Significance
Synthetic biology and genetic engineering would greatly benefit from engineered genetic elements that are orthogonal to the host in which they operate. Two-component signaling pathways are the prevalent signal processing modality in prokaryotes that is also found in low eukaryotes and plants but absent from vertebrate cells. Here we investigate whether the elements of prokaryotic two-component pathways are operational in mammalian cells. We find that the core biochemical processes are maintained, whereas the capacity to sense chemical ligands is diminished or obscured. We use the pathways for multiinput gene regulation and show that they can serve as a rich source of orthogonal building blocks for gene expression control in mammalian cells. Our findings open new avenues in synthetic circuit design.
Keywords: synthetic biology, two-component signaling, biological computing
Abstract
Signaling pathway engineering is a promising route toward synthetic biological circuits. Histidine–aspartate phosphorelays are thought to have evolved in prokaryotes where they form the basis for two-component signaling. Tyrosine-serine–threonine phosphorelays, exemplified by MAP kinase cascades, are predominant in eukaryotes. Recently, a prokaryotic two-component pathway was implemented in a plant species to sense environmental trinitrotoluene. We reasoned that “transplantation” of two-component pathways into mammalian host could provide an orthogonal and diverse toolkit for a variety of signal processing tasks. Here we report that two-component pathways could be partially reconstituted in mammalian cell culture and used for programmable control of gene expression. To enable this reconstitution, coding sequences of histidine kinase (HK) and response regulator (RR) components were codon-optimized for human cells, whereas the RRs were fused with a transactivation domain. Responsive promoters were furnished by fusing DNA binding sites in front of a minimal promoter. We found that coexpression of HKs and their cognate RRs in cultured mammalian cells is necessary and sufficient to strongly induce gene expression even in the absence of pathways’ chemical triggers in the medium. Both loss-of-function and constitutive mutants behaved as expected. We further used the two-component signaling pathways to implement two-input logical AND, NOR, and OR gene regulation. Thus, two-component systems can be applied in different capacities in mammalian cells and their components can be used for large-scale synthetic gene circuits.
Signaling pathways are the cell’s major interface with the environment. Their “purpose” is to deliver environmental signals either to the nucleus to modulate gene expression, or to other cellular proteins for posttranscriptional responses. Membrane receptors respond to external signals and transduce them downstream, where interesting phenomena are observed including signal amplification (1), noise filtering (2), response sharpening (3), adaptation (4), etc. Virtually all pathways use protein phosphorylation by ATP followed by phosphoryl transfer between proteins to transmit information, resulting in fast transmission timescales that translate to fast overall responses when gene expression is not required, e.g., in chemotaxis. Substantial modularity allows tens if not hundreds of pathways to operate simultaneously, but it is complemented with interpathway cross-talk and a plethora of one-to-many or many-to-one input–output relationships (5–7). In the context of synthetic biology (8–15), signaling represents a challenging case because pathway modification requires protein reengineering or de novo design. Both are hard tasks, but progress has been steady (1, 16–19).
An attractive paradigm in genetic engineering and synthetic biology is what can be termed “cross-kingdom pathway transplantation,” which, as the name implies, involves transplantation of a pathway from an evolutionarily distinct donor into the host cell. Redesign of a native pathway often carries a risk of unwanted interactions with the other pathways. However, introduction of a completely foreign pathway from the donor that is as divergent as possible from the recipient can minimize this risk. Indeed, the application of phage-derived recombinases (20) and prokaryotic-based transcription regulators (21) were breakthrough developments in genetic engineering in animals and plants alike.
