Significance
Both overexpression of wild-type fused in sarcoma (FUS) protein and missense mutations can be pathogenic in a group of related neurodegenerative disorders that includes amyotrophic lateral sclerosis and frontotemporal lobar degeneration. It is unclear how FUS overexpression and missense mutations cause disease in human patients. In this work, we generated novel transgenic mouse models expressing low levels of wild-type and mutant human FUS, both of which recapitulate aspects of the human diseases. We found a profound difference in the underlying mechanisms by which missense mutation and wild-type overexpression cause disease. Overexpression of wild-type FUS protein alters its nuclear function at the level of gene expression. In contrast, missense mutation disrupts activity-dependent synaptic homeostasis to gain a toxic function at dendritic spines.
Keywords: FUS, frontotemporal lobar degeneration, amyotrophic lateral sclerosis, metabotropic glutamate receptors, synaptic homeostasis
Abstract
The RNA-binding protein fused-in-sarcoma (FUS) has been associated with amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD), two neurodegenerative disorders that share similar clinical and pathological features. Both missense mutations and overexpression of wild-type FUS protein can be pathogenic in human patients. To study the molecular and cellular basis by which FUS mutations and overexpression cause disease, we generated novel transgenic mice globally expressing low levels of human wild-type protein (FUSWT) and a pathological mutation (FUSR521G). FUSWT and FUSR521G mice that develop severe motor deficits also show neuroinflammation, denervated neuromuscular junctions, and premature death, phenocopying the human diseases. A portion of FUSR521G mice escape early lethality; these escapers have modest motor impairments and altered sociability, which correspond with a reduction of dendritic arbors and mature spines. Remarkably, only FUSR521G mice show dendritic defects; FUSWT mice do not. Activation of metabotropic glutamate receptors 1/5 in neocortical slices and isolated synaptoneurosomes increases endogenous mouse FUS and FUSWT protein levels but decreases the FUSR521G protein, providing a potential biochemical basis for the dendritic spine differences between FUSWT and FUSR521G mice.
Amyotrophic lateral sclerosis (ALS) is characterized by the degeneration of upper and lower motor neurons, leading to muscle weakness, paralysis, and death within 3–5 y of onset. Interestingly, ∼10–15% of ALS patients have clinical features of frontotemporal lobar degeneration (FTLD), marked by a decline in decision-making, behavioral control, emotion, and language, and as many as half have mild-to-moderate cognitive or behavioral abnormalities (1). FTLD comprises a group of heterogeneous diseases characterized by progressive neurodegeneration of the frontal and temporal lobes and clinically by frontotemporal dementia (FTD) with or without motor neuron disease. There is no cure or effective therapy for those who suffer from ALS or FTLD, and the mechanisms by which these diseases occur are not well understood.
The clinical, pathological, and genetic overlap between ALS and FTLD suggests that there are mechanisms shared by these diseases. The RNA-binding proteins fused in sarcoma (FUS) and transactive response DNA-binding protein-43 (TDP-43) are the major protein components of inclusions that are characteristic of ALS and FTLD-U (FTLD with ubiquitinated inclusions) (2). More than 50 genetic FUS mutations have been identified in these related neurodegenerative disorders (3). Similarly, more than 40 dominant mutations in the TDP-43 gene have been linked to ALS cases and, to a lesser extent, to FTLD (4). The identification of mutations in the FUS and TDP-43 genes has provided insights for uncovering the disease mechanisms for ALS and FTLD.
FUS is a ubiquitously expressed RNA-binding protein that exists in dynamic ribonucleoprotein complexes involved in pre-mRNA splicing, mRNA stability, and mRNA transport. FUS is a member of the FET family of proteins that bind RNAs (5) and contains an RNA recognition motif, three arginine-glycine-glycine (RGG) boxes, and a zinc finger (ZnF) (6). RGG2-ZnF-RGG3 is the major RNA-binding domain, which has a preference for GU-rich sequences (7, 8). The N terminus of FUS contains a low-complexity sequence domain involved in RNA granule formation (9). Nucleocytoplasmic shuttling of FUS occurs by a nonclassical proline-tyrosine nuclear localization signal (PY-NLS) and a nuclear export signal (NES) (10). Methylation of the C-terminal RGG3 domain of FUS is necessary for transportin 1 interaction and nuclear localization (11).
The majority of clinical ALS/FTLD-associated FUS mutations occur in its C-terminal PY-NLS sequence (12), which is believed to enhance the cytoplasmic localization and aggregation propensity of the protein and reduce its ability to bind nuclear RNAs. In response to various stressors, FUS localizes into cytoplasmic stress granules (13). In neurons, there is more immunodetectable FUS at dendritic spines in response to metabolic glutamate receptor (mGluR) agonists (14). Moreover, neurons cultured from FUS-knockout mice have abnormal spine morphology and spine density (14). It is unclear whether pathological FUS mutations disrupt activity-dependent synaptic structure or function.
Besides missense mutations at the C terminus of FUS protein, overexpression of wild-type FUS caused by mutations in its 3′ UTR also has been linked to ALS (15), suggesting that overexpression of wild-type FUS is pathogenic under certain circumstances. Indeed, pathogenic effects of increased levels of wild-type proteins are common in other neurodegenerative disorders, as exemplified by increased gene dose or overexpression of wild-type TDP-43, α-synuclein, and amyloid β precursor protein (APP) in ALS/FTLD, Parkinson disease, and Alzheimer’s disease (16–18). However, in all these cases (including FUS), it is unclear whether protein overexpression and missense mutations contribute to neurodegenerative disorders via common or distinct mechanisms. In this work, we developed novel FUS transgenic mice expressing low levels of human wild-type FUS (FUSWT) and an ALS-associated missense mutation (FUSR521G), which is located in the PY-NLS, to investigate the pathological consequences and molecular mechanisms of FUS overexpression and missense mutation.
Results
Development of Cre-Inducible Transgenic Mice Globally Overexpressing Low Levels of Human FUSWT and FUSR521G.
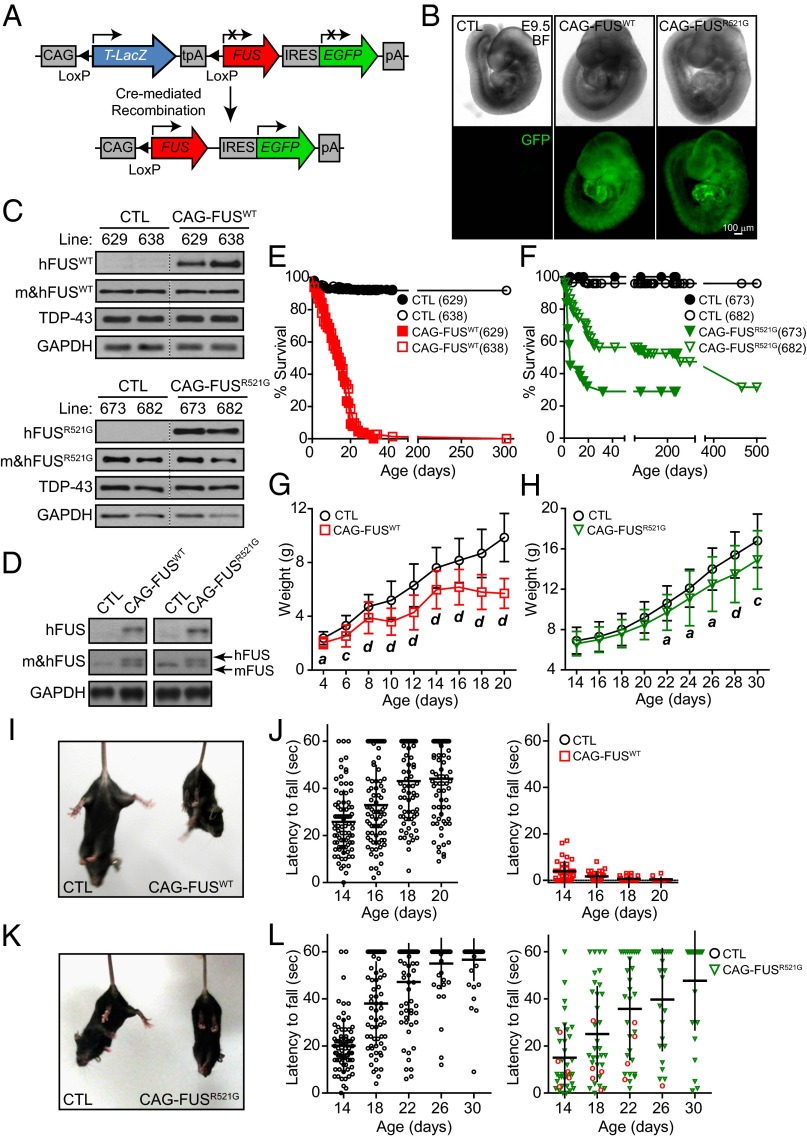
We generated transgenic mice expressing human wild-type FUS or the R521G mutation under the control of the cytomegalovirus immediate early enhancer-chicken β-actin hybrid (CAG) promoter (Fig. 1A), referred to as “CAG-FUSWT” and “CAG-FUSR521G,” respectively. In this study, we chose to overexpress the FUS transgenes globally, starting from germ line and at low levels to recapitulate more closely the expression profile and FUS levels in patients with ALS and FTLD, because FUS is ubiquitously expressed in human tissues, and human patients carry FUS mutations (or overexpressed wild-type protein) all their lives, starting from germ line. Accordingly, two mouse lines harboring CAG-Z-FUSWT-IRES-EGFP or CAG-Z-FUSR521G-IRES-EGFP were crossed with the germ-line Meox2Cre mice. Cre recombinase excises the LacZ DNA sequence flanked by loxP sequences, allowing translation of FUS and GFP (Fig. 1 B and C). Transgenic CAG-FUSWT mice (lines 629 and 638) and CAG-FUSR521G mice (lines 673 and 682) were born at normal Mendelian ratios (SI Appendix, Table S1). Analysis of total brain lysates from CAG-FUSWT (line 638) and CAG-FUSR521G (line 682) mice showed that the level of human FUS expression was similar to that of endogenous mouse FUS (Fig. 1D).
