Significance
Streptococcus mutans adhesin P1 is a target of protective immunity and a vaccine candidate. P1’s complex structure dictates its function and makes it of interest from a protein folding perspective as well. An interaction between N- and C-terminal sequences contributes to antigenicity, adherence behavior, and stability. This is now explained by the identification of a previously unidentified fold in which the N terminus forms a stabilizing scaffold at the base of P1’s helical stalk to physically lock it in place via interactions with the C terminus. Disruption of this intramolecular lock not only negatively affects stability, but also prevents proper folding of the purified full-length protein.
Keywords: X-ray crystallography, protein folding, adhesin, Streptococcus, intramolecular lock
Abstract
The cariogenic bacterium Streptococcus mutans uses adhesin P1 to adhere to tooth surfaces, extracellular matrix components, and other bacteria. A composite model of P1 based on partial crystal structures revealed an unusual complex architecture in which the protein forms an elongated hybrid alpha/polyproline type II helical stalk by folding back on itself to display a globular head at the apex and a globular C-terminal region at the base. The structure of P1’s N terminus and the nature of its critical interaction with the C-terminal region remained unknown, however. We have cocrystallized a stable complex of recombinant N- and C-terminal fragments and here describe a previously unidentified topological fold in which these widely discontinuous domains are intimately associated. The structure reveals that the N terminus forms a stabilizing scaffold by wrapping behind the base of P1’s elongated stalk and physically “locking” it into place. The structure is stabilized through a highly favorable ΔGsolvation on complex formation, along with extensive hydrogen bonding. We confirm the functional relevance of this intramolecular interaction using differential scanning calorimetry and circular dichroism to show that disruption of the proper spacing of residues 989–1001 impedes folding and diminishes stability of the full-length molecule, including the stalk. Our findings clarify previously unexplained functional and antigenic properties of P1.
Streptococcus mutans is a recognized cause of human dental caries (cavities), the most common infectious disease worldwide (1). Identifying how S. mutans interacts with host components at the molecular level is essential to fully understand its virulence properties. The sucrose-independent adhesin P1 (AgI/II, antigen B, PAc) is localized on the surface of this oral pathogen, along with many other streptococci (2–7). In the oral cavity, S. mutans P1 interacts with the salivary agglutinin glycoprotein complex composed predominantly of scavenger receptor gp340/DMBT1 (2, 3, 5–10). Without a complete structural model, the mechanisms by which P1 binds to host components have not yet been fully characterized.
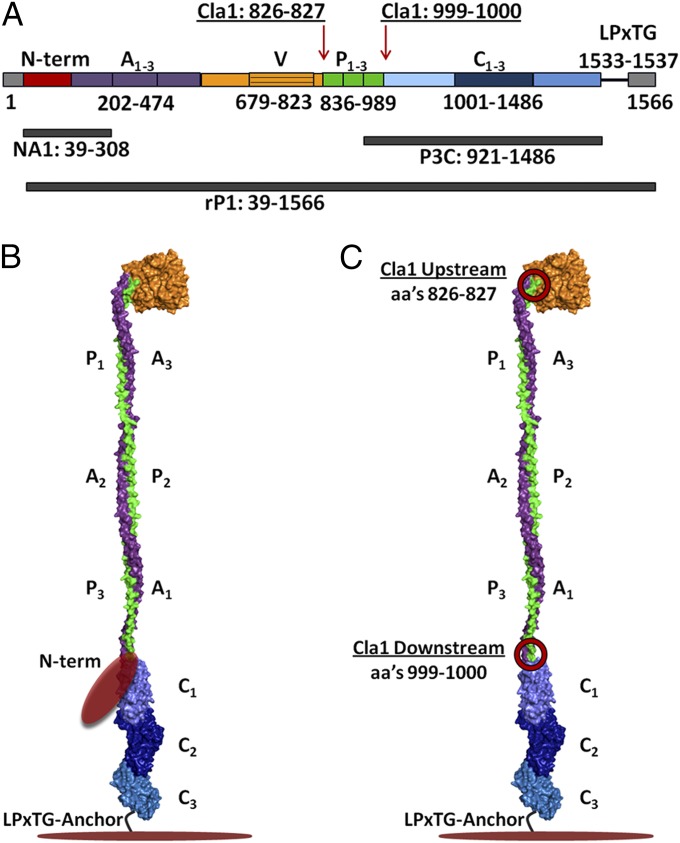
P1’s primary structure (Fig. 1A) contains a 38-residue signal sequence, the heretofore uncharacterized N-terminal region, three alanine-rich repeats (A1–3), a central domain containing a so-called variable (V) region (11), three proline-rich repeats (P1–3), a C-terminal region consisting of three domains (C1–3), an LPxTG sortase-recognition motif, and wall- and membrane-spanning regions (12, 13). Recent partial X-ray crystal structure and velocity centrifugation studies of the intact protein unveiled a unique architecture in which the ∼185-kDa (1,561-aa) protein folds back on itself to form a ∼50-nm elongated hybrid helical stalk that separates two independent adherence domains, with a globular head at the apex and a globular C-terminal region at the base (13–15) (Fig. 1B).
Fig. 1.
Schematic representation of the primary and modeled tertiary structure of P1. (A) Primary structure of P1 and location of polypeptides used in this study. (B) Proposed tertiary model of P1 based on velocity centrifugation and crystal structures of A3VP1 and C123 fragments (13). (C) Diagram showing the locations of the two engineered Cla1 sites (circled in red) that added isoleucine and aspartic acid residues to either side of the proline-rich region (20).
The crystal structure of the third alanine-rich repeat through the first proline-rich repeat first revealed the unusual interaction between the A and P regions to form a hybrid alpha/polyproline type II helix (14). In this model, a globular β super sandwich domain sits at the apex of the molecule (15). At the other end of the hybrid helix, the three contiguous domains of the C-terminal region each adopt a DE-variant Ig-like (DEv-IgG) fold stabilized by isopeptide bonds (13, 16). Despite this recent progress, the structure of the ∼20-kDa N terminus of P1 remained unknown, however (Fig. 1B). We previously demonstrated that proper folding and function of P1 on the surface of S. mutans requires an interaction between N- and C-terminal segments (17), thus increasing the imperative to elucidate the structure of the N terminus in complex with its intramolecular binding partner.
