Abstract
Bacteria and archaea have evolved sophisticated adaptive immune systems, known as CRISPR–Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) systems, which target and inactivate invading viruses and plasmids. Immunity is acquired by integrating short fragments of foreign DNA into CRISPR loci, and following transcription and processing of these loci, the CRISPR RNAs (crRNAs) guide the Cas proteins to complementary invading nucleic acid, which results in target interference. In this Review, we summarize the recent structural and biochemical insights that have been gained for the three major types of CRISPR–Cas systems, which together provide a detailed molecular understanding of the unique and conserved mechanisms of RNA-guided adaptive immunity in bacteria and archaea.
Viruses, including those that infect bacteria (known as bacteriophages) and archaea, are the most abundant biological agents on our planet1. In response to viral predation, bacteria and archaea have evolved a range of defence mechanisms, and many of these protective systems, such as restriction–modification systems (R–M systems), abortive infection and the modification of virus receptors, provide innate immunity2. However, the genomes of almost all archaea and of about one-half of the bacteria contain CRISPR–Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins)3 loci, which are responsible for adaptive immunity. The sequences and lengths of CRISPR arrays vary, but they all have a characteristic pattern of alternating repeat and spacer sequences. In addition, CRISPR arrays are usually located adjacent to the cas genes (FIG. 1).
Figure 1. Overview of the CRISPR–Cas system.
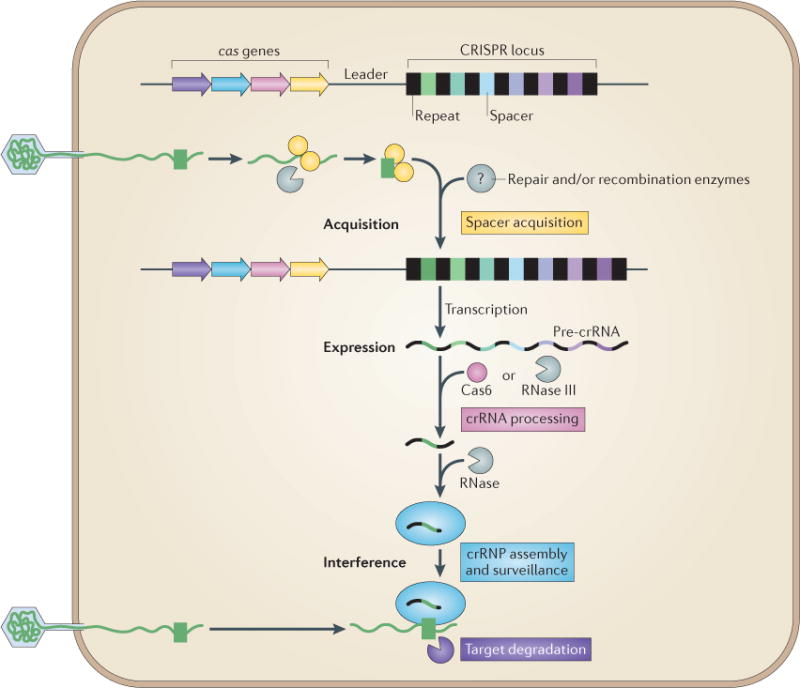
Adaptive immunity by CRISPR–Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) systems is mediated by CRISPR RNAs (crRNAs) and Cas proteins, which form multicomponent CRISPR ribonucleoprotein (crRNP) complexes. The cas genes are coloured according to function, as indicated by the four functional categories in coloured boxes: spacer acquisition (yellow); crRNA processing (pink); crRNA assembly and surveillance (blue); and target degradation (purple). Involvement of non-Cas components (grey) is indicated, either when experimentally demonstrated (for example, RNase III processing in type II systems) or when anticipated (for example, the potential involvement of housekeeping repair and/or recombination enzymes). The first stage is known as acquisition, which occurs following the entry of an invading mobile genetic element (in this case, a viral genome). The invading DNA is fragmented and a new protospacer (green) is selected, processed and integrated as a new spacer at the leader end of the CRISPR array. During the second stage, which is known as expression, the CRISPR locus is transcribed and the pre-crRNA is processed into small crRNAs by CRISPR-associated (Cas6) and/or housekeeping ribonucleases (such as RNase III). The mature crRNAs and Cas proteins assemble to form a crRNP complex. During the final stage of interference, the crRNP scans invading DNA for a complementary nucleic acid target and on successful recognition, the target is eventually degraded by Cas nucleases.
In 2005, three groups recognized that the sequences of some CRISPR spacers were identical to sequences from mobile genetic elements (MGEs), including viruses and conjugative plasmids4–6. In addition, a positive correlation was found between the possession of virus-derived spacers and resistance to the corresponding virus4,5, which suggested that CRISPR loci might participate in a nucleic acid-based immune system. This hypothesis was tested by phage-challenge experiments, which revealed that CRISPR loci acquire fragments of invading DNA and that these new spacers result in sequence-specific resistance to the corresponding phage. Moreover, it was found that the cas genes are required for this process7. Subsequent research has shown that CRISPR-mediated adaptive immunity occurs in three stages: the recruitment of new spacers (known as the acquisition stage), transcription of the CRISPR array and subsequent processing of the precursor transcript into smaller CRISPR RNAs (crRNAs) (known as the expression stage), and crRNA-directed cleavage of invading DNA by the Cas nucleases or other nucleases (known as the interference stage) (FIG. 1). In this Review, we discuss the recent mechanistic insights that have been gained from structural and functional analyses of Cas proteins and CRISPR ribo nucleoprotein (crRNP) complexes, which emphasize both conserved and unique features of adaptive immunity in bacteria and archaea.
CRISPR–Cas diversity
CRISPR–Cas systems are highly diverse, which is probably due to the rapid evolution of immune systems as a result of the dynamic selective pressures that are imposed by invading MGEs. Initial comparative analyses of CRISPR loci revealed that there are major differences in CRISPR repeat sequences8, in cas gene sequences and in the architecture of the cas operons9–11. On the basis of these differences, CRISPR–Cas systems have been classified into three main types and several subtypes12 (FIG. 2; Supplementary information S1 (table)). Each type has a specific ‘signature’ Cas protein: type I systems all contain the Cas3 nuclease–helicase, type II systems are defined by the Cas9 nuclease, and type III systems all have Cas10, which is a large protein of unknown function12 (FIG. 2; Supplementary information S1 (table)). Type I and type III systems seem to be distantly related, whereas type II systems are phylogenetically and structurally distinct13. In order to target and cleave invading nucleic acid, crRNAs and Cas proteins form crRNP complexes, the nomenclature of which is defined by their composition12. Type I-A to type I-F crRNP complexes are known as Cascade (CRISPR-associated complex for antiviral defence), whereas all crRNPs in type II systems (that is, type II-A, type II-B and type II-C systems) are known as Cas9 complexes. In addition, type III-A crRNP complexes are known as Csm complexes, whereas those that belong to type III-B systems are known as Cmr complexes.
Figure 2. Diversity of CRISPR–Cas systems.
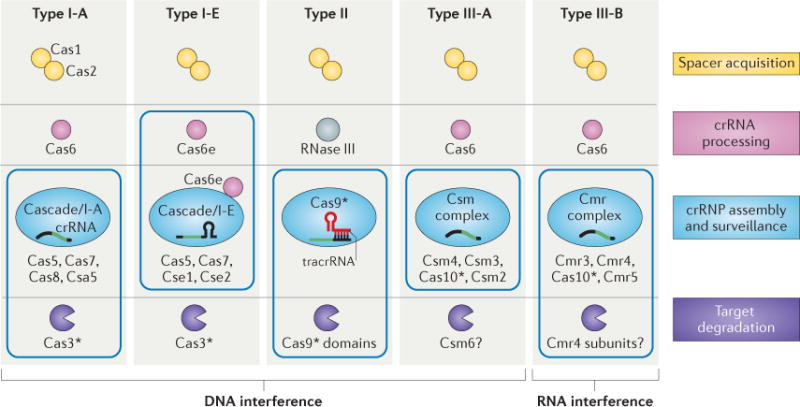
The CRISPR-associated (Cas) proteins can be divided into distinct functional categories as shown. The three types of CRISPR–Cas systems are defined on the basis of a type-specific signature Cas protein (indicated by an asterisk) and are further subdivided into subtypes. The CRISPR ribonucleoprotein (crRNP) complexes of type I and type III systems contain multiple Cas subunits, whereas the type II system contains a single Cas9 protein. Boxes indicate components of the crRNP complexes for each system. The type III-B system is unique in that it targets RNA, rather than DNA, for degradation.
Type I and type III CRISPR–Cas systems are found in various combinations among phylogenetically diverse bacteria and archaea, whereas the distinct type II systems (sometimes in combination with other CRISPR–Cas types) are restricted to bacteria10,14. Interestingly, CRISPR–Cas systems have also been found in viral genomes and plasmids15–19, which is consistent with phylogenetic studies that suggest that these systems are frequently exchanged via horizontal gene transfer11. Despite their diversity, all Cas proteins can be grouped into four functional categories (FIGS 1,2): nucleases and/or recombinases, which are involved in spacer acquisition; ribonucleases, which catalyse the processing of crRNA guides; proteins that assemble with the RNA guides to form the crRNP complexes for target surveillance; and nucleases, which are responsible for degradation of the DNA or RNA targets.