Fig. 1.
Generation of CAG-FUS transgenic mice. (A) Schematic of the CAG-Z-FUS-IRES-EGFP transgenic construct. Mice carrying the CAG-Z-FUS-IRES-EGFP transgene are crossed with germ-line Meox2Cre mice. Cre recombinase excises the DNA sequence flanked by loxP sequences, allowing translation of FUS (red) and EGFP (green). (B) Embryos at E9.5 (bright field, BF) that have both Meox2Cre and CAG-Z-FUSWT-IRES-EGFP (CAG-FUSWT) or CAG-Z-FUSR521G-IRES-EGFP (CAG-FUSR521G) have global GFP expression. (C) Immunoblot of lysates from P0 mouse brains from CAG-FUSWT (lines 629 and 638) and CAG-FUSR521G (lines 673 and 682) transgenic lines for human (h)FUS, total mouse and human (m&h)FUS (shown is the FUS Santa Cruz antibody), TDP-43, and GAPDH. Samples were pooled from three pups from each line. (D) Immunoblot of P20 mouse whole brain showing hFUS and resolved endogenous mouse and exogenous (m&h)FUS proteins (shown is the FUS Sigma antibody) from CAG-FUSWT (638) and CAG-FUSR521G (682) mice. (E and F) Survival curves of CAG-FUSWT (E) and CAG-FUSR521G (F) mice. (G and H) Body weight curves of CAG-FUSWT (638) (P4–P20) (G) and CAG-FUSR521G (682) (P14-P30) (H) mice. (I and K) CAG-FUSWT (I) and CAG-FUSR521G (K) mice display hindlimb curl. (J and L) Grip test of CAG-FUSWT mice (638), postnatal stages (P14–P20) (n = 21 litters) (J) and of CAG-FUSR521G mice (682), postnatal stages (P14–P30) (n = 17 litters) (L). Red circles indicate CAG-FUSR521G mice that had severe deficits in motor function and early lethality. Quantification is shown in SI Appendix, Table S2. (G and H) a, P < 0.05; b, P < 0.01; c, P < 0.005; d, P < 0.001 (one-way repeated measures ANOVA and post hoc Tukey test). Error bars represent SD of the mean.
Mice from CAG-FUSWT (629 and 638) and CAG-FUSR521G (673 and 682) transgenic lines were found to have reduced lifespan (Fig. 1 E and F): Nearly 100% of CAG-FUSWT (lines 629 and 638) mice die before postnatal day (P)30, and ∼70% of the CAG-FUSR521G (line 673) and ∼50% of CAG-FUSR521G (line 682) mice have early lethality before P30. In monitoring the body weights of these mice from birth, we observed that the body weights of CAG-FUSWT mice are significantly different from their littermates starting at ∼P4 (Fig. 1G and SI Appendix, Fig. S2A). The weight differences between CAG-FUSR521G mice and their littermates are less obvious (Fig. 1H and SI Appendix, Fig. S2B). CAG-FUSWT mice developed gait abnormalities at P10. By P14 their grip strength and righting ability were reduced, and hindlimb clasping was present (Fig. 1 I and J and SI Appendix, Table S2). Animals at this stage either died or were euthanized. Compared with the CAG-FUSWT mice, the CAG-FUSR521G mice that die early display similar but less severe impairments in locomotion in terms of gait, grip strength, righting ability, and hindlimb clasping (Fig. 1 K and L and SI Appendix, Table S2). The CAG-FUSR521G mice that escaped early lethality had somewhat reduced body mass and displayed subtle motor impairment (SI Appendix, Fig. S2 C–E).
Defective Neuromuscular Synapses and Neuroinflammation in Juvenile CAG-FUSWT and CAG-FUSR521G Mice with Severe Motor Impairment.
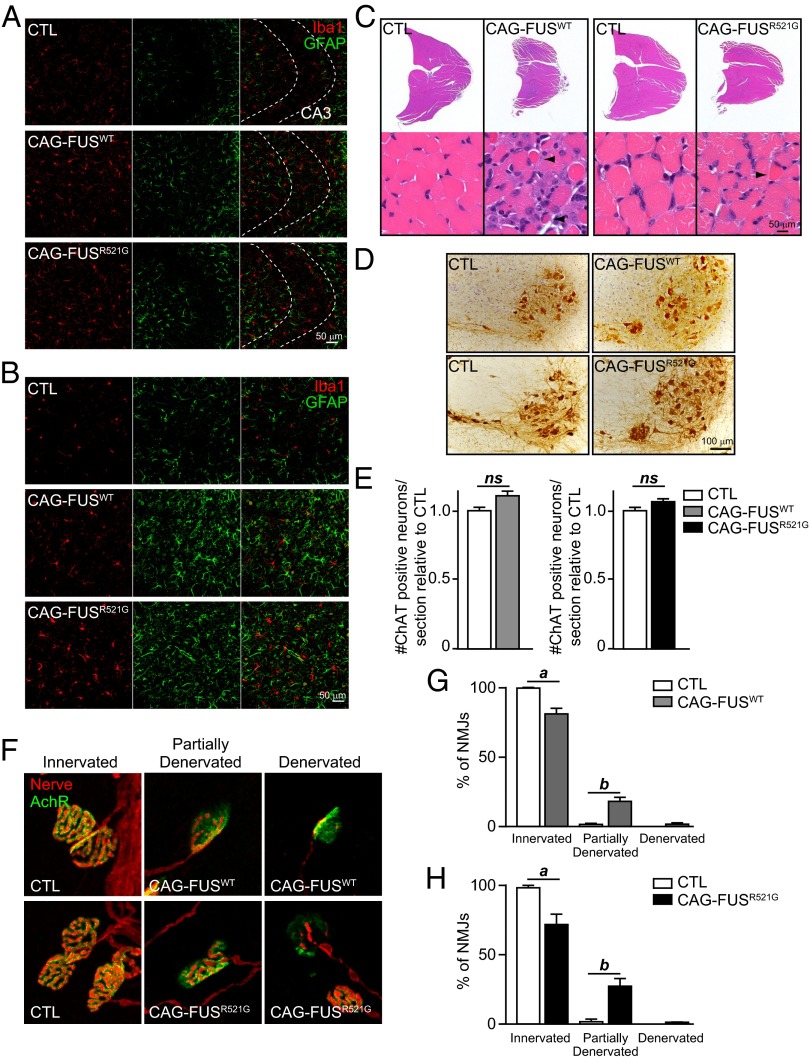
Inflammation, degeneration of motor neurons, and FUS aggregation are present in patients with ALS-FUS (3, 19, 20). We examined brains and spinal cords of end-stage CAG-FUSWT and CAG-FUSR521G mice and found no detectible cytoplasmic FUS localization or protein aggregates (SI Appendix, Fig. S1 E–H). Using immunohistochemistry, we then examined the activation of microglia and astrocytes as markers for neuroinflammation in the brains and spinal cords of end-stage CAG-FUSWT and CAG-FUSR521G mice. We found evidence of activation of astrocytes and microglia in all regions of the brain and spinal cord (Fig. 2 A and B and SI Appendix, Fig. S3 A and B). In contrast, CAG-FUSR521G mice that escaped early lethality did not have these markers of neuroinflammation (SI Appendix, Fig. S3 C and D).
Fig. 2.