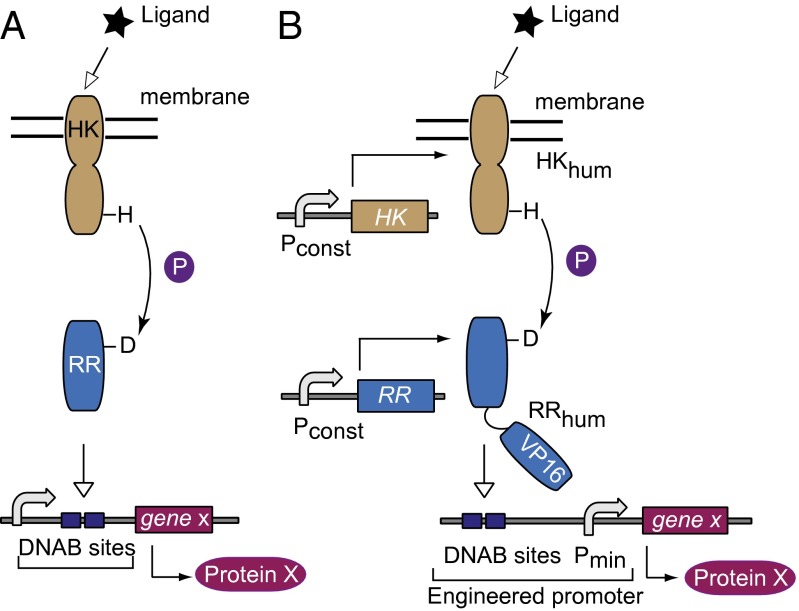
Two-component signaling (TCS) pathways are ubiquitous in prokaryotes, comprising in their simplest form a sensor and a regulator component (22, 23). The membrane sensor histidine kinase (HK) typically has extracellular and cytoplasmic domains linked via a transmembrane domain. Upon ligand binding to the extracellular domain and subsequent conformational change, autophosphorylation of the conserved histidine residue in the cytoplasmic domain takes place. The phosphate is then transferred to the aspartic residue on the response regulator (RR), inducing a conformation change that usually facilitates DNA binding and modulation of gene expression (Fig. 1A). In other cases such as chemotaxis, TCS results in posttranslational modifications of downstream proteins (24). The phosphoryl transfer from a histidine to aspartate residue is what sets these pathways apart. Similar biochemistry is encountered in plants (25) and lower eukaryotes, whereas it is absent in vertebrates (26), where tyrosine-serine–threonine phosphorelay is the rule (27). Thus, implementation of prokaryotic signaling in mammalian cells would amount to cross-kingdom pathway transplantation mentioned above.
Fig. 1.
Schematics of native and transplanted two-component signaling pathways. (A) The native pathway consists of a receptor histidine kinase protein, which senses and propagates the signal to a cognate response regulator that regulates gene expression. (B) The envisioned adaptation to the mammalian host. Subscript “hum” indicates human-optimized codon sequence. DNAB, DNA binding. Pconst, constitutive mammalian promoter. P, phosphate; Pmin, minimal mammalian promoter; VP16, VP16 transactivator domain.
Small size and modularity of these pathways have already made them a target for rewiring and synthetic application in prokaryotic cells (28–30) as well as Arabidopsis (31). There was also a report on functional complementation by a plant-derived HK in yeast (32). However, the adaptation of TCS system to mammalian cells has not, to the best of our knowledge, been shown yet. Here, we report on such an adaptation and show that the internal core of the pathways and their specific interactions are functional in mammalian cells. We have used the transplanted modules to implement complex logical programs of gene expression. In addition, we show that the pathway genes can serve as a rich source of orthogonal building blocks for engineering genetic circuits in mammalian cells.
Methods
Plasmid Construction.
Plasmid design was done with SnapGene (GSL Biotech LLC). Escherichia coli DH5α competent cells were used for cloning. Transformed cells were grown in LB Broth Miller Difco (BD) supplemented with selective antibiotics. PCR was done with Phusion High-Fidelity DNA polymerase (NEB). Primers were purchased from Sigma-Aldrich. Synthesis of gene constructs was done by DNA 2.0 and Genscript. gBlocks were purchased from IDT. Plasmids were purified with PureYield Midiprep System (Promega), followed by endotoxin removal (Norgen Biotek Corp), quantification with NanoDrop ND-2000 and gel electrophoresis, and sequencing (Microsynth AG). Restriction enzymes (NEB) were used as per manufacturer protocols. Ligation was performed with T4 DNA Ligase (NEB). Cloning steps are described in SI Appendix and primer list is in SI Appendix, Table S19. Maps of plasmids not published elsewhere are available on European Nucleotide Archive, www.ebi.ac.uk/ena.
Cell Culture and Transfection.
HEK293 cell line (Life Technologies, #11631–017) was maintained at 37 °C, 5% CO2 in RPMI-1640 medium (Life Technologies), supplemented with 10% (vol/vol) FBS (Sigma-Aldrich) and 500 µg/mL Penicillin-streptomycin Solution (Tecomedical AG). HeLa cell line (ATCC, #CCL-2) was maintained in DMEM/GlutaMAX medium (Life Technologies) with the same supplements. Passaging of up to 20 times occurred at 70–80% confluency roughly every 2–3 d. Twelve-well culture plates (NUNC) were used in transfection experiments. Wells were seeded with 1.4 × 105 cells per well 24 h pretransfection. For transfection, DNA was resuspended in OPTI-MEM without serum (Life Technologies) in combination with Lipofectamine 2000 (Life Technologies) at a 1:2 ratio of DNA to lipofectamine. The cells were incubated for 48 h before characterization.
Fluorescence Microscopy.