For this, we used the recombinant N-terminal (NA1) and C-terminal (P3C) P1 fragments (Fig. 1A), which have been shown to form a stable high-affinity and functionally active complex (17). The NA1/P3C protein complex was copurified and then cocrystallized for X-ray diffraction data collection to 2.0-Å resolution. We observed that the N terminus adopts a previously unidentified fold that serves as an intramolecular scaffold connecting it to C-terminal portions of the molecule, thus locking P1’s hybrid helical stalk into place. We validated this model experimentally using differential scanning calorimetry (DSC) and circular dichroism to demonstrate decreased thermal stability and altered secondary structure in a P1 mutant containing two extra amino acids within the region that normally reacts with the N-terminal intramolecular scaffold. Our X-ray crystallography model and stability measurements agree well with biophysical data characterizing the NA1/P3C complex (17) and provide mechanistic insight into why the N-terminal segment supports the proper folding, function, and stability of the full-length P1 protein. This information contributes to our ability to interpret data regarding protective and nonprotective immune responses and preventative therapies, and will inform future studies evaluating bacterial adhesion and biofilm formation by S. mutans and related organisms.
Results
Crystallization and Structure Determination.
Recombinant P1 polypeptides NA1 and P3C (Fig. S1A) were copurified and crystallized together. A complete dataset was collected from a single crystal that diffracted to 2.0-Å resolution and was assigned to the space group C2 with unit cell parameters a = 197.5, b = 68.9, c = 81.3 Å, and β = 96.8°, consistent with the protein’s elongated structure. Data collection and refinement statistics are provided in Table 1. In the structural overlay, the C-terminal portion of our model (C123) is nearly identical to that described previously (13), with an rmsd of 0.52 Å (Fig. 2C). The sole difference is a lack of ordered electron density corresponding to a surface loop containing residues 1342–1363 within C3. Two calcium ions were coordinated at identical positions within C2, as reported previously (13). The calcium ion observed in C3 of previously published C-terminal structures (13, 16) was not observed. We were able to build our model beginning from the 13th residue after the P1 signal sequence cleavage site. Ordered electron density was observed for residues 52–172 within NA1 and residues 980–1486 within P3C; thus, these regions were modeled in the structure.
Table 1.
Crystallography data collection and refinement statistics
| Parameter | NA1/P3C |
| Temperature, K | 93 |
| PDB ID code | 4TSH |
| Space group | C2 |
| Unit-cell parameters, Å; ° | a = 197.5, b = 68.9, c = 81.3; β = 96.8 |
| Resolution, Å | 35.9–2.0 (2.07–2.00)* |
| Rsym, %† | 9.0 (64.9) |
| I/σ, I | 18.5 (2.0) |
| Completeness, % | 99.7 (97.0) |
| Average redundancy | 3.7 (3.6) |
| Number of unique reflections | 73,048 |
| Rcryst‡/Rfree§, % | 17.1/20.8 |
| No. of atoms, protein | 4,692 |
| Water | 545 |
| Ions | 4 |
| B-factors, Å2 | |
| Main chain/side chains/solvent/ion | 39.0/48.9/49.6/29.5 |
| rmsd, bond (Å)/angle | 0.012/1.22 |
| Ramachandran statistics, % | |
| Most favored, allowed | 98.5, 1.5 |
Values in parentheses represent the highest-resolution bin.
Rsym = (Σ |I − <I>|/ Σ <I>) × 100.
Rcryst = (Σ |Fo| − |Fc|/ Σ |Fo|) × 100.
Rfree is calculated in same manner as Rcryst, except that it uses 2.7% of the reflection data (2,000 reflections) excluded from refinement.
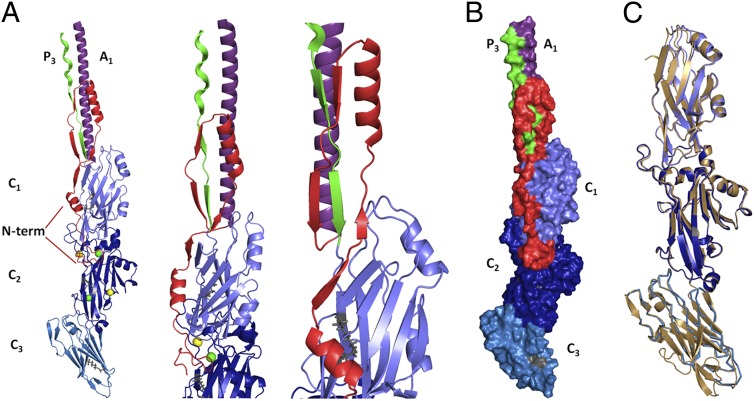
Fig. 2.
Multiple views of the N terminus in complex with the C-terminal region of P1 and comparison with the previously published structure of the C terminus of S. mutans AgI/II. (A) Three separate views displaying the interaction of the N terminus with the C-terminal region. The N terminus interacts with the C terminus at the C1/C2 interface (amino acids 1114–1158) and immediately upstream of the C2/C3 interface (amino acids 1321–1323). This segment continues past the C1 domain to form a previously unidentified fold that wraps behind the base of P1’s hybrid helical stalk. The N terminus is shown in red, the alanine-rich region is in purple, the proline-rich and immediate downstream region is in green, and each domain of the C terminus is in a different shade of blue. Calcium ions identified within the structure are shown in yellow. Magnesium ions are shown in green. (B) Space-filling model displaying the interface of the N terminus (red) with the C-terminal domains. (C) Structure of the C123 domains within the NA1/P3C complex (blue) compared with the previously published C terminus (gold) of S. mutans Ag I/II (13) superimposed with an rmsd of 0.52 Å.
Overall Structure.
