Significance
Application of epoxide hydrolases in synthesizing chiral drug compounds has been hindered by their limited substrate range. The enzymatic production of bulky epoxides has proven remarkably challenging. In this work, we identified an active tunnel for substrate access and product release of an epoxide hydrolase with unusual (R)-enantioselectivity. Mutagenesis targeted to unblock the steric hindrance in the active pocket or the potential product release site resulted in variants with much higher activity toward α-naphthyl glycidyl ether, the precursor of β-adrenergic receptor blocking drug (S)-propranolol. The strategy presented here may be a useful alternative choice for rational design of enzymes toward bulky substrates.
Keywords: epoxide hydrolase, X-ray crystallography, protein engineering, product release, bulky substrate
Abstract
Optically pure epoxides are essential chiral precursors for the production of (S)-propranolol, (S)-alprenolol, and other β-adrenergic receptor blocking drugs. Although the enzymatic production of these bulky epoxides has proven difficult, here we report a method to effectively improve the activity of BmEH, an epoxide hydrolase from Bacillus megaterium ECU1001 toward α-naphthyl glycidyl ether, the precursor of (S)-propranolol, by eliminating the steric hindrance near the potential product-release site. Using X-ray crystallography, mass spectrum, and molecular dynamics calculations, we have identified an active tunnel for substrate access and product release of this enzyme. The crystal structures revealed that there is an independent product-release site in BmEH that was not included in other reported epoxide hydrolase structures. By alanine scanning, two mutants, F128A and M145A, targeted to expand the potential product-release site displayed 42 and 25 times higher activities toward α-naphthyl glycidyl ether than the wild-type enzyme, respectively. These results show great promise for structure-based rational design in improving the catalytic efficiency of industrial enzymes for bulky substrates.
Optically pure epoxides and the corresponding vicinal diols are valuable chiral building blocks for the production of pharmaceutically active compounds and other fine chemicals (1). Existing approaches for preparing enantiopure epoxides and diols include the asymmetric epoxidation or dihydroxylation of olefin substrates and the resolution of racemic epoxides. These reactions can be accomplished with either chemical catalysts such as chiral salen cobalt complexes and porphyrin manganese adducts or biocatalysts such as monooxygenases and epoxide hydrolases (EHs) (2–4). In the past two decades, EHs have received much attention because they are cofactor-independent enzymes that are “easy to use” for catalyzing the hydrolysis of racemic epoxides to yield highly enantiopure epoxides and vicinal diols (1, 5, 6). However, application of EHs in laboratory and industry was often hindered by their narrow substrate scope, low enantioselectivity, and regioselectivity, or product inhibition (7, 8).
Many protein-engineering efforts have been made to overcome these drawbacks (9, 10). For example, directed evolution by error-prone PCR or DNA shuffling has been used to enhance the activity and enantioselectivity of EHs (11–13). Structure-guided mutagenesis also generated a few EH variants with improved catalytic performance (14–16). The strategy of iterative Combinatorial Active Site-Saturation Test (CAST) combines the rational approach and directed evolution to yield high-quality and small focused mutant libraries for screening EHs with better enantioselectivity (7, 17). By mutating residues at the substrate-binding site, the substrates of EHs have been expanded to include cyclic meso-epoxides, phenyl glycidyl ether (PGE) derivatives, and other styrene oxide-like analogs (18, 19). However, the catalytic efficiency of EH is still not satisfactory for bulky epoxide substrates including precursors of (S)-propranolol, (S)-alprenolol, and other β-adrenergic receptor blocking drugs (20, 21).
In this work, we select BmEH, an EH cloned from Bacillus megaterium ECU1001, to expand its substrate scope for bulky pharmaco substrate α-naphthyl glycidyl ether (NGE). This enzyme is a potential industrial biocatalyst because it has unusual (R)-enantioselectivity and resolves ortho-substituted PGEs and para-nitrostyrene oxide with excellent enantiomeric ratios (E > 200) (22). We first identified the active tunnel of BmEH by solving its crystal structure complexed with a substrate analog phenoxyacetamide (POA) and analyzing the routes of substrate entry and product release by mass spectrum analysis. Alanine scanning experiments targeted to the potential product-release site of BmEH resulted in two variants, F128A and M145A, with efficient bioresolution abilities on NGE. Further kinetic measurements and structural analysis showed that M145A has much higher activity for the transition state intermediate formation, whereas both mutants exhibited expanded product-release site. The M145A BmEH variant has been successfully applied for the preparation of (S)-propranolol on a gram scale. The engineering of the potential product-release site described herein should have great promise for structure-based rational design of better industrial enzymes.
Results
Structures of BmEH and the BmEH–POA Complex.
