Abstract
Hematopoietic stem/progenitor cells (HSPCs) function to give rise to mature blood cells. Effective DNA damage response (DDR) and maintenance of genomic stability are crucial for normal functioning of HSPCs. Mammalian target of rapamycin (mTOR) integrates signals from nutrients and growth factors to control protein synthesis, cell growth, survival and metabolism, and has been shown to regulate DDR in yeast and human cancer cells through the p53/p21 signaling cascade. Here, we show that gene targeting of mTOR in HSPCs causes a defective DDR due to a variety of DNA damage agents, mimicking that caused by deficient FANCD2, a key component of the Fanconi anemia (FA) DDR machinery. Mechanistically, mTOR −/− HSPCs express drastically reduced FANCD2. Consistent with these genetic findings, inactivation of mTOR in human lymphoblast cells by pp242 or Torin 1, mTOR kinase inhibitors, suppresses FANCD2 expression and causes a defective DDR that can be rescued by reconstitution of exogenous FANCD2. Further mechanistic studies show that mTOR deficiency or inactivation increases phosphorylation and nuclear translocation of nuclear factor (NF)-κB, which results in an enhanced NF-kB binding to FANCD2 promoter to suppress FANCD2 expression. Thus, mTOR regulates DDR and genomic stability in hematopoietic cells through a noncanonical pathway involving NF-κB-mediated FANCD2 expression.
Keywords: mTOR, FANCD2, NF-κB, DNA damage response, hematopoietic cells
INTRODUCTION
Maintenance of genomic stability is crucial for cell survival and tissue homeostasis.1 Mammalian cells have evolved a complex network of DNA damage response (DDR) to sense and respond to genotoxic agents.2 Activation of DDR leads to the repair of DNA damage and genomic restoration, with cell death being an alternative outcome.3 A major DDR network consists of the tumor suppressor p53 and its downstream target p21.4,5 Another important DDR mechanism involves Fanconi anemia (FA) DNA repair pathway; mutation of each of the 15 known FANC genes causes FA syndrome in human, which is often manifested by bone marrow failure and/or progression to leukemia.6–8 It has recently been proposed that p53/p21 may function downstream of FA pathway in DDR of hematopoietic stem/progenitor cells (HSPCs) from FA patients.9 HSPCs give rise to multilineage mature blood cells. Normal functioning of HSPCs requires a faithful DDR. Indeed, a variety of hematopoietic diseases can be attributed to deficiency of the DDR signaling circuitry.10–12
Mammalian target of rapamycin (mTOR) is a serine/threonine kinase and has a critical role in cell growth, survival and metabolism.13 mTOR is known to function through two cellular complexes: mTOR complex 1 (mTORC1) and 2 (mTORC2).13 mTOR has been suggested to regulate DDR in yeast and human cancer cells through the p53/p21 pathway.14,15 It also has been suggested that inhibiting the mTOR pathway may sensitize cancer cells to chemotherapy and radiotherapy;16–18 however, the molecular mechanism by which this occurs remains largely unknown.
Here, we have investigated the role and underlying molecular mechanism of mTOR in DDR of HSPCs using mouse gene-targeting approaches. We found that mTOR deficiency sensitized HSPCs to chemotherapy- and radiotherapy-induced DNA damage in vitro and in vivo. Significantly, we discovered that mTOR deficiency, but not that of the mTORC1 component Raptor or the mTORC2 component Rictor, suppressed the expression of FANCD2, a critical component of FA pathway.6 The down-regulation of FANCD2 was due to an activation of transcriptional factor nuclear factor (NF)-κB that in turn inhibited FANCD2 promoter function. These results indicate that mTOR regulates DDR of HSPCs by modulating a NF-κB/FANCD2 signaling cascade.
MATERIALS AND METHODS
Mice and reagents
Conditional gene-targeted mTORloxp/loxp mice were generated as described previously.19 The flox allele contains loxP sites flanking exon 1–5 of mTOR gene. To delete mTOR in vivo in hematopoietic stem cells, mTORloxp/loxp; Mx-Cre+ mice were generated by breeding mTORloxp/loxp mice with Mx-Creþ transgenic mice carrying a bacteriophage Cre recombinase driven by an interferon-α-inducible Mx1 promoter. The expression of Cre was induced by 6–8 intraperitoneal injections of 10 mg/g of body weight polyinosine–polycytidine (Amersham Pharmacia Biotech, Piscataway, NJ, USA) into the Mx-Cre+ mice at 2-day intervals. The use of mice was in strict accordance with the guidelines for the Care and Use of Laboratory Animals of the Cincinnati Children's Hospital Research Foundation. The animal protocol was approved by the Committee on the Ethics of Animal Experiments of the Cincinnati Children's Hospital Research Foundation and consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association.
mTOR inhibitors pp242 and Torin 1 were purchased from Chemdea (Ridgewood, NJ, USA) and TOCRIS Bioscience (Bristol, UK), respectively. NF-κB inhibitor JSH-23, melphalan and mitomycin (MMC) were purchased from Sigma (St Louis, MO, USA).