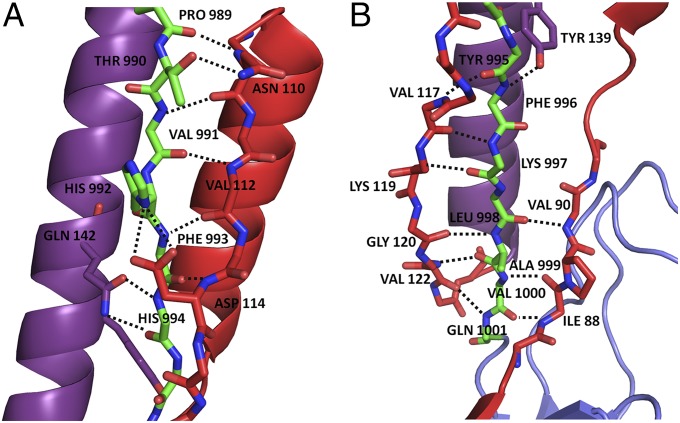
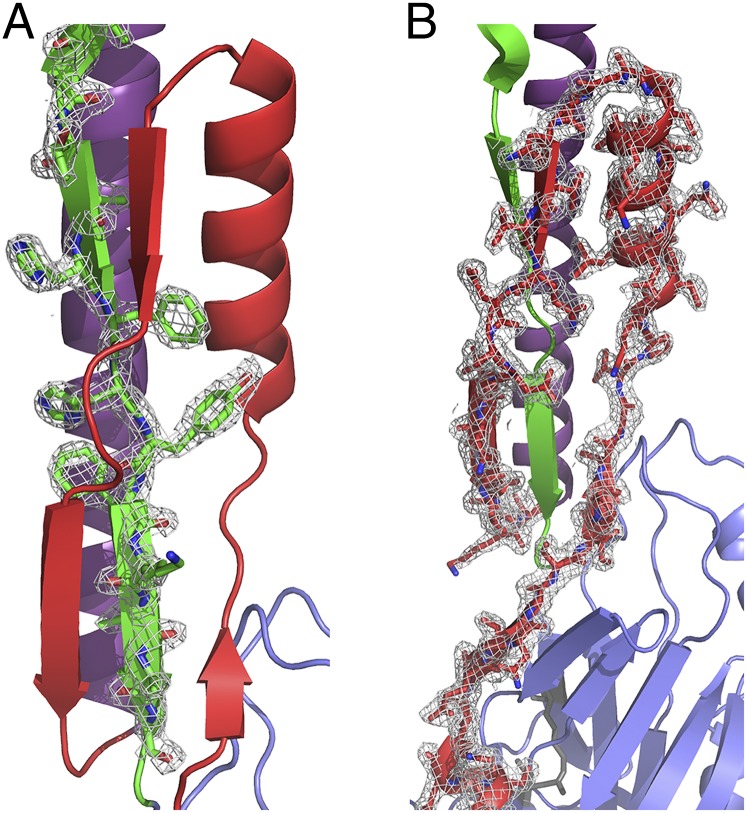
The structure of the N terminus of P1 was determined in complex with the C terminus. This structure displays a previously unidentified topological fold in which an extensive loop runs parallel to the base of P1’s elongated hybrid helical stalk and acts to support the stalk (Fig. 2A). An omit map displays a single polypeptide chain of the N terminus forming a loop around the post-P3 region, as modeled into our structure (Fig. S2). Within the modeled structure, the N terminus forms two alpha helices (amino acids 72–79 and 95–106) and four β-strands (amino acids 84–86, 88–90, 110–113, and 117–122) (Table S1). These secondary structure elements form numerous contacts with C2 and C1, as well as with the scaffolding loop visible immediately downstream of the proline-rich region at the base of the stalk (Fig. 2A). The interface between the N- and C-terminal segments continues along C1/C2, ending just before the junction of C2/C3 (Fig. 2B). As estimated by PISA (Protein Interfaces, Surfaces, and Assemblies) (18), the interactions between NA1 and P3C are stabilized by a highly energetically favorable ∆Gsolvation of −32.5 kcal/mol, along with 39 hydrogen bonds and three salt bridges. Twenty-three of the 42 stabilizing bonds are formed between the N terminus and the region immediately C-terminal to the proline-rich region (amino acids 989–1001) (Fig. 3 A and B), revealing the specific molecular nature of the previously observed high-affinity interaction (17). A string of bulky residues, HFHYFK (amino acids 992–997), lies in alternating directions directly within the “eye” of the loop formed by the N terminus (Fig. 4) and may provide the necessary steric constraints to promote the formation of stabilizing interactions within the N terminus and help position it in place around the base of the stalk. Ten of the remaining 19 H bonds that bridge NA1 and P3C lie at the interface of C1/C2 (amino acids 1114–1158); the others occur within C1 (amino acids 1023–1054) or just before the interface of C2/C3 (amino acids 1321–1323).
Fig. 3.
Illustration of the stabilizing bonds, represented by dotted black lines, between residues 989–1001 of P1’s postproline-rich region and the N-terminal intramolecular scaffold. The N terminus is shown in red, the alanine-rich region is in purple, and the postproline-rich region is in green. Side chains not participating in stabilizing interactions have been deleted for clarity. (A) Top portion of the N-terminal scaffold, stabilizing residues 989–994 of the postproline region. (B) Bottom portion of the N-terminal scaffold, stabilizing residues 995–1001 of the postproline-rich region.
Fig. 4.
A 2Fo-Fc map contoured at 2σ, displaying the observed electron density of the postproline-rich region and the N terminus. The N terminus is shown in red, the alanine-rich region is in purple, and the postproline-rich region is in green. (A) The postproline-rich region displays a high number of stabilizing bonds with the N-terminal intramolecular scaffold. The orientation of HFHYFK residues (amino acids 992–997) with side chains lying in alternating directions within the “eye” of the N-terminal topological loop are illustrated. (B) The N-terminal scaffold intimately interacts with and wraps behind the postproline-rich region.
Thermal Stability of P1 Mutants.
