Missense mutations reported | 63 | 30 | 14 | 19 | 25 | 12 | 15 | |||
| Non-sense mutations reported | 21 | 21 | 5 | 7 | 10 | 0 | 2 | |||
Mutation discovery for Mendelian traits in non-laboratory animals
Initial success
The molecular revolution began with the discovery of the structure of DNA by Watson & Crick (1953). The subsequent development of methods for manipulating and sequencing DNA was at the heart of this revolution. By the time of the award of the 1980 Nobel Prize for chemistry to Berg (recombinant DNA) and Gilbert and Sanger (DNA sequencing) (http://www.nobelprize.org/nobel_prizes/chemistry/laureates/1980/), sufficient molecular tools were in place for a concerted effort to determine the molecular basis of Mendelian traits. The most straightforward challenges were disorders with an obvious candidate gene.
Familial goitre in cattle (OMIA 000424-9913) was the first success story in non-laboratory animals, the obvious candidate molecule being the peptide thyroglobulin (TG). Tellingly, all authors of the three key papers (Tassi et al. 1984; Ricketts et al. 1985, 1987) were working in human medical research laboratories (in Naples, Cape Town and Brussels). The first of these papers reported a 10- to 15-fold decrease in the concentration of TG mRNA in affected cattle; the second paper reported the use of S1 nuclease assays and electron microscopy of RNA/DNA hybridisation to narrow down the location of the mutation to near the junction of exon 9 and intron 9 of the TG gene; and the third (1987) paper reported the cloning and sequencing of TG cDNA from an affected animal, and the cloning and sequencing of genomic DNA from the TG exon-9–intron-9 junction of an affected and a normal animal, leading to discovery of the causal mutation as a non-sense mutation: a C>T transition towards the end of exon 9 of TG, resulting in p.Arg697Ter. It is interesting to note that this 1987 discovery predates by four years the first report of a mutation in the TG gene causing familial goitre in humans (Ieiri et al. 1991) and that the last author of this human report is also the last author of the 1987 bovine report. Judging from the text of the human paper, it is evident that the bovine research substantially informed the research leading to the discovery of the first human TG mutation.
The next success story, citrullinaemia in cattle (OMIA 000194-9913), was published in 1989. This disorder also has an obvious candidate gene, in this case the gene encoding the enzyme argininosuccinate synthetase (ASS1). Armed with DNA samples from affected and normal Australian cattle, animal researchers Julie Dennis and Peter Healy crossed the Pacific Ocean to the Baylor College of Medicine (Houston) laboratory of Arthur L. Beaudet and William E. O'Brien, who were by then just about to announce the first characterisation of mutations in the ASS1 gene causing human citrullinaemia (Jackson et al. 1989). Very quickly, the mutation in the bovine cases was shown to be (coincidentally) exactly the same type of non-sense mutation as for familial goitre: a C>T transition resulting in p.Arg86Ter (Dennis et al. 1989). Incidentally, this discovery was greatly aided by the use of the then still-novel polymerase chain reaction (PCR).
Two months after the publication of the bovine citrullinaemia mutation came the first report of the molecular basis of a Mendelian disorder in dogs. As with the bovine cases, this disorder (haemophilia B; OMIA 000438-9615) had an obvious candidate molecule, namely coagulation factor IX (F9). Once again, the research was conducted by (human) medical researchers, in this case a team led by the pioneer haemophilia researcher Kenneth M. Brinkhous, who maintained colonies of haemophilic dogs at the University of North Carolina at Chapel Hill (the haemophilia A colony since 1947; the haemophilia B colony since 1966). In the early 1970s, matings between dogs from these two colonies enabled Brinkhous et al. (1973) to show that the loci for canine haemophilias A and B are both on the X chromosome, but far apart. By 1989, a team from Brinkhous's laboratory was able to use PCR to amplify the entire F9 cDNA from affected animals and to show by sequencing that haemophilia B in this colony of dogs is due to a missense mutation (c.1477G>A), replacing a highly conserved glycine (p.Gly379Glu) (Evans et al. 1989).
An indication of the importance of these first three discoveries is that each was published in Proceedings of the National Academy of Sciences of the United States of America (PNAS).
The next year, 1990, saw the publication of two newly identified non-sense mutations, each in a likely candidate gene, as the cause of X-linked tremor in dogs (OMIA 000770-9615; Nadon et al. 1990) and maple syrup urine disease in cattle (OMIA 000627-9913; Zhang et al. 1990).
Two new molecular characterisations were reported in 1991. The first of these, Henny feathering in chickens (OMIA 000452-9031), became the first documented case of an insertion mutation in non-laboratory animals, in this case the insertion of the terminal repeat sequence of a retrovirus into the 5′ promoter region of the gene encoding the candidate molecule aromatase (CYP19A1) (Matsumine et al. 1991).
The second trait was malignant hyperthermia (MH) in pigs (OMIA 000621-9825), which had been the subject of extensive research in many laboratories around the world for several decades. Its importance arose from its manifestation as porcine stress syndrome (PSS) and pale, soft, exudative (PSE) meat. For reasons that will become evident, it is worth retelling the MH story in some detail.
It had long been realised that MH in humans is triggered by an adverse reaction to the anaesthetic halothane. This gave rise to the development of the halothane test in pigs (Webb & Jordan 1978), by which hundreds of thousands of pigs around the world were phenotyped for PSS. So extensive was the use of the halothane test that the porcine gene for PSS became known as Hal. By the mid-1980s, Hal was known to belong to a linkage group comprising six loci (reviewed by Archibald & Imlah 1985). By the late 1980s, this linkage group had been physically mapped to pig chromosome SSC6p12–q22 (Davies et al. 1988). In the same year, Mickelson et al. (1988) presented strong evidence to suggest that the physiological fault in porcine MH muscle may reside in the so-called ryanodine receptor, which is actually the Ca2+-release channel of a muscle organelle known as sarcoplasmic reticulum and hence has central involvement in muscle contraction, a key feature of MH. This sparked considerable interest in the ryanodine receptor: the peptide was isolated from rabbits and sequenced by Takeshima et al. (1989), who used this information to isolate, clone and sequence the corresponding rabbit cDNA. This sequence was then used to isolate, clone and hence physically map the ryanodine receptor gene (RYR1) in humans (MacLennan et al. 1989) and pigs (Harbitz et al. 1990). In humans, it maps to HSA19cen–q13.2, which (tantalisingly) contains a gene (GPI) that also occurs in the porcine Hal linkage group; in pigs, it maps (just as tantalisingly) to exactly the same (by then refined) location as the Hal linkage group, that is, SSC6p11–q21. At the same time, it was becoming increasingly evident that there is strongly conserved synteny between these two regions, that is, between SSC6p11–q21 and (the narrowed region) HSA19q12–q13.2 (Chowdhary et al. 1989; Yerle et al. 1990). Making use of this information, human geneticists checked to see whether the then unmapped MH in humans is linked to HSA19q12–13.2 markers and, sure enough, it is (MacLennan et al. 1990; McCarthy et al. 1990). This is the first case of a human Mendelian disorder being mapped on the basis of information gleaned from non-laboratory animals. In the same paper, MacLennan et al. (1990) reported that the ryanodine receptor shows zero recombination with MH in humans. The race was then on to search for causal mutations in this candidate gene in humans and in pigs& It turned out to be a Herculean task: the coding sequence, alone, is 15.2 kb in length. The race was won by MacLennan's team (Fujii et al. 1991) who showed that the porcine Hal mutation involves yet another C>T transition (c.1843C>T in RYR1), resulting in p.Arg615Cys. Thus, the smallest possible change (a single base substitution causing a single amino acid substitution) in a very large molecule comprising 5035 amino acids was shown to be the cause of a disorder that had been a major financial burden for the global pig industry for several decades. Intriguingly, when MacLennan's team searched for the same mutation in human families segregating MH, they discovered the corresponding mutation (c.1840C>T; p.Arg614Cys) in one family, and this became the first human causal MH mutation to be reported (later that same year, by Gillard et al. 1991).
The p.Arg615Cys mutation is responsible for almost all cases of porcine MH throughout the world, suggesting that the origin of this mutation predates breed formation or that it has been spread among breeds due to introgression followed by strong positive selection. The positive selection arose from the fact that the p.Arg615Cys allele is associated with leanness (reviewed by Sellier 1998), a trait increasingly favoured in the last 50 years. Thus, the p.Arg615Cys allele is both favourable (increasing leanness) and unfavourable (MH susceptible when homozygous). The inevitable result was selection favouring heterozygotes (overdominance) at this locus, which explains why this harmful allele was so frequent in pig populations throughout the world. On the reasonable conclusion that its disadvantages (PSS and PSE) outweigh its advantages, DNA tests based on the c.1843C>T mutation were used to remove the allele from most pig populations in the decade or so following the allele's discovery. In contrast to the situation with pigs, MH in humans has a wide range of different causal mutations (all at relatively low frequency) in more than one gene (see OMIM 145600, OMIM 601887).