Fluorescent protein (FP) images, not including localization study, were taken with Nikon Eclipse Ti microscope using a Hamamatsu ORCA-R2 camera. AmCyan FP was visualized with CFP HC-filter set (HC 438/24, HC 483/32, BS 458) at 300-ms exposure, and DsRed with TRITC HC-filter set (HC 543/22, HC 593/40, BS 562) at 200-ms exposure. Images were uniformly processed with ImageJ software using lookup table (LUT) values 0–33,000 for DsRed and AmCyan channels, with the exception of Fig. 3 B and C, where an LUT range of 3,000–6,000 was used for AmCyan to visualize lower expression levels. To measure localization of HK::mCherry fusion proteins, CellMask Deep Red Plasma Membrane stain (Life Technologies) was used according to the manufacturer's protocol at a 1:1,000 dilution and incubated with cells for 20 min at 37 °C and 5% CO2 before changing the medium. The cells were imaged using a Nikon Confocal A1R Microscope with a Plan Apo VC 100× (oil) objective. The mCherry and the CellMask stain were visualized by excitation at 561 nm and emission at 595 nm, and excitation at 641 nm and emission at 700 nm, respectively. Colocalization was analyzed using Imaris software (33).
Fig. 3.
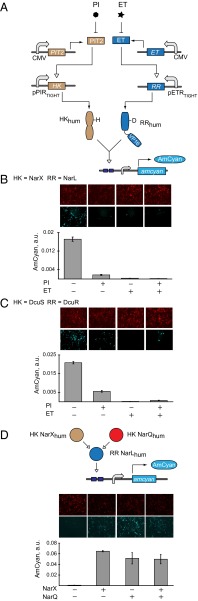
Logic with TCS. (A) NOR-gate circuit schematics comprising antibiotic-regulated HK and RR genes. (B) Quantitative data for antibiotic-regulated NarXL pathway. (C) Quantitative data for antibiotic-regulated DcuSR pathway. PI and ET are at 10 µg/mL and 4 µg/mL, respectively. Plasmid composition and output values are in SI Appendix, Tables S13 and S14. (D) (Top) Schematic representation of an OR gate between NarX and NarQ. (Bottom) Quantitative data and representative images. Plasmid composition and output values are in SI Appendix, Tables S15 and S16. In all panels the images are shown with red pseudocolor indicating DsRed transfection marker, and cyan pseudocolor indicating AmCyan output. The resultant data are presented as mean ± SD of biological triplicates.
Flow Cytometry.
Cell analysis was done with BD LSR Fortessa. Cells were trypsinized with 0.5% Trypsin-EDTA (Life Technologies). DsRed was measured using 561-nm laser and 586/15 BP emission filter with a photomultiplier tube (PMT) voltage of 270 or equivalent. AmCyan was measured with 445-nm laser and 473/10 BP emission filter with PMT voltage of 253 or equivalent. Due to hardware modifications in the course of the project, PMT voltages were adjusted using calibration beads to keep numerical values comparable across experiments.
Data Analysis.
Flow cytometry data analysis was done with FlowJo software (Tree Star). Quantification of AmCyan output in arbitrary expression units (a.u.) is as follows: (i) Live cells gating using forward and side scatter. (ii) Gating of cells positive for a particular FP and calculating freq(FP+), the fraction of FP-positive cells among live cells, and mean(FP+), the mean expression of this FP among positively gated cells. (iii) Calculating AmCyan level: AmCyan (a.u.) = [mean(AmCyan+)*freq(AmCyan+)]/[(mean(DsRed+)*freq(DsRed+)], where DsRed is a transfection control.
Results
TCS Is Partially Functional in Mammalian Cells.
Transplanting prokaryotic TCS pathways to mammalian cell lines requires a number of adaptations (Fig. 1B). Three TCS pathways from E. coli were selected, including EnvZ-OmpR, NarXL, and DcuSR. Each gene coding for a TCS protein was codon-optimized for expression in human cells and placed under the control of a constitutive CMV immediate early promoter. The amino acid sequence of each HK and RR was preserved, whereas the RR sequences were augmented with three minimal VP16 transactivating domains at the C terminus (31). Nuclear localization signal (NLS) was not fused to RRs as we reasoned that NLS would preclude RR from interacting with nonnuclear HK. In addition, a few prokaryotic RRs without NLS were shown to translocate to the nucleus in plants (34). The responsive promoter driving a fluorescent reporter AmCyan was created by fusing known DNA binding sites of different RRs in front of a minimal promoter. For brevity, we also call these sites response elements (REs). Specifically, pairs of consensus REs for OmpR (35) and NarL (36) separated by a 10-bp spacer and a single DcuR response element from the dctA gene (37) were cloned upstream of the TATA box in the minimal promoter developed in our group (SI Appendix, Table S1).