To experimentally validate the stabilizing contacts observed in our crystal model, we took advantage of a previously constructed mutant polypeptide in which amino acid spacing immediately downstream of the P region was placed out of register by the addition of intervening isoleucine and aspartic acid residues introduced via Cla1 restrictions sites used to reintroduce DNA encoding the P region into a deletion construct used in an earlier study (19, 20). This complemented mutant construct was unstable when reintroduced into S. mutans via a shuttle vector and was not properly localized to the cell surface (20). In addition, the complemented recombinant polypeptide exhibited altered antigenicity and no longer interacted with immobilized salivary agglutinin (17). These results now make sense in light of the NA1/P3C crystal structure.
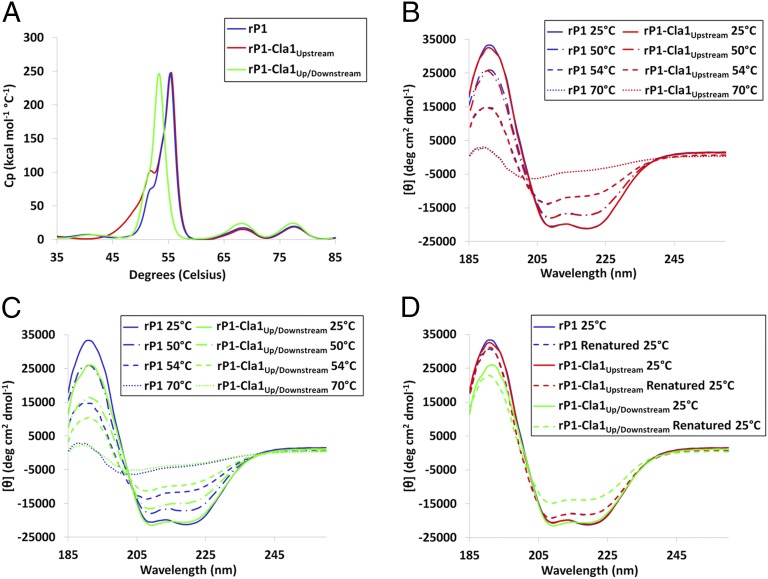
To confirm that these amino acid additions interfered with stabilizing interactions between the N terminus and C terminus, we performed thermal denaturation experiments using full-length rP1 and two different mutants, the original PC967 complementation polypeptide (20) containing an added Cla1 site on either side of the P region, referred to here as rP1-Cla1Up/Downstream, as well another polypeptide in which the downstream Cla1 site was eliminated, referred to as rP1-Cla1Upstream. Recombinant P1/Cla1Upstream contains added isoleucine and aspartic acid residues at positions 826 and 827 immediately upstream of the P region near the apex of the molecule, whereas rP1-Cla1Up/Downstream contains these residues as well as added isoleucine and aspartic acid residues at positions 999–1000 downstream of the P region and immediately before the C1 domain (Fig. 1C and Fig. S1B).
Based on our crystal model, the isoleucine and aspartic acid residues added at positions 999–1000 fall directly within the portion of P1 that is intimately associated with the N-terminal scaffold at the base of the protein’s hybrid helical stalk. Using DSC, we compared the thermal denaturation profiles of purified rP1, rP1-Cla1Upstream, and rP1-Cla1Up/Downstream to measure the ΔH of unfolding between 30 and 85 °C (Fig. 5A). The melting curves of rP1 and rP1-Cla1Upstream were essentially identical, both displaying large transitions occurring at the same temperature between 50 and 60 °C, whereas rP1-Cla1Up/Downstream displayed a shift of approximately −2.2 °C for the largest transition compared with rP1 or rP1-Cla1Upstream. The lower transition temperature for the largest unfolding event between 50 and 60 °C in rP1-Cla1Up/Downstream compared with WT rP1 or rP1-Cla1Upstream indicates that the insertion of two additional amino acids directly within the region of the C terminus found to interact with the stabilizing loop formed by the N terminus in fact has a negative affect on the stability of the full-length P1 molecule. In contrast, the addition of extra isoleucine and aspartic acid residues at the opposite end of the folded stalk has no effect on protein stability (Fig. 5A).
Fig. 5.
Thermal stability and refolding after thermal denaturation of rP1-Cla1Upstream and rP1-Cla1Up/Downstream compared with rP1. (A) The melting curves of rP1-Cla1Upstream, rP1-Cla1Up/Downstream, and full-length rP1 during thermal denaturation as measured by DSC. (B and C) Changes in circular dichroism spectra during thermal denaturation of rP1-Cla1Upstream compared with rP1 (B) and rP1-Cla1Up/Downstream compared with rP1 (C). Far-UV CD spectra were measured from 260 to 185 nm over a temperature range of 25–70 °C. Representative spectra are shown at specific temperatures during thermal denaturation. (D) Circular dichroism evaluation of refolded rP1-Cla1Upstream and rP1-Cla1Up/Downstream compared with rP1 after thermal denaturation. Far-UV CD spectra were measured from 260 to 185 nm at 25 °C to compare the polypeptides with and without previous thermal denaturation at 70 °C.
Changes in Secondary Structure and Refolding Following Thermal Denaturation.
CD spectroscopy was used to gain insight into which regions of rP1-Cla1Up/Downstream are less stable than rP1-Cla1Upstream and rP1, by measuring the CD spectrum of each protein from 25 °C to 70 °C to estimate changes in secondary structure owing to thermal denaturation. The initial CD spectra of rP1-Cla1Up/Downstream compared with rP1 and rP1-Cla1Upstream at 25 °C displayed slight but observable differences in local minima and maxima, suggesting differences in the secondary structure of the proteins (Fig. S3), but subsequent deconvolution of these spectra did not identify the exact differences (Fig. S3, Inset). On heating, however, the difference in stability of rP1-Cla1Up/Downstream compared with rP1 and rP1-Cla1Upstream became readily apparent (Fig. S4). The CD spectra indicated that rP1-Cla1Up/Downstream melted out significantly faster than either rP1 or rP1-Cla1Upstream (Fig. 5 B and C). Deconvolution of the CD spectra confirmed that rP1-Cla1Up/Downstream denatures at ∼2 °C lower than either rP1 or rP1-Cla1Upstream (Table S2). Much of the unfolding of P1 that occurs on heating appears to be attributable to a loss of helical character between 50 and 60 °C, which would correlate with the largest transition observed in the DSC spectra.