Acquisition of spacers
The acquisition of new invader-derived spacers generally proceeds in a polarized manner at the leader-end of the CRISPR locus6,7,20 (FIGS 1,3a), which results in a chronological record of previously encountered foreign nucleic acid. The most recent experimental data support the following model for the step-wise acquisition of novel spacers (FIG. 3a). The recognition and fragmentation of invading DNA is likely to be the first step in the process. A recent study reported functional synergy between an R–M system and CRISPR–Cas in Streptococcus thermophilus21, which suggests that fragments of invader DNA that are generated by the R–M system might be potential substrates for spacer acquisition. The CRISPR–Cas system selects suitable spacers by the detection of a specific protospacer adjacent motif (PAM)22–24 (BOX 1), followed by processing of the DNA substrates into spacer precursors of a defined size25. After the opening of the leader-end repeat by the nicking of both strands at opposite sides of the repeat26, the new spacer is integrated in a specific, PAM-dependent orientation25,27 (FIG. 3a). In support of this model, the leader-end repeat is duplicated during spacer acquisition28. In addition to DNA that is derived from MGEs (that is, ‘non-self’ DNA), fragments of chromosomal DNA (that is, ‘self’ DNA) are occasionally integrated as novel CRISPR spacers. However, as these self-targeting spacers are associated with cytotoxicity29, their presence in the genome is typically associated with a modified PAM or an inactivated CRISPR–Cas system30. In the absence of Cas proteins that are essential for target cleavage, the acquisition of chromosome-derived spacers has indeed been observed, but it occurs at least 100-fold less frequently than the acquisition of plasmid-derived spacers28. This suggests that CRISPR–Cas systems can distinguish invading, non-self DNA from self DNA (BOX 1) — either directly, by an unknown mechanism, or indirectly, by interacting with other defence systems (such as R–M systems).
Figure 3. CRISPR spacer acquisition.
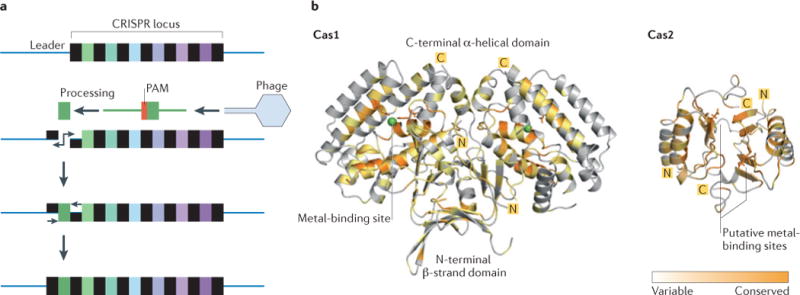
a | Proposed stages of CRISPR spacer acquisition: fragmentation of invading DNA (in this case, phage DNA), selection of the protospacer by recognition of the protospacer adjacent motif (PAM), processing of the pre-spacer, nicking of the leader-end repeat in the CRISPR locus, integration of the new spacer and duplication of the flanking repeat. Both type I and type II systems rely on PAM recognition for spacer integration, whereas the type III systems do not. b | Crystal structures of Cas1 (from Pseudomonas aeruginosa; Protein Databank (PDB) accession 3GOD)31 and Cas2 (from Desulfovibrio vulgaris, PDB accession 3OQ2)36, which are the two main endonucleases that are involved in spacer acquisition. Cas1 is a metal-dependent, dimeric endonuclease (DNase) with a unique three-dimensional fold that consists of an amino-terminal β-strand domain and a carboxy-terminal α-helical domain. Sequence conservation (indicated by colour intensity) of Cas1 shows that the metal ion-binding site is highly conserved among Cas1 family proteins. Cas2 is a metal-dependent, dimeric endonuclease (RNase and/or DNase), with a metal-binding site at the interface of the two subunits (which is composed of RAMP domains). The conservation model was generated using Consurf and the figure was made using PyMol.
Box 1. ‘Self’ versus ‘non-self’ discrimination by CRISPR–Cas systems.
All immune systems must efficiently distinguish ‘self’ from ‘non-self’ to avoid autoimmunity. In DNA-targeting CRISPR–Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) systems, the mechanism of discrimination occurs during CRISPR surveillance. The protospacer itself cannot be used for discrimination, as the crRNA spacer is also complementary to its template in the CRISPR locus on the host chromosome. Instead, in silico analyses of sequences that flank the protospacers recognized by CRISPR–Cas type I and type II systems have revealed that type-specific short sequences (of 2–3 nucleotides), which are collectively known as protospacer adjacent motifs (PAMs)22,23, are necessary for discrimination. The most important feature of the PAM is that it differs from the corresponding sequence of the CRISPR repeat24, which enables discrimination between a non-self target and a self non-target. Indeed, experimental analyses of CRISPR interference by type I (REFS 54,99,101,103) and type II (REFS 76,77) systems have confirmed an important role for the PAM motif. Moreover, studies of CRISPR adaptation in these systems22,25,28,49 indicate that the PAM is also important for spacer acquisition. This makes sense, as only functional protospacers (that is, those that provide immunity) are selected for integration into the CRISPR array.
Type I and type II systems use a ‘non-self activation’ strategy that involves protein-mediated detection of a PAM that is located adjacent to the targeted protospacers in the invading DNA. This eventually results in the ‘switching on’ of interference, most probably by a conformational change that triggers either the recruitment of a nuclease to the crRNP complex (for example, Cas3 in type I systems) or the induction of intrinsic crRNP nuclease activity (for example, Cas9 in type II systems)84,108,125. In type I systems, PAMs are located downstream (at the 3′ end) of the protospacer on the target strand, whereas PAMs of type II systems are located upstream (at the 5′ end) of the protospacer22,23,126. Recognition of PAMs may occur in a single-stranded conformation, which either exclusively involves the strand that base pairs with the crRNA (in type I systems)84,103 or the displaced strand (in type II systems)76.
Type III systems seem to lack the PAM-based system; instead, the type III-A system uses a ‘self inactivation’ strategy that involves base pairing between the 5′ handle of the crRNA (as part of the Csm complex) and the repeat sequence in the CRISPR locus on the host chromosome. Base paring in this region of the crRNA signals binding to the chromosomal CRISPR array (self DNA), which seems to trigger the ‘switching off’ of the interference process, possibly by preventing the recruitment of the nuclease111.
The involvement of Cas1 and Cas2
The strict conservation of Cas1 and Cas2 in all CRISPR–Cas systems12, together with the finding that Cas1 and Cas2 are required for the integration of new spacers28, suggests that the basic mechanism of CRISPR adaptation is conserved (FIG. 3). Although the simultaneous expression of both Cas1 and Cas2 enables spacer acquisition28, their precise functions in the adaptation process remain elusive. Cas1 is a metal-dependent endonuclease that catalyses the cleavage of double-stranded DNA (dsDNA), single-stranded DNA (ssDNA) and branched DNA in a sequence-independent manner31,32. Crystal structures of the homodimeric Cas1 protein have shown that it consists of an amino-terminal β-strand domain and a carboxy-terminal α-helical domain31,32 (FIG. 3b). The C-terminal domain contains a conserved binding site for a divalent metal ion31, which is crucial for DNA degradation in vitro and spacer acquisition in vivo28. The metal-binding site is surrounded by a cluster of basic residues that form a positively charged strip across the surface of the C-terminal domain. This surface has been implicated in DNA binding and might be involved in the positioning of substrates close to the metal ion in the active site31,33.
Cas2 is a metal-dependent nuclease that contains a RAMP-like fold34 with a typical β1α1β2β3α2β4 arrangement, in which the two α-helices are positioned together on one face of a four-stranded antiparallel β-sheet35–37 (FIG. 3b). The β-sheets from two Cas2 protomers form a β-sandwich, and conserved amino acids are positioned along the dimer interface. The substitution of a conserved aspartic acid residue in each protomer, located at the dimer interface, does not affect their assembly (FIG. 3b), but it perturbs the binding of a metal ion and disrupts nuclease activity37. Although several studies have reported that Cas2 proteins are endoribonucleases35, other Cas2 proteins mainly catalyse the cleavage of dsDNA, which indicates that they are deoxyribonucleases37. Differences in the loop regions might explain differences in substrate preference; for example, Cas2 proteins that have a long loop connecting α2 to β4 have a relatively narrow substrate-binding cleft and correspond to ribonucleases. By contrast, Cas2 proteins that have long β1–α1 loops contain wider substrate-binding clefts and show deoxyribonuclease activity37. A recent study128 has revealed that Cas1 and Cas2 from E. coli form a stable complex that interacts with the CRISPR locus. The data show that an intact Cas1–Cas2 complex is essential for spacer acquisition in vivo. Importantly, although Cas1 activity is required for protospacer processing and/or spacer integration, Cas2 activity is not needed for spacer acquisition.
Other factors involved in spacer acquisition
In addition to the participation of Cas1 and Cas2, there are indications that a variable set of accessory factors might be involved in spacer acquisition. Pulldown assays have shown that Cas1 of Escherichia coli interacts with RecBCD and RuvB, which are housekeeping proteins that are involved in general DNA repair and recombination32. Moreover, several cases of gene fusion and conserved gene clustering12,38 suggest that CRISPR acquisition might require additional Cas proteins, such as Csn2, Cas4, Csa1 and Cas3 (Supplementary information S1 (table)). Attempts have been made to verify the putative roles of some of these proteins in CRISPR adaptation, as discussed below.