The following year (1992) saw the first publication of a causal mutation in horses, namely a missense mutation in SCN4A resulting in periodic paralysis II (OMIA 000785-9796). This was identified by Rudolph et al. (1992b), following discovery that SCN4A, a strong comparative candidate gene, co-segregates with the disorder (Rudolph et al. 1992a). Interestingly, this disorder provides another example of heterozygote advantage: the mutant allele confers increased muscularity, which is favoured by show judges (Naylor 1994). Exactly the same comparative positional candidate gene strategy was used by Klungland et al. (1995) to identify the first reported molecular basis of coat colour mutations in any non-laboratory animal (dominant black and recessive red in cattle; OMIA 001199-9913) and by Johansson Moller et al. (1996) to show that mutations in KIT are responsible for dominant white spotting in pigs (OMIA 000209-9825).
As more and more single-locus traits were characterised at the DNA level in humans and in laboratory animals (especially mice), the comparative candidate gene approach proved to be very productive for the identification of the molecular basis of homologous traits in non-laboratory animals. By the end of the 1990s, this strategy had led to the publication of causal mutations for 73 single-locus traits in non-laboratory animals.
There were, however, many single-locus traits with no obvious candidate gene, comparative or otherwise. One obvious strategy for tackling such traits was to map them in the relevant species and then to utilise the ever-increasing knowledge of comparative maps between non-laboratory animal species and very well-mapped species, such as humans and mice, to identify positional comparative candidate genes. The first step in this strategy required sufficient polymorphic markers to cover all regions of all chromosomes. Microsatellites filled that bill.
The utility of genome-wide microsatellite linkage maps
The power of microsatellites was shown by three pioneering studies in 1993. Using 233 microsatellites in cattle, Michel Georges and colleagues identified markers linked to the Weaver locus (OMIA 000827-9913; Georges et al. 1993a) and the polled locus (OMIA 000483-9913; Georges et al. 1993b). In sheep, Montgomery et al. (1993) identified random microsatellite markers and an RFLP linked to the Booroola fecundity locus (OMIA 000383-9940). An indication of the importance of these discoveries at that time is that, even though each of these papers went no further than reporting (loose) linkage to markers, two of them were published in Nature Genetics and the other in PNAS& As the numbers of markers increased, Charlier et al. (1996) showed how homozygosity (identity-by-descent) mapping on a relatively small number of affected animals (e.g. 12) and on a pool of DNA from control animals could be a powerful first step in mapping a single-locus trait, in their case narrowing down the location of syndactyly in cattle (OMIA 000963-9913) to three candidate regions, each comprising sizeable proportions of a different bovine chromosome. Subsequent conventional linkage analysis (involving additional animals genotyped for additional markers in the candidate regions) by the same authors identified closely linked markers in one of the candidate regions.
By the end of the 1990s, reasonably comprehensive microsatellite linkage maps had been published for cattle (Barendse et al. 1994, 1997; Bishop et al. 1994; Ma et al. 1996; Kappes et al. 1997), pigs (Ellegren et al. 1994; Rohrer et al. 1994, 1996; Archibald et al. 1995; Marklund et al. 1996; Mikawa et al. 1999), sheep (Crawford et al. 1995; de Gortari et al. 1998), chickens (Cheng et al. 1995; Crooijmans et al. 1996; Groenen et al. 1998), goats (Vaiman et al. 1996), dogs (Lingaas et al. 1997; Mellersh et al. 1997), horses (Lindgren et al. 1998; Guérin et al. 1999) and cats (Menotti-Raymond et al. 1999). At the same time, by integrating these maps with physical maps, linkage groups were being allocated to particular chromosomes: it was now possible to ‘scan’ all regions of all chromosomes. This development set the stage for the widespread use of genome scans followed by linkage analysis or homozygosity/autozygosity mapping, followed by comparative positional candidate gene analysis.
The first trait in non-laboratory animals for which a causal mutation was identified from the combined results of a genome scan and a subsequent comparative positional candidate gene strategy was muscular hypertrophy (double muscling) in cattle (OMIA 000683-9913; Charlier et al. 1995; Grobet et al. 1997; Kambadur et al. 1997; McPherron & Lee 1997). This combined strategy became increasingly popular and productive in the first half of the 2000s.
In some cases, mapping was so precise that, even in the absence of any comparative candidate genes, sequencing of the fine-mapped candidate region in the species of interest revealed a new gene directly relevant to the trait, closely followed by discovery of a causal mutation within that gene. The first published application of this (positional cloning) strategy was the discovery of the PRKAG3 gene for the Rendement Napole meat quality trait by Milan et al. (2000); OMIA 001085-9825). Other early examples include discovery of the non-coding RNA gene PISRT1 for polled/intersex syndrome in goats (OMIA 000483-9925; Pailhoux et al. 2001) and LIMBIN (now EVC2) for chondrodysplasia in cattle (OMIA 000187-9913; Takeda et al. 2002).
The next revolution: genome assemblies and SNP chips
At the same time as genome scans were beginning to yield causal mutations in non-laboratory animals, the first draft genome sequence assemblies were published for humans (Lander et al. 2001; Venter et al. 2001) and mice (Waterston et al. 2002), ushering in a new revolution. Draft genome sequence assemblies soon became available for the main non-laboratory animals, and genome-assembly publications followed for chicken (International Chicken Genome Sequencing Consortium 2004), dog (Lindblad-Toh et al. 2005), cat (Pontius et al. 2007), sheep (Dalrymple et al. 2007), cattle (Elsik et al. 2009; Liu et al. 2009; Zimin et al. 2009), horse (Wade et al. 2009) and pig (Archibald et al. 2010; Groenen et al. 2012). The continual improvements in the quality and the extent of annotation of these genome assemblies saw a consequential decrease in the need for comparative mapping analysis. Coincident with the initial creation of draft genome sequence assemblies came the recognition that single nucleotide polymorphisms (SNPs), detectable from comparisons of sequence data, could be powerful DNA markers for research and application in animal genetics (Vignal et al. 2002), especially given that their exact location in the sequence, and hence in the genome, is (in principle) automatically known.
The generation of raw sequence data for genome assemblies and for SNP discovery was itself revolutionised by the advent in the mid-2000s of next-generation sequencing methods, which dramatically reduced the cost of sequencing (reviewed by Schuster 2008). The 2000s can therefore be dubbed the decade of SNP discovery and utilisation. In next to no time, or so it seemed, tens (and later, hundreds) of thousands of more-or-less equally spaced SNPs were available on arrays (‘chips’), providing very dense coverage across an entire genome: for chicken (Wong et al. 2004; Muir et al. 2008; Groenen et al. 2011), dog (Lindblad-Toh et al. 2005), cat (Pontius et al. 2007; Mullikin et al. 2011), cattle (Charlier et al. 2008; Van Tassell et al. 2008; Eck et al. 2009; Gibbs et al. 2009; Matukumalli et al. 2009; Stothard et al. 2011), horse (Wade et al. 2009; McCue et al. 2012), pig (Amaral et al. 2009; Ramos et al. 2009) and sheep (International Sheep Genomics Consortium et al. 2010).
The immense combined power of dense SNP chips and increasingly better-annotated genome assemblies for each of the major non-laboratory animal species was well illustrated in two landmark papers by Karlsson et al. (2007) and Charlier et al. (2008), who showed that with these tools, single-locus traits could be fine-mapped with amazingly small numbers of affecteds and controls (around 10 of each) to a sufficiently small region of a chromosome by either association analysis or homozygosity/autozygosity mapping, respectively; and that the candidate region could be sufficiently small so as to provide a manageable list of positional candidate genes for sequencing.
Single nucleotide polymorphisms chips can sometimes be used on pooled DNA, thereby dramatically reducing the cost of a project. Wells et al. (2012), for example, fine-mapped scaleless in chickens (OMIA 000889-9031) to a 1.25-Mb region using just two 60K SNP chips: one on DNA pooled from 86 chickens homozygous for scaleless and the other on a pool from 120 heterozygous sibs. Sequencing within that region enabled the same authors to identify the causal mutation as a non-sense mutation in FGF20.
Detecting mutations that cause abortions and/or stillbirths
The existence of mutants which when homozygous result in embryonic lethality (i.e. ‘natural’ abortion) has been known for many decades, e.g. Hutt (1934), Eaton (1937) and Lerner (1944). Typically, such mutants have been discovered because they result in a distinctive phenotype when heterozygous, i.e., their distinctive phenotype is dominant (or partially dominant) and the lethality is recessive. It has long been recognised, however, that there must be many recessive lethal mutants that have no identifiable effect in heterozygotes. How does one detect such mutants? Occasionally, luck can play a part. As part of a nutrition study at the University of Illinois, the level of orotic acid was measured in the milk of cows in the university herd. Some cows had exceptionally high levels of this acid, and one possible explanation was that they were deficient in uridine monophosphate synthetase (UMPS), the enzyme responsible for converting orotic acid to uridine monophosphate. Subsequent biochemical tests showed that these cows had only 50% of the normal activity of this enzyme (Robinson et al. 1983). Given that UMPS catalyses the last two steps of pyrimidine synthesis, it was expected (and subsequently reported by Shanks & Robinson 1989) that homozygosity for the deficiency results in embryonic lethality. The causal mutation for deficiency of UMPS in cattle, namely a non-sense mutation in UMPS, was reported by Schwenger et al. (1993) (DUMPS; OMIA 000262-9913).