NF-κB essential modulator (NEMO) peptide (acetyl-RRRRRRRLKAQADIYK ADFQAERHAREKLVEKK EYLQEQLEQLQREFNKL-amide) and control peptide (acetyl-RRRRRRRLKAQADIYKADFQAERHAR EKLVEKKEYPQEQLEQPQREFNK L-amide) were synthesized by GenScript (Piscataway, NJ, USA). NF-kB p65 −/− and IKKa −/− mouse embryonic fibroblasts (MEFs) were kindly provided by Dr Ying Xia (University of Cincinnati, Cincinnati, OH, USA) and generated from NF-κB p65 −/− and IKKα−/− fetuses as previously described.20,21
Cell apoptosis analysis
Apoptotic cell population was determined by annexin V staining and analyzed by flow cytometry.
Western blot
Whole-cell lysates or nuclear extracts were prepared and separated by 10% SDS-polyacrylamide gel electrophoresis. The expression or activation (phosphorylation) of mTOR, p53, S6K, FANCD2, NF-κB p65 and p50, and IKKα was probed by using corresponding antibodies (Cell Signaling Technology, Danvers, MA, USA).
Quantitative real-time RT-PCR
Total RNA was isolated by using RNeasy Micro Kit (Qiagen, Hilden, Germany) and recommended protocol. Reverse transcription was performed with random hexamers and Superscript II RT (Invitrogen, Grand Island, NY, USA) and was carried out at 42 °C for 60 min and stopped at 95 °C for 5 min. Real-time quantitative PCR was carried out in an ABI Prism 7900 Sequence Detector by using SYBR Green PCR Master Mix reagent (Applied Biosystems, Grand Island, NY, USA). Primer sequences used in this study are: mouse FANCA, forward 5’-TGAGCACCTCTCATGTCCTG-3’, reverse 5’-ACATCTGGATCCACCTCTGG-3’; mouse FANCC, forward 5’-GAGACCCCAG ACCACCTGTA-3’, reverse 5’-AAGAAGAAGGCGTCTGACCA-3’; mouse ATM, forward 5’-TTACGATGGCAACAGCAGAG-3’, reverse 5’-AGAACCTTGCCAACG AGAGA-3’; mouse ATR, forward 5’-AAAGGAGCTTCGCCAGTGTA-3’, reverse 5’-TGGGCAGGAGTAATTCTTGG-3’; mouse FANCD2, forward 5’-AGGCTCAAT GCTGCTTTTGT-3’, reverse 5’-CGTATTTGCTGAGGGGACAT-3’; human FANCD2, forward 5’-GCATTGTGTTTTGCCATCAC-3’, reverse 5’-TGAGGAAC TTGCTCGTCCTT-3’.
Alkaline comet assay
Lin− cells were isolated from bone marrow by MACS beads using a lineage depletion kit (Miltenyi, Auburn, CA, USA) and cultured with a combination of cytokines stem cell factor (100 ng/ml), granulocyte colony-stimulating factor (100 ng/ml), and thrombopoietin (100 ng/ml) for 24 h. The cells were then incubated with or without melphalan for 16 h, or treated with or without ionizing radiation, followed by 7-h incubation. For in vivo induction of DNA damage, mice were injected i.p. with or without MMC and killed after 72 h, and Lin− cells were purified from bone marrow. Human JY lymphoblasts, PD20 cells derived from human FA patient, or FANCD2-reconstituted PD20 cells were treated with or without pp242, rapamycin and/or MMC for 16 h. Damaged DNA content in Lin− or human cells was then determined by comet assay using a Kit from Trevigen (Gaithersburg, MD, USA) per the manufacturer's instructions. Images were captured using a Zeiss fluorescence microscope with an Axiovision camera driven by Axiovision software (Carl Zeiss, Oberkochen, Germany). Images were saved as bitmap files and olive tail moments calculated using TriTek CometScore Freeware v1.5 (TriTek Corp, Sumerduck, VA, USA).
Immunofluorescence
Cells were plated onto 100 mg/ml poly-L-lysine (Sigma)-coated coverslips and fixed with 2% paraformaldehyde. Coverslips were incubated in 0.2% Triton X-100 for 3 min, blocked with 4% bovine serum albumin and incubated with antibody against γH2AX (Upstate, Billerica, MA, USA) for 1 h. Coverslips were incubated in fluorescence-conjugated secondary antibodies (Invitrogen) for 30 min, and mounted onto glass slides with DAPI Vector Vectashield mounting media (Vector Laboratories, Burlingame, 2041 CA, USA). Images were taken on a Zeiss fluorescence microscope with an Axiovision camera driven by Axiovision software.
