Abstract
The field of plasmid biology has historically focused on bacteria growing in liquid culture. Surface attached communities of bacterial biofilms have recently been understood to be the normal environment of bacteria in the natural world. Thus, studies examining plasmid replication, maintenance, and transfer in biofilms are essential for a true understanding of bacterial plasmid biology. This chapter reviews the current knowledge of the interplay between bacterial biofilms and plasmids, focusing on the role of plasmids in biofilm development and the role of biofilms in plasmid maintenance, copy number control, and transfer. The studies examined herein highlight the importance of biofilms as an important ecological niche in which bacterial plasmids play an essential role.
Introduction
The natural state for many bacteria is not growth in liquid culture but rather, living as a community attached to a surface. These bacterial communities, termed biofilms, exist in the natural world as well as in the human host. The Centers for Disease Control and the National Institutes of Health have estimated that approximately 65-80% of human infections are biofilm-related. A recent burgeoning area of research has examined the role of plasmids in biofilms including the effect of conjugative plasmid transfer on biofilm formation as well as the role of biofilms in plasmid dissemination. In addition, heterogeneity in the biofilm population in terms of plasmid carriage has also been demonstrated. Most published studies of plasmid biology and conjugation in biofilms have focused on gram negative spp. such as Pseudomonas aeruginosa and Escherichia coli. In this chapter, we will review these studies in relation to recent work focusing on effects of biofilm growth on plasmid-related functions such as gene transfer and antimicrobial resistance in gram positive pathogens such as Enterococcus faecalis and Staphylococcus aureus.
Formation of bacterial biofilms involves three steps (Figure 1). Initially, individual cells growing planktonically attach to a surface. Following surface adherence, additional cells may bind to previously attached cells. As the attached cells grow and divide, they produce an extracellular polymeric substance known as the biofilm matrix that stabilizes attachment of the cells to one another and to the surface. The biofilm matrix components may differ between species but frequently contain DNA (1), proteins (2), and polysaccharides (3) as well as other nutrients and cellular components (recently reviewed (4)). During and following formation of a fully structured biofilm, individual cells or even large pieces of the biofilm may break away. These cells may then revert back to a planktonic lifestyle or may attach to a surface elsewhere and seed a new biofilm (Figure 1).
Figure 1. Formation of a bacterial biofilm.
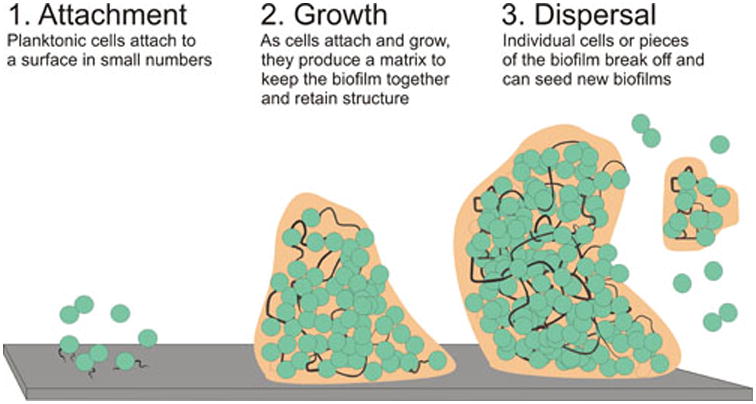
Bacterial biofilm development involves three stages. 1. Initial attach of single or small groups of bacteria to a surface, often aided by attachment structures such as pili. 2. Growth of these attached cells as well as attachment of additional cells increases the biomass of the biofilm. Concurrently, the bacteria produce an extracellular matrix made of up various components including DNA, protein and polysaccharides which help the biofilm retain its structure and keeps the biofilm cells attached to the surface and to each other. 3. During and after the formation of a large biofilm individual cells or even large pieces of the biofilm may break off. These detached cells may go on to live a planktonic life style or seed new biofilms. Dark lines indicate components of the matrix used to attach cells to each other and the surface such as eDNA (1) while orange extracellular material indicates other matrix components used to retain biofilm structure and surface attachment.
Many of the seminal studies of biofilm development that led to developmental models like the one shown in Figure 1 utilized rod-shaped motile bacteria such as E. coli, P. aeruginosa and Bacillus subtilis (5-9). In such bacteria, sensing of surface attachment and transition from planktonic to biofilm growth may involve components of the motility machinery and is accompanied by loss of motility, whereas the dispersal phase can involve reactivation of motility. It is interesting that non-motile genera including staphylococci, streptococci and enterococci show very similar patterns of biofilm development and dispersal, including characteristic cellular architecture and extracellular matrix in the biofilm structure. As biofilms are believed to be highly heterogenous in terms of nutrient, pH, and oxygen gradients, it is likely that they are composed of heterogenous populations of cells that differ in terms of metabolic potential and phenotypic characteristics.