Gliosis, muscle atrophy, and denervated NMJs in CAG-FUSWT and CAG-FUSR521G mice. (A and B) Immunofluorescence staining of the CA3 region of the hippocampus (A) and of the ventral horn of the spinal cord (B) for Iba1 (microglia; red) and GFAP (astrocytes; green) of end-stage mice. Images are representative of three animals per genotype. (C) H&E staining of the gastrocnemius muscle of CAG-FUSWT and CAG-FUSR521G mice at end stage show wasting of the muscle and scattered and grouped muscle atrophy, characteristic of motor axon degeneration. Arrowheads indicate pyknotic myofibers. Images are representative of three animals per genotype. (D) Immunostaining for ChAT highlights motor neurons in the cervical spinal cord of CAG-FUSWT and CAG-FUSR521G mice. (E) Quantification of spinal motor neuron numbers in the cervical spinal cord of CAG-FUSWT and CAG-FUSR521G mice show no evidence of neuron loss compared with control littermates. n = 3 CAG-FUSWT mice and littermate controls, and n = 4 CAG-FUSR521G mice and littermate controls. ns, not significant (Student t test). (F) Costaining for presynaptic terminals (nerve; red) and with bungarotoxin for postsynaptic terminals (AchR; green) shows that NMJs are denervated in P20–P24 CAG-FUSWT and CAG-FUSR521G mice at end stage compared with littermate controls. (G and H) Quantification of innervated NMJs. a, P < 0.05; b, P < 0.01 (Student t test). Error bars represent SD of the mean.
ALS patients develop muscle atrophy caused by the degeneration of spinal motor neurons, together with axonal degeneration and sclerosis of the later columns of the spinal cord, which contain the corticospinal tracts. Examination of the lumbar region of the spinal cord revealed no degeneration of axons in the dorsal corticospinal tract or lateral columns or in the dorsal or ventral roots (SI Appendix, Fig. S4 B and C), indicating that descending motor axons were not altered in CAG-FUSWT or CAG-FUSR521G mice. Muscle histology from end-stage CAG-FUSWT and CAG-FUSR521G mice showed scattered and grouped atrophic muscle fibers (Fig. 2C), a characteristic of denervation in muscle from patients with ALS. CAG-FUSWT muscle showed more severe abnormalities, as observed by the presence of pyknotic myofibers (Fig. 2C, Left), whereas muscle abnormalities in CAG-FUSR521G mice were less severe (Fig. 2C, Right). Quantification of spinal motor neuron numbers in the cervical spinal cord of CAG-FUSWT and CAG-FUSR521G mice showed no evidence of neuron loss as compared with control littermates (Fig. 2 D and E). Importantly, there were abnormalities in the neuromuscular junctions (NMJs) of end-stage animals (SI Appendix, Fig. S5), and analysis of the NMJs revealed significant denervation (Fig. 2 F–H). Our results indicate that degeneration of NMJs and muscle atrophy contribute to loss of motor function in CAG-FUSWT and CAG-FUSR521G mice.
FUS has been implicated in transcriptional and posttranscriptional regulation of gene expression (21–26). We therefore asked whether changes in gene-expression patterns in CAG-FUSWT and CAG-FUSR521G mice explain both the similarities and differences in the behavioral and cellular phenotypes of the wild-type and mutant transgenic mice. To do so, we generated paired-end RNA sequencing (RNA-seq) libraries from total RNA isolated from spinal cords of CAG-FUSWT and CAG-FUSR521G mice and their littermate controls. To avoid secondary effects of end-stage mice on gene expression, we selected transgenic mice that had not yet shown severe deficits in motor function and did not meet our end-stage criteria. To this end, we used P20 mice with a health score between 1 and 2 (as described in Materials and Methods). We carefully selected these mice to be phenotypically similar. Additionally, the samples for each RNA-seq library (n = 2 for each genotype) were pooled from three individual mice, to take into account any phenotype variability (see SI Appendix, Supplemental Experimental Procedures for details). The analysis from CAG-FUSWT mice revealed 185 differentially expressed genes (with adjusted P value <0.05) (SI Appendix, Fig. S6A and Table S3). Genes with increased expression are enriched with Gene Ontology (GO) terms related to immune response: “DNA replication, recombination and repair” and “regulation of cell proliferation.” Genes with decreased expression show GO terms related to lipid and sterol biosynthesis. In contrast, CAG-FUSR521G mice had very few genes that were differentially expressed (with adjusted P value <0.05) (SI Appendix, Fig. S6B and Table S3), yielding no significant GO terms. The transcriptome profiles of these mice are consistent with the phenotypic differences observed between the CAG-FUSWT and CAG-FUSR521G transgenic models, wherein altering wild-type FUS levels is more deleterious than expression of FUSR521G.
Impaired Motor Function and Sociability in Adult FUSR521G Transgenic Mice.
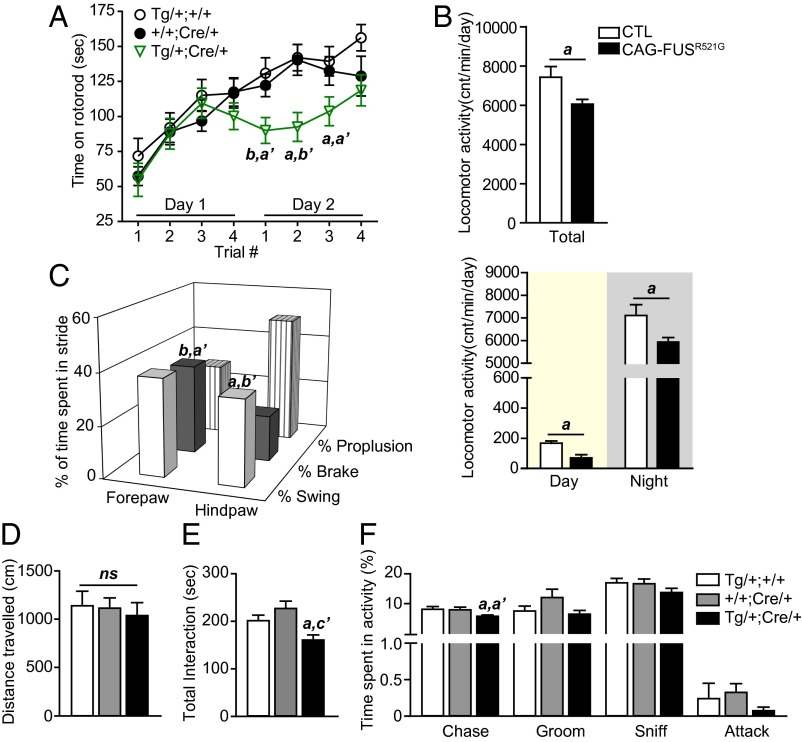
CAG-FUSR521G mice that escape early lethality were monitored further. They showed persistently lower body weight (SI Appendix, Fig. S2 C and D) with no obvious deficits in locomotion (Fig. 3D and SI Appendix, Fig. S7A) and displayed subtle behavioral differences as compared with their littermates. The motor function of CAG-FUSR521G mice was assessed on a rotorod over a 2-d period. On day 1 of rotorod testing, CAG-FUSR521G mice performed as well as their littermate controls, but on day 2 they had impaired motor function (Fig. 3A). CAG-FUSR521G mice were monitored on voluntary running wheels and showed less activity over a 9-d period (Fig. 3B). Despite the reduction in overall locomotor activity, food intake was not altered significantly (SI Appendix, Fig. S7 B and C). Gait analysis of the CAG-FUSR521G mice revealed that the braking phase was greater in the forelimbs (Fig. 3C and SI Appendix, Table S4), and the swing phase was reduced in the hindlimbs (Fig. 3C and SI Appendix, Table S5). Results from the ladder-walking test indicate that the forelimbs have more errors in stepping with few deficits in the hindlimbs (SI Appendix, Fig. S7 E–H). These data indicate that deficits in the motor function of the CAG-FUSR521G mice are modest and are more prominent when their motor function is challenged.
Fig. 3.
Decline in motor function and social interaction in CAG-FUSR521G transgenic mice that escape early lethality. (A) Rotorod performance of 2-mo-old CAG-FUSR521G mice showing motor impairment on day 2. (B) Total running-wheel activity of 2-mo-old CAG-FUSR521G mice (Upper) and their activity during the day and night (Lower). cnt, wheel revolutions. (C) Gait analysis of 2-mo-old CAG-FUSR521G mice showing that the braking phase is greater in the forepaws and the swing phase is greater in the hindpaws. (D) Open field test in 8-mo-old mice shows no differences in total distance traveled. (E and F) The resident/intruder test in 8-mo-old CAG-FUSR521G mice age shows significant reduction in total interactions (E) and particularly in chasing behavior (F). (A, C, and D–F) Three-way statistical comparisons with littermate controls used one-way ANOVA. a, P < 0.05; b, P < 0.01. (′ compares +/+;Cre/+ with Tg/+;Cre/+). ns, not significant. (B) a, P < 0.05; b, P < 0.01; c, P < 0.001 (Student t test). Error bars represent SEM.