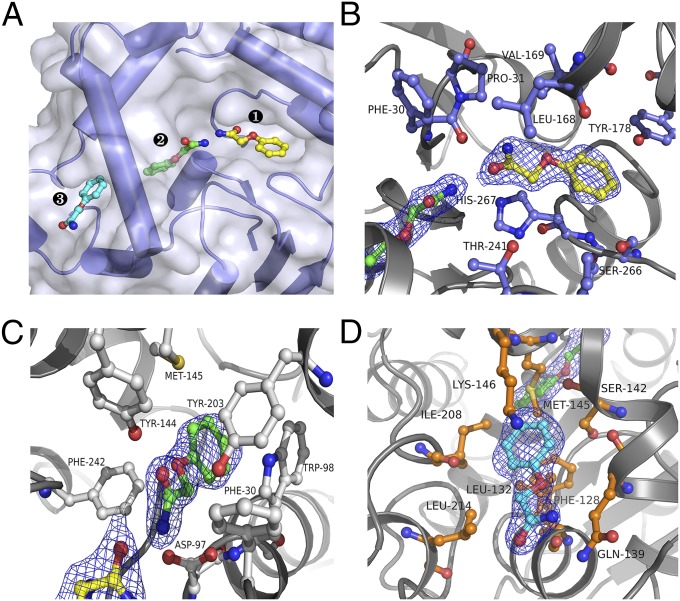
The overall structure of BmEH, which is similar to the structures of other α/β-hydrolases, consists of a catalytic α/β domain and a lid domain that caps the active site (SI Appendix, Fig. S1). Key residues involved in the catalytic triad (Asp-97, -239, and His-267), binding motif (Tyr-144 and -203), and oxyanion hole (Phe-30 and Trp-98) are highly conserved in BmEH, HssEH (23), MmsEH (24), MtEH (25), AnEH (26), ArEH (27), and StEH (28) (SI Appendix, Table S1). In the BmEH–POA complex structure, three POA molecules were observed in the catalytic zones (Fig. 1A and SI Appendix, Fig. S2). One POA molecule (Fig. 1B) was located in a deep cleft (zone 1) surrounded by residues Leu-168, Tyr-178, Pro-240, Thr-241, Ser-266, and His-267. Another POA molecule (Fig. 1C) was deeply buried in the catalytic cavity (zone 2), which was identified on the basis of structural similarity with the catalytic centers of other EHs. The amide nitrogen of this POA was hydrogen-bonded to the side chain of Asp-97 (N–O distance 2.7 Å), which is responsible for the nucleophilic attack on epoxide in the catalytic mechanism, and the amide oxygen and the ether oxygen were hydrogen-bonded to binding motif residues Tyr-144 and -203, respectively. The substrate analog may have been further stabilized by π–π stacking of the phenyl rings of POA and Trp-98. The third POA (Fig. 1D, cyan) was located on a bowl-like surface (zone 3) underneath zone 2; Met-145, Phe-128, Ile-208, and Ser-142 made up the base of the bowl, and Gln-139, Leu-132, Phe-209, Lys-146, and Leu-214 made up the side walls.
Fig. 1.
Three POA binding sites found in the active tunnel of the BmEH–POA complex. (A) POA molecules in zones 1 (yellow), 2 (green), and 3 (cyan). (B–D) The POA binding pattern in zones 1 (B), 2 (C), and 3 (D). The residues of BmEH interacting directly with POA are shown as ball-and-stick models.
Identification of the Active Tunnel of BmEH.
Comparison of the BmEH–POA complex structure with those of other EHs showed that there is an active tunnel in BmEH consisting of zones 1, 2, and 3. Zone 2 is conserved in all EH structures and serves as the catalytic center. It is accessible from zone 1 and very close to, but detached from, zone 3 (SI Appendix, Fig. S3). Analysis of the B factor for each zone in BmEH revealed that the atomic fluctuations of residues 125–156 and 204–225, located between zones 2 and 3, were significant (SI Appendix, Fig. S4). This fluctuation was confirmed by root mean square (rms) fluctuation and rms deviation (rmsd) statistics from an 80-ns molecular dynamics (MD) simulation (SI Appendix, Fig. S5). These results suggested that such fluctuations might promote the transfer of substrate or product between zones 2 and 3. Indeed, the M145F mutant designed specifically to block the tunnel between zones 2 and 3 lost ∼80% activities for PGE and NGE (SI Appendix, Table S2).
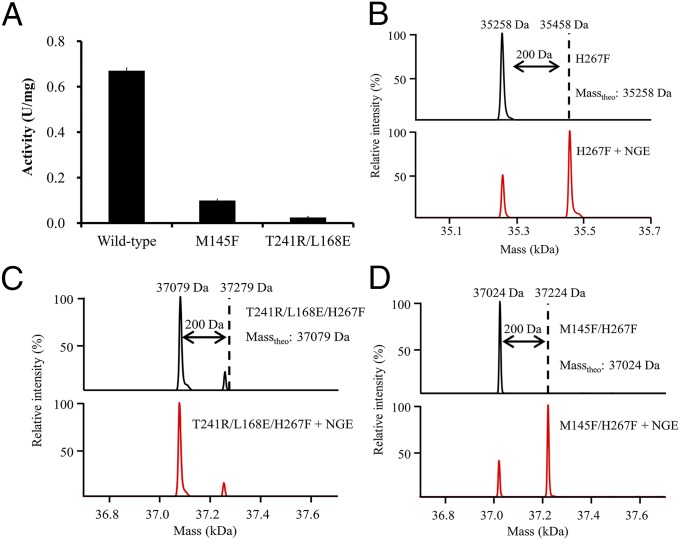
To verify whether zones 1 and 3 are responsible for substrate access in BmEH catalysis, we developed a mass spectrum method for capturing the transition state intermediates (SI Appendix, Fig. S6). The T241R/L168E BmEH variant was constructed by increasing the steric hindrance between zones 1 and 2. The catalytically dead mutation H267F was introduced into T241R/L168E and M145F to prevent the covalent transition state intermediates from being hydrolyzed. After the BmEH variants were incubated with the bulky substrate NGE for 10 min, the reaction mixture was separated by a chromatographic capillary column and further subjected to mass spectrum analysis. As shown in Fig. 2, incubation of H267F or M145F/H267F with NGE generated a peak with molecular mass increase of 200 Da, which exactly matches the theoretical size of NGE. However, we did not detect any corresponding peaks in the reaction mixture of T241R/L168E/H167F and NGE, suggesting that the tunnel between zones 1 and 2 plays an essential role in substrate access.