Csn2 is encoded by all type II-A systems and has been shown to be involved in CRISPR adaptation in Streptococcus thermophilus7. Several structural studies have revealed that Csn2 forms a tetrameric ring-shaped complex with a positively charged central cavity that binds to, and slides along, DNA fragments39–43. The apparent lack of Csn2 catalytic activity suggests that it might have an accessory role during spacer acquisition (such as stabilizing the double-strand break during spacer integration) or that it might be involved in the recruitment of additional factors39.
Cas4 and Csa1 share amino acid sequence similarity with RecB- and AddB-type nuclease–helicases11,44. The Cas4 protein of Sulfolobus solfataricus is a ring-shaped decamer that has DNA-targeting 5′ to 3′ exonuclease activity44,45. In addition, some Cas4 homologues have been reported to have endonuclease activity as well as helicase activity44,45. Fusions of Cas4 and Cas1 occur in several bacterial and archaeal type I and type III systems, which indicates that the two proteins are functionally related12,38. Cas4 from Thermoproteus tenax has been shown to form a complex in vitro with a Cas1–Cas2 fusion protein and Csa1 (REF. 46). However, such complexes have not yet been isolated from a natural system, which may indicate that the proteins interact only transiently in vivo. Furthermore, it is likely that fusion proteins (such as Cas4–Cas1 and Cas1–Cas2) might contribute to stabilizing these complexes11,46.
Cas3 is a multidomain nuclease–helicase that is fused to Cas2 in type I-F systems47 (Supplementary information S1 (table)). In the type I-F system of Pectobacterium atrosepticum, a direct interaction between Cas1 and the Cas2–Cas3 fusion protein has been observed, which suggests that Cas3 has a dual role, functioning during CRISPR interference as well as during spacer acquisition48. The proposed role for Cas3 during both acquisition and interference might be related to a phenomenon that is known as ‘primed spacer acquisition’ (REFS 25,49). Priming refers to the positive-feedback loop that accelerates the acquisition of new spacers from previously encountered genetic elements50. In the type I-E system, this process requires Cas1, Cas2, Cas3 and an RNP complex that is composed of crRNA and multiple Cas proteins (that is, Cascade), which suggests that many proteins participate in this process. However, the mechanism of primed spacer acquisition is currently unknown.
Processing of crRNA guides
Transcription of the CRISPR array generates a long precursor transcript (known as a pre-crRNA) (FIG. 4a). Primary processing of the pre-crRNA involves endoribo-nucleolytic cleavage within the repeat sequences, either by Cas6 homologues (FIG. 4b) or by RNase III (FIG. 4c).
Figure 4. Biogenesis of crRNAs.
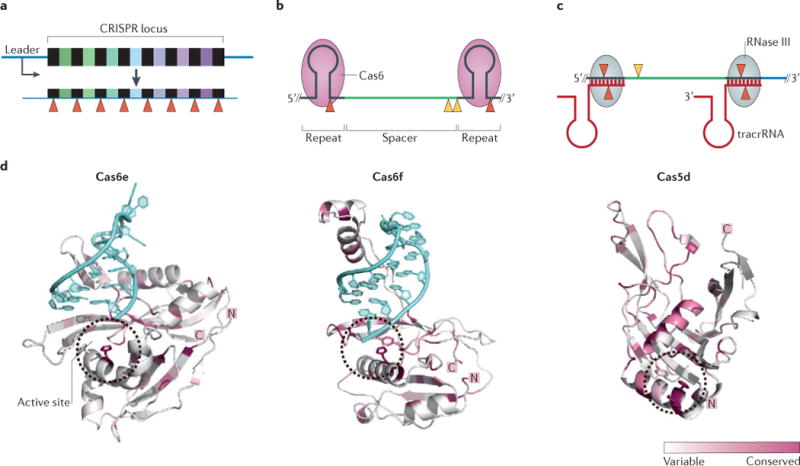
a | The CRISPR array is transcribed to produce a pre-CRISPR RNA (pre-crRNA) transcript, the primary processing of which occurs by cleavage (red triangles) within the repeat sequences, producing crRNAs in which spacers are flanked by repeat-derived handles. b | Generation of CRISPR guide RNAs in type I and type III CRISPR–Cas systems. Primary processing of the pre-crRNA is catalysed by Cas6, which typically results in a crRNA with a 5′ handle of 8 nucleotides, a central spacer sequence and a longer 3′ handle. In some subtypes, the 3′ handle forms a stem–loop structure, in other systems, secondary processing of the 3′ end of crRNA (yellow triangles) is catalysed by unknown ribonucleases. c | In type II CRISPR–Cas systems, the repeat sequences of the pre-crRNA hybridize with complementary sequences of transactivating CRISPR RNA (tracrRNA). The double-stranded RNA is cleaved by RNase III (red triangles) and further trimming of the 5′ end of the spacer is carried out by additional nucleases (yellow triangle). d | Crystal structures of CRISPR-associated ribonucleases that catalyse primary processing of pre-crRNA. Cas6e (from the type I-E system; Protein Databank (PDB) accession 4C9D) and Cas6f (from the type I-F system; PDB accession 4AL7) are shown complexed to the hairpin of the crRNA (blue). In type I-C systems, a Cas5 variant (known as Cas5d) substitutes for Cas6 and is involved in pre-crRNA processing (PDB accession 4F3M). For all three structures, the location of the active site (which contains a catalytic histidine residue) is indicated with a circle. Sequence conservation is indicated by colour intensity. The conservation model was generated using Consurf and the figure was made using PyMol.
Type I and type III systems
In type I and type III systems, Cas6-like nucleases are responsible for the primary processing of the pre-crRNA (FIG. 4b), which has been extensively reviewed elsewhere34. Although Cas6 homologues generally consist of two RAMP domains, a considerable level of structural variation has been described34,51.
Despite the structural variability, Cas6 enzymes specifically cleave the pre-crRNA by hydrolysing a single phosphodiester bond in the repeat sequences of the transcript. This typically results in crRNAs that have a repeat-derived 5′ handle of 8 nucleotides, followed by the complete spacer sequence and a repeat-derived 3′ handle of variable size that forms a hairpin structure in some systems (FIG. 4b). Cas6 variants are metal-independent endoribonucleases that generate crRNAs that have a 5′ hydroxyl group and either a 3′ phosphate or a cyclic 2′–3′ phosphate52–55. This suggests that Cas6 has a general acid–base catalytic mechanism, in which the deprotonated hydroxyl at the 2′ position of the ribose functions as a nucleophile. The catalytic sites of all characterized Cas6-like enzymes are composed of an invariant histidine residue, a tyrosine or serine residue and, in some cases, a lysine residue. However, the relative positions of these residues are poorly conserved (FIG. 4d), which might explain the observed functional variations in Cas6 activity (see below)33,56,57.
In the type I-E and type I-F systems, the Cas6 proteins (which are known as Cas6e and Cas6f, respectively) have a high affinity for the cleaved crRNA product, which results in single-turnover reaction kinetics51. This is consistent with the observation that Cas6e and Cas6f are core components of Cascade complexes, in which they remain firmly associated with the hairpin at the 3′ handle of the crRNA51,55,56,58–65 (FIG. 4d). By contrast, the Cas6 variants of other type I systems and all type III systems function as stand-alone nucleases that deliver primary crRNAs to the respective crRNP complexes55,66 (FIG. 1). After transfer of the crRNAs to these complexes, the 3′ handles are accessible for nucleolytic trimming67. This secondary processing seems to result in short 3′ handles in type I-A, type I-B, type I-C and type I-D systems. In type III systems, differential trimming typically results in the production of two mature crRNA species that differ by 6 nucleotides68–70 (FIG. 4b).
In type I-C systems, Cas6 is substituted by a Cas5 variant (known as Cas5d; FIG. 4d) that cleaves the pre-crRNA to form the mature crRNA71–73. The resulting crRNA contains an 11 nucleotide 5′ handle (rather than the 8 nucleotide handle that is generated by Cas6) and a 21–26 nucleotide 3′ handle. Similarly to Cas6, Cas5d is a RAMP protein with an active site that is composed of a catalytic triad (containing tyrosine, lysine and histidine), and like Cas6e and Cas6f, Cas5d remains associated with its crRNA product and assembles with other Cas proteins to form the multisubunit Cascade complex72. In all other type I systems, catalytically inactive Cas5 homologues are a subunit of Cascade and have been proposed to interact with the 5′ handle of the crRNA74 (see below).
Type II systems
In type II systems, processing of pre-crRNAs relies on a completely different mechanism (FIG. 4c). In addition to a cas operon and a CRISPR array, the CRISPR locus of these systems includes a gene that encodes a transactivating crRNA (tracrRNA)75. The tracrRN A contains a 25 nucleotide sequence that is complementary to the repeat region of the pre-crRNA transcript. Base pairing between these two RNAs results in a double-stranded region that is recognized and cleaved by the housekeeping ribonuclease RNase III (FIG. 4c). Cas9 is required for primary crRNA processing75,76, most probably for binding and positioning the RNA molecules for cleavage by RNase III (REF. 14). After initial processing by RNase III, the crRNA–tracrRNA hybrid remains firmly associated with Cas9 (REFS 76,77). The 5′ end of the crRNA spacer (which is 24–27 nucleotides in length) is trimmed by an unknown nuclease, which typically results in a spacer that is 20 nucleotides long75. The mature crRNA–tracrRNA hybrid is required for target interference76 — probably for proper anchoring and positioning of the crRNA in Cas9 — in a way that might be analogous to the binding of the crRNA hairpin in Cascade. In addition, recently obtained crystal structures of Cas9 reveal that major domain rearrangements occur following the binding of target nucleic acids (see below).
