Significance
Tumor cells are heterogeneous, and much variation occurs at the single-cell level, which may contribute to therapeutic response. Here, we studied drug resistance dynamics in a model of tolerance with a metastatic breast cancer cell line by leveraging the power of single-cell RNA-Seq technology. Drug-tolerant cells within a single clone rapidly express high cell-to-cell transcript variability, with a gene expression profile similar to untreated cells, and the population reacquires paclitaxel sensitivity. Our gene expression and single nucleotide variants analyses suggest that equivalent phenotypes are achieved without relying on a unique molecular event or fixed transcriptional programs. Thus, transcriptional heterogeneity might ensure survival of cancer cells with equivalent combinations of gene expression programs and/or single nucleotide variants.
Keywords: single cell, paclitaxel, tumor heterogeniety, drug resistance, RNA-Seq
Abstract
The acute cellular response to stress generates a subpopulation of reversibly stress-tolerant cells under conditions that are lethal to the majority of the population. Stress tolerance is attributed to heterogeneity of gene expression within the population to ensure survival of a minority. We performed whole transcriptome sequencing analyses of metastatic human breast cancer cells subjected to the chemotherapeutic agent paclitaxel at the single-cell and population levels. Here we show that specific transcriptional programs are enacted within untreated, stressed, and drug-tolerant cell groups while generating high heterogeneity between single cells within and between groups. We further demonstrate that drug-tolerant cells contain specific RNA variants residing in genes involved in microtubule organization and stabilization, as well as cell adhesion and cell surface signaling. In addition, the gene expression profile of drug-tolerant cells is similar to that of untreated cells within a few doublings. Thus, single-cell analyses reveal the dynamics of the stress response in terms of cell-specific RNA variants driving heterogeneity, the survival of a minority population through generation of specific RNA variants, and the efficient reconversion of stress-tolerant cells back to normalcy.
A major barrier to successful cancer treatment is the recurrence of cancer cells with acquired resistance to chemotherapy (1–3). However, the molecular events underlying cancer cell evolution toward a drug-resistant phenotype are largely unknown. Recent studies using next-generation sequencing (NGS) systems have attempted to identify the genetic changes that drive tumorigenesis and resistance to treatments (4, 5). These sequencing studies have revealed that many of the resistance-imparting mutations identified are different from tumor to tumor. In addition to heterogeneity across tumors from different patients, intratumor heterogeneity adds another level of complexity. Minor subpopulations of cancer cells can harbor aberrations that are associated with resistance to therapy and tumor progression (6–8). Thus, treatments may be effective against the majority of the tumor, but a small population of resistant cells can cause the persistence, recrudescence, or recurrence of cancer that is refractory to further treatment. Sequencing studies on bulk tumor tissue can only identify mutations present in subpopulations of a heterogeneous tumor in a limited capacity. By sequencing the transcriptome of single cells in depth, low abundance mutations can be detected that will facilitate identifying the drivers of drug resistance.
Recent advances have enabled the analysis of DNA and RNA within a single cell. The coupling of whole genome amplification and DNA sequencing have allowed multiple groups to study the genetics of single cells, but not without significant amplification biases (9–11). Moreover, single-cell exome sequencing confirmed the clonal heterogeneity of a solid tumor identifying key mutations across much of the genome (12). DNA sequencing can identify mutations across the genome, but is unable to illuminate expressional differences that can contribute significantly to drug resistance. Multiplexed single-cell quantitative PCR (qPCR) assays allow expression-based analysis of up to 96 targets in a single experiment (13). Recently, a few groups have demonstrated that RNA-Seq of single cells using NGS technology is feasible, reproducible, and usable for gene expression-based classification of cell subpopulations (14–17). A major advantage of RNA-Seq in single-cell studies is that the entire transcriptome can be surveyed, rather than a limited number of genes. DNA and RNA methodologies are not mutually exclusive and can be combined to generate more biologically significant information.
Paclitaxel (Taxol) is a chemotherapy drug commonly used to treat solid cancers including breast tumors (18). This toxin targets microtubules to interfere with the mitotic spindle, resulting in cell cycle arrest and ultimately apoptosis. Paclitaxel treatment kills most tumor cells but, for the residual cancer cells, the mechanisms of resistance are unclear (18). An important question is whether mutations that drive drug resistance are common in a population or arise from unique mutations in individual cells.
Here we leverage the power of single-cell RNA-Seq to identify single nucleotide variants (SNVs) and gene expression at the single-cell level in an insightful drug-tolerance experimental paradigm. We evaluated three groups of cells from the human breast carcinoma cell line, MDA-MB-231: untreated cells, stressed cells that had been exposed to paclitaxel treatment for 5 d plus 1 d drug free, and drug-tolerant cells from a small (n < 64) clonal population of cells that resumed proliferation after paclitaxel treatment. In addition to sequencing the mRNA of single cells, we also performed DNA sequencing of a population of untreated cells and RNA-Seq of a population from each of the three groups to facilitate the identification of SNVs and RNA variants. We also performed differential gene expression profiling for single cells and population cells of the three groups to identify the transcriptional stress response and cytotoxic effects of paclitaxel on gene expression.
Results
Generation of a Paclitaxel Tolerance Paradigm in Metastatic Human Cancer Cells and Isolation of Single Cells.
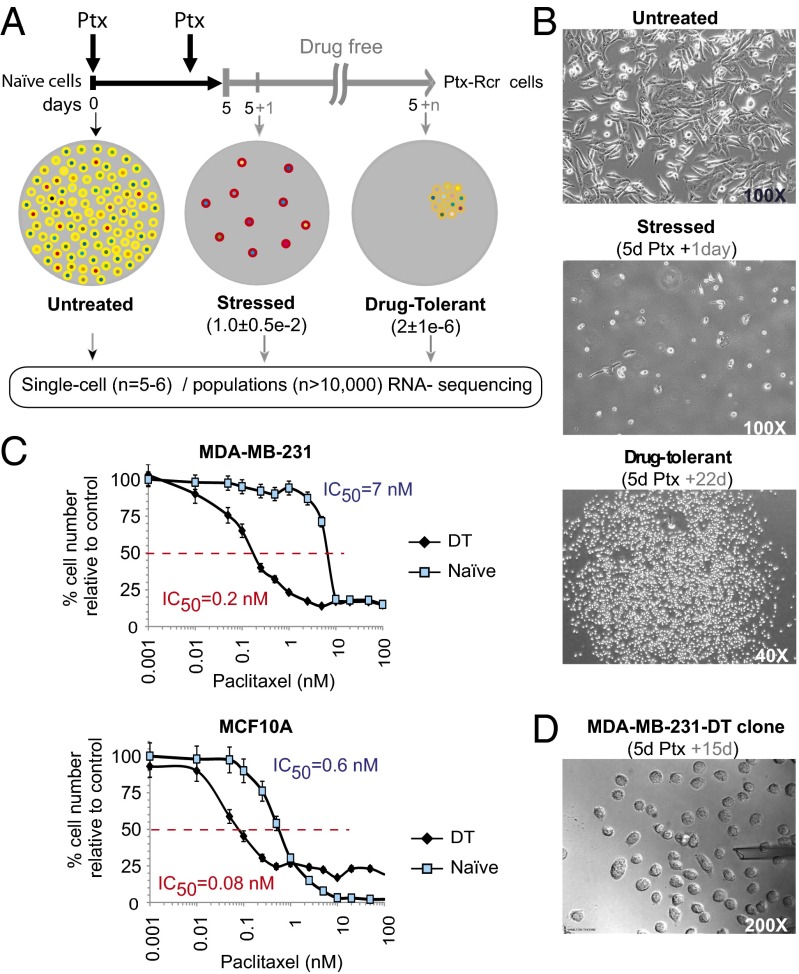
To investigate the molecular events associated with the response of cancer cells to drug-treatment followed by drug withdrawal that may be potentially associated with drug tolerance, we exposed the paclitaxel-sensitive (IC50 < 10 nM) (18) metastatic human breast cancer cell line MDA-MB-231 to paclitaxel (100 nM) according to the regimen diagrammed in Fig. 1A. After 5 d of drug exposure, most cells had died. Residual cells alive 1 d after paclitaxel removal were considered to be a stressed cell population, and the majority of these cells underwent apoptosis within 2–4 wk. A small number of residual stressed cells resumed proliferation and established clones, and such cells were considered to be drug-tolerant cells (Fig. 1B). A drug–toxicity curve was also constructed using a range of paclitaxel concentrations (Fig. 1C). The IC50 was ∼7 nM with ∼20% viable cells. Interestingly, reexposure of the drug-tolerant cell population to paclitaxel resulted in these cells becoming much more drug sensitive, with an IC50 of ∼0.2 nM. This apparent increase in sensitivity was also found in precancerous MCF10A cells (Fig. 1C, Lower), indicating that this is not an exclusive phenomenon of fully transformed cells and that drug-tolerant cell populations retain a cellular memory of the prior drug exposure. This result suggests that smaller paclitaxel doses would be more effective in killing drug-tolerant cells. However, we observed that higher doses of paclitaxel reduced the growth rate of tolerant cells to a similar (MDA-231) or lesser (MCF10A) extent than in naïve cells, indicating that high dosage of paclitaxel is equally effective for drug-tolerant cells. These results suggested that (i) phenotypic heterogeneity is reestablished after expansion of these cells; (ii) drug tolerance is reversible; and (iii) the new IC50 may reflect a protective preconditioning effect by stopping cell growth even at lower concentrations of paclitaxel on the drug-tolerant cells.