Fig. 2.
Mass-spectrometric analysis of the covalent intermediates in the BmEH-catalyzed NGE hydrolysis reaction. (A) Relative activities of mutant M145F and T241R/L168E compared with the wild-type BmEH. (B–D) Comparison of the molecular mass changes of the H267F (B), T241R/L168E/H267F (C), and M145F/H267F (D) BmEH variants before (black) and after (red) reacting with NGE. The theoretical molecular mass increase of the enzymes is 200 Da. Mutations at residue Met-145 severely blocked the formation of the enzymatic covalent intermediates.
Mutations Expanded the Product Release Site of BmEH.
Although the tunnel between zones 2 and 3 disfavors the substrate access, it is important for BmEH. As mentioned earlier, blocking this area by mutating Met-145 to Phe resulted in an ∼80% activity decrease for PGE and NGE (SI Appendix, Table S2). It is reasonable to propose that this area may be responsible for product release, and blocking the product exit caused severe activity loss. Another possibility is that mutation in this area might induce conformational changes and thereafter affect the catalysis in zone 2. This finding offers prospects for generating and screening BmEH variants with better bioresolution efficiency toward bulky epoxide substrates. Among all constructed variants, the activities of L132A, M145A, and F128A were 13, 25, and 42 times higher toward NGE, respectively. The kinetic data of M145A and F128A showed that the enhanced activities were mainly due to their higher kcat values (57 and 32 times the kcat of wild-type; Table 1). As a control, mutations of the residues in zones 1 and 2 (L168A, L206A, L219A, F220A, and F242A) did not yield enzymes with increased catalytic activity toward NGE.
Table 1.
Kinetic parameters for the wild-type, M145A, and F128A BmEH toward the substrate rac-PGE or -NGE
| Enzyme | rac-PGE | rac-NGE | ||||
| kcat, s−1 | KM, mM | kcat/KM, s−1⋅mM−1 | kcat, s−1 | KM, mM | kcat/KM, s−1⋅mM−1 | |
| Wild type | >400* | >50† | N.A. | 0.60 ± 0.06 | 1.3 ± 0.3 | 0.45 ± 0.12 |
| M145A | >9.5* | >50† | N.A. | 19.0 ± 2.7 | 1.5 ± 0.5 | 12 ± 5 |
| F128A | 4.4 ± 0.2 | 12 ± 1 | 0.37 ± 0.05 | 34.1 ± 0 0.4 | 0.75 ± 0.24 | 45 ± 15 |
The PGE and NGE concentrations were varied in the ranges of 3–50 and 0.3–10 mM, respectively. N.A., not available.
Calculated based on the highest rate detected.
No saturation was observed. The KM values were beyond the concentration range of substrate. The concentration of PGE for activity determination was limited by its low solubility.
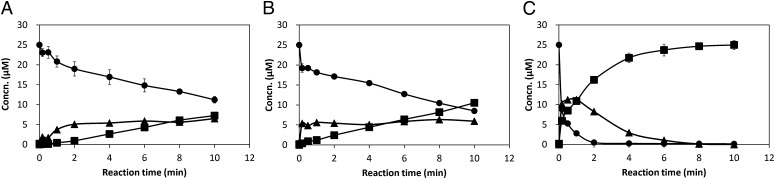
To interpret the kinetic aspects of F128A and M145A, the stopped-flow method was used to examine the fluorescence signal change during the hydrolysis reaction of NGE. Unfortunately, because of the low signal/noise level, no significant burst-phase for the pre-steady-state kinetics analysis could be detected from the observed kinetic trace of reactions (SI Appendix, Fig. S7). Nevertheless, the stopped-flow fluorescence experiment showed that the H267F mutation caused relatively slow intermediate formation and hydrolysis (SI Appendix, Fig. S7). This finding enabled us to detect the subtle difference between the substrate-consumption rate and the product-releasing rate of BmEH variants by HPLC in minute scale. As shown in Fig. 3, the F128A/H267F mutant had a similar pattern of intermediate formation to that of H267F BmEH, whereas the M145A/H267F mutant behaved differently and proceeded at a higher intermediate formation rate. This result suggested that the M145A mutation could accelerate the formation of enzymatic intermediate and thereby increase its catalytic activity toward the bulky NGE.
Fig. 3.
The enzyme–substrate intermediate formation curve of H267F (A), F128A/H267F (B), and M145A/H267F (C). ●, Concentration of substrate (NGE); ■, concentration of product (NPD); ▲, concentration of intermediate. Values were calculated by deduction of the remaining NGE and NPD concentrations from the inputs (25 μM). Reaction conditions were as follows: Each variant at a concentration of 25 μM was mixed with 25 μM (R)-NGE in potassium phosphate buffer (100 mM, pH 7.0) containing 10% DMSO at 30 °C. Samples were withdrawn at different intervals, mixed with methanol for termination, and analyzed by RP-HPLC (C18 column). The concentration of NGE and NPD was quantified by the area of corresponding peaks on HPLC.