In recent years, systematic strategies have been developed for detecting mutants causing abortions and/or stillbirths. In the first of these, Flisikowski et al. (2010) described a Finnish Ayrshire bull with an incidence of almost 50% of late abortion/stillbirth in his progeny. A half-sib linkage analysis with the bovine 50K SNP chip implicated the maternally imprinted PEG3 domain on chromosome BTA18. Close examination of this region disclosed that this bull was heterozygous for a 110-kb deletion in the MIMT1 LincRNA gene (OMIA 001565-9913). All of his offspring receive a maternally imprinted (and hence non-expressed) version of this gene from their dam. The 50% of his offspring that receive the deletion from the bull will therefore have no functional MIMT1 gene. The vast majority of these offspring die in late pregnancy, resulting in late abortion/stillbirth.
A powerful strategy was described by VanRaden et al. (2011), namely searching for common haplotypes that are never homozygous in live animals. By using genotype data from tens of thousands of North American Holsteins, Jerseys and Brown Swiss cattle, each genotyped with the bovine 50K SNP chip, these authors identified and (automatically) fine-mapped five new recessive lethal haplotypes in cattle (OMIA 001825-9913, OMIA 000001-9913, OMIA 001823-9913, OMIA 001824-9913 and OMIA 001697-9913). They also confirmed the lethal or partially lethal effects of eight previously documented mutants. The lethal mutation responsible for one of these (OMIA 000001-9913) was reported by Adams et al. (2012). Four more of these lethal haplotypes were characterised at the DNA level in 2013 (see OMIA 001826-9913, OMIA 001697-9913, OMIA 001827-9913 and OMIA 001828-9913).
Having discovered the causal mutation for brachyspina in cattle (OMIA 000151-9913) and having noticed that a large proportion of affected calves die in utero, Charlier et al. (2012) suggested a different strategy for detecting lethals, namely exome sequencing of 100–200 animals per breed to detect ‘disruptive’ (candidate) mutations, followed by genotyping around 5000 animals in relevant breeds for these mutations, and then searching for those mutations that are common (say, >0.03) but significantly deficient as homozygotes.
Mutation detection in a candidate region: sequence the region or the whole genome?
An obvious strategy for detecting a causal mutation in a candidate region is to ‘capture’ DNA from the region via a ‘tiling’ array of oligonucleotides and then to (deep) sequence it. The first published example of this strategy in non-laboratory animals reported the discovery of a causal mutation of arachnomelia in Brown Swiss cattle (OMIA 000059-9913; Drögemüller et al. 2010) as a single base insertion in the SUOX gene. Interestingly, this same disorder in Simmental cattle (OMIA 001541-9913) is due to a (2-bp deletion) mutation in a different gene, namely MOCS1 (Buitkamp et al. 2011). As noted by Buitkamp et al. (2011), this heterogeneity makes sense when it is realised that MOCS1 encodes two peptides (MOCS1A and MOCS1B) that are involved in the synthesis of molybdenum cofactor (Moco), which in turn is involved in the synthesis of sulphite oxidase, the gene for which is SUOX. Thus, mutations in either SUOX or MOCS1 are expected to result in the same biochemical deficiency and hence the same clinical signs.
Given the extraordinary decreases in the cost of sequencing, to the extent that in October 2013 the cost of sequencing an entire mammalian genome to an average coverage depth of 30X was less than US$5000 (J. Taylor and R. Schnabel, pers. comm.), it has been argued (e.g. Wade 2012; Schnabel et al. 2013) that whole-genome sequencing of an affected animal is a feasible alternative to ‘capturing’ and sequencing a region in that animal, especially because each sequenced affected animal can be re-used as a control for comparison with whole-genome-sequenced animals that are affected with different disorders. Of course, the storage and analysis of the vast data from whole-genome sequencing imposes its own challenges, but these are quickly being overcome with the increasing availability of capacity for storing sequence data and the increasing power of bioinformatics tools for analysis, for example Nordström et al. (2013). As sequencing costs continue to decrease, whole-genome sequencing alone may well become the strategy of choice for simultaneously mapping traits and revealing their causal mutations (Schnabel et al. 2013; Willet et al. 2013).
Another sequencing strategy for mutation detection
An alternative to genome sequencing is so-called mRNA sequencing from a tissue that is central to the trait. A dramatic example of the power of this strategy was provided by Forman et al. (2012), who discovered a causal mutation for cerebellar cortical degeneration in dogs (OMIA 000175-9615) by sequencing the cDNA from the cerebellum of just one affected dog and then comparing that sequence with the sequence of human genes known to be implicated in homologous disorders.
The mutation-discovery timeline
The time scale of mutation detection for Mendelian traits in non-laboratory animals is shown in Fig.1. It is evident that the rate of discovery has increased quite markedly over time. For the first six years after the initial 1987 discovery, the combined rate for the eight most studied species (listed separately in Fig.1) averaged around two per year. As genome-wide microsatellite maps became available and were published (1994–1999), the average combined rate for those eight species increased to nearly 10 per year and to around 15 per year for the ensuing five years (2000–2004). The following five years (2005–2009) witnessed the availability and publication of SNP chips and an average combined discovery rate of around 25 per year. The final three years shown in Fig.1, with an average of around 45 per year, illustrate the fruits of SNP chips and annotated genome assemblies. As can be seen from the solid line in Fig.1, by the end of 2012, the discovery rate across all non-laboratory animal species was more than one per week.
Figure 1.
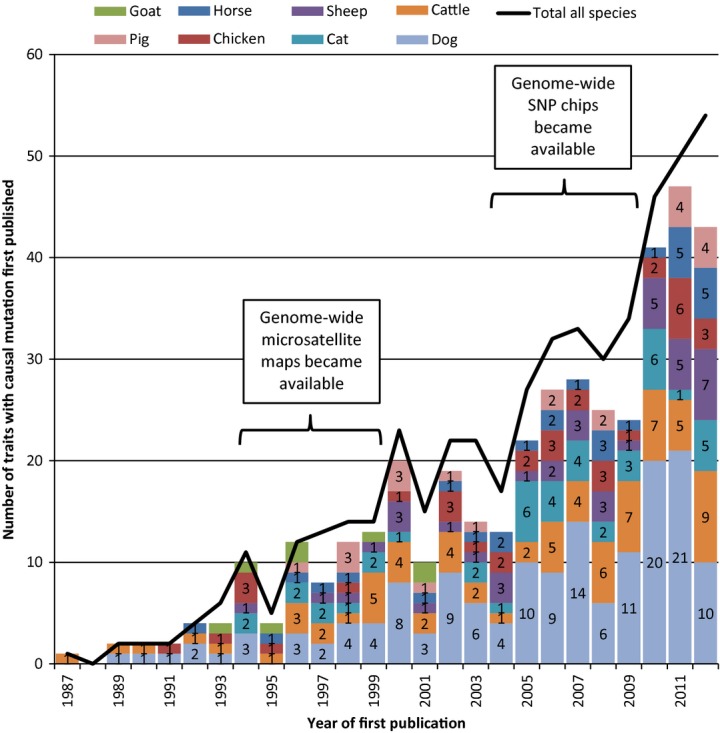
The time scale of discovery of causal mutations for Mendelian traits in non-laboratory animals, from the discovery of a causal mutation for familial goitre in cattle in 1987 up to 2012. Results are shown for the eight major species (in stacked bars) and for all species (solid line). Also indicated are the periods during which genome-wide sets of mapped microsatellites and genome-wide single-nucleotide polymorphism (SNP) chips became available.
Landmarks along the mutation-discovery timeline
The above discussion has provided a summary of the evolution of strategies by which causal mutations have been detected in non-laboratory animals. In concentrating on strategies, some discoveries of major importance have not been mentioned. Those that (in the opinion of the authors) deserve mention are listed chronologically in Table2.
Table 2.