Fig. 1.
Reversible phenotypic equilibrium in response to paclitaxel in cancer cells. (A) Regimen for expansion of paclitaxel (Ptx) stress-tolerant cells. Highly metastatic MDA-MB-231 naïve (yellow) cells were treated with Ptx (100 nM) on day 1 and day 3. After 5 d, Ptx was removed, and cells were left in a drug-free culture. Most stressed cells arrested (red) and ultimately died, whereas rare drug-tolerant cells (orange) resumed proliferation after 10–15 d, and clones were expanded. Five single cells per group were analyzed either before treatment, 1 d after Ptx removal, as well as from recently established (n < 64) or long-term expanded, drug-tolerant clones. Populations were analyzed from long-term expanded clones. Frequencies of surviving stressed and drug-tolerant cells observed are indicated between parentheses. Cell-to-cell heterogenous RNA content is indicated with varying colors. (B) Bright field images of untreated, stressed, and drug-tolerant cells at the indicated times after drug removal. Total magnification is indicated. (C) Paclitaxel toxicity assays on naïve or drug-tolerant MDA-MB-231 cells (Upper) and MCF10A cells (Lower). Growth inhibitory concentrations 50% (IC50) are indicated. Data shown are the mean ± SEM from a quadruplicated representative experiment. (D) Bright-field image of an MDA-MB-231-Ptx-tolerant clone (n < 64) during single cell collection by micromanipulation. The number of days in drug-free culture is indicated at the top. The opening of the micropipette of roughly 20 μm is shown. Total magnification is indicated.
Early Drug Tolerance Dynamics Analysis at the Single-Cell Level.
To better understand the early events occurring soon after the onset of proliferation of the rare drug-tolerant cells, we conducted whole transcriptome sequencing analyses at the single cell level for untreated, stressed and drug-tolerant (collected from a proliferating clone at less than six cell divisions) populations. Five single cells were isolated from each treatment group by picking single cells with glass needles using micromanipulators over an inverted microscope and immediately placing each cell in lysis buffer (Fig. 1D). For whole population analyses, >10,000 pooled cells were collected from each group. We used a linear RNA amplification system for the whole transcriptome sequencing (19). The use of such a system prevents reproduction of an error introduced in earlier amplification cycles, a concern in exponential amplification systems.
We generated similar average numbers of sequencing reads for individual single cells and each cell population, 77 million reads and 100 million reads, respectively (SI Appendix, Table S1). With a somewhat similar number of sequencing reads, RNA-Seq from single cells generated a much greater sequencing depth than it did for cell populations. On average, we had 117 times coverage for single cells and 23 times coverage for cell populations. By contrast, RNA sequencing reads of the cell populations covered 5.4 times more genomic regions compared with that of a single cell (SI Appendix, Table S2). This result indicates that with a comparable number of reads generated, the single-cell RNA sequencing generates less coverage than the cell population RNA sequencing, with the consideration that each individual cell may be expressing only a fraction of the genes that are expressed in the bulk population. Indeed, the fraction of genes expressed above 1 RPKM (or 1 adj-RPKM; Experimental Procedures) in single cells compared with their bulk populations is only 20%, whereas pooling and mapping the reads from each cell within the same group resulted in a much greater approximation of the number of genes expressed above the same threshold (SI Appendix, Table S3).
Low-Abundance Novel RNA Variants in Single Cells Are Not Detectable in Cell Populations.
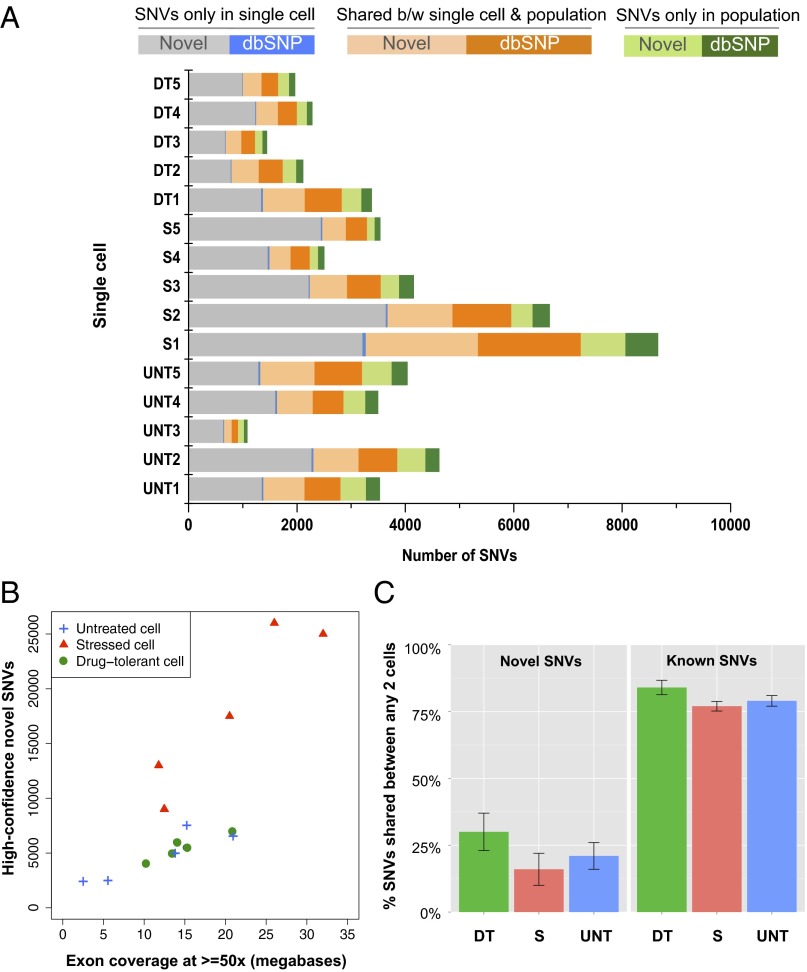
SNVs consist of somatic mutations propagated from DNA and other RNA variants that are introduced by processes such as RNA editing or errors in transcription. RNA variants in our data are supported by sufficient evidence that they are only present in the RNA sequencing reads and not in any of the DNA sequencing reads. Novel SNVs are variants that are not present in dbSNP (The SNP Database) (20). Variants in dbSNP are common SNPs that are found in at least 1% of the human population; therefore, they are not rare variants. Most of the novel SNVs identified in single cells were not detected at the cell population level, despite the fact that there were 2–10 times more total SNVs found in cell populations than in single cells (SI Appendix, Table S4) and that SNVs detected in the cell populations cover more genomic regions than those from single cells, as indicated above. Within comparable genomic regions where there was at least 10 times RNA read coverage, there were about 6 times fewer SNVs detected at the population level than in single cells. In most cases, the novel variants in single cells were not the major variants in the cell populations, whereas most dbSNP variants were shared between single cells and population cells (Fig. 2A). Because the number of RNA variants called could be directly related to the depth of sequencing, we compared the amount of SNVs detected in both single cells and cell populations at various depth of read coverage thresholds (SI Appendix, Fig. S1). The number of SNVs was normalized by the number of genomic bases with the corresponding depth of read coverage to ensure that the difference in the number of SNVs was not due to differences in genome coverage breadth. Strikingly, most SNVs were found at genomic regions with less than 60 times read coverage, whereas the genomic regions with deeper read coverage do not present more SNVs. Moreover, more novel (non-dbSNP) variants were detected from single cells than from populations regardless of the depth of coverage. In contrast, more dbSNP variants were detected at the population-cell level compared with the single-cell level at all depths of coverage. Additionally, we observed that with a similar number of uniquely mapped reads for single-cells vs. cell populations; the latter have only slightly more transcriptomic bases with reads at lower depth of coverage, but this difference is minimal or even reverts in regions with higher depth of coverage (SI Appendix, Fig. S2). Thus, with a similar amount of resources and effort, RNA sequencing from single cells has increased sensitivity to detect novel SNVs that are not apparent from RNA-Seq from cell populations.