We also compared the refolding of rP1-Cla1Up/Downstream with that of rP1 and rP1-Cla1Upstream following thermal denaturation. On renaturation, rP1-Cla1Up/Downstream displayed a substantially different CD spectrum than rP1 or rP1-Cla1Upstream (Fig. 5D). Deconvolution of the CD spectra again revealed that much of the difference in secondary structure is the result of decreased helical content (Table S3). Because the majority of P1’s helical character lies within its hybrid helical stalk, these data are consistent with the assumption that this region of rP1-Cla1Up/Downstream is less stable, and thus less amenable to proper refolding, compared with the full-length molecule or rP1-Cla1Upstream. Taken together, these results suggest that the introduction of two extra amino acids in the region of P1 immediately before C1 disrupts the register of residues that interact directly with the “eye” of the N-terminal scaffolding loop and prevents formation of the molecular lock identified in the crystal model.
Discussion
Recent crystal structures have enabled construction of a nearly complete tertiary model of S. mutans P1 (13–16); however, the N terminus and nature of its integral association with the C-terminal region remained uncharacterized. Our crystal structure lends insight into why the N-terminal segment of P1 has such a pronounced impact on its immunogenicity, antigenicity, folding, stability, and adherent function (17, 21–26), The NA1/P3C complex contains an elongated alpha/polyproline type II hybrid helix similar to that identified in the crystallized A3VP1 fragment (14), except that in this case it comprises the A1 and P3 repeats rather than the A3 and P1 repeats. In addition, the pre-A region N terminus is found in immediate juxtaposition with the post-P region sequence and extends into the globular C-terminal domains. The entire structure is secured by a previously unidentified topological fold formed by the N terminus that wraps behind the base of P1’s hybrid helix to physically lock it into place.
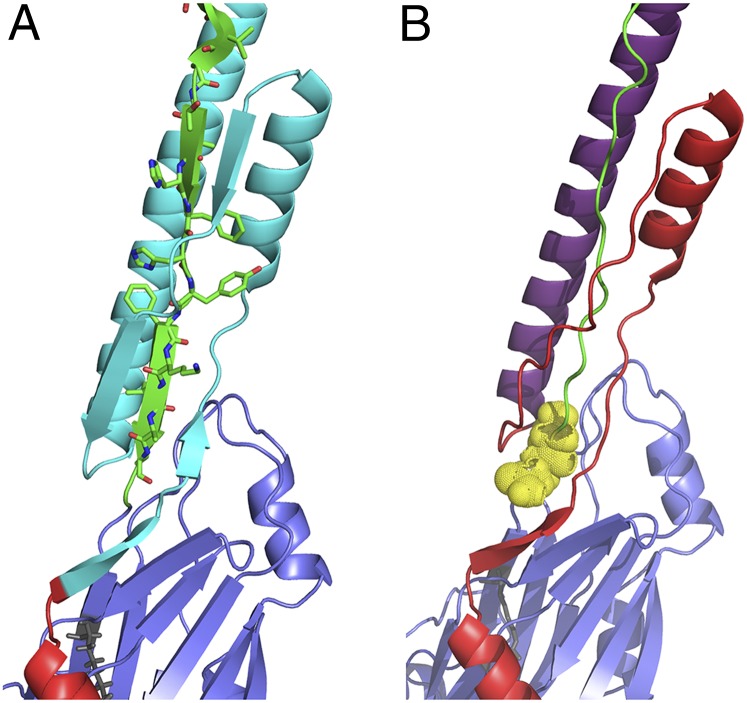
Our crystal structure demonstrates why the deletion of P1 residues 84–190 decreases protein stability and impedes refolding after denaturation (17). Despite mapping to widely spaced locations within the amino acid sequence, the N-terminal deletion polypeptide (25) and the rP1-Cla1Up/Downstream construct evaluated in this work have similar properties. This is explained by the close proximity of the two different mutations within the tertiary model. The N-terminal deletion eliminates the scaffolding loop that supports P1’s helical stalk (Fig. 6A), whereas the two additional amino acids encoded by the downstream Cla1 site throw out of register the post-P region residues that form H bonds with the N terminus to stabilize the structure (Fig. 6B).
Fig. 6.
Illustrations of the regions within the tertiary structure of P1 affected by two different mutations that exhibit instability, impeded folding, and inability to adhere to immobilized salivary agglutinin when expressed in S. mutans (17). The N terminus is shown in red, the alanine-rich region is in purple, and the postproline-rich region is in green. (A) The region deleted within the previously characterized NR7 N-terminal deletion polypeptide (31) is highlighted in cyan and includes the intramolecular scaffold formed around the postproline-rich region. (B) The two added isoleucine and aspartic acid residues encoded by the engineered downstream Cla1 site within polypeptide PC967 (20) correspond to positions 999 and 1000 and are highlighted in yellow. The insertion of extra residues at this location apparently perturbs the alignment and subsequent interactions between postproline-rich region residues and the N-terminal intramolecular scaffold.
It has been reported that C1/C2 compose the majority of the adherent capability of P1’s C terminus (13). In addition, an adhesion epitope was mapped to residues 1025–1044, now known to fall within C1, and a synthetic peptide corresponding to these residues was found to inhibit S. mutans adhesion in vivo (27). Interestingly, the N terminus of P1 interacts intimately with both C1 and C2, with the strongest interactions directly along their interface. It also forms a loop within the cleft formed at the N-terminal end of C2. In this cleft, several glucose molecules were found within the original C-terminal structure, which is believed to represent a sugar-binding domain and site of interaction with gp340 (8, 13). Although we did not identify glucose molecules within our structure, the proximity of this site to the C1/C2 interface likely explains why the N terminus not only contributes to the stability of P1’s elongated helical stalk, but also enhances the adherence of the C terminus to salivary agglutinin when NA1 and P3C interact to form a complex (17).