Chromosome breakage assay
Cells were treated with 0.05 μg/ml Colcemid (Gibco, Grand Island, NY, USA) for 90 min, followed by 0.4% KCl hypotonic solution at 37 °C for 20 min, fixed with methanol and acetic acid at 4 °C for 15 min, and dropped onto microscope slides. The cells were then rinsed with isoton, stained with Giemsa for 5 min and rinsed with Gurr Buffer (CTL Scientific, Deer Park, NY, USA) and Milli-Q-filtered deionized water. A total of 50 cells from each sample were scored for chromosome breaks.
Electrophoretic mobility shift assay
Nuclear extracts were prepared from human JY lymphoblasts. Oligonucleotide probes corresponding to canonical NF-κB consensus sequence (5’-TAGTTGAGGGGACTTTCCCAG-3’) or FANCD2-specific consensus NF-κB-binding sites (5’-TTCAGACAGGGGCTCTCCCATTGCAA-3’ (probe I); 5’-TTTCCCCAGGAAACCCCAATTTGCAA-3’ (probe II); 5’-TTAATATACTAAAAA ACCCTGAATAA-3’ (probe III); and 5’-TTTGAAGTGGGGCTTCCCAGACTGAA-3’ (probe IV))20 were labeled with γ-[32P]ATP using T4 polynucleotide kinase and purified in Bio-Spin chromatography columns (Bio-Rad, Hercules, CA, USA). A NF-1-binding probe (5’-CTTATTTTGGATTGAAGCCAATAT-3’) was used to assay NF-1 DN- binding activity as loading control.22 Ten micrograms of nuclear protein were preincubated with electrophoretic mobility shift assay buffer (12 mmol/l HEPES, pH 7.9, 4 mmol/l Tris-HCl, pH 7.9, 25 mmol/l KCl, 5 mmol/l MgCl2, 1 mmol/l EDTA, 1 mmol/l dithiothreitol, 50 ng/ml poly [d(I-C)], 12% glycerol v/v and 0.2 mmol/l phenylmethylsulfonyl fluoride) on ice for 10 min before addition of the radiolabeled oligonucleotide for an additional 10 min. To determine the binding specificity of NF-κB, supershift assay was performed by co-incubating nuclear extracts with NF-κB-specific antibodies against p65 or p50 before addition of the radiolabeled oligonucleotide. Protein–nucleic acid complexes were resolved using a nondenaturing polyacrylamide gel consisting of 5% acrylamide (29:1 ratio of acrylamide:bisacrylamide) and run in 0.5 TBE (45 mmol/l Tris-HCl, 45 mmol/l boric acid, 1 mmol/l EDTA) for 1 h at constant current (30 mA). Gels were transferred to Whatman 3M paper, dried under a vacuum at 80 °C for 1 h and exposed to a photographic film at –70 °C with an intensifying screen.
Statistical analysis
All experimental data were analyzed and compared for statistically significant differences by two-tailed Student's t-test. Data are presented as the averaged values±s.d., where applicable. The following values were considered significant: *P<0.05, and **P<0.01.
RESULTS AND DISCUSSION
mTOR regulates DDR of HSPCs
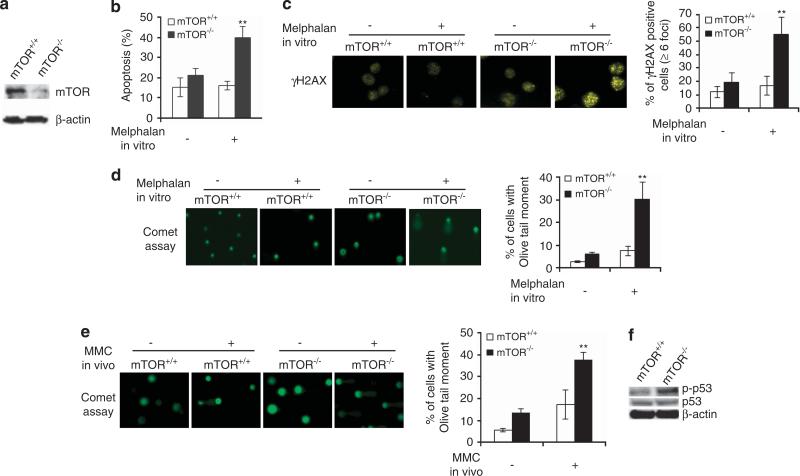
To explore the role of mTOR in DDR of HSPCs, we ablated mTOR gene from adult bone marrow by crossing polyinosine– polycytidine-inducible Mx1-Cre recombinase mice with conditional mTOR knockout mice (mTORloxp/loxp), and inducing Cre expression in the compound mTORloxp/loxp;Mx1-Creþ mice by injection of polyinosine–polycytidine (Figure 1a). Lineage-negative (Lin− ) cells that are enriched in HSPCs were isolated from the mouse bone marrow. Interestingly, mTOR −/− HSPCs grew in culture similar to that of mTORþ/þ cells, and did not display a significant defect in survival (Figure 1b). Under the treatment of the DNA damage agent, melphalan, however, mTOR −/− Lin− cells were more susceptible to apoptosis (Figure 1b). DNA damage agents induce phosphorylation of histone H2AX and foci formation of γH2AX at or around DNA damage sites.4,23 A significant increase in γH2AX foci formation was detected in melphalan-treated mTOR −/− Lin− cells (Figure 1c). When we directly measured DNA damage by employing the comet assay using a Fpg-FLARE (fragment length analysis using repair enzymes) assay kit,24,25 a marked accumulation of DNA damage manifested by tail DNA contents— olive tail moment in melphalan-treated mTOR −/− Lin− cells was observed (Figure 1d). Similar defects in DDR were also detected in γ-ionizing radiation-treated mTOR −/− cells (Supplementary Figures S1a and b). To examine DDR of the HSPCs in vivo, we treated mTOR −/− and mTOR+/+ mice with MMC, and probed for DDR in Lin− cells of the bone marrow. We found that mTOR deficiency enhanced olive tail moment in vivo induced by MMC (Figure 1e). Consistently with a defect in DDR, we found that phospho-p53 was elevated in mTOR −/− Lin cells (Figure 1f). Thus, mTOR regulates DDR of HSPCs.