Fig. 2.
SNVs identified at the single cell level. (A) The percentage of SNVs shared between single cells and their corresponding population was small. The majority of novel variants in single cells were unique (gray bars) and not detected at the population level. Most of the common variants that were present in a single cell and its corresponding population were known variants cataloged in dbSNP (orange bars). A relatively small number of unique SNVs were detected in the population in genomic regions that had read coverage in both the single cell and population (green bars). Also see SI Appendix, Table S4. (B) There were highly disparate variant rates within the different treatment conditions. Single cells from the stressed cell group (red triangles) contained about two times more novel SNVs than did single untreated cells or single drug-tolerant cells. Drug-tolerant cells had a variant rate similar to that of untreated cells. (C, Left) Cell-to-cell novel SNVs comparison. Novel variants are the ones that are not present in the dbSNP database. We only considered SNVs in genomic regions where both single cells have at least 10 reads in the cell-to-cell comparison; therefore, the differences between two cells are not due to the differing amount of coverage. The bar plot shows the average percent of novel shared variants between any two single cells for each group. (Right) Most known variants present in one cell were present in another cell of any different group. The bar plot shows the average percent of known (dbSNP) shared variants between any two single cells for each group. Also see SI Appendix, Fig. S3.
RNA Variation Is Similar in Drug-Tolerant Cells and Other Cancer Cells.
We measured the number of novel variants shared between any two cells in the genomic regions where each single cell has at least 10 times read coverage. Before paclitaxel treatment, untreated single cells shared about 30% of novel variants with any other untreated cell. On exposing the cells to 5 d of paclitaxel treatment, the stressed cells appeared to have accumulated additional novel variants that were not previously present in the untreated cells (Fig. 2B). On average, stressed cells shared 24% of novel variants with each other, but fewer than 20% novel variants in stressed cells were found in any single cell in either the untreated or drug-tolerant group (Fig. 2C and SI Appendix, Fig. S3). Although drug-tolerant cells were clonal, they shared similar percentages of novel SNVs among themselves compared with untreated single cells: about 25–45%. Drug-tolerant cells and untreated cells shared about 30% of novel variants. Overall, most novel variants in one cell were unique (Fig. 2C and SI Appendix, Fig. S3). Furthermore, all single cells shared more than 75% of the known dbSNP variants with any other cell of any of the three groups (Fig. 2C). This result shows that those variants cataloged in the dbSNP are not rare even at the single cell level.
To compare our RNA-Seq variant calls and variant calls using other single cell RNA-Seq datasets that used normal human cells and other cancer cells, we performed SNV analysis with two other published single cell RNA-Seq datasets using the same SNV analysis method described in Experimental Procedures, including single cells collected from human early embryos (21), human embryonic stem cells (hESCs) (14, 21), and human melanoma cells (14). Single-cell RNA-Seq from human early embryos including oocytes, two-cell embryos, four-cell embyros, and hESCs (passage 0) shows that as cells go through each cell division, the number of novel variants progressively increases (SI Appendix, Table S5). Importantly, the frequency of total cell-specific SNVs, i.e., those not present in any other cell of the drug-tolerant group, is about 4.7e-4/bp (∼7.8e-5 SNVs/bp/cell division; SI Appendix, Table S6). Meanwhile, the polymorphism frequencies from the other two datasets were 3.3e-4/bp for hES cells, 7.0e-4/bp for cancer cells (14), 1.4e-4/bp for two-cell stage human embryos, and 4.7e-4/bp for four-cell stage human embryos, with 2.4e-4 SNVs/bp/cell division (21). Moreover, although neither untreated cells nor long-term stressed cells are monoclonal, the frequency of cell-specific SNVs in the latter is greater, suggesting either that cellular stress may increase errors in transcription such as when RNA polymerase inserts the wrong base into the transcript, or RNA editing events, or those cells that persisted in a permanent arrest state are rare cells preexisting in the heterogeneous untreated population.
To identify the RNA editing events, one needs to compare DNA and RNA from the same cell. However, it is not yet possible to sequence whole genome DNA and whole transcriptome RNA simultaneously from a single cell. To gain insight into whether our single-cell RNA variants result from RNA editing, we compared the base substitution patterns between our single-cell RNA variants and other previously published RNA editing events.
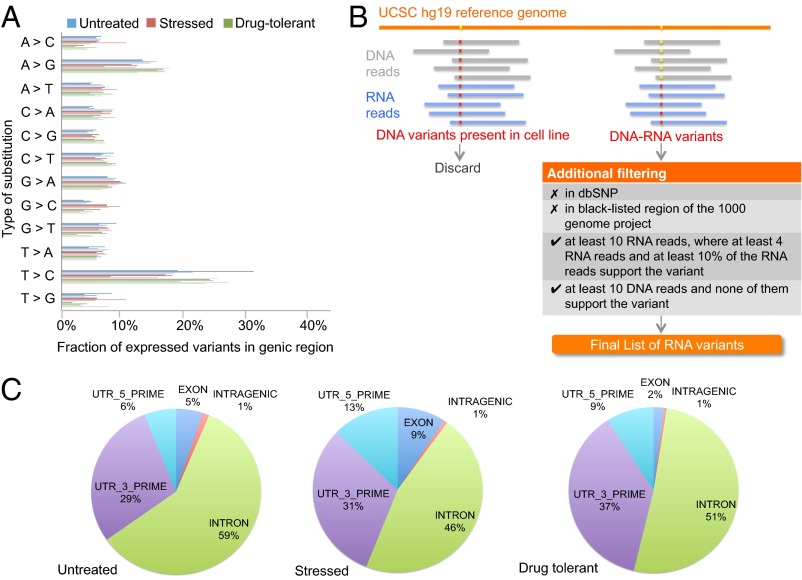
A-to-G substitution is typically the most frequently occurring RNA variants detected in other RNA editing studies (22–25). Curiously, we found that after T-to-C, A-to-G substitutions were the more frequent substitutions identified in our study (Fig. 3A). In addition, most of the A-to-G RNA variants observed in the single cells occurred in the intronic regions and UTRs (Fig. 3C), in agreement with previous A-to-G RNA editing studies (24, 26, 27).
Fig. 3.
RNA variants identified at the single-cell level. (A) The distribution of the 12 RNA variants at the single-cell level showed that the most abundant types of RNA variants were A-to-G and T-to-C. (B) Multiple filters were applied to identify candidate RNA variants. Any SNV in the DNA of the cell line that did not match the human genome reference was considered to be a DNA variant specific to the MDA-MB-231 cell line. DNA-RNA variants were those in which the base call in the single cell RNA reads differed from that in the cell line DNA reads, and the base call in the reference genome agreed with that in the cell line DNA. The DNA-RNA variants were first subjected to two filters that removed all known variants and variants that were not within the accessible genome defined by the 1000 Genome Project Consortium. Two filters were applied to the rest of the DNA-RNA variants to ensure that the RNA variants have enough sequencing reads to support an RNA variant call. (C) The distribution of A-to-G RNA variants relative to gene boundaries. The majority of the A-to-G RNA variants were clustered in noncoding regions including introns and 5′ and 3′ UTRs. Also see SI Appendix, Tables S7 and S8.
RNA Variants Found Only in Drug-Tolerant Cells Are Involved in Microtubule Stabilization and Organization.