Assembly of crRNP complexes
Mature crRNAs associate with Cas proteins to form stable crRNP complexes. Type I systems form multisubunit surveillance complexes that are called Cascade (FIG. 2). Although the proteins associated with the type III crRNPs are phylogenetically distinct from those in the type I system, recent structural studies have shown that there are striking architectural similarities between the type III and type I crRNPs69,78,79. By contrast, the type II crRNP complex (in which Cas9 is the only protein component) is fundamentally different76,77,80,81 (FIG. 2; Supplementary information S1 (table)).
Type I crRNP complexes
The first crRNP complex to be identified was the Cascade complex of the E. coli type I-E system. Owing to striking structural similarities, the crRNP complexes of other type I variants are generally referred to as Cascade34,82. The type I-E crRNP (Cascade/I-E) from E. coli is composed of a core complex (Cas5, Cas6, Cas7 and a single 61 nucleotide crRNA) and two less tightly associated subunits (Cse1 and Cse2)52,83,84 (FIG. 2). The complete complex has an uneven subunit stoichiometry: (Cse1)1–(Cse2)2–(Cas5)1–(Cas7)6–(Cas6)1 (REFS 52,85) (FIG. 5a), which is a typical feature of all type I and type III crRNP complexes and is controlled by differential translation of the encoding polycistronic mRNA86. The overall architectures of Cascade complexes of type I-C72 and type I-F systems53 share a helical backbone structure that is composed of Cas7 (known as Csy3 in type I-F systems), Cas8 (known as Csy1 in type I-F systems), Cas5 (known as Csy2 in type I-F systems) and a crRNA (FIG. 5a). Interestingly, in vitro assembly of Cascade/I-A has shown that, in addition to Cas7, Cas8, Cas5 and Csa5 (known as the small subunit; Supplementary information S1 (table)), the truncated domains of Cas3 (the helicase domain, which is known as Cas3′; and the nuclease domain, which is known as Cas3″) form part of the complex46 (FIG. 5a). Thus, despite many similarities, the occurrence of structural differences suggests that there are minor functional variations.
Figure 5. Architecture of crRNP complexes.
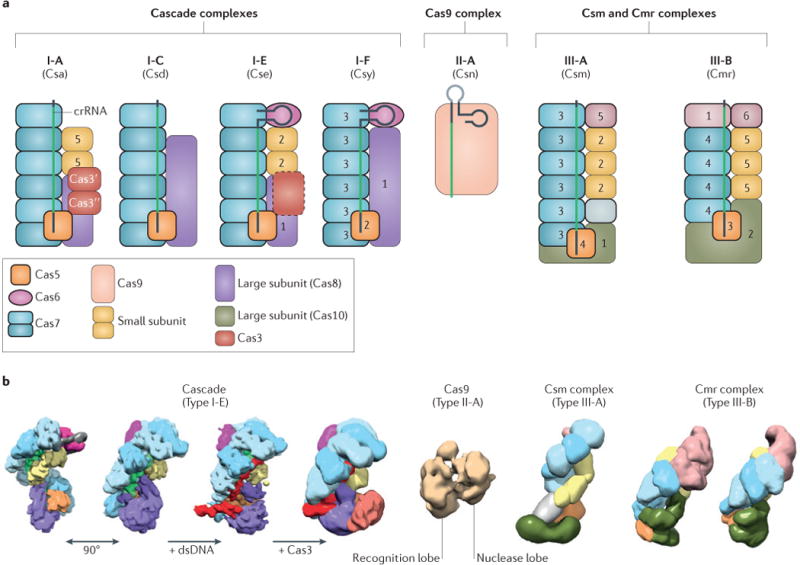
a | Schematic representation of the subunit composition of different CRISPR ribonucleoprotein (crRNP) complexes from all three CRISPR–Cas types. The colours indicate homology with conserved Cas proteins or defined components of the complexes, as shown in the key. The numbers refer to protein names that are typically used for individual subunits of each subtype (for example, subunit 5 of the type I-A (Csa) complex refers to Csa5, whereas subunit 2 of the type I-E (Cse) complex refers to Cse2, and so on). The CRISPR RNA (crRNA) is shown, including the spacer (green) and the flanking repeats (grey). Truncated Cas3 domains (Cas3′ and Cas3″) have been suggested to be part of the type I-A complex127, and fusions of Cas3 with Cascade subunits (for example, with Cse1 (REF. 103)) have been found in some type I-E systems (shown as a dashed Cas3 homologue). Cas9 is depicted in complex with single-guide RNA (sgRNA), with an artificial linker (light grey) between the crRNA and the tracrRNA. Subunits with a RAMP (that is, an RNA-recognition motif (RRM)) fold are shown with a bold outline. The grey subunit in the type III-A Csm complex has been proposed to be a Cas7 homologue78. b | Structural comparison of crRNP complexes (colours as in part a): cryo- electron microscopy (cryo-EM) structures of Escherichia coli Cascade/I-E bound to a crRNA (two views after 90 ° rotation; Electron Microscopy Data Bank (EMDB) accession 5314; 8.8 Å)74, with additional double-stranded DNA (dsDNA) target (9 Å)89 and with additional Cas3 (20 Å)89. Cryo-EM structure of Streptococcus pyogenes Cas9 (of the type II-A system) bound to a single-guide RNA (sgRNA; not shown) and a 20 nucleotide target single-stranded DNA (ssDNA; not shown) (EMDB accession 5860; 21 Å), revealing a recognition lobe and a nuclease lobe, with a cleft in which the crRNA–DNA hybrid is located (see crystal structure; Supplementary information S2 (figure)). Cryo-EM structure of type III crRNP complexes: Sulfolobus solfataricus Csm complex (EMDB accession 2420; 30 Å)78, and Cmr complexes from Pyrococcus furiosus (EMDB accession 5740; 12 Å)79 and Thermus thermophilus69.
A major advance in our understanding of crRNA-guided surveillance came from two cryo-electron microscopy (cryo-EM) structures (of 8–9 Å resolution) of E. coli Cascade/I-E74 (FIG. 5b). These structures revealed an overall seahorse-shaped architecture in which the 3′ and 5′ handles of the crRNA are anchored at opposite ends of the complex and the 32 nucleotide spacer sequence is displayed along the helical backbone, which is composed of six Cas7 proteins. This is a typical feature of Cascade complexes, in which a string of Cas7 subunits provides a backbone that has an elongated binding cleft for the crRNA guide (FIG. 5a). Cas7 of Cascade/I-A has a crescent-shaped structure that contains a central RAMP domain87. Although the Cas7 RAMP domain resembles a typical RNA-recognition motif (RRM), it seems to lack some of the conserved aromatic residues that are responsible for RNA binding by canonical RRMs87,88. By mapping the highly conserved residues onto the three-dimensional structure, two conserved clusters were identified on the concave surface of the Cas7 structure87 that is involved in binding to the crRNA74,89.
Type II crRNP complexes
The recently established high-resolution structures of Cas9 have been a major break-through in the field (REFS 80,81). Crystal structures have been obtained of two Cas9 proteins (from the type II-A system of Streptococcus pyogenes and the II-C system of Actinomyces naeslundii) in the absence of nucleic acids80. In a second study, the structure of Cas9 from a type II-A system (from S. pyogenes) was solved, with a single-guide RNA (sgRNA; which is an engineered functional fusion of crRNA and tracrRNA76) hybridized to a 20 nucleotide DNA target81 (Supplementary information S2 (figure)). These structures show that Cas9 has a conserved architecture that consists of two distinct lobes: the α-helical recognition lobe, which is primarily involved in coordinating the guide RNA, and the nuclease lobe, which is responsible for PAM recognition and subsequent cleavage of the target DNA (see below) (Supplementary information S2 (figure)). In the apo state, the lobes are oriented in a position that would preclude the binding and cleavage of target DNA80. Complementary single-particle electron microscopy reconstructions show that the two structural lobes undergo a reorientation following binding of the sgRNA, which results in the formation of a central channel that allows for the binding of DNA substrates80 (FIG. 5b). This is in agreement with the structure of Cas9–sgRNA with a single-stranded DNA target, in which the RNA–DNA heteroduplex is located in the positively charged groove at the interface of the two lobes81 (FIG. 5b; Supplementary information S2 (figure)). The observation that the catalytic sites of the two nuclease domains — HNH and RuvC — are not positioned properly for cleavage81 may suggest that an R-loop configuration is required to reach the ultimate cleavage-competent state of Cas9.
Type III crRNP complexes, and similarities with type I systems
Structures of the type III-A crRNP complex from S. solfataricus (known as the Csm complex78; FIG. 5b) and two type III-B crRNP complexes from Pyrococcus furiosus and Thermus thermophilus (known as Cmr complexes; FIG. 5b) were recently determined by electron microscopy69,79. Type III complexes have a multicopy backbone (which is composed of Csm3 in type III-A systems and Cmr4 in type III-B systems; FIG. 5a, b) that is morphologically similar to the Cas7 backbone of type I Cascade complexes (FIG. 5a, b). Indeed, the crystal structure of Csm3 has revealed that it is a structural homologue of Cas7 (REF. 90) (FIG. 5a, b). Moreover, cryo-EM structures of the Cmr complex from P. furiosus have shown that the crRNA is positioned along the backbone, similarly to the crRNA in Cascade79.