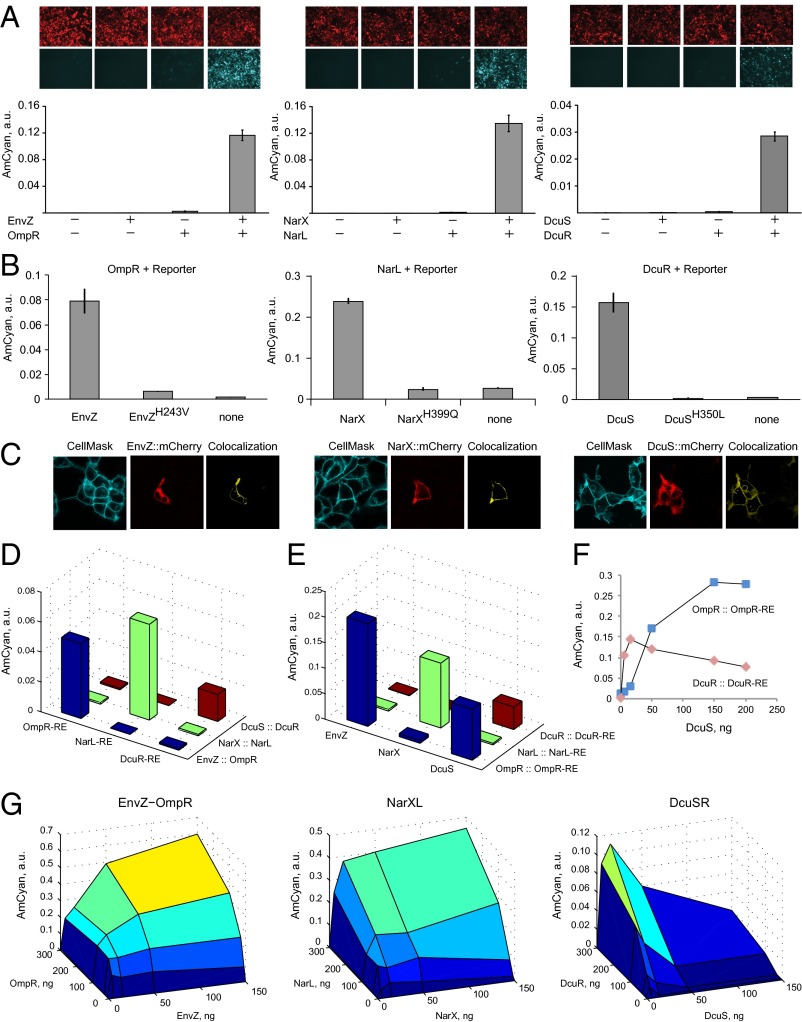
First we inquired whether coexpression of the pathway genes in cultured human cells would result in gene activation. We transiently transfected HEK293 cells with (i) none of the TCS cassettes; (ii) HK or RR cassettes alone; and (iii) both HK and RR. All three pathways elicited strong expression of the reporter gene when both the HK and RR were present but not when either component was missing (Fig. 2A). The induction with DcuSR was weaker due to a single DNA binding site in the promoter. The same trends were observed in HeLa cells (SI Appendix, Fig. S1). We cloned DcuR-inducible promoters with two DcuR sites and varying spacer length (SI Appendix, Table S1) and found that the activation in both HEK and HeLa increased dramatically in both cases (SI Appendix, Fig. S2). We further mutated the catalytic histidine in all three HKs and observed loss of induction to the background level (Fig. 2B), with slightly elevated activity in EnvZ mutant likely due to experimental variation. Lastly, we fused mCherry protein to all three HKs. We confirmed their activity in the pathways (SI Appendix, Fig. S3) and found that they are capable of localizing to the plasma membrane (Fig. 2C). Fluorescence was observed in other compartments as well, probably due to embedding in other membrane components as well as due to turnover in proteasome.
Fig. 2.
Characterization of the transplanted TCS pathways. (A) Component requirements for RE activation. Each component combination is accompanied by representative microscopy pictures with red pseudocolor indicating DsRed transfection marker, and cyan pseudocolor indicating pathway-induced AmCyan protein output. The bar charts display AmCyan level in a.u. as mean ± SD of independent biological triplicates. Plasmid composition and output values are in SI Appendix, Tables S2 and S3. (B) Loss of function via mutating catalytic histidine residues of HKs (references 2, 8, and 9 in SI Appendix). Each bar chart shows, left to right, the readout with wild-type HK, mutated HK, and without HK gene. Plasmid composition and output values are in SI Appendix, Tables S4 and S5. (C) Cellular localization of the HK::mCherry fusion proteins. CellMask membrane stain is shown in cyan, Cherry is shown in red, and colocalized region (yellow) was calculated using Imaris software. (D) Cross-talk of HK::RR pairs with noncognate response elements. Plasmid composition and output values (including SD) are in SI Appendix, Tables S6 and S7. (E) Cross-talk of noncognate HKs with RR::RE pairs, indicating that DcuS phosphorylates OmpR. Plasmid composition and output values (including SD) are in SI Appendix, Tables S8 and S9. (F) Detailed look into the DcuS-OmpR cross-talk. DcuS-OmpR interaction is displayed using blue markers and cognate DcuS-DcuR is shown with red markers. Data points are duplicate averages. Plasmid composition and output values (including SD) are in SI Appendix, Tables S10 and S11. (G) The relationship between the amounts of HKs and RRs and the reporter level. For each surface plot, all pairwise combinations of HK and RR were measured with HK at 0, 5, 50, and 150 ng per transfection in a 12-well plate and RR at 0, 10, 30, 100, and 300 ng. Filler DNA was added to keep the total DNA amount constant. All data points are duplicate averages. Output values are in SI Appendix, Table S12. Different colors represent output intensity from dark blue (low) to yellow (high).