In any single cell, there were ∼5,000 cell line-specific SNVs that were different from the human reference genome (hg19), and about 63,000 cell line-specific SNVs were found in the population cells of all three groups. After removing all cell line-specific variants, the remaining DNA-RNA variants that passed all additional filters were considered to be RNA variants (Fig. 3B). The accuracy of the variant calls was validated through pyrosequencing (SI Appendix, Table S7). We validated the SNVs at 10 different loci on a new set of single cells from independent groups of untreated MDA-MB-231 cells and different drug-tolerant clones. All of the SNVs identified by pyrosequencing agreed with the ones detected using Illumina HiSEq. 2000. To estimate the percentage of base calls generated due to sequencing/amplification errors, we calculated the false-positive SNV rate by comparing the fraction of known (dbSNP) variants that were found only in a single cell but not in the population in comparable genomic regions (where both the population and the single cell have at least 10 times read coverage). In most single cells, there are less than 5% false-positive variant calls (SI Appendix, Table S4).
There were 38 RNA variants in at least three of five drug-tolerant cells that were not present in any untreated or stressed cells and were not detected in the population cells (SI Appendix, Table S8). These RNA variants are identical at the nucleotide level in the drug-tolerant cells. One of the variants present in all five drug-tolerant single cells was located on chromosome 8 (chr8:17885150) and represented a missense mutation in the PCM1 (pericentriolar material 1) gene that encodes a protein essential for anchoring microtubules to the centrosome (28, 29). PCM1 is involved in microtubule stabilization and assembly of centrosomal proteins (29). Centrosome function is essential for completion of interphase and mitosis (30), and aberrant centrosomal activity has been implicated in tumor progression (31, 32). Three other RNA variants unique to the drug-tolerant single cells were found in genes that were involved in microtubule organization and stabilization during mitosis: RAPGEF4, NUDCD3, and KIAA1671 (Table 1). RAPGEF4 (Rap guanine nucleotide exchange factor 4) was previously shown to interact with protein complexes that were involved in microtubule polymerization and organization (33, 34). RAPGEF4 protein is also known as exchange protein directly activated by cAMP 2 (EPAC2) and is one of the binding partners of MAP1A (microtubule-associated protein 1A) (33). MAP1A is known to promote elongation and nucleation of tubulin (35). Depletion of RAPGEF4 showed a significant increase in paclitaxel-induced microtubule stabilization in paclitaxel-resistant A549-T12 lung carcinoma cells and partially restored paclitaxel sensitivity in a previous study (36). The gene NUDCD3 encodes the NudCL (nuclear distribution gene C-like) protein. NudCL has been shown to interact with the dynein complex, a minus-end-directed microtubule motor (37), and is required for mitosis and cytokinesis (38). Depletion of NudCL causes loss of dynein function, which leads to insufficient recruitment of γ-tubulin to spindle poles and mislocalization of the dynein complex during mitosis (37). The protein encoded by KIAA1671 is involved in mitosis and chromosome segregation (39, 40). Antibodies against this protein were found in sera of breast cancer patients that had developed autoantibodies (41). We also analyzed the presence of SNVs in other genes known for their role in paclitaxel resistance, including RAPGEF4. Most of these genes showed variable depth of coverage (SI Appendix, Table S9), and although other SNVs were called in some of these genes, only RAPGEF4 presented a SNV in at least three of the drug-tolerant cells (SI Appendix, Table S10). Interestingly, three different missense variants were found in three different survivor cells in the same gene, RTN4. RTN4 protein was previously shown to sequester antiapoptotic proteins BCL2 and BCL-xL in the endoplasmic reticulum and prevent them from entering mitochondria (42). The missense mutations in RTN can potentially alter its binding affinity for BCL2 and BCL-xL and increase the concentration of the antiapoptotic proteins in the mitochondria, which could prevent cells from entering apoptosis.
Table 1.
List of RNA variants present in at least three of five of the drug-tolerant single cells and not found in untreated or stressed cells
| RNA variants in drug-tolerant cells only | Function | Mutation | Locus | Δaa* | Ref. | |
| RAPGEF4 | Rap guanine nucleotide exchange factor (GEF) 4 | Paclitaxel resistance | Missense | chr2: 173916571 | L1785M | (35) |
| AMOTL1 | Angiomotin like 1 | 3′-UTR | chr11: 94607183 | — | (42, 43) | |
| PCM1 | Pericentriolar material 1 | Microtubule organization and stabilization | Missense | chr8: 17885150 | G227R | (27, 28) |
| NUDCD3 | NudC domain containing 3 | Missense | chr7: 44425714 | H131D | (36) | |
| KIAA1671 | Uncharacterized protein KIAA1671 | 3-UTR | chr22: 25592835 | — | (38, 39) | |
Amino acid change.
Stressed Cells Undergo a Paclitaxel-Induced Transcription Response That Is Not Apparent in Drug-Tolerant Cells.
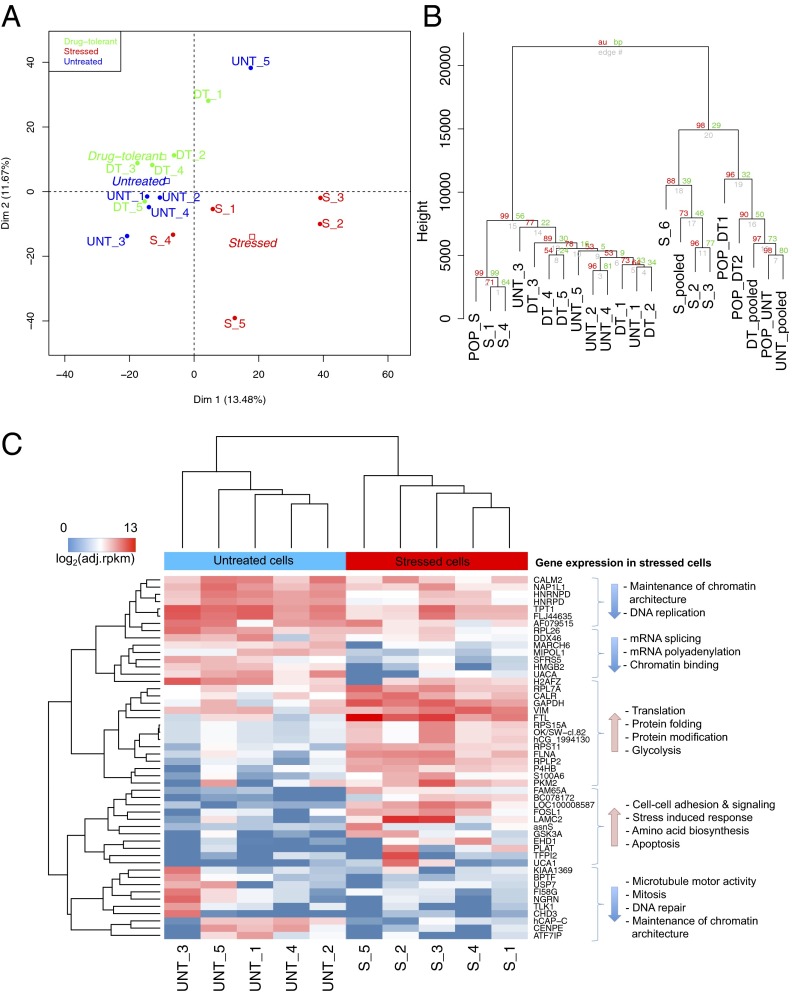
Although acute changes in gene expression (4–24 h) have been extensively analyzed for a number of stressors including chemotherapeutic compounds, the gene expression profiles in long-term stressed cells are largely unknown. Single cells from our long-term stressed cell group exhibited distinct gene expression patterns. We characterized the cell type using the adjusted reads per kilobase per million (RPKM) (Experimental Procedures) for 15 single cells using principal component analysis (PCA). The first and second PCA components clearly separated these stressed single cells from the untreated and drug-tolerant cells by their gene expression profile (Fig. 4A). We also performed hierarchical cluster analysis using the adjusted RPKM, and the same clustering pattern was observed (SI Appendix, Fig. S4A). The differentially expressed genes showed a long-term stress-induced response and the effects of paclitaxel on microtubules and mitosis in stressed cells including down-regulation of genes involved in maintenance of chromatin architecture, microtubule motor activity, mitosis, DNA repair, mRNA splicing, mRNA polyadenylation, and chromatin binding. In addition, gene expression was up-regulated in the following functional areas: cell–cell adhesion and signaling, stress-induced response, apoptosis, glycolysis, amino acid biosynthesis, translation, protein folding, and protein modification (Fig. 4C). The differential gene expression analysis between stressed and drug-tolerant cells showed a reversed trend in up- and down-regulation compared with that observed between untreated and stressed cells (SI Appendix, Fig. S4A). Differential gene expression analysis between untreated and drug-tolerant cells showed that microtubule motor activity, microtubule binding, and protein kinase activity were up-regulated in drug-tolerant cells. Furthermore, genes involved in mRNA splicing, mRNA transcription factor activity, translation, and cell adhesion were down-regulated in drug-tolerant cells. Interestingly, we find that expression of ITGA6 (integrin α6), histone demethylase KDM5A, and IGF1R (IGF1 receptor) were each up-regulated in drug-tolerant cells but not in untreated or stressed cells (SI Appendix, Fig. S4 A and B). Expression of these genes was observed in the majority of single cells as well as the population. Importantly, our data are consistent with studies by Sharma et al. (8) that implicated IGF1R signaling and an altered chromatin state conferred, in part, by KDM5A as being required to maintain a dynamic drug-tolerant phenotype. Thus, despite apparent heterogeneity in single cells, our analysis captured known features of drug tolerance conversion paths previously described with cell population studies.