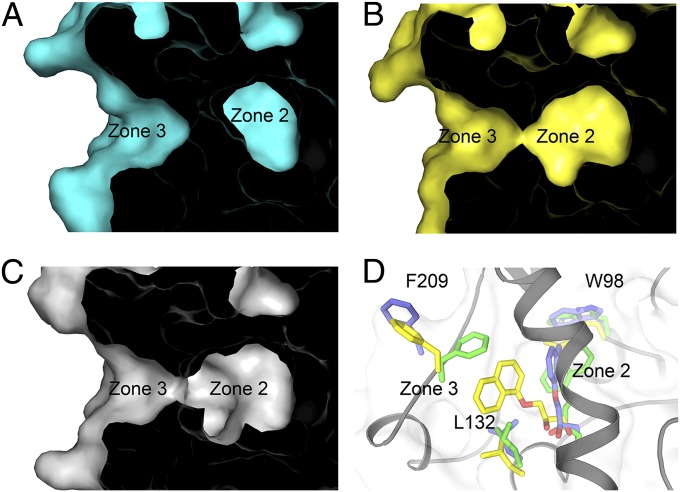
We further solved the crystal structure of BmEHM145A and BmEHF128A–(R)-NPD complex [NPD, the hydrolyzed product of (R)-NGE] at resolutions of 1.70 and 2.90 Å, respectively (SI Appendix, Table S3). Although the overall structures of M145A and F128A are highly similar to that of the native BmEH (with rmsd values of 0.186 and 0.239 Å, respectively), the mutations expanded the cavity of zone 2 and made it more accessible for zone 3 (Fig. 4 A–C). In the complex structure of BmEHF128A–(R)-NPD, the diol moiety of (R)-NPD is located near the boundary between zones 2 and 3, whereas the naphthyl moiety of (R)-NPD is in the active cleft of zone 2 but very close to zone 1. However, based on a structural alignment of other EHs with the BmEH–POA complex, the epoxide ring should be hydrolyzed around the area between zones 1 and 2 (mainly by residues Asp-97, Tyr-144, and -203), whereas the naphthyl ring would sit in the cavity of zone 2 close to zone 3 and be stabilized by π–π stacking of Trp98. This finding suggested that there was a 90° conformational rotation of the hydrolyzed product (R)-NPD when it was released from the catalytic center. We further used the BmEHF128A–(R)-NPD complex as the initial model to perform 80-ns MD simulation calculations to predict the product-release pathways in the active tunnel of BmEH. As shown in Fig. 4D and Movie S1, the product (R)-NPD penetrated zone 2 to reach zone 3 for exit. This process was accompanied by dramatic conformational change of residues Trp-98, Leu-132, and Phe-209. Collectively, these findings indicated a possible catalytic route for BmEH: The substrate enters through zone 1, then undergoes hydrolysis in zone 2, reaches zone 3, and, finally, exits to the solvent.
Fig. 4.
The M145A and F128A mutations made zone 2 expand and attach to zone 3. (A−C) Surface presentation of zones 2 and 3 in wild-type BmEH (cyan), M145A (yellow), and F128A (gray). (D) Snapshots of MD simulation on the BmEHF128A–(R)-NPD complex at 0 ns (green), 42 ns (blue), and 80 ns (yellow). Significant conformation changes took place in residues Trp-98, Leu-132, and Phe-209 around zones 2 and 3.
Gram-Scale Synthesis of (S)-Propranolol with M145A.
With mutants F128A and M145A in hand, we turned our attention to NGE bioresolution. Because F128A was expressed mainly as inclusion bodies and had lower enantioselectivity (E = 45) than M145A (E > 100), we chose M145A for organic synthesis. The kinetic parameters of BmEH WT and M145A toward (R)- and (S)-NGE were measured (Table 2). The wild-type BmEH and mutant M145A had similar KM values for the preferred (R)-NGE and less preferred (S)-NGE substrates. However, in the wild-type BmEH, the substrate specificity constant (kcat/KM) for the preferred (R)-NGE was ∼30 times that for the less preferred (S)-NGE; in mutant M145A, the kcat/KM for the preferred (R)-NGE was ∼200 times that for the less preferred (S)-NGE. Mutation of Met-145 to alanine increased both the catalytic activity and the enantioselectivity for NGE. Indeed, when racemic NGE was bioresolved at a substrate load of 20 g⋅L−1, (S)-NGE and (R)-diol were obtained in 99.4% ee and 91.1% ee, respectively. Compared with that of wild-type BmEH, the E value of mutant M145A was markedly improved from 25 to 131. Subsequently, a simple reaction of (S)-NGE with isopropylamine afforded the desired (S)-propranolol in 36% yield (1.86 g).
Table 2.
Kinetic parameters of the wild-type and M145A BmEH catalyzed reactions of (R)- and (S)-NGE
| Enzyme | (S)-NGE | (R)-NGE | E [=(kcat/KM)R/(kcat/KM)S] | ||||
| kcat, s−1 | KM, mM | kcat/KM, s−1⋅mM−1 | kcat, s−1 | KM, mM | kcat/KM, s−1⋅mM−1 | ||
| Wild type | 0.039 ± 0.005 | 1.40 ± 0.42 | 0.028 ± 0.009 | 0.75 ± 0.08 | 0.87 ± 0.32 | 0.86 ± 0.33 | 31 |
| M145A | 0.23 ± 0.02 | 1.62 ± 0.32 | 0.14 ± 0.03 | 29.1 ± 3.1 | 1.01 ± 0.36 | 28.8 ± 10.7 | 206 |
Reactions were performed in 0.1 M potassium phosphate buffer at pH 7.0 with 10% DMSO as cosolvent and 0.02% Tween 80, and the (R)- and (S)-NGE concentrations were varied in the range of 0.3–10 mM.