Conjugative plasmids in biofilms
It is well established that the biofilm is an important niche for horizontal gene transfer (HGT) by transformation in naturally competent bacteria (10, 11), and that biofilm development and competence are mediated and regulated by many of the same gene products (10, 12, 13). HGT, the transferring genetic material between cells in which reproduction does not play a role, includes the processes of conjugation, transformation, and transduction. Increasingly, new studies have examined the interplay between conjugation and biofilm development (14-20). The seminal paper by Ghigo describing the role of plasmids in biofilm formation described the effects of the well-studied conjugative F plasmid of E. coli biofilms (15). These experiments demonstrated that addition of the F plasmid to E. coli cells greatly increased their ability to form biofilms a conjugation-independent and plasmid-encoded pilus-dependent fashion (15). This report documented an increase in biofilm formation by other gram-negative bacteria when grown with pilus-encoding natural conjugative plasmids. In the case of F and other plasmids like it that express pili and other conjugation functions constitutively, the presence of the plasmid was associated with increased biofilm formation. Monocultures of donor strains carrying repressed plasmids such as R1 did not exhibit increased biofilm development. However in biofilms formed from donor/recipient mixtures, or in recipient biofilms subsequently exposed to planktonic donor cells, Ghigo observed enhanced biofilm development. Ghigo hypothesized that a small number of spontaneously-depressed pilus-producing bacteria in the planktonic donor cultures could adhere to the recipient biofilms,and transfer their plasmids, followed by a period of “epidemic spread” through the entire biofilm. The increase in pilus-expressing bacteria could then aid in formation of a large bacterial biofilm (15). The top portion of Figure 2 illustrates the model proposed by Ghigo. The lower panels depict a variation on the theme of induction of conjugation and surface adhesins in a biofilm context for Enterococcus faecalis, where the expression of conjugative functions in donor cells can be activated by a peptide mating pheromone produced by recipients, which is further discussed below. Both models illustrate how activation of conjugation in a biofilm context can lead to both plasmid transfer and increased biofilm biomass.
Figure 2. The interplay between conjugation and biofilm development.
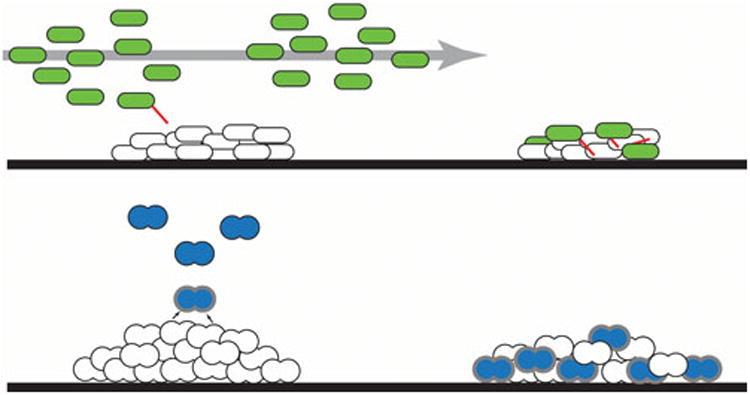
Top panel: A model proposed by Ghigo (15), based on his analysis of conjugation between E. coli strains. Planktonic populations of donor cells (green), carrying plasmids such as R1, whose conjugation functions are normally repressed, contain a few spontaneously depressed individuals. When these depressed cells encounter a biofilm containing recipient cells (white) they can attach via their sex pili (red) and transfer the plasmid. In newly generated transconjugants, there is a transient period where repression of conjugation is not operative. This can be followed by “epidemic spread” of the plasmid through the biofilm population, and the associated production of sex pili also can increase the biofilm biomass directly. Lower panel: In E. faecalis, expression of conjugation is regulated by peptide mating pheromones produced by recipient cells. In the scenario depicted on the left, the pheromone produced by recipient cells (white) in a biofilm turns on expression of conjugation in planktonic donor cells (blue) in close proximity, and the resulting synthesis of pheromone-induced surface adhesins (thick, gray layer) promotes both an increase in biofilm resulting from increased attachment of planktonic cells, and also leads to plasmid transfer within the biofilm. In the right panel, development of a mixed biofilm as a result of attachment of both donors and recipients to the same surface may allow for signaling and conjugation between sessile donor and recipient cells in close proximity (34).