Important discoveries of the molecular basis of Mendelian traits not mentioned elsewhere in this review, arranged chronologically.
| Important discovery | OMIA ID | References† |
|---|---|---|
| First report of a mutation causing increased ovulation rate in sheep [fecundity, Inverdale, FecX(I)] | 000386-9940 | Galloway et al. (2000) |
| Discovery of a causal mutation for the Booroola fecundity gene in sheep | 000383-9940 | Wilson et al. (2001); Mulsant et al. (2001); Souza et al. (2001) |
| Discovery of a causal mutation for adverse reaction to a range of important drugs in dogs | 001402-9615 | Mealey et al. (2001) |
| Discovery of a causal mutation for the callipyge mutation in sheep | 001354-9940 | Freking et al. (2002); Smit et al. (2003) |
| Discovery of a causal mutation for the ‘trademark’ Siamese and Burmese phenotypes in cats | 000202-9685 | Lyons et al. (2005); Schmidt-Küntzel et al. (2005) |
| First report of an inherited disorder in non-laboratory animals being due to an expanded repeat | 000690-9615 | Lohi et al. (2005) |
| Discovery of a coat colour polymorphism in the woolly mammoth | 001199-37349 | Rompler et al. (2006) |
| First report of an animal disorder due to a spontaneous mutation in mitochondrial DNA | 001130-9615 | Li et al. (2006) |
| Discovery of the coding sequence for yellow skin in chickens, which also showed that present-day poultry evolved not only from the red jungle fowl (as Darwin had correctly surmised), but also from the grey jungle fowl, which was the source of the yellow-skin allele | 001449-9031 | Eriksson et al. (2008) |
| Discovery of a most unusual causal mutation for the iconic white fleece phenotype of sheep | 000201-9940 | Norris & Whan (2008) |
| Discovery of the causal mechanism for a classic horse phenotype, namely greying with age | 001356-9796 | Rosengren Pielberg et al. (2008) |
| Discovery of the causal mutation for the classic feline tabby coat colour pattern | 001429-9685 | Kaelin et al. (2012) |
| Discovery of the most unusual molecular basis of colour sidedness in cattle | 001576-9913 | Durkin et al. (2012) |
| An explanation of the molecular basis of rose comb in chickens that also provides a molecular explanation for the very first case of epistasis ever reported (by Bateson & Punnett 1905), namely the interaction between the rose comb and pea comb loci, resulting in walnut comb | 000884-9031 | Imsland et al. (2012) |
| Identification of a candidate causal mutation for polledness in non-Holstein cattle | 000483-9913 | Medugorac et al. (2012) |
| Discovery of a mutation that plays a major role in determining modes of locomotion in mammals | 001715-9796 | Andersson et al. (2012) |
OMIA, Online Mendelian Inheritance in Animals.
These and other ‘landmark’ papers are presented in an annotated list under a tab labelled ‘Landmarks, Reviews, Maps’, accessible from the OMIA home page (http://omia.angis.org.au).
The rose comb and polled discoveries listed in Table2 are notable also because they represent the last of the six original Mendelian traits reported by Bateson (1902) and Bateson & Saunders (1902) to be characterised at the molecular level. Thus, 110 years after they were first shown to be inherited in a Mendelian manner, all six of the original Bateson/Saunders animal Mendelian traits now have a known causal mutation.
Complete lists of Mendelian traits with known causal mutations
As shown in Table1, the total number of traits in non-laboratory animals with known causal mutations was 499 at the end of 2012. A complete list of Mendelian traits with known causal mutations, up to 2012, is presented in Tables S1 and S2. In Table S1, the traits are listed alphabetically; in Table S2, the listing is by gene symbol. Up-to-date versions of each table are freely available from the ‘Download’ tab of the OMIA home page (http://omia.angis.org.au).
Types of mutations
For many years, the Human Gene Mutation Database (HGMD; http://www.hgmd.org) has been recording all published human mutations and categorising them into the 10 classes shown on the X-axis of Fig.2. With over 141 000 mutations documented in more than 5700 different genes, the HGMD distribution of the 10 classes of mutation provides a very accurate indication of the frequency of different types of mutations in humans. The number of mutations published for non-laboratory animals is far more modest: 556 in 312 different genes in OMIA up to 2012. (The number of mutations is greater than the total number of traits with a known causal mutation in OMIA, because more than one mutation has been published for some OMIA traits.) Although small by human standards, this number is sufficient for a comparison of the frequencies of mutation types between non-laboratory animals (OMIA) and humans (HGMD), which is illustrated in Fig.2. For most classes, the frequencies are quite similar. The stand-out result is the predominance of missense/non-sense mutations (i.e. mutations involving a single base substitution): around half of published mutations in humans and in non-laboratory animals are missense/non-sense. The next most common type of mutation is small deletions (defined by HGMD as 20 bp or less), followed by splicing mutations and gross deletions. Interestingly, insertions are less frequent than deletions in both humans and non-laboratory animals. Although the two distributions do have considerable similarity, a chi-squared contingency test on the actual numbers indicates a significant difference between the two distributions: χ2 = 92.74; df = 9; P < 0.0001. By far the greatest difference in distributions comes from gross (>20 bp) insertions, which are far more frequent in non-laboratory animals (5.8%) than in humans (1.7%). A close inspection of the data (not shown) indicates that this discrepancy is primarily in dogs, in which gross insertions account for 10.5% of published mutations in that species, compared with 3.8% across all other non-laboratory animal species with known causal mutations. Consistent with the ideas of Andersson (2012, 2013), this difference between humans and animals may reflect the fact that, unlike the HGMD list, the OMIA list includes single-locus traits that have been favoured by human selection. Such traits are more likely to be due to gain-of-function mutations arising from structural rearrangement (e.g. caused by insertions) of regulatory elements.
Figure 2.
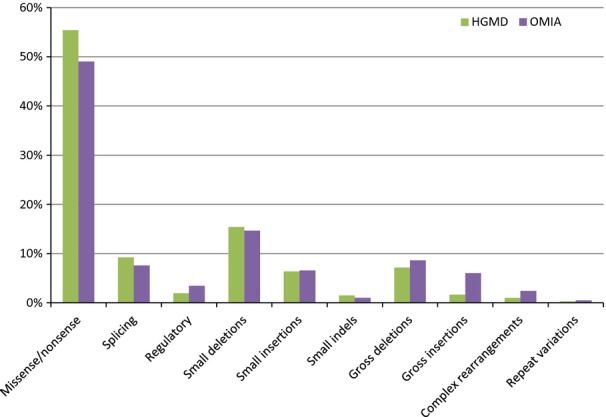
The distribution of types of mutation in humans (HGMD) and non-laboratory animals (OMIA). HGMD data obtained from their public web site (http://www.hgmd.org) on 3 July 2013. Definitions of HGMD's 10 types of mutations are provided at the same web site. HGMD, Human Gene Mutation Database.
Although HGMD combines missense and non-sense mutations together, these two types of mutations are recorded separately in OMIA. Across all 556 mutations in the OMIA data set up to 2012, 193 are missense and 78 are non-sense, indicating that missense mutations are 2.5 times as frequent as are non-sense mutations. Of the 61 non-stop codons in the genetic code, 21 are ‘near-stop’, that is, just one base substitution away from a stop codon. Consequently, if all non-stop codons occur equally frequently, and if base substitutions occur randomly, the expected ratio of missense to non-sense mutations is 40:21. The observed ratio of 193:78 is consistent with this expectation (χ2 = 3.82; df = 1; P = 0.05). It is realised, of course, that non-stop codons are not equally frequent and neither is base substitution completely random. However, it is interesting to see that the observed relative frequencies of missense and non-sense mutations in non-laboratory animals are consistent with those arising from the simplest of expectations, that is, that the observed proportion of non-sense mutations in non-laboratory animals (78/271 = 0.29) is in the same ball-park as the proportion of near-stop codons (21/61 = 0.34).
Finally, an analysis among the seven animal species with the greatest number of published causal mutations (dog, cattle, cat, sheep, pig, horse and chicken) indicates homogeneity in the ratio of missense to non-sense mutations (data are presented in the last two rows of Table1; χ2 = 11.76; df = 6; P = 0.07).
Conclusion
Most of the information in this review is freely available from the OMIA web site (http://omia.angis.org.au). The authors of this review are very much aware that the information in OMIA is incomplete and that some information is incorrect. In stark contrast to the situation faced by the pioneering cataloguers of the pre-computer era, information in OMIA can easily be updated/corrected from anywhere in the world where there is Internet access. Even better, updates/corrections are automatically and immediately made freely available on the web site. The OMIA home page includes a list of colleagues who are registered as OMIA curators. There is an open invitation to anyone who is aware of anything in OMIA that needs updating/correcting to contact Frank Nicholas, who will be very pleased to register new OMIA curators and to thereby enable readers to contribute to this knowledge base. The authors are also very keen to hear from anyone who has the time and patience to take responsibility for any section of OMIA.
Some of the information included in this review is not yet available from the OMIA web site, for example details of individual mutations in tabular form and graphs like Figures1 & 2. As soon as resources are found for the necessary software development, this information, and more, will be made freely available.
The term ‘non-laboratory animal’ used in this review embraces all species of animals except those that are well covered by phenotypic databases for their own species, such as mice, rats and zebrafish.