Discussion
EHs hydrolyze the racemic epoxides to yield highly enantiopure epoxides and vicinal diols. However, their application was greatly hindered by limited substrate scope and low enantioselectivity and regioselectivity. Previous protein-engineering efforts on EHs including direct evolution, rational design, and CAST strategy have generated several mutants with improved enantioselectivities. However, the catalytic efficiency of engineered EHs on bulky epoxide substrates is still not satisfying. In this work, we have identified an active tunnel in an EH with unusual (R)-enantioselectivity and modified it suitably for efficient bioresolution of bulky pharmaco NGE. Although the wild-type BmEH exhibits higher affinity for NGE (KM = 1.3 ± 0.3 mM) than for less bulky PGE (KM > 50 mM) (Table 1), its efficiency on NGE hydrolysis was much lower, with an activity decreased by two orders of magnitude and an E value reduced to ∼25. We found that this result might be caused by the jammed active tunnel. The F128A and M145A variants, designed to remove the steric hindrance in the product-release site, were found to successfully expand zone 2 and enable it to more openly communicate with zone 3. Interestingly, the M145A mutant was also found to be beneficial for the formation of transition state intermediate. The kcat/KM values of F128A and M145A toward NGE were enhanced by 57 and 32 times that of wild-type BmEH, respectively. Moreover, we can carry out gram-scale synthesis of (S)-propranolol with the M145A variant of BmEH. Further studies to generate an EH library by semisaturation mutations on the identified hot spots has broadened the substrate scope to more drug precursors in our laboratory (29).
As illustrated in the modified Michaelis–Menten equation, the catalytic efficiency (kcat) of the enzymatic reaction is determined by both k2 of the hydrolysis step and k3 of the product-release step (SI Appendix, Fig. S8). When the product dissociation is presumed as the rate-limiting step, expanding the product-release site of active tunnel would increase k3 and thereby enhance kcat. The release of product from enzyme is the last step in the catalytic cycle, which is very important for the catalytic performance, and the substrate specificity might be partially controlled by this step (30). Much effort has been made to identify and engineer the product-release sites of various enzymes. In most cases, the product release and substrate access share the same tunnel in proteins (31–33). However, the product-release tunnel in enzymes such as haloalkane dehalogenases is distinct from the substrate access tunnel, which sometimes makes the product release step a rate-limiting bottleneck, thereby providing a new target for efficient protein engineering (34–36). Previously, there was no report of an independent product-release site in EHs. In this work, we have identified a potential product-release site in BmEH and successfully used this information to engineer mutants with substrate specificity toward bulky epoxides (Fig. 5). Considering of the high structural similarity among the superfamily of α/β-hydrolases, this strategy might become an alternative choice for expanding the substrate scope of enzymes.
Fig. 5.
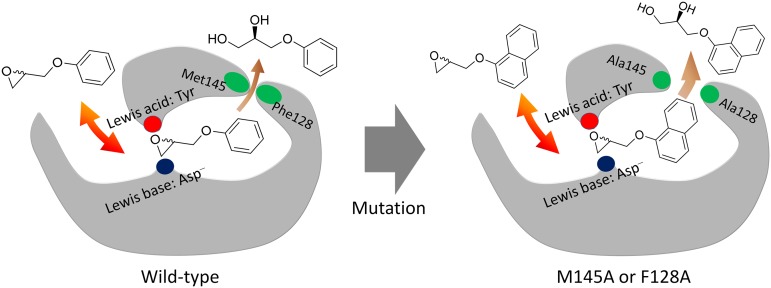
A proposed model for bulky (R)-diol production by the M145A and F128A BmEH variants. The M145A or F128A mutation expanded the predicted product release site of BmEH, which would facilitate the hydrolysis of bulky NGE. Tyrosines (Tyr-144 and -203) and aspartic acid (Asp-97) were served as Lewis acid and Lewis base, respectively.
Compared with the existing methods for rational protein engineering, targeting the substrate-access or the transition-state sites (37, 38), the “two birds with one stone” approach described herein may might be advantageous for increasing the catalytic activity of the target enzyme and for reducing its product inhibition. With the rapid growth of new enzyme structures and development of molecular modeling dynamics software, more and more valuable enzymes might be modified by this approach.
Materials and Methods
Chemical Reagents.
PGE was purchased from TCI. The 2-phenoxyacetamide was bought from Alfa Aesar. Racemic NGE was prepared by using corresponding phenols and epichlorohydrin as reported (22). (R)- and (S)-NGE were prepared with a preparative-scale HPLC (Waters) equipped with an OD-H 10 × 250 mm (Daicel) column. (R)-NPD {(R)-3-[1]-naphthyloxy-propane-1,2-diol}, the hydrolyzed product of (R)-NGE, was prepared in the laboratory of J.-H.X. All other chemicals were obtained commercially.
Expression and Purification of BmEH.
The wild-type and mutant BmEH from B. megaterium strain ECU1001 were expressed as N-terminal His-tagged proteins in Escherichia coli BL21(DE3) as described (22). All mutations were generated by PCR using the QuikChange method (Strategene) and confirmed by DNA sequencing. Proteins were purified by using one Ni-NTA columns, followed by thrombin protease treatment, and one gel filtration column (Superdex 75 Hiload 16/60; GE Healthcare). Details are described in SI Appendix. Selenomethionine-substituted (SeMet) BmEH was expressed via the methionine inhibitory pathway (39) and purified similarly to the wild-type protein.
Crystallization, Data Collection, and Structure Determination.