Transmission of a conjugative F plasmid also induces biofilm formation by a mixed population of laboratory and wild isolates of E. coli and plays a role in the overall structure of the biofilm (5, 20). Addition of an F-like conjugative plasmid, R1drd19 which, like F, constitutively synthesizes pili, was also shown to induce greater biofilm formation in E. coli cultures (19). Interestingly, the presence of the R1drd19 plasmid also increased expression of numerous chromosomal genes including those related to envelope stress, motility, and other genes known to be involved in biofilm formation. It was also demonstrated that F pilus production caused increased colonic acid and curli (thin fimbriae) production (21). Curli have previously been shown to stimulate attachment of E. coli cells to surfaces (22). Other conjugative plasmids of E. coli including pOLA52 and pMAS2027 have been shown to enhance biofilm formation through type 3 fimbriae (14, 17). The pOLA52 plasmid can also be transferred to a variety of organisms and retains its ability to induce biofilm formation in Salmonella typhimurium, Kluyvera sp., and Enterobacter aerogenes (14).
Studies on the TOL conjugative plasmid of Pseudomonas putida demonstrated that carriage of conjugative plasmids could also increase biofilm formation by increasing the amount of extracellular DNA (eDNA), thus aiding the formation of the biofilm matrix (16). Although not fully understood, the mechanism of increased eDNA does not appear to be caused by increased cell lysis and may, alternatively, be due to increased DNA secretion. These results demonstrate that the conjugative pili and fimbriae are not the only factors involved in plasmid-mediated enhancement of biofilm formation.
Co-culture studies with P. putida, E. coli, and Kluyvera spp. also demonstrated the impact of conjugative plasmids on biofilm development. In these experiments, the conjugative plasmid pKJK5, an IncP-1 plasmid (23), altered biofilm development during growth in co-culture. Co-cultures of the three bacteria produced increased biofilms when compared to any of the strains alone. Interestingly, when P. putida carried pKJK5, biofilm formation of a mixed co-culture was decreased (24). In this case, the presence of a conjugative plasmid had a negative effect on biofilm formation rather than enhancing it. It is apparent from these studies that the role of conjugative plasmids in biofilm development is likely complex and varied and could depend strongly on host as well as plasmid-encoded factors.
The interplay between conjugative plasmids and biofilm formation is not a one-way street. Conjugative plasmids influence the development of biofilms and, in turn, biofilms affect horizontal transfer of conjugative plasmids. Biofilm promotion of higher levels of HGT has been demonstrated for a variety of systems (18, 25-27). One of the first papers outlining HGT rates in biofilms used a GFP-tagged broad host-range plasmid pRK415 (28) requiring a separate plasmid containing the cognate conjugation system, pRK2013. Using the GFP reporter and a TRITC counterstain, researchers were able to determine approximate conjugation rates in biofilms (25). Similarly, it was determined that transfer of the F-like plasmid R1drd19 in E. coli occurred at significantly higher rates in a biofilm and transfer kinetics were similar between laboratory biofilms and growth in a mouse intestine (29). Another study looking at conjugative transfer of plasmid RP4 between Pseudomonas species demonstrated high transfer frequencies in biofilms and showed that shear force affected HGT, suggesting that altering various biofilm parameters affects rates of gene transfer (30).
A recent study attempted to determine the various factors involved in conjugative transfer of antibiotic resistance plasmids in E. coli biofilms in a comprehensive fashion (31). The transfer efficiency of 19 drug-resistant plasmids was measured using biofilms of different ages and different times of exposure of biofilms to plasmid donor cells. Wide variation was observed in transfer efficiencies between different plasmids and efficiencies depended on conditions such as biofilm thickness and age with older and thicker biofilms having a smaller proportion of transconjugants. Not surprisingly, allowing donor cells to incubate for longer periods of time with established biofilms also allowed for increased conjugation. Transfer was not only dependent on the particular plasmid and biofilm conditions; transfer efficiency was also increased when the donor strain background had increased ability to attach to recipient cells, although this was also seen in liquid cultures (31).
HGT in gram positive bacterial biofilms has also been studied, although to a lesser extent. S. aureus, an important gram positive human pathogen, is known to form biofilms related to infections such as endocarditis, indwelling device-associated infections, and osteomyelitis. One report demonstrated that biofilm growth in S. aureus also facilitates higher levels of HGT with increased conjugation and mobilization frequencies leading to the spread of antibiotic resistance (27).