To denote potential structural and/or functional similarities between components of type I and type III crRNP complexes, the terms ‘large subunits’ and ‘small subunits’ have been introduced91. In most type I systems, the large subunits are Cas8 homologues (such as Cas8a, Cas8b, Cas8c, Cse1 and Csy1), whereas in type III systems, the large subunits are Cas10 homologues (such as Csm1 and Cmr2) (Supplementary information S1 (table)). The large subunit in type I Cascade complexes is positioned adjacent to Cas5, close to the 5′ handle of the crRNA (FIG. 5a, b). Similarly, native mass spectrometry of a type III-A Csm complex revealed that the large subunit Csm1 (which is a Cas10 homologue) interacts with Csm4 (which is a Cas5 homologue)78 (FIG. 5a, b). In addition, evidence for a similar interaction was obtained from a crystal structure of a partial Cmr complex that consisted of Cmr2 (a Cas10 homologue) and Cmr3 (a Cas5 homologue)92,93 (FIG. 5a, b). Comparison of the crystal structures of the type I-E subunit Cse1 (REFS 84,94) (which is a Cas8 homologue) and the type III-B subunit Cmr2 (REFS 95,96) (which is a Cas10 homologue) showed that these two proteins do not share obvious structural similarity34. However, the conserved position of the large subunits in type I and type III crRNP complexes, as well as the apparent substitution of Cas8 by Cas10 in type I-D crRNP complexes (Supplementary information S1 (table)), suggests that they have analogous roles.
The small subunits are Csa5 and Cse2 in type I systems and Csm2 and Cmr5 in type III systems (FIG. 5a; Supplementary information S1 (table)). The absence of genes that encode small subunits in certain cas operons (for example, types I-B, I-C, I-D and I-F) has been proposed to be compensated for by extensions of the large subunits91,97. A structural comparison of small subunits has shown that there is structural conservation between Cmr5 (from the type III-B Cmr complex) and the N-terminal domain of Cse2 (from Cascade/I-E), and between Csa5 (from Cascade/I-A) and the C-terminal domain of Cse2 (from Cascade/I-E)97. In Cascade/I-E, the Cse2 dimer constitutes a protein bridge that connects Cas6 in the head of the complex to Cse1 in the tail of the complex (FIG. 5a, b). Moreover, electron microscopy structures of Csm and Cmr complexes have shown that the small subunits constitute a second helical string of subunits, which run parallel to the Cas7 backbone and form a solid bridge that connects the ‘bottom’ (large subunit and the Cas5-like proteins Csm4 or Cmr3) to the head (Csm5 or Cmr1 and Cmr6) in all type I and type III complexes. The set of small subunits in type III complexes consists of three copies of Csm2 in the type III-A complex78 and three copies of Cmr5 in the type III-B complexes69,79 (FIG. 5a).
Target surveillance and interference
The targeting of invading MGEs by the different crRNP complexes seems to proceed in a stepwise manner33,82,84,98,99 (FIG. 6). Finding a protospacer sequence that is complementary to the crRNA involves scanning of the invader DNA, discriminating self from non-self (BOX 1) and base pairing between the 7–8 nucleotide seed region of the spacer and the complementary protospacer, followed by extended base pairing between the spacer and protospacer, which eventually results in complete strand displacement82,84,100. Hybridization of crRNA to the target strand generates an R-loop structure, which — at least in some cases — has been shown to trigger a conformational change in the crRNP complex52,74,79,89. This structural transition may function as a signal that recruits a trans-acting nuclease (for type I and type III-A systems) or lead to the activation of intrinsic nuclease activity (for type II and type III-B systems) (FIG. 6).
Figure 6. Surveillance and interference by crRNP complexes.
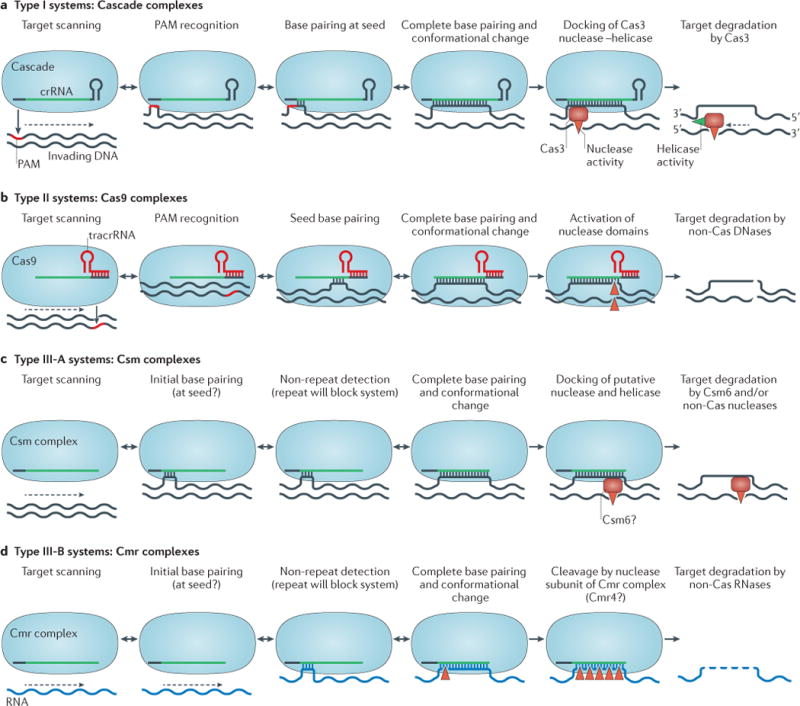
Proposed mechanisms of targeting for the three different types of CRISPR–Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) systems. a | In type I systems, the Cascade complex searches for a complementary protospacer in the invader DNA via target scanning. The large subunit (Cse1 or Cas8) of the complex recognizes the protospacer adjacent motif (PAM) sequence by a ‘non-self activation’ strategy (BOX 1), which is followed by hybridization between the seed sequence and the target DNA. If these initial criteria are met, complete base pairing results in R-loop formation and a simultaneous conformational change in the Cascade complex, which probably triggers Cas3 recruitment and subsequent degradation of the displaced target DNA strand (red triangles indicate endonucleolytic cleavage). The dashed arrow indicates processivity by the concerted helicase (green triangle) and exonuclease activities in the 3′ to 5′ direction. b | In type II systems, the Cas9 complex, bound to the CRISPR RNA (crRNA)–transactivating crRNA (tracrRNA) duplex, follows a similar mechanism of PAM-dependent recognition of invading DNA. However, unlike type I systems, the PAM is located upstream (at the 5′ end) of the protospacer and both target DNA strands are cleaved by Cas9-mediated nuclease activity. c | In type III-A systems, the crRNA-bound Csm complex targets DNA in a PAM-independent process, using a ‘self inactivation’ strategy (BOX 1). The stand-alone nuclease that is responsible for DNA degradation has been proposed to be Csm6 (also known as Csx1) (FIG. 2; Supplementary information S1 (table)). d | In type III-B systems, the crRNA-guided Cmr complex targets invading RNA in a PAM-independent process. After recognition and hybridization of crRNA and a complementary target RNA sequence, cleavage of this target occurs at multiple sites (red triangles). The nuclease that is responsible for RNA degradation has been proposed to be a subunit of the Cmr complex (Cmr4; Supplementary information S1 (table)). With the exception of type I systems, in which Cas3 mediates target degradation (part a), all other systems (parts b–d), are thought to involve non-Cas nucleases for complete target degradation.
Type I systems
Scanning of invader DNA seems to be strongly dependent on nonspecific interactions between the crRNP complex and the invading DNA. In type I-E systems, the Cse1 subunit of Cascade is important for nonspecific association with DNA and also has a key role in preventing autoimmunity (the targeting of chromosomal sequences)52,84,101. Structural and biochemical studies suggest that a flexible loop in Cse1 interacts with the 3 nucleotide PAM motif84,94. The PAM is an antigenic signature, and PAM recognition by Cse1 might destabilize the DNA duplex, thereby enabling the crRNA to access the target DNA for hybridization84. For successful interference, base pairing between the seed region of the crRNA spacer and the complementary target protospacer is essential at positions closest to the PAM. In Cascade/I-E, the seed region of the crRNA includes nucleotides 1–5 and 7–8 at the 5′ boundary of the spacer99. Mutated targets, in which base pairing within this seed region is imperfect, generally escape detection by Cascade99,102. In the case of PAM recognition and successful seed base pairing, crRNA-guided strand invasion of the dsDNA proceeds in an ATP-independent manner, which generates an R-loop that might be stabilized by the positively charged surface of the two small subunits (Cse2)74,98.
The formation of a complete R-loop coincides with a major conformational change of the Cascade/I-E complex52,74 and local bending of the target DNA98,103. In type I systems, these structural changes seem to trigger the recruitment of the Cas3 nuclease–helicase (REF. 103). Single-particle EM reconstructions of dsDNA-bound Cascade have recently shown that Cascade positions the kinked DNA duplex in such a way that the PAM motif contacts the Cse1 subunit89. Interestingly, this study showed that docking of Cas3 on Cascade occurs at a site that is provided by Cse1, in close proximity to Cas5 (FIG. 5b). Although the observed density for Cas3 in this reconstruction only corresponds to part of Cas3 (REF. 89), its binding site on Cascade is consistent with the occurrence of some natural Cas3 fusions, such as Cas3–Cse1 in type I-E systems103 and Cas5–Cas3 in type I-B systems (J.v.d.O, unpublished observations).