Building on the long-standing convention of Online Mendelian Inheritance in Man (OMIM; http://omim.org/), each trait in non-laboratory animals has been allocated an OMIA ID comprising a six-digit trait ID and (separated by a hyphen) the NCBI taxonomy ID (see http://www.ncbi.nlm.nih.gov/guide/taxonomy/). At the first mention of any trait in this review, the relevant OMIA ID is given. Researchers are encouraged to include the OMIA ID in any publication relating to a trait in OMIA. If a trait of interest is not yet in OMIA, please contact Frank Nicholas who will be pleased to create a new entry. The URL for any OMIA entry is http://omia.angis.org.au/OMIAxxxxxx/yyyy/, where xxxxxx is the 6-digit OMIA trait ID and yyyy is the NCBI species taxonomy ID (usually four digits, but sometimes longer).
Acknowledgments
The authors are very grateful to John James for his help with the analysis and interpretation of the data illustrated in Fig.2 and for his very helpful feedback on a draft. They are also very grateful to Tosso Leeb, Leif Andersson and an anonymous reviewer for detailed, constructive feedback which greatly improved the manuscript. Thanks are also due to Alan Archibald, David Cooper, Bianca Haase, Donna Maglott, Hamutal Mazrier, Thomas Peterson, Bob Schnabel, Jerry Taylor, Claire Wade and Cali Willet for useful feedback. The analyses of mutation types were stimulated by the assembly of OMIA mutation data by Thomas Peterson and Maricel Kann. The authors are grateful to Sue Tok and Chris Moran for the time they devoted to our attempts to incorporate hyperlinks in the text.
Supporting Information
Additional supporting information may be found in the online version of this article.
An alphabetical list of Mendelian traits/disorders for which a causal mutation has been reported, up to the end of 2012.
Genes (listed alphabetically by symbol) in which mutations have been shown to result in Mendelian traits/disorders in non-rodent animals, up to the end of 2012.
References
- Adams HA, Sonstegard T, VanRaden PM, Null DJ, Van Tassell C. Lewin H. Identification of a nonsense mutation in APAF1 that is causal for a decrease in reproductive efficiency in dairy cattle. Plant and Animal Genome (PAG) XX. 2012 doi: 10.3168/jds.2015-10517. Abstract P0555. Available at: https://pag.confex.com/pag/xx/webprogram/Paper3932.html. [DOI] [PubMed] [Google Scholar]
- Amaral AJ, Megens HJ, Kerstens HH, Heuven HC, Dibbits B, Crooijmans RP, den Dunnen JT. Groenen MA. Application of massive parallel sequencing to whole genome SNP discovery in the porcine genome. BMC Genomics. 2009;10:374. doi: 10.1186/1471-2164-10-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L. How selective sweeps in domestic animals provide new insight into biological mechanisms. Journal of Internal Medicine. 2012;271:1–14. doi: 10.1111/j.1365-2796.2011.02450.x. [DOI] [PubMed] [Google Scholar]
- Andersson L. Molecular consequences of animal breeding. Current Opinion in Genetics & Development. 2013;23:295–301. doi: 10.1016/j.gde.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Andersson LS, Larhammar M, Memic F, et al. et al. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature. 2012;488:642–6. doi: 10.1038/nature11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald AL. Imlah P. The halothane sensitivity locus and its linkage relationships. Animal Blood Groups and Biochemical Genetics. 1985;16:253–63. doi: 10.1111/j.1365-2052.1985.tb01478.x. [DOI] [PubMed] [Google Scholar]
- Archibald AL, Haley CS, Brown JF, et al. The PiGMaP consortium linkage map of the pig (Sus scrofa. Mammalian Genome. 1995;6:157–75. doi: 10.1007/BF00293008. [DOI] [PubMed] [Google Scholar]
- Archibald AL, Bolund L, Churcher C, et al. Pig genome sequence – analysis and publication strategy. BMC Genomics. 2010;11:438. doi: 10.1186/1471-2164-11-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barendse W, Armitage SM, Kossarek LM, et al. A genetic linkage map of the bovine genome. Nature Genetics. 1994;6:227–35. doi: 10.1038/ng0394-227. [DOI] [PubMed] [Google Scholar]
- Barendse W, Vaiman D, Kemp SJ, et al. A medium-density genetic linkage map of the bovine genome. Mammalian Genome. 1997;8:21–8. doi: 10.1007/s003359900340. [DOI] [PubMed] [Google Scholar]
- Bateson W. Experimental studies in the physiology of heredity. Part II. Poultry. Reports to the Evolutionary Committee of the Royal Society. 1902;1:87–124. [Google Scholar]
- Bateson W. Facts limiting the theory of heredity. Science. 1907;26:649–60. doi: 10.1126/science.26.672.649. [DOI] [PubMed] [Google Scholar]
- Bateson W. Mendel's Principles of Heredity. London: Cambridge University Press; 1909. [Google Scholar]
- Bateson W. Saunders ER. The facts of heredity in the light of Mendel's discovery. Reports to the Evolution Committee of the Royal Society. 1902;1:125–60. [Google Scholar]
- Bateson W. Punnett RC. Experimental studies in the physiology of heredity. Poultry. Reports to the Evolutionary Committee of the Royal Society. 1905;2:99–119. [Google Scholar]
- Bishop MD, Kappes SM, Keele JW, et al. A genetic linkage map for cattle. Genetics. 1994;136:619–39. doi: 10.1093/genetics/136.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkhous KM, Davis PD, Graham JB. Dodds WJ. Expression and linkage of genes for X-linked hemophilias A and B in the dog. Blood. 1973;41:577–85. [PubMed] [Google Scholar]
- Buitkamp J, Semmer J. Götz KU. Arachnomelia syndrome in Simmental cattle is caused by a homozygous 2-bp deletion in the molybdenum cofactor synthesis step 1 gene (MOCS1. BMC Genetics. 2011;12:11. doi: 10.1186/1471-2156-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle WE. Heredity in Relation to Evolution and Animal Breeding. New York, NY: D. Appleton and Co; 1911. [Google Scholar]
- Charlier C, Coppieters W, Farnir F, et al. The MH gene causing double-muscling in cattle maps to bovine chromosome 2. Mammalian Genome. 1995;6:788–92. doi: 10.1007/BF00539005. [DOI] [PubMed] [Google Scholar]
- Charlier C, Farnir F, Berzi P, Vanmanshoven P, Brouwers B, Vromans H. Georges M. Identity-by-descent mapping of recessive traits in livestock: application to map the bovine syndactyly locus to chromosome 15. Genome Research. 1996;6:580–9. doi: 10.1101/gr.6.7.580. [DOI] [PubMed] [Google Scholar]
- Charlier C, Coppieters W, Rollin F, et al. Highly effective SNP-based association mapping and management of recessive defects in livestock. Nature Genetics. 2008;40:449–54. doi: 10.1038/ng.96. [DOI] [PubMed] [Google Scholar]
- Charlier C, Agerholm JS, Coppieters W, et al. A deletion in the bovine FANCI gene compromises fertility by causing fetal death and brachyspina. PLoS ONE. 2012;7:e43085. doi: 10.1371/journal.pone.0043085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HH, Levin I, Vallejo RL, Khatib H, Dodgson JB, Crittenden LB. Hillel J. Development of a genetic map of the chicken with markers of high utility. Poultry Science. 1995;74:1855–74. doi: 10.3382/ps.0741855. [DOI] [PubMed] [Google Scholar]
- Chowdhary BP, Harbitz I, Makinen A, Davies W. Gustavsson I. Localization of the glucose phosphate isomerase gene to the p12–q21 segment of chromosome 6 in pig by in situ hybridization. Hereditas. 1989;111:73–8. doi: 10.1111/j.1601-5223.1989.tb00378.x. [DOI] [PubMed] [Google Scholar]
- Crawford AM, Dodds KG, Ede AJ, et al. An autosomal genetic linkage map of the sheep genome. Genetics. 1995;140:703–24. doi: 10.1093/genetics/140.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooijmans RP, van Oers PA, Strijk JA, van der Poel JJ. Groenen MA. Preliminary linkage map of the chicken (Gallus domesticus) genome based on microsatellite markers: 77 new markers mapped. Poultry Science. 1996;75:746–54. doi: 10.3382/ps.0750746. [DOI] [PubMed] [Google Scholar]
- Dalrymple BP, Kirkness EF, Nefedov M, et al. Using comparative genomics to reorder the human genome sequence into a virtual sheep genome. Genome Biology. 2007;8:R152. doi: 10.1186/gb-2007-8-7-r152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies W, Harbitz I, Fries R, Stranzinger G. Hauge JG. Porcine malignant hyperthermia carrier detection and chromosomal assignment using a linked probe. Animal Genetics. 1988;19:203–12. doi: 10.1111/j.1365-2052.1988.tb00809.x. [DOI] [PubMed] [Google Scholar]