P1 is a known target of protective immunity (28). Certain anti-P1 monoclonal antibodies (mAbs) act as beneficial immunomodulatory agents to redirect the host immune response when bound to the surface of S. mutans before mucosal or parenteral immunization (22–24, 26, 29, 30). It has been hypothesized that this defect stems from destabilization of the protein and exposure of cryptic epitopes, resulting in a more effective adherence-inhibiting antibody response (22, 30). Two of the mAbs map to P1’s hybrid helical stalk (22, 31, 32), whereas epitopes of the remaining three mAbs (23, 31) are contributed to by the same N-terminal sequence that we now identify as corresponding to the stabilizing loop at the base of the stalk. Our structural model provides a mechanistic explanation for the earlier immunization results, showing that the mAbs bind to P1 regions critical to the maintainence of structural integrity. Thus, perturbing stability would allow the formation of protective antibodies against epitopes that would not otherwise be immunogenic in the locked structure. In more recent experiments, immunization of mice with P3C alone elicited antibodies capable of inhibiting adherence of S. mutans to immobilized human salivary agglutinin; however, immunization with the P3C/NA1 complex was significantly less effective (21). Taken together, these previous studies suggest that the association between the N- and C-terminal regions of P1 not only stabilizes the protein structure and enhances the adherent function of the C-terminal region, but also acts to mask protective epitopes from the immune system.
The fold exhibited by P1’s N terminus in which a stabilizing loop forms around the base of an elongated helical structure appears to be unique. The Dali server (33) was used to compare the tertiary structure of NA1 in complex with P3C with other known structures within the Protein Data Bank, revealing no significant similarities. Bacterial adhesins, such as the Escherichia coli Ig-binding proteins (34), Yersinia enterocolitica adhesin A (35), and other trimeric autotransporter adhesins (36), also exhibit an elongated helical stalk and globular β-rich head domain; however, these structures differ from P1 in their lack of an identifiable stabilizing loop. The topological loop formed by the N terminus of P1 is reminiscent of an amino terminal slipknot (37–39), but is not a true knot. It is hypothesized that true knots have been systematically discriminated against by nature, except in cases in which they preserve a functional advantage that outweighs the complex folding challenges (40). True knotted structures are quite rare and found in far fewer proteins than would be predicted based on random folding algorithms. The protein knots identified to date are found within compact globular enzymes (41, 42) or within the context of the final assembled form of viral capsids (43–45). In the case of P1, the stabilizing loop can better be categorized as a type of mechanical clamp in which interdomain contacts play an essential role in stabilizing a knot loop within a multidomain single-chain protein with a patch of H bonds fastening the ends (46, 47).
Although complex topologies within proteins are beginning to be recognized, and it has been speculated that these may play a role in stability and resistance to protease degradation (37, 39, 43, 48–50), there are few experimental examples addressing their biological and biophysical significance (37, 48, 49). The seminal findings of Anfinsen (51) established that all information required for folding of a protein is inherent within its amino acid sequence. In the case of P1, the key positioning and simple addition of only two residues within its 1,561-aa sequence precluded complete and proper folding. Our experimental data are consistent with the view that the N terminus of P1 functions as an intramolecular chaperone and serves as a critical internal building block to achieve a thermodynamically stable state during folding (52) and lock the full-length adhesin into its functional conformation (17).
The complex and unusual structure of S. mutans P1 has presented an experimental challenge since its discovery in the late 1970s (3, 4, 53). The identification of its hybrid alpha-PPII helical stalk (14) explained some, but not all, of its puzzling functional and immunological features (28). Identification of the topological loop adopted by P1’s N terminus further explains historical data and brings to light the integral role of this protein segment in facilitating folding, enhancing stability and function, and influencing both immunogenicity and antigenicity. This information broadens our mechanistic understanding of a key virulence factor and will aid in the rational development of therapeutic strategies to interfere with S. mutans adhesion to host tissues.
Methods
Crystallization, cloning, protein expression, purification, DSC, and circular dichroism studies are described in detail in SI Methods.
Crystallization and Data Collection.
The NA1/P3C complex was dialyzed into 25 mM Tris, 150 mM sodium chloride (pH 7.4) and concentrated to 17 mg/mL using an Amicon filter (Millipore). Initial crystallization trials were performed by sitting-drop vapor diffusion in 96-well plates using an Art Robbins Gryphon automated pipetting robot (Rigaku) and standard crystal screening kits (Hampton Research). NA1/P3C protein crystals were optimized from condition 44 of the Hampton PEG/Ion HT screen. Crystals were routinely grown at room temperature using hanging-drop vapor diffusion and a reservoir solution of 24% polyethylene glycol 4000 and 150 mM ammonium phosphate (pH 7.5). Crystals were soaked in a well solution augmented with 25% glycerol before flash-cooling in liquid nitrogen and data collection. X-ray diffraction data were collected on the Cornell High-Energy Synchrotron Source beamline A1 at 93 K under a gaseous N2 stream with an ADSC Quantum-210 CCD detector, at a wavelength of 0.976 Å, an exposure time of 5.0 s, an oscillation angle of 1.0°, and a crystal-to-detector distance of 120 mm. Diffraction data were integrated and scaled with the HKL-2000 program (54).
Structure Determination and Refinement.
Molecular replacement was carried out using the full C terminus (amino acids 1000–1486) of AgI/II (PDB ID code 3QE5) as the input model in PHASER (55). Molecular replacement yielded a single solution, and the model was completed using iterative rounds of manual model building in Coot (56) and refinement using phenix.refine (57).
Supplementary Material
Acknowledgments
We thank Dr. Shyamasri Biswas for help with crystallization screening, Dr. Steve Hagen for use of his CD spectrometer, and Dr. William McArthur and the late Dr. Arnold Bleiweis for valuable discussions. This work was supported by a University of Florida Alumni Fellowship, National Institute for Dental and Craniofacial Research (NIDCR) Predoctoral Fellowship T90 DE021990-03, and NIDCR Grants R01 DE08007 and R01 DE21789.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4TSH).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413018111/-/DCSupplemental.