We also examined the social interactions of CAG-FUSR521G mice with intruder/novel juvenile and adult mice. We found that the interaction with juvenile mice was significantly reduced at 4 mo of age (SI Appendix, Fig. S7I). When introduced to intruder adult mice, CAG-FUSR521G mice did not show any significant deficits before 8 mo of age (Fig. 3E and Movies S1 and S2). We analyzed the types of social interactions of 8-mo-old CAG-FUSR521G mice with an intruder adult and found that chasing behavior was reduced in CAG-FUSR521G mice (Fig. 3F). No alterations in cognitive function or olfaction were detected in the CAG-FUSR521G mice (SI Appendix, Fig. S7 J and K).
Altered Dendritic Branching in Spinal Motor Neurons and Sensorimotor Neurons of CAG-FUSR521G Mice.
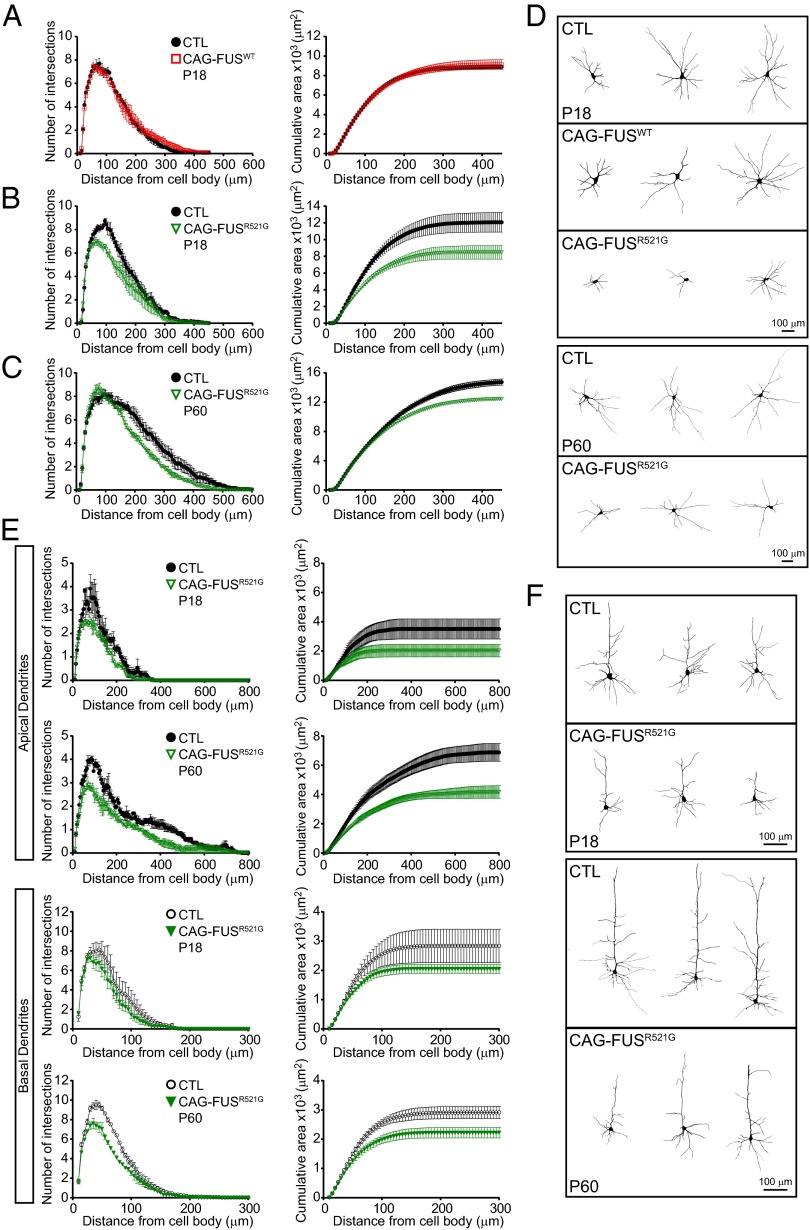
FUS is found in RNA granules at dendritic spines, and immunodetectable FUS at synapses increases in response to group 1 mGluR stimulation (14, 27). Hippocampal cultures from FUS-knockout mice have altered dendritic branching and reduced mature spines (14), suggesting that FUS has an important role at the synapse. We hypothesized that deficits in motor function and sociability in the CAG-FUSR521G “escapers” could be caused by alterations in dendrites or dendritic spines in the motor neurons and/or sensory motor cortex. We first examined the dendrites of spinal motor neurons in P18 FUS transgenic mice and found no reduction in the number of dendritic intersections or cumulative area in CAG-FUSWT mice (Fig. 4 A and D). In contrast, the dendritic intersections and cumulative area of dendrites were reduced significantly in spinal motor neurons in CAG-FUSR521G mice of the same age (Fig. 4 B and D). We then examined the CAG-FUSR521G escapers at age 2 mo (P60). Even though the distribution of the numbers of intersections and cumulative area of dendrites were slightly different in P18 and P60 mice, we found significant and persistent deficits in the dendritic branches in spinal motor neurons (Fig. 4 C and D). Moreover, analysis of apical and basal dendrites in neurons in sensorimotor cortex layers IV–V in CAG-FUSR521G mice showed fewer intersections and reduced cumulative area in the apical and basal dendrites of P18 and P60 mice (Fig. 4 E and F).
Fig. 4.
Reduced dendritic branching in spinal motor neurons and sensorimotor neurons of CAG-FUSR521G mice. (A) Sholl analyses show no reduction in the number of dendritic intersections or cumulative area of dendrites in spinal motor neurons in CAG-FUSWT mice. (B and C) In contrast, the dendritic intersections and cumulative area of dendrites show significant reductions of spinal motor neurons in P18 (B) and in 2-mo-old (P60) (C) CAG-FUSR521G mice. (D) Representative images of Neurolucida tracing of the dendrites of spinal motor neurons in control (CTL), CAG-FUSWT, and CAG-FUSR521G mice. A total of 36 spinal motor neurons were analyzed in CAG-FUSWT and CAG-FUSR521G mice and corresponding littermate controls. (E, Upper) Sholl analyses show reduced intersections and cumulative area in the apical dendrite within 50–250 μm from the cell body of cortical neurons of P18 and P60 CAG-FUSR521G mice. (Lower) Similar reductions in the dendritic intersections and cumulative surface areas are also identified in the basal dendrites of CAG-FUSR521G neurons. A total of 24 neurons from cortical layers IV–V were analyzed in CAG-FUSR521G mice and corresponding littermate controls. (F) Representative images of Neurolucida tracing of the apical and basal dendrites in neurons from layers IV–V in the sensorimotor cortex in control and CAG-FUSR521G mice. For each group three or four animals were analyzed. (A–C and E) P < 0.0001 (two-way repeated measures ANOVA). Error bars represent SEM.
Activity-Dependent Reduction of FUSR521G Protein Levels at Synapses.
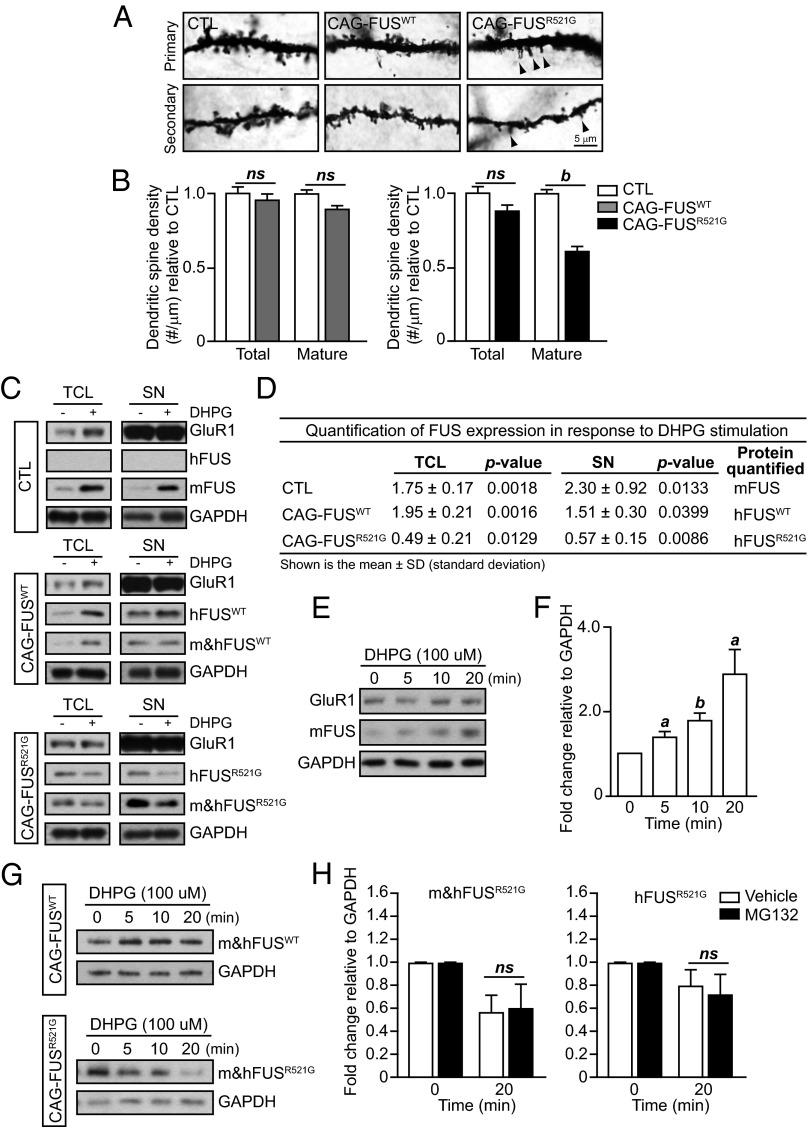
Cultured neurons from FUS-knockout mice have abnormal spine morphology as well as spine density (14). Therefore we decided to examine whether our transgenic models had alterations in the number of mature spines. We found that there was no difference in the total number of mature spines in the CAG-FUSWT mice, but CAG-FUSR521G mice had a significant decrease in the number and density of mature spines (Fig. 5 A and B).