Rhombohedral crystals of BmEH were grown at 20 °C by using the sitting-drop vapor diffusion method by mixing the protein (20 mg/mL) with an equal volume of reservoir solution containing 0.5 M LiCl, 0.1 M Tris⋅HCl (pH 6.5), and 25% (wt/vol) PEG 6000. The SeMet BmEH was crystallized under the same condition. Crystals of the BmEH–POA complex were prepared by cocrystallization of 5 mM POA in the protein solution and grown under the condition of 0.2 M Li2SO4, 0.1 M Tris⋅HCl (pH 7.0), and 1.6 M (NH4)2SO4. The M145A mutant was crystallized under the condition of 0.2 M Li2SO4, 0.1 M Tris⋅HCl (pH 8.5), 34% (wt/vol) PGE 3000. The complex of F128A and (R)-NPD was made by cocrystallizing the mutant enzyme with 5 mM (R)-NGE under the condition of 0.2 M Li2SO4, 0.1 M Tris⋅HCl (pH 7.5), and 2.0 M (NH4)2SO4. The hydrolysis of (R)-NGE in the crystallization process afforded the product (R)-NGE. Before data collection, crystals were soaked in Paratone-N (Hampton Research) and then flash-cooled in liquid nitrogen.
Diffraction data of the native and SeMet BmEH were collected at the wavelength of 0.9791 Å by using a MX-225 CCD detector and an ADSC Quantum 315r detector, respectively, at beamline BL17U of the Shanghai Synchrotron Radiation Facility. Diffraction data of the BmEH–POA complex, BmEHM145A, and BmEHF128A–(R)-NPD complex were collected at the wavelength of 1.5418 Å on a RaxisIV++ imaging plate detector. All datasets were indexed, integrated, and scaled by using the HKL2000 package (40). The single-wavelength anomalous diffraction phases were calculated with SHELX C/D/E (41). A model of BmEH was built automatically by ARP/wARP (42) and manually adjusted by using COOT (43). Rounds of automated refinement were performed with PHENIX (44). The structure of the BmEH–POA complex was solved by molecular replacement method using the program PHASER (45). The atomic models of BmEH, BmEH–POA complex, BmEHM145A, and the BmEHF128A–(R)-NPD complex have been refined to 1.85, 1.7, 1.95, 1.7, and 2.9 Å, separately. Crystallographic statistics are summarized in SI Appendix, Table S3. All figures of the protein models were prepared with Pymol (www.pymol.org/).
Activity Assay.
The specific activities of BmEH proteins were measured by monitoring the conversion of substrate by HPLC. Details of the measurements are provided in SI Appendix. Determination of the enantiomeric excesses of epoxides and diols were performed as reported (22).
Mass Spectrometric Analyses.
The BmEH variants with H267F mutation at a concentration of 1 mg/mL (0.028 mM) were incubated with substrate NGE (0.1 mM) for 10 min. The BmEH protein solution was then loaded onto a homemade capillary column (150 μm inner diameter, 3 cm long) packed with Poros R2 medium (AB-Sciex). The BmEH proteins were eluted by an Agilent 1100 binary pump system with the following solvent gradient: 0–100% B in 60 min (A = 0.1 M acetic acid in water; B = 0.1 M acetic acid/40% acetonitrile/40% isopropanol). The eluted proteins were sprayed into a QSTAR XL mass spectrometer (AB-Sciex) equipped with a Turbo Electrospray ion source. The instrument was acquired in MS mode under 5K volts spray voltage. The protein charge envelop was averaged across the corresponding protein elution peaks and deconvoluted into noncharged forms by the BioAnalyst software provided by the manufacturer.
Stopped-Flow Data Acquisition and Analysis.
The stopped-flow experiments were conducted on an Applied Photophysics model SX20 stopped-flow spectrofluorimeter fitted with a Xenon lamp. Samples were excited at 310 nm, and the fluorescence difference of NGE and NPD was observed at 380 nm through a monochromator. All reactions were performed in 100 mM potassium phosphate buffer (pH 7.0) containing 10% DMSO at 25 °C with enzyme concentrations of 25 μM and substrate concentrations of 125 μM. Volumes of 80 μL were injected from each syringe, and the reported concentrations are those in the reaction chamber. A path length of 1 cm was used throughout the stopped-flow experiments.
Chemoenzymatic Synthesis of (S)-Propranolol.
Gram-scale bioresolution of NGE was conducted by mixing crude enzyme extracted from ∼5 g of fresh cell pellets and 4 g of racemic NGE in a 500 mL conical flask. The reaction was terminated at ∼16 h, and the mixture was saturated by adding sodium chloride followed by extracting with ethyl acetate (150 mL × 3). The resultant mixture with ees of 99% and eep of 91% was purified by using a silica gel column, affording the enantiomerically pure epoxide (99% ee) and optically enriched diol (88% ee). The obtained enantiopure (S)-NGE was refluxed in isopropylamine for 48 h, producing 1.86 g of (S)-propranolol (36% yield after recrystallization).
1H NMR (400 MHz, CDCl3) δ 1.15 (6H, d, J = 6.3 Hz), 2.84–3.01 (3H, m), 3.03 (2H, d, J = 12.4), 4.15–4.23 (3H, m), 6.83 (1H, d, J = 7.4 Hz), 7.40–7.55 (4H, m), 7.84 (1H, m), 8.32 (1H, m).
Computational Modeling of the BmEH Active Tunnel.
Structures of the BmEH–POA complex and the BmEHF128A–(R)-NPD complex were imported into the Maestro program (Version 9.2) (46). All MD simulations were performed by using the Schrodinger 2011-based Desmond package (47). Eighty nanoseconds for the BmEH–POA complex and 80 ns for the BmEHF128A–(R)-NPD complex production MD on each system were executed. A total of 100 structures of BmEH extracted throughout the whole MD simulation trajectories were used for CAVER (48) input files. Output from CAVER was finally visualized and rendered in Pymol. Details of computational modeling are provided in SI Appendix.