These results indicate that major pathway features, such as protein folding, membrane insertion, phosphorelay, and differential DNA binding and transactivation, are operational in human cells. However, gene induction occurs in the apparent absence of cognate external stimulus. EnvZ and NarX respond to generic inputs that could be present in the medium or the cytoplasm. DcuS is responsive to fumarate that is present in cell cytoplasm at low concentration, e.g., 40 μM in HEK293 cells (38), and absent from the medium. Efforts to further induce the pathway with fumarate in both HEK293 and HeLa cells failed to elicit a significant effect. We speculate that cytoplasmic and/or media components induce the HKs, and in-depth study of HK induction will be the subject of future work.
TCS Orthogonality and Cross-Talk in Human Cells.
TCS operate without extensive cross-talk in their endogenous milieu. We asked whether the same holds in the mammalian host. Being able to operate multiple pathways in parallel would allow facile scaling up of genetic circuits (39). First, we kept the canonical HK::RR pairs together but varied the response elements (Fig. 2D). The activation is highly specific to the correct responsive promoter. In the second experiment, we combined the RRs with their cognate responsive promoters and varied the upstream HKs. We found that in one case, an HK DcuS activates a noncognate RR OmpR whereas in all other cases the phosphorelay is specific (Fig. 2E). We measured the dose–response of the cognate as well as noncognate DcuS activity (Fig. 2F) and saw that the correctly assembled pathway exhibits sharp dose–response that decreases at high DcuS levels, whereas cross-reactivity increased slowly and monotonously. Thus, operating at the optimal DcuS level results in high selectivity toward the cognate pathway. Note that the interaction between DcuS and OmpR was not reported in a large cross-reactivity study (40). Our findings suggest that human cell host could be an attractive in vivo model for studying cross-talk in TCS pathways without the need to purify proteins for biochemical assays.
The nonlinear dose–response (Fig. 2F) prompted us to map the input–output relationship of the transplanted pathways. We kept the response plasmid constant and varied the amounts of both HK- and RR-encoding plasmids (Fig. 2G). All pathways show marked sensitivity to the amount of HK, which is unsurprising given its enzymatic nature. The dependency on the amount of RR is gradual; however, half the maximum induction is reached with very low amounts of both HK and RR relative to the response gene. The input–output map of DcuSR pathway reproduces the nonmonotonous behavior observed in Fig. 2F. The reason for this phenomenon will be the subject of future investigation.
TCSs as Building Blocks for Genetic Logic Circuits.
The experiments above show that the response requires expression of both pathway components. This can be described as AND logic (12, 41–43) between HK and RR genes as inputs. Controlling these genes with external stimuli can generate additional logic behaviors. To exemplify this possibility, we cloned the components of NarXL and DcuSR pathways in vectors controlled by engineered transactivators PIT2 (44) and ET (45), modulated, respectively, with antibiotics pristinamycin 1A (PI) and erythromycin (ET). Because both cofactors inhibit DNA binding of their cognate transactivators, the underlying AND logic translates into NOR logic (46) between antibiotic inputs (Fig. 3 A–C). The resultant logic behavior is consistent with expectation (highest output in the absence of both inputs, low output otherwise). The NarXL pathway displays the largest dynamic range, whereas with DcuSR pathway the leakage in one of the OFF states is likely due to very high sensitivity of this pathway to minute amounts of the DcuS HK and residual expression of DcuS in the presence of PI. The low level of induction in the ON state is due to multiple plasmid cotransfection. Similar experiments in HeLa cells showed comparable trends (SI Appendix, Fig. S4).
Natural TCS cross-talk provides additional types of logic control. For example, HK NarQ activates NarL almost as efficiently as NarX (47). We constructed a pathway in which NarL is controlled by both NarX and NarQ and measured almost-perfect OR behavior as the function of NarX and NarQ presence (Fig. 3D).