Fig. 5.
Activity-dependent reduction of FUSR521G in response to mGluR stimulation. (A and B) Golgi images of dendritic spines in the apical and basal dendrites of control (CTL), CAG-FUSWT, and CAG-FUSR521G mice. Spine density was analyzed in a total of 30 neurons from cortical layers IV–V in P18 CAG-FUSWT and CAG-FUSR521G mice and their corresponding littermate controls (n = 3 mice per group). In CAG-FUSWT mice the density of mature dendritic spines does not differ in apical and basal dendrites. However, in CAG-FUSR521G mice the density of mature dendritic spines is reduced in the apical and secondary dendrites compared with littermate controls. (We have defined “mature” spines as being mushroom-shaped.) Arrowheads indicate immature spines. (C) Acute cortical tissue slices from littermate P18CTL, CAG-FUSWT, or CAG-FUSR521G mice (n = 3 mice per group) were pretreated with AMPA (20 μM 6,7-dinitroquinoxaline-2,3-dione, DNQX) and NMDA [5 μM 3-(2-carboxypiperazin-4-yl)propyl-1-phosphonic acid, CPP] inhibitors, followed by treatment with 100 μM DHPG for 10 min or no DHPG treatment. Total cell lysates (TCL) and synaptoneurosomes (SNs) were immunoblotted for human FUS (hFUS), mouse FUS (mFUS), total FUS (m&hFUS), GAPDH, and GluR1. GluR1 is enriched in the synaptoneurosome fraction. (D) Quantification of FUS expression from total cell lysates and synaptoneurosomes from acute cortical tissue slices treated with DHPG relative to untreated groups. Immunoblots are representative of three or four separate experiments. P values were obtained by Student t test. (E) Isolated synaptoneurosomes from CTL mice were treated with 100 μM DHPG for the indicated time and were immunoblotted for mFUS, GAPDH, and GluR1. (F) Quantification of FUS relative to GAPDH indicates a significant increase in expression in response to DHPG treatment. (G) Isolated synaptoneurosomes from CAG-FUSWT and CAG-FUSR521G mice were treated with 100 μM DHPG for the indicated time and were immunoblotted for total m&hFUS and GAPDH. Immunoblots are representative of two separate experiments. (H) Isolated synaptoneurosomes from CAG-FUSR521G mice were pretreated with 25 μM MG132 or vehicle (DMSO) before stimulation with 100 μM DHPG for 20 min. Synaptoneurosome lysates were immunoblotted, and m&hFUS and hFUS levels were quantified relative to GAPDH. MG132 did not inhibit the decrease in FUS expression. (B, D, F, and H) ns, not significant. a, P < 0.05; b, P < 0.01 (Student t test). The FUS Santa Cruz antibody was used for all blots for m&hFUS. Three animals were used in each experimental group. (B and H) Error bars represent SEM; (F) Error bars represent SD.
Activation of group 1 mGluRs in hippocampal neurons has been shown to affect spine shape in a protein synthesis-dependent manner (28). Given what is known about the existence of FUS at synapses and its response to mGluR signaling, we hypothesized that deficits in dendritic branching and spine formation may stem from altered responses of FUSR521G protein to mGluR activation. To test this hypothesis, we determined whether FUSR521G protein displayed an altered synaptic expression upon activation of mGluRs. Using acute cortical tissue slices, we demonstrated that endogenous mouse FUS and human FUSWT protein levels are increased in total cell lysates and in synaptoneurosome fractions after treatment with the group 1 mGluR agonist (R,S)-3,5-dihydroxyphenylglycine (DHPG) (Fig. 5 C and D). This result is consistent with an increase in immunodetectable FUS at synapses in response to mGluR stimulation previously reported in dissociated hippocampal cultured neurons (14, 27). In contrast, FUSR521G protein levels were reduced in response to DHPG treatment in acute cortical slices (Fig. 5 C and D). These results indicate that mutant FUS does not respond properly to mGluR activation, and the reduced FUS levels may lead to the altered dendritic branching and spines.
The decrease in FUSR521G could be caused by a deficit in the synthesis, trafficking, and/or degradation of FUS proteins. To test whether FUS’s response to DHPG stimulation is a local event at synapses, we isolated synaptoneurosomes using a discontinuous Percoll–sucrose gradient and treated them with DHPG in vitro. We found that FUS expression is induced significantly in the synaptoneurosomes of control mice (Fig. 5 E and F), suggesting that local synthesis of the protein does occur. We then performed these same in vitro experiments in synaptoneurosomes from CAG-FUSWT and CAG-FUSR521G mice. We found that both endogenous mouse FUS and exogenous human FUSWT increase in the CAG-FUSWT samples and decrease in the CAG-FUSR521G samples (Fig. 5G), as is consistent with our observation in the acute cortical tissue slice model (Fig. 5 C and D). We then tested whether the decrease in FUSR521G could be blocked using a proteasome inhibitor, MG132, and found that inhibiting the proteasome does not prevent a decrease in FUS expression (Fig. 5H). Together, these observations suggest that the alterations of FUS levels in response to mGluR activation are local synaptic events, likely related to protein synthesis.
Discussion
Cre-Inducible Transgenic Mice Expressing Low Levels of FUS as Novel Models of ALS and FTLD.
The pathological and genetic association of FUS with ALS and FTLD suggests that dysregulation of FUS may lead to neurodegenerative diseases. However, the mechanism by which FUS aggregation or mutations cause ALS and FTLD is not known. To study the role of FUS in neurodegeneration, we generated Cre-inducible FUS transgenic mice that express low levels of wild-type (FUSWT) or mutant (FUSR521G) proteins. Under control of the CAG promoter, the human FUS transgene is expressed ubiquitously in the germ line of CAG-FUSWT and CAG-FUSR521G mice (Fig. 1). CAG-FUSWT and CAG-FUSR521G mice that develop severe deficits in motor function have denervation of the NMJs, muscle atrophy, neuroinflammation, and early lethality (Figs. 1 and 2). The phenotypes observed in our transgenic models phenocopy aspects of adult cases of ALS. However, the onset of phenotypes in the mouse models is earlier, more closely reflecting FUS-linked juvenile ALS (29–31).
A portion of CAG-FUSR521G mice that escape early lethality have impairments in motor function and sociability (Figs. 1 I–L and 3), which are likely linked to the alterations in dendritic branches and spines in the upper and lower motor neurons (Figs. 4 and 5). Adult CAG-FUSR521G mice do not perform as well on the rotorod and they are less active on a running wheel (Fig. 3 A and B). Specifically, the forelimbs of these mice are impaired (SI Appendix, Fig. S7 E and F). Upper or lower limb weakness is common in both ALS and FTLD with motor function deficits (32–35). Changes in social interactions also are a common clinical feature of patients with FTLD and in ALS patients with dementia (1). Similar to progranulin (Grn)-knockout mice, a model of familial FTLD that has deficits in social interaction (36), CAG-FUSR521G mice have deficits in social interactions with intruder/novel juvenile and adult mice (Fig. 3 E and F and SI Appendix, Fig. S7I).
Differences and Commonalities in FUS Overexpression and Missense Mutations.