Plasmid copy number control and maintenance in biofilms
Biofilm growth does not only affect plasmid transfer but it also appears to play a role in plasmid maintenance and copy number control. In 1995, Davies and Geesey examined the regulation of alginate biosynthesis in P. aeruginosa biofilms using a reporter plasmid pNZ63 carrying a β-lactamase marker. During the course of their experiments they noted that although the presence of the β-lactam antibiotic did not affect the average plasmid copy number in the population, biofilm growth coincided with an increase in plasmid number by ~1.5 fold (32). They also determined that the change in copy number was not due to plasmid loss as most of the cells retained antibiotic resistance. Additionally, it was shown that the copy number of pBR322, a plasmid carrying resistance genes against ampicillin and tetracycline, was increased approximately two-fold in E. coli cells growing in a biofilm compared to planktonic copy numbers (33). Interestingly, an E. coli strain containing the pBR322 plasmid formed less biofilm than the plasmid-free cells, which the authors attributed to the presence of the bla ampicillin resistance gene. Addition of tetracycline or a combination of tetracycline and ampicillin at sub-inhibitory concentrations induced higher biofilm formation by E. coli cells harboring the plasmid but not plasmid-free cells (33). This study also found that the addition of antibiotics to planktonic populations caused an increased in plasmid copy number to levels seen in biofilm cells, supporting the hypothesis that increased plasmid copy number correlates with increased antibiotic resistance (33). Importantly, these studies examined average copy number in biofilms which could significantly underestimate the copy number heterogeneity that may exist in subpopulations of biofilm cells.
In 2011, a report by Cook et al. demonstrated that the copy number of the conjugative plasmid pCF10 of E. faecalis was increased during growth in a biofilm as compared to planktonic growth (34). Not only was the average copy number of pCF10 increased in biofilm populations but copy number heterogeneity in the population was also highly increased, supporting the model of biofilms as complex communities in which not all cells are identical. While the average number of pCF10 copies/chromosome in biofilm cells was about twice that of planktonic cells, fluorescence-activated cell sorting experiments showed the existence of sub-populations of biofilm cells containing as many as 5 times as many plasmid copies as planktonic cells (34).
In the case of pCF10, an increase in plasmid copy number causes a subsequent increase in plasmid-borne negative regulators of conjugation including an inhibitory peptide, iCF10 and the negative regulator of conjugation, PrgX. This results in tighter control of conjugation induction; thus, biofilm cells carrying more copies of pCF10 require a higher concentration of inducer peptide, cCF10, to turn on conjugation. These same cells may show a significantly increased level of conjugation gene expression once the threshold inducing concentration is exceeded, leading to a stronger response to peptide. It is hypothesized that this plasmid copy number control allows restrictive regulation of conjugation, preventing the energetically costly process to occur when potential plasmid recipient cells are not in the immediate vicinity (34). This is especially important in the case of E. faecalis as the non-motile bacteria are fixed in the biofilm without the ability to migrate towards potential recipients.
Following this study, further analysis was completed on the copy number of various enterococcal plasmids to determine whether increased copy number was a pCF10-specific phenomenon. It was found that at least four other plasmids showed an increase in both plasmid copy number and copy number heterogeneity when cells were grown in a biofilm (35). These four plasmids were not conjugative, were unrelated to pCF10, had different native copy numbers, and included both rolling circle and theta replicating elements. In this study, increased plasmid copy number was also correlated with increased expression of plasmid-borne antibiotic resistance genes as well as increased plating efficiency on high concentrations of antibiotics (35). It was not possible to determine the heterogeneity of plasmid copy number in biofilm cells with all the plasmids analyzed, but the available data indicates that the heterogeneity observed with pCF10 could also exist for other, unrelated replicons.