Supplementary Material
Acknowledgments
We are grateful for access to beamline BL17U1 at Shanghai Synchrotron Radiation Facility and thank the beamline staff for technical support. We also thank Dr. Peter K. Park and Profs. Zhihong Guo and Ran Hong for helpful discussions. This work was supported by National Program on Key Basic Research of China Grant 2011CB710800 (to J.-H.X. and J.Z.), National Grand Project for Medicine Innovation Grant 2012ZX10002006 (to J.Z.), National Natural Science Foundation of China Grant 21276082 (to J.-H.X.), and a grant from the Open Fund from the State Key Laboratory of Bioreactor Engineering (to J.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4G00, 4G02, 4INZ, and 4IO0).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404915111/-/DCSupplemental.
References
- 1.Archelas A, Furstoss R. Synthetic applications of epoxide hydrolases. Curr Opin Chem Biol. 2001;5(2):112–119. doi: 10.1016/s1367-5931(00)00179-4. [DOI] [PubMed] [Google Scholar]
- 2.Tokunaga M, Larrow JF, Kakiuchi F, Jacobsen EN. Asymmetric catalysis with water: Efficient kinetic resolution of terminal epoxides by means of catalytic hydrolysis. Science. 1997;277(5328):936–938. doi: 10.1126/science.277.5328.936. [DOI] [PubMed] [Google Scholar]
- 3.Schmid A, Hofstetter K, Feiten HJ, Hollmann F, Witholt B. Integrated biocatalytic synthesis on Gram scale: The highly enantioselective preparation of chiral oxiranes with styrene monooxygenase. Adv Synth Catal. 2001;343:732–737. [Google Scholar]
- 4.de Vries EJ, Janssen DB. Biocatalytic conversion of epoxides. Curr Opin Biotechnol. 2003;14(4):414–420. doi: 10.1016/s0958-1669(03)00102-2. [DOI] [PubMed] [Google Scholar]
- 5.Choi WJ, Choi CY. Production of chiral epoxides: Epoxide hydrolase-catalyzed enantioselective hydrolysis. Biotechnol Bioprocess Eng. 2005;10:167–179. [Google Scholar]
- 6.Sareen D, Kumar R. Prospecting for efficient enantioselective epoxide hydrolases. Indian J Biotechnol. 2011;10:161–177. [Google Scholar]
- 7.Reetz MT, Zheng H. Manipulating the expression rate and enantioselectivity of an epoxide hydrolase by using directed evolution. ChemBioChem. 2011;12(10):1529–1535. doi: 10.1002/cbic.201100078. [DOI] [PubMed] [Google Scholar]
- 8.Gurell A, Widersten M. Modification of substrate specificity resulting in an epoxide hydrolase with shifted enantiopreference for (2,3-epoxypropyl)benzene. ChemBioChem. 2010;11(10):1422–1429. doi: 10.1002/cbic.201000185. [DOI] [PubMed] [Google Scholar]
- 9.Lee EY, Shuler ML. Molecular engineering of epoxide hydrolase and its application to asymmetric and enantioconvergent hydrolysis. Biotechnol Bioeng. 2007;98(2):318–327. doi: 10.1002/bit.21444. [DOI] [PubMed] [Google Scholar]
- 10.Solares LF, Mateo C. Improvement of the epoxide hydrolase properties for the enantioselective hydrolysis of epoxides. Curr Org Chem. 2013;17(7):744–755. [Google Scholar]
- 11.Rui L, Cao L, Chen W, Reardon KF, Wood TK. Protein engineering of epoxide hydrolase from Agrobacterium radiobacter AD1 for enhanced activity and enantioselective production of (R)-1-phenylethane-1,2-diol. Appl Environ Microbiol. 2005;71(7):3995–4003. doi: 10.1128/AEM.71.7.3995-4003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Loo B, et al. Directed evolution of epoxide hydrolase from A. radiobacter toward higher enantioselectivity by error-prone PCR and DNA shuffling. Chem Biol. 2004;11(7):981–990. doi: 10.1016/j.chembiol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 13.Reetz MT, et al. Enhancing the enantioselectivity of an epoxide hydrolase by directed evolution. Org Lett. 2004;6(2):177–180. doi: 10.1021/ol035898m. [DOI] [PubMed] [Google Scholar]
- 14.van Loo B, et al. Improved enantioselective conversion of styrene epoxides and meso-epoxides through epoxide hydrolases with a mutated nucleophile-flanking residue. Enzyme Microb Technol. 2009;44:145–153. [Google Scholar]
- 15.Choi SH, Kim HS, Lee EY. Multiple sequence alignment-inspired mutagenesis of marine epoxide hydrolase of Mugil cephalus for enhancing enantioselective hydrolytic activity. J Ind Eng Chem. 2012;18:72–76. [Google Scholar]
- 16.Thomaeus A, Naworyta A, Mowbray SL, Widersten M. Removal of distal protein-water hydrogen bonds in a plant epoxide hydrolase increases catalytic turnover but decreases thermostability. Protein Sci. 2008;17(7):1275–1284. doi: 10.1110/ps.034173.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reetz MT, et al. Directed evolution of an enantioselective epoxide hydrolase: Uncovering the source of enantioselectivity at each evolutionary stage. J Am Chem Soc. 2009;131(21):7334–7343. doi: 10.1021/ja809673d. [DOI] [PubMed] [Google Scholar]
- 18.Choi WJ. Biotechnological production of enantiopure epoxides by enzymatic kinetic resolution. Appl Microbiol Biotechnol. 2009;84(2):239–247. doi: 10.1007/s00253-009-2110-9. [DOI] [PubMed] [Google Scholar]
- 19.Hwang S, Choi CY, Lee EY. Bio- and chemo-catalytic preparations of chiral epoxides. J Ind Eng Chem. 2010;16(1):1–6. [Google Scholar]
- 20.Cordato DJ, Mather LE, Herkes GK. Stereochemistry in clinical medicine: A neurological perspective. J Clin Neurosci. 2003;10(6):649–654. doi: 10.1016/j.jocn.2002.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Zelaszczyk D, Kieć-Kononowicz K. Biocatalytic approaches to optically active beta-blockers. Curr Med Chem. 2007;14(1):53–65. doi: 10.2174/092986707779313480. [DOI] [PubMed] [Google Scholar]