The CAG-FUSWT and CAG-FUSR521G mouse models demonstrate that increased expression of FUS alone can cause cellular toxicity. This result is consistent with the recent finding that mutations in the 3′ UTR of FUS increase FUS expression levels and cause ALS (15). This result is also in agreement with the observations that increased gene dose or overexpression of APP, α-synuclein, and TDP-43 can cause Alzheimer’s disease, Parkinson disease, and ALS/FTLD (16–18). On the other hand, we did not detect overt motor neuron loss or apparent ubiquitin-positive aggregation and mislocalization of FUS in neurons and glia of either of our transgenic mouse models (SI Appendix, Fig. S1 E–H), suggesting that permanent FUS mislocalization, aggregation, and motor neuron loss are not necessary for disease onset but might be end-stage pathological markers or outcomes in human patients. Moreover, our studies suggest that peripheral and central synapses are more vulnerable than axons and cell bodies and that synaptic defects precede axonal and neuronal degeneration.
The cellular phenotypes observed in our animal models may represent cellular events occurring before FUS mislocalization, aggregation, and neuronal death that are the key neuropathological features of ALS/FTD. Although FUSR521G is not overtly mislocalized in our animals, it is likely that the shuttling dynamics of FUSR521G are altered, because FUS R521 residue is located at PY-NLS. On the other hand, PY-NLS has very high binding affinity (kd = 9.5 nM) for karyopherinβ2 (Kapβ2, also known as “transportin”), which mediates FUS nuclear shuttling (37). Although ALS mutations in PY-NLS reduce Kapβ2-binding affinities by several fold (37), the mutant FUS proteins still have nanomolar affinity and thus are expected to be translocated efficiently to the nucleus unless the nuclear import machinery is overwhelmed (such as when wild-type or mutant FUS proteins are massively overexpressed). Therefore it is not surprising that there is no overt FUS mislocalization in our transgenic mice with low FUS expression.
Some of our CAG-FUSR521G mutant mice escaped early lethality, but none of the CAG-FUSWT mice survived to adulthood. The similar phenotypes we observe in both of the CAG-FUSWT and CAG-FUSR521G founding lines reduces the possibility that these observations are caused by insertional effects. Also, these transgenic lines have very low copy numbers of transgenes with single genomic insertion (SI Appendix, Fig. S1C). Interestingly, in ALS patients the age of disease onset and clinical phenotypes are variable, with incomplete penetrance for TDP-43 and FUS mutations. It is possible that other factors, such as genetics and environment, have an impact on whether an individual can escape the consequences of these autosomal dominant mutations. Incomplete penetrance has also has been observed in carriers of the APOE4 allele, 50% of whom develop Alzheimer’s disease but the remainder do not. Another argument for incomplete penetrance of FUS mutations has to do with the possibility that mutations in the PY-NLS are also partial loss-of-function mutations, presumably in gene expression. This is supported by the finding that the ALS-associated FUS mutant proteins R521G and H517Q have reduced binding to intronic sequences of its nuclear RNA targets (24).
We note that the CAG-FUSWT mice readily recapitulated human diseases caused by increased levels of wild-type FUS. Although CAG-FUSR521G mice can model the toxic gain of functions of FUS (which is highly relevant to the studies of ALS/FTLD), modeling loss of function is more difficult in the mutant mice. Nevertheless, results from our parallel studies of the wild-type and mutant animals are consistent with a model wherein overexpression of FUSWT alters the nuclear function of endogenous FUS at the level of gene expression, but FUSR521G mutation has both a partial loss of function in RNA regulation and gene expression and a partial toxic gain of function in disrupting synapses. This model is supported by our transcriptome analysis of the spinal cords of the transgenic mice, which revealed that the gene-expression pattern is altered in CAG-FUSWT mice but not in CAG-FUSR521G mice (SI Appendix, Fig. S6). This result also potentially explains why an increase in wild-type FUS level is more deleterious than the overexpression of FUSR521G (Fig. 1 and SI Appendix, Fig. S6 and Table S2). At the steady state, both FUSWT and FUSR521G stay mainly in the nucleus, but FUSR521G has no apparent effect on gene expression. This result supports the view that overexpressed FUSR521G has a diminished ability to alter the nuclear function of endogenous FUS in gene expression. As discussed in more detail in the next section, although FUSR521G does not affect gene expression, it has a toxic gain of function disrupting synaptic homeostasis at dendritic spines. In contrast, FUSWT does not affect synaptic homeostasis.
In other published animal models (38–40), overexpression of wild-type FUS has been reported as being less toxic than overexpression of mutant FUS. In addition, a recently developed FUSWT transgenic mouse line showed no deficits until crossed to homozygosity, wherein these mice displayed progressive hindlimb paralysis and neuromuscular denervation (41). These studies contrast with our observation that overexpression of FUSWT is more toxic than overexpression of FUSR521G in terms of early lethality. The reason for the different degree of toxicity in different FUS transgenic models needs to be examined further and will yield important insight into disease progression. In this regard, we note that in our models FUS proteins are expressed globally at low levels and during early embryonic development. Because ALS is thought to be a non–cell-autonomous disease (42), it will be important to examine the contribution of different cell types, particularly astrocytes and microglia, to the phenotypes observed in our transgenic models. The Cre-inducible transgenic system developed herein will allow temporal and spatial expression of FUS proteins in glial cells to test the hypothesis that ALS and FTLD are non–cell-autonomous. Similarly, it will be informative to use Cre-lines specific for motor neurons and cortical neurons to examine separately FUS’s contributions to ALS and FTLD.
Mutant-Specific Disruption of mGluR-Dependent Synaptic Homeostasis.
Primary hippocampal cultures from FUS-knockout mice have altered dendritic branching and reduced mature spines (14). Based on this information and the behavioral phenotypes in the CAG-FUSR521G mice, we examined dendritic branching in the motor neurons and sensorimotor cortex and found significant reductions in dendritic intersections and in the cumulative area of dendrites. Moreover, the density of mature dendritic spines is reduced in the apical and secondary dendrites in the mutant mice (Fig. 5). These data are consistent with transgenic mice harboring the R521C mutation under control of the Syrian hamster prion promoter (43). However, Qiu et al. (43) reported only transgenic mice for FUS mutant R521C, without comparable wild-type transgenic animals. Therefore it was unclear whether the phenotypes in their studies were caused by simple overexpression of FUS protein or were specific to the FUS mutation. In our studies we observed persistent dendritic defects in the spinal motor neurons and cortical neurons in FUSR521G mice at P18 and P60, suggesting that the negative impacts of FUSR521G on dendritic morphology can occur at young age. Importantly, we did not see the same alterations in the CAG-FUSWT mice, indicating that although certain aspects of the CAG-FUSWT and CAG-FUSR521G models are similar, the alterations in synaptic homeostasis resulting in alterations in dendritic branches and spines are specific to the R521G mutation. It is likely that disruption of synaptic homeostasis at dendritic spines contributes to the alterations in motor function and social interaction of the mutant transgenic animals.
Interestingly, Grn-knockout mice display similar alterations in dendritic branching and spine maturation, which correspond with deficits in social interaction (36). FTLD and ALS share common clinical and pathological features including loss of cognition, motor impairment, and TDP-43– or FUS-positive inclusions. Although the FUS R521G mutation is associated with familial ALS, the rare FUS mutations P106L, G206S, and M254V are linked to familial FTLD (3). Indeed, our CAG-FUSR521G mice phenocopy aspects of the loss of motor function observed in ALS. They also show changes in social interactions resembling those observed in FTLD. On the other hand, alterations in dendritic branching and spines have not been documented for ALS or FTLD, but the findings from our CAG-FUSR521G mice suggest that these alterations might exist in human patients.
FUS also localizes to RNA granules at the synapse (27) and copurifies with the NMDA receptor (44). In response to mGluR5 stimulation, there is more immunodetectable FUS at dendritic spines (14). Fujii et al. (14) did not assess whether the increase in immunodetectable FUS at the synapse was caused by local translation of FUS mRNA or by localization to the synapse. Using an in vitro assay to assess local protein translation in isolated synaptoneurosomes, we demonstrate that the increase in FUS expression at the synapse in response to mGluR activation is a local event (Fig. 5 E and F). The acute increase in synaptic FUS expression in response to mGluR activation as demonstrated by Fujii et al. (14) and in our study strongly suggests that FUS participates in the regulation of mRNAs important to synaptic function and serves as an important synaptic RNA-binding protein. This finding is reproducible in our studies using acute cortical tissue slices, where FUSWT protein is increased but FUSR521G protein is reduced in response to mGluR activation (Fig. 5 C and D). We also found that inhibiting the proteasome does not prevent the activity-dependent decrease of FUS expression in synaptoneurosomes isolated from CAG-FUSR521G mice. Together, these observations suggest that the alterations of FUS levels in response to mGluR activation are local synaptic events that are likely to be related to protein synthesis. Future studies will test whether dysregulation of synaptic FUS in response to mGluR activation contributes to the altered dendritic branching and maturation of spines in the CAG-FUSR521G mice and perhaps also in human patients with ALS or FTLD.