- Dennis JA, Healy PJ, Beaudet AL. O'Brien WE. Molecular definition of bovine argininosuccinate synthetase deficiency. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:7947–51. doi: 10.1073/pnas.86.20.7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drögemüller C, Tetens J, Sigurdsson S, Gentile A, Testoni S, Lindblad-Toh K. Leeb T. Identification of the bovine Arachnomelia mutation by massively parallel sequencing implicates sulfite oxidaseSUOX) in bone development. PLoS Genetics. 2010;6:e1001079. doi: 10.1371/journal.pgen.1001079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin K, Coppieters W, Drögemüller C. Serial translocation by means of circular intermediates underlies colour sidedness in cattle. Nature. 2012;482:81–4. doi: 10.1038/nature10757. [DOI] [PubMed] [Google Scholar]
- Eaton ON. A summary of lethal characters in animals and man. Journal of Heredity. 1937;28:320–6. [Google Scholar]
- Eck SH, Benet-Pages A, Flisikowski K, Meitinger T, Fries R. Strom TM. Whole genome sequencing of a single Bos taurus animal for single nucleotide polymorphism discovery. Genome Biology. 2009;10:R82. doi: 10.1186/gb-2009-10-8-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Chowdhary BP, Johansson M, Marklund L, Fredholm M, Gustavsson I. Andersson L. A primary linkage map of the porcine genome reveals a low rate of genetic recombination. Genetics. 1994;137:1089–100. doi: 10.1093/genetics/137.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, Tellam RL, Worley KC, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324:522–8. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J, Larson G, Gunnarsson U, et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genetics. 2008;4:e1000010. doi: 10.1371/journal.pgen.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JP, Brinkhous KM, Brayer GD, Reisner HM. High KA. Canine hemophilia B resulting from a point mutation with unusual consequences. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:10095–9. doi: 10.1073/pnas.86.24.10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flisikowski K, Venhoranta H, Nowacka-Woszuk J, et al. A novel mutation in the maternally imprinted PEG3 domain results in a loss of MIMT1 expression and causes abortions and stillbirths in cattle (Bos taurus) PLoS ONE. 2010;5:e15116. doi: 10.1371/journal.pone.0015116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman OP, De Risio L, Stewart J, Mellersh CS. Beltran E. Genome-wide mRNA sequencing of a single canine cerebellar cortical degeneration case leads to the identification of a disease associated SPTBN2 mutation. BMC Genetics. 2012;13:55. doi: 10.1186/1471-2156-13-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freking BA, Murphy SK, Wylie AA, Rhodes SJ, Keele JW, Leymaster KA, Jirtle RL. Smith TPL. Identification of the single base change causing the callipyge muscle hypertrophy phenotype, the only known example of polar overdominance in mammals. Genome Research. 2002;12:1496–506. doi: 10.1101/gr.571002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii J, Otsu K, Zorzato F, de Leon S, Khanna VK, Weiler JE, O'Brien PJ. MacLennan DH. Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science. 1991;253:448–51. doi: 10.1126/science.1862346. [DOI] [PubMed] [Google Scholar]
- Galloway SM, McNatty KP, Cambridge LM, et al. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nature Genetics. 2000;25:279–83. doi: 10.1038/77033. [DOI] [PubMed] [Google Scholar]
- Georges M, Dietz AB, Mishra A, et al. Microsatellite mapping of the gene causing weaver disease in cattle will allow the study of an associated quantitative trait locus. Proceedings of the National Academy of Sciences of the United States of America. 1993a;90:1058–62. doi: 10.1073/pnas.90.3.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges M, Drinkwater R, King T, et al. Microsatellite mapping of a gene affecting horn development in Bos taurus. Nature Genetics. 1993b;4:206–10. doi: 10.1038/ng0693-206. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Taylor JF, Van Tassell CP, et al. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science. 2009;324:528–32. doi: 10.1126/science.1167936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillard EF, Otsu K, Fujii J, Khanna VK, de Leon S, Derdemezi J, Britt BA, Duff CL, Worton RG. MacLennan DH. A substitution of cysteine for arginine 614 in the ryanodine receptor is potentially causative of human malignant hyperthermia. Genomics. 1991;11:751–5. doi: 10.1016/0888-7543(91)90084-r. [DOI] [PubMed] [Google Scholar]
- de Gortari MJ, Freking BA, Cuthbertson RP, Kappes SM, Keele JW, Stone RT, Leymaster KA, Dodds KG, Crawford AM. Beattie CW. A second-generation linkage map of the sheep genome. Mammalian Genome. 1998;9:204–9. doi: 10.1007/s003359900726. [DOI] [PubMed] [Google Scholar]
- Grobet L, Martin LJ, Poncelet D, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nature Genetics. 1997;17:71–4. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- Groenen MA, Crooijmans RP, Veenendaal A, Cheng HH, Siwek M. van der Poel JJ. A comprehensive microsatellite linkage map of the chicken genome. Genomics. 1998;49:265–74. doi: 10.1006/geno.1998.5225. [DOI] [PubMed] [Google Scholar]
- Groenen MA, Megens HJ, Zare Y, Warren WC, Hillier LW, Crooijmans RP, Vereijken A, Okimoto R, Muir WM. Cheng HH. The development and characterization of a 60K SNP chip for chicken. BMC Genomics. 2011;12:274. doi: 10.1186/1471-2164-12-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen MA, Archibald AL, Uenishi H, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491:393–8. doi: 10.1038/nature11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérin G, Bailey E, Bernoco D, et al. Report of the International Equine Gene Mapping Workshop: male linkage map. Animal Genetics. 1999;30:341–54. doi: 10.1046/j.1365-2052.1999.00510.x. [DOI] [PubMed] [Google Scholar]
- Harbitz I, Chowdhary B, Thomsen PD, Davies W, Kaufmann U, Kran S, Gustavsson I, Christensen K. Hauge JG. Assignment of the porcine calcium release channel gene, a candidate for the malignant hyperthermia locus, to the 6p11-q21 segment of chromosome 6. Genomics. 1990;8:243–8. doi: 10.1016/0888-7543(90)90278-3. [DOI] [PubMed] [Google Scholar]
- Hurst CC. Experiments with poultry. Reports to the Evolutionary Committee of the Royal Society. 1905;2:131–54. [Google Scholar]
- Hutt FB. Inherited lethal characters in domestic animals. Cornell Veterinarian. 1934;24:1–25. [Google Scholar]
- Ieiri T, Cochaux P, Targovnik HM, Suzuki M, Shimoda S, Perret J. Vassart G. A 3′ splice site mutation in the thyroglobulin gene responsible for congenital goiter with hypothyroidism. The Journal of Clinical Investigation. 1991;88:1901–5. doi: 10.1172/JCI115513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imsland F, Feng C, Boije H, et al. The Rose-comb mutation in chickens constitutes a structural rearrangement causing both altered comb morphology and defective sperm motility. PLoS Genetics. 2012;8:e1002775. doi: 10.1371/journal.pgen.1002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- International Sheep Genomics Consortium. Archibald AL, Cockett NE, et al. The sheep genome reference sequence: a work in progress. Animal Genetics. 2010;41:449–53. doi: 10.1111/j.1365-2052.2010.02100.x. [DOI] [PubMed] [Google Scholar]
- Jackson MJ, Kobayashi K, Beaudet AL. O'Brien WE. Analysis of deletions at the human argininosuccinate synthetase locus. Molecular Biology & Medicine. 1989;6:179–86. [PubMed] [Google Scholar]
- Johansson Moller M, Chaudhary R, Hellmen E, Hoyheim B, Chowdhary B. Andersson L. Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mammalian Genome. 1996;7:822–30. doi: 10.1007/s003359900244. [DOI] [PubMed] [Google Scholar]
- Kaelin CB, Xu X, Hong LZ, et al. Specifying and sustaining pigmentation patterns in domestic and wild cats. Science. 2012;337:1536–41. doi: 10.1126/science.1220893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambadur R, Sharma M, Smith TP. Bass JJ. Mutations in myostatinGDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Research. 1997;7:910–6. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- Kappes SM, Keele JW, Stone RT, McGraw RA, Sonstegard TS, Smith TP, Lopez-Corrales NL. Beattie CW. A second-generation linkage map of the bovine genome. Genome Research. 1997;7:235–49. doi: 10.1101/gr.7.3.235. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nature Genetics. 2007;39:1321–8. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Klungland H, Vage DI, Gomez-Raya L, Adalsteinsson S. Lien S. The role of melanocyte-stimulating hormone (MSH) receptor in bovine coat color determination. Mammalian Genome. 1995;6:636–9. doi: 10.1007/BF00352371. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lerner IM. Lethal and sublethal characters in farm animals: a checklist and proposed numbering system. Journal of Heredity. 1944;35:219–24. [Google Scholar]