The published studies of biofilm growth of E. faecalis, and its effect on plasmid copy number, plasmid transfer and antibiotic resistance can be used to generate hypothetical mechanistic models that may inform the design of future experiments. Figure 3 illustrates three possible mechanisms, all of which assume that a subpopulation of plasmid-carrying cells in a biofilm display a significant increase in copy number. It has recently been shown that during early biofilm growth of E. faecalis, there is substantial secretion of eDNA into the extracellular matrix by a mechanism that does not require cell lysis, and may be carried out by a fraction of cells in the monospecies biofilm community (1). As shown in Figure 3A, if chromosomal (rather than plasmid) DNA was a specific substrate for this novel secretion pathway, the relative intracellular concentration of plasmid would be increased. This could also indirectly increase transcription of plasmid genes relative to chromosomal genes, by increasing the fraction of the cellular pool of RNA polymerase available to transcribe plasmid genes. Figure 3B depicts a scenario where the global physiological changes in a fraction of biofilm cells disrupts initiation of chromosome replication, even when nutrients are not limiting and the overall growth rate of the community is high. In both 2A and 2B, the increased ratio of plasmids actually results from a decrease in replication and reduced numbers of chromosomes/cell, and in some cases concomitant “runaway replication” (36) of plasmids residing in the same cell could occur. Runaway replication of plasmids has been demonstrated to occur in the presence of environmental factors such as antibiotics, nutrient levels, and temperature (36-38). In the previously published work (34, 35), significant changes in total DNA per cell were not found in the pooled biofilm cells, but the methods used may not have detected changes in subpopulations. In the model illustrated in Figure 3C, the altered physiology of biofilm cells is postulated to increase initiation of vegetative replication, or conjugative nicking, both of which result in generation of new copies of the plasmid. The models depicted in Figure 3 all involve actively-growing cells in early biofilm formation, when there is no a significant level of cell death or lysis. For cells in older biofilms experiencing nutrient limitation and other stresses, there are likely additional factors that could have significant impacts on plasmid maintenance and copy control, as noted below.
Figure 3. Three mechanistic models that could account for increased ratios of plasmids/chromosome in a fraction of E. faecalis biofilm cells.
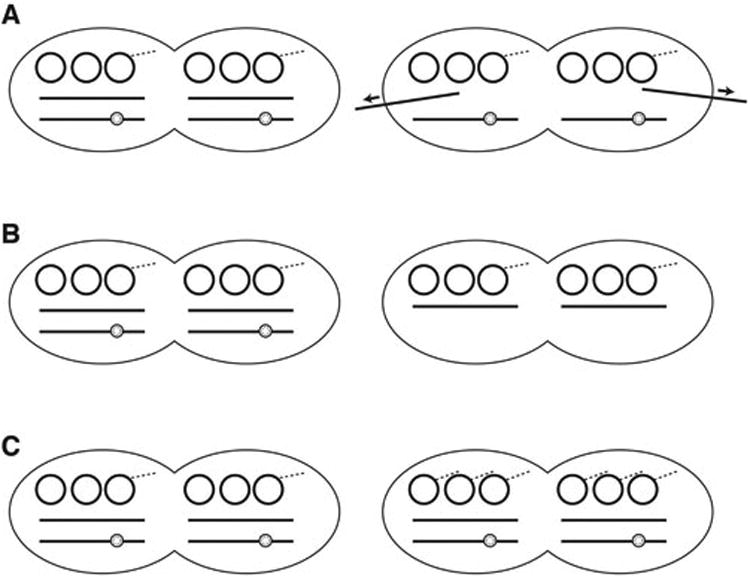
Figures depict actively growing “planktonic-like” diplococcal cells prior to cell division. Each cell half contains two copies of the chromosome (represented by thick lines), where a new round of replication has initiated on one chromosome (indicated by the “bubble”). Each cell half also contains 3 copies of a plasmid, with the dashed lines indicating rolling circle replication initiated by either a single-stranded replication initiator protein or a conjugative relaxase. (It should be noted that some plasmids showing increased copy number in biofilms actually use a “theta” mode of replication similar to the chromosome (35)). Each diagram contains one diplococcal cell (left) with a similar plasmid copy number to that of planktonic cells and a second cell with altered copy number, representing a subpopulation of the biofilm community. The three diagrams illustrate 3 possible mechanisms by which the plasmid/chromosome ratio could be increased in these subpopulations. A. Secretion of eDNA into the biofilm matrix by a fraction of cells, reduces the ratio of chromosome/plasmid. B. Biofilm growth reduces chromosomal copy number in subpopulations by inhibiting initiation of chromosome replication. C. In a subpopulation of biofilm cells, changes in cellular physiology could disrupt normal mechanisms limiting conjugative nicking or vegetative plasmid replication initiation, leading to increases in copy number. Additionally for some well-studied plasmids, such as ColE1, blockage of chromosome replication initiation (B) can lead to “runaway” plasmid replication (36). Based on published results (1, 34) all of these models are postulated to operate in the early stages of biofilm development, when the adherent cells are still growing, and there is no significant death or lysis.