- 22.Zhao J, et al. An unusual (R)-selective epoxide hydrolase with high activity for facile preparation of enantiopure glycidyl ethers. Adv Synth Catal. 2011;353(9):1510–1518. [Google Scholar]
- 23.Gomez GA, Morisseau C, Hammock BD, Christianson DW. Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis. Biochemistry. 2004;43(16):4716–4723. doi: 10.1021/bi036189j. [DOI] [PubMed] [Google Scholar]
- 24.Argiriadi MA, Morisseau C, Hammock BD, Christianson DW. Detoxification of environmental mutagens and carcinogens: Structure, mechanism, and evolution of liver epoxide hydrolase. Proc Natl Acad Sci USA. 1999;96(19):10637–10642. doi: 10.1073/pnas.96.19.10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biswal BK, et al. The molecular structure of epoxide hydrolase B from Mycobacterium tuberculosis and its complex with a urea-based inhibitor. J Mol Biol. 2008;381(4):897–912. doi: 10.1016/j.jmb.2008.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou J, et al. Structure of Aspergillus niger epoxide hydrolase at 1.8 A resolution: Implications for the structure and function of the mammalian microsomal class of epoxide hydrolases. Structure. 2000;8(2):111–122. doi: 10.1016/s0969-2126(00)00087-3. [DOI] [PubMed] [Google Scholar]
- 27.Nardini M, et al. The x-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1. An enzyme to detoxify harmful epoxides. J Biol Chem. 1999;274(21):14579–14586. [PubMed] [Google Scholar]
- 28.Mowbray SL, Elfström LT, Ahlgren KM, Andersson CE, Widersten M. X-ray structure of potato epoxide hydrolase sheds light on substrate specificity in plant enzymes. Protein Sci. 2006;15(7):1628–1637. doi: 10.1110/ps.051792106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong XD, Ma Q, Zhou J, Zeng BB, Xu JH. A smart library of epoxide hydrolase variants and the top hits for synthesis of (S)-β-blocker precursors. Angew Chem Int Ed Engl. 2014;53(26):6641–6644. doi: 10.1002/anie.201402653. [DOI] [PubMed] [Google Scholar]
- 30.Ma B, Nussinov R. Enzyme dynamics point to stepwise conformational selection in catalysis. Curr Opin Chem Biol. 2010;14(5):652–659. doi: 10.1016/j.cbpa.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lüdemann SK, Lounnas V, Wade RC. How do substrates enter and products exit the buried active site of cytochrome P450cam? 1. Random expulsion molecular dynamics investigation of ligand access channels and mechanisms. J Mol Biol. 2000;303(5):797–811. doi: 10.1006/jmbi.2000.4154. [DOI] [PubMed] [Google Scholar]
- 32.Köhler A, et al. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol Cell. 2001;7(6):1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 33.Mendes KR, Kantrowitz ER. The pathway of product release from the R state of aspartate transcarbamoylase. J Mol Biol. 2010;401(5):940–948. doi: 10.1016/j.jmb.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klvana M, et al. Pathways and mechanisms for product release in the engineered haloalkane dehalogenases explored using classical and random acceleration molecular dynamics simulations. J Mol Biol. 2009;392(5):1339–1356. doi: 10.1016/j.jmb.2009.06.076. [DOI] [PubMed] [Google Scholar]
- 35.Wen Z, Baudry J, Berenbaum MR, Schuler MA. Ile115Leu mutation in the SRS1 region of an insect cytochrome P450 (CYP6B1) compromises substrate turnover via changes in a predicted product release channel. Protein Eng Des Sel. 2005;18(4):191–199. doi: 10.1093/protein/gzi023. [DOI] [PubMed] [Google Scholar]
- 36.Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE. Quantitative NMR spectroscopy of supramolecular complexes: Dynamic side pores in ClpP are important for product release. Proc Natl Acad Sci USA. 2005;102(46):16678–16683. doi: 10.1073/pnas.0507370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazlauskas RJ, Bornscheuer UT. Finding better protein engineering strategies. Nat Chem Biol. 2009;5(8):526–529. doi: 10.1038/nchembio0809-526. [DOI] [PubMed] [Google Scholar]
- 38.Morley KL, Kazlauskas RJ. Improving enzyme properties: When are closer mutations better? Trends Biotechnol. 2005;23(5):231–237. doi: 10.1016/j.tibtech.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229(1):105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology, ed Carter, Jr CW (Academic, New York), Vol 276, pp 307–326.
- 41.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58(10-2):1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 42.Mooij WTM, Cohen SX, Joosten K, Murshudov GN, Perrakis A. “Conditional restraints”: Restraining the free atoms in ARP/wARP. Structure. 2009;17(2):183–189. doi: 10.1016/j.str.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 45.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banks JL, et al. Integrated modeling program, applied chemical theory (IMPACT) J Comput Chem. 2005;26(16):1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shan Y, et al. A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc Natl Acad Sci USA. 2009;106(1):139–144. doi: 10.1073/pnas.0811223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chovancová E, et al. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.