Activity-dependent down-regulation of the FUSR521G protein at the synapse and its potential role in disrupting the formation or maintenance of dendritic spines provides a tantalizing mechanism for FUS regulation at the synapse. In this context, we note that our finding that FUS dysfunction disrupts synaptic homeostasis at dendritic spines somewhat parallels observations for another RNA-binding protein, fragile X mental retardation protein (FMRP). FMRP has been shown to regulate spine shape in a protein synthesis-dependent manner. In response to mGluR signaling, FMRP regulates local translation of mRNAs at the synapse (45). Loss-of-function mutations in the FMR1 gene cause fragile X mental retardation syndrome, in which a deficit in spine maturation is thought to underlie the autism-like symptoms in individuals with the syndrome (46). In the future, it would be important to test how deficits in the synthesis of mutant FUS proteins lead to the disruption of synaptic homeostasis. Moreover, it would be of interest to examine whether disruption of synaptic homeostasis caused by dysfunction of RNA metabolism represents a common theme of brain disorders.
Materials and Methods
For more details, see SI Appendix, Supplemental Experimental Procedures.
Generation of FUS Transgenic Mice.
Wild-type or mutant R521G human FUS cDNAs were inserted into the CAG-Z-IRES-EGFP vector (provided by Yuji Mishina, University of Michigan, Ann Arbor, MI). The CAG-Z-FUS-IRES-EGFP construct was digested with AflII and SpeI to remove the vector sequence and then was injected into fertilized oocytes from C57BL/6 female mice and implanted into pseudopregnant ICR mice. Mice carrying the transgene were identified by PCR analysis and β-galactosidase activity as previously reported (47). To induce global overexpression of human FUS, the CAG-Z-FUS-IRES-EGFP mice were bred to Meox2-Cre mice to yield CAG-FUSWT or CAG-FUSR521G mice. CAG-FUSWT and CAG-FUSR521G pups were monitored daily and scored as follows: 0 = healthy; 1 = limp tail or hindlimb weakness; 2 = limp tail and hindlimb weakness; 3 = moderate hindlimb weakness and/or unilateral hindlimb paralysis; 4 = bilateral, complete hindlimb paralysis; and 5 = moribund state accompanied by complete hindlimb paralysis with forelimb weakness. Mice with a score of 3 or higher or that had a loss of total body weight >20, were considered to have reached end stage and were euthanized. All experimental procedures involving animals in this study were reviewed and approved by the University of Texas Southwestern Institutional Animal Care and Use Committee.
Behavior Testing.
Grip test.
Mice were placed on a 15.5 × 15.5-cm wire grid. The grid then was inverted and secured 42 cm above a padded surface. Latency to fall was measured, with a maximum trial time of 1 min. The latency to fall is reported for P12–P30 CAG-FUSWT (n = 21 litters) and CAG-FUSR521G (n = 17 litters) transgenic mice and their littermate controls.
Rotorod.
Mice were placed on a stationary rotorod (IITC Life Science Inc.). The rod then was accelerated from 5–45 rpm over 5 min. The time that each mouse fell from the rod was recorded. Mice that held onto the rod for two complete rotations were scored as if they had fallen from the rod. Each mouse was tested four times a day for two consecutive days with an intertrial interval of at least 15 min. CAG-FUSR521G (Tg/+;Cre/+, n = 18) and littermate controls (Tg/+;+/+, n = 19 and +/+;Cre/+, n = 19) were tested.
Running wheel.
Mice were housed individually in running-wheel cages with access to food ad libitum. Wheel-running activity was recorded continuously for 14 d using the ClockLab data collection system (Actimetrics). Locomotor activity was assessed in the final 9 d of recording, allowing acclimation to the running wheel. CAG-FUSR521G (n = 6) and littermate controls (n = 4) were tested at 5 mo of age.
Digigait.
Mice were placed onto the Digigait (Mouse Specifics) and allowed to explore and habituate for 2–3 min. Then the treadmill was started at 10 cm/s, and the speed was increased rapidly to 20 cm/s. A camera located below the transparent treadmill collected the images, and the data were analyzed automatically by the software. Approximately 10 steps at a constant pace were recorded. CAG-FUSR521G (Tg/+;Cre/+, n = 18) and littermate controls (Tg/+;+/+, n = 19 and +/+;Cre/+, n = 19) were tested at 2 mo of age.
Resident/intruder test.
Adult test mice were housed singly in clean cages for 24 h before testing. An adult intruder/novel mouse (age 3–8 mo, same sex as the test mouse and not heavier) was introduced into the cage containing the resident/test mouse. Each trial duration was 10 min, and active behaviors were recorded using the Noldus Observer program. Active behaviors were defined as attack (resident biting, pinning, and kicking of the hind limbs), chase (resident closely following the intruder), grooming (resident climbing on intruder, tugging its hair or tail, or rubbing its snout on the intruder’s body), and sniffing (resident sniffing the intruder’s anogenital region or other body part). Intruder mice were used only twice. Acclimation and testing were conducted under red light to minimize any stress and anxiety. CAG-FUSR521G (Tg/+;Cre/+, n = 17) and littermate controls (Tg/+;+/+, n = 15 and +/+;Cre/+, n = 16) were tested at 8 mo of age.
Histology Staining.
Tissues stained with H&E were fixed in 10% (wt/vol) formalin fixative for 48 h, paraffin embedded, and sectioned to 8-μm thickness.
Immunostaining.
Brains, spinal cords, and NMJs from whole mounts of triangularis sterni muscles from mice (age P18–P25) were prepared and immunostained as previously described (47–49). Tissue sections were immunostained with primary antibodies: GFAP (AB5541; Millipore), IBA1 (019-19741; Wako), GFP (Aves 1020), human FUS (B327D), and To-Pro3 (T3605; LifeTechnologies). Triangularis sterni muscles were immunostained with Alexa Fluor 647 α-bungarotoxin (AchR; Invitrogen) or Syntaxin 1 (a gift from Thomas Südhof, Stanford University, Stanford, CA). Primary antibodies were incubated overnight at 4 °C, and Alexa Fluor-conjugated secondary antibodies (Invitrogen) were incubated for 2 h at room temperature. Three animals from each genotype (n = 3) were analyzed, and a minimum of 400 NMJs per genotype were assessed.
Western Blot Analysis.
Lysates from tissues were processed, and equal proteins were resolved by SDS/PAGE as previously reported (47). Primary antibodies used were glutamate receptor 1 (GluR1; MAB2263; Millipore), GAPDH (G9545; Sigma), FUS (HPA008784; Sigma), FUS (sc-47711; Santa Cruz), GFP (1020; Aves); human FUS antibody was a gift from Hongxia Zhou and Xu-Gang Xia, Thomas Jefferson University, Philadelphia (40) and human FUS peptide antibody B327D (SYGQPQSGSYSQQPS) was generated in rabbits as previously described (47). Immunodetected proteins were quantified by densitometry using the NIH ImageJ software. We were able to estimate the amounts of human and mouse FUS in the transgenic animals visually by using larger SDS/PAGE gels (Fig. 1D) but were unable to quantify the human FUS level accurately because of the similar molecular weights of human FUS (526 amino acids) and mouse FUS (518 amino acids) (SI Appendix, Fig. S1D).
Golgi Staining for the Analysis of Dendrites in Cortical Neurons and Cervical Spinal Motor Neurons.
Both male and female CAG-FUSWT and CAG-FUSR521G mice and their littermate controls were used for Golgi staining, and samples were analyzed used the Neurolucida software (MicroBrightField) as previously reported (43). Three animals from each genotype (n = 3), with 12 cervical spinal motor neurons (from the ventral horn region) and 10 cortical neurons (from layers IV–V in the sensorimotor cortex) were traced and analyzed.
Choline Acetyltransferase Staining of Spinal Cord and Quantification of Spinal Motor Neurons.
Tissues from the cervical spinal cord were processed, immunostained, and analyzed as previously reported (43). Anti-choline acetyltransferase (ChAT) (1:300; Millipore), biotinylated rabbit anti-goat IgG antibody (1:100; Vector Labs); and the VECTASTAIN Elite ABC Kit (Vector Labs) were used for staining motor neurons. ChAT-positive cells in the ventral horn region were quantified using at least 12 images per animal. CAG-FUSR521G (n = 3) and CAG-FUSWT (n = 4) transgenic mice and their littermate controls (n = 3 or 4) were analyzed.
Acute Treatment of Cortical Tissue Slices with DHPG.
After DHPG treatment of acute cortical tissue slices, total cell lysates were collected, and the remaining homogenate was passed through two 100-μm filters and then through one 10-μm filter. Synaptoneurosomes were pelleted after 10-min centrifugation at 1,000 × g. Total cell lysates, supernatants, and synaptoneurosomes were lysed as previously reported (50). Experimental replicates (n = 3) were analyzed for each genotype.