Although plasmid copy number was increased in biofilms for numerous plasmids listed above, researchers have conversely demonstrated that, for some plasmids, plasmid loss is increased in biofilm populations, especially at specific foci within the biofilm. The TOL plasmid of P. putida, known for carrying genes allowing degradation of organic compounds, is used in bioremediation and the ability of this plasmid to be transferred stably in an environmental setting such as a biofilm was examined. It was found that the probability of plasmid loss was increased in biofilms compared to planktonic cells. As was shown with plasmid copy number, plasmid loss was also heterogenous within the biofilm with the outside layers experiencing up to 80 times higher plasmid loss than the populations in the middle of the biofilm (39). Loss of plasmids in P. putida biofilm populations was attributed to segregational loss. In this study, actual copy number of plasmids/cell was not examined, only loss of the plasmid based on selective plating. It is important to remember that the P. putida biofilms that were examined ranged from 1-7 days old whereas plasmid copy number analyses in E. faecalis were done on “young” biofilms at 4 or 24 hours after initial inoculation. The age of the biofilm could greatly impact the growth rate of the cells and thus affect plasmid copy number control or plasmid loss.
Results of these studies demonstrate that the heterogeneous traits of biofilm cells extend to plasmid carriage. Upregulation of plasmid copy number presents a novel possible mechanism of the high level of antibiotic resistance observed in biofilms. From the available data, testable models (Figure 3) can be generated to facilitate the design of new experiments focused on the mechanisms for alteration of copy number in biofilm growth. It has yet to be determined whether this phenomenon may be found in other human pathogens and whether it might impact antibiotic susceptibility of bacterial biofilm infections clinically.
To examine plasmid maintenance theoretically, Imran et al. derived a mathematical model to determine the interplay between the cost and benefits of plasmid carriage (40). This model assumed that the plasmid in question was beneficial for biofilm formation itself which may not be the case for most plasmids but is applicable to some of the conjugative plasmids discussed above. Although this model does not provide definitive data on plasmid carriage in biofilms, it provides a good beginning framework for future work on this topic.
Non-conjugative plasmids and biofilm formation
The majority of the studies examining plasmids and their relation to biofilm development have focused on conjugative plasmids but recent research has demonstrated the influence of non-conjugative plasmids in biofilm formation as well. Teodosio et al. found that addition of small non-conjugative plasmids pET28 and pUC8 to E. coli increased the concentration of cells growing in a biofilm compared to non-transformed cells (41). Additionally, horizontal transfer of non-conjugative plasmids and genetic competence have also been demonstrated to occur in E. coli systems (42-44). Using DNase I to degrade DNA in F- E. coli biofilms, it was demonstrated that some horizontal transfer (likely through transformation) occurred in biofilm cells at a higher level than in planktonically growing cells (45).
Conclusion
The role of plasmids and plasmid copy number in bacterial biofilm populations is likely much more complex than what is observed in homogenous batch cultures. Plasmid carriage in biofilms is probably highly heterogenous and may allow for the development of subgroups within the biofilm population that are specifically poised to react to different environments and stimuli. Development of improved methods for quantitative determination of plasmid copy number in individual cells, as well as improved visualization of individual conjugation events in real time will be necessary for a full understanding of this topic. Recently, improved fluorescent reporter proteins, and highly sensitive and specific reagents for fluorescent staining were used to demonstrate localization of the competence machinery and visualize individual genetic transformation events in Streptococcus pneumoniae. (46). Similar approaches hold great promise for analysis of plasmid copy number and conjugation in enterococci and staphylococci growing in biofilms.
The research summarized in this review detail the importance of plasmids in the development of biofilms and that both single and multi-species biofilms are important ecological niches for HGT of plasmids by conjugation. Recent studies have also revealed that the physiological changes associated with the transition from planktonic to biofilm growth can impact the control of plasmid copy number, antibiotic resistance and conjugative transfer (34, 35). A better understanding of these processes will be essential for development of improved methods to prevent and control biofilm related infections by resistant organisms as well as for manipulation of biofilms to enhance the use of bacteria for biotechnological applications.
Acknowledgments
Our biofilm and plasmid research was supported by NIH grants 1RO1AI58134 and 1RO1GM49530. LCC was a predoctoral trainee under T32GM008347 from NIGMS (2007-2009), and a fellowship from the American Academy of University Women (2010-2011). We thank Tim Leonard for making figures 2 & 3.