Cas3 consists of a HD-nuclease domain104,105 (Supplementary information S3 (figure)) fused to an SF2-helicase domain (superfamily 2 helicase domain)47. In Cascade/I-E, it has been shown that the aforementioned loop in Cse1 is not only involved in PAM recognition but is also required for Cascade-associated Cas3 nuclease activity89. The ATP-dependent helicase activity of Cas3, combined with its metal-dependent nuclease activity, mediates complete degradation of the target DNA54,103. After initial endonucleolytic cleavage of the displaced strand of the R-loop54, exonucleolytic degradation proceeds in the 3′ to 5′ direction54,103,104 (FIG. 6a). The other DNA strand undergoes endonucleolytic and exonucleolytic degradation54,103–106.
Type II systems
The interference mechanism of type II systems is completely different from that of type I and type III systems (FIG. 6b). In type II systems, interference is mediated by the Cas9–RNP complex that consists of Cas9 and two RNAs (that is, crRNA and tracrRNA)75–77,107 (FIG. 5a, b). Targeting of a complementary DNA fragment by Cas9 has recently been shown to proceed in a step-wise manner108. As mentioned above, loading of the sgRNA triggers a structural rearrangement that leads to the formation of a central channel that binds to the target DNA80. Next, the Cas9–RNA complex scans the DNA for a PAM motif (BOX 1; FIG. 6b). Scanning and identification of the target DNA by type II systems seems to be a mirror image of the initial steps of type I interference; the PAM motif resides on the displaced strand of the target DNA and is located close to the 3′ handle of the crRNA guide. Two tryptophan-containing flexible loops in the C-terminal domain of the Cas9 nuclease lobe are involved in PAM recognition, and mutation of these residues affects both the binding and cleavage of target DNA80. Interaction with the PAM motif is required for DNA binding, and subsequent DNA strand displacement and R-loop formation initiate at the PAM. Base pairing progresses over a 12 nucleotide seed sequence towards the distal end of the target sequence108, with little room for mismatch109 (FIG. 6b). During the final stage of interference, Cas9 — which has adopted a cleavage-competent state (as described above) — uses its intrinsic nucleolytic activity to cleave the DNA. Endonuclease activity is catalysed by two active sites at separate locations in the nuclease lobe. The HNH-like nuclease domain cleaves the DNA strand that base pairs with the crRNA, and the RuvC-like nuclease domain cleaves the displaced DNA strand76,80. Cas9-mediated nuclease activity results in a blunt double-stranded end at a specific site, which is typically 3 nucleotides from the 3′ end of the protospacer107,110 (FIG. 6b). Cas9 has been reported to be a single-turnover enzyme108.
Type III systems
Csm complexes of type III-A systems typically consist of at least five distinct proteins (Csm1–Csm5) (FIG. 2; Supplementary information S1 (table)) and crRNAs with conserved 8 nucleotide 5′ handles and variable 3′ ends67,78. The mechanistic details underlying the targeting of complementary protospacers by type III crRNP complexes remain elusive, but some information is emerging. Type III-A systems discriminate self DNA from non-self DNA in a PAM-independent manner111 (BOX 1), with the possible involvement of the Cas10-like protein Csm1 (that is, the large subunit)112. Similarly to some type I systems, the 5′ end of the spacer region of the crRNA has been proposed to contain a seed sequence112 (FIG. 6c). The type III-A system has been shown to target DNA in vivo113; however, no in vitro DNA-degrading activity has so far been reported for a purified Csm complex78,111. Genetic analysis suggests that an additional protein, Csm6 (also known as Csx1), is required for interference114. It is tempting to speculate that Csm6 is a helicase and/or nuclease that is recruited for DNA interference, analogous to Cas3 in type I systems (FIG. 6a, c), but this remains to be determined.
Cmr complexes of type III-B systems consist of at least six distinct proteins (Cmr1–Cmr6) (FIG. 2; Supplementary information S1 (table)). They are unique among CRISPR–Cas systems in that they target RNA rather than DNA115. It has recently been shown that the RNA target is cleaved by the T. thermophilus Cmr complex at 4–5 distinct sites69,115 (FIG. 5d). Cleavage of the target RNA occurs processively, in the 3′ to 5′ direction, in 6 nucleotide intervals69. This sequential slicing of the target RNA has been confirmed for two additional Cmr complexes (M. Terns and S. Bailey, personal communication). Whether or not there is a seed-like sequence at the 5′ end of the spacer remains to be determined. The regularly spaced cleavage pattern of Cmr complexes indicates that there are multiple catalytic sites along the backbone of the complex69, which suggests that Cmr4 might be the catalytic subunit, as discussed below.
Two Cmr complexes (Cmr-α and Cmr-β) are encoded by different gene clusters in Sulfolobus islandicus. Unexpectedly, the Cmr-α complex (which is composed of Cmr1–Cmr6) has been reported to target plasmid DNA in vivo, using a Csx1-dependent mechanism that requires the transcription of its target114; this seems to be a functional analogue of the aforementioned Csm complex of III-A systems. By contrast, the S. solfataricus Cmr-β complex (which is composed of Cmr1–Cmr7) targets RNA in vitro68; however, the reported catalytic mechanism differs substantially from that which has been described for the III-B systems of P. furiosus and T. thermophilus69,79. These studies suggest that there is further mechanistic and/or functional diversity among type III systems114.
Heteroduplex formation
Among the different CRISPR–Cas types, the molecular details of crRNA binding by crRNP complexes differ substantially. As mentioned above, Cas5 of Cascade/I-E is most probably involved in binding the 5′ handle of the crRNA. At the other end of the crRNA, the 3′ hairpin (which consists of a 6 base pair stem and a 4 nucleotide loop) is firmly bound by the Cas6e nuclease subunit56,59,74. These stable interactions at both ends of the mature crRNA constrain base pairing with target nucleic acids to 5–6 helical segments that are each separated by short non-helical sequences74 (Supplementary information S4 (figure)). This type of crRNA–target interaction is supported by biochemical analysis74,84 and genetic data, which indicate that some nucleotides in the spacer do not base pair and, as such, are not essential for target recognition50,116. Interestingly, this base-pairing pattern is reminiscent of the DNA–DNA interaction that is mediated by the RecA protein, which forms a nucleoprotein filament during homologous recombination. Strings of RecA molecules, which are complexed with a ssDNA template, invade a dsDNA helix. After the displacement of the non-complementary strand, the newly formed hybrid of complementary DNA strands is globally underwound and stretched but is locally allowed to adopt a classic B-form conformation117, which resembles the crRNA–target hybrid configuration of the Cascade R-loop (Supplementary information S5 (figure)).
In the Cascade complexes of type I-A and type I-C systems, as well as in the crRNP complexes of type II and type III systems, crRNAs are bound by only a single handle. This probably results in increased flexibility of the crRNA and possibly leads to a different structure of the crRNA–target heteroduplex. Indeed, the structure of Cas9 reveals that there is complete base pairing between the sgRNA and the 20 nucleotide target DNA81 (Supplementary information S2 (figure)). In RNA targeting by type III-B systems, the situation might resemble eukaryotic RNA interference (RNAi), in which an Argonaute nuclease typically uses a 21 nucleotide guide RNA with a firmly anchored 5′ end to recognize a complementary mRNA target. Interactions between the Argonaute complex and a complementary target nucleic acid trigger the release of the 3′ end of the guide RNA, which results in the formation of at least 15 contiguous base pairs in an A-form conformation118.
Conclusions and outlook
Since the publication of the landmark paper by Barrangou et al.7, which describes the discovery of the CRISPR–Cas system as a prokaryotic adaptive immune system, impressive progress has been made with respect to understanding many of the unique mechanistic features that are associated with these remarkable systems. Although in silico studies initially showed that there was overwhelming variation in CRISPR–Cas systems, subsequent comparative sequence analyses resulted in the identification of three major types12 and in potential scenarios for the evolution of CRISPR–Cas variants91. Moreover, recent structural analyses of Cas proteins and crRNP complexes have revealed that there are unanticipated similarities between the type I and type III crRNP complexes. In contrast to these multisubunit complexes, the single-protein Cas9–crRNP complex from type II systems is structurally unrelated to other crRNP complexes, and there are major mechanistic differences at the levels of CRISPR expression and interference.
Outstanding gaps in our mechanistic understanding of CRISPR–Cas functionality include the molecular details of the spacer acquisition process, particularly the role of Cas1 and Cas2, the involvement of Cas3 and/or Cas4 and assistance by general repair and/or recombination enzymes. At the level of CRISPR interference, the general picture has become clear, but several relevant details regarding self versus non-self discrimination, target degradation by trans-acting nucleases (such as Cas3) and interference by allosteric regulation of the nuclease domain (for example, by Cas9 and the Cmr complex) remain poorly understood. Molecular insights rely heavily on the availability of high-resolution crystal structures. A steadily growing number of structures are available for individual Cas proteins, but a major challenge is to obtain atomic-resolution models (such as crystal structures and cryo-EM structures) of the RNP complexes that are involved in CRISPR acquisition, expression and interference. Important progress has recently been made for all three types of CRISPR–Cas system (FIG. 5a,b), but future breakthroughs are required to address some of the outstanding questions.