Figure 1.
mTOR deficiency leads to defective DDR. (a) Western blotting of mTOR protein in bone marrow cells. (b–d) DDR of mTOR −/− and mTOR+/+ Lin− cells in vitro. Lin− cells were stimulated with or without melphalan (0.3 mg/ml). After 16 h, gH2AX foci formation was visualized by immunofluorescence and quantified (c). The images are representative of at least 30 cells (c). The percentage of positive cells (X6 gH2AX foci) was assessed from 30–50 nuclei (c). Damaged DNA was visualized by comet assay, and olive tail moment in at least 50 cells was calculated (d). After 48 h, the apoptotic cells were measured by FACS analysis of annexin V+ cells (b). (e) DDR of mTOR −/− and mTOR+/+ Lin− cells in vivo. Mice were injected i.p. with MMC (1 mg/kg body weight) and killed after 72 h. Lin− cells were isolated and analyzed for damaged DNA by comet assay. (f) p53 activity in mTOR −/− and mTOR+/+ Lin− cells. Phospho- and total p53 in Lin− cells was detected by Western blot. Data are represented as mean±s.d. **P<0.01.
mTOR regulates DDR of hematopoietic cells by regulating FANCD2 expression
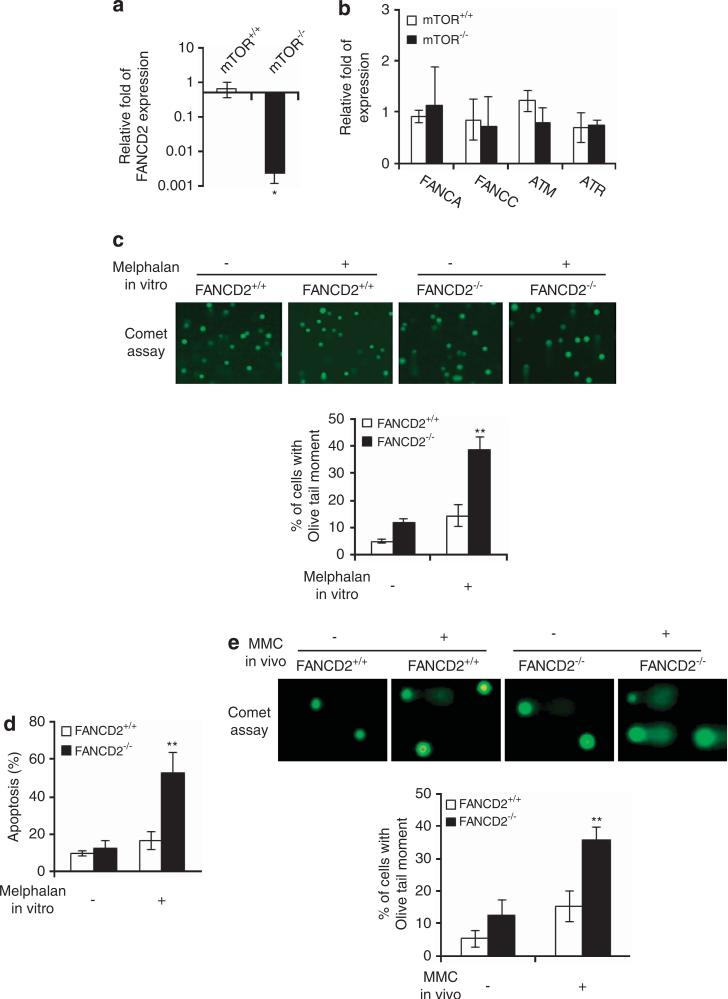
Inhibition of mTOR signaling in MCF-7 cells has been shown to increase the rate of DNA damage-induced radial chromosomes, a hallmark of the chromosome aberration of the cells with defects in FA signaling pathway.26,27 Moreover, the FA pathway has been shown to regulate DDR through p53.9 These observations led us to hypothesize that mTOR signaling may regulate DDR of HSPCs by affecting FA signaling. In support of this, we detected an over 100-fold decrease in FANCD2 mRNA expression, but not that of the FANCD2-related FANCA and FANCC nor the mTOR-related ATM/ATR kinases in mTOR −/− LSK (Lin scal+c-kit+) HSPCs (Figures 2a and b). Furthermore, FANCD2 −/− Lin− cells recapitulated the DDR phenotypes of mTOR −/− Lin− cells in vitro and in vivo, which are manifested by increased DNA damage and apoptosis after melphalan treatment (Figures 2c–e). These genetic results suggest that mTOR signaling to DDR may be mediated through the FANCD2 pathway.