References
- 1.Barnes AM, Ballering KS, Leibman RS, Wells CL, Dunny GM. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. MBio. 2012;3:e00193–00112. doi: 10.1128/mBio.00193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 4.Flemming HC, Wingender J. The biofilm matrix. Nature Reviews Microbiology. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 5.Reisner A, Haagensen JA, Schembri MA, Zechner EL, Molin S. Development and maturation of Escherichia coli K-12 biofilms. Mol Microbiol. 2003;48:933–946. doi: 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- 6.Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Molecular Microbiology. 30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 7.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 9.Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Molecular Microbiology. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aspiras MB, Ellen RP, Cvitkovitch DG. ComX activity of Streptococcus mutans growing in biofilms. FEMS Microbiol Lett. 2004;238:167–174. doi: 10.1016/j.femsle.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 11.Li T, Lau P, Lee J, Ellen R, Cvitkovitch D. Natural genetic transformation of Streptococcus mutans growing in biofilms. Journal of Bacteriology. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li YH, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol. 2002;184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton RE, Jacques NA. Deletion of competence-induced genes over-expressed in biofilms caused transformation deficiencies in Streptococcus mutans. Mol Oral Microbiol. 2010;25:406–417. doi: 10.1111/j.2041-1014.2010.00589.x. [DOI] [PubMed] [Google Scholar]
- 14.Burmolle M, Bahl MI, Jensen LB, Sorensen SJ, Hansen LH. Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology. 2008;154:187–195. doi: 10.1099/mic.0.2007/010454-0. [DOI] [PubMed] [Google Scholar]
- 15.Ghigo J. Natural conjugative plasmids induce bacterial biofilm development. Nature. 2001;412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 16.D'Alvise PW, Sjoholm OR, Yankelevich T, Jin Y, Weurtz S, Smets BF. TOL plasmid carriage enhances biofilm formation and increases extracellular DNA content in Pseudomonas putida KT2440. FEMS Microbiology Letters. 2010;312:84–92. doi: 10.1111/j.1574-6968.2010.02105.x. [DOI] [PubMed] [Google Scholar]
- 17.Ong CL, Beatson SA, McEwan AG, Schembri MA. Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Appl Environ Microbiol. 2009;75:6783–6791. doi: 10.1128/AEM.00974-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induced enhanced stabilisation of the biofilm structure. Current Opinion in Biotechnology. 2003;14:255–261. doi: 10.1016/s0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 19.Yang X, Ma Q, Wood TK. The R1 Conjugative Plasmid Increases Escherichia coli Biofilm Formation through an Envelope Stress Response▽†. Appl Environ Microbiol. 2008;74:2690–2699. doi: 10.1128/AEM.02809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reisner A, Holler BM, Molin S, Zechner EL. Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. J Bacteriol. 2006;188:3582–3588. doi: 10.1128/JB.188.10.3582-3588.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.May T, Okabe S. Escherichia coli harboring a natural IncF conjugative F plasmid develops complex mature biofilms by stimulating synthesis of colanic acid and Curli. J Bacteriol. 2008;190:7479–7490. doi: 10.1128/JB.00823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castonguay MH, van der Schaaf S, Koester W, Krooneman J, van der Meer W, Harmsen H, Landini P. Biofilm formation by Escherichia coli is stimulated by synergistic interactions and co-adhesion mechanisms with adherence-proficient bacteria. Res Microbiol. 2006;157:471–478. doi: 10.1016/j.resmic.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Bahl MI, Hansen LH, Goesmann A, Sorensen SJ. The multiple antibiotic resistance IncP-1 plasmid pKJK5 isolated from a soil environment is phylogenetically divergent from members of the previously established alpha, beta and delta sub-groups. Plasmid. 2007;58:31–43. doi: 10.1016/j.plasmid.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Roder HL, Hansen LH, Sorensen SJ, Burmolle M. The impact of the conjugative IncP-1 plasmid pKJK5 on multispecies biofilm formation is dependent on the plasmid host. FEMS Microbiol Lett. 2013 doi: 10.1111/1574-6968.12175. epub. [DOI] [PubMed] [Google Scholar]
- 25.Hausner M, Wuertz S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Applied and Environmental Microbiology. 1999;65:3710–3713. doi: 10.1128/aem.65.8.3710-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. Studying plasmid horizontal transfer in situ: a critical review. Nat Rev Microbiol. 2005;3:700–710. doi: 10.1038/nrmicro1232. [DOI] [PubMed] [Google Scholar]