Fig. 4.
Gene expression analyses. (A) The clustering gene expression data in adjusted-RPKM using PCA showed that gene expression profiles of stressed cells (red) were much different from those of untreated (green) and drug-tolerant cells (black). (B) Hierarchical clustering of gene expression data for single cells, pooled cells, and population cells in Euclidean distance with 1,000 bootstrap replications. Values at branches are approximately unbiased (AU) P values (Left), bootstrap probability (BP) values (Right), and single-cell labels (Lower). (C) Hierarchical clustering of gene expression data for the 50 most significant differentially expressed genes between untreated and stressed single cells: P < 0.001, false discovery rate < 0.025. Also see SI Appendix, Fig. S4.
The averaged patterns of gene expression between untreated and stressed or drug-tolerant groups of cells show a large number of transcripts with diverging expression (SI Appendix, Fig. S6A). However, a similar if not higher degree of divergence was found between single cells from one drug-tolerant clone (SI Appendix, Fig. S6B). The extent of divergence cannot be attributed solely to possible differences in cell cycle stage as the same extent was seen with stress-arrested single cells (SI Appendix, Fig. S6C).
Gene Expression Profiles of Single Cells Are Distinct from That of the Population.
To determine whether the gene expression profile of the population is representative of that in a single cell, we performed PCA and hierarchical clustering with the gene expression data from single cells, pooled single cells (combing all paired-end reads of five single cells and treated them as one sample for read mapping and gene expression analysis for each cell group), and populations. The hierarchical clustering showed that pooled cells clustered closer to the population cells in the untreated and drug-tolerant groups (Fig. 4B). The single cells did not cluster with their corresponding population, except for the stressed cells. However, each stressed cell appeared to have distinct gene expression patterns; therefore, they did not cluster together as tightly as untreated and drug-tolerant single cells (Fig. 4B and SI Appendix, Fig. S5B). The distance between the stressed cell population and the untreated population was farther than that between the untreated and drug-tolerant populations, which agreed with the pattern observed at the single-cell level. Moreover, great divergence of gene expression was found between any drug-tolerant single cell and the averaged expression of the five-cell group. These results indicated that gene expression in a single cell is not reflected by the average expression found in the five-cell group or in a large population of cells (Fig. 4B and SI Appendix, Fig. S5B). These results underscore the value of single-cell RNA-Seq to enhance the resolution of gene expression analysis otherwise masked by averaged values of gene expression in a bulk population.
The higher variability of single-cell gene expression compared with bulk measurements makes it more difficult to find clear patterns of differential gene expression in single cells, particularly for those that are highly variable, as has been recently described when performing RNA-Seq from normal single nuclei from human neurons (43). Moreover, a much larger sample size would empower a better examination for those transcripts that are consistently highly variable.
Discussion
It has become technically and economically feasible to sequence RNA from single cells, which enables highly sensitive detection of rare SNVs and single cell-specific gene expression programs. Such technologies will be critical for examining individual cells from tissue biopsies of heterogeneous populations. For example, we recently identified that TGF-β1 signaling represses the gene expression program induced by DNA damage, and our immunohistochemistry studies revealed heterogeneity of this phenomenon in different cancer cells present in the same tumor (44). Although not all rare variants are relevant to personalized cancer treatment, some have the potential to drive drug resistance or serve as biomarkers of therapeutic success. Thus, the ability to detect rare SNVs and specific gene expression profiles distinguishing drug terminally arrested vs. drug-tolerant single cells at the very early onset of recurrence offers extremely valuable information. This information may potentially provide diagnostic/prognostic value to assess success or failure of cancer chemotherapies shortly after administration and guide the selection of appropriate treatments that will ultimately increase therapeutic efficacy. Of note, performing single-cell transcroptomic analysis involves its own challenges and limitations from the sample acquisition, data generation, data analysis, and interpretation perspectives, which is now starting to gain attention (17, 45).
Here, by using a single-cell RNA-Seq approach, we interrogated both the RNA variants and expression levels present at the very early onset of evolution of a monoclonal population of drug-tolerant cells. We demonstrated that the majority of novel RNA variants in a single cell were unique to that cell. Most of the RNA variants shared among single cells or between populations and any single cell were SNPs cataloged in the dbSNP. The statistics of single-cell RNA-Seq for SNV identification is summarized in Table 2. There were more SNVs detected in stressed cells compared with untreated and drug-tolerant cells. This finding could be the result of paclitaxel-associated down-regulation of DNA repair that was detected in the differential gene expression analysis.
Table 2.
Statistics of SNV detection using single-cell RNA-Seq
| Single-cell statistics | Average |
| Percent transcriptomic coverage of single-cell RNA-Seq | 8.3% |
| Percent of gene expressed in the population detected at the single-cell level | 20% |
| Percent of non-dbSNP variants shared between any two single cells | 30% |
| Percent of dbSNP variants shared between any two single cells | 75% |
| Number of variants only present in the RNA but not in the DNA in a single-cell of MDA-MB-231 cell line | ∼500 |
Although it would be more rigorous to measured mutation frequency at the DNA level, our data provide an indirect approximate estimation of the maximum effective mutation rate of cancer cells from individual cells at the RNA level, with less than 5% of false-positive variant calls. Interestingly, the cell-specific polymorphism frequencies we found in the datasets from Ramsköld et al. (14) and Yan et al. (21) were very similar to ours and among themselves, regardless of cells being normal or cancer cells and the different protocols used in each study (SI Appendix, Fig. S7). This result suggests that our single-cell RNA-Seq appears to show equivalent if not superior base call fidelity than those previously published.
We identified drug-tolerant-specific RNA variants that were not found in untreated or stressed cells. Two of them resided in genes that were previously reported to be involved in paclitaxel resistance. One was a missense mutation located in RAPGEF4 that encodes a protein involved in microtubule polymerization and organization (33, 34). The other was found in the 3′ UTR region of AMOTL1. AMOTL1 protein interacts with the Hippo pathway component TAZ, which is implicated in paclitaxel resistance in breast cancer cells (46, 47). Four drug tolerant-specific RNA variants were found in genes involved in microtubule stabilization and organization, including RAPGEF4, PCM1, NUDCD3, and KIAA1671. Interestingly, expression of all these genes is still quite variable between cell to cell of any group with the sole exception of RAPGEF4, which is almost completely undetected in all untreated or stressed cells compared with drug-tolerant cells (SI Appendix, Fig. S9).
The PCA and hierarchical clustering analysis differentiates distinct gene expression profiles of stressed cells from that of untreated and drug-tolerant cells. This information could be valuable in predicting the clinical significance of residual cancer cells having either profile after chemotherapy. In fact, untreated and drug-tolerant cells have quite similar gene expression profiles. The 50 most significant differentially expressed genes between untreated cells and stressed cells were those involved in the stress response. Genes that are up-regulated in stressed cells are involved in apoptosis, glycolysis, protein synthesis, cell-to-cell adhesion, and signaling. Genes required for chromatin architecture, DNA repair, DNA replication, mRNA splicing, mRNA polyadenylation, microtubule motor activity, and mitosis are down-regulated in stressed cells, and their expression levels are similar between untreated- and drug-tolerant cells. We observed that expression of ITGA6, the histone demethylase KDM5A, and IGF1R was each up-regulated in drug-tolerant cells but not in untreated or stressed cells.