The unique capacity of sequence-specific DNA targeting and cleavage allows for the application of CRISPR–Cas components as novel tools for genome editing. The detailed characterization of the Cas9–guide complex and its interference mechanism in 2012 (REFS 72,73) has led to a revolution of CRISPR-based genetic engineering119,120, including directed recombination in bacteria109,121, transcriptional activation and repression of synthetic regulons122,123 and genome editing in eukaryotic cells, ranging from yeast to plant and from zebrafish to human124. In addition, RNA targeting by type III-B systems (as well as by Cas9) may function as an alternative system for the directed silencing of gene expression124. In terms of applications of CRISPR-associated nucleases in general, and Cas9 in particular, the sky seems to be the limit. However, even for Cas9, there is still room for improvement — for example, by lowering the stringency of its PAM dependence and reducing its off-target cleavage. We anticipate that fundamental details of CRISPR–Cas structure and function will not only further improve our understanding of these unique defence systems but will also be crucial for optimizing and further expanding the applicability of CRISPR–Cas systems.
Supplementary Material
Acknowledgments
The authors thank members of the van der Oost laboratory (R. Staals and S. Brouns) and the Wiedenheft laboratory (specifically M.-C. Rollins and S. Golden) for discussions and critical reading of this manuscript. This work was supported by a Netherlands Organisation for Scientific Research (NWO) Earth and Life Sciences (ALW-TOP) grant (grant number 854.10.003) and NWO ALW Open competition to J.O. (grant number 820.02.003). E.R.W. received funding from the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007–2013) under REA grant agreement number 327606. Postdoctoral training for R.N.J. is supported by the US National Institutes of Health (grant number R01GM108888). B.W. is supported by the US National Institutes of Health (grant numbers P20GM103500 and R01GM108888), US National Science Foundation EPS-CoR (grant number EPS-110134), the M.J. Murdock Charitable Trust and the Montana State University Agricultural Experimental Station.
Glossary
- Restriction–modification systems
(R–M systems). Innate defence systems in bacteria and archaea that enable the discrimination of ‘non-self’ DNA from ‘self’ DNA. These systems typically consist of an endonuclease that specifically recognizes and cleaves a short palindromic sequence motif in invading DNA and a methyltransferase that methylates a nucleotide within the same motif in the genomic DNA of the host cell, thereby protecting self DNA from degradation.
- Cascade
(CRISPR-associated complex for antiviral defence). A multisubunit Cas (CRISPR-associated protein) complex that associates with a CRISPR RNA (crRNA) in type I CRISPR–Cas systems. Recent insights have revealed that the Cascade core is conserved in type III CRISPR ribonucleoprotein (crRNP) complexes.
- Leader
The DNA sequence upstream of a CRISPR locus it contains the promoter and sequence elements that drive polarized repeat duplication and spacer acquisition.
- Protospacer adjacent motif
(PAM). A short signature sequence (of 2–5 nucleotides) that flanks the protospacer in invading DNA. Recognition of the PAM by type I and type II CRISPR–Cas systems triggers interference.
- RAMP
A protein fold that resembles the widely distributed RNA-recognition motif (RRM); it is also referred to as a ferredoxin fold.
- Protospacer
The target DNA sequence that is complementary to the spacer of the CRISPR RNA.
- RNase III
A dimeric endoribonuclease that cleaves double-stranded RNA; it typically generates products that have a 2 nucleotide overhang at the 3′ end.
- HNH
One of two nucleolytic domains in Cas9; it is related to the nucleolytic domain of McrA-like restriction endonucleases.
- RuvC
One of two nucleolytic domains in Cas9; it is homologous to nucleases that are involved in recombination.
- R-loop
A structure that is formed by the hybridization of an RNA strand with double-stranded DNA. The RNA base pairs with a complementary sequence in one of the DNA strands, which causes the displaced DNA strand to form a loop.
- Seed
A short sequence within the CRISPR RNA that is required for perfect base pairing with the target sequence. This short stretch of 7–8 nucleotides is most probably the site of initial hybridization with the complementary target strand, resulting in R-loop formation and CRISPR interference.
- HD-nuclease domain
The domain of Cas3 that is responsible for the nucleolytic degradation of double-stranded DNA targets. In many Cas3 proteins, this nuclease domain is fused to a helicase domain.
- SF2-helicase domain
(Superfamily 2-helicase domain). The multidomain component of Cas3 that is responsible for unwinding double-stranded DNA targets. Proteins of the SF2-type helicase superfamily consist of two RecA-like domains (with an ATP-binding site at their interface) and a flexible carboxy-terminal domain.
- B-form
The classic right-handed DNA double helix (established by Watson and Crick), which is the predominant DNA conformation under physiological conditions.
- Argonaute
A nuclease that is involved in eukaryotic RNA interference and bacterial and archaeal DNA interference. Argonaute contains an RNaseH nucleolytic domain that is homologous to RuvC.
- A-form
A conformation of the DNA helix that is more compact than the B-form; it is often present in double-stranded RNA and in DNA–RNA hybrids.
Footnotes
Competing interests statement
The authors declare no competing interests.
DATABASES
Electron Microscopy Data Bank (EMDB):
http://www.ebi.ac.uk/pdbe/emdb
5314 | 5860 | 2420 | 5740
Protein Data Bank (PDB):
http://www.rcsb.org/pdb/home/home.do
3GOD | 3OQ2 | 4C9D | 4AL7 | 4F3M
SUPPLEMENTARY INFORMATION
See online article: S1 (table) | S2 (figure) | S3 (figure) | S4 (figure) | S5 (figure)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Suttle CA. Marine viruses — major players in the global ecosystem. Nature Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 2.Samson JE, Magadan AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nature Rev Microbiol. 2013;11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 3.Makarova KS, Wolf YI, Koonin EV. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. This paper reports the discovery that CRISPR spacers are homologous to fragments of mobile genetic elements, which led to the hypothesis that the system is an adaptive immune system. [DOI] [PubMed] [Google Scholar]
- 5.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 6.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 7.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. This study provides the first experimental evidence that the CRISPR–Cas system is an adaptive immune system. [DOI] [PubMed] [Google Scholar]
- 8.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. This paper reports the discovery of the link between repetitive arrays (that is, CRISPR sequences) and CRISPR-associated (cas) genes. [DOI] [PubMed] [Google Scholar]
- 10.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makarova KS, et al. Evolution and classification of the CRISPR–Cas systems. Nature Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. This article describes a revised CRISPR–Cas classification system, in which the three major system types are proposed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heidrich N, Vogel J. Same same but different: new structural insight into CRISPR–Cas complexes. Mol Cell. 2013;52:4–7. doi: 10.1016/j.molcel.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 14.Fonfara I, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR–Cas systems. Nucleic Acids Res. 2013;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–729. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- 16.Lillestol RK, et al. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009;72:259–272. doi: 10.1111/j.1365-2958.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- 17.Seed KD, Lazinski DW, Calderwood SB, Camilli A. A bacteriophage encodes its own CRISPR–Cas adaptive response to evade host innate immunity. Nature. 2013;494:489–491. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minot S, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minot S, et al. Rapid evolution of the human gut virome. Proc Natl Acad Sci USA. 2013;110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–207. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- 21.Dupuis ME, Villion M, Magadan AH, Moineau S. CRISPR–Cas and restriction–modification systems are compatible and increase phage resistance. Nature Commun. 2013;4:2087. doi: 10.1038/ncomms3087. [DOI] [PubMed] [Google Scholar]
- 22.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. This paper reports the discovery of the PAM motif. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 24.Shah SA, Erdmann S, Mojica FJ, Garrett RA. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. CRISPR interference directs strand specific spacer acquisition. PLoS ONE. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diez-Villasenor C, Guzman NM, Almendros C, Garcia-Martinez J, Mojica FJ. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR–Cas I-E variants of Escherichia coli. RNA Biol. 2013;10:792–802. doi: 10.4161/rna.24023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erdmann S, Garrett RA. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol. 2012;85:1044–1056. doi: 10.1111/j.1365-2958.2012.08171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vercoe RB, et al. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013;9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiedenheft B, et al. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 32.Babu M, et al. A dual function of the CRISPR–Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol. 2011;79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 34.Reeks J, Naismith JH, White MF. CRISPR interference: a structural perspective. Biochem J. 2013;453:155–166. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beloglazova N, et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samai P, Smith P, Shuman S. Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1552–1556. doi: 10.1107/S1744309110039801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam KH, et al. Double-stranded endonuclease activity in Bacillus halodurans clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas2 protein. J Biol Chem. 2012;287:35943–35952. doi: 10.1074/jbc.M112.382598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Arslan Z, et al. Double-strand DNA end-binding and sliding of the toroidal CRISPR-associated protein Csn2. Nucleic Acids Res. 2013;41:6347–6359. doi: 10.1093/nar/gkt315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nam KH, Kurinov I, Ke A. Crystal structure of clustered regularly interspaced short palindromic repeats (CRISPR)-associated Csn2 protein revealed Ca2+-dependent double-stranded DNA binding activity. J Biol Chem. 2011;286:30759–30768. doi: 10.1074/jbc.M111.256263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellinger P, et al. The crystal structure of the CRISPR-associated protein Csn2 from Streptococcus agalactiae. J Struct Biol. 2012;178:350–362. doi: 10.1016/j.jsb.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Koo Y, Jung DK, Bae E. Crystal structure of Streptococcus pyogenes Csn2 reveals calcium-dependent conformational changes in its tertiary and quaternary structure. PLoS ONE. 2012;7:e33401. doi: 10.1371/journal.pone.0033401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee KH, et al. Identification, structural, and biochemical characterization of a group of large Csn2 proteins involved in CRISPR-mediated bacterial immunity. Proteins. 2012;80:2573–2582. doi: 10.1002/prot.24138. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J, Kasciukovic T, White MF. The CRISPR associated protein Cas4 is a 5′ to 3′ DNA exonuclease with an iron–sulfur cluster. PLoS ONE. 2012;7:e47232. doi: 10.1371/journal.pone.0047232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemak S, et al. Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe–4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus. J Am Chem Soc. 2013;135:17476–17487. doi: 10.1021/ja408729b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plagens A, Tjaden B, Hagemann A, Randau L, Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson RN, Lavin M, Carter J, Wiedenheft B. Fitting CRISPR-associated Cas3 into the helicase family tree. Curr Opin Struct Biol. 2014;24C:106–114. doi: 10.1016/j.sbi.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richter C, Gristwood T, Clulow JS, Fineran PC. In vivo protein interactions and complex formation in the Pectobacterium atrosepticum subtype I-F CRISPR/Cas System. PLoS ONE. 2012;7:e49549. doi: 10.1371/journal.pone.0049549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Datsenko KA, et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nature Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 50.Fineran PC, et al. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci USA. 2014;111:E1629–E1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niewoehner O, Jinek M, Doudna JA. Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases. Nucleic Acids Res. 2014;42:1341–1353. doi: 10.1093/nar/gkt922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jore MM, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nature Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 53.Wiedenheft B, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinkunas T, et al. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nature Struct Mol Biol. 2011;18:680–687. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 57.Reeks J, et al. Structure of a dimeric crenarchaeal Cas6 enzyme with an atypical active site for CRISPR RNA processing. Biochem J. 2013;452:223–230. doi: 10.1042/BJ20130269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ebihara A, et al. Crystal structure of hypothetical protein TTHB192 from Thermus thermophilus HB8 reveals a new protein family with an RNA recognition motif-like domain. Protein Sci. 2006;15:1494–1499. doi: 10.1110/ps.062131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nature Struct Mol Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 60.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Przybilski R, et al. Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol. 2011;8:517–528. doi: 10.4161/rna.8.3.15190. [DOI] [PubMed] [Google Scholar]
- 62.Haurwitz RE, Sternberg SH, Doudna JA. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 2012;31:2824–2832. doi: 10.1038/emboj.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–672. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc Natl Acad Sci USA. 2011;108:21218–21222. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hatoum-Aslan A, Samai P, Maniv I, Jiang W, Marraffini LA. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J Biol Chem. 2013;288:27888–27897. doi: 10.1074/jbc.M113.499244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staals RH, et al. Structure and activity of the RNA-targeting Type III-B CRISPR–Cas complex of Thermus thermophilus. Mol Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hale CR, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garside EL, et al. Cas5d processes pre-crRNA and is a member of a larger family of CRISPR RNA endonucleases. RNA. 2012;18:2020–2028. doi: 10.1261/rna.033100.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nam KH, et al. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR–Cas system. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koo Y, Ka D, Kim EJ, Suh N, Bae E. Conservation and variability in the structure and function of the Cas5d endoribonuclease in the CRISPR-mediated microbial immune system. J Mol Biol. 2013;425:3799–3810. doi: 10.1016/j.jmb.2013.02.032. [DOI] [PubMed] [Google Scholar]
- 74.Wiedenheft B, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. This study reports cryo-EM structures of Cascade, which is the archetype for type I and type III crRNP complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. This study shows that crRNA maturation in type II systems requires RNase III and tracrRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. This study characterizes the mechanism of action of Cas9 and reveals a requirement for both the crRNA and tracrRNA (or an sgRNA fusion) for target cleavage. In addition, it presents the first experimental evidence for Cas9-directed genome editing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rouillon C, et al. Structure of the CRISPR interference complex CSM reveals key similarities with Cascade. Mol Cell. 2013;52:124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spilman M, et al. Structure of an RNA silencing complex of the CRISPR–Cas immune system. Mol Cell. 2013;52:146–152. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jinek M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. This paper reports the crystal structures of Cas9 endonucleases and cryo-EM structures that demonstrate major conformational changes in Cas9 following binding of the sgRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nishimasu H, et al. Crystal structure of cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. This paper reports the crystal structure of Cas9 in complex with an sgRNA and an ssDNA target. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Westra ER, et al. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 83.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. This study reports the discovery of the Cascade complex that, together with Cas3, is responsible for CRISPR-based DNA interference in type I systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Duijn E, et al. Native tandem and ion mobility mass spectrometry highlight structural and modular similarities in clustered-regularly-interspaced shot-palindromic-repeats (CRISPR)-associated protein complexes from Escherichia coli and Pseudomonas aeruginosa. Mol Cell Proteom. 2012;11:1430–1441. doi: 10.1074/mcp.M112.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quax TE, et al. Differential translation tunes uneven production of operon-encoded proteins. Cell Rep. 2013;4:938–944. doi: 10.1016/j.celrep.2013.07.049. [DOI] [PubMed] [Google Scholar]
- 87.Lintner NG, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 89.Hochstrasser ML, et al. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci USA. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hrle A, et al. Structure and RNA-binding properties of the Type III-A CRISPR-associated protein Csm3. RNA Biol. 2013;10:1670–1678. doi: 10.4161/rna.26500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR–Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Osawa T, Inanaga H, Numata T. Crystal structure of the Cmr2–Cmr3 subcomplex in the CRISPR–Cas RNA silencing effector complex. J Mol Biol. 2013;425:3811–3823. doi: 10.1016/j.jmb.2013.03.042. [DOI] [PubMed] [Google Scholar]
- 93.Shao Y, et al. Structure of the Cmr2–Cmr3 subcomplex of the Cmr RNA silencing complex. Structure. 2013;21:376–384. doi: 10.1016/j.str.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mulepati S, Orr A, Bailey S. Crystal structure of the largest subunit of a bacterial RNA-guided immune complex and its role in DNA target binding. J Biol Chem. 2012;287:22445–22449. doi: 10.1074/jbc.C112.379503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhu X, Ye K. Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR–Cas systems. FEBS Lett. 2012;586:939–945. doi: 10.1016/j.febslet.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 96.Cocozaki AI, et al. Structure of the Cmr2 subunit of the CRISPR–Cas RNA silencing complex. Structure. 2012;20:545–553. doi: 10.1016/j.str.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Makarova KS, Wolf YI, Koonin EV. The basic building blocks and evolution of CRISPR–Cas systems. Biochem Soc Trans. 2013;41:1392–1400. doi: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Westra ER, et al. Cascade-mediated binding and bending of negatively supercoiled DNA. RNA Biol. 2012;9:1134–1138. doi: 10.4161/rna.21410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Semenova E, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 101.Westra ER, et al. Type I-E CRISPR–Cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013;9:e1003742. doi: 10.1371/journal.pgen.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kunne T, Swarts DC, Brouns SJ. Planting the seed: target recognition of short guide RNAs. Trends Microbiol. 2014;22:74–83. doi: 10.1016/j.tim.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 103.Westra ER, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beloglazova N, et al. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30:4616–4627. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mulepati S, Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) J Biol Chem. 2011;286:31896–31903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sinkunas T, et al. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 108.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR–Cas systems. Nature Biotech. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Magadan AH, Dupuis ME, Villion M, Moineau S. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3–Cas system. PLoS ONE. 2012;7:e40913. doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. This study characterizes self versus non-self discrimination in type III-A systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hatoum-Aslan A, Maniv I, Samai P, Marraffini LA. Genetic characterization of antiplasmid immunity through a type III-A CRISPR–Cas system. J Bacteriol. 2014;196:310–317. doi: 10.1128/JB.01130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Deng L, Garrett RA, Shah SA, Peng X, She Q. A novel interference mechanism by a type IIIB CRISPR–Cmr module in Sulfolobus. Mol Microbiol. 2013;87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- 115.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA–Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. This study provides evidence that RNA (rather than DNA) is targeted by type III-B systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stoll B, et al. Requirements for a successful defence reaction by the CRISPR–Cas subtype I-B system. Biochem Soc Trans. 2013;41:1444–1448. doi: 10.1042/BST20130098. [DOI] [PubMed] [Google Scholar]
- 117.Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 118.Sheng G, et al. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc Natl Acad Sci USA. 2014;111:652–657. doi: 10.1073/pnas.1321032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pennisi E. The CRISPR craze. Science. 2013;341:833–836. doi: 10.1126/science.341.6148.833. [DOI] [PubMed] [Google Scholar]
- 120.Wilkinson R, Wiedenheft BA. CRISPR method for genome engineering. F1000Prime Rep. 2014;6:3. doi: 10.12703/P6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gomaa AA, et al. Programmable removal of bacterial strains by use of genome-targeting CRISPR–Cas systems. mBio. 2014;5:e00928–13. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bikard D, et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR–Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sampson TR, Weiss DS. Exploiting CRISPR/Cas systems for biotechnology. Bioessays. 2014;36:34–38. doi: 10.1002/bies.201300135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Almendros C, Guzman NM, Diez-Villasenor C, Garcia-Martinez J, Mojica FJ. Target motifs affecting natural immunity by a constitutive CRISPR–Cas system in Escherichia coli. PLoS ONE. 2012;7:e50797. doi: 10.1371/journal.pone.0050797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Horvath P, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Plagens A, et al. In vitro assembly and activity of an archaeal CRISPR–Cas type I-A Cascade interference complex. Nucleic Acids Res. 2014;42:5125–5138. doi: 10.1093/nar/gku120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nuñez JK, et al. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nature Struct Mol Biol. 2014 doi: 10.1038/nsmb.2820. http://dx.doi.org/10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