- Li FY, Cuddon PA, Song J, Wood SL, Patterson JS, Shelton GD. Duncan ID. Canine spongiform leukoencephalomyelopathy is associated with a missense mutation in cytochrome b. Neurobiology of Diseases. 2006;21:35–42. doi: 10.1016/j.nbd.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–19. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lindgren G, Sandberg K, Persson H, Marklund S, Breen M, Sandgren B, Carlsten J. Ellegren H. A primary male autosomal linkage map of the horse genome. Genome Research. 1998;8:951–66. doi: 10.1101/gr.8.9.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingaas F, Sorensen A, Juneja RK, et al. Towards construction of a canine linkage map: establishment of 16 linkage groups. Mammalian Genome. 1997;8:218–21. doi: 10.1007/s003359900393. [DOI] [PubMed] [Google Scholar]
- Liu Y, Qin X, Song XZ, et al. Bos taurus genome assembly. BMC Genomics. 2009;10:180. doi: 10.1186/1471-2164-10-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohi H, Young EJ, Fitzmaurice SN, et al. Expanded repeat in canine epilepsy. Science. 2005;307:81. doi: 10.1126/science.1102832. [DOI] [PubMed] [Google Scholar]
- Lyons LA, Imes DL, Rah HC. Grahn RA. Tyrosinase mutations associated with Siamese and Burmese patterns in the domestic cat (Felis catus) Animal Genetics. 2005;36:119–26. doi: 10.1111/j.1365-2052.2005.01253.x. [DOI] [PubMed] [Google Scholar]
- Ma RZ, Beever JE, Da Y, et al. A male linkage map of the cattle (Bos taurus) genome. Journal of Heredity. 1996;87:261–71. doi: 10.1093/oxfordjournals.jhered.a022999. [DOI] [PubMed] [Google Scholar]
- MacLennan DH, Zorzato F, Fujii J, et al. Cloning and localisation of the human calcium release channel (ryanodine receptor) gene to the proximal long arm (Cen -> q13.2) of human chromosome 19. American Journal of Human Genetics. 1989;45(Suppl):A205. Abstract 0803. [Google Scholar]
- MacLennan DH, Duff C, Zorzato F, Fujii J, Phillips M, Korneluk RG, Frodis W, Britt BA. Worton RG. Ryanodine receptor gene is a candidate for predisposition to malignant hyperthermia. Nature. 1990;343:559–61. doi: 10.1038/343559a0. [DOI] [PubMed] [Google Scholar]
- Marklund L, Johansson Moller M, Hoyheim B, Davies W, Fredholm M, Juneja RK, Mariani P, Coppieters W, Ellegren H. Andersson L. A comprehensive linkage map of the pig based on a wild pig-Large White intercross. Animal Genetics. 1996;27:255–69. doi: 10.1111/j.1365-2052.1996.tb00487.x. [DOI] [PubMed] [Google Scholar]
- Matsumine H, Herbst MA, Ou SH, Wilson JD. McPhaul MJ. Aromatase mRNA in the extragonadal tissues of chickens with the henny-feathering trait is derived from a distinctive promoter structure that contains a segment of a retroviral long terminal repeat. Functional organization of the Sebright, Leghorn, and Campine aromatase genes. Journal of Biological Chemistry. 1991;266:19900–7. [PubMed] [Google Scholar]
- Matukumalli LK, Lawley CT, Schnabel RD, et al. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE. 2009;4:e5350. doi: 10.1371/journal.pone.0005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy TV, Healy JM, Heffron JJ, Lehane M, Deufel T, Lehmann-Horn F, Farrall M. Johnson K. Localization of the malignant hyperthermia susceptibility locus to human chromosome 19q12-13.2. Nature. 1990;343:562–4. doi: 10.1038/343562a0. [DOI] [PubMed] [Google Scholar]
- McCue ME, Bannasch DL, Petersen JL, et al. A high density SNP array for the domestic horse and extant Perissodactyla: utility for association mapping, genetic diversity, and phylogeny studies. PLoS Genetics. 2012;8:e1002451. doi: 10.1371/journal.pgen.1002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron AC. Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:12457–61. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealey KL, Bentjen SA, Gay JM. Cantor GH. Ivermectin sensitivity in collies is associated with a deletion mutation of the MDR1 gene. Pharmacogenetics. 2001;11:727–33. doi: 10.1097/00008571-200111000-00012. [DOI] [PubMed] [Google Scholar]
- Medugorac I, Seichter D, Graf A, Russ I, Blum H, Göpel KH, Rothammer S, Förster M. Krebs S. Bovine polledness - an autosomal dominant trait with allelic heterogeneity. PLoS ONE. 2012;7:e39477. doi: 10.1371/journal.pone.0039477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellersh CS, Langston AA, Acland GM, Fleming MA, Ray K, Wiegand NA, Francisco LV, Gibbs M, Aguirre GD. Ostrander EA. A linkage map of the canine genome. Genomics. 1997;46:326–36. doi: 10.1006/geno.1997.5098. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Lyons LA, Schaffer AA, Tomlin JF, Hutton MK. O'Brien SJ. A genetic linkage map of microsatellites in the domestic cat (Felis catus. Genomics. 1999;57:9–23. doi: 10.1006/geno.1999.5743. [DOI] [PubMed] [Google Scholar]
- Mickelson JR, Gallant EM, Litterer LA, Johnson KM, Rempel WE. Louis CF. Abnormal sarcoplasmic reticulum ryanodine receptor in malignant hyperthermia. Journal of Biological Chemistry. 1988;263:9310–5. [PubMed] [Google Scholar]
- Mikawa S, Akita T, Hisamatsu N, et al. A linkage map of 243 DNA markers in an intercross of Gottingen miniature and Meishan pigs. Animal Genetics. 1999;30:407–17. doi: 10.1046/j.1365-2052.1999.00493.x. [DOI] [PubMed] [Google Scholar]
- Milan D, Jeon JT, Looft C, et al. A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science. 2000;288:1248–51. doi: 10.1126/science.288.5469.1248. [DOI] [PubMed] [Google Scholar]
- Montgomery GW, Crawford AM, Penty JM, et al. The ovine Booroola fecundity gene (FECB) is linked to markers from a region of human chromosome 4q. Nature Genetics. 1993;4:410–4. doi: 10.1038/ng0893-410. [DOI] [PubMed] [Google Scholar]
- Muir WM, Wong GK, Zhang Y, et al. Review of the initial validation and characterization of a chicken 3K SNP array. World's Poultry Science Journal. 2008;64:219–26. [Google Scholar]
- Mullikin JC, Hansen NF, Shen L, et al. Light whole genome sequence for SNP discovery across domestic cat breeds. BMC Genomics. 2011;11:406. doi: 10.1186/1471-2164-11-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulsant P, Lecerf F, Fabre S, et al. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5104–9. doi: 10.1073/pnas.091577598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadon NL, Duncan ID. Hudson LD. A point mutation in the proteolipid protein gene of the ‘shaking pup’ interrupts oligodendrocyte development. Development. 1990;110:529–37. doi: 10.1242/dev.110.2.529. [DOI] [PubMed] [Google Scholar]
- Naylor JM. Selection of Quarter horses affected with hyperkalemic periodic paralysis by show judges. Journal of the American Veterinary Medical Association. 1994;204:926–8. [PubMed] [Google Scholar]
- Nordström KJ, Albani MC, James GV, Gutjahr C, Hartwig B, Turck F, Paszkowski U, Coupland G. Schneeberger K. Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nature Biotechnology. 2013;31:325–30. doi: 10.1038/nbt.2515. [DOI] [PubMed] [Google Scholar]
- Norris BJ. Whan VA. A gene duplication affecting expression of the ovine ASIP gene is responsible for white and black sheep. Genome Research. 2008;18:1282–93. doi: 10.1101/gr.072090.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pailhoux E, Vigier B, Chaffaux S, et al. A 11.7-kb deletion triggers intersexuality and polledness in goats. Nature Genetics. 2001;29:453–8. doi: 10.1038/ng769. [DOI] [PubMed] [Google Scholar]
- Pontius JU, Mullikin JC, Smith DR, et al. Initial sequence and comparative analysis of the cat genome. Genome Research. 2007;17:1675–89. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punnett RC. Mendelism. Cambridge: Macmillan and Bowes; 1905. [Google Scholar]