Drug-tolerant cells present gene expression profiles more similar to untreated cells than to long-term stressed cells. These cells could be either cells that became stressed and then resolved the stress or cells that had been in a preexisting condition and were never engaged in a stress response. However, these cells are more sensitive to a second round of paclitaxel (Fig. 1C) and present some SNVs in genes related to tubulin metabolism showing a cellular memory of the damage previously encountered. This result would suggest that these cells have been stressed, but obviously they enacted a rare program that ensured survival. Drug-tolerant cells might originate from stressed cells. When we performed hierarchical clustering on expressed genes that are involved in cell cycle, DNA damage signaling, senescence, drug resistance, metabolism, and apoptosis, we observed that stressed cells are clustered together, but single cells within the stressed group exhibit very distinct expression patterns compared with each other (SI Appendix, Fig. S8). Thus, a single molecular mechanism for drug tolerance might not be needed because the diversity will ensure at any time a given cancer cell containing the right gene expression and/or RNA variant will be able to overcome massive stress.
Single-cell RNA-Seq not only allows us to detect cell-to-cell transcript heterogeneity at the single nucleotide level, but it can also identify gene expression heterogeneity among single cells. By contrast, analyzing cell populations only generates an averaged gene expression level in all cells.
Interestingly, it has been recently reported that to resolve the stochastic gene expression heterogeneity found between single cells, pooling the single cell RNA sequencing results from 30 to 100 single cells was able to reconstitute the averaged gene expression given by an entire population of cells (17). This observation might help to explain at least partially why pooling the data will cluster closer to the actual population than with only five cells (although our findings do not contradict these data but agree with their proposed idea, because when comparing one cell to another, they still find a high degree of variability). Our data suggest that pooling five cells from different treatments or biological conditions is not sufficient to accurately reconstitute the averaged expression of the populations.
Here we demonstrated that single-cell gene expression profiles differ from profiles of their corresponding populations in significant and illuminating ways. These data reveal logical connections to genes involved in stress, cell proliferation, and cell death. Importantly, we were able to trace the regeneration of cancer cell heterogeneity in as few as six cell divisions after resuming proliferation from one founder drug-tolerant cell. This colony led to the reestablishment of a heterogeneous response to paclitaxel on further expansion, indicating that clonal evolution of cancer cells can regenerate drug-tolerant and drug-sensitive subpopulations. Use of this technique in a deeper examination may further illuminate the underlying basis of such phenotypic switching, as well as lead to the identification of genes not yet implicated in cancer, cancer treatment, or other disease states.
Experimental Procedures
Cell Culture, Drug Treatments, and the Paclitaxel Paradigm.
MDA-231 cells were obtained from the Princeton Physical Sciences Oncology Center tissue biorepository and routinely cultured in DMEM supplemented with 10% (vol/vol) FBS. Taxol (paclitaxel; Sigma) was prepared as a 5 mM stock solution in ethanol, and serial dilutions were prepared for toxicity assays.
The paclitaxel treatment paradigm was established as indicated in the diagram of Fig. 1A. Briefly, 1 × 106 cells were plated in 100-mm culture dishes for 24 h and then treated with 100 nM paclitaxel. After 3 d, fresh 100 nM paclitaxel-containing media was added for another 2 d, totaling 5 d of paclitaxel treatment. Cells were then rinsed with PBS and maintained in drug-free culture with media replacement every 48 h, and clones of drug-tolerant cells were expanded by the ring cloning technique. Cells still alive 1 d after paclitaxel removal were considered residual cells undergoing a stress response, most of which died within the next 2–4 wk. The clones of cells that resumed proliferation are considered recurrent drug-tolerant cells. To calculate the frequencies of stressed and drug-tolerant cells, the number of the counted stressed or drug-tolerant founder cells was divided by the total number of cells submitted to drug treatment.
Paclitaxel Toxicity Assays.
Cell growth of naïve or expanded recurrent drug-tolerant cells was determined as follows. Briefly, 25,000 cells were plated in each well of 12-well plates and after 24 h were treated with vehicle-ethanol or up to 100 nM paclitaxel-containing media. After 4 d, cells were fixed with 10% formaldehyde, and the IC50 was established by Giemsa staining (44). Cell number was plotted as a percent of cells relative to vehicle control with SE from four replicated wells from a representative experiment.
Cell Analysis Experimental Design.
For the single cells, we performed RNA sequencing on the RNAs from five naïve, five stressed (day 5 + 1 d drug free), and five drug-tolerant cells from one clone at early growth (5 d paclitaxel + 15 d drug free). Thus, the RNA-Seq was on five drug-tolerant cells from a unique clone. The clone shown in Fig. 1B is a clone that was ultimately expanded from an individual cell up to >8 million cells (>23 population doublings). This clone was used to generate data in Fig. 1C, and the results were similar to three other clones. For population RNA-Seq, we used 10,000 naïve (untreated) MDA-231 cells, 10,000 stressed cells (day 5 + 1 drug free, nonclonal), and 10,000 cells from three independent, new drug-tolerant clones expanded as explained above to render various millions of cells per clone. Finally, we focused on SNVs that would be present in different drug-tolerant clones rather than in clone-specific ones. For that reason, we performed pyrosequencing from additional single cells from different drug-tolerant clones, as well as from additional untreated single cells obtained as described above.
Isolation of Single Cells and Cell Populations and cDNA Synthesis.
Typically, five single cells from populations of untreated, stressed, or proliferating drug-tolerant cells obtained from single clones as indicated in Fig. 1 were collected as follows. Media were removed and replaced by PBS at room temperature. Single cells were picked within the next 10 min with <20-µm-diameter glass needles using Narshige MO-188 and MN-188 hydraulic micromanipulators over an inverted microscope, washed, and immediately lysed in Prelude Direct Lysis Module (NuGEN Technologies) on glass-mounted microdroplets. For population analyses, >10,000 pooled untreated, stressed, or drug-tolerant cells were lysed. Snap frozen lysates were stored at −80 °C. cDNA was generated for each single cell lysate using the Ovation RNA-Seq system (NuGEN Technologies) per the manufacturer’s recommended protocols and as described previously in Tariq et al. (48). Briefly, total RNA of cell lysate was reverse-transcribed to first-strand cDNA using a combination of random hexamers and poly-T chimeric primers and then converted to double-stranded (ds) DNA using fragmentation and RNA-dependent DNA polymerase. Finally, the ds cDNA was amplified linearly using a single-primer isothermal amplification process and purified by using MyOne carboxilic acid-coated superparamagnetic beads (Invitrogen). The cDNA was prepared for 15 individual single cells for library preparation. The quality and quantity of single-cell cDNA were evaluated using the Agilent Bioanalyzer 2100 DNA High Sensitivity chip (Agilent).
RNA-Seq Library Preparation and Sequencing.
For paired-end whole transcriptome library preparation, ∼0.5–1.0 µg cDNA of each sample was sheared to a size ranging between 200 and 300 bp using the Covaris-S2 sonicator (Covaris) according to the manufacturer’s recommended protocols. Fragmented cDNA samples were used for the preparation of RNA-Seq libraries using TruSeq v1 Multiplex Sample Preparation kit (Illumina). Briefly, cDNA fragments were end-repaired, dA-tailed, and ligated to multiplex adapters according to the manufacturer's instructions. After ligation, DNA fragments smaller than 150 bp were removed with AmPure XP beads (Beckman Coulter Genomics). The purified adapter ligated products were enriched using PCR (14 cycles). The final amplified libraries were resolved on 2.0% agarose gel and manually size-selected in the range of 350–380 bp. The final single cell RNA-Seq libraries were quantitated using the Agilent bioanalyzer 2100 and pooled together in equal concentration for sequencing. The pooled multiplexed libraries were sequenced in five independent sequencing runs, with eight lanes per run, and generated 50-bp paired-end reads on HiSEq. 2000 (Illumina).