Known Mutant Behaviors Are Recapitulated in the Mammalian Host.
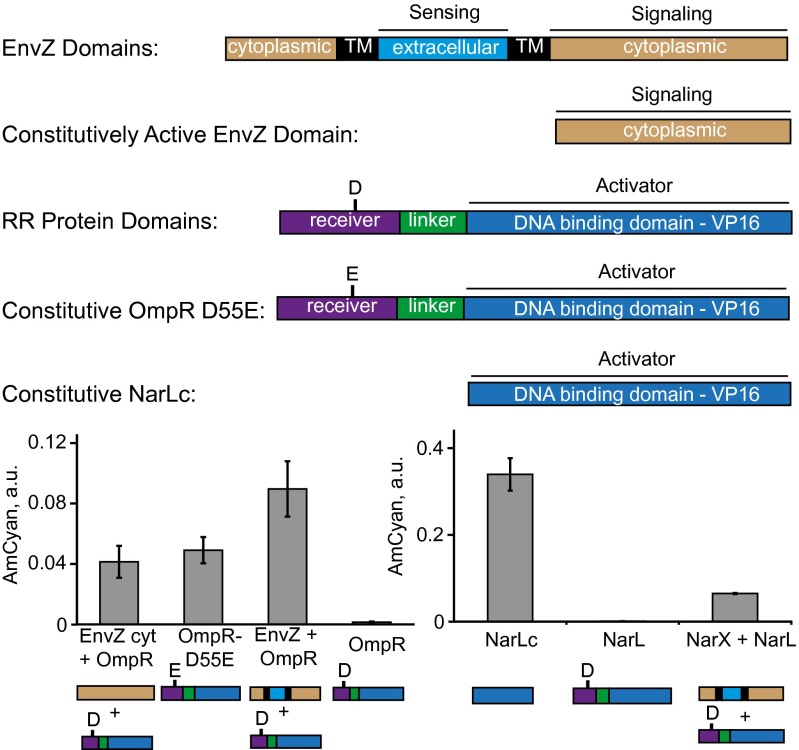
TCS research uncovered a large number of mutants that possess certain qualities that could be of use in synthetic pathways (Fig. 4). For example, cytoplasmic domains of HKs are known to support constitutive signaling and we found that cytoplasmic domain of EnvZ (EnvZ cyt) (48) supports constitutive signaling via OmpR. Mutant RRs OmpR D55E (49) and C-terminal domain of NarL (NarLc) (48) were reported to be constitutive activators, and we found them to be constitutive inducers of their respective REs in mammalian cells. These data suggest that the findings made in the native prokaryotic setting translate into the humanized system. They also imply that TCS-encoding genes and their variants can be used as a huge source of “biological parts” for mammalian gene circuits.
Fig. 4.
Activity of mutant TCS components. Full-length, truncated, and mutant TCS genes are shown with each protein domain color-coded and labeled. Experimental data are given below. Each bar represents mean ± SD of a biological triplicate. The output values for NarX + NarL and NarL are from Fig. 3B; they are displayed again for side-by-side comparison. Plasmid composition and output values are in SI Appendix, Tables S17 and S18. TM, transmembrane domain.
Discussion
Histidine–aspartate phosphorelay is absent from vertebrate cells whereas it is found in plants, yeast, lower eukaryotes, and most commonly in prokaryotes. The lack of homologous genes in vertebrates suggests prokaryotic TCS pathways to be orthogonal signal processing modules in mammalian cells for circuit engineering. However, the preservation of the basic biochemical processes during mammalian “transplantation” is by no means guaranteed, as it required three conditions. First, the internal operation of a pathway has to be preserved as much as possible. Second, the pathway components should not affect the host nonspecifically. Third, the host should not interfere with the pathway components.
Our results show that the phosphorelays between HK and RR, and differential DNA binding by the RR followed by gene induction, occur in mammalian cells. The presence of a cognate ligand does not seem to be necessary, and the elucidation of ligand induction will be addressed in future work. We speculate that cytoplasmic metabolites or media components can be responsible for HK activation; if this is the case, there is an intriguing possibility to use HKs as biosensors for cytoplasmic metabolites. However, the fact that full-length HK genes are functional suggests that they are properly folded, as supported by their observed localization to plasma membrane and other membrane-bound organelles. With respect to pathway effect on the host cells, no gross adverse effects were observed. The individual RR DNA binding sequences tested in this work vary in size from 16 to 26 bp, and a tandem repeat of those is required for strong induction. Such sequences are not expected to occur frequently in the human genome. Finally, the response elements were silent on their own, meaning that no endogenous activator bound to these sites. RRs in combination with REs generated only low background expression, likely due to residual DNA binding of the nonphosphorylated RRs rather than due to phosphoryl transfer to the RR by endogenous kinases.