Figure 2.
FANCD2 deficiency recapitulated DDR phenotypes of mTOR deficiency. (a) Quantitative RT-PCR analysis of FANCD2 expression in LSK cells. (b) Quantitative RT-PCR analysis of FANCA, FANCC, ATM and ATR expression in LSK cells. (c, d) DDR of FNACD2 −/− and FANCD2+/+ Lin cells in vitro. Lin cells were stimulated with or without melphalan (0.3 μg/ml). After 16 h, damaged DNA was visualized by comet assay, and olive tail moment in at least 50 cells was calculated (c). After 48 h, the apoptotic cells were measured by FACS analysis of annexin V+ cells (d). (e) DDR of FNACD2 −/− and FANCD2+/+ Lin cells in vivo. Mice were injected i.p. with MMC (1 mg/kg body weight) and killed after 72 h. Lin cells were isolated and analyzed for damaged DNA by comet assay. Data are represented as mean±s.d. *P<0.05; **pP<0.01.
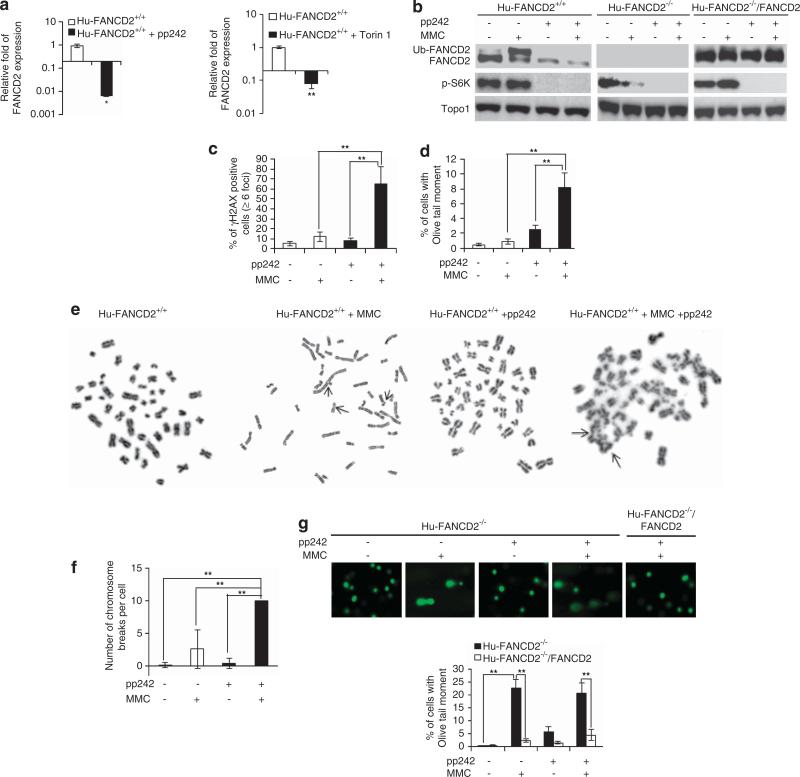
The mTOR-FANCD2 DDR signaling axis also exists in human cells. Treatment of human B lymphoblasts (hereafter referred to as Hu-FANCD2+/+) with pp242 or Torin 1, inhibitors of mTOR kinase activity, markedly decreased FANCD2 mRNA transcript expression (Figure 3a). Treatment of Hu-FANCD2+/+ cells with pp242 also inhibited MMC-induced FANCD2 monoubiquitination (Figure 3b), the biochemical hallmark of activation of the FA repair pathway.6–9 Notably, although pp242 was effective in inhibiting endogenous FANCD2 expression, it did not suppress the expression of a FANCD2 cDNA driven by a retroviral promoter (Figure 3b), suggesting that mTOR may exert its regulatory effect on the promoter of the FANCD2. Consequently, Hu-FANCD2+/+ cells treated with pp242 displayed a dampened DDR in response to MMC, leading to increased γH2AX foci and DNA damage (Figures 3c and d). Cytogenetic analysis revealed that pp242-treated cells responded to MMC-induced DNA damage in producing significantly increased chromosome breaks and abnormalities, including radial structure similar to that of FANCD2-deficient PD20 cells derived from a FA patient (hereafter referred to as Hu-FANCD2 −/− ) (Figures 3e and f and Supplementary Figures S2a and b). In parallel, Hu-FANCD2 −/− cells exhibited a defect in MMC-induced DDR, but did not show an additive effect to MMC after pp242 treatment (Figure 3g). Furthermore, the MMC-elicited DDR of Hu-FANCD2 −/− cells with or without pp242 could be rescued by retrovirally expressed FANCD2 (Figures 3b and g). Thus, mTOR regulates DDR in a FANCD2-dependent manner. Interestingly, deletion of the mTORC1 component Raptor or the mTORC2 component Rictor had no effect on FANCD2 expression (Supplementary Figures S3a and b), suggesting that mTOR regulates FANCD2-mediated DDR independently from the canonical mTOR signaling of mTORC1 or mTORC2.