Whole Genome DNA Sequencing of Naïve MDA-MB-231 Cells.
For high-throughput sequencing, high-molecular-weight genomic DNA (gDNA) was obtained from MDA-MB-231 cells (Princeton PSOC). For the DNA library prep, 1 µg of gDNA was first sheared down to 200–300 bp using the Covaris S2 per the manufacturer’s recommendations. A target insert size of 200–250 bp was then size-selected using the automated electrophoretic DNA fractionation system LabChip XT (Caliper Life Sciences). Paired-end sequencing libraries were prepared using Illumina’s TruSeq DNA Sample Preparation Kit. Following DNA library construction, samples were quantified using the Agilent Bioanalyzer per the manufacturer’s protocol. DNA libraries were sequenced using the Illumina HiSEq. 2000 in two flow cell lanes with sequencing paired-end read length at 2 × 100 bp. Reads were demultiplexed using CASAVA (version 1.8.2).
Sequencing Reads Quality Control and Mapping.
Reads were subjected to a series of preprocessing steps. First, the sequencing adapter sequences were removed from the reads using SeqPrep (github.com/jstjohn/SeqPrep). The first nine bases from the 5′ end of the reads were trimmed due to nucleotide biases introduced during cDNA synthesis. The quality of the preprocessed reads was evaluated with FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). The preprocessed reads were mapped as paired-end reads with the Tophat package (v.1.3.2), using default parameters (49), against the UCSC (University of California, Santa Cruz) hg19 human DNA reference. Duplicates reads were removed using the rmdup option of samtools (50). Uniquely mapped reads were used for all of the analyses in this paper. These reads were extracted from the bam file generated by Tophat with tag “NH:i:1” using GNU fgrep package (www.gnu.org/s/grep/).
SNV Calling.
The SNVs in single-cell RNA were called using BamBam (51). For all data analyses, we only used variants that passed the strand bias filter and read quality filter and had a genotype accuracy likelihood score greater than 30. We identified two types of SNVs: known SNVs (those cataloged in dbSNP Build ID: 137), and novel SNVs (those not present in the dbSNP). BamBam uses a Bayesian mutation caller and can be run in either single-sample mode or in a two-sample mode in which we used both DNA and RNA. Each mutation was appointed a confidence score according to the genotype accuracy likelihood. Variants with supporting reads only in the first or last third of a read’s data were removed.
Determination of the SNV Rate.
The SNVs were filtered to find those within the exons of UCSCs known gene canonical transcripts, where the exon’s average mapped read coverage in the cell line was greater than 50. Each cell line’s variant rate was calculated by summing the total number of variants that pass this filter and dividing by the total number of bases in exons with average coverage greater than 50.
Identification of Common and Unique SNVs.
To compare the SNVs between a single cell and its corresponding population cells, we first identified the comparable genomic regions where both the single cell and the population cells of the same group have at least 10 times RNA-Seq read coverage. We then identified the common and different SNVs between the single cell and the population cells that are within the comparable genomic regions. For identifying common and unique SNVs between any two single cells, we performed all pairwise comparisons of single cells. We first identified comparable genomic regions where both single cells have at least 10 times RNA-Seq read coverage. Then, we identified common and different SNVs between the two single cells that are within the comparable genomic regions.
Detection of DNA-RNA Variants.
DNA-RNA variants are the SNVs that are only present in the RNA but not in the DNA. We identified the DNA-RNA variants by comparing single-cell RNA-Seq data with the DNA sequencing data from cell populations. DNA-RNA variants were detected for each of the three single cell groups (untreated, stressed, and drug tolerant). We first identified the DNA variants that were specific to the cell line by comparing the population DNA to the human reference genome (UCSC hg19). The population DNA was from two sources: one is from our whole genome DNA sequencing (20 times), and the other is from ultra-deep exome sequencing (200 times) (52). Next, we compared untreated RNA to population DNA and the human reference and declared all differences as DNA-RNA variants and those with sufficient evidence as RNA variants. An event has sufficient evidence if there are enough reads in the RNA to support the variant and enough coverage in the DNA to determine that the variant seen in the RNA is not a DNA variant that is specific to the cell line. RNA variants must be covered by at least 10 reads, and at least 4 of the reads need to support the variant. In addition, at least 10% of the RNA reads must support the variant. We also require 10 or more reads in the population DNA, and none of the reads can support the RNA variant. We identified the high-confidence RNA variants by requiring at least 100 reads in the cell line DNA at those loci, and none of the reads can support the RNA variant. We continued to determine candidate RNA variants that were newly emerged in the stressed single cells by comparing them to the population DNA, human reference, and the untreated RNA. Lastly, we identified RNA variants that had only occurred in the drug-tolerant cells by comparing them to the population DNA, human reference, untreated RNA, and stressed single cells. We performed additional filtering steps for all of the RNA variants and removed all of the RNA variants that overlapped with SNP sites in dbSNP. We only considered RNA variants that were within the accessible genome defined by the 1000 Genome Project Consortium (30). To eliminate false-positive RNA variants, we used BLAT (BLAST-like alignment tool) to ensure unique mapping of reads that support any RNA variant (53).
Gene Expression Analysis.
The number of reads per transcript was measured using RSeQC (54). Whole transcriptome gene expression was calculated by normalizing the read counts per transcript by the kilobases of the transcript per million mapped reads that has at least 10 times coverage denoted as adjusted RPKM (55). We characterized the single cells using gene expression data for all 15 single cells and population cells from the three groups using PCA and hierarchical clustering with 1,000 bootstrapping replications. A matrix was generated with adjusted RPKM for each gene in each single cell sample. Only those genes with RPKM > 0 in at least one sample were retained for further analysis. The PCA was performed using FactoMineR (56) to cluster the gene expression data. We used the Pvclust package in R to perform the hierarchical clustering analysis using the Ward’s method with distance measured in Euclidean distance (57). Differential expression analysis was performed on single cells and population cells using DEGSeq that uses a nonparametric approach with resampling to account for the different sequencing depths (58). Functional classification of the differential expressed genes was performed using the PANTHER Classification System, version 8 (www.pantherdb.org). Hierarchical clustering of differentially expressed genes was performed using the heatmap function in R (version 2.15.1) (59).
PCR Amplification for Targeted SNV Pyrosequencing.
Ten SNVs were selected for validation with pyrosequencing using cDNA from nine different single cells: two untreated cells, two stressed cells, and five drug-tolerant cells. PCR primers and internal sequencing primers were designed using Pyrosequencing Assay Design Software (Biotage) and were synthesized by IDT. Amplicons were designed to be 100–200 bp long. Amplicons used in pyrosequencing were amplified from cDNA used to generate HiSeq libraries. Each PCR in a 50-µL volume contained the following: 5 ng cDNA, 0.1 µM of each forward and reverse primer, 2.0 mM MgCl2, 200 µM dNTPs, 75 mM Tris⋅HCl (pH 8.0), and 1.5 U of Titanium Taq polymerase (Clontech Laboratories). The amplification was performed in a Gene Amp PCR System 9700 Thermal Cycler (Applied Biosystems) under the following conditions: 95 °C for 5 min, followed by 25 cycles of denaturing at 95 °C for 30 s and annealing at the primer specific annealing temperature for 30 s, and a final extension at 72 °C for 4 min.
Validating SNVs with Pyrosequencing.
Fifty microliters of biotinylated PCR amplicons was immobilized onto streptavidin-coated paramagnetic beads (Dynabeads M-280-streptavidin; Dynal AS) in 2× binding wash buffer (10 mM Tris⋅HCl, pH 7.5, 1 mM EDTA, and 2 M NaCl) and incubated at room temperature for 15 min. The immobilized PCR product was treated with 100 µL of 20 mM NaOH for 5 min to denature into single-stranded DNA. Single-stranded DNA attached to the beads was washed twice with 1× annealing buffer (200 mM magnesium acetate and 0.1 M Tris-acetate, pH 7.75). Immobilized single-stranded DNA was resuspended in 20 µL of 1× annealing buffer and 5 µL of sequencing primer at 10 µM. The sequencing primer was annealed to single-stranded template at 95 °C for 2 min and then 50 °C for 8 min. Primed single-stranded template was sequenced using the PyroMark Q24 system (Qiagen). SNV quantification was performed using the PyroMark Q24 1.010 software (Qiagen).