Among themselves, the pathways exhibited an impressive lack of cross-talk. We reproduced a known cross-reactivity between NarQ and NarL and uncovered an unexpected interaction between DcuS and OmpR. This assay might present an attractive approach to study TCS biochemistry in vivo on the clean background devoid of interference.
The RR genes in this study did not include NLS, and their activity can be due to their small size of about 30 kDa, which is lower than the size cutoff of a nuclear pore (40–60 kDa). In plants, RR translocation to the nucleus was shown upon ligand activation (34), but whether or not this is also true in mammalian cells can only be tested when we achieve selective pathway induction by an environmental ligand. Even in the absence of response to external ligands, TCS can support complex logic signal integration in mammalian cells. We implemented AND, NOR, and OR gates using constitutive and inducible HKs and RRs. Given the difficulty to implement AND-like gene activation in mammalian cells, adapted TCS pathways are an attractive source of such control elements. In addition, constitutive mutants were shown to act consistently with their behavior in prokaryotes; these mutants can be a rich source of mutually orthogonal building blocks in large gene circuits.
Supplementary Material
Acknowledgments
We thank M. Fussenegger for the plasmids encoding PIT2 and ET and their DNA binding sites. We thank M. Laub for discussions; the Y.B. laboratory, and especially N. Lapique, for help and advice; and T. Horn, V. Jaeggin, E. Montani, and A. Ponti from the Single Cell Unit for help with imaging and flow cytometry. The research was funded by Eidgenössische Technische Hochschule Zürich (ETH Zurich).
Footnotes
Conflict of interest statement: The results of this paper have been filed as a priority application with European Patent Office.
This article is a PNAS Direct Submission.
Data deposition: The plasmid maps reported in this paper have been deposited in the European Nucleotide Archive, www.ebi.ac.uk/ena (accession no. LN613236–LN613260).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406482111/-/DCSupplemental.
References
- 1.O’Shaughnessy EC, Palani S, Collins JJ, Sarkar CA. Tunable signal processing in synthetic MAP kinase cascades. Cell. 2011;144(1):119–131. doi: 10.1016/j.cell.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews BW, Yi TM, Iglesias PA. Optimal noise filtering in the chemotactic response of Escherichia coli. PLOS Comput Biol. 2006;2(11):e154. doi: 10.1371/journal.pcbi.0020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrell JE., Jr Tripping the switch fantastic: How a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci. 1996;21(12):460–466. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- 4.Shimizu TS, Tu Y, Berg HC. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol Syst Biol. 2010;6:382. doi: 10.1038/msb.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kashtan N, Alon U. Spontaneous evolution of modularity and network motifs. Proc Natl Acad Sci USA. 2005;102(39):13773–13778. doi: 10.1073/pnas.0503610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3(10):e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Procaccini A, Lunt B, Szurmant H, Hwa T, Weigt M. Dissecting the specificity of protein-protein interaction in bacterial two-component signaling: orphans and crosstalks. PLoS ONE. 2011;6(5):e19729. doi: 10.1371/journal.pone.0019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss R, Homsy GE, Knight TF. Toward in vivo digital circuits. In: Landweber LF, Winfree E, editors. Evolution as Computation: DIMACS Workshop. Springer; Berlin: 1999. pp. 275–295. [Google Scholar]
- 9.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403(6767):339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 10.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403(6767):335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 11.Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21(7):796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 12.Kramer BP, Fischer C, Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol Bioeng. 2004;87(4):478–484. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- 13.Endy D. Foundations for engineering biology. Nature. 2005;438(7067):449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 14.Kitney R, Freemont P. Synthetic biology - the state of play. FEBS Lett. 2012;586(15):2029–2036. doi: 10.1016/j.febslet.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Benner SA, Sismour AM. Synthetic biology. Nat Rev Genet. 2005;6(7):533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dueber JE, Yeh BJ, Chak K, Lim WA. Reprogramming control of an allosteric signaling switch through modular recombination. Science. 2003;301(5641):1904–1908. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- 17.Bashor CJ, Helman NC, Yan S, Lim WA. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319(5869):1539–1543. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 18.Regot S, et al. Distributed biological computation with multicellular engineered networks. Nature. 2011;469(7329):207–211. doi: 10.1038/nature09679. [DOI] [PubMed] [Google Scholar]
- 19.Galloway KE, Franco E, Smolke CD. Dynamically reshaping signaling networks to program cell fate via genetic controllers. Science. 2013;341(6152):1235005. doi: 10.1126/science.1235005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Backman K, Oconnor MJ, Maruya A, Erfle M. Use of synchronous site-specific recombination in vivo to regulate gene-expression. Bio-Technology. 1984;2(12):1045–1049. [Google Scholar]