Figure 3.
mTOR inhibition mimics FANCD2 deficiency with defective DDR in human cells. (a, b) FANCD2 expression in human cells upon mTOR inhibition. (a) Human JY B lymphoblasts (Hu-FANCD2+/+) were treated overnight with or without mTOR kinase inhibitor pp242 (2.4 μM) (left) or Torin 1 (100 nM) (right) and analyzed by quantitative RT-PCR for FANCD2 expression. (b) Hu-FANCD2+/+ cells, human Fanconi anemia patient-derived PD20 cells deficient for FANCD2 (Hu-FANCD2 −/− ), and FANCD2-reconstituted Hu-FANCD2 −/− cells (Hu-FANCD2 −/− / FANCD2) were preincubated with or without pp242 for 1 h and then cultured overnight in the presence or absence of MMC (100 ng/ml). The cells were subjected to western blotting for FANCD2 and phopho-S6K. Topo1 was blotted as a loading control. Ub-FANCD2: ubiquitinized FANCD2. (c, d) DDR of Hu-FANCD2+/+ cells after a treatment with pp242 and/or MMC. (e, f) Chromosome breaks and radial formation of Hu-FANCD2+/+ cells after a treatment with pp242 and/or MMC. Hu-FANCD2+/+ cells were preincubated with or without pp242 for 1 h and then cultured for 2 days in the presence or absence of MMC. Chromosomes were then subjected to cytogenetic analysis. A total of 50 cells from each sample were examined and scored. Arrowheads point to chromosome breaks in the Hu-FANCD2+/+ + MMC cells or to radial structures in the Hu-FANCD2+/+ + MMC + pp242 cells. As too many breaks were observed in Hu-FANCD2+/+ + MMC + pp242 cells, the number of chromosome breaks per cell in these cells was artificially set to 10. (g) DDR of Hu-FANCD2 −/− and Hu-FANCD2−/− /FANCD2 cells after a treatment with pp242 and/or MMC. Data are represented as mean±s.d. *P<0.05; **P<0.01.
mTOR promotes FANCD2 expression by suppressing NF-κB activity
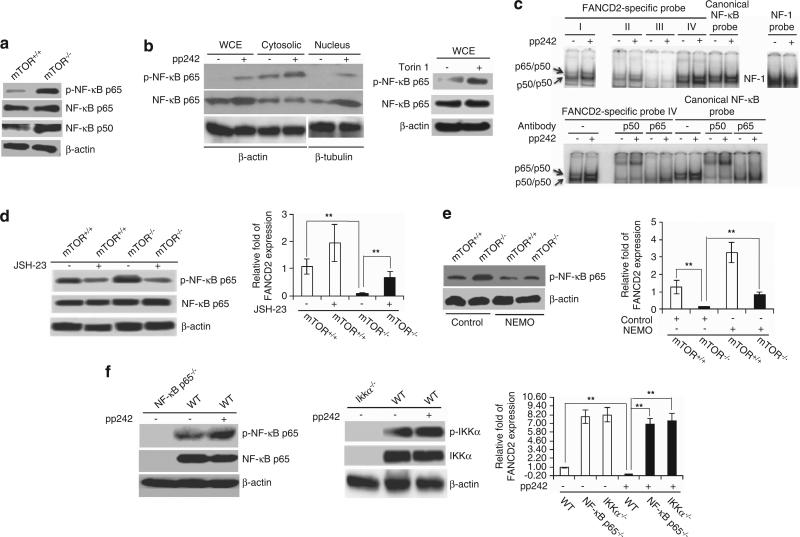
Next, we sought to determine the molecular link between mTOR signaling and the FANCD2 pathway. As FANCD2 gene promoter region contains four putative NF-κB binding sites and mTOR has been suggested to regulate NF-κB pathway in cancer cell lines,22,28 we first examined the activities of NF-κB in mTOR −/− Lin− bone marrow cells and pp242- or Torin 1-treated Hu-FANCD2+/+ cells. These cells showed significantly increased levels of phospho-p65 and p50 of NF-κB in comparison with mTOR+/+ Lin− cells and non-pp242- or non-Torin 1-treated cells, respectively (Figures 4a and b). We also observed an enhanced nuclear translocation of NF-κB p65 subunit, and DNA binding activity of NF-κB p65/p50 heterodimer and p50/p50 homodimer to canonical NF-κB binding site as well as the NF-κB binding sites of the FANCD2 promoter, particularly the binding site IV, in pp242-treated Hu-FANCD2+/+ cells (Figures 4b and c). The binding specificity of NF-κB p65/p50 and p50/p50 to canonical NF-κB binding site, as well as the NF-κB binding sites of the FANCD2 promoter was detected by a supershift assay with anti-p65 and anti-p50 antibodies (Figure 4c). Consistent with an elevated NF-κB activity upon mTOR suppression, pp242-treated HEK 293T cells showed an augmented generic NF-κB luciferase reporter activity, and mTOR −/− HSPCs (LSK cells) displayed an increased expression of a number of NF-κB target genes, including IL-1R2 and IGFR as revealed by quantitative real-time PCR (Supplementary Figures S4a and b). These data are indicative of an increase in canonical NF-κB transcriptional activity upon mTOR inhibition or deficiency. Importantly, FANCD2 expression could be partially restored by treatment of mTOR −/− Lin− cells with a NF-κB inhibitor, JSH-23 (Figure 4d), or a NF-κB inhibitory peptide, NEMO (Figure 4e). A restoration of FANCD2 expression by JSH-23 rescued defective DDR in mTOR −/− Lin cells as shown by decreased γH2AX foci formation in DDR assays (Supplementary Figures S5a–c). In MEFs, genetic deletion of NF-κB p65 or NF-κB upstream activator IKKa was able to fully reverse mTOR inhibitor (pp242)-induced FANCD2 downregulation (Figure 4f). Together, these data indicate that mTOR positively regulates FANCD2 expression by suppressing NF-κB.
Figure 4.
NF-κB mediates mTOR-dependent FANCD2 expression. (a) Western blotting of phospho- and total NF-κB p65 and p50 in Lin− cells. (b) Western blotting of phospho- and total NF-κB p65 in whole-cell extracts (WCE), cytosolic and nuclear fractions of Hu-FANCD2+/+ cells treated overnight with or without pp242 (2.4 μM) (left) or Torin 1 (100 nM) (right). (c) Electrophoretic mobility shift assay (EMSA) on DNA binding activity of NF-κB to canonical, as well as four FANCD2-specific NF-κB consensus sites by using pp242-treated Hu-FANCD2+/+ cells (upper). Supershift assays were performed to determine the binding specificity of NF-κB by using NF-κB-specific antibodies against p65 or p50 (lower). NF-1 DNA binding activity was assayed as a control. (d) Left hand panel: western blotting of phospho- and total NF-κB p65 in Lin− cells treated overnight with or without NF-κB inhibitor JSH-23 (10 μM). Right hand panel: quantitative RT-PCR analysis of FANCD2 expression in JSH-23-treated Lin− cells. (e) Left hand panel: western blotting of phospho-NF-κB p65 in Lin− cells treated overnight with or without NF-κB inhibitory peptide NEMO (15 μM) or control peptide (15 μM). Right hand panel: quantitative RT-PCR analysis of FANCD2 expression in NEMO-treated Lin cells. (f) Left hand panel: western blotting of phospho- and total NF-κB p65 in NF-κB p65+/+ (WT) and NF-κB p65 −/− MEFs treated for 12 h with or without pp242. Middle panel: western blotting of phospho- and total IKKα in IKKα+/+ (WT) and IKKα −/− MEFs treated for 12 h with or without pp242. Right hand panel: quantitative RT-PCR analysis of FANCD2 expression in pp242-treated MEFs. Data are represented as mean±s.d. **P<0.01.
As mTOR was reported to bind to IKKa in prostate cancer cells,28 we further examined if inhibition of NF-κB by mTOR could be attributed to mTOR binding to IKKa and a sequestration of IKKa from activating NF-κB. IKKα co-immunoprecipitated with mTOR in Hu-FANCD2+/+ cells, and a treatment with pp242 abolished the co-immunoprecipitation (Supplementary Figure S6a). Blockade of the interaction between mTOR and IKKα by pp242 correlated with an increased binding of IKKa to its upstream activator TAK1 (Supplementary Figure S6b). Thus, it appears that mTOR negatively regulates NF-κB activity by binding to IKK, and that mTOR knockout or inhibition disrupts the mTOR–IKK complex to release IKK for binding to and activation by TAK1, leading to increased NF-κB nuclear activity.