References
- 1.Hamada S, Slade HD. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980;44(2):331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hajishengallis G, Koga T, Russell MW. Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J Dent Res. 1994;73(9):1493–1502. doi: 10.1177/00220345940730090301. [DOI] [PubMed] [Google Scholar]
- 3.Russell MW, Bergmeier LA, Zanders ED, Lehner T. Protein antigens of Streptococcus mutans: Purification and properties of a double antigen and its protease-resistant component. Infect Immun. 1980;28(2):486–493. doi: 10.1128/iai.28.2.486-493.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell RR. Wall-associated protein antigens of Streptococcus mutans. J Gen Microbiol. 1979;114(1):109–115. doi: 10.1099/00221287-114-1-109. [DOI] [PubMed] [Google Scholar]
- 5.Russell MW, Mansson-Rahemtulla B. Interaction between surface protein antigens of Streptococcus mutans and human salivary components. Oral Microbiol Immunol. 1989;4(2):106–111. doi: 10.1111/j.1399-302x.1989.tb00107.x. [DOI] [PubMed] [Google Scholar]
- 6.Bleiweis AS, Oyston PC, Brady LJ. Molecular, immunological and functional characterization of the major surface adhesin of Streptococcus mutans. Adv Exp Med Biol. 1992;327:229–241. doi: 10.1007/978-1-4615-3410-5_25. [DOI] [PubMed] [Google Scholar]
- 7.Brady LJ, Piacentini DA, Crowley PJ, Oyston PC, Bleiweis AS. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect Immun. 1992;60(3):1008–1017. doi: 10.1128/iai.60.3.1008-1017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purushotham S, Deivanayagam C. The calcium-induced conformation and glycosylation of Gp340's SRCR domains influences the high-affinity interaction with antigen I/II homologs. J Biol Chem. 2014;289(32):21877–87. doi: 10.1074/jbc.M114.565507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kishimoto E, Hay DI, Gibbons RJ. A human salivary protein which promotes adhesion of Streptococcus mutans serotype c strains to hydroxyapatite. Infect Immun. 1989;57(12):3702–3707. doi: 10.1128/iai.57.12.3702-3707.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bikker FJ, et al. Identification of the bacteria-binding peptide domain on salivary agglutinin (gp-340/DMBT1), a member of the scavenger receptor cysteine-rich superfamily. J Biol Chem. 2002;277(35):32109–32115. doi: 10.1074/jbc.M203788200. [DOI] [PubMed] [Google Scholar]
- 11.Brady LJ, et al. Restriction fragment length polymorphisms and sequence variation within the spaP gene of Streptococcus mutans serotype c isolates. Infect Immun. 1991;59(5):1803–1810. doi: 10.1128/iai.59.5.1803-1810.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly C, et al. Sequencing and characterization of the 185-kDa cell surface antigen of Streptococcus mutans. Arch Oral Biol. 1990;35(Suppl):33S–38S. doi: 10.1016/0003-9969(90)90128-w. [DOI] [PubMed] [Google Scholar]
- 13.Larson MR, et al. Crystal structure of the C-terminus of Streptococcus mutans antigen I/II and characterization of salivary agglutinin adherence domains. J Biol Chem. 2011;286(24):21657–66. doi: 10.1074/jbc.M111.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larson MR, et al. Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices. Proc Natl Acad Sci USA. 2010;107(13):5983–5988. doi: 10.1073/pnas.0912293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Troffer-Charlier N, Ogier J, Moras D, Cavarelli J. Crystal structure of the V-region of Streptococcus mutans antigen I/II at 2.4-A resolution suggests a sugar preformed binding site. J Mol Biol. 2002;318(1):179–188. doi: 10.1016/S0022-2836(02)00025-6. [DOI] [PubMed] [Google Scholar]
- 16.Nylander A, Forsgren N, Persson K. Structure of the C-terminal domain of the surface antigen SpaP from the caries pathogen Streptococcus mutans. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67(Pt 1):23–26. doi: 10.1107/S174430911004443X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heim KP, Crowley PJ, Brady LJ. An intramolecular interaction involving the N terminus of a streptococcal adhesin affects its conformation and adhesive function. J Biol Chem. 2013;288(19):13762–13774. doi: 10.1074/jbc.M113.459974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Brady LJ, et al. Deletion of the central proline-rich repeat domain results in altered antigenicity and lack of surface expression of the Streptococcus mutans P1 adhesin molecule. Infect Immun. 1998;66(9):4274–4282. doi: 10.1128/iai.66.9.4274-4282.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crowley PJ, et al. Requirements for surface expression and function of adhesin P1 from Streptococcus mutans. Infect Immun. 2008;76(6):2456–2468. doi: 10.1128/IAI.01315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinette RA, et al. Alterations in immunodominance of Streptococcus mutans AgI/II: Lessons learned from immunomodulatory antibodies. Vaccine. 2014;32(3):375–382. doi: 10.1016/j.vaccine.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinette RA, Oli MW, McArthur WP, Brady LJ. A therapeutic anti-Streptococcus mutans monoclonal antibody used in human passive protection trials influences the adaptive immune response. Vaccine. 2011;29(37):6292–6300. doi: 10.1016/j.vaccine.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isoda R, Robinette RA, Pinder TL, McArthur WP, Brady LJ. Basis of beneficial immunomodulation by monoclonal antibodies against Streptococcus mutans adhesin P1. FEMS Immunol Med Microbiol. 2007;51(1):102–111. doi: 10.1111/j.1574-695X.2007.00279.x. [DOI] [PubMed] [Google Scholar]
- 24.Oli MW, Rhodin N, McArthur WP, Brady LJ. Redirecting the humoral immune response against Streptococcus mutans antigen P1 with monoclonal antibodies. Infect Immun. 2004;72(12):6951–6960. doi: 10.1128/IAI.72.12.6951-6960.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhodin NR, Cutalo JM, Tomer KB, McArthur WP, Brady LJ. Characterization of the Streptococcus mutans P1 epitope recognized by immunomodulatory monoclonal antibody 6-11A. Infect Immun. 2004;72(8):4680–4688. doi: 10.1128/IAI.72.8.4680-4688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhodin NR, Van Tilburg ML, Oli MW, McArthur WP, Brady LJ. Further characterization of immunomodulation by a monoclonal antibody against Streptococcus mutans antigen P1. Infect Immun. 2004;72(1):13–21. doi: 10.1128/IAI.72.1.13-21.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly CG, et al. A synthetic peptide adhesion epitope as a novel antimicrobial agent. Nat Biotechnol. 1999;17(1):42–47. doi: 10.1038/5213. [DOI] [PubMed] [Google Scholar]
- 28.Brady LJ, et al. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol Microbiol. 2010;77(2):276–286. doi: 10.1111/j.1365-2958.2010.07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brady LJ, van Tilburg ML, Alford CE, McArthur WP. Monoclonal antibody-mediated modulation of the humoral immune response against mucosally applied Streptococcus mutans. Infect Immun. 2000;68(4):1796–1805. doi: 10.1128/iai.68.4.1796-1805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinette RA, Oli MW, McArthur WP, Brady LJ. Beneficial immunomodulation by Streptococcus mutans anti-P1 monoclonal antibodies is Fc independent and correlates with increased exposure of a relevant target epitope. J Immunol. 2009;183(7):4628–4638. doi: 10.4049/jimmunol.0803300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McArthur WP, et al. Characterization of epitopes recognized by anti-Streptococcus mutans P1 monoclonal antibodies. FEMS Immunol Med Microbiol. 2007;50(3):342–353. doi: 10.1111/j.1574-695X.2007.00260.x. [DOI] [PubMed] [Google Scholar]
- 32.van Dolleweerd CJ, Kelly CG, Chargelegue D, Ma JK. Peptide mapping of a novel discontinuous epitope of the major surface adhesin from Streptococcus mutans. J Biol Chem. 2004;279(21):22198–22203. doi: 10.1074/jbc.M400820200. [DOI] [PubMed] [Google Scholar]
- 33.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(web server issue):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leo JC, et al. The structure of E. coli IgG-binding protein D suggests a general model for bending and binding in trimeric autotransporter adhesins. Structure. 2011;19(7):1021–1030. doi: 10.1016/j.str.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 35.Koretke KK, Szczesny P, Gruber M, Lupas AN. Model structure of the prototypical non-fimbrial adhesin YadA of Yersinia enterocolitica. J Struct Biol. 2006;155(2):154–161. doi: 10.1016/j.jsb.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 36.Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VA. Trimeric autotransporter adhesins: Variable structure, common function. Trends Microbiol. 2006;14(6):264–270. doi: 10.1016/j.tim.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 37.Sułkowska JI, Rawdon EJ, Millett KC, Onuchic JN, Stasiak A. Conservation of complex knotting and slipknotting patterns in proteins. Proc Natl Acad Sci USA. 2012;109(26):E1715–E1723. doi: 10.1073/pnas.1205918109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor WR. Protein knots and fold complexity: Some new twists. Comput Biol Chem. 2007;31(3):151–162. doi: 10.1016/j.compbiolchem.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 39.King NP, Yeates EO, Yeates TO. Identification of rare slipknots in proteins and their implications for stability and folding. J Mol Biol. 2007;373(1):153–166. doi: 10.1016/j.jmb.2007.07.042. [DOI] [PubMed] [Google Scholar]
- 40.Sułkowska JI, et al. Knotting pathways in proteins. Biochem Soc Trans. 2013;41(2):523–527. doi: 10.1042/BST20120342. [DOI] [PubMed] [Google Scholar]
- 41.Potestio R, Micheletti C, Orland H. Knotted vs. unknotted proteins: Evidence of knot-promoting loops. PLOS Comput Biol. 2010;6(7):e1000864. doi: 10.1371/journal.pcbi.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lua RC, Grosberg AY. Statistics of knots, geometry of conformations, and evolution of proteins. PLOS Comput Biol. 2006;2(5):e45. doi: 10.1371/journal.pcbi.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeates TO, Norcross TS, King NP. Knotted and topologically complex proteins as models for studying folding and stability. Curr Opin Chem Biol. 2007;11(6):595–603. doi: 10.1016/j.cbpa.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Virnau P, Mirny LA, Kardar M. Intricate knots in proteins: Function and evolution. PLOS Comput Biol. 2006;2(9):e122. doi: 10.1371/journal.pcbi.0020122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wikoff WR, et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289(5487):2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 46.Valbuena A, et al. On the remarkable mechanostability of scaffoldins and the mechanical clamp motif. Proc Natl Acad Sci USA. 2009;106(33):13791–13796. doi: 10.1073/pnas.0813093106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sikora M, Cieplak M. Mechanical stability of multidomain proteins and novel mechanical clamps. Proteins. 2011;79(6):1786–1799. doi: 10.1002/prot.23001. [DOI] [PubMed] [Google Scholar]
- 48.Soler MA, Faísca PF. Effects of knots on protein folding properties. PLoS ONE. 2013;8(9):e74755. doi: 10.1371/journal.pone.0074755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sayre TC, Lee TM, King NP, Yeates TO. Protein stabilization in a highly knotted protein polymer. Protein Eng Des Sel. 2011;24(8):627–630. doi: 10.1093/protein/gzr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sułkowska JI, Sulkowski P, Szymczak P, Cieplak M. Stabilizing effect of knots on proteins. Proc Natl Acad Sci USA. 2008;105(50):19714–19719. doi: 10.1073/pnas.0805468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 52.Ma B, Tsai CJ, Nussinov R. Binding and folding: In search of intramolecular chaperone-like building block fragments. Protein Eng. 2000;13(9):617–627. doi: 10.1093/protein/13.9.617. [DOI] [PubMed] [Google Scholar]
- 53.Russell MW, Lehner T. Characterisation of antigens extracted from cells and culture fluids of Streptococcus mutans serotype c. Arch Oral Biol. 1978;23(1):7–15. doi: 10.1016/0003-9969(78)90047-x. [DOI] [PubMed] [Google Scholar]
- 54.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.