Supplementary Material
Acknowledgments
We thank Kikuye Koyano, James Perrott, Rameet Brar, and Ms. Ruo Huang for technical assistance. We thank Dr. Christopher Benner for bioinformatics training and expert advice to F.J.L.-D. We thank Dr. Joe Gray for sharing his unpublished MDA-MB-231 exome sequencing data. This work was supported in part by National Cancer Institute Grant U54CA143803, National Institutes of Health Grant P01-35HG000205, the Chambers Medical Foundation, the GemCon Family Foundation, the Olive Tupper Foundation, and Cancer Center Grant P30CA014195 (to B.M.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequencing short reads data have been deposited in the Sequence Read Archive at the National Center for Biotechnology Information, www.ncbi.nlm.nih.gov/sra (accession no. SRP040309).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404656111/-/DCSupplemental.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Bock C, Lengauer T. Managing drug resistance in cancer: Lessons from HIV therapy. Nat Rev Cancer. 2012;12(7):494–501. doi: 10.1038/nrc3297. [DOI] [PubMed] [Google Scholar]
- 3.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 4.Mardis ER. A decade’s perspective on DNA sequencing technology. Nature. 2011;470(7333):198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 5.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 6.Saunders NA, et al. Role of intratumoural heterogeneity in cancer drug resistance: Molecular and clinical perspectives. EMBO Mol Med. 2012;4(8):675–684. doi: 10.1002/emmm.201101131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauri A, et al. Assessment of MDA efficiency for genotyping using cloned embryo biopsies. Genomics. 2013;101(1):24–29. doi: 10.1016/j.ygeno.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Lao K, Xu NL, Straus NA. Whole genome amplification using single-primer PCR. Biotechnol J. 2008;3(3):378–382. doi: 10.1002/biot.200700253. [DOI] [PubMed] [Google Scholar]
- 11.Zhou HG, Zhang C. [Study on application of the whole genome amplification in LCN] Fa Yi Xue Za Zhi. 2006;22(1):43–44, 47. [PubMed] [Google Scholar]
- 12.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148(5):886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalerba P, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29(12):1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramsköld D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cann GM, et al. mRNA-Seq of single prostate cancer circulating tumor cells reveals recapitulation of gene expression and pathways found in prostate cancer. PLoS ONE. 2012;7(11):e49144. doi: 10.1371/journal.pone.0049144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Islam S, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21(7):1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marinov GK, et al. From single-cell to cell-pool transcriptomes: Stochasticity in gene expression and RNA splicing. Genome Res. 2014;24(3):496–510. doi: 10.1101/gr.161034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauer JA, et al. RNA interference (RNAi) screening approach identifies agents that enhance paclitaxel activity in breast cancer cells. Breast Cancer Res. 2010;12(3):R41. doi: 10.1186/bcr2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurn N, et al. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin Chem. 2005;51(10):1973–1981. doi: 10.1373/clinchem.2005.053694. [DOI] [PubMed] [Google Scholar]
- 20.Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9(8):677–679. [PubMed] [Google Scholar]
- 21.Yan L, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20(9):1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 22.Bahn JH, et al. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 2012;22(1):142–150. doi: 10.1101/gr.124107.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinman CL, Adoue V, Majewski J. RNA editing of protein sequences: A rare event in human transcriptomes. RNA. 2012;18(9):1586–1596. doi: 10.1261/rna.033233.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park E, Williams B, Wold BJ, Mortazavi A. RNA editing in the human ENCODE RNA-seq data. Genome Res. 2012;22(9):1626–1633. doi: 10.1101/gr.134957.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng Z, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. 2012;30(3):253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 26.Gu T, et al. Canonical A-to-I and C-to-U RNA editing is enriched at 3’UTRs and microRNA target sites in multiple mouse tissues. PLoS ONE. 2012;7(3):e33720. doi: 10.1371/journal.pone.0033720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge X, Frank CL, Calderon de Anda F, Tsai LH. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron. 2010;65(2):191–203. doi: 10.1016/j.neuron.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159(2):255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81(2):261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- 31.Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: Implications for genomic stability and cell polarity. Proc Natl Acad Sci USA. 1998;95(6):2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brinkley BR, Goepfert TM. Supernumerary centrosomes and cancer: Boveri’s hypothesis resurrected. Cell Motil Cytoskeleton. 1998;41(4):281–288. doi: 10.1002/(SICI)1097-0169(1998)41:4<281::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 33.Magiera MM, et al. Exchange protein directly activated by cAMP (EPAC) interacts with the light chain (LC) 2 of MAP1A. Biochem J. 2004;382(Pt 3):803–810. doi: 10.1042/BJ20040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sehrawat S, Cullere X, Patel S, Italiano J, Jr, Mayadas TN. Role of Epac1, an exchange factor for Rap GTPases, in endothelial microtubule dynamics and barrier function. Mol Biol Cell. 2008;19(3):1261–1270. doi: 10.1091/mbc.E06-10-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedrotti B, Islam K. Purified native microtubule associated protein MAP1A: Kinetics of microtubule assembly and MAP1A/tubulin stoichiometry. Biochemistry. 1994;33(41):12463–12470. doi: 10.1021/bi00207a013. [DOI] [PubMed] [Google Scholar]
- 36.Ahmed AA, et al. Modulating microtubule stability enhances the cytotoxic response of cancer cells to Paclitaxel. Cancer Res. 2011;71(17):5806–5817. doi: 10.1158/0008-5472.CAN-11-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou T, Zimmerman W, Liu X, Erikson RL. A mammalian NudC-like protein essential for dynein stability and cell viability. Proc Natl Acad Sci USA. 2006;103(24):9039–9044. doi: 10.1073/pnas.0602916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou T, Aumais JP, Liu X, Yu-Lee LY, Erikson RL. A role for Plk1 phosphorylation of NudC in cytokinesis. Dev Cell. 2003;5(1):127–138. doi: 10.1016/s1534-5807(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 39.Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3(104):ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 40.Dephoure N, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105(31):10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernández-Madrid F, et al. Autoantibodies to Annexin XI-A and Other Autoantigens in the Diagnosis of Breast Cancer. Cancer Res. 2004;64(15):5089–5096. doi: 10.1158/0008-5472.CAN-03-0932. [DOI] [PubMed] [Google Scholar]
- 42.Tagami S, Eguchi Y, Kinoshita M, Takeda M, Tsujimoto Y. A novel protein, RTN-XS, interacts with both Bcl-XL and Bcl-2 on endoplasmic reticulum and reduces their anti-apoptotic activity. Oncogene. 2000;19(50):5736–5746. doi: 10.1038/sj.onc.1203948. [DOI] [PubMed] [Google Scholar]
- 43.Grindberg RV, et al. RNA-sequencing from single nuclei. Proc Natl Acad Sci USA. 2013;110(49):19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.López-Díaz FJ, et al. Coordinate transcriptional and translational repression of p53 by TGF-β1 impairs the stress response. Mol Cell. 2013;50(4):552–564. doi: 10.1016/j.molcel.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saliba AE, Westermann AJ, Gorski SA, Vogel J. Single-cell RNA-seq: Advances and future challenges. Nucleic Acids Res. 2014;42(14):8845–8860. doi: 10.1093/nar/gku555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011;71(7):2728–2738. doi: 10.1158/0008-5472.CAN-10-2711. [DOI] [PubMed] [Google Scholar]
- 47.Chan SW, et al. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem. 2011;286(9):7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tariq MA, Kim HJ, Jejelowo O, Pourmand N. Whole-transcriptome RNAseq analysis from minute amount of total RNA. Nucleic Acids Res. 2011;39(18):e120. doi: 10.1093/nar/gkr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanborn JZ. 2012. Tumor versus matched-normal sequencing analysis and data integration. PhD dissertation (California Digital Library, Santa Cruz, CA)
- 52.Daemen A, et al. Modeling precision treatment of breast cancer. Genome Biol. 2013;14(10):R110. doi: 10.1186/gb-2013-14-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Wang S, Li W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 55.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 56.Lê S, Josse J, Husson F. FactoMineR: An R package for multivariate analysis. J Stat Softw. 2008;25(1):18. [Google Scholar]
- 57.Suzuki R, Shimodaira H. Pvclust:Aan R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22(12):1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 58.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 59.Team RC. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.