Synaptoneurosome Isolation, in vitro Treatment with DHPG.
Synaptoneurosomes used for in vitro DHPG stimulation experiments were isolated as previously reported (51). Synaptoneurosomes were equilibrated to room temperature for 10 min before stimulation with DHPG (100 μM). Pretreatment with DMSO (vehicle) or 25 μM MG132 (Tocris Biosciences) was performed at room temperature for 10 min before DHPG stimulation.
Supplementary Material
Acknowledgments
We thank Jim Richardson and John Shelton of the University of Texas Southwestern (UTSW) Histology Core Facilities for assistance with histology staining and analysis; Shari Birnbaum and Laura Peca of the UTSW Behavior Core Facility for assistance with rotorod, Digigait, social interaction, olfactory, and Y-test testing; Erik Plautz and Sherry Rovinsky of the UTSW Neuro-Models Facility, with support from the Haggerty Center for Brain Injury and Repair, for assistance with ladder-rung testing and analysis; Leighton Stein of the Roswell Park Cancer Institute for assistance with FISH analysis of copy number in mouse embryonic fibroblasts; Weichun Lin for advice on NMJ studies; Vincent Zimmern (Ecole Polytechnique Federale de Lausanne) and Pradipta Ray (University of Texas at Dallas) for contributions to the initial RNA-Seq analysis; and Paul A. Dutchak for experimental discussion and critical feedback on the manuscript. This work was supported by Alzheimer's Association, Consortium for Frontotemporal Dementia Research, Friends of the Alzheimer's Disease Center of UT Southwestern Medical Center, National Institutes of Health, National Natural Science Foundation of China, Ministry of Science and Technology of China, US Department of Veterans Affairs Biomedical Laboratory Research and Development Merit and Pilot Awards, and Muscular Dystrophy Association.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406162111/-/DCSupplemental.
References
- 1.Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59(7):1077–1079. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 2.Mackenzie IR, et al. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: An update. Acta Neuropathol. 2010;119(1):1–4. doi: 10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dormann D, Haass C. Fused in sarcoma (FUS): An oncogene goes awry in neurodegeneration. Mol Cell Neurosci. 2013;56:475–486. doi: 10.1016/j.mcn.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Lee EB, Lee VM, Trojanowski JQ. Gains or losses: Molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci. 2012;13(1):38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan AY, Manley JL. The TET family of proteins: Functions and roles in disease. J Mol Cell Biol. 2009;1(2):82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 7.Lerga A, et al. Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem. 2001;276(9):6807–6816. doi: 10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- 8.Iko Y, et al. Domain architectures and characterization of an RNA-binding protein, TLS. J Biol Chem. 2004;279(43):44834–44840. doi: 10.1074/jbc.M408552200. [DOI] [PubMed] [Google Scholar]
- 9.Kato M, et al. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149(4):753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee BJ, et al. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126(3):543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dormann D, et al. Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 2012;31(22):4258–4275. doi: 10.1038/emboj.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dormann D, Haass C. TDP-43 and FUS: A nuclear affair. Trends Neurosci. 2011;34(7):339–348. doi: 10.1016/j.tins.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Bentmann E, et al. Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43) J Biol Chem. 2012;287(27):23079–23094. doi: 10.1074/jbc.M111.328757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujii R, et al. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005;15(6):587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 15.Sabatelli M, et al. Mutations in the 3′ untranslated region of FUS causing FUS overexpression are associated with amyotrophic lateral sclerosis. Hum Mol Genet. 2013;22(23):4748–4755. doi: 10.1093/hmg/ddt328. [DOI] [PubMed] [Google Scholar]
- 16.Gitcho MA, et al. TARDBP 3′-UTR variant in autopsy-confirmed frontotemporal lobar degeneration with TDP-43 proteinopathy. Acta Neuropathol. 2009;118(5):633–645. doi: 10.1007/s00401-009-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rovelet-Lecrux A, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38(1):24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 18.Chartier-Harlin MC, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364(9440):1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 19.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 20.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz JC, et al. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 2012;26(24):2690–2695. doi: 10.1101/gad.204602.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishigaki S, et al. Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions. Sci Rep. 2012;2:529. doi: 10.1038/srep00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogelj B, et al. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci Rep. 2012;2:603. doi: 10.1038/srep00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoell JI, et al. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol. 2011;18(12):1428–1431. doi: 10.1038/nsmb.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagier-Tourenne C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15(11):1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon I, et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155(5):1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belly A, Moreau-Gachelin F, Sadoul R, Goldberg Y. Delocalization of the multifunctional RNA splicing factor TLS/FUS in hippocampal neurones: Exclusion from the nucleus and accumulation in dendritic granules and spine heads. Neurosci Lett. 2005;379(3):152–157. doi: 10.1016/j.neulet.2004.12.071. [DOI] [PubMed] [Google Scholar]
- 28.Vanderklish PW, Edelman GM. Dendritic spines elongate after stimulation of group 1 metabotropic glutamate receptors in cultured hippocampal neurons. Proc Natl Acad Sci USA. 2002;99(3):1639–1644. doi: 10.1073/pnas.032681099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conte A, et al. P525L FUS mutation is consistently associated with a severe form of juvenile amyotrophic lateral sclerosis. Neuromuscul Disord. 2012;22(1):73–75. doi: 10.1016/j.nmd.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Belzil VV, et al. Novel FUS deletion in a patient with juvenile amyotrophic lateral sclerosis. Arch Neurol. 2012;69(5):653–656. doi: 10.1001/archneurol.2011.2499. [DOI] [PubMed] [Google Scholar]
- 31.Zou ZY, et al. De novo FUS gene mutations are associated with juvenile-onset sporadic amyotrophic lateral sclerosis in China. Neurobiol Aging. 2013;34(4):1312.e1–1312.e8. doi: 10.1016/j.neurobiolaging.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Lee EB, et al. Topography of FUS pathology distinguishes late-onset BIBD from aFTLD-U. Acta Neuropathologica Comm. 2013;1(9):1–11. doi: 10.1186/2051-5960-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taieb G, et al. R521C mutation in the FUS/TLS gene presenting as juvenile onset flail leg syndrome. Muscle Nerve. 2013;48(6):993–994. doi: 10.1002/mus.23956. [DOI] [PubMed] [Google Scholar]
- 34.Rademakers R, et al. Fus gene mutations in familial and sporadic amyotrophic lateral sclerosis. Muscle Nerve. 2010;42(2):170–176. doi: 10.1002/mus.21665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corrado L, et al. Mutations of FUS gene in sporadic amyotrophic lateral sclerosis. J Med Genet. 2010;47(3):190–194. doi: 10.1136/jmg.2009.071027. [DOI] [PubMed] [Google Scholar]
- 36.Petkau TL, et al. Synaptic dysfunction in progranulin-deficient mice. Neurobiol Dis. 2012;45(2):711–722. doi: 10.1016/j.nbd.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Zhang ZC, Chook YM. Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS) Proc Natl Acad Sci USA. 2012;109(30):12017–12021. doi: 10.1073/pnas.1207247109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanson NA, Jr, et al. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet. 2011;20(13):2510–2523. doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murakami T, et al. ALS mutations in FUS cause neuronal dysfunction and death in Caenorhabditis elegans by a dominant gain-of-function mechanism. Hum Mol Genet. 2012;21(1):1–9. doi: 10.1093/hmg/ddr417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang C, et al. FUS transgenic rats develop the phenotypes of amyotrophic lateral sclerosis and frontotemporal lobar degeneration. PLoS Genet. 2011;7(3):e1002011. doi: 10.1371/journal.pgen.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitchell JC, et al. Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age- and dose-dependent fashion. Acta Neuropathol. 2013;125(2):273–288. doi: 10.1007/s00401-012-1043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187(6):761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu H, et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014;124(3):981–999. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3(7):661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 45.Darnell JC, Klann E. The translation of translational control by FMRP: Therapeutic targets for FXS. Nat Neurosci. 2013;16(11):1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irwin SA, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex. 2000;10(10):1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- 47.Sephton CF, et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285(9):6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Sugiura Y, Lin W. The role of synaptobrevin1/VAMP1 in Ca2+-triggered neurotransmitter release at the mouse neuromuscular junction. J Physiol. 2011;589(Pt 7):1603–1618. doi: 10.1113/jphysiol.2010.201939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dewey CM, et al. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol. 2011;31(5):1098–1108. doi: 10.1128/MCB.01279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sephton CF, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2011;286(2):1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westmark PR, Westmark CJ, Jeevananthan A, Malter JS. Preparation of synaptoneurosomes from mouse cortex using a discontinuous percoll-sucrose density gradient. J Vis Exp. 2011 doi: 10.3791/3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.