- 27.Savage VJ, Chopra I, O'Neill AJ. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob Agents Chemother. 2013;57:1968–1970. doi: 10.1128/AAC.02008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keen NT, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 29.Licht TR, Christensen BB, Krogfelt KA, Molin S. Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology. 1999;145(Pt 9):2615–2622. doi: 10.1099/00221287-145-9-2615. [DOI] [PubMed] [Google Scholar]
- 30.Ehlers LJ, Bouwer EJ. Rp4 plasmid transfer among species of Pseudomonas in a biofilm reactor. Water Sci Tech. 1999;39:163–171. [Google Scholar]
- 31.Krol JE, Wojtowicz AJ, Rogers LM, Heuer H, Smalla K, Krone SM, Top EM. Invasion of E. coli biofilms by antibiotic resistance plasmids. Plasmid. 2013;70:110–119. doi: 10.1016/j.plasmid.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies D, Geesey G. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl Environ Microbiol. 1995;61:860–867. doi: 10.1128/aem.61.3.860-867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.May T, Ito A, Okabe S. Induction of Multidrug Resistance Mechanism in Escherichia coli Biofilms by Interplay between Tetracycline and Ampicillin Resistance Genes. Antimicrob Agents Chemother. 2009;53:4628–4639. doi: 10.1128/AAC.00454-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cook L, Chatterjee A, Barnes A, Yarwood J, Hu WS, Dunny G. Biofilm growth alters regulation of conjugation by a bacterial pheromone. Molecular Microbiology. 2011;81:1499–1510. doi: 10.1111/j.1365-2958.2011.07786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cook LC, Dunny GM. Effects of Biofilm Growth on Plasmid Copy Number and Expression of Antibiotic Resistance Genes in Enterococcus faecalis. Antimicrobial Agents and Chemotherapy. 2013;57:1850–1856. doi: 10.1128/AAC.02010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clewell DB. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972;110:667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Togna PA, Shuler ML, Wilson DB. Effects of Plasmid Copy Number and Runaway Plasmid Replication on Overproduction and Excretion of Beta- Lactamase from Escherichia coli. Biotechnology Progress. 1993;9:31–39. doi: 10.1021/bp00019a005. [DOI] [PubMed] [Google Scholar]
- 38.Uhlin BE, Nordström K. A runaway-replication mutant of plasmid R1drd-19: Temperature-dependent loss of copy number control. Molecular and General Genetics. 1978;165:167–179. doi: 10.1007/BF00269904. [DOI] [PubMed] [Google Scholar]
- 39.Ma H, Katzenmeyer KN, Bryers JD. Non-invasive in situ monitoring and quantification of TOL plasmid segregational loss within Pseudomonas putida biofilms. Biotechnol Bioeng. 2013 doi: 10.1002/bit.24953. epub ahead. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imran M, Jones D, Smith H. Biofilms and the plasmid maintenance question. Mathematical Biosciences. 2005;193:183–204. doi: 10.1016/j.mbs.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Teodosio JS, Simoes M, Mergulhao FJ. The influence of nonconjugative Escherichia coli plasmids on biofilm formation and resistance. Journal of Applied Microbiology. 2012;113:373–382. doi: 10.1111/j.1365-2672.2012.05332.x. [DOI] [PubMed] [Google Scholar]
- 42.Baur B, Hanselmann K, Schlimme W, Jenni B. Genetic transformation in freshwater: Escherichia coli is able to develop natural competence. Appl Environ Microbiol. 1996;62:3673–3678. doi: 10.1128/aem.62.10.3673-3678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsen SD, Fang SS, Chen MJ, Chien JY, Lee CC, Tsen DH. Natural plasmid transformation in Escherichia coli. J Biomed Sci. 2002;9:246–252. doi: 10.1007/BF02256071. [DOI] [PubMed] [Google Scholar]
- 44.Maeda S, Sawamura A, Matsuda A. Transformation of colonial Escherichia coli on solid media. FEMS Microbiol Lett. 2004;236:61–64. doi: 10.1016/j.femsle.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 45.Maeda S, Ito M, Ando T, Ishimoto Y, Fujisawa Y, Takahashi H, Matsuda A, Sawamura A, Kato S. Horizontal transfer of nonconjugative plasmids in a colony biofilm of Escherichia coli. FEMS Microbiol Lett. 2006;255:115–120. doi: 10.1111/j.1574-6968.2005.00072.x. [DOI] [PubMed] [Google Scholar]
- 46.Berge MJ, Kamgoue A, Martin B, Polard P, Campo N, Claverys JP. Midcell Recruitment of the DNA Uptake and Virulence Nuclease, EndA, for Pneumococcal Transformation. PLoS Pathog. 2013;9:e1003596. doi: 10.1371/journal.ppat.1003596. [DOI] [PMC free article] [PubMed] [Google Scholar]