- Ramos AM, Crooijmans RP, Affara NA, et al. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS ONE. 2009;4:e6524. doi: 10.1371/journal.pone.0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts MH, Pohl V, de Martynoff G, Boyd CD, Bester AJ, Van Jaarsveld PP. Vassart G. Defective splicing of thyroglobulin gene transcripts in the congenital goitre of the Afrikander cattle. EMBO Journal. 1985;4:731–7. doi: 10.1002/j.1460-2075.1985.tb03690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts MH, Simons MJ, Parma J, Mercken L, Dong Q. Vassart G. A nonsense mutation causes hereditary goitre in the Afrikander cattle and unmasks alternative splicing of thyroglobulin transcripts. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:3181–4. doi: 10.1073/pnas.84.10.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JL, Drabik MR, Dombrowski DB. Clark JH. Consequences of UMP synthase deficiency in cattle. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:321–3. doi: 10.1073/pnas.80.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer GA, Alexander LJ, Keele JW, Smith TP. Beattie CW. A microsatellite linkage map of the porcine genome. Genetics. 1994;136:231–45. doi: 10.1093/genetics/136.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer GA, Alexander LJ, Hu Z, Smith TP, Keele JW. Beattie CW. A comprehensive map of the porcine genome. Genome Research. 1996;6:371–91. doi: 10.1101/gr.6.5.371. [DOI] [PubMed] [Google Scholar]
- Rompler H, Rohland N, Lalueza-Fox C, Willerslev E, Kuznetsova T, Rabeder G, Bertranpetit J, Schoneberg T. Hofreiter M. Nuclear gene indicates coat-color polymorphism in mammoths. Science. 2006;313:62. doi: 10.1126/science.1128994. [DOI] [PubMed] [Google Scholar]
- Rosengren Pielberg G, Golovko A, Sundstrom E, et al. A cis-acting regulatory mutation causes premature hair graying and susceptibility to melanoma in the horse. Nature Genetics. 2008;40:1004–9. doi: 10.1038/ng.185. [DOI] [PubMed] [Google Scholar]
- Rudolph JA, Spier SJ, Byrns G. Hoffman EP. Linkage of hyperkalaemic periodic paralysis in quarter horses to the horse adult skeletal muscle sodium channel gene. Animal Genetics. 1992a;23:241–50. doi: 10.1111/j.1365-2052.1992.tb00136.x. [DOI] [PubMed] [Google Scholar]
- Rudolph JA, Spier SJ, Byrns G, Rojas CV, Bernoco D. Hoffman EP. Periodic paralysis in quarter horses: a sodium channel mutation disseminated by selective breeding. Nature Genetics. 1992b;2:144–7. doi: 10.1038/ng1092-144. [DOI] [PubMed] [Google Scholar]
- Schmidt-Küntzel A, Eizirik E, O'Brien SJ. Menotti-Raymond M. Tyrosinase and tyrosinase related protein 1 alleles specify domestic cat coat color phenotypes of the albino and brown loci. Journal of Heredity. 2005;96:289–301. doi: 10.1093/jhered/esi066. [DOI] [PubMed] [Google Scholar]
- Schnabel R, Mhlanga-Mutangadura T, Gilliam DH, Decker J, O'Brien DP, Coates JR, Taylor J. Johnson GS. Application of whole genome sequencing to identify Mendelian disease candidates in dogs using a 1-case + n-control design. Plant and Animal Genome (PAG) 2013 Abstract W 140. Available at: https://pag.confex.com/pag/xxi/webprogram/Paper7104.html. [Google Scholar]
- Schuster SC. Next-generation sequencing transforms today's biology. Nature Methods. 2008;5:16–8. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- Schwenger B, Schober S. Simon D. DUMPS cattle carry a point mutation in the uridine monophosphate synthase gene. Genomics. 1993;16:241–4. doi: 10.1006/geno.1993.1165. [DOI] [PubMed] [Google Scholar]
- Sellier P. Genetics of meat and carcass traits. In: Ruvinsky A, editor; Rothschild MF, editor. The Genetics of the Pig. Wallingford, UK: CAB International; 1998. pp. 463–510. [Google Scholar]
- Shanks RD. Robinson JL. Embryonic mortality attributed to inherited deficiency of uridine monophosphate synthase. Journal of Dairy Science. 1989;72:3035–9. doi: 10.3168/jds.S0022-0302(89)79456-X. [DOI] [PubMed] [Google Scholar]
- Smit M, Segers K, Carrascosa LG, Shay T, Baraldi F, Gyapay G, Snowder G, Georges M, Cockett N. Charlier C. Mosaicism of Solid Gold supports the causality of a noncoding A-to-G transition in the determinism of the callipyge phenotype. Genetics. 2003;163:453–6. doi: 10.1093/genetics/163.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza CJ, MacDougall C, MacDougall C, Campbell BK, McNeilly AS. Baird DT. The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor, type 1BBMPR1B) gene. Journal of Endocrinology. 2001;169:R1–6. doi: 10.1677/joe.0.169r001. [DOI] [PubMed] [Google Scholar]
- Stothard P, Choi JW, Basu U, Sumner-Thomson JM, Meng Y, Liao X. Moore SS. Whole genome resequencing of black Angus and Holstein cattle for SNP and CNV discovery. BMC Genomics. 2011;12:559. doi: 10.1186/1471-2164-12-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H, Takami M, Oguni T, et al. Positional cloning of the gene limbin responsible for bovine chondrodysplastic dwarfism. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10549–54. doi: 10.1073/pnas.152337899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Nishimura S, Matsumoto T, Ishida H, Kangawa K, Minamino N, Matsuo H, Ueda M, Hanaoka M, Hirose T. Numa S. Primary structure and expression from complementary DNA of skeletal muscle ryanodine receptor. Nature. 1989;339:439–45. doi: 10.1038/339439a0. [DOI] [PubMed] [Google Scholar]
- Tassi VP, Di Lauro R, Van Jaarsveld P. Alvino CG. Two abnormal thyroglobulin-like polypeptides are produced from Afrikander cattle congenital goiter mRNA. Journal of Biological Chemistry. 1984;259:10507–10. [PubMed] [Google Scholar]
- Vaiman D, Schibler L, Bourgeois F, Oustry A, Amigues Y. Cribiu EP. A genetic linkage map of the male goat genome. Genetics. 1996;144:279–305. doi: 10.1093/genetics/144.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tassell CP, Smith TP, Matukumalli LK, Taylor JF, Schnabel RD, Lawley CT, Haudenschild CD, Moore SS, Warren WC. Sonstegard TS. SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nature Methods. 2008;5:247–52. doi: 10.1038/nmeth.1185. [DOI] [PubMed] [Google Scholar]
- VanRaden PM, Olson KM, Null DJ. Hutchison JL. Harmful recessive effects on fertility detected by absence of homozygous haplotypes. Journal of Dairy Science. 2011;94:6153–61. doi: 10.3168/jds.2011-4624. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vignal A, Milan D, SanCristobal M. Eggen A. A review on SNP and other types of molecular markers and their use in animal genetics. Genetics, Selection, Evolution: GSE. 2002;34:275–305. doi: 10.1186/1297-9686-34-3-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade CM. International Society for Animal Genetics (ISAG) 2012. Efficiency of light Illumina HiSeq 2000 whole-genome sequence for mutation detection, Cairns, Australia. Abstract S0120.
- Wade CM, Giulotto E, Sigurdsson S, et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326:865–7. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Watson JD. Crick FH. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953;171:737–8. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Webb AJ. Jordan CHC. Halothane sensitivity as a field test for stress susceptibility in the pig. Animal Production. 1978;26:157–68. [Google Scholar]
- Wells KL, Hadad Y, Ben-Avraham D, Hillel J, Cahaner A. Headon DJ. Genome-wide SNP scan of pooled DNA reveals nonsense mutation in FGF20 in the scaleless line of featherless chickens. BMC Genomics. 2012;13:257. doi: 10.1186/1471-2164-13-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterstrand KA. 2013. DNA Sequencing costs: data from the NHGRI Genome Sequencing Program (GSP). URL Available at: www.genome.gov/sequencingcosts.
- Willet C, Haase B. Wade CM. 2013. Better identification of candidate functional SNPs from whole genome sequence. Tenth International Equine Gene Mapping Workshop, Azores, Portugal.
- Wilson T, Wu XY, Juengel JL, et al. Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biology of Reproduction. 2001;64:1225–35. doi: 10.1095/biolreprod64.4.1225. [DOI] [PubMed] [Google Scholar]
- Wong GK, Liu B, Wang J, et al. A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature. 2004;432:717–22. doi: 10.1038/nature03156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerle M, Archibald AL, Dalens M. Gellin J. Localization of the PGD and TGF beta-1 loci to pig chromosome 6q. Animal Genetics. 1990;21:411–7. doi: 10.1111/j.1365-2052.1990.tb01985.x. [DOI] [PubMed] [Google Scholar]
- Zhang B, Healy PJ, Zhao Y, Crabb DW. Harris RA. Premature translation termination of the pre-E1 alpha subunit of the branched chain alpha-ketoacid dehydrogenase as a cause of maple syrup urine disease in polled Hereford calves. Journal of Biological Chemistry. 1990;265:2425–7. [PubMed] [Google Scholar]
- Zimin AV, Delcher AL, Florea L, et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biology. 2009;10:R42. doi: 10.1186/gb-2009-10-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An alphabetical list of Mendelian traits/disorders for which a causal mutation has been reported, up to the end of 2012.
Genes (listed alphabetically by symbol) in which mutations have been shown to result in Mendelian traits/disorders in non-rodent animals, up to the end of 2012.