The current studies reveal a previously unappreciated role of mTOR in DDR of hematopoietic cells that is mediated through a NF-κB/FANCD2 signaling cascade dependent upon the kinase activity of mTOR. Neither mTORC1 nor mTORC2 signaling alone could account for the observed mTOR regulation of FANCD2 expression, because gene deletion of the Raptor or Rictor is insufficient to yield an effect on FANCD2 expression (Supplementary Figures S3a and b). Although mTOR has been suggested to activate NF-κB in prostate cancer cells,28 our genetic data show that mTOR suppresses, rather than activating, NF-κB activity in hematopoietic cells. Thus, mTOR may regulate NF-κB activity in a cell type-specific manner. One generic role of NF-κB is to promote gene transcription. Indeed, activation of NF-κB in mTOR-deficient HSPCs correlates with increased mRNA expression of NF-κB target genes IL-1R2 and IGFR (Figure S4b), as well as a NF-κB reporter (Figure S4a). Furthermore, NF-κB seems to promote FANCD2 gene expression in multiple myeloma cells.22 However, our data demonstrate that in the mTOR signaling context, NF-κB serves as a suppressor in regulating FANCD2 expression in normal hematopoietic cells (Figure 4), suggesting that the mTOR-mediated role of NF-κB in governing FANCD2 expression is distinct from that of canonical NF-κB pathways. In summary, our studies unveil a noncanonical pathway of mTOR signaling in DDR of hematopoietic cells. The findings suggest that a DNA repair-targeted therapy can come by mTOR kinase inhibition to sensitize cancer cells to DNA damage agents and enhance anticancer potential.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Markus Grompe (Oregon Health & Sciences University) for FANCD2+/− mice, Dr Alan D'Andrea (Harvard Medical School) for the pMMP-Puro and pMMPFANCD2 retroviral vectors. This work was supported by NIH grants R01 HL076712 and T32 HL091805.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 4.Milyavsky M, Gan OI, Trottier M, Komosa M, Tabach O, Notta F, et al. A distinctive DNA damage response in human hematopoietic stem cell reveals an apoptosis-independent role of p53 in self-renewal. Cell Stem Cell. 2010;7:186–197. doi: 10.1016/j.stem.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 5.Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Beau MM, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kee Y, D'Andrea AD. Molecular pathogenesis and clinical management of Fanconi anemia. J Clin Invest. 2012;122:3799–3806. doi: 10.1172/JCI58321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitao H, Takata M. Fanconi anemia: a disorder defective in the DNA damage response. Int J Hematol. 2011;93:417–424. doi: 10.1007/s12185-011-0777-z. [DOI] [PubMed] [Google Scholar]
- 8.Leguit RJ, van den Tweel JG. The pathology of bone marrow failure. Histopathology. 2010;57:655–670. doi: 10.1111/j.1365-2559.2010.03612.x. [DOI] [PubMed] [Google Scholar]
- 9.Ceccaldi R, Parmar K, Mouly E, Delord M, Kim JM, Regairaz M, et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11:36–49. doi: 10.1016/j.stem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 11.Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 12.Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 13.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2005;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beuvink I, Boulay A, Fumagalli S, Zilbermann F, Ruetz S, O'Reilly T, et al. The mTOR inhibitord RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120:747–759. doi: 10.1016/j.cell.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 15.Shen C, Lancaster CS, Shi B, Guo H, Thimmaiah P, Bjornsti MA. TOR signaling is a determinant of cell survival in response to DNA damage. Mol Cell Biol. 2007;27:7007–7017. doi: 10.1128/MCB.00290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol. 2009;27:2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yap TA, Garrett MD, Walton MI, Raynaud F, de Bono JS, Workman P. Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls, and promises. Curr Opin Pharmacol. 2008;8:393–412. doi: 10.1016/j.coph.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz JF, et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol. 2004;24:9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 21.Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, et al. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKa subunit of IkB kinase. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- 22.Yarde DN, Oliveira V, Mathews L, Wang X, Villagra A, Boulware D, et al. Targeting the Fanconi anemia/BRCA pathway circumvents drug resistance in multiple myeloma. Cancer Res. 2009;69:9367–9375. doi: 10.1158/0008-5472.CAN-09-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fairbairn DW, Olive PL, O'Neill KL. The comet assay: a comprehensive review. Mutat Res. 1995;339:37–59. doi: 10.1016/0165-1110(94)00013-3. [DOI] [PubMed] [Google Scholar]
- 25.Olive PL, Banáth JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. Radiat Res. 1990;122:86–94. [PubMed] [Google Scholar]
- 26.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross) linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Venkitaraman AR. Tracing the network connecting BRCA and Fanconi anaemia proteins. Nat Rev Cancer. 2004;4:266–276. doi: 10.1038/nrc1321. [DOI] [PubMed] [Google Scholar]
- 28.Dan HC, Cooper MJ, Cogswell PC, Duncan JA, Ting JP, Baldwin AS. Akt-dependent regulation of NF-kB is controlled by mTOR and Raptor in association with IKK. Genes Dev. 2008;22:1490–1500. doi: 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.