- 21.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slauch JM, Russo FD, Silhavy TJ. Suppressor mutations in rpoA suggest that OmpR controls transcription by direct interaction with the alpha subunit of RNA polymerase. J Bacteriol. 1991;173(23):7501–7510. doi: 10.1128/jb.173.23.7501-7510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laub MT, Goulian M. Specificity in Two-Component Signal Transduction Pathways. Annual Review of Genetics. Vol 41. Annual Reviews; Palo Alto, CA: 2007. pp. 121–145. [DOI] [PubMed] [Google Scholar]
- 24.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. The two-component signaling pathway of bacterial chemotaxis: A molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413(6854):383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- 26.Klumpp S, Krieglstein J. Reversible phosphorylation of histidine residues in vertebrate proteins. Biochim Biophys Acta. 2005;1754(1-2):291–295. doi: 10.1016/j.bbapap.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 27.Karin M, Hunter T. Transcriptional control by protein phosphorylation: Signal transmission from the cell surface to the nucleus. Curr Biol. 1995;5(7):747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- 28.Levskaya A, et al. Synthetic biology: Engineering Escherichia coli to see light. Nature. 2005;438(7067):441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 29.Skerker JM, et al. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133(6):1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabor JJ, et al. A synthetic genetic edge detection program. Cell. 2009;137(7):1272–1281. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antunes MS, et al. Programmable ligand detection system in plants through a synthetic signal transduction pathway. PLoS ONE. 2011;6(1):e16292. doi: 10.1371/journal.pone.0016292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urao T, et al. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11(9):1743–1754. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costes SV, et al. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86(6):3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antunes MS, et al. Engineering key components in a synthetic eukaryotic signal transduction pathway. Mol Syst Biol. 2009;5:270. doi: 10.1038/msb.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda S, et al. Activation of the osmoregulated ompC gene by the OmpR protein in Escherichia coli: A study involving synthetic OmpR-binding sequences. J Biochem. 1991;110(3):324–327. doi: 10.1093/oxfordjournals.jbchem.a123579. [DOI] [PubMed] [Google Scholar]
- 36.Darwin AJ, Tyson KL, Busby SJW, Stewart V. Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement. Mol Microbiol. 1997;25(3):583–595. doi: 10.1046/j.1365-2958.1997.4971855.x. [DOI] [PubMed] [Google Scholar]
- 37.Abo-Amer AE, et al. DNA interaction and phosphotransfer of the C4-dicarboxylate-responsive DcuS-DcuR two-component regulatory system from Escherichia coli. J Bacteriol. 2004;186(6):1879–1889. doi: 10.1128/JB.186.6.1879-1889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koivunen P, et al. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: Possible links between cell metabolism and stabilization of HIF. J Biol Chem. 2007;282(7):4524–4532. doi: 10.1074/jbc.M610415200. [DOI] [PubMed] [Google Scholar]
- 39.Stanton BC, et al. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat Chem Biol. 2014;10(2):99–105. doi: 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto K, et al. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem. 2005;280(2):1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- 41.Wang B, Kitney RI, Joly N, Buck M. Engineering modular and orthogonal genetic logic gates for robust digital-like synthetic biology. Nat Commun. 2011;2:508. doi: 10.1038/ncomms1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohmueller JJ, Armel TZ, Silver PA. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 2012;40(11):5180–5187. doi: 10.1093/nar/gks142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahdavi A, et al. A genetically encoded and gate for cell-targeted metabolic labeling of proteins. J Am Chem Soc. 2013;135(8):2979–2982. doi: 10.1021/ja400448f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fussenegger M, et al. Streptogramin-based gene regulation systems for mammalian cells. Nat Biotechnol. 2000;18(11):1203–1208. doi: 10.1038/81208. [DOI] [PubMed] [Google Scholar]
- 45.Weber W, et al. Macrolide-based transgene control in mammalian cells and mice. Nat Biotechnol. 2002;20(9):901–907. doi: 10.1038/nbt731. [DOI] [PubMed] [Google Scholar]
- 46.Ran T, Douek Y, Milo L, Shapiro E. A programmable NOR-based device for transcription profile analysis. Sci Rep. 2012;2:641. doi: 10.1038/srep00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabin RS, Stewart V. Either of two functionally redundant sensor proteins, NarX and NarQ, is sufficient for nitrate regulation in Escherichia coli K-12. Proc Natl Acad Sci USA. 1992;89(18):8419–8423. doi: 10.1073/pnas.89.18.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forst S, Delgado J, Inouye M. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci USA. 1989;86(16):6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lan CY, Igo MM. Differential expression of the OmpF and OmpC porin proteins in Escherichia coli K-12 depends upon the level of active OmpR. J Bacteriol. 1998;180(1):171–174. doi: 10.1128/jb.180.1.171